Abstract
The relative replicative fitness of human immunodeficiency virus type 1 (HIV-1) mutants selected by different protease inhibitors (PIs) in vivo was determined. Each mutant was compared to wild type (WT), NL4-3, in the absence of drugs by several methods, including clonal genotyping of cultures infected with two competing viral variants, kinetics of viral antigen production, and viral infectivity/virion particle ratios. A nelfinavir-selected protease D30N substitution substantially decreased replicative capacity relative to WT, while a saquinavir-selected L90M substitution moderately decreased fitness. The D30N mutant virus was also outcompeted by the L90M mutant in the absence of drugs. A major natural polymorphism of the HIV-1 protease, L63P, compensated well for the impairment of fitness caused by L90M but only slightly improved the fitness of D30N. Multiply substituted indinavir-selected mutants M46I/L63P/V82T/I84V and L10R/M46I/L63P/V82T/I84V were just as fit as WT. These results indicate that the mutations which are usually initially selected by nelfinavir and saquinavir, D30N and L90M, respectively, impair fitness. However, additional mutations may improve the replicative capacity of these and other drug-resistant mutants. Hypotheses based on the greater fitness impairment of the nelfinavir-selected D30N mutant are suggested to explain observations that prolonged responses to delayed salvage regimens, including alternate PIs, may be relatively common after nelfinavir failure.
Human immunodeficiency virus type 1 (HIV-1) evolution under chemotherapeutic selection pressure in vivo involves a complex interplay between an increasing magnitude of drug resistance and changes in viral replicative capacity. The first mutants to be selected when HIV-1 replication is ongoing during therapy are generally those with single-amino-acid substitutions that confer low-level resistance to protease inhibitors (PIs) or reverse transcriptase (RT) inhibitors. Several such resistant mutants with single-amino-acid substitutions selected in vivo have been shown to reduce viral replicative capacity in vitro in the absence of an inhibitor (2, 12, 27, 28, 36, 38, 50, 51, 57). Further replication during drug therapy may allow these mutants to accumulate additional drug-selected mutations. These may be selected in vivo because they increase the level of phenotypic resistance (10, 11, 29, 32, 39, 41, 46, 48) and/or because they cause compensatory increases in viral replicative capacity even in the absence of the drug (4, 12, 28, 35, 36, 43, 55, 56, 61).
Further understanding of this process may contribute to an explanation for prolonged responses to relatively late initiation of salvage regimens, including alternate PIs, in the majority of patients for whom nelfinavir therapy fails with a D30N nelfinavir-resistant mutant (6, 58, 62) but in a minority of patients for whom therapy with other PIs fails (10, 11, 14, 44, 62). These differences cannot be explained entirely by direct cross-resistance because neither the primary saquinavir-selected L90M mutation, the nelfinavir-selected because neither the primary saquinavirus-selected L90M mutation, the nelfinavir-selected D30N mutation, nor any single indinavir-selected mutation confers cross-resistance to other PIs in vitro as single mutations (10, 11, 29, 41, 45, 46, 48).
Effects of drug-selected mutations on the relative replicative capacities of HIV-1 mutants in vitro in the absence of an inhibitor have not been as well characterized as effects of mutations on susceptibility to different drugs. Methods for characterization of replicative capacity have included comparisons of enzyme catalytic activities or substrate-inhibitor binding (2, 7, 12, 23, 38, 39, 51, 56), kinetic analyses of virus production in cell cultures infected with one virus (2, 12, 28, 36, 55, 57), infectivity/virion particle ratios (57), and competition between two different virus variants in a single mixed culture (26, 27, 32, 43, 57). We used several of these methods, including a quantitative analysis of outcomes of competition between two viruses, to determine if protease mutants with single substitutions had impaired replication and if additional protease mutations accumulating during PI failure could fully restore replicative capacity to the level of that of wild-type virus.
Single-substitution mutants most commonly selected in vivo by either saquinavir (L90M) (29) or nelfinavir (D30N) (45, 46) were each studied in the genetic context of 63L or 63P. This allowed evaluation of possible compensation by L63P, which is of interest because it is a common polymorphism in pretreatment isolates (31, 33), it can be selected during failure of several PIs, and it does not alter PI susceptibility (11). L63P has been reported to improve the replicative capacity of the impaired double mutant V82F/I84V virus (36), and also, indinavir-selected M46L/L63P improves protease enzyme catalytic efficiency without changing indinavir binding (56). Two multiple-substitution protease mutants selected by indinavir in vivo were also studied. These mutants (M46I/L63P/V82T/I84V and L10R/M46I/L63P/V82T/I84V) evolved by accumulation of additional mutations in a V82T/I84V mutant genome during failure of indinavir therapy (11). We determined the relative fitness of the mutants selected by different PIs in vivo by three different experimental approaches to confirm that the conclusions were independent of a particular methodology.
MATERIALS AND METHODS
Cells and viruses.
Peripheral blood mononuclear cells (PBMCs) and the MT2 and P815 cell lines were used. PBMCs from HIV-seronegative blood donors were obtained by Ficoll-Hypaque density gradient centrifugation of citrate phosphate dextrose adenine (CPDA-1)-treated venous blood. The cells were stimulated with 2 μg of phytohemagglutinin (PHA; Difco Laboratories)/ml for 3 days and maintained in RPMI 1640 (Cellgro) supplemented with 20% fetal calf serum (Sigma), 2 mM l-glutamine (Cellgro), 10 mM HEPES buffer (Cellgro), interleukin-2 (IL-2) (100 U/ml), 50 μg of penicillin/ml and 50 μg of streptomycin/ml (both from Cellgro) prior to in vitro infection (these cells are referred to below as prestimulated PBMCs). An aliquot of HIV-seronegative donor PBMCs was also cultured in the absence of PHA or IL-2 for 3 days prior to infection (these cells are referred to below as unstimulated cells). MT2 (24, 25) and P815 cells were maintained as described above for PBMCs, except that 10% fetal calf serum was included and IL-2 was not.
Viral stocks were prepared in PHA- and IL-2-prestimulated PBMCs. Viruses included wild-type (WT) NL4-3 (1), two HIV-1 protease mutants with indinavir-selected protease mutations in the NL4-3 genetic background (M46I/L63P/V82T/I84V and L10R/M46I/L63P/V82T/I84V [10, 11]), and other mutants constructed by site-directed mutagenesis for this study.
Site-directed mutagenesis.
Site-directed mutagenesis was performed to construct D30N, L90M, L63P/L90M, and D30N/L63P mutant viruses by a modification of recombinant PCR (54, 60). For the single mutants D30N and L90M, an 852-bp fragment that included the 3′ end of Pr55gag (p7/p1/p6), protease, and part of RT was amplified from NL4-3-infected cell DNA in two fragments, one 5′- and one 3′-PCR product. Two primer pairs were used for each mutant, including overlapping, internal mutagenic primers which had dUMP incorporated in place of dTMP (mutations are underlined in the primer sequences below; the numbers in the names of primers refer to nucleotides in the NL4-3 sequence [1]). The primer pair which amplified the 5′-PCR product to construct D30N was 5′-CUA CUA CUA CUA ACA TAG CCA IAA ATT GCA GGG CCC CTA G-3′ (Apa1988UDG) and 5′-ACU GUA UTA UCU GCU CCU GUA UCT AAT AGA-3′ (D30N-2312L36). The primer pair which amplified the 3′-PCR product to construct D30N was 5′-ACA GGA GCA GAU AAU ACA GUA UTA GAA GAA-3′ (D30N-2328U33) and 5′-CAU CAU CAU CAU CTG ATT TTT TCT GTT TTA ACC CTG CAG GAT G-3′ (Sse2835UDG). The overlapping internal mutagenic primers for introducing L90M were 5′-CUG AGU CAU CAG ATT TCT TCC AAT TAT GTT-3′ (L90M-2499L30) and 5′-CUG AUG ACU CAG AUT GGC TGC ACT TTA AAT T-3′ (L90M-2517U31), used with the outer Apa1988UDG and Sse2835UDG primers, respectively. Thirty cycles of PCR amplification (XL DNA PCR kit; Perkin-Elmer) were performed with 1 mM Mg(OAc)2, 0.2 mM (each) deoxynucleoside triphosphate (dNTP), 0.8 μM (each) primer, and 2 U of recombinant Tth DNA polymerase (at 94°C for 10 s, 60°C for 45 s, followed by an extension step at 72°C for 10 min).
PCR products were mixed with pAMP1 vector DNA (Clone-Amp; Gibco-BRL) and digested with uracil deglycosylase (Gibco-BRL) for 20 min at 37°C followed by 10 min at 65°C. This allowed annealing of undigested single strands complementary to the internal mutagenic primers and of the undigested flanking single-stranded ends of each PCR product to complementary single strands of pAMP1 vector DNA (7, 53, 60). Escherichia coli DH10B electrocompetent cells (Gibco-BRL) were transformed according to the manufacturer’s protocol, plated on Luria broth plates containing ampicillin, and incubated overnight at 37°C. Infectious mutant proviral clones were reconstructed by subcloning the 833-bp ApaI-Sse8387I fragment, containing the mutagenized protease gene, into the NL4-3 5′-half-genome plasmid (pJM11ΔGPR), with a protease gene deleted (37), using standard ligase-mediated methods. All PCR-derived segments, including the complete protease gene, were verified by DNA sequencing of plasmid clones. Infectious virus was produced by cotransfection of MT2 cells with the mutant proviral 5′-half-genome plasmid and the NL4-3 3′-half-genome plasmid (p83-10) (20). Supernatant fluids were collected when HIV-1 p24 antigen was positive (by enzyme-linked immunosorbent assay [Dupont]). Virus stock was generated by one passage in PBMCs stimulated with PHA and IL-2.
The double mutants D30N/L63P and L63P/L90M were constructed as described above, except that DNA from PBMCs infected with the cloned mutants D30N and L90M served as templates for recombinant PCR. The internal mutagenic primers for introducing L63P were 5′-GAU CAG AUA CCC AUA GAA AUC UGC GGA CAU AAA GC-3′ (L63P-2424L35) and 5′-GUC CGC AGA UTU CUA UGG GUA UCU GAU CAU ACT G-3′ (L63P-2430U34).
Viral growth kinetics in parallel cultures.
The 50% tissue culture-infective dose (TCID50) of each virus stock was determined by endpoint dilution in PBMCs (30). Inocula of either 5,000 or 3,000 TCID50 of each virus were used to infect 5 × 106 PHA–IL-2-prestimulated PBMCs (multiplicity of infection [MOI], 0.001 and 0.0006, respectively) in 1 ml of RPMI 1640 medium supplemented with 10% fetal calf serum. After incubation for 1 h at 37°C, the cells were resuspended at a concentration of 106/ml and reincubated. The cells were fed with medium containing 100 U of IL-2/ml on days 4, 11, and 17 after infection and with PHA- and IL-2-stimulated allogeneic PBMCs on days 7 and 14 after infection (7). Cell-free supernatant fluids were assayed by enzyme-linked immunosorbent assay for HIV-1 p24 antigen every 3 or 4 days after infection. Each experiment compared at least three different viruses in separate virus-infected cultures of the same donor’s PBMCs in the absence of drugs. The mean p24 antigen values were determined and plotted over time.
In separate experiments, an inoculum of 3,000 TCID50 of each virus was used to infect unstimulated PBMCs, which were maintained in the absence of PHA until day 7 after infection. These cultures were fed with medium containing PHA (2μg/ml) and IL-2 (10%) on days 7, 10, 14, 17, 21, and 24. No allogeneic cells were added at any time to cells stimulated after infection.
After viral growth kinetics experiments were completed, total genomic DNA was extracted from infected cell pellets with the Puregene DNA isolation kit (Gentra). HIV-1 DNA was amplified with primers Apa1988VDG and Sse2835VDG, and the PCR products were purified with the Qiaquick PCR purification kit (Qiagen). An uncloned amplicon containing the codons for cleavage sites p7/p1 and p1/p6 in Pr55gag (15, 61), the complete protease coding region, and the first 99 amino acids in the RT gene was sequenced directly, in bulk, using cycle sequencing with fluoresceinated primers (ThermoSequenase; Amersham) and electrophoresis in an automated sequencer (ALF; Pharmacia).
Infectivity/virion particle ratios.
TCID50 values (30) and HIV-1 p24 antigen concentrations (in nanograms per milliliter) were obtained in triplicate for a single clarified stock supernatant of each HIV-1 variant. Ratios of TCID50 to p24 antigen indicate infectivity per unit of virion antigen; lower ratios imply less infectivity per virion.
Growth competition assays.
All experiments were performed in the absence of drugs in MT2 cells except where PBMCs are indicated. Infections were initiated with unequal amounts of two competing virus variants, typically 20 and 80%, based on infectivity titrations in corresponding cells (MT2 cells or PBMCs). Unequal ratios were used based on the rationale that an increase in proportion of the initially less abundant virus suggests a relatively better replicative capacity than the competing, initially more abundant virus, although random factors cannot be excluded with either unequal or equal starting ratios. A 2-ml inoculum of 18,000 TCID50 was made up of a mixture of 3,600 and 14,400 TCID50 of each virus variant. Two 1-ml aliquots were adsorbed to 9 × 106 cells each (MOI = 0.001) for 1 h at 37°C. Subsequently, the cells were pooled, centrifuged, washed with cold phosphate-buffered saline, resuspended in medium at a final concentration of 106/ml, and incubated at 37°C. The cells were counted and the cultures were fed with medium every 3 or 4 days. Cells were added every 7 days as needed to maintain a concentration of 106/ml. Although the total cell number remained constant over the entire course of each experiment, the number of viable cells decreased at and after day 7, presumably because of increasing virus production. Aliquots were removed from the cultures at days 0, 1, 7, 14 (or 17), and 21 and centrifuged; total genomic DNA was extracted from the cells for genotypic analysis; and supernatants were stored at −80°C. The day 0 sample was obtained at the conclusion of the 1-h adsorption period. MT2 cells were also separately infected with mutants in the absence of WT virus to assess possible spontaneous reversion of the mutated codons.
Three independent amplifications of each infected cell lysate were done with primers Apa1988UDG and Sse2835UDG for 35 cycles of PCR. Amplicons from each sample were pooled to minimize possible bias introduced by PCR, cloned into pAMP1 (53), and used to transform E. coli. Recombinant plasmids were purified from ampicillin-resistant colonies with Qiawell (Qiagen). An average of 15 plasmid clones were used per time point to determine the proportion of each virus in the mixed competitive cultures. For the competitive culture comparing L10R/M46I/L63P/V82T/I84V and WT, the same PCR products used for cloning were also sequenced directly as a bulk population with at least two fluoresceinated primers, each hybridizing to a different strand.
Genotyping was performed on 10 μg of cloned PCR product plasmid DNA by the HIV-1 site-specific sequencing (HIV-SSS) method (37). In this assay a multiplex of several oligonucleotide primers of different lengths hybridizes to different positions on a template of an HIV-1 amplicon plasmid DNA clone. Then T7 polymerase (Sequenase; Amersham) extends each primer by a single base by incorporating one of four fluorescein-labeled dideoxynucleoside triphosphates (ddNTPs; NEN Life Science Products). These single-base-extended products are separated in a denaturing polyacrylamide gel electrophoresis with an automated DNA sequencer (ALF). The products are classified by protease or RT codon number according to length and the genotype that is determined based on the fluoresceinated ddNTP that has been incorporated. The primers used here detected nucleotide changes in codons 30 (first position), 46 (third position), 63 (second position), 82 (first and second positions), and 90 (first position) in the protease gene. Additional primers testing changes in codons 46 (first position) in protease, 41 (first position) in RT, and 74 (first position) in RT served as controls in the same multiplex mix because nucleotide bases at these positions were identical in the two competing viruses. In some reactions, additional control primers were used to analyze specific codons in cleavage sites between p7/p1 and p1/p6 in Pr55gag.
Controls were performed to determine if the HIV-1 PCR products genotyped on day 0 accurately reflected the initial infectious inoculum. Stocks of L90M mutant virus were incubated with the murine tumor cell line P815 and then lysed as described for day 0 samples from the mixed-infection cultures. HIV-1 virions cannot enter P815 murine CD8+ CD4− T lymphocytes, and HIV-1 DNA was not amplified from this control. This indicated that noninfectious HIV-1 DNA was not artifactually carried over in the cell-free virus inoculum and that DNA amplified from the mixed competitive cultures on day 0 was derived from intracellular HIV-1 DNA. The proportional composition of the competitive cultures at early time points was also supported by another line of evidence. The proportion of clones determined by the clonal genotyping assay correlated with the input TCID50 ratio of the two different viral variants, each determined in triplicate (R2 = 0.93; P = 0.002). In some experiments, the ratio of input TCID50 was the only measure of the proportion of each virus in the starting population.
Statistical methods.
Nonparametric tests were used. The statistical significance of differences among viral strains at each time point of viral growth kinetics experiments in parallel cultures was determined by either the Kruskal-Wallis or Mann-Whitney U tests for two or three viruses in the same experiment, respectively. In the mixed-competitive-culture experiments, Fisher’s exact test was used to determine if the difference between the baseline proportion of the two competing viruses and the proportion at a later time point was statistically significant.
RESULTS
The replicative fitness of the nelfinavir-selected mutants D30N and D30N/L63P, the saquinavir-selected mutants L90M and L63P/L90M, and the indinavir-selected mutants M46I/L63P/V82T/I84V and L10R/M46I/L63P/V82T/I84V was determined by using kinetics of HIV-1 p24 antigen production, infectivity/virion particle ratios, and clonal genotypic analysis of mixed competitive cultures.
Fitness of mutants with a single substitution.
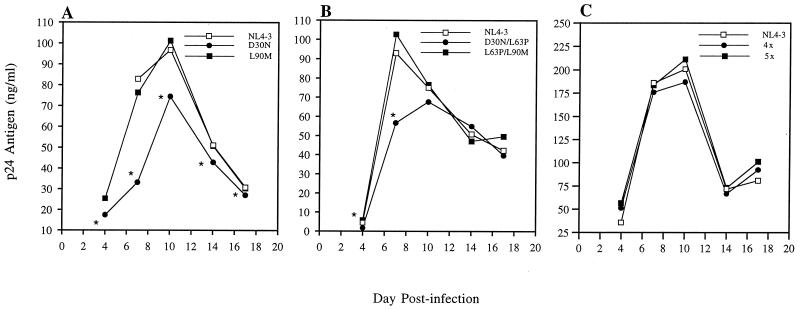
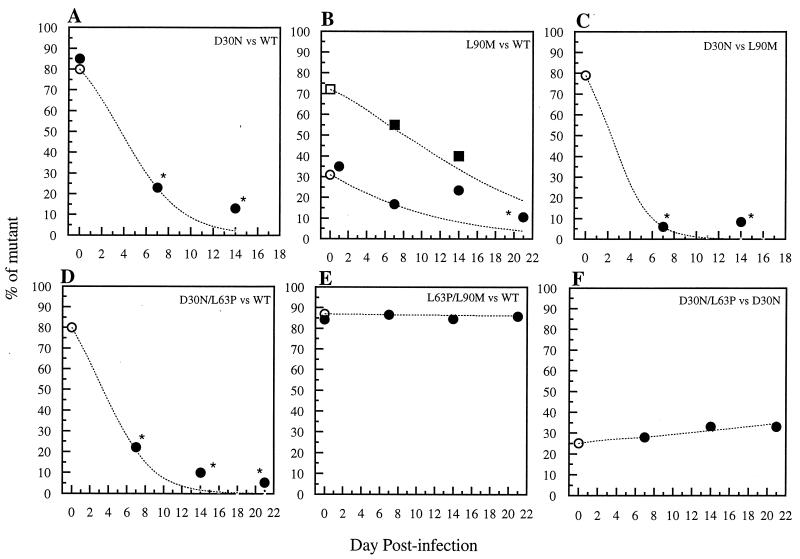
The nelfinavir-selected D30N mutant virus replicated less well than WT (NL4-3) virus in each of the assay systems. Analyses of replication kinetics in parallel cultures showed that significantly less p24 antigen was produced from cells infected with the D30N mutant than from cells infected with WT virus at all time points (Fig. 1A). This mutant also had a decreased TCID50/p24 antigen ratio compared to WT, which was the lowest infectivity-to-viral particle ratio observed here (Table 1). In a mixed infection starting with 80% D30N mutant and 20% WT virus (Fig. 2A), a sharp decrease in the proportion of D30N mutant virus occurred at days 7 and 14. This decrease in the proportion of D30N mutant, and increase in WT, was statistically significant (P < 0.02). Sequencing of the D30N mutant clones from the end of the competition did not identify any other mutation in these clones in either the protease coding region or in the p7/p1 or p1/p6 cleavage sites in Pr55gag. The sequencing also confirmed that protease D30N was correctly identified by the HIV-SSS clonal genotyping assay in each case. In the culture infected with only the D30N mutant virus, there was no reversion to WT at protease codon 30 (AAT to WT GAT) among nine clones obtained at day 7. Therefore, the observed decrease in D30N mutant virus by day 7 of the competitive culture is most likely due to WT virus outcompeting it rather than to reversion.
FIG. 1.
Replication kinetics of PI-resistant HIV-1 mutants in parallel cultures. One thousand TCID50 of each virus was used per 106 PHA-prestimulated PBMCs (MOI = 0.001). Virus production was monitored at days 4, 7, 10, 14, and 17 by measuring HIV-1 p24 antigen levels in supernatant fluids of triplicate cultures of each virus. The means of p24 antigen values are depicted for each virus over time. (A) D30N versus L90M versus WT; (B) D30N/L63P versus L63P/L90M versus WT; (C) L10R/M46I/L63P/V82T/I84V (5×) versus M46I/L63P/V82T/I84V (4×) versus WT. An asterisk indicates a statistically significant difference (P < 0.05; Mann-Whitney U test).
TABLE 1.
HIV-1 infectivity-to-virion particle ratiosa
| Protease variant | TCID50/ml | p24 antigen (ng/ml) | TCID50/p24 antigen ratio |
|---|---|---|---|
| NL4-3 (WT) | 45,275 (12,999) | 455 (54) | 100 |
| D30N | 8,947 (1,253) | 305 (18) | 29 |
| D30N/L63P | 30,667 (15,858) | 266 (4) | 115 |
| L90M | 32,700 (8,250) | 453 (26) | 72 |
| L63P/L90M | 32,467 (16,267) | 408 (10) | 79 |
| L10R/M46I/L63P/V82T/I84V | 64,967 (33) | 498 (13) | 130 |
Infectivity was measured by determining TCID50, and virion particles were estimated by HIV-1 p24 antigen measurement from a single clarified supernatant (virus stock) of each HIV variant. All assays were performed in triplicate, and the standard error is in parentheses.
FIG. 2.
Fitness dynamics based on clonal analysis of mixed competitive infections of MT2 cells with drug-resistant HIV-1 mutants selected by nelfinavir or saquinavir in vivo. Coinfections were carried out with nonequivalent amounts of PI-resistant mutants versus either WT (NL4-3) or another mutant. The proportion of mutant virus, or the first mutant virus listed in the inset caption for each panel, is plotted over time. Two experiments, each initiated with a different proportion of the two viruses, are plotted in panel B; one experiment is plotted in each of the other panels. A mean of 15 clones was analyzed at each time point. (A) D30N versus WT starting with 85% D30N mutant, depicting the percentage of D30N mutant over time; (B) L90M versus WT starting with either 31% (circles) or 72% (squares) L90M mutant, depicting the percentage of L90M mutant over time; (C) D30N versus L90M starting with 80% D30N mutant, depicting the percentage of D30N mutant over time; (D) D30N/L63P versus WT starting with 80% D30N/L63P mutant, depicting the percentage of D30N/L63P mutant over time; (E) L63P/L90M versus WT starting with 87% L63P/L90M mutant, depicting the percentage of L63P/L90M mutant over time; (F) D30N/L63P versus D30N starting with 25% D30N/L63P mutant, depicting the percentage of D30N/L63P over time. Open symbols on day 0 refer to input TCID50 proportions (mutant TCID50/total TCID50). Solid symbols on day 0 refer to proportions of clones determined by clonal genotyping (number of mutant clones/total number of clones). The dotted lines show the percentage of each mutant predicted by a formula that modeled the effects of selection at a single locus in an asexual haploid population during continuous replication in overlapping generations (42) and which has been used in other studies of relative fitness of drug-resistant HIV-1 mutants (16, 22, 57); the relative fitness determined at day 7 was used to solve for proportions at other time points. The time points after day 7 do not agree perfectly with the model’s predictions in cases where the variants differ markedly, suggesting this model is inadequate. The asterisks indicate a statistically significant difference from day 0 (P < 0.05; Fisher’s exact test).
The saquinavir-selected L90M mutant had minimally decreased replicative fitness compared to WT, which was only apparent when analyzed by infectivity-to-particle ratios and the mixed-competitive-culture method. Mutant L90M virus produced levels of p24 antigen similar to those of WT over the entire time course of the growth kinetics experiment (Fig. 1A). However, its TCID50/p24 antigen ratio was slightly less than that of WT (Table 1). The competitive culture also demonstrated a decrease in the proportion of L90M mutant virus, whether the experiment was initiated with 70 or 30% of L90M mutant (Fig. 2B). In the competitive culture starting with 30% L90M, the proportion of L90M decreased significantly from 0 to day 21 (P < 0.05). L90M, in the absence of competing virus, was also stable and did not revert in any of 24 clones derived from day 7 of the culture and infected with only L90M mutant virus.
The L90M mutant virus also outcompeted the D30N mutant in a separate competitive culture, confirming that the D30N mutant was less fit than L90M. The proportion of D30N mutant virus was significantly decreased from baseline on days 7 and 14 (Fig. 2C), and the proportion of L90M was significantly increased. Up to 12% of genotyped PCR product clones were recombinant; half were doubly mutant (D30N/L90M). The recombinant double mutants were not selected over time.
Fitness of mutants with multiple substitutions.
The double mutant D30N/L63P virus had a reduced replication capacity compared to that of either WT or L63P/L90M mutant virus when assayed by comparative kinetics of p24 antigen production in parallel PBMC cultures in triplicate (Fig. 1B). The decrease, relative to WT or L63P/L90M mutant virus, was statistically significant at days 4 and 7 (P < 0.05). A second experiment comparing triplicate PBMC cultures of D30N/L63P and WT confirmed significantly decreased p24 antigen levels of the D30N/L63P mutant virus at days 4, 7, 10, and 14 (P = 0.049 at each time point; data not shown). Likewise, the proportion of D30N/L63P mutant clones significantly decreased between baseline and day 7 in a competitive culture with WT (Fig. 2D). The stability of the D30N/L63P double mutant in cultures containing only this mutant again supported competition rather than reversion. There was a minimal increase in the proportion of D30N/L63P mutant over time in a competitive culture with the D30N single mutant (Fig. 2F), and the TCID50/p24 antigen ratio for D30N/L63P was similar to that of WT rather than reduced, as was observed for the D30N mutant (Table 1).
In contrast, the double mutant L63P/L90M had a replication capacity similar to that of WT virus when assayed by kinetics of p24 antigen (Fig. 1B), infectivity-to-particle ratios (Table 1), and growth competition experiments (Fig. 2E). In the competitive culture with WT, the proportion of L63P/L90M mutant clones did not significantly decrease during the time of the study (Fig. 2E). Six percent of PCR product plasmid clones were single-mutant recombinants, but none were selected over time.
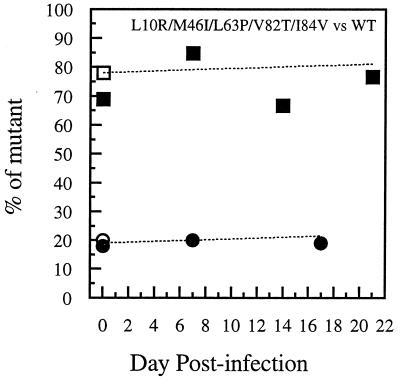
Kinetics of p24 antigen production of the two indinavir-selected, multiply mutant viruses (M46I/L63P/V82T/I84V and L10R/M46I/L63P/V82T/I84V) were indistinguishable from parental WT virus, NL4-3, in parallel cultures of PBMCs stimulated with PHA before infection at either of the two low MOIs used (0.001 and 0.0006). Data from six replicate infections with each virus were averaged (Fig. 1C). Sequencing of PCR products amplified from infected, prestimulated PBMC DNA after 17 days of culture confirmed the stability of the protease mutations and documented the fact that no additional, potentially compensatory mutations developed during the experiment, either in the protease coding region or in codons of the cleavage sites gag p7/p1, gag p1/p6, gag/PR, or PR/RT. In addition, no differences in p24 antigen production were observed between WT and L10R/M46I/L63P/V82T/I84V infections of quiescent PBMCs stimulated by mitogen 7 days after infection in the absence of drugs. This multiple mutant had a TCID50/p24 antigen ratio that was slightly higher than that of WT. Competitive cultures in MT2 cells showed little change in the proportion of mutant virus, starting with either more or less than 50% of the mutant L10R/M46I/L63P/V82T/I84V relative to WT (Fig. 3). Similar results were seen for competition experiments in prestimulated PBMCs. Eleven percent of the genotyped clones were recombinant; none was selected over time.
FIG. 3.
Fitness dynamics based on clonal analysis of mixed competitive infections of MT2 cells with a drug-resistant HIV-1 mutant selected by indinavir in vivo versus WT. Coinfections were carried out with nonequivalent amounts of PI-resistant L10R/M46I/L63P/V82T/I84V mutant versus WT (NL4-3). The proportion of mutant virus is plotted over time. Two experiments, each initiated with a different proportion of the two viruses, are plotted. A mean of 15 clones was analyzed at each time point of the culture of L10R/M46I/L63P/V82T/I84V mutant versus WT starting with either 20% (circles) or 80% (squares) of the L10R/M46I/L63P/V82T/I84V mutant; the percentage of L10R/M46I/L63P/V82T/I84V mutant is depicted over time. Open symbols on day 0 refer to input TCID50 proportions (mutant TCID50/total TCID50). Solid symbols on day 0 refer to proportions of clones determined by clonal genotyping (number of mutant clones/total number of clones). The dotted lines show the percentage of the mutant predicted by a formula (42) used in several other studies of the relative fitness of drug-resistant HIV-1 mutants (16, 22, 57); the relative fitness determined at day 7 was used to solve for proportions at other time points.
The same PCR products used for molecular cloning and clonal genotyping were also directly sequenced at several time points of the competitive culture which was initiated with 20% L10R/M46I/L63P/V82T/I84V mutant virus and 80% WT (Fig. 3). Differences in relative peak heights of the mixed bases were noted from each of two primers used for direct sequencing of the same PCR product. Recombination between the two input viruses was not detected by sequencing uncloned PCR products.
DISCUSSION
In the present study, several HIV-1 protease mutations selected by different PIs in vivo were documented to either impair or restore replicative fitness in the absence of drugs. The resistance mutations initially selected by nelfinavir and saquinavir impaired fitness relative to WT and will be discussed before those associated here with compensatory increases in fitness.
In this study the protease D30N mutant virus was the least fit of the HIV-1 mutants compared to the parental WT virus. This was observed in comparisons of kinetics of HIV-1 p24 antigen production and of infectivity-to-particle ratios to WT in parallel cultures, as well as in experiments directly competing D30N mutant virus against WT in mixed infections. The saquinavir-selected L90M mutant virus also showed a consistent decrease in replicative capacity when competed in two mixed cultures with NL4-3 with different input virus concentrations. Its loss of fitness relative to WT was of a smaller magnitude than that of D30N. The impairment in fitness of L90M was evident in the competitive culture versus WT, but not in the less-sensitive comparison of kinetics of viral antigen production. Direct competition between D30N and L90M mutant viruses confirmed that the D30N mutant virus was also less fit than the L90M mutant virus.
Several substitutions commonly selected in patients because they confer resistance to antiretroviral drugs have previously been shown to impair HIV-1 replicative fitness in the absence of drugs in vitro. These include mutations in protease codons 82 and 84, which are usually selected early during failure of ritonavir or indinavir (16, 36, 43, 50, 55). An earlier report noted incidentally that the protease L90M mutant was outcompeted by WT virus in the absence of drugs, but no data were presented (39). The same protease mutant may be less fit in some HIV-1 genetic backgrounds, including NL4-3, than in others; this is poorly understood (55). RT resistance mutations previously documented to impair fitness include several selected relatively early in virologic failure, such as zidovudine (ZDV)-selected RT T215Y (5, 8), lamivudine-selected RT M184V (2, 32), and didanosine-selected L74V (57), as well as the ZDV resistance mutation RT L210W, which is selected later (27).
Using the same mathematical model (42) employed for quantitative estimates of fitness in earlier studies of RT mutants in codons 41 (21), 74 (57), 210 (27), and 215 (16, 21, 22) and a PR mutant in codon 82 (16) and assuming continuous HIV-1 replication with a generation time of 2.6 days (52), the changes in proportions of viruses between days 0 and 7 of the current study’s competitive cultures indicate that D30N mutant virus is 37% less fit than WT and the L90M mutant virus is 10% less fit than WT. However, in some cases the data points do not perfectly fit the theoretical mathematical model (Fig. 2A to D). This mathematical model, developed for asexual haploid populations (42), may not adequately quantify relative growth rates of different HIV-1 mutants; the time dependence of the number of infected cells is not specified in the model or measured in our experiments. Indeed, the best comparison of different viruses is a relative ranking by direct experimentation, as performed here for WT versus each of the mutants and for D30N versus L90M or D30N/L63P. The purpose of this attempt at a more quantitative estimate of the magnitude of fitness loss relative to WT is to illustrate that the impairments in replicative capacity of D30N and L90M mutants documented in the present study appear to be of a magnitude similar to or greater than those of other resistant HIV-1 mutants studied earlier with the same model (16, 21, 22, 27, 57).
Relative fitness in the absence of drugs, as measured here, may be related to the catalytic efficiency of the mutant enzyme. The substantial fitness loss for the nelfinavir-selected D30N mutant virus documented by the experimental data in the present study agrees with a prior study of the protease enzyme function of this mutant (34). A relatively impaired replicative capacity of a particular drug-selected mutant virus in the absence of the drug will likely limit its potential for persistence after withdrawal of the selecting antiretroviral agent, transmission to another person, and persistence in a newly infected host in the absence of drug selection pressure. The present study suggests that differences in fitness impairment among PI-resistant mutants are great enough to cause appreciable variation in their potential transmissibility and persistence in the absence of selection pressure; this hypothesis requires clinical testing.
Relative fitness need not be similar in the presence and absence of drugs. Further study of these mutants will be needed to directly measure their relative fitness in the presence of drugs and whether it correlates with measures of their biochemical “vitality,” a multiplicative combination of relative catalytic efficiency and Ki ratio which has been correlated with selection of mutations in the presence of drugs (23). However, if a marked reduction in fitness of D30N mutants extends to situations where a drug is present, it may explain the rapid loss of the D30N mutant in favor of a different mutant during one experiment that selected for nelfinavir resistance in vitro (48).
Mutations may accumulate during drug failure in vivo because they compensate for a replication defect conferred by an initial mutation in the absence of drugs (7, 28, 43, 61) and/or increase the level of phenotypic drug resistance. The possibility that L63P could compensate for the replication impairment caused by D30N or L90M was studied here because L63P is seen in many patients who are naive to PI therapy (31, 33), it can be selected during PI therapy (10, 11), it does not alter 50% inhibitory concentration to PIs by itself (10, 11), and it compensates in vitro for the impaired replication caused by mutations in protease codons 82 and 84 (36, 56). L63P did completely restore viral replicative fitness to WT levels when it was linked to L90M (Fig. 1B and Table 1; also compare Fig. 2B and E), consistent with observations that these mutations are often both selected during saquinavir therapy in vivo (29). However, the addition of L63P to D30N did not restore HIV p24 antigen production to WT levels (Fig. 1B) or substantially alter the results of a losing competition with WT (compare Fig. 2A and D). The competitive culture between the D30N/L63P double mutant and the D30N single mutant (Fig. 2F) suggests minimal improvement in fitness of the D30N/L63P mutant over the D30N mutant, although this double mutant was still less fit than WT (Fig. 2D). These data on fitness in the absence of drugs, in conjunction with the lack of increase in nelfinavir resistance of the D30N/L63P mutant over that of the D30N mutant (data not shown), suggest the testable hypothesis that there may be minimal selection pressure for a linkage between D30N and L63P during nelfinavir therapy. Some in vivo data support this prediction (46, 47).
Compensatory mutations for PI-resistant mutants other than D30N or L90M have been defined in the Pr55gag codons for gag p7/p1 and gag p1/p6 cleavage sites (15, 61), as well as in additional protease codons, including 10, 46, 47, and 71 (4, 12, 28, 36, 51, 55). Our results demonstrated that the replication capacity of mutant L10R/M46I/L63P/V82T/I84V was identical to that of WT virus, as measured with each of the methods used here, including viral antigen production kinetics (Fig. 1C), competitive growth in either MT2 cells (Fig. 3) or PBMCs, and infectivity/particle ratios. These results, and the L63P/L90M mutant results, indicate that HIV-1 replicative fitness in the absence of drugs can be fully restored to the WT level by additional protease mutations, in the absence of any cleavage site mutations in Pr55gag (61).
Our results also indirectly suggest which protease mutations may compensate for impairment in replication observed in earlier studies to be caused by I84V and V82T (43, 49, 55): specifically, M46I and/or L63P. This is in agreement with previous observations that M46I and L63P improved enzyme catalytic activity impaired by the V82T and I84V active site mutations (56). Although L10I has been reported to rescue impaired growth of NL4-3 with G48V/A71T/V82A substitutions (55), we were unable to demonstrate such a role for L10R in the present study (Fig. 1C), although this cannot be excluded given that p24 antigen kinetics may not discriminate small differences in fitness. Others have reported that the impaired replication of a protease V82T and I84V mutant, initially selected by ritonavir, was improved by subsequent acquisition of A71V and K20R/A71V (43). Additional mutations (e.g., A71V and N88D) have been associated with selection of nelfinavir-resistant D30N mutant virus (45–48), and the present data showing impaired fitness of D30N mutant virus support the merit of future experimental investigation to determine if they compensate for the effect of D30N on replication in the presence or absence of drugs.
The fit protease mutant with multiple indinavir-selected substitutions (L10R/M46I/L63P/V82T/I84V) did not replicate better than WT in the absence of drugs in cells stimulated after infection. In an earlier study, we found that certain ZDV-selected viruses with multiple RT substitutions had even better replicative fitness than WT virus in infections of quiescent PBMCs which were stimulated with PHA 7 days after infection, although their replication rates were comparable to that of WT in prestimulated PBMCs (7). This was thought to occur because lower levels of dNTPs were available during reverse transcription in quiescent cells, providing an advantage to the mutant, more processive RT over WT only in such cells. The similar fitness of PR L10R/M46I/L63P/V82T/I84V to that of WT in the present study is consistent with an RT-specific explanation for the improved fitness of the ZDV-resistant mutant studied previously (7).
Compensatory restoration of replicative fitness to the level of WT virus caused by an accumulation of multiple mutations is likely to contribute, along with cross-resistance, to generally poorer virologic responses to a second (i.e., salvage) agent in the same mechanistic class (7, 13). The single-resistance mutations usually initially selected by PIs do not confer cross-resistance to other PIs by themselves. It is reasonable to expect that additional protease mutations would accumulate in linkage with a mutation initially selected by any PI because of selection for increased resistance and cross-resistance and/or restored replicative capacity in the absence or presence of drugs. Indeed, selection of broadly cross-resistant, fit HIV-1 with multiple protease mutations has been commonly observed during indinavir failure (10, 11), and it has been felt that this is likely applicable to all PIs. In fact, poor response rates have been reported for relatively delayed salvage after failure of initial regimens containing saquinavir (hard gel capsule) (44) as well as regimens containing indinavir (10, 11, 14).
However, several studies suggest that prolonged maximal HIV-1 suppression may be common among patients who are treated with a second regimen containing PI following a prolonged time on a failing regimen containing nelfinavir (6, 58, 62). Switching early during failure of a first regimen containing PI including either indinavir or nelfinavir, presumably before several mutations accumulate, led to good responses to a salvage regimen including ritonavir plus saquinavir (18, 19). Taken together, the studies now available suggest that patients may have better responses to relatively delayed salvage after nelfinavir failure, although the lack of a mechanistic explanation has made this interpretation somewhat contentious. The results of the present study suggest the hypothesis that a relatively less fit virus (e.g., nelfinavir-selected D30N mutant virus) may require a larger number of compensatory mutations to increase its replicative capacity to WT levels. In order to determine if the observations in this study are relevant to this clinical issue, it will be necessary to compare the relative fitness of mutant viruses from patients for whom therapy with nelfinavir and other PIs has failed, both in the presence and absence of drugs.
There are also several methodological implications of these data. We measured replicative fitness by three different approaches: kinetics of viral antigen production of independent cultures, virion infectivity/particle ratios, and virus competition assays in the absence of drugs. Differences among the methods were noted. The competitive-culture approach was more quantifiable and more able to discriminate small differences than the other methods. The consistency of results when competitions were repeated with different input ratios (Fig. 2B and 3), as well as the corroboration of relative fitness by the other methods, suggest that systematic differences between viruses were more important than random factors in determining the outcome of the competitions. We measured the proportion of mutant versus WT molecular clones of PCR products over time instead of analyzing electropherograms of PCR product population sequences, as earlier studies had done (26, 27, 57). Mixed base ratios in the direct PCR product sequence differed somewhat from those of each of two primers in the present study; proportions of clones with either WT or mutant sequence were unambiguous and therefore preferable. Clonal analysis also has the potential to improve detection of minorities. If a large enough number of clones is analyzed so that sampling error is small, the sensitivity to detect minorities will improve to less than 20%, the lower limit detectable by direct PCR product sequencing in our hands. We have not determined the number of clones needed to improve minority detection compared to that with direct PCR product sequencing. However, we have shown that sequence linkage and, therefore, rare recombination events between competing variants can be determined by sequencing clones but not by directly sequencing PCR products. The infrequent recombination noted here could have occurred either during virus growth in culture or during PCR (40); this could not be differentiated in the present study. Finally, the analysis of clones in the present study was speeded by using HIV-SSS instead of complete nucleotide sequencing; this facilitates analysis of a larger number of clones than sequencing. The base at each position in the codons of interest can be read directly as the gel electrophoresis is running without the need to perform the postrun alignment required for analysis of even a short protease sequence. This advantage would be greater if mutations distributed over a longer segment (e.g., protease and RT) were genotyped.
The comparison of kinetics of viral antigen production in parallel cultures of different variants was less able to discriminate small fitness differences than the competitive cultures (e.g., L90M versus WT). It may be limited for PI-resistant viruses by the impaired efficacy of gag and gag-pol precursor cleavage by the mutant protease (59), since cleavage intermediates present in nonreplicative virions have been reported to be detectable by p24 antibody (17). In the present study, it appeared that the viral infectivity-to-particle ratios were less able to discriminate differences than the other methods. The D30N/L63P mutant had a higher infectivity-to-particle ratio than WT (Table 1), although both the competitive-culture and kinetics of p24 production experiments indicated it was relatively less fit than WT.
The proportion of the less fit virus seemed to decrease more slowly after day 7 in some, but not all, competitive cultures (Fig. 2A, C, and D). The different patterns in different competitions with identical culture conditions suggest an explanation related to differences among HIV-1 competitions. Mutations developing during culture which compensated for effects of D30N were sought as a potential explanation of the observed data. However, no additional mutations with possible compensatory effects were identified in the protease coding region or in codons for cleavage sites between gag p7/p1 and gag p1/p6 in Pr55gag at the latest times in cultures studied here.
The biology of different drug-selected mutants of HIV-1 can only be fully understood by characterizing their replicative capacities and the likelihood of acquiring additional compensatory mutations, in addition to drug susceptibilities. A report that long-term nonprogressors harbor relatively less fit non-syncytium-inducing viruses than progressors also emphasizes the potential clinical relevance of viral fitness (3). The possibility of selection of compensatory mutations suggests that it would be best to change antiretroviral drugs before such mutants evolve.
ACKNOWLEDGMENTS
We thank Joan Kaplan for many helpful discussions and critical manuscript review, as well as M. P. DePasquale, G. Hanna, B. Clotet, D. Merrill, and M. Hirsch for their input. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3 from Malcolm Martin (NIAID), MT2 cell line from Douglas Richman (UCSD), and protease M46I/L63P/V82T/I84V and L10R/M46I/L63P/V82T/I84V mutant HIV-1 from Emilio Emini and William Schleif (Merck). We also thank Margot Coppelman and Mark Pasternack for their gift of P815 cells.
This work was supported by NIH grants AI-29193, a subcontract of AI-27659, and CA-12464. J.M.-P. was supported by a postdoctoral fellowship from the Spanish Ministry of Education.
REFERENCES
- 1.Adachi A, Gendelman H E, Koening S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and non-human cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back N K, Nijhuis M, Keulen W, Boucher C A, Oude Essink B O, van Kuilenburg A B, van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 3.Blaak H, Brouwer M, Ran L J, Wolf F, Schuitemaker H. In vitro replication kinetics of human immunodeficiency virus type 1 (HIV-1) variants in relation to virus load in long-term survivors of HIV-1 infection. J Infect Dis. 1998;177:600–610. doi: 10.1086/514219. [DOI] [PubMed] [Google Scholar]
- 4.Borman A M, Paulous S, Clavel F. Resistance of human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J Gen Virol. 1996;77:419–426. doi: 10.1099/0022-1317-77-3-419. [DOI] [PubMed] [Google Scholar]
- 5.Boucher C A, van Leeuwen R, Kellam P, Schipper P, Tijnagel J, Lange J M, Larder B A. Effects of discontinuation of zidovudine treatment on zidovudine sensitivity of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1525–1530. doi: 10.1128/aac.37.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks J T, Vliet M V, Sax P E. Abstracts of the 12th World AIDS Conference. 1998. Viral load reduction after changing from nelfinavir- to indinavir-containing regimens and associated resistance mutations, abstract 12342. [Google Scholar]
- 7.Caliendo A M, Savara A, An D, DeVore K, Kaplan J C, D’Aquila R T. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J Virol. 1996;70:2146–2153. doi: 10.1128/jvi.70.4.2146-2153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow Y K, Hirsch M S, Merrill D P, Bechtel L J, Eron J J, Kaplan J C, D’Aquila R T. Use of evolutionary limitations of HIV-1 multidrug resistance to optimize therapy. Nature. 1993;361:650–654. doi: 10.1038/361650a0. [DOI] [PubMed] [Google Scholar]
- 9.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 10.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelsi L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a HIV-1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 12.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of HIV-1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Aquila R T, Johnson V A, Welles S L, Japour A J, Kuritzkes D R, DeGruttola V, Reichelderfer P S, Coombs R W, Crumpacker C S, Kahn J O, et al. Zidovudine resistance and HIV-1 disease progression during antiretroviral therapy. AIDS Clinical Trials Group Protocol 116B/117 Team and the Virology Committee Resistance Working Group. Ann Intern Med. 1995;122:401–408. doi: 10.7326/0003-4819-122-6-199503150-00001. [DOI] [PubMed] [Google Scholar]
- 14.Deeks S, Grant R, Horton N, Simmonds N, Follansbee S, Eastman S. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Virologic effect of ritonavir plus saquinavir in subjects who have failed indinavir, abstr. I-205. [Google Scholar]
- 15.Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D. Second locus involved in HIV-1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eastman P S, Mittler J, Kelso R, Gee C, Boyer E, Kolberg J, Urdea M, Leonard J M, Norbeck D W, Mo H, Markowitz M. Genotypic changes in human immunodeficiency virus type 1 associated with loss of suppression of plasma viral RNA levels in subjects treated with ritonavir (Norvir) monotherapy. J Virol. 1998;72:5154–5164. doi: 10.1128/jvi.72.6.5154-5164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson-Viitanen S, Manfredi J, Viitanen P, Tribe D E, Tritch R, Hutchison C A D, Loeb D D, Swanstrom R. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res Hum Retroviruses. 1989;5:577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 18.Gallant J E, Barnett S, Raines C, Hall C. Abstracts of the 12th World AIDS Conference. 1998. Efficacy and durability of ritonavir/saquinavir as salvage therapy after failure of initial protease inhibitor regimen, abstr. 12330. [Google Scholar]
- 19.Gallant J E, Hall C, Barnett S, Raines C. Abstracts of the 5th Conference on Retroviruses and Opportunistic Infections. 1998. J. E., C. Hall, S. Barnett, and C. Raines. 1998. Ritonavir/saquinavir (RTV/SQV) as salvage therapy after failure of initial protease inhibitor (PI) regimen, abstr. 510. [Google Scholar]
- 20.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res Hum Retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 21.Goudsmit J, de Ronde A, de Rooij E, de Boer R. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J Virol. 1997;71:4479–4484. doi: 10.1128/jvi.71.6.4479-4484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goudsmit J, de Ronde A, Ho D D, Perelson A S. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J Virol. 1996;70:5662–5664. doi: 10.1128/jvi.70.8.5662-5664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulnik S V, Suvorov L I, Liu B, Yu B, Anderson B, Mitsuya H, Erickson J W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 24.Haertle T, Carrera C J, Wasson D B, Sowers L C, Richman D D, Carson D A. Metabolism and anti-human immunodeficiency virus-1 activity of 2-halo-2′,3′-dideoxyadenosine derivatives. J Biol Chem. 1988;263:5870–5875. [PubMed] [Google Scholar]
- 25.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 26.Harrigan P R, Bloor S, Larder B A. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J Virol. 1998;72:3773–3778. doi: 10.1128/jvi.72.5.3773-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrigan P R, Kinghorn I, Bloor S, Kemp S D, Najera I, Kohli A, Larder B A. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J Virol. 1996;70:5930–5934. doi: 10.1128/jvi.70.9.5930-5934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho D D, Toyoshima T, Mo H, Kempf D J, Norbeck D, Chen C-M, Wideburg N E, Burt S K, Erickson J W, Singh M K. Characterization of HIV-1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobsen H, Hänggi M, Ott M, Duncan I B, Owen S, Andreoni M, Vella S, Mous J. In vivo resistance to a HIV-1 proteinase inhibitor: mutations, kinetics, and frequencies. J Infect Dis. 1996;173:1379–1387. doi: 10.1093/infdis/173.6.1379. [DOI] [PubMed] [Google Scholar]
- 30.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J M, Lane J, Black R J, Reichelderfer P S, D’Aquila R T, et al. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. The RV-43 Study Group, the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 32.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 33.Lech W J, Wang G, Yang Y L, Chee Y, Dorman K, McCrae D, Lazzeroni L C, Erickson J W, Sinsheimer J S, Kaplan A H. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loeb D D, Swanstrom R, Everitt L, Manchester M, Stamper S E, Hutchison C A. Complete mutagenesis of the HIV-1 protease. Nature. 1989;340:397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- 35.Maeda Y, Venzon D J, Mitsuya H. Altered drug sensitivity, fitness, and evolution of human immunodeficiency virus type 1 with pol gene mutations conferring multi-dideoxynucleoside resistance. J Infect Dis. 1998;177:1207–1213. doi: 10.1086/515282. [DOI] [PubMed] [Google Scholar]
- 36.Markowitz M, Mo H, Kempf D J, Norbeck D W, Bhat T N, Erickson J W, Ho D D. Selection and analysis of HIV-1 variants with increased resistance to ABT-538, a novel protease inhibitor. J Virol. 1995;69:701–706. doi: 10.1128/jvi.69.2.701-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Picado J, Sutton L, Helfant A H, Savara A, D’Aquila R T. Abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication. 1997. Novel clonal analyses of resistance to HIV-1 RT and protease inhibitors, abstr. 45. [Google Scholar]
- 38.Maschera B, Darby G, Palu G, Wright L L, Tisdale M, Myers R, Blair E D, Furfine E S. Human immunodeficiency virus. Mutations in the viral protease that confer resistance to saquinavir increase the dissociation rate constant of the protease-saquinavir complex. J Biol Chem. 1996;271:33231–33235. doi: 10.1074/jbc.271.52.33231. [DOI] [PubMed] [Google Scholar]
- 39.Maschera B, Furfine E, Blair E D. Analysis of resistance to HIV-1 protease inhibitors by using matched bacterial expression and proviral infection vectors. J Virol. 1995;69:5431–5436. doi: 10.1128/jvi.69.9.5431-5436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyerhans A, Vartanian J P, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H-M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A B, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 42.Nagylaki T. Introduction to theoretical population genetics. Berlin, Germany: Springer-Verlag KG; 1992. pp. 25–27. [Google Scholar]
- 43.Nijhuis M, Schuurman R, de Jong D, Schipper P, Danner S, Boucher C. Abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication. 1997. Selection of HIV-1 variants with increased fitness during ritonavir therapy, abstr. 92. [Google Scholar]
- 44.Para M F, Coombs R, Collier A, Glidden D, Bassett R, Duff F, Boucher C, Leavitt R, Condra J, Pettinelli C the ACTG 333 Study Team. Abstracts of the 5th Conference on Retroviruses and Opportunistic Infections. 1998. Relationship of baseline genotype to RNA response in ACTG 333 after switching from long term saquinavir to indinavir or saquinavir soft gel capsule, abstr. 511. [Google Scholar]
- 45.Patick A K, Duran M, Cao Y, Ho T, Pei A, Keller M R, Peterkin J, Chapman S, Anderson B, Ho D, Markowitz M. Abstracts of the 5th International Workshop on HIV Drug Resistance. 1996. Genotypic and phenotypic characterization of HIV-1 variants isolated from in vitro selection studies and from patients treated with the protease inhibitor, nelfinavir, abstr. 29. [Google Scholar]
- 46.Patick A K, Duran M, Cao Y, Shugarts D, Keller M R, Mazabel E, Knowles M, Chapman S, Kuritzkes D R, Markowitz M. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob Agents Chemother. 1998;42:2637–2644. doi: 10.1128/aac.42.10.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patick A K, Kuritzkes D, Johnson V A, Shugarts D, Bakhtiari M, Potts K E, Farnsworth A, Anderson R, Koel J L, Hazelwood J D, Nail C D, Duran M, Markowitz M, Ho D, Richman D. Abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication. 1997. Genotypic and phenotypic analyses of HIV-1 variants isolated from patients treated with nelfinavir and other HIV-1 protease inhibitors, abstr. 18. [Google Scholar]
- 48.Patick A K, Mo H, Markowitz M, Appelt K, Wu B, Musick L, Kalish V, Kaldor S, Reich S, Ho D, Webber S. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob Agents Chemother. 1996;40:292–297. doi: 10.1128/aac.40.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patick A K, Rose R, Greytok J, Bechtold C M, Hermsmeier M A, Chen P T, Barrish J C, Zahler R, Colonno R J, Lin P F. Characterization of a human immunodeficiency virus type 1 variant with reduced sensitivity to an aminodiol protease inhibitor. J Virol. 1995;69:2148–2152. doi: 10.1128/jvi.69.4.2148-2152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paul M, Logue K, Bacheler L T. Abstracts of the 4th Conference on Retroviruses and Opportunistic Infections. 1997. Fitness of protease inhibitor resistant viruses, abstr. 119. [Google Scholar]
- 51.Pazhanisamy S, Stuver C M, Cullinan A B, Margolin N, Rao B G, Livingston D J. Kinetic characterization of human immunodeficiency virus type-1 protease-resistant variants. J Biol Chem. 1996;271:17979–17985. doi: 10.1074/jbc.271.30.17979. [DOI] [PubMed] [Google Scholar]
- 52.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 53.Rashtchian A, Buchman D M, Schuster D M, Beringer M S. Uracil DNA glycosilase-mediated cloning of PCR-amplified DNA: application to genomic and cDNA cloning. Anal Biochem. 1992;206:91–97. doi: 10.1016/s0003-2697(05)80015-6. [DOI] [PubMed] [Google Scholar]
- 54.Rashtchian A, Thorton C G, Heidecker G. A novel method for site-directed mutagenesis using PCR and uracil DNA glycosylase. PCR Methods Appl. 1992;2:124. doi: 10.1101/gr.2.2.124. [DOI] [PubMed] [Google Scholar]
- 55.Rose R E, Gong Y-F, Greytok J A, Bechtold C M, Teryy B J, Robinson B S, Alam M, Colonno R J, Lin P-F. HIV-1 viral background plays a major role in development of resistance to protease inhibitors. Proc Natl Acad Sci USA. 1996;93:1648–1653. doi: 10.1073/pnas.93.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schock H B, Garsky V M, Kuo L C. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. J Biol Chem. 1996;271:31957–31963. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 57.Sharma P L, Crumpacker C S. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J Virol. 1997;71:8846–8851. doi: 10.1128/jvi.71.11.8846-8851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tebas P, Kane E, Klebert M, Simpson J, Powderly W G, Henry K. Abstracts of the 5th Conference on Retroviruses and Opportunistic Infections. 1998. Virologic responses to a ritonavir/saquinavir containing regimen in patients who have previously failed nelfinavir, abstr. 510. [Google Scholar]
- 59.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang D, Caliendo A M, Eron J J, DeVore K M, Kaplan J C, Hirsch M S, D’Aquila R T. Resistance to 2′,3′-dideoxycytidine conferred by a mutation in codon 65 of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1994;38:282–287. doi: 10.1128/aac.38.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y-M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during indinavir therapy is caused by mutations in the protease gene and its gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zolopa A R, Shafer R W, Warford A, Montoya J G, Katzenstein D, Merigan T C. Abstracts of the 2nd International Workshop on Drug Resistance and Treatment Strategies. 1998. Predictors of antiviral response to saquinavir/ritonavir therapy in a clinical cohort who failed prior protease inhibitors: a comparison of clinical characteristics, antiretroviral drug history and HIV genotype, abstr. 54. [Google Scholar]