PURPOSE
Optimized strategies for risk classification are essential to tailor therapy for patients with biologically distinctive disease. Risk classification in pediatric acute myeloid leukemia (pAML) relies on detection of translocations and gene mutations. Long noncoding RNA (lncRNA) transcripts have been shown to associate with and mediate malignant phenotypes in acute myeloid leukemia (AML) but have not been comprehensively evaluated in pAML.
METHODS
To identify lncRNA transcripts associated with outcomes, we evaluated the annotated lncRNA landscape by transcript sequencing of 1,298 pediatric and 96 adult AML specimens. Upregulated lncRNAs identified in the pAML training set were used to establish a regularized Cox regression model of event-free survival (EFS), yielding a 37 lncRNA signature (lncScore). Discretized lncScores were correlated with initial and postinduction treatment outcomes using Cox proportional hazards models in validation sets. Predictive model performance was compared with standard stratification methods by concordance analysis.
RESULTS
Training set cases with positive lncScores had 5-year EFS and overall survival rates of 26.7% and 42.7%, respectively, compared with 56.9% and 76.3% with negative lncScores (hazard ratio, 2.48 and 3.16; P < .001). Pediatric validation cohorts and an adult AML group yielded comparable results in magnitude and significance. lncScore remained independently prognostic in multivariable models, including key factors used in preinduction and postinduction risk stratification. Subgroup analysis suggested that lncScores provide additional outcome information in heterogeneous subgroups currently classified as indeterminate risk. Concordance analysis showed that lncScore adds to overall classification accuracy with at least comparable predictive performance to current stratification methods that rely on multiple assays.
CONCLUSION
Inclusion of the lncScore enhances predictive power of traditional cytogenetic and mutation-defined stratification in pAML with potential, as a single assay, to replace these complex stratification schemes with comparable predictive accuracy.
INTRODUCTION
Iterative refinements in risk stratification have been a cornerstone for improved outcomes in children with leukemia. The criteria used in stratifying pediatric acute myeloid leukemia (pAML) have evolved substantially in recent years, driven by better recognition of recurrent molecular changes that modify biology and response to chemotherapy (Data Supplement, online only).1-5 Inclusion of minimal residual disease (MRD) assessment of induction response6 and recent identification of high-risk immunophenotypes7 have added additional features for stratification and prognostication.
CONTEXT
Key Objective
Does long noncoding RNA (lncRNA) expression in pediatric acute myeloid leukemia offer improved performance compared with conventional clinical risk classification criteria?
Knowledge Generated
We define and validate the lncScore, a 37-gene–based lncRNA expression classifier that yields comparable predictive performance to traditional cytogenetic and molecular testing, while also uncovering new predictive information not available in current techniques.
Relevance (S. Bhatia)
The lncScore as a single assay carries the potential to replace the current complex stratification schemes without losing predictive accuracy.*
*Relevance section written by JCO Associate Editor Smita Bhatia, MD, MPH, FASCO.
Prognostic classification by coding gene expression has been studied in acute myeloid leukemia (AML) for nearly 20 years.8-12 Although such studies have been important for better understanding of AML biology, stratification by gene expression patterns has been slow to penetrate clinical practice and is not widely used in pAML. Among several reasons for this failure, including challenges in reproducibility and ease of assay performance, a key problem has been that these classifiers have not yielded additional information compared with traditional testing.13
More recent efforts have focused on broadening the definition of gene expression to include nonprotein coding transcripts, including both micro-RNA and long noncoding RNA (lncRNA) expression.14 lncRNAs are defined as transcripts longer than 200 base pairs that lack protein coding potential.15 They are widely expressed in eukaryotes and increasingly recognized as critical mediators of diverse processes in normal development and differentiation including transcriptional regulation, chromatin architectural reorganization, modulation of translation, and post-translational modifications.16 Altered lncRNA expression is implicated in a variety of disease processes, including neoplasia.17 lncRNA are characteristically differentially expressed during normal and malignant hematopoiesis.18-21 Recent studies have demonstrated novel prognostic potential using lncRNA expression in adult AML22-27; however, their relevance in pediatric disease is not well established.
We investigated the expression of lncRNA in childhood AML, testing its utility for risk characterization. We define a lncRNA-based expression classifier that, as a single assay, has comparable predictive performance to complex modern, multiassay-based stratification procedures, while also adding novel prognostic information uncaptured by current techniques. This work demonstrates that lncRNA-based risk stratification could augment or replace current stratification schemes to yield less complex and more precise risk stratification of childhood AML.
METHODS
AML Cohorts
We assayed 68 normal bone marrow and 1,299 AML cases from Children's Oncology Group (COG) studies (CCG-2961,28 AAML03P1,29 AAML0531,30 and AAML103131) by RNA-seq. Most samples (1,060, 82%) come from AAML1031, representing all cases from that trial where high-quality RNA was available; the remainder were selected from prior studies and were enriched for high-risk features. An additional unselected set comprising 96 adult AML specimens treated on SWOG Cancer Research Network trials S9031,32 S9333,33 S0112,34 and S010635 (ClinicalTrials.gov identifier: NCT01503541) was examined for further validation, representing all cases with rRNA depleted RNA-seq data available for study. Written informed consent for biological correlative studies was obtained from participants during enrollment in the parent clinical trials, which were conducted in accordance with the Declaration of Helsinki. The Fred Hutchinson Cancer Research Center Institutional Review Board and the COG Myeloid Biology Committee approved and oversaw the conduct of this study.
Methods for RNA sequencing, transcript quantification, revised risk classification, training/validation set randomization, and lncScore model generation are outlined in the Data Supplement.
Outcome Analyses
Analyses of the association between lncScore and survival outcomes were performed using Kaplan-Meier estimates and the log-rank test. Hazard ratios (HRs) and associated CI were estimated in single and multivariable models by Cox proportional hazards regression. Predictive performance was assessed by concordance index,36 with submodel comparisons performed using the method of Uno.37 P-values < 0.05 were considered significant. Event-free survival (EFS) was defined as the time from enrollment to first event (relapse, induction failure, or death) or last follow-up. Overall survival (OS) was defined as the time from study enrollment to death or last follow-up. Relapse rate (RR) was defined as the time from end of induction to relapse or last follow-up. Post-induction disease-free survival (DFS) and OS were defined starting from the end of induction cycle 1 through these end points, respectively.
RESULTS
lncRNA Is Differentially Expressed Between pAML and Normal Bone Marrow
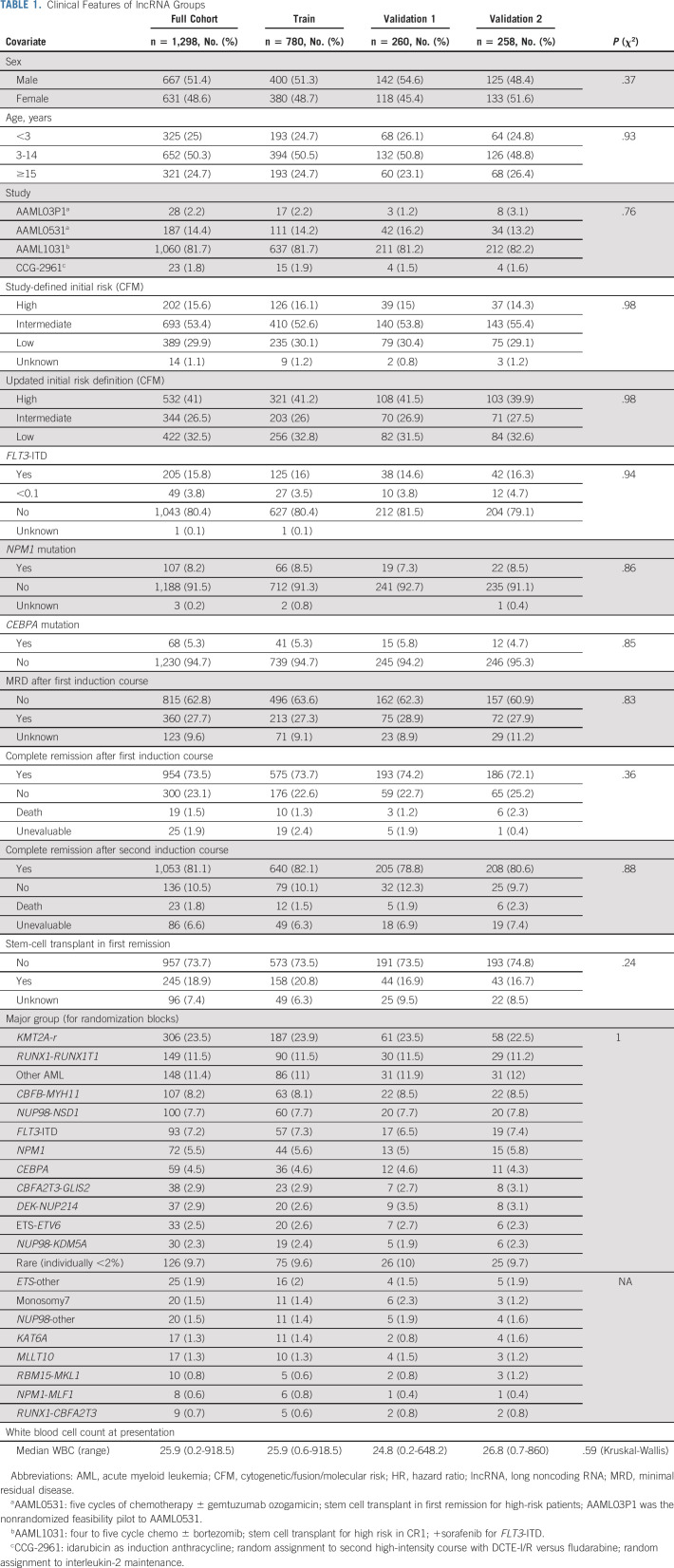
We assessed the relationship between lncRNA expression and outcomes in pAML specimens sequenced as a part of the Target Pediatric AML Initiative.5,14,38,39 The cohort initially consisted of 1,299 pediatric patients with de novo AML treated on four COG phase III trials. Patient characteristics are summarized in Table 1. Revision of risk classification from prior study definitions to current standards defined a large proportion of high-risk patients (Data Supplement). Overall outcomes for this cohort, according to a modern definition of presenting risk by cytomolecular features (cytogenetic/fusion/molecular risk, CFM) and by final risk (FR), a definition that incorporates presenting risk and postinduction MRD determination,31 are illustrated in Figures 1A-1D.
TABLE 1.
Clinical Features of lncRNA Groups
FIG 1.
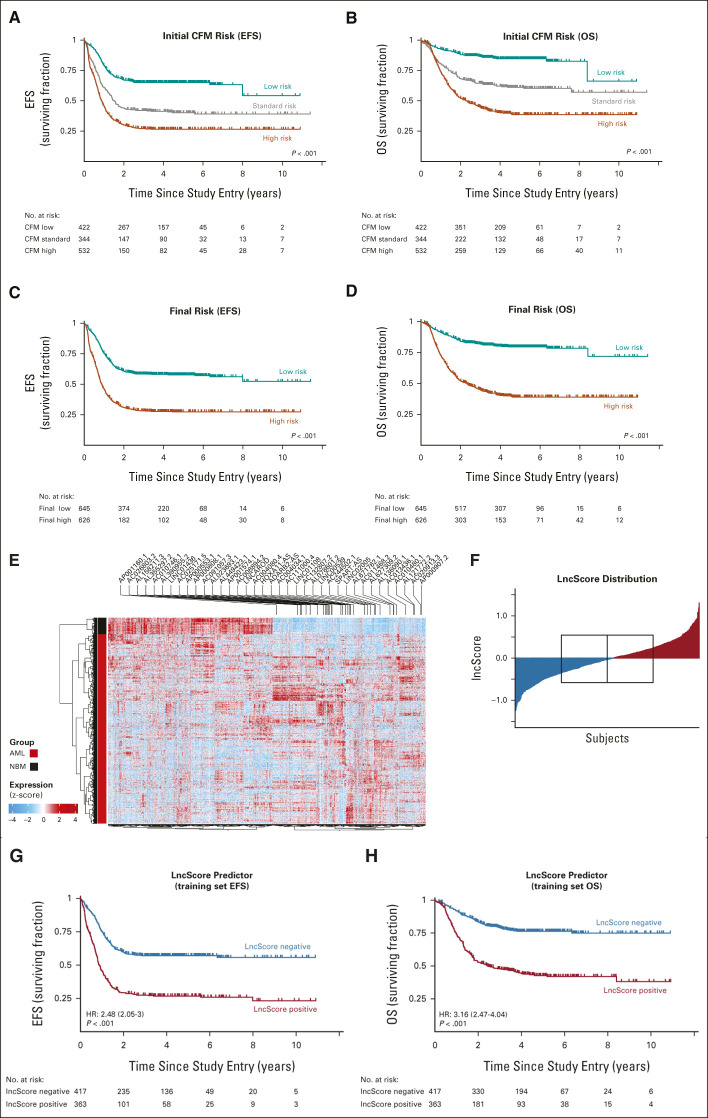
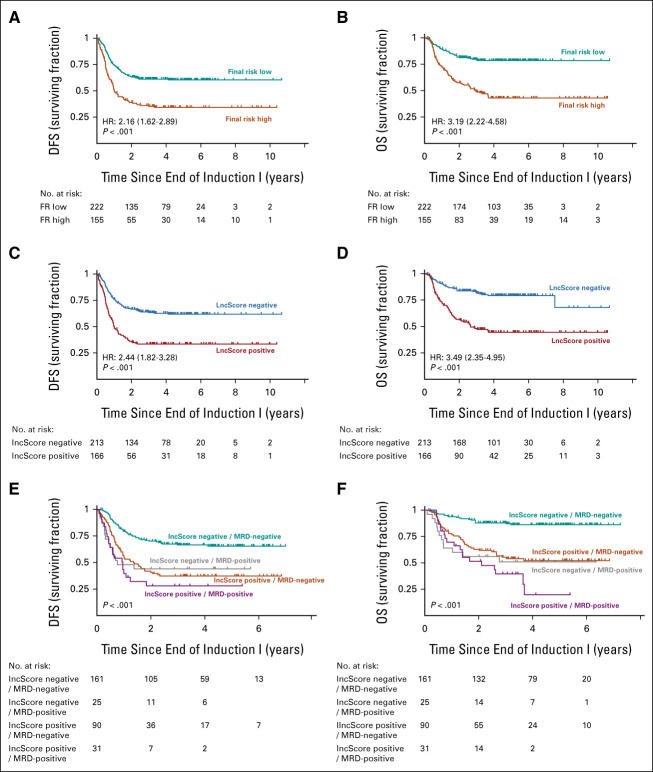
Identification of a lncRNA signature associated with outcome in pediatric AML. Overall treatment outcomes of the 1,298-subject study cohort after reclassification to a current schema for initial risk ([A] CFM EFS; [B] CFM OS) and postinduction risk determination ([C] EFS for FR; [D] OS by FR classification). (E) Differential analysis of annotated lncRNA expression in pAML training set data compared with normal bone marrow–identified 1,346 lncRNAs with altered expression (adjusted P-value of <0.05, >2-fold change in average expression). We trained a Cox regression model for EFS on the aberrantly upregulated training set lncRNAs (right half of dendrogram) from training set data to identify 37 lncRNAs (labeled above) with significant effect on EFS. (F) The lncScore was calculated as the weighted sum after applying model coefficients to lncRNA expression estimates. In the training cohort set, lncScores ranged from –1.24 to 1.31 and were nearly centered at 0 (F; red, positive lncScore; blue, negative lncScore; boxed region indicates interquartile range and median). lncScores were strongly correlated with (G) event-free survival and (H) overall survival, with the HR for a positive score of 2.48 (P < .001) and 3.16 (P < .001), respectively. AML, acute myeloid leukemia; CFM, cytogenetic/fusion/molecular risk; EFS, event-free survival; FR, final risk; HR, hazard ratio; lncRNA, long noncoding RNA; NBM, normal bone marrow; OS, overall survival; pAML, pediatric acute myeloid leukemia.
The study population was divided into training (n = 781), validation 1 (n = 260), and validation 2 (n = 258) groups using a randomization scheme blocked for key molecular features. There were no significant differences in the distribution of these features among the groups (Table 1). After derivation of the lncScore, one subject in the training set was determined to be ineligible and was removed from subsequent outcome analyses. A diagram for the investigations reported here is illustrated in the Data Supplement.
Identification and Validation of a 37-Gene lncRNA Score for pAML Outcome
We defined differentially expressed lncRNAs by comparison of AML data from the training set to bulk normal control bone marrow. Differential expression analysis of annotated lncRNA revealed 1,346 transcripts, with 647 upregulated in pAML (Fig 1E and Data Supplement). To determine whether lncRNA expression at the time of diagnosis was predictive of treatment outcome, we selected these upregulated lncRNAs for inclusion in a regularized Cox proportional hazards regression model of EFS. We limited analysis to upregulated genes in AML to best allow identification of lncRNAs that may be associated with disease progression and development, and whose expression may be identifiable as a detectable biomarker, rather than finding absence of expression in downregulated lncRNAs between normal and AML samples.
This approach defined a set of 37 lncRNAs (Data Supplement). We applied the trained model coefficients to the normalized lncRNA expression data (Data Supplement), producing a weighted sum of expression for each patient to create an expression score, which we term the lncScore. The distribution of lncScores revealed approximately equal numbers of patients with positive and negative scores in the training cohort, with values ranging from –1.24 to +1.31 (Fig 1F). lncScore was significantly predictive of both OS and EFS as a continuous variable (training set HR for OS, 3.67; 95% CI, 2.69 to 5.02; HR for EFS, 4.1; 95% CI, 3.27 to 5.14; all P < .001) and when discretized by quartile (Data Supplement). Since the median lncScore was close to 0, and clinical decision making revolves around identifying patients for early intensification by bone marrow transplantation, we dichotomized the training set cohort into those with positive or negative lncScores for further analysis. Comparison of these groups revealed positive lncScores had an EFS of 27% ± 5% at 5 years from diagnosis compared with 57% ± 5% for those with negative scores (HR, 2.48; 95% CI, 2.05 to 3; P < .001, Fig 1G and Data Supplement). lncScore was similarly predictive of OS (5-year OS, 43% ± 6% v 76% ± 4%; HR, 3.16; 95% CI, 2.47 to 4.04; P < .001; Fig 1H and Data Supplement).
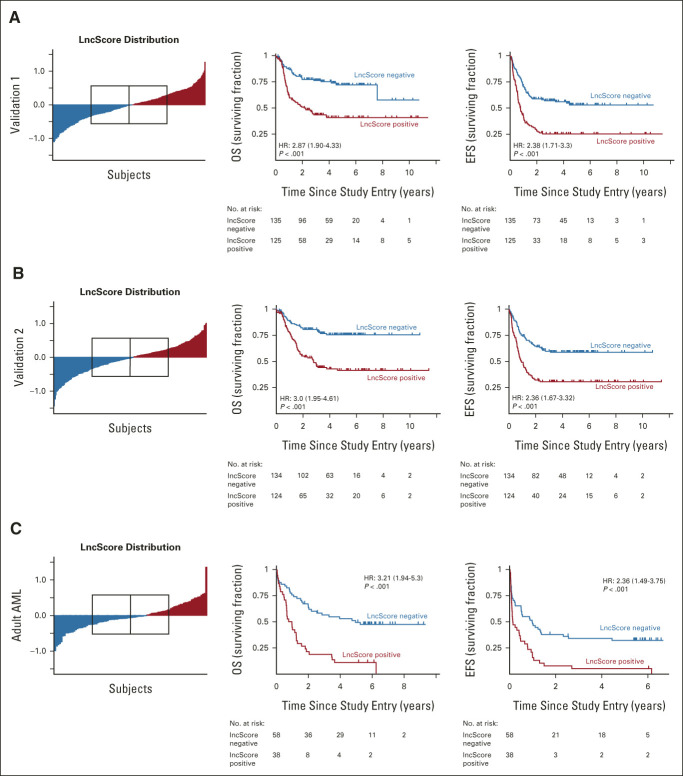
We validated the association of lncScore with survival measures in two pediatric validation sets not used during lncRNA selection or survival model training, with one set reserved for potential model revision. As in the training cohort, both validation 1 and validation 2 cohorts showed similar distribution of lncScores across samples (median, –0.011 and –0.026; range, –1.10 to 1.27 and –1.44 to 1.01, respectively). The magnitude and significance level of predictive effect was comparable with the training set for both OS (HR, 2.87 and 3, respectively; P < .001) and EFS (HR, 2.38 and 2.36, respectively; P < .001; Figs 2A and 2B). These results suggested adequate predictive performance for the lncScore as initially derived from the training set. Without a need to hold an additional validation set in reserve for optimization, we subsequently combined the pAML validation 1 and 2 groups (n = 518) for increased power in multivariable analyses (Data Supplement).
FIG 2.
Validation of the prognostic significance of lncScore in independent pediatric and adult data sets. Independent univariable analysis in both (A) validation 1 and (B) validation 2 sets showed a comparable distribution of lncScores to that observed in the training set, with median scores nearly centered on 0 (red, positive lncScore; blue, negative lncScore; boxed region in the left-hand panels indicates interquartile range and median). Estimates of the HR for OS and EFS were comparable with those seen in the training set and remained highly significant (P < .001). (C) Application of lncScores to an independent adult RNA-seq data set demonstrated a similar distribution across samples, with slightly lower percentage of positive cases. Comparison of adult cases with positive versus negative lncScore shows a significant difference in both OS (HR, 3.21; 95% CI, 1.94 to 5.3; P < .001) and EFS (HR, 2.36; 95%, CI, 1.49 to 3.75; P < .001). AML, acute myeloid leukemia; EFS, event-free survival; HR, hazard ratio; OS, overall survival.
To establish generalizability, we tested whether the lncScore was predictive of adult AML outcomes in a technically and clinically distinctive data set. Evaluation of lncScores in 96 adult AML cases also showed a significant association for EFS (HR, 2.36; 95% CI, 1.49 to 3.75; P < .001) and OS (HR, 3.21; 95% CI, 1.94 to 5.3; P < .001; Fig 2C and Data Supplement). These results suggest that the lncScore is a robust and reproducible predictor of outcome with potential relevance across the AML age spectrum.
lncScore Is an Independent Predictor of pAML Outcome With Accuracy Comparable With Established Initial Risk Markers
To examine whether lncScore provides independent prognostic information, we performed multivariable analysis on OS and EFS from study enrollment and on RR from end induction. Including those factors defined at diagnosis that were identified as significant by univariable Cox regression in both training and validation sets (lncScore, CFM risk class, and presenting WBC count; Data Supplement), lncScore retained prognostic significance in the training set (HR of 2.07, 1.84, and 1.82 for OS, EFS, and RR from EOI1, respectively; all at P < .001; Data Supplement). Concordance statistics exceeded 66% for each survival metric (OS, 0.7; EFS, 0.667; RR, 0.679). Comparable results were observed in the combined validation group (HR OS, 1.75; P = .001; C-stat, 0.672; EFS, 1.56; P = .001; C-stat, 0.671; RR, 1.67; P = .004; C-stat, 0.689). Estimates of the concordance difference between submodels containing CFM classification compared with lncScore showed negligible differences, with large P-values for these comparisons in both validation and training sets. The full model containing lncScore, CFM, and WBC count slightly outperformed either submodel, but this effect was only statistically significant against the CFM comparison in the training group and lncScore comparison in validation set (Data Supplement). These results suggested that the lncScore provides comparable accuracy to traditional pretherapy classification metrics while having potential to add additional prognostic information.
lncScore Is Informative in Heterogeneous Cytogenetic Subgroups
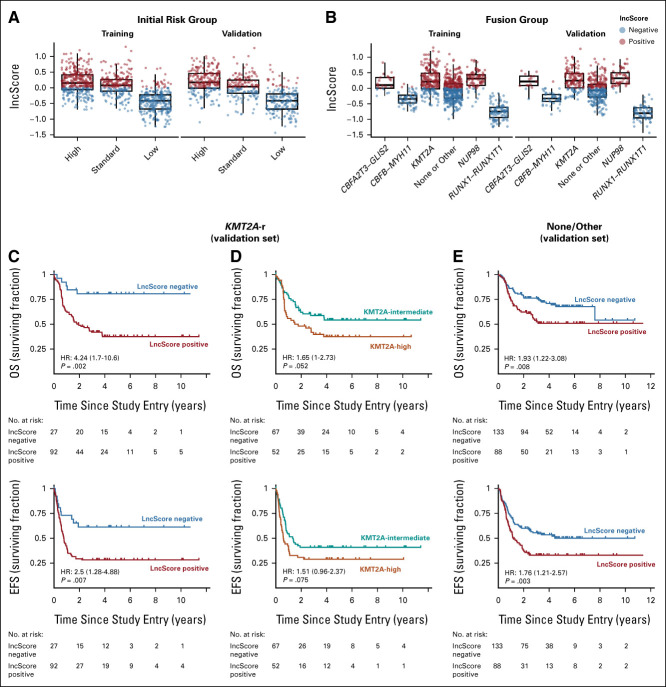
To determine the distribution and prognostic contribution of lncScores across key presenting features, we analyzed lncScores in the context of upfront CFM classification and key fusions: CBFA2T3-GLIS2, CBFB-MYH11, KMT2A-r, NUP98-r, RUNX1-RUNX1T1, and those lacking a cytogenetic change or with rare fusions (none/other). Positive lncScores were most common among CFM high- and intermediate-risk group patients but infrequent among patients with low-risk disease (Fig 3A). lncScores similarly tracked with cytogenetic markers: few positive scores were detected among favorable translocations, but predominantly positive scores were seen in unfavorable groups such as NUP98-rearranged and CBFA2T3-GLIS2 fusions. By contrast, lncScores were widely distributed in heterogeneous subgroups including cases with KMT2A fusions and those with rare or lacking a detectable fusion (none/other; Fig 3B).
FIG 3.
lncScore is informative in heterogeneous subgroups. (A) In both the training and validation pediatric data sets, unfavorable lncScores were most common in high initial-risk cases, mixed in intermediate-risk, but infrequent among low-risk cases. (B) When separated by fusion class, cases with higher-risk fusions including CBFA2T3-GLIS2 and NUP98 fusions showed nearly exclusively positive lncScores, whereas favorable-risk translocations showed the converse. Heterogeneous fusion groups including KMT2A-r and those with rare or lacking an identifiable fusion showed high levels of variability in lncScores. (C) Outcome differences in the KMT2A-r validation set were dramatic, with 81% versus 37% and 61% versus 28% 5-year survival for OS and EFS respectively (HR, 4.25; 95% CI, 1.7 to 10.6; P < .001; and 2.51; 95% CI, 1.29 to 4.89; P < .001). (D) Outcomes for these KMT2A-r validation set patients as stratified by fusion partner risk group are illustrated as contrast. (E) Cases with rare recurrent fusions or lacking a detectable fusion also showed significant outcome differences when stratified by lncScore (5-year OS, 67.7% v 48.4%; HR, 1.9; P = .002; 5-year EFS, 50.6% v 31.9%; HR, 1.8; P = .001). Training set cases with KMT2A-r or in the none/other fusion category showed similar findings, as illustrated in the Data Supplement. EFS, event-free survival; HR, hazard ratio; OS, overall survival.
We evaluated treatment outcomes in the KMT2A-rearranged and none/other groups to further examine the relationship between lncScore and subgroup outcomes. Survival analysis in KMT2A fusion cases in the validation cohort (N = 119) confirmed a marked separation observed in the training set (validation set 5-year OS, 81% ± 16% v 37% ± 10%; HR, 4.24; P = .002; 5-year EFS, 61% ± 19% v 28% ± 9%; HR, 2.5; P = .007; Figs 3C and 4A-4C Data Supplement) that was substantially better than the current standard for allocating to high- or intermediate-risk groups on the basis of fusion partner (Fig 3D and Data Supplement). Cases with rare fusions or lacking a fusion also showed a significant outcome association when stratified by lncScore (validation set 5-year OS, 68% ± 9% v 51% ± 11%; HR, 1.93; P = .008; 5-year EFS, 50% ± 10% v 33% ± 10%; HR, 1.76; P = .003; Fig 3E and Data Supplement). In aggregate, these data suggest that the lncScore may recapitulate prognostic information available from known high- and low-risk cytogenetic classes but adds additional information to the current standard classification model, particularly in heterogeneous groups presently classified as intermediate risk.
FIG 4.
Assessment of lncScores with postinduction prognostic factors. Outcomes in the validation set by FR category as defined by current (AAML1831) stratification guidelines show (A) 59.1% versus 38.7% 5-year DFS (HR, 2.16; 95% CI, 1.62 to 1.89; P < .001) and (B) 78.4% versus 46.1% for OS (HR, 3.19; 95% CI, 2.22 to 4.58; P < .001). Comparison of cases binarized by lncScore shows comparable results, (C) with a 5-year DFS of 65.7% versus 35% (HR, 2.44; 95% CI, 1.82 to 3.28; P < .001) and (D) with a 5-year OS of 82% versus 45.8% (HR, 3.41; 95% CI, 2.35 to 4.95; P < .001; Data Supplement). Patients with negative lncScore and absence of detectable end-induction MRD comprise over half of cases and show historically excellent outcomes, with (E) a DFS of 65% ± 8% and (F) an OS of 86% ± 6% in the validation cohort. Patients with one or both markers positive showed poor outcomes, with DFS ranging from 28% to 44% and OS from 20% to 52%. DFS, disease-free survival; EFS, event-free survival; FR, final risk; HR, hazard ratio; MRD, minimal residual disease; OS, overall survival.
Comparison and Integration of lncScore With Postinduction Prognostic Criteria
To test whether lncScores could replace or augment a modern stratification scheme, we compared lncScore predictions with updated FR determination. FR grouping within the context of current (AAML1831, ClinicalTrials.gov identifier: NCT04293562) and recent COG studies is a complex criterion incorporating cytologic and molecular classification at the time of presentation with induction response by assessment of MRD to determine length and intensity of consolidative therapy.31,40
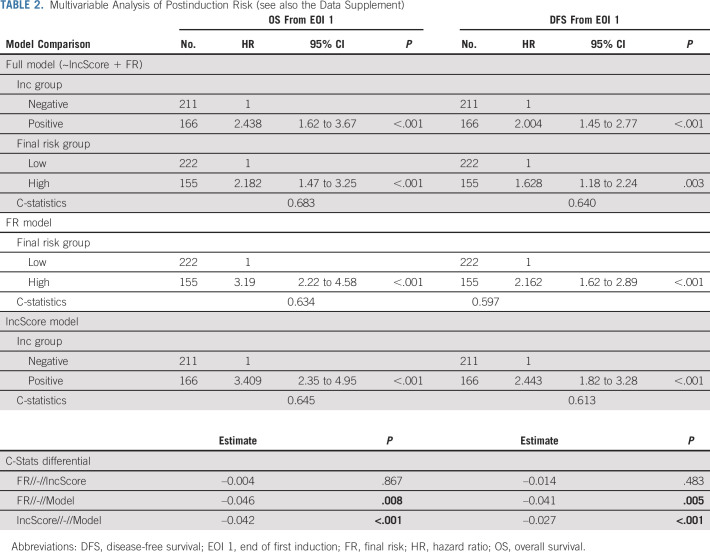
Because FR is established after induction, we assessed OS and DFS after first induction course in a multivariable model including FR and lncScore, again comparing with single-term submodels. lncScore and FR class were both significant at P < .001 in multivariable Cox models of training and validation sets for both outcome measures (Table 2; Figs 4A-4C; Data Supplement). Point estimates for HR were slightly higher by lncScore than FR for all but training set OS, with all CI showing substantial overlap. Comparison of single term submodels of lncScore versus FR in both training and validation groups showed similar trends, slightly favoring lncScore, but with nonsignificant concordance differential estimates (validation set HR for OS, 3.41 v 3.19; C-index diff, –0.004; P = .867; validation set DFS HR 2.44 v 2.16; C-index diff, –0.014; P = .483; training set OS, 3.13 v 4.21; C-index diff, –0.016; P = .7; training set DFS HR, 2.5 v 2.29; C-index diff, –0.028; P = .168). As with initial risk features, a full model containing FR and lncScore outperformed either FR or lncScore alone, with significant P-values in both possible concordance model comparisons for DFS and OS in the validation set (validation P-value range 0.008-<0.001; Table 2; Data Supplement). Together, these data suggest that lncScore, as a single assay, has comparable predictive performance to the currently used complex stratification scheme, while addition of lncScore to current standards outperforms either approach alone.
TABLE 2.
Multivariable Analysis of Postinduction Risk (see also the Data Supplement)
Since lncScore does not include the induction response information that is nested in FR class, we tested inclusion of MRD with lncScore for postinduction outcome prediction. lncScores remained highly significant in both training and validation sets by multivariable regression (validation HR for DFS and OS, 2.19 and 2.83, respectively; both P < .001; Data Supplement). The effect of MRD was significant but more modest (validation set DFS HR, 1.89; P < .001; OS HR, 2.5; P ≤ 0.001). The complete model including lncScore and MRD outperformed either factor alone, suggesting that inclusion of MRD indeed improves predictive accuracy of the lncScore. Practically, this approach defined a large group of patients (approximately 50%) with both negative lncScore and without MRD demonstrating historically excellent outcomes in pAML (validation set OS and DFS of 85% and 65%, respectively), while those with either or both markers positive showed markedly inferior outcomes (Figs 4E and 4F; Data Supplement).
DISCUSSION
In this study, we identify and validate a 37-gene lncRNA-based classification system that improves upon state-of-the art predictive strategies to better differentiate pAML into lower- and higher-risk categories at risk for treatment failure. Our findings generally corroborate the results from studies of adult patients with AML in illustrating the relevance of lncRNA expression to outcome prediction.22-27 While developed for pediatric disease, our study also suggests the lncScore may be predictive beyond pediatric AML to adult disease, although the adult sample examined here is limited in size.
Notably, none of the lncRNAs identified in the signature have been previously implicated in AML outcomes. Several factors may explain this discrepancy including unique biological differences of pediatric versus adult AML (including a substantially higher proportion of oncofusion-driven disease),5 the relatively large size of the training data set used here, the sequencing chemistry—which used stranded sequencing after rRNA depletion rather than poly(A) selection, allowing for the detection of both polyadenylated (poly-A) and non–poly-A lncRNAs41 as well as antisense transcripts—and generally high coverage transcriptome sequencing, potentially allowing for better detection of weakly expressed lncRNAs. As in prior lncRNA studies, it is challenging to determine on the basis of available data whether individual lncRNA components serve as direct mediators of therapy resistance or are passengers marking a broader transcriptional milieu of resistance.24 Consistent with the generally incomplete state of lncRNA annotation, many of the lncScore transcripts presently lack any functional annotation. Several, however, have potentially important direct roles in AML biology through WNT42 signaling, HOXA cluster expression,43 and stem-cell maintenance.44 The marked improvement in predicting KMT2A-r case outcomes on the basis of lncScore, compared with fusion partner identity, is likely partially due to passenger effects, since transformed cell of origin has been demonstrated to significantly influence transcriptional patterns in KMT2A-r leukemia.45 On the basis of the dramatic differences between these two predictive methods in KMT2A-r pAML, it seems plausible that lncScore encodes partner-gene and cell-of-origin information that is unavailable from KMT2A partner definition alone.
Several features of the lncScore make it favorable for future development and clinical application. Our selection of transcripts overexpressed relative to normal bone marrow leaves the assay less susceptible to sensitivity issues in partially diluted marrow samples. In four cohorts (training, validation 1, validation 2, and adult), the median lncScore score lay extremely close to 0, thus motivating our selection of 0 as an absolute cutpoint for dichotomization. This approach obviates the need for large reference data sets or concurrent controls for median definitions.
In addition to improving on state-of-the art prognostics, this study suggests that the lncScore offers comparable performance to modern stratification methods in a single assay. Both findings are of potential importance since prognostic classification schemes presently used in pAML leave room for predictive strengthening while posing substantial practical hurdles to execution; successful stratification is presently a logistical and interpretive challenge. For example, the schema currently used by COG requires bone marrow testing for numerous targeted gene assessments by Sanger sequencing and fragment length analysis technology, g-banded cytogenetics, interphase fluorescent in situ hybridization against numerous targets, immunophenotyping, as well as targeted DNA and RNA short-read sequencing to define rare fusion partners, potentially supplemented with directed quantitative polymerase chain reaction assays. This complexity is costly and burdensome even in high-volume centers and in patients with ample bone marrow for testing but becomes particularly problematic where limited marrow is available, in lower-volume centers that infrequently encounter patients with pAML, or in settings of limited medical resources. Hence, there is motivation to improve and simplify predictive testing. The inclusion of lncScore, as a standalone assay, or in combination with other sequencing analyses, is fully compatible with a broader move toward next-generation sequencing as an upfront diagnostic modality in AML.46,47
Jason E. Farrar
Research Funding: Novartis
Megan Othus
Consulting or Advisory Role: Glycomimetics, Cascadia Labs, Merck, Daiichi Sankyo, Biosight
Other Relationship: Celgene, Glycomimetics
Tiffany Hylkema
Stock and Other Ownership Interests: Moderna Therapeutics, Quest Diagnostics
Sneha Challa
Employment: Fred Hutchinson Cancer Research Center
Research Funding: Fred Hutchinson Cancer Research Center
Timothy I. Shaw
Patents, Royalties, Other Intellectual Property: Timothy Shaw reports a patent application in CAR T cell therapy US20220267425A1, Timothy Shaw reports a pending patent application on software for survival analysis
Timothy J. Triche
Patents, Royalties, Other Intellectual Property: Inventor on patent licensed to Adela Bio (Inst)
Alan S. Gamis
Consulting or Advisory Role: Novartis
Richard Aplenc
Expert Testimony: Vorys, Sater, Seymour, and Pease LLP
E. Anders Kolb
This author is a member of the Journal of Clinical Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Travel, Accommodations, Expenses: Roche/Genentech
Derek L. Stirewalt
Stock and Other Ownership Interests: Yatiri Bio
No other potential conflicts of interest were reported.
See accompanying article on page 3059
PRIOR PRESENTATION
Presented at the American Society of Hematology meeting, Orlando, FL, December 6-10, 2019.
SUPPORT
Research reported in this publication was supported by the Children's Oncology Group and the SWOG Cancer Research Network through the National Cancer Institute of the National Institutes of Health under award numbers U10CA180899, U10CA180888, U10CA180886, U10CA180819, U24CA196173, as well as R01CA114563 (S.M.), R01CA160872 (D.L.S.), R01CA190661 (D.L.S.), P20GM121293 (J.E.F.), Target Pediatric AML (S.M., E.A.K.), the Andrew McDonough B+ Foundation (S.M.), St Baldrick's Foundation (J.E.F., B.H., S.M.), the COG Foundation (S.M., J.E.F.), the Rally Foundation (J.E.F.), the Jeffrey Pride Foundation (J.E.F.), Project Stella (S.M.), Hyundai Hope on Wheels (S.M., J.E.F.), Alex's Lemonade Stand Foundation (S.M.), and the Arkansas Biosciences Institute (J.E.F.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
DATA SHARING STATEMENT
The covariates, outcomes, and lncRNA expression data supporting the development of this biomarker are included in the supplementary data files. The complete transcriptomic data are available in dbGaP as outlined in the Data Supplement methods section.
AUTHOR CONTRIBUTIONS
Conception and design: Jason E. Farrar, Jenny L. Smith, Benjamin J. Huang, Alan S. Gamis, E. Anders Kolb, Soheil Meshinchi
Financial support: Jason E. Farrar, Soheil Meshinchi
Administrative support: Jason E. Farrar, Soheil Meshinchi
Provision of study materials or patients: Richard Aplenc, Soheil Meshinchi
Collection and assembly of data: Jason E. Farrar, Jenny L. Smith, Megan Othus, Rhonda Ries, Tiffany Hylkema, Era L. Pogosova-Agadjanyan, Amanda Leonti, Alan S. Gamis, Richard Aplenc, Todd A. Alonzo
Data analysis and interpretation: Jason E. Farrar, Jenny L. Smith, Benjamin J. Huang, Yi-Cheng Wang, Rhonda Ries, Era L. Pogosova-Agadjanyan, Sneha Challa, Timothy I. Shaw, Alan S. Gamis, Richard Aplenc, E. Anders Kolb, Xiaotu Ma, Derek L. Stirewalt, Todd A. Alonzo, Soheil Meshinchi
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Long Noncoding RNA Expression Independently Predicts Outcome in Pediatric Acute Myeloid Leukemia
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jason E. Farrar
Research Funding: Novartis
Megan Othus
Consulting or Advisory Role: Glycomimetics, Cascadia Labs, Merck, Daiichi Sankyo, Biosight
Other Relationship: Celgene, Glycomimetics
Tiffany Hylkema
Stock and Other Ownership Interests: Moderna Therapeutics, Quest Diagnostics
Sneha Challa
Employment: Fred Hutchinson Cancer Research Center
Research Funding: Fred Hutchinson Cancer Research Center
Timothy I. Shaw
Patents, Royalties, Other Intellectual Property: Timothy Shaw reports a patent application in CAR T cell therapy US20220267425A1, Timothy Shaw reports a pending patent application on software for survival analysis
Timothy J. Triche
Patents, Royalties, Other Intellectual Property: Inventor on patent licensed to Adela Bio (Inst)
Alan S. Gamis
Consulting or Advisory Role: Novartis
Richard Aplenc
Expert Testimony: Vorys, Sater, Seymour, and Pease LLP
E. Anders Kolb
This author is a member of the Journal of Clinical Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Travel, Accommodations, Expenses: Roche/Genentech
Derek L. Stirewalt
Stock and Other Ownership Interests: Yatiri Bio
No other potential conflicts of interest were reported.
REFERENCES
- 1.Zwaan CM, Meshinchi S, Radich JP, et al. : FLT3 internal tandem duplication in 234 children with acute myeloid leukemia: Prognostic significance and relation to cellular drug resistance. Blood 102:2387-2394, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Brown P, McIntyre E, Rau R, et al. : The incidence and clinical significance of nucleophosmin mutations in childhood AML. Blood 110:979-985, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard JA, Alonzo TA, Gerbing RB, et al. : Prevalence and prognostic significance of KIT mutations in pediatric patients with core binding factor AML enrolled on serial pediatric cooperative trials for de novo AML. Blood 115:2372-2379, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tarlock K, Meshinchi S: Pediatric acute myeloid leukemia: Biology and therapeutic implications of genomic variants. Pediatr Clin North America 62:75-93, 2015 [DOI] [PubMed] [Google Scholar]
- 5.Bolouri H, Farrar JE, Triche T, Jr, et al. : The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 24:103-112, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loken MR, Alonzo TA, Pardo L, et al. : Residual disease detected by multidimensional flow cytometry signifies high relapse risk in patients with de novo acute myeloid leukemia: A report from Children's Oncology Group. Blood 120:1581-1588, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eidenschink Brodersen L, Alonzo TA, Menssen AJ, et al. : A recurrent immunophenotype at diagnosis independently identifies high-risk pediatric acute myeloid leukemia: A report from Children's Oncology Group. Leukemia 30:2077-2080, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yagi T, Morimoto A, Eguchi M, et al. : Identification of a gene expression signature associated with pediatric AML prognosis. Blood 102:1849-1856, 2003 [DOI] [PubMed] [Google Scholar]
- 9.Bullinger L, Dohner K, Bair E, et al. : Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med 350:1605-1616, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Ross ME, Mahfouz R, Onciu M, et al. : Gene expression profiling of pediatric acute myelogenous leukemia. Blood 104:3679-3687, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Marcucci G, Maharry K, Whitman SP, et al. : High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B Study. J Clin Oncol 25:3337-3343, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Ng SWK, Mitchell A, Kennedy JA, et al. : A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 540:433-437, 2016 [DOI] [PubMed] [Google Scholar]
- 13.Huang BJ, Smith JL, Farrar JE, et al. : Integrated stem cell signature and cytomolecular risk determination in pediatric acute myeloid leukemia. Nat Commun 13:5487, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim EL, Trinh DL, Ries RE, et al. : MicroRNA expression-based model indicates event-free survival in pediatric acute myeloid leukemia. J Clin Oncol 35:3964-3977, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapranov P, Cheng J, Dike S, et al. : RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316:1484-1488, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Yao RW, Wang Y, Chen LL: Cellular functions of long noncoding RNAs. Nat Cell Biol 21:542-551, 2019 [DOI] [PubMed] [Google Scholar]
- 17.Schmitt AM, Chang HY: Long noncoding RNAs in cancer pathways. Cancer Cell 29:452-463, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarzer A, Emmrich S, Schmidt F, et al. : The non-coding RNA landscape of human hematopoiesis and leukemia. Nat Commun 8:218, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Dominguez JR, Lodish HF: Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood 130:1965-1975, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paralkar VR, Weiss MJ: Long noncoding RNAs in biology and hematopoiesis. Blood 121:4842-4846, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo M, Jeong M, Sun D, et al. : Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell; 16:426-438, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garzon R, Volinia S, Papaioannou D, et al. : Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A 111:18679-18684, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mer AS, Lindberg J, Nilsson C, et al. : Expression levels of long non-coding RNAs are prognostic for AML outcome. J Hematol Oncol 11:52, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck D, Thoms JAI, Palu C, et al. : A four-gene LincRNA expression signature predicts risk in multiple cohorts of acute myeloid leukemia patients. Leukemia 32:263-272, 2018 [DOI] [PubMed] [Google Scholar]
- 25.Tsai CH, Yao CY, Tien FM, et al. : Incorporation of long non-coding RNA expression profile in the 2017 ELN risk classification can improve prognostic prediction of acute myeloid leukemia patients. EBioMedicine 40:240-250, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bill M, Papaioannou D, Karunasiri M, et al. : Expression and functional relevance of long non-coding RNAs in acute myeloid leukemia stem cells. Leukemia 33:2169-2182, 2019 [DOI] [PubMed] [Google Scholar]
- 27.Papaioannou D, Nicolet D, Ozer HG, et al. : Prognostic and biologic relevance of clinically applicable long noncoding RNA profiling in older patients with cytogenetically normal acute myeloid leukemia. Mol Cancer Ther 18:1451-1459, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lange BJ, Smith FO, Feusner J, et al. : Outcomes in CCG-2961, a children's oncology group phase 3 trial for untreated pediatric acute myeloid leukemia: A report from the children's oncology group. Blood 111:1044-1053, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper TM, Franklin J, Gerbing RB, et al. : AAML03P1, a pilot study of the safety of gemtuzumab ozogamicin in combination with chemotherapy for newly diagnosed childhood acute myeloid leukemia: A report from the Children's Oncology Group. Cancer 118:761-769, 2012 [DOI] [PubMed] [Google Scholar]
- 30.Gamis AS, Alonzo TA, Meshinchi S, et al. : Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: Results from the randomized phase III Children's Oncology Group trial AAML0531. J Clin Oncol 32:3021-3032, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aplenc R, Meshinchi S, Sung L, et al. : Bortezomib with standard chemotherapy for children with acute myeloid leukemia does not improve treatment outcomes: A report from the Children's Oncology Group. Haematologica 105:1879-1886, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Godwin JE, Kopecky KJ, Head DR, et al. : A double-blind placebo-controlled trial of granulocyte colony-stimulating factor in elderly patients with previously untreated acute myeloid leukemia: A Southwest oncology group study (9031). Blood 91:3607-3615, 1998 [PubMed] [Google Scholar]
- 33.Anderson JE, Kopecky KJ, Willman CL, et al. : Outcome after induction chemotherapy for older patients with acute myeloid leukemia is not improved with mitoxantrone and etoposide compared to cytarabine and daunorubicin: A Southwest Oncology Group study. Blood 100:3869-3876, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Hiddemann W, Kern W, Schoch C, et al. : Management of acute myeloid leukemia in elderly patients. J Clin Oncol 17:3569-3576, 1999 [DOI] [PubMed] [Google Scholar]
- 35.Petersdorf SH, Kopecky KJ, Slovak M, et al. : A phase 3 study of gemtuzumab ozogamicin during induction and postconsolidation therapy in younger patients with acute myeloid leukemia. Blood 121:4854-4860, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrell FE, Jr., Califf RM, Pryor DB, et al. : Evaluating the yield of medical tests. JAMA 247:2543-2546, 1982 [PubMed] [Google Scholar]
- 37.Uno H, Cai T, Pencina MJ, et al. : On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med 30:1105-1117, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrar JE, Schuback HL, Ries RE, et al. : Genomic profiling of pediatric acute myeloid leukemia reveals a changing mutational landscape from disease diagnosis to relapse. Cancer Res 76:2197-2205, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maxson JE, Ries RE, Wang YC, et al. : CSF3R mutations have a high degree of overlap with CEBPA mutations in pediatric AML. Blood 127:3094-3098, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper TM, Ries RE, Alonzo TA, et al. : Revised risk stratification criteria for children with newly diagnosed acute myeloid leukemia: A report from the Children's Oncology Group. Blood 130:407, 2017 [Google Scholar]
- 41.St Laurent G, Wahlestedt C, Kapranov P: The landscape of long noncoding RNA classification. Trends Genet 31:239-251, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ntini E, Louloupi A, Liz J, et al. : Long ncRNA A-ROD activates its target gene DKK1 at its release from chromatin. Nat Commun 9:1636, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sessa L, Breiling A, Lavorgna G, et al. : Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. RNA 13:223-239, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng SY, Johnson R, Stanton LW: Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J 31:522-533, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krivtsov AV, Figueroa ME, Sinha AU, et al. : Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia 27:852-860, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arindrarto W, Borras DM, de Groen RAL, et al. : Comprehensive diagnostics of acute myeloid leukemia by whole transcriptome RNA sequencing. Leukemia 35:47-61, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duncavage EJ, Schroeder MC, O'Laughlin M, et al. : Genome sequencing as an alternative to cytogenetic analysis in myeloid cancers. N Engl J Med 384:924-935, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The covariates, outcomes, and lncRNA expression data supporting the development of this biomarker are included in the supplementary data files. The complete transcriptomic data are available in dbGaP as outlined in the Data Supplement methods section.