Abstract
The Asian mouse Mus castaneus is resistant to infection by the polytropic mink cell focus-inducing (MCF) subgroup of murine leukemia viruses (MuLVs). Genetic crosses showed this recessive resistance to be governed by a single gene that maps at or near the gene encoding the polytropic viral receptor, Rmc1. To investigate this resistance, we mated M. castaneus with mice carrying the wild mouse Sxv variant of the Rmc1 receptor that allows infection by xenotropic as well as polytropic virus. Unlike other F1 hybrids of M. castaneus, these F1 mice were resistant to both xenotropic and polytropic classes of MuLVs. Analysis of backcrossed progeny of the F1 hybrids mated to Sxv mice indicates that resistance is due to inheritance of two M. castaneus genes. Cells from individual backcross mice were also examined for cell surface antigen by fluorescence-activated cell sorter analysis with monoclonal antibodies reactive with xenotropic or MCF virus env glycoproteins. A correlation was observed between virus resistance and antigen, suggesting that virus resistance is due to expression of endogenous viral envelope genes that interfere with infection by exogenous virus. Since the inbred strain Rmc1 receptor remains functional in the presence of these M. castaneus genes, and since M. castaneus contains multiple copies of xenotropic MuLV env genes, we suggest that these resistance genes control expression of xenotropic env glycoprotein that interferes with exogenous virus in cells containing the Sxv variant of Rmc1.
The various host range subgroups of murine leukemia viruses (MuLVs) infect cells by interacting with distinct cell surface receptors. Ecotropic viruses, which infect only mouse cells, use as their receptor the cationic amino acid transporter CAT1 (1). Amphotropic MuLVs employ Pit2, and the recombinant amphotropic virus 10A1 can use either Pit2 or Pit1 (Glvr1) (17), both of which function as phosphate transporters. Mice also have a fourth receptor, termed the Rmc1 receptor, used by polytropic mink cell focus-inducing (MCF) viruses (11).
Some differences in susceptibility to exogenous MuLV infection have been noted among the inbred strains and wild mouse species, and these differences are mediated at the level of the virus-cell receptor interaction. Resistance of Mus dunni cells to Moloney ecotropic virus has been attributed to allelic variation of the ecotropic CAT1 receptor (6). Resistance to all ecotropic viruses in Mus castaneus, Mus museulus molossinus, and Lake Casitas mice of California is due to the presence of the Fv4 endogenous ecotropic env, expression of which interferes with binding of exogenous virus (22). Although all common strains of laboratory mice are resistant to xenotropic viruses, one allelic variant of the MCF Rmc1 receptor, termed Sxv, can also function as an inefficient receptor for xenotropic MuLVs in many wild mouse species (12). Two different receptor-mediated resistance phenomena have been described for polytropic MCF viruses. First, DBA/2 and some CBA mouse substrains are partially resistant to MCF virus due to the presence of a dominant allele at Rmcf that is thought to control expression of endogenous MCF virus envelope glycoprotein (2, 10, 21). More recently, we have shown that the Asian mouse M. castaneus is resistant to infection with MCF MuLVs. This resistance is controlled by a single recessive gene, and genetic studies have mapped this gene to distal chromosome 1 at or very near the position of the Rmc1 receptor (16). This recessive inheritance together with the map location suggested that M. castaneus carries an allelic variation of Rmc1 that lacks receptor function.
In order to further characterize the phenotypic variants that have been attributed to the Rmc1 receptor, we have now done additional genetic studies to examine the function of the M. castaneus receptor in crosses with mice carrying the Sxv variant of Rmc1. Analysis of the progeny of these genetic crosses indicates that M. castaneus contains additional genetic factors that interact with the Sxv receptor to produce the resistance phenotype.
MATERIALS AND METHODS
Viruses, cells, and virus assays.
The viruses used in the infectivity assays were originally obtained from J. Hartley (National Institute of Allergy and Infectious Diseases, Bethesda, Md.). Three MCF virus isolates (Akv1 MCF, AKR13 MCF, and Moloney MCF-HIX [5, 8]) and one amphotropic virus (4070A [9]), were used. Wild mouse xenotropic virus was isolated from M. castaneus mice following treatment of spleen cells with bacterial lipopolysaccharide and 5-iododeoxyuridine as previously described (14). All viruses were grown in mink lung cells (Mv-1-Lu; ATCC-CCL64).
Primary cultures of tail biopsy tissue were established from 7- to 20-day old mice as previously described (15). Subconfluent cultures were infected with 0.2 ml of each virus dilution in the presence of Polybrene (4 μg/ml; Aldrich, Milwaukee, Wis.). Cultures from each mouse were infected with amphotropic virus, Moloney MCF-HIX, and one of two other MCF virus isolates. The culture medium was changed the next day, and the cells were maintained for 4 to 5 days and then lethally irradiated. MCF MuLV-infected cultures were overlaid with 2 × 105 mink lung cells or 6 × 105 mink S+L− cells (18), and amphotropic MuLV-infected cultures were overlaid with mink S+L− cells. Cultures were examined for foci after 6 to 8 days.
Mice.
CAST/Ei mice were obtained from The Jackson Laboratory, Bar Harbor, Maine. DBA/2N and NFS/N mice were originally obtained from the Small Animal Section, National Institutes of Health, Bethesda, Md. M. castaneus mice were obtained from random-bred colonies maintained at PerImmune, Rockville, Md. (National Cancer Institute contract N01-CB-21075), and kindly provided by M. Potter (National Cancer Institute, Bethesda, Md.).
Two congenic stocks were developed in our laboratory on the NFS/N genetic background. NFS/N-Bxv1/Bxv1 mice carry the Bxv1 locus of BALB/c, and NFS/N-Sxv/Sxv mice carry the Sxv locus of M. musculus domesticus (formerly Mus praetextus) (12). Both congenic stocks were constructed by backcrossing to NFS/N for 10 generations, followed by brother-sister mating for over 10 years.
F1 hybrids and backcross mice were bred in our laboratory. Specifically, M. castaneus males and females were bred with various inbred and congenic stocks to obtain F1 hybrids. Backcross progeny were generated by mating (NFS/N-Sxv/Sxv × M. castaneus)F1 mice with NFS/N-Sxv/Sxv mice.
Antibodies.
Seven antibodies were used to screen for cell surface retroviral envelope glycoprotein. The derivation and description of these antibodies have been reported previously, and all were kindly provided by John Portis (Rocky Mountain Laboratory, National Institute of Allergy and Infectious Diseases, Hamilton, Mont.). The monoclonal antibodies (MAbs) and their reactivities are listed in Table 1.
TABLE 1.
Reactivity of anti-env MAbs with M. castaneus xenotropic virus
| Antibody | Reactivity | Reference | Reactivity with M. castaneus xenotropic MuLVa |
|---|---|---|---|
| 83A25 | Most MuLVs | 7 | + |
| 24-6 | Most xenotropic and some MCF viruses | 20 | + |
| 603 | Some xenotropic viruses | 19 | + |
| 19-1 | Some xenotropic viruses | 20 | + |
| 18-6 | Some xenotropic and MCF viruses | 20 | − |
| 613 | Some xenotropic viruses | 19 | − |
| 514 | MCF viruses | 4 | − |
MAbs were tested by fluorescence-activated cell sorter analysis for reactivity with mink lung cells and mink cells infected with M. castaneus xenotropic virus. For MAbs identified as positive, mean fluorescence with infected cells was increased 3- to 20-fold.
Flow cytometry assays for viral env.
Cell surface env was detected by staining approximately 106 trypsinized cells with 100 μl of antibody to MuLV env diluted 1:1,000 in Hanks buffered saline solution with 0.1% bovine serum albumin and 0.1% sodium azide for 30 min on ice. After being washed, the cells were incubated with 40 μl of a 1:100 dilution of rat anti-mouse immunoglobulin G conjugated to fluorescein isothiocyanate (Life Technologies, Gaithersburg, Md.) for 30 min on ice. The cells were washed two or three times and then analyzed with an EPICS profile cytometer (Coulter, Hialeah, Fla.).
RESULTS AND DISCUSSION
Infectivity of M. castaneus and F1 hybrid mice with nonecotropic virus.
Previous studies had demonstrated that the genes encoding the wild mouse susceptibility to xenotropic virus (Sxv) (12) and the M. castaneus resistance to polytropic MCF virus (16) map to the site on distal chromosome 1 that encodes the cell surface receptor for polytropic MCF virus (11). The MCF virus resistance of M. castaneus was determined to be recessive in crosses with various inbred strains (16). In contrast, the Sxv-mediated susceptibility to xenotropic virus characteristic of many wild mouse species is dominant in crosses with the common inbred strains (12). These previous observations predict that F1 hybrids between M. castaneus and Sxv-bearing mice would be susceptible to both polytropic and xenotropic MuLVs due to the presence of the dominantly expressed, fully functional Sxv receptor.
We tested this prediction by mating M. castaneus with a congenic stock bearing the wild mouse Sxv allele on the NFS/N strain genetic background. Tail cultures established from individual F1 mice were tested for susceptibility to xenotropic, polytropic, and amphotropic virus (Table 2). These F1 hybrids were susceptible to amphotropic virus to the same extent as the parental mice. However, these cells were resistant to infection by MCF virus as well as xenotropic MuLVs. Cell cultures established from five different F1 mice consistently showed resistance to both classes of virus. Identical results were obtained for F1 mice produced by using M. castaneus stocks obtained from two different sources (PerImmune and The Jackson Laboratory). In contrast, parental NFS/N-Sxv/Sxv cells were susceptible to both xenotropic and MCF MuLVs, and inbred NFS/N-Bxv1/Bxv1 and DBA/2N mice and their F1 progeny with M. castaneus were susceptible only to MCF virus, although the titers were reduced in DBA/2N mice due to the presence of the Rmcf resistance allele. These results indicate that the F1 progeny between M. castaneus and Sxv mice is phenotypically equivalent to the virus-resistant parent. The different results obtained for the F1 mice with NFS/N-Sxv/Sxv and NFS/N-Bxv1/Bxv1 mice confirm the role of the receptor in this resistance.
TABLE 2.
Virus susceptibility of parental mice and F1 hybrids
| Micea | Log10 virus titer
|
||
|---|---|---|---|
| MCF | Xenotropic | Amphotropic | |
| Parental | |||
| M. castaneus | NDb | ND | 4.1 |
| NFS/N-Bxv1/Bxv1 | 3.8 | ND | 3.8 |
| DBA/2N | 2.2 | ND | 4.0 |
| NFS/N-Sxv/Sxv | 4.0 | 1.5 | 4.2 |
| Hybrids | |||
| (DBA/2N × M. castaneus)F1 | 2.1 | ND | 3.9 |
| NFS/N-Bxv1/Bxv1 × M. castaneus)F1 | 3.9 | ND | 3.9 |
| NFS/N-Sxv/Sxv × NFS/N)F1 | 4.0 | 1.7 | 4.4 |
| NFS/N-Sxv/Sxv × M. castaneus)F1 | ND | ND | 4.3 |
Titers of MCF and amphotropic virus were reported previously for NFS/N-Bxv1/Bxv1, DBA/2N, (NFS/N-Bxv1/Bxv1 × M. castaneus)F1, and M. castaneus (16).
ND, not detectable.
Analysis of backcross mice.
Because of the unexpected resistance of the (NFS/N-Sxv/Sxv × M. castaneus)F1 mice to MCF virus, additional crosses were done to determine the genetic basis of this phenotype. We mated the virus-resistant F1 mice with the susceptible parent, NFS/N-Sxv/Sxv. Tail cultures were established from 140 individual backcross mice and challenged with MCF and amphotropic virus. Mice were classed as susceptible to MCF virus if 100 or more foci were obtained and resistant if fewer than 20 foci were detected on mink S+L− cells. Seventeen of the 140 mice were excluded from further analysis due to poor replication of amphotropic virus or intermediate titers of MCF virus. The majority of the remaining backcross mice (87 of 123 [71%]) were typed as resistant to infection. This pattern is consistent with inheritance of two genes from the M. castaneus parent, either one of which is capable of producing the resistance phenotype (χ2 = 1.2, P = 0.27). These results suggest that M. castaneus contains two genes that block virus infection in crosses with Sxv mice.
Immunological detection of viral env gene expression.
The inheritance pattern observed in this backcross is reminiscent of other systems in which expression of specific endogenous retroviral env genes is responsible for resistance to virus of the same host range (Fv4 and Rmcf) (2, 10, 21, 22). This suggests that the two genes detected in this backcross may represent endogenous M. castaneus env genes. In order to provide more direct evidence for this model, we examined resistant and susceptible backcross progeny for expression of viral envelope.
We tested seven MAbs known to be reactive with MCF and/or xenotropic envelope glycoproteins. Because endogenous MLV env genes of M. castaneus are predominantly xenotropic (13), the MAbs were first tested by fluorescence-activated cell sorter analysis on mink cells infected with a virus of xenotropic host range isolated from the spleen of an M. castaneus mouse (Table 1). Four of the MAbs tested (83A25, 24-6, 19-1, and 603) identified cell surface antigen on the infected cells. All four of these MAbs were then tested to determine if they could identify viral glycoprotein on uninfected M. castaneus cells, but only 83A25 consistently reacted with these cells. With 83A25, mean fluorescence was, on average, fourfold higher than mean fluorescence obtained with the secondary antibody alone.
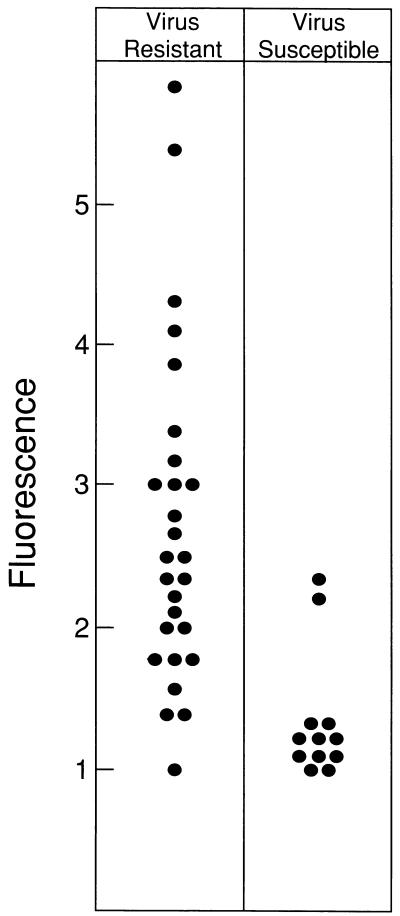
Duplicate cultures of tail biopsy tissue from 39 backcross mice were tested for virus susceptibility and reactivity with MAb 83A25. Of these mice, 27 were resistant to virus and 12 were susceptible. Cells from the different mice varied in their reactivity with 83A25, and comparison of this reactivity with virus susceptibility is shown in Fig. 1. Among the resistant mice, the mean fluorescence ratio (determined as fluorescence with 83A25 divided by control fluorescence with the secondary antibody alone) was 2.7 compared to 1.35 for the susceptible mice. Comparison of the levels of fluorescence of susceptible and resistant groups of backcross mice by the two-tailed t test, assuming equal variance shows a significant difference (P < 0.0001). The assumption of unequal variance does not alter this result. These results indicate a significant correlation between cell surface envelope glycoprotein and resistance in this backcross. These results are consistent with the proposal that this resistance may be due to expression of endogenous retroviral env genes in M. castaneus.
FIG. 1.
Flow cytometry analysis of nonecotropic env glycoprotein on the surface of cells cultured from tail biopsies taken from mice of the backcross (NFS/N-Sxv/Sxv × M. castaneus) × NFS/N-Sxv/Sxv. Fluorescence is given as the ratio of the mean fluorescence with antibody 83A25 over the mean fluorescence with only the secondary antibody.
The genetic basis of virus resistance in M. castaneus.
The common inbred strains and most wild mouse species are susceptible to infection with MCF virus, whereas M. castaneus is not. This virus susceptibility of common inbred strains is maintained in F1 hybrids with virus-resistant M. castaneus mice, and our previous genetic studies implicated the M. castaneus Rmc1 viral receptor allele in this resistance. In the present study, we demonstrate that Sxv receptor-mediated MCF and xenotropic MuLV susceptibility is extinguished in crosses with M. castaneus. Genetic analysis of this phenomenon demonstrates that this loss of susceptibility is due to inheritance of either of two dominantly expressed M. castaneus genes. Immunological assays further showed that resistance is associated with expression of MuLV env glycoprotein. This suggests a mechanism in which expression of one of two MuLV env genes blocks receptor interaction with exogenous virus. This interference mechanism resembles that proposed for the virus resistance genes Fv4 and Rmcf (2, 10, 21, 22). In this case, however, there are at least two alleles at Rmc1; the Sxv allele is susceptible to this env-mediated resistance, but the common inbred strain allele is not. This explains why the M. castaneus hybrids with one Rmc1 allele from common inbred strains are virus susceptible, whereas hybrids with the Sxv allele are not.
The nature of the interfering env is not suggested by the known reactivity of the MAb 83A25, which reacts with virus of the ecotropic, xenotropic, polytropic, and amphotropic host ranges (7). However, previous studies have indicated that the predominant MuLV env type in M. castaneus is xenotropic (13). These mice carry more than 50 xenotropic env genes, but only 1 or 2 MCF virus env copies. It is therefore tempting to speculate that the interfering env genes are xenotropic, since this would explain why the Sxv receptor is affected, but that of inbred strains is not.
It is not clear from these experiments whether M. castaneus carries a defective Rmc1 receptor, as suggested by our earlier studies (16), or whether these mice have a functional Rmc1 allele that is blocked by endogenous env gene expression. To determine whether M. castaneus mice contain a functional Rmc1 receptor, we are in the process of breeding the M. castaneus allele into a laboratory mouse genetic background to isolate it from the endogenous M. castaneus proviruses. If M. castaneus is found to contain a functional receptor, its inactivation by interference would suggest that it is more closely related to the Sxv receptor than to that of the inbred laboratory strains. This possibility is consistent with the fact that virtually all other Asian mice tested contain the Sxv receptor variant (12). These questions may be resolved soon, since several laboratories have recently reported cloning the Rmc1 receptor (5a, 23). The isolation of a molecular clone of the M. castaneus gene should make it possible to evaluate it directly for receptor function and to compare its sequence with that of the inbred strain and Sxv mouse alleles.
M. castaneus and its naturally occurring hybrid, M. musculus molossinus, are the only wild mouse species known to harbor infectious virus of both the ecotropic and xenotropic host ranges. These mice have multiple endogenous copies of both virus subgroups (13), and infectious virus of both subgroups can be isolated from these mice (3). Despite the damage that can be caused by these viruses through somatic and germ line mutation, M. castaneus mice show no ill effects, since these mice are resistant to both virus subgroups. As shown previously, M. castaneus mice contain Fv4 (22), which is responsible for resistance to ecotropic virus. The present study indicates that a similar mechanism is responsible for resistance to polytropic and/or xenotropic viruses. That the same type of interference mechanism is responsible for the resistance to multiple viruses in this species, as well as for virus resistance in other mouse and avian systems, suggests that it represents a common and powerful method of ensuring survival in the presence of persistent viral infection.
ACKNOWLEDGMENT
We thank Jonathan Silver for helpful discussions.
REFERENCES
- 1.Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 2.Bassin R H, Ruscetti S, Ali I, Haapala D K, Rein A. Normal DBA/2 mouse cells synthesize a glycoprotein which interferes with MCF virus infection. Virology. 1982;123:139–151. doi: 10.1016/0042-6822(82)90301-4. [DOI] [PubMed] [Google Scholar]
- 3.Chattopadhyay S K, Oliff A I, Linemeyer D L, Lander M R, Lowy D R. Genomes of murine leukemia viruses isolated from wild mice. J Virol. 1981;39:777–791. doi: 10.1128/jvi.39.3.777-791.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chesebro B, Britt W, Evans L, Wehrly K, Nishio J, Cloyd M. Characterization of monoclonal antibodies with murine leukemia viruses: use in analysis of strains of Friend MCF and Friend ecotropic murine leukemia virus. Virology. 1983;127:134–148. doi: 10.1016/0042-6822(83)90378-1. [DOI] [PubMed] [Google Scholar]
- 5.Cloyd M W, Hartley J W, Rowe W P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980;151:542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Cunningham, J. M. Personal communication.
- 6.Eiden M V, Farrell K, Warsowe J, Mahan L C, Wilson C A. Characterization of a naturally occurring ecotropic receptor that does not facilitate entry of all ecotropic murine retroviruses. J Virol. 1993;67:4056–4061. doi: 10.1128/jvi.67.7.4056-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans L H, Morrison R P, Malik F G, Portis J, Britt W J. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J Virol. 1990;64:6176–6183. doi: 10.1128/jvi.64.12.6176-6183.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischinger P J, Nomura S, Bolognesi D P. A novel murine oncornavirus with dual ecotropic and xenotropic properties. Proc Natl Acad Sci USA. 1975;77:531–535. doi: 10.1073/pnas.72.12.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartley J W, Rowe W P. Naturally occurring murine leukemia viruses in wild mice: characterization of a new “amphotropic” class. J Virol. 1976;19:19–25. doi: 10.1128/jvi.19.1.19-25.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartley J W, Yetter R A, Morse H C., III A mouse gene on chromosome 5 that restricts infectivity of mink cell focus-forming recombinant murine leukemia viruses. J Exp Med. 1983;158:16–24. doi: 10.1084/jem.158.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozak C A. Genetic mapping of a mouse chromosomal locus required for mink cell focus-forming virus replication. J Virol. 1983;48:300–303. doi: 10.1128/jvi.48.1.300-303.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak C A. Susceptibility of wild mouse cells to exogenous infection with xenotropic leukemia viruses: control by a single codominant locus on chromosome 1. J Virol. 1985;55:690–695. doi: 10.1128/jvi.55.3.690-695.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak C A, O’Neill R R. Diverse wild mouse origins of xenotropic, mink cell focus-forming and two types of ecotropic proviral genes. J Virol. 1987;61:3082–3088. doi: 10.1128/jvi.61.10.3082-3088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak C A, Rowe W P. Genetic mapping of xenotropic murine leukemia virus-inducing loci in five mouse strains. J Exp Med. 1980;152:219–228. doi: 10.1084/jem.152.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lander M R, Moll B, Rowe W P. A procedure for culture of cells from mouse tail biopsies: brief communication. J Natl Cancer Inst. 1978;60:477–478. [PubMed] [Google Scholar]
- 16.Lyu M S, Kozak C A. Genetic basis for resistance to polytropic murine leukemia viruses in the wild mouse species Mus castaneus. J Virol. 1996;70:830–833. doi: 10.1128/jvi.70.2.830-833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller D G, Miller A D. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peebles P T. An in vitro focus-induction assay for xenotropic murine leukemia viruses, feline leukemia virus C, and the feline-primate viruses RD114/CCC/M-7. Virology. 1975;67:288–291. doi: 10.1016/0042-6822(75)90427-4. [DOI] [PubMed] [Google Scholar]
- 19.Portis J L, McAtee F J. Monoclonal antibodies derived during graft-versus-host reaction. II. Antibodies detect unique determinants common to many MCF viruses. Virology. 1983;126:96–105. doi: 10.1016/0042-6822(83)90464-6. [DOI] [PubMed] [Google Scholar]
- 20.Portis J L, McAtee F J, Cloyd M W. Monoclonal antibodies to xenotropic and MCF murine leukemia viruses derived during the graft-versus-host reaction. Virology. 1982;118:181–190. doi: 10.1016/0042-6822(82)90331-2. [DOI] [PubMed] [Google Scholar]
- 21.Ruscetti S, Davis L, Feild J, Oliff A. Friend murine leukemia virus-induced leukemia is associated with the formation of mink cell focus-inducing viruses and is blocked in mice expressing endogenous mink cell focus-inducing xenotropic viral envelope genes. J Exp Med. 1981;154:907–920. doi: 10.1084/jem.154.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki S. Fv-4: a new gene affecting the splenomegaly induction by Friend leukemia virus. Jpn J Exp Med. 1975;45:473–478. [PubMed] [Google Scholar]
- 23.Tailor C S, Nouri A, Lee C G, Kozak C, Kabat D. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci USA. 1999;96:927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]