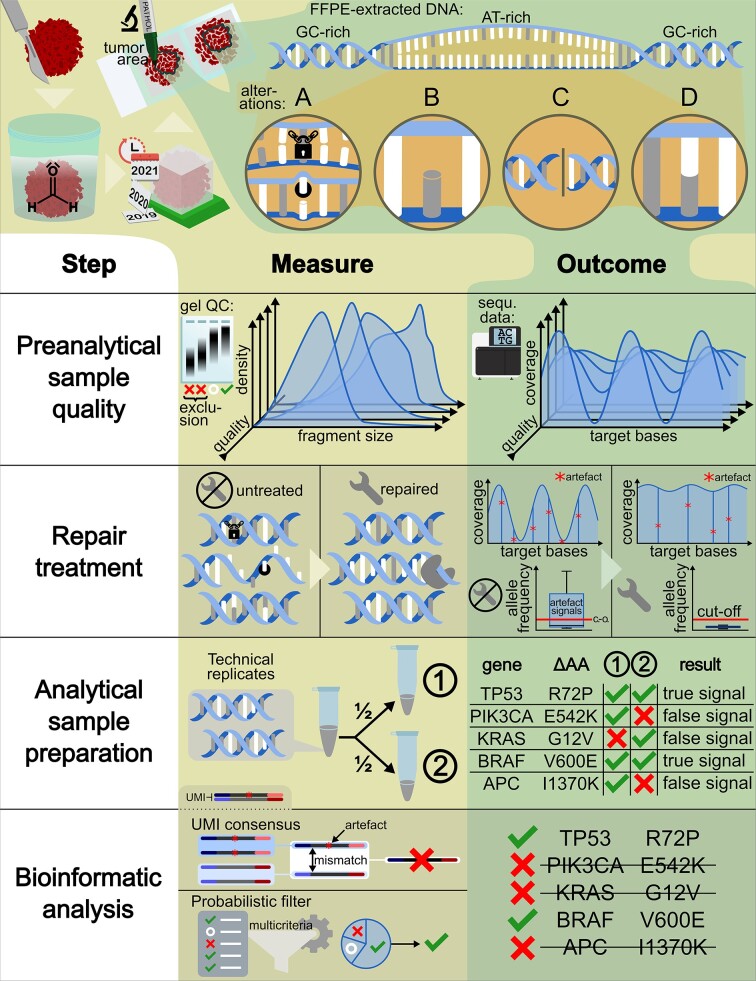
Abstract
In the late 19th century, formalin fixation with paraffin-embedding (FFPE) of tissues was developed as a fixation and conservation method and is still used to this day in routine clinical and pathological practice. The implementation of state-of-the-art nucleic acid sequencing technologies has sparked much interest for using historical FFPE samples stored in biobanks as they hold promise in extracting new information from these valuable samples. However, formalin fixation chemically modifies DNA, which potentially leads to incorrect sequences or misinterpretations in downstream processing and data analysis. Many publications have concentrated on one type of DNA damage, but few have addressed the complete spectrum of FFPE-DNA damage. Here, we review mitigation strategies in (I) pre-analytical sample quality control, (II) DNA repair treatments, (III) analytical sample preparation and (IV) bioinformatic analysis of FFPE-DNA. We then provide recommendations that are tested and illustrated with DNA from 13-year-old liver specimens, one FFPE preserved and one fresh frozen, applying target-enriched sequencing. Thus, we show how DNA damage can be compensated, even when using low quantities (50 ng) of fragmented FFPE-DNA (DNA integrity number 2.0) that cannot be amplified well (Q129 bp/Q41 bp = 5%). Finally, we provide a checklist called ‘ERROR-FFPE-DNA’ that summarises recommendations for the minimal information in publications required for assessing fitness-for-purpose and inter-study comparison when using FFPE samples.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Formalin, an aqueous solution of formaldehyde, was introduced as a fixative for the preservation of biological tissue specimens in the late 19th century (1), frequently in combination with paraffin embedding. Historically, formalin fixation was utilised to conserve the tissue's cellular morphology, but it also conserves protein epitopes, enabling pathologists to stain histological sections for morphological and immunohistochemical analyses. Due to the low handling and maintenance costs, formalin is still the most widely used fixative in medical sciences (2). Since the practice was introduced, millions of FFPE specimens have been preserved, some of which for more than a century, so that nowadays FFPE specimens are available from almost every disease, often paired with detailed pathological and clinical documentation (3,4).
As nucleic acids are preserved in FFPE specimens, they are a rich source for nucleotide sequence analysis of samples of various types and ages. Based on a report by Ferlay et al. (5) we estimate that for solid tumours alone, globally between 50 and 80 million FFPE specimens are potentially suitable for next-generation sequencing (NGS) analysis. Their wide availability and clinical diversity, in combination with modern DNA sequencing applications, offer a tremendous resource for biomedical research (6).
However, over time formalin fixation introduces a variety of chemical modifications of the DNA that poses technical challenges and compromises accurate sequencing. These challenges include analytical sample preparation failure from FFPE-DNA, i.e. insufficient library yield, and FFPE-induced chemical modifications of the DNA potentially leading to incorrect base identification (7). The latter can have serious consequences, for instance, detection of false positive variants. False positives observed in FFPE-DNA are particularly problematic for variant-based signatures or patterns (8,9) and for somatic mutations of lower variant allele frequency (VAF) in cancer specimens (10).
Here, we first review the chemical alterations found in FFPE-DNA and their effects on sequencing and single nucleotide variant identification. For application of NGS to FFPE samples there are four critical parameters that most affect sequencing results: (I) pre-analytical sample quality and its specifications, (II) optional application of DNA repair treatment, (III) analytical sample preparation and (IV) bioinformatic analysis. Each of these is briefly reviewed here, after which published solutions to mitigate frequently occurring problems are presented, backed up with experimental data to illustrate their individual effects. We demonstrated the importance of each parameter by generating DNA sequences from older FFPE samples and compared this to DNA from fresh frozen (FF) tissue. Certain problems can be specified but not controlled, while others can be managed. Therefore, we give recommendations on the minimal amount of information that scientific publications on sequences derived from FFPE-DNA should include. Finally, we indicate the remaining challenges that will need to be overcome in order to fully exploit the use of FFPE-DNA for future research.
FORMALIN-INDUCED ALTERATIONS TO DNA
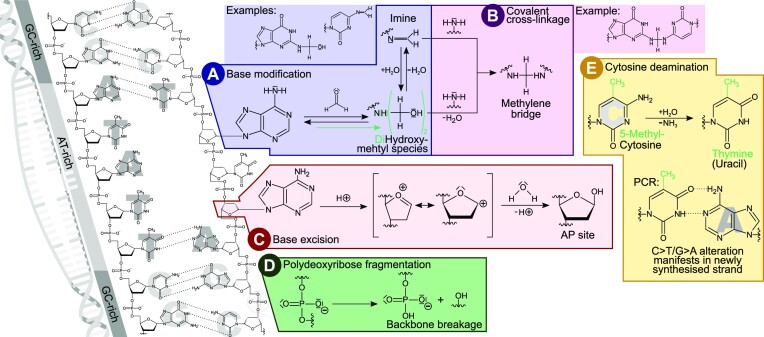
Typical formaldehyde-induced chemical alterations of DNA are summarised in Figure 1. All of these alterations are initial steps, leading to double-strand denaturation and base unstacking mainly in AT-rich genomic regions (11). The process is then magnified due to local strand separation increasing the chance for further modifications, leading to a vicious cycle resulting in increased DNA modifications in AT-rich regions and their flanks.
Figure 1.
Summary of DNA modifications typically observed in FFPE samples. DNA instability is initiated by double strand denaturation and base unstacking, especially in AT-rich regions (far left). Modifications influencing base pairing then induce further local double strand denaturation and accelerate base modifications, leading to local hot spots of alterations. (A) Base modification caused by the nucleophilic attack of a base's amino group towards the electrophilic carbon of formaldehyde. The resulting hydroxymethyl can condensate to form an imine (altering base pairing) or further react to a dihydroxymethyl species. (B) Methylene bridges can form a covalent crosslink with another nucleophilic group of, e.g. a base or a protein, both leading to DNA polymerase blockage. (C) Base excision by hydrolysis of the N-glycosylic bond leaves a 2-deoxy-d-ribose AP site in the phosphate backbone. A transition state can form as an intermediate containing a highly reactive cyclic oxocarbenium ion that reacts with water. (D) Formaldehyde conservation also promotes the slow hydrolysis of phosphodiester bonds that breaks the phosphate backbone and fractures the DNA. (E) As glycosylase repair enzymes are inactivated by the fixation, spontaneous cytosine deamination converting cytosine to uracil is no longer corrected. In case of 5-methylcytosine this conversion results in thymine. Either way, the base will now pair with adenine instead of the original C/G base pair at that location.
The formalin-induced alterations described in the literature can be classified into five different mechanistic processes.
A chemical addition reaction of formaldehyde to a nucleophilic group such as an amino group of a DNA base results in a modified base species with altered base pairing abilities (Figure 1A) (12,13).
Such a modified base can further react to form, via methylene bridges, a covalent cross-link with another nucleophilic group in its proximity (Figure 1B) (13). During sequencing library preparation, such modifications can locally alter base pairing characteristics, leading to the incorporation of non-complementary nucleotides in daughter strands. Alternatively, they lead to blockage of DNA polymerase during amplification of the template strand (14).
In addition, formaldehyde fixation accelerates the cleavage of glycosidic bonds and the generation of apurinic/apyrimidinic (AP) sites within the double strand (Figure 1C) (15). While DNA remains relatively stable under physiological conditions, these AP sites are more susceptible to damage and fragmentation (16,17) and to incorporation of alternative nucleotides (18). DNA polymerases generally have low bypass efficacies for such AP sites (19). Therefore, such DNA molecules may not be amplified sufficiently for sequencing. This means a reduced diversity of functional sequencing library molecules, termed as lower ‘library complexity’, resulting in an information loss.
Moreover, polydeoxyribose fragmentation, the cleavage of the backbone of the DNA macromolecule into separate segments, is widely observed in FFPE-DNA (Figure 1D) (15,20). Samples that were fixed in unbuffered formalin, yielding formic acid over time, are particularly sensitive to increased DNA degradation, because under acidic conditions, AP-sites form more easily by hydrolysis of protonated purines (21).
The most frequently encountered chemical alteration of FFPE-DNA is due to spontaneous deamination of cytosine. In living cells this is repaired by glycosylases, however, such events accumulate in formalin-fixed tissues (22) due to enzyme inactivation by the fixation. Deaminated cytosine results in uracil, which pairs with adenine instead of guanine; when cytosine is methylated (5-methylcytosine) its deamination leads to thymine that also pairs with adenine. Either case leads to the base pair alteration C>T/G>A (Figure 1E) (23). Other types of single base substitution artefacts in FFPE-extracted DNA have also been reported in the literature, but they cannot easily be attributed to a single chemical mechanism (23–26).
In contrast to the alteration mechanisms in Figure 1A–D that all result in the loss or underrepresentation of original sequence information, i.e. in reduced library complexity, the mechanism in Figure 1E introduces false signals. The combination of false signals within regions of diminished true sequences leads to high VAF of these false signals.
The effects of the chemical alterations summarised in Figure 1 propagate into downstream applications and consequently into sequencing results. One of the first reported downstream effects of formalin fixation is polymerase chain-reaction (PCR) amplification failure (27–29). Dropouts of FFPE-DNA amplicons (30,31) or sequencing libraries (32,33) exacerbate the outcome for NGS applications.
Nevertheless, many studies have fallen into the pitfalls of non-rigorous interpretation of the complex consequences of formalin fixation, especially in the context of NGS. For example, NGS artefacts were often addressed by merely trying to reduce the absolute artefact count, rather than maximising the amount of usable DNA from the sample. Moreover, most previous work was limited to deamination artefacts (C>T/G>A), presumably because here the mechanism was obvious and addressable. In contrast, the term ‘FFPE artefacts’ refers here to the sum of all false positives that are observed in FFPE-DNA, as they can be misinterpreted as true variants independent of their individual causes.
CONSEQUENCES OF FORMALIN FIXATION
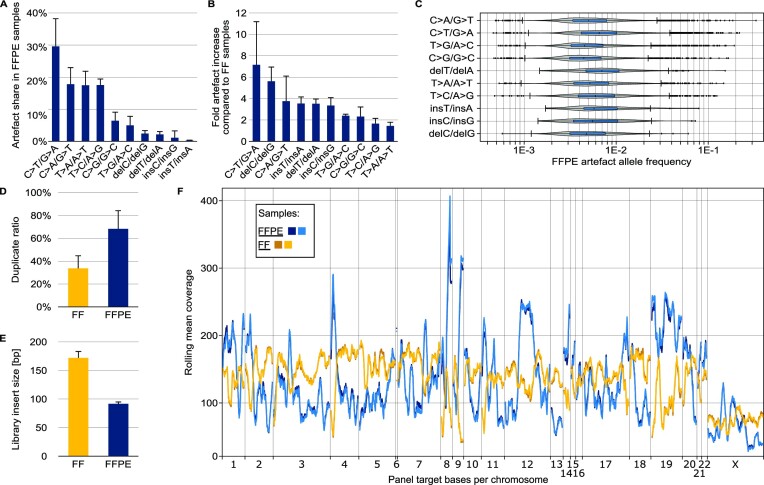
The consequences of formalin fixation are even more complex than previously summarised from the literature. Figure 2 shows the differences between DNA sequences obtained from FF and FFPE specimens, which the authors investigated, as part of the EASI-Genomics consortium. They included a 13-year-old sample with a case-matched FF sample, analysed in a large number of replicates by applying a diverse set of in vitro and in silico strategies. This approach showed the effect of formalin fixation storage on the resulting sequences, as well as the type of alterations and the relative frequencies of these artefacts.
Figure 2.
Characterisation of differences in NGS of FFPE-DNA and FF-DNA. FF-DNA was taken from the same tissue sample as FFPE-DNA. Experimental details are described in the online methods section. (A) Proportion of each artefact type in a set of five different FFPE samples of varying qualities and preparation workflows. (B) Fold increase in artefact number in FFPE-DNA compared to FF-DNA sequences. FFPE and FF read files were appropriately down-sampled before comparison. (C) Allelic frequency of artefacts in a typical FFPE sample of low quality. (D) Sequence duplicate ratios for low-quality FFPE-DNA and matching FF-DNA samples. (E) Insert sizes for the sample pairs used in (D). (F) Systematic coverage bias typical for targeted sequencing of FFPE samples. The plot shows the rolling mean coverage over the target region of a hybridisation capture bait panel. The reads were randomly down-sampled so that the mean unique coverage over the target bases was identical in all four libraries.
Figure 2A shows the repertoire of potential artefacts. The two most prevalent artefact types in FFPE-extracted DNA reported in the literature are C>T/G>A caused by cytosine deamination and C>A/G>T that mostly results from base oxidation (34). Other single base substitution artefacts such as T>A/A>T and T>C/A>G changes are also known (26,35). These were equally prevalent in the 13-year-old sample and contributed to its total artefact repertoire.
Figure 2B exemplifies the increase of the most frequently encountered artefacts in FFPE-DNA samples compared to their case-matched FF-DNA. The highest, 7-fold increase was observed for C>T/G>A. However, a large number of FFPE artefacts can be filtered bioinformatically if its VAF is lower than a threshold of interest, e.g. lower than 5%.
The distribution of artefact allele frequencies (AAF), some of which exceeded 10% in the analysed samples, is shown in Figure 2C. FFPE-DNA artefacts with high AAFs are particularly located in regions of low sequencing coverage (36), i.e. low information. The low coverage is a direct result of many genomic fragments of that region being severely damaged, not amplified, and therefore not sequenced. Those genomic fragments that are not so severely damaged may result in artefact-bearing sequences that are overrepresented. Consequently, artefacts reaching high AAFs may not only be related to mechanisms shown in Figure 1 but could also stem from any other root cause such as oxidation or sequencing errors. For example, in the 13-year-old FFPE specimen, the highest AAF was explained by its low sequence coverage and not obtained for a ‘typical’ C>T/G>A artefact but for a C>A/G>T change.
Figure 2D–F shows the three main mechanisms of information loss in FFPE-DNA sequencing: First, the sequence duplication ratio is higher in FFPE-DNA compared to FF-DNA (33,37,38), which increases sequencing cost for unique coverage. Unique coverage represents the sequences derived from original genomic molecules after correction for PCR duplication. The duplicated sequences can be identified by bioinformatic analysis and then eliminated; here, this revealed the average true unique coverage in FFPE libraries to be half as high as for FF libraries (Figure 2D). Consequently, the information content per sequence was half as high.
Second, the severely fragmented FFPE-DNA leads to reduced library insert sizes (38,39), which are approximately half of the FF libraries (Figure 2E). In Illumina-based sequencing, library molecules are usually sequenced from the adapters at both ends (paired-end sequencing). Small genomic inserts between those adapters reduce unique coverage because their paired-end reads may overlap (fewer unique bases sequenced per read). In addition to overlap, the reduced length makes unique bioinformatic mapping, the alignment of the sequence to a reference sequence, more difficult. This is caused by ambiguities since shorter sequences map to more genomic loci than longer sequences.
Third, FFPE-DNA leads to decreased coverage uniformity (i.e. evenness of coverage) (36). Figure 2F shows decreased coverage uniformity in FFPE compared to FF replicates. Of note, the rolling mean coverage in the FFPE replicates was generally more volatile and especially high in low-coverage regions of FF replicates and vice versa. Such findings have been described before: a systematic relationship with sequence context was observed by the 100 000 Genomes Project, reporting dropouts in FFPE versus FF in AT-rich DNA regions (36). Also, Xiao et al. (10) confirmed this relationship for targeted sequencing of FFPE-DNA, contradicting an earlier report by others (40). In our example, the mean coverage in AT-rich regions in FFPE replicates was lower than that of FF replicates (Supplementary Figure S1), in line with the 100 000 Genomes Project and Xiao et al. Taken together, against the background of generally non-uniform and locally extremely low coverage, artefacts observed in FFPE may achieve such high allelic frequencies that they might ultimately be mistaken as biological variants, despite deep sequencing (40).
PARAMETER I: PRE-ANALYTICAL SAMPLE SPECIFICATIONS AND QUALITY CONTROL CRITERIA
Pre-analytical sample specifications of quality and quantity are particularly important for FFPE-DNA extracted from old (2) or small specimens, such as needle biopsies (41). Of paramount importance in the pre-analytical procedure is the specimen collection and the fixation procedure. The specimen quality is impaired by tissue dehydration, and delayed, too short, or prolonged fixation (42–46).
The quality of extracted FFPE-DNA critically depends on the formalin concentration and pH, fixation temperature, thickness of the sample and fixation time, and the specimen storage conditions (13,46,47). Specimens without documented collection and fixation protocols should be prepared with all applicable precautions and be interpreted accordingly. When preparing the nucleic acid extractions from these specimens, air-exposed sections from the FFPE block surface should be discarded (7) because tissue areas at the block surface are prone to oxidation. The cells of interest (e.g. tumour cells) should ideally be enriched in the sections (or in the punched-out material). Deparaffinisation is often performed using agitation, but more gentle approaches without agitation should be preferred. An overnight proteinase K digestion in aqueous solution should be included in the extraction protocol. Of note is that the performance of FFPE-DNA extraction kits varies (38,48). If the resulting FFPE-DNA eluate needs to be concentrated, no additional heat should be applied, as this leads to further DNA degradation. Instead, lyophilisation (freeze drying) is a better concentration method.
Generally, FFPE-DNA should be prepared with care (e.g. gentle mixing, avoidance of unnecessary freeze-thaw cycles) to optimally preserve its integrity. To facilitate reproducibility and quality assurance, the international standard ISO/FDIS 20166-3:2018 (49) provides general guidelines and specifications for specimen collection, formalin fixation, DNA extraction, storage, and documentation.
Specifically, for NGS, criteria and thresholds for the adequacy of FFPE-extracted DNA have been defined in the literature, such as the preferred use of low-concentration (4% v/v, formaldehyde), neutral-buffered formalin (50) for fixation, specimen age below eight years, amplifiability (51,52), and a DNA integrity number (DIN) of >2.05 (30). The amplifiability is defined by the ratio of longer amplicons to shorter amplicons in qPCR. In severely impaired FFPE-DNA, longer amplicons drop out, resulting in a smaller ratio. For example, a Q129 bp/Q41 bp ratio is recommended to be >10% (51) or even 40% (52). The minimal amount of DNA required for NGS as specified by most laboratories is 50 ng (4,46), but sometimes a requirement of 10 ng for amplifiable DNA fragments is set (4). As discussed above, a major consequence of poor FFPE-DNA quality is the low availability of amplifiable DNA fragments of appropriate sizes (2). Low availability of such templates leads to a low library conversion rate, resulting in a high ratio of PCR duplicates (33) and the non-uniform coverage distribution that is typical for FFPE-extracted samples (36).
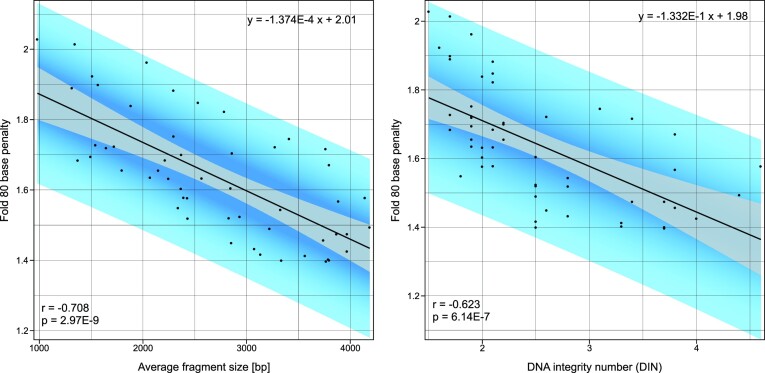
Due to the challenges induced by formalin fixation, many studies have tried to identify easy-to-determine metrics that correlate with DNA quality, so that the outcome can be predicted and unsuitable samples can be identified and excluded from further analysis. As illustrated in the previous section, one of the largest differences between high-quality FF-DNA and low-quality FFPE-DNA is the non-uniformity of read coverage (Figure 2F). The coverage uniformity can be represented in general terms by a fold-N base penalty, defined as the factor of sequencing required, so that the mean sequencing depth is fulfilled in N% of the targeted genomic region. A cutoff of 80% is deemed practical (53), therefore the fold-80 base penalty (F80BP) is usually assessed for the sequencing quality control. With an appropriate set of samples and data, it can be evaluated whether coverage uniformity, as a quality metric for FFPE-DNA, correlates with DNA fragmentation.
Figure 3 shows a correlation for the average FFPE-DNA fragment size, as measured by electrophoresis (P = 2.97E−9, r = –0.708) with coverage uniformity represented by F80BP, and also with the DNA integrity number (P = 6.14E−7, r = –0.623). This exemplifies that DNA fragment size provides a useful quality control measure for NGS of FFPE-DNA. Hence, to avoid wasting resources, large-scale sequencing studies can exclude low-quality specimens based on FFPE-DNA fragmentation that can be assessed by electrophoresis.
Figure 3.
FFPE-DNA fragment size (left) and DIN (right) correlate with NGS coverage uniformity (Fold 80 base penalty). DNA fragment size and DIN were determined on a gel electrophoresis system. Fold 80 base penalty was determined bioinformatically after sequence alignment. This correlation is based on 53 identically prepared whole exome sequencing libraries. Perfect coverage uniformity is defined by Fold 80 base penalty value of 1.
To summarise, meaningful sequence output can be achieved provided that specific conditions are met, even from decade-old FFPE specimens. Importantly, the use of neutral-buffered formalin in fixation and the use of as high as possible input amounts of FFPE-DNA is recommended. Given the many variables affecting sample quality, it makes sense to explore the suitability of a particular sample collection with a small proof-of-principle pilot study before investing in a large-scale study. Inclusion of FFPE reference material may also be considered for a pilot study, as long as the reference material is of a similar quality as the study samples. Some caution is needed here: when we compared fragmentation profiles of typical clinical FFPE samples from our laboratories to commercial reference material, we observed that the reference was less degraded than clinical samples and therefore not suitable for testing the NGS workflow (Supplementary Figure S2). If these commercial reference samples are not representative for a real-life study cohort or clinical samples, the outcome of many NGS-based data analyses will be worthless.
PARAMETER II: FFPE-DNA REPAIR TREATMENTS
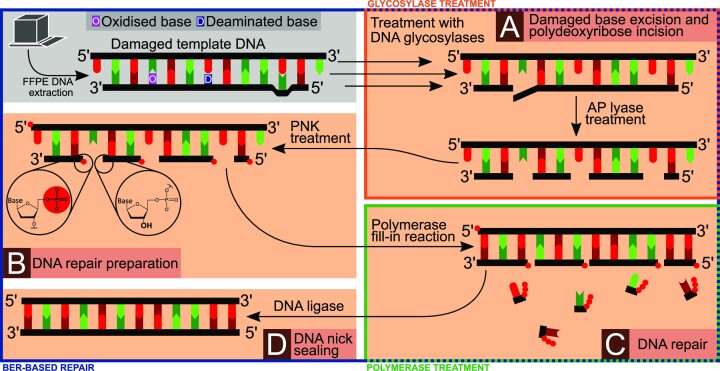
DNA treatments improving the performance in downstream analysis can be based on three different principles: (i) heat treatment, (ii) single-step enzyme treatment or (iii) multi-step enzyme treatment. Figure 4 summarises the main subprocesses of the in vivo base excision repair (BER) pathway (54) that comprises all common in vitro FFPE-DNA repair principles. While some of these commonly used repair principles constitute only individual steps of the BER pathway, other repair principles replicate more subprocesses of this pathway.
Figure 4.
Principles of enzymatic FFPE-DNA repair treatments. The grey panel shows template DNA extracted from FFPE tissue containing oxidised, deaminated and mismatched bases. The original, unaltered sequence is represented as the top strand. (A) Altered base species can be excised by DNA glycosylases leaving an AP site or, in the case of bifunctional glycosylases, producing a 5′-phosphate and a 4-hydroxy-5-phospho-2-pentenal on the 3′-end. AP lyase activity of the respective enzymes excises the pentanal species, leaving a 5′-phosphate and a 3′-hydroxy end. (B) In the next repair step, these ends are processed by DNA polynucleotide kinase (PNK) that phosphorylates all 5′-ends and dephosphorylates any 3′-ends. (C) Next, DNA polymerase fills in complementary nucleotides into the double strand gaps. In this step different polymerases can be used that have a higher tolerance for altered base species or that generate blunt ends. (D) Finally, DNA ligase seals the double strand nicks. The blue frame indicates a BER-based approach, the orange frame simple glycosylase treatment, and the green frame simple polymerase treatment.
Heat treatments (e.g. exposure to 95°C (55,56)) can help to break any methylene interstrand cross-links that would otherwise block polymerases from amplifying the template strands. This results in fewer truncated PCR products, hence in improved library complexity and fewer duplicate sequences.
-
The simplest single-enzyme based repair uses specific polymerases that are more tolerant to modified base species to produce complementary strands prior to PCR amplification (green frame in Figure 4). Although this results in a higher yield of amplifiable fragments and longer DNA fragments (21,57), it may increase artefacts caused by altered base pairs.
Another form of (ii) is glycosylase treatment (orange frame in Figure 4), such as the treatment with uracil-DNA glycosylase (UDG) that can excise deaminated bases (58). Alternatively, formamidopyrimidine-DNA glycosylase (FPG) treatment excises a broader range of oxidised bases (59). The sole application of UDG repair methods (58), as referred to in the ISO standard (49), is often disadvantageous: excision by the glycosylase generates an AP-site, increasing DNA fragmentation (17), especially in combination with ultrasonication, consequently lowering library complexity. The same applies to isolated use of bifunctional glycosylases, which lead to phosphate-ribose backbone cleavage. With such enzyme treatments alone, the original DNA strand and its information are not restored.
A more extensive enzymatic repair treatment (blue frame in Figure 4) consists of multiple steps involving base excision and backbone incision by different glycosylases and AP lyases (Figure 4A), polynucleotide kinase treatment (Figure 4B), DNA polymerase fill-in (Figure 4C) and nick sealing with DNA ligase (Figure 4D), which in combination mimic physiological BER (60). The advantage of a BER-based approach is that the DNA fragment is restored using the information of the complementary undamaged template strand.
To illustrate how BER-based DNA repair treatments remove artefacts, two such approaches were experimentally compared. As a benchmark, we used a commercially available FFPE-DNA repair mix (‘NEBrepair’, New England Biolabs). This was compared to a sequential BER-based repair approach that uses different glycosylases, In vitro Sequential Base Excision repair (‘IQBErepair’), with details described in Supplementary Figure S3. The protocol can be found in the online methods. It evolved from existing protocols (60), as it restores damaged DNA fragments by a sequential treatment of glycosylases and it was modelled on physiological base excision repair steps, as reviewed in (61). In contrast to other approaches suppressing artefacts, IQBErepair increases the molecular diversity and hence elevates the unique coverage and improves the coverage uniformity from low DNA input amounts. Thymine–DNA glycosylase (TDG) and N-methylpurine-DNA glycosylase (MPG) treatments address potential base modifications (62,63).
We then challenged the perception that FFPE-DNA with quality metrics Q129 bp/Q41 bp ratio <10% (51) or DIN <2.05 would be unsuitable for cancer somatic mutation detection (30). Such somatic mutation detection is particularly confounded by artefacts. To illustrate that these artefacts can be managed in practice, we used a low input amount (50 ng) of degraded DNA extracted from the 13-year-old healthy liver sample that had been fixed with buffered formalin. The quality metrics of the FFPE-DNA were an average fragment size of 1490 bp, a DIN of 2.0, and a Q129 bp/Q41 bp ratio of 5%. In order to assess the reproducibility of the findings, sequencing and data analysis were performed in replicates in two sequencing centres (Supplementary Figure S4, experimental data are given in the online methods. An overview of all samples, libraries, and replicates prepared can be found in the supplementary data).
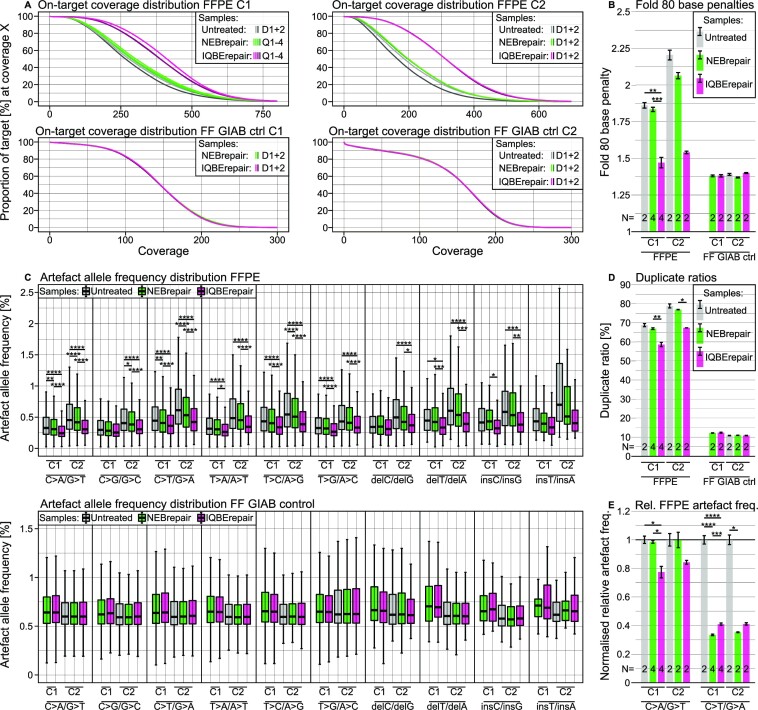
When comparing mitigation strategies for sequencing artefacts it is important to remove true biological variants and generate a set of pure artefacts. Therefore, the variants detected in sequencing data must be filtered to remove all potential true biological positives from the dataset, e.g. with the help of replicate experiments and FF-DNA from the same tissue sample. The subsequent figures illustrate artefacts only, indicating the individual effects of mitigation strategies. FFPE-DNA samples were aliquoted from a single DNA isolate from a 13-year-old FFPE tissue specimen and sent to participating centres. Various in vitro or in silico strategies are shown and how well they performed in removing these artefacts. As a first step, DNA repair treatments were assessed with the aim of analysing the effects of the BER-based repair approaches. Fresh-frozen Genome-in-a-Bottle (GIAB) DNA, a reference standard DNA sample, was repaired as a negative control.
IQBErepair resulted in a significantly higher coverage in both centres, with 53% and 80% more unique bases compared to untreated DNA (Figure 5A). Furthermore, the coverage uniformity metric F80BP was improved (Figure 5B). The AAF was significantly reduced for most artefact types, and the median AAF was lowest following IQBErepair for all artefact types, while no significant differences could be observed for the GIAB control (Figure 5C). IQBErepair also significantly reduced the sequence duplicate ratios in FFPE samples (Figure 5D) compared to NEBrepair. The main advantage of IQBErepair was the improved coverage uniformity, resulting in higher coverage at otherwise low-covered regions and hence in less artefacts with very high AAF. Therefore, for the most common FFPE-artefact types, namely C>T/G>A and C>A/G>T (cf. Figure 2A), the number of artefacts per sequenced base, normalised by the respective untreated libraries, was decreased (Figure 5E).
Figure 5.
Effect of FFPE-DNA repair on the on-target sequence coverage and artefacts. Untreated DNA (grey) is compared to DNA treated with BER-mixes, NEBrepair (green) and IQBErepair (magenta), and FF-DNA as a negative control, in two centres (C1, C2). (A) In the coverage curves, the y-axis shows the percentage of target region with coverage of at least x reads. For FFPE-DNA, the magenta and grey curves represent the most and least uniform coverage, respectively. The FF-DNA curves are concordant. The number of replicates is shown in the inset legend with D: duplicate, Q: quadruplicate. (B) Coverage uniformity metric F80BP for FFPE-DNA and FF GIAB DNA. F80BP of FFPE-DNA is improved by repair treatments, especially by IQBErepair. The number of libraries (N) is given in the lower region of the bar chart. (C) Artefact allele frequencies of FFPE-DNA and FF GIAB control DNA. Improved coverage (cf. panels A–C) and reduced artefact occurrence (cf. panel E) lower the median AAF, generally leading to significant differences for repaired FFPE-DNA, regardless of artefact type. (D) Sequence duplication ratios. The restauration of damaged genomic fragments lowers the duplicate ratios for repaired FFPE-DNA. (E) Normalised relative artefact frequency, i.e. the number of artefacts per sequenced base in the repaired DNA, normalised by the untreated DNA. The frequency of deamination C>T/G>A artefacts is considerably reduced by DNA repair, while oxidisation C>A/G>T artefacts are only mitigated by IQBErepair.
Challenges in FFPE-DNA and its repair encompass chimeric reads, i.e. observed reads that include sequences from two distant genomic loci, falsely implying genomic fusions such as fusion genes or structural variation. FFPE-DNA chimeras are most likely caused by spontaneous priming of randomly reverse complementary fragments (64). In our example, a higher number of chimeric reads were observed in IQBErepair that, however, could be removed by appropriate bioinformatic filtering. Another challenge is mechanical ultrasonication DNA fragmentation in some NGS protocols. While IQBErepair improved results for low-input (50 ng) enzymatically prepared tagmentase (Tn5-transposase (65)) libraries, it did not have an effect on the coverage distribution of DNA sheared by ultrasonication in a high-input (200 ng) protocol (Supplementary Figure S5). Lastly, Figure 5 shows that these BER-based repair methods did not lead to significant differences in the negative control (GIAB FF) compared to untreated FF-DNA.
We conclude that repair treatment of damaged FFPE-DNA is an option for specimens that are small and irreplaceable. Such treatment can be considered especially for precious historical samples or for focused, hypothesis-driven studies of rare clinical conditions. In such cases, instead of a sole glycosylase treatment, it is recommended to use a BER-based repair protocol that restores fragments based on the complementary strand. As shown by the experimental example, the inherently low availability of intact fragments in FFPE-DNA leads to poor uniformity of coverage with locally low-covered regions (Figure 2F), where artefacts result in more intense signals. This is the key hurdle to overcome for correct analysis and interpretation of mutation profiles in severely damaged FFPE samples. The high number of alterations, whether mechanistically associated with FFPE treatment or formed by unknown mechanisms, has been observed in many studies (22,23,35,60,66,67), and these can be better addressed by the recovery of additional DNA fragments than through the sole excision of damaged bases.
PARAMETER III: OPTIMISING ANALYTICAL SAMPLE PREPARATION
The steps of library preparation from FFPE-DNA and optional enrichment for target fragments are collectively considered here as analytical sample preparation steps. It is difficult to prepare FFPE-DNA for NGS on a high throughput scale, due to sample variability (damage, fragmentation, etc.). Fragmentation by shearing is usually performed to generate required DNA sizes for NGS. FFPE-DNA requires gentler shearing settings than in standard protocols to achieve these desired fragment sizes. Ideally, fragmentation parameters should be fine-tuned for individual samples to avoid over-fragmentation, or, when the DNA is already extremely fragmented, the fragmentation step can be skipped (68). Even when FFPE-DNA is relatively intact, it is quite fragile due to the presence of single-strand breaks and AP sites, so it should be treated extremely gently.
The question whether ultrasonication or enzymatic approaches should be used for FFPE-DNA is debatable. Ultrasonication fragmentation appears to cause irreversible DNA damage which, in our experience, could not be remedied using the tested FFPE-DNA repair treatments. However, ultrasonication also has certain advantages, for instance it allows better control of fragment size, and it serves well to remove compromised DNA molecules from the pool. When ultrasonication is applied, the introduction of additional oxidative base alterations should be avoided, for which ultrasonication is best carried out in Tris-EDTA buffer (69).
End repair of nucleotide overhangs and dA-tailing, commonly performed in ultrasonication protocols, constitutes another critical step whereby artefacts can get entrenched (70). Tagmentation (71) or other enzymatic fragmentation techniques (68) are mild alternatives to ultrasonication fragmentation. Tagmentase libraries have been reported to be input-efficient (10) and to produce results for high-quality FFPE-DNA that are comparable to FF-DNA (72). Finally, for high-quality FF-DNA, the measured input mass is almost equal to the usable amount of DNA. In contrast, the input mass measurement of FFPE-DNA typically overestimates the usable fraction of DNA, so that the amounts should be adjusted accordingly.
General analytical measures
A number of other options may be considered alone or in combination to improve sample preparation. Targeted enrichment has been standard practice to increase the coverage in genomic regions of interest and for decreasing FFPE-caused noise (40). When enough material is available, technical replicates from the same sample can be prepared, as these greatly reduce false positives (73). Library preparation approaches that use hairpin adapters (74), which are cleaved by UDG and Endonuclease VIII, may help to increase the library conversion rate (75) and hence library complexity. Library protocols that leverage single-stranded DNA (ssDNA) present in FFPE-DNA can further increase library complexity (76,77) and hence improve the output. As ssDNA suffers from elevated levels of artefacts, it is advised to suppress their contribution, for instance by application of dedicated glycosylase treatments (78), since the original genomic sequence cannot be restored due to the lack of a complementary template strand.
Using a single library approach, DNA repair could reduce the number of artefacts by 20–40% compared to untreated DNA (Figure 6, inset table). Multi-library approaches may be considered to improve these results. These can be simple or optimised: replicates can be used to remove artefacts by only keeping variants detected in each library (79). For this approach it can be advantageous to use untreated DNA for one library preparation, and repaired DNA for the library preparation of the replicate, or, alternatively, to use two different DNA repair protocols. Pseudorandomised artefacts (80) cancel each other out in replicate combinations, making technical replicates especially useful in NGS of FFPE-DNA (10,81,82).
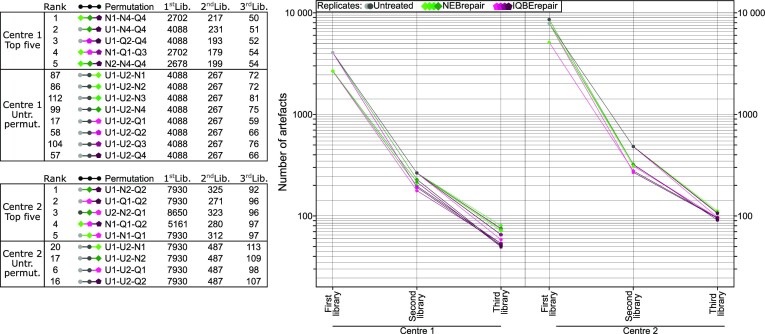
Figure 6.
Permutation analysis to identify the top library replicate strategies. Artefacts were bioinformatically filtered by their presence in library replicates of untreated and repaired FFPE-DNA. The choice of library combination in a multi-library approach can lead to a different number of remaining artefacts. Here, all possible permutations of libraries were bioinformatically tested. The top permutations for artefact removal are depicted in the graph and the tables for FFPE-DNA replicates processed in two sequencing centres. In addition, all permutations of untreated libraries are included. Untreated FFPE-DNA (U, grey), NEBrepaired FFPE-DNA (N, green), and IQBErepaired FFPE-DNA (Q, magenta) libraries were used. For this combined analysis a 1% VAF detection threshold was applied and artefacts that did not pass this VAF filter in all libraries of the doublets or triplets, respectively, were removed.
Figure 6 illustrates the reduction in number of artefacts for the five best permutations of multi-libraries, based on the pure datasets of artefacts. The number of artefacts was reduced by approximately 94% when two libraries were combined. Adding a third library only marginally improved the filtering, to a 98% total reduction of number of artefacts. Replicates that combined two different DNA repair protocols, or untreated and repaired DNA, consistently performed best to filter out artefacts, whereas library replicates based on identical DNA treatment resulted in less effective filtering. Compared to untreated FFPE-DNA library doublets, doublets involving repaired FFPE-DNA reduced artefacts by 15–45%, with a stronger reduction by IQBErepair than NEBrepair. The combination of untreated and repaired FFPE-DNA to prepare a library doublet appears to be a good compromise for practical purposes. The number of unfiltered artefacts depends on the VAF filter threshold. In the presented example we applied a 1% VAF filter threshold, in order to demonstrate that even highly resolved data can be obtained from old and degraded FFPE samples, as required for many cancer NGS applications.
In NGS, multiple libraries are sequenced in a pool on a single flow cell. Therefore, it is important to equally pool and sequence all samples, avoiding unequal sequencing of just one or few samples. In the case of FFPE-DNA, unequal sequencing output is commonly observed (83), resulting in inferior coverages or even total sample drop-out. Target enrichment is commonly performed in pools of multiple libraries (multiplex enrichment) before sequencing. Multiplex enrichments are inexpensive, however, they exacerbate the problem of unequal sequencing output. This can be mitigated by pooling libraries of similar fragment sizes to the same pool. On the other hand, single-plex target enrichment can allow individual balancing of the final product. Furthermore, using single-plex enrichments, an underrepresented sample can easily be rebalanced and repeated in a second NGS run. Optimal balancing before sequencing can be achieved by qPCR quantification, which however adds an additional step compared to balancing by DNA mass alone.
If sequencing costs need to be optimised, then a sequencing kit with fewer cycles (read length) but higher output (number of reads) can be chosen after the median library sizes have been measured. Finally, small, low-cost pilot experiments to fine-tune all conditions are strongly encouraged prior to large-scale sequencing projects with FFPE-DNA, as each different FFPE-DNA source may require different optimal parameters. While these are current workarounds, it will be exciting to see which developments emerge in the coming years to improve the challenges of FFPE library preparation and balancing.
UMIs and their analytical use with FFPE-DNA
In contrast to replicate strategies that can be used to remove artefacts on the library level, unique molecular identifiers (UMIs) enable bioinformatic filtering at the molecular level. UMIs are a set of random (or sufficiently unique) nucleotides that are typically introduced into one or both sequencing adapters and label an individual source DNA or RNA molecule (84), making it distinct from other DNA or RNA fragments with the same sequence. Originally, the first UMI-based NGS library preparation protocols were developed with the aim of counting the exact absolute numbers of molecules (85), for error correction of the fidelity limitations in NGS (86,87) or for needle-in-a-haystack applications, the search for variants at the NGS detection limit (88–92). Later, UMIs were adapted and used to overcome artefacts resulting from formalin (81).
UMIs collate PCR progenies to their original source molecule so that their read family can ultimately be collapsed bioinformatically (89,93). The simplest form of collapsing is to select one representative read of a family (Figure 7, UMI dedup), e.g. randomly picking a specific read with the highest base quality sum. Single-UMI methods cannot trace back the source molecule to the original template strand. However, dual UMIs can be used to assign whether their source molecule originated from the Watson or Crick strand. State-of-the-art dual-UMI approaches featuring this functionality are Duplex (89) and SaferSeqS (94). Such dual-UMI approaches enable more complex bioinformatic processing strategies.
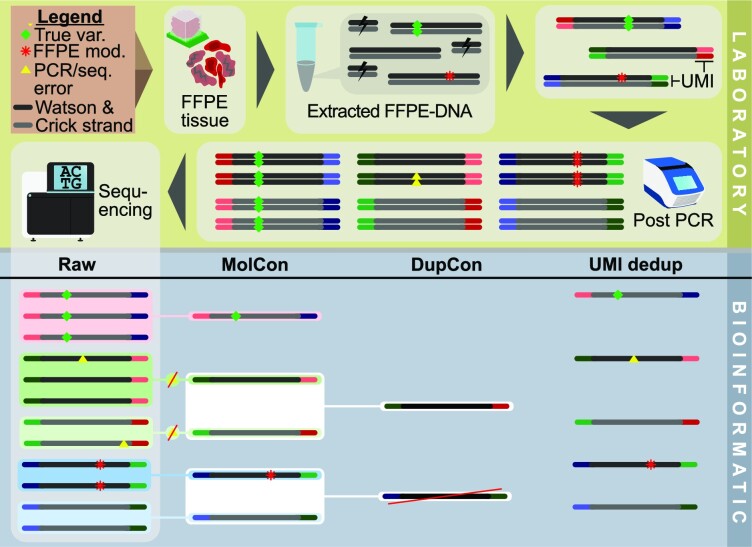
Figure 7.
Analytical use of dual UMIs in the context of FFPE-DNA sequencing. In the laboratory (top part), extraction of FFPE-DNA from formalin impaired tissue results in a low diversity of functional molecules. In general, true variants (green diamond) occur in both strands whereas FFPE modifications (red asterisk) are theoretically restricted to one strand. During library preparation, adapters containing UMI sequences are ligated to both strands. The product is amplified by PCR. During PCR and sequencing, additional errors occur (yellow triangles). Only a fraction of the library's diversity is analysed during sequencing. Overrepresentation of molecules that are preferentially amplified affect the read diversity. Bioinformatic processing (bottom part) of raw reads can group reads belonging to a read family to build the molecular consensus (MolCon) using a statistical model with error removal. The duplex consensus (DupCon) combines both molecular consensuses of the Watson and Crick strands. DupCon allows single-stranded FFPE modifications to be detected and removed, as the molecular consensuses of the single strands (MolCon) are contradictory. However, true variants get suppressed (red raw read group) if the complementary molecule is not sequenced. In the right column, deduplication using UMI (UMI dedup) randomly picked a read from each family. Compared to UMI dedup, consensus approaches reduce errors, although they also result in lower coverage.
In combination with appropriate bioinformatic tools, dual UMIs can be used to error-correct the read families on a strand level. Variants that are observed only in a fraction of redundant reads from a given read family (e.g. caused by polymerase or sequencing errors) are suppressed. Consensus reads can be generated on the molecular, single strand level, where they represent the consensus of all redundant reads derived from one single strand source molecule (Figure 7, MolCon). Alternatively, two of these single strand consensuses can in turn be used to compute a consensus sequence representing the information of the complementary double strand source molecules (Figure 7, DupCon) (89,95). This approach is often referred to as a Duplex (89) UMI method.
Duplex consensus (DupCon) approaches confer the ability to reduce artefacts resulting from formalin modifications, as reads can be discarded in case the complementary sequences originating from one double strand do not match. Figure 7 illustrates a complete overview on the analytical use and an exemplary logic of UMI filtering in context of FFPE. Suppression of formalin-induced artefacts is exemplified by the blue raw read groups, where the formalin artefact is present on the Watson strand reads and not present on the Crick strand reads. In this example, the result of the DupCon is the rejection of the read.
However, due to the nature of the UMI approach that leverages read redundancies, such approaches require significantly deeper sequencing efforts per sample than conventional approaches; the data loss and computational resources required to calculate the consensus can be considerable (96,97). Despite using an UMI (MolCon) approach, Bhagwate et al. (81) experienced a very high number of variant calls in FFPE samples and only their additional application of a 5% VAF filter could remove 92% of FFPE related artefacts (81). These authors therefore recommended the use of FFPE-FF sample pairs when possible, or at least the inclusion of FFPE replicates.
In general, bioinformatic processing involves splitting a respective read into the template and the UMI sequence. The latter is then added to a tag of the specific read in the alignment file. After alignment of the reads and merging of the read's tag and the alignment information, the reads can be grouped. Subsequently, the desired consensus sequence from each family can be generated. Additional filters can be applied on the consensus data, such as a minimal number of reads for the consensus call, a maximal read or base error rate of the read family, or a minimal consensus base quality phred score. Alternatively, there are bioinformatic tools that do not require an alignment of the reads (98). Some library kits with UMI deliberately reduce the diversity of UMIs with a set of predefined UMI sequences that are less sensitive to sequencing errors within the UMI (84,99).
In conclusion, for sequencing studies, we recommend various workflow adaptations compared to FF-DNA such as gentler and sample-individual preparation and shearing conditions, the use of higher input amounts of FFPE-DNA, the application of targeted enrichment in single-plex reactions, and the use of replicate experiments. Replicate libraries improve the specificity of bioinformatic variant calling, especially when low allele frequency mutations are expected in a specimen. The replicate library approach is recommended for tumour mutation burden analysis when tumour mutation allele frequencies are typically low (73), for intra-tumoral heterogeneity, for low tumour content in a FFPE tissue sample from which the FFPE-DNA was isolated, or for detection of sub-clonal mutations with metastatic potential and clinical actionability. The use of library preparation kits with dual UMIs is particularly suitable for severely impaired FFPE-DNA of low diversity, however, to leverage their potential in bioinformatic filtering, significantly deeper sequencing is required.
PARAMETER IV: BIOINFORMATIC CONSIDERATIONS
Bioinformatic analyses are designed to identify the most relevant information from the flood of generated sequence data. As already illustrated in the previous sections, the data derived from FFPE-DNA are distinctly different from FF-DNA data, and typically suffer from low-coverage regions, short insert sizes, and changes in the artefact repertoire. Therefore, it is necessary to optimise the bioinformatic analysis for FFPE samples to correct for this as best as possible, while at the same time sensitivity and specificity must be maintained.
General bioinformatic measures
Bioinformatic filtering is the application of computational inclusion or exclusion criteria that may use a single criterion (e.g. variant quality score filtering) or multiple criteria of arbitrary complexity (e.g. variant quality and gene of interest). In FFPE-DNA sequence analysis, the exclusion of detected variants with VAF < 5% or 10% is commonplace, which may exclude true variants of importance or interest. Therefore, Do et al. suggested that such excluded variants of interest with VAF <10% may be manually re-analysed (23). The allelic frequency threshold depends on the research question: for germline variants a 20% threshold may be used. In the context of somatic variant calling in tumour material the desirable threshold may be as low as 1%, depending on the expected tumour content.
The mapping quality of aligned sequences can be bioinformatically filtered to remove chimeric reads from datasets. These chimeric reads are commonly observed in FFPE-DNA (64) and in repaired DNA. Removal of reads with low mapping quality does not generally affect the variant calling sensitivity. To demonstrate this, after alignment with a Burrows-Wheeler algorithm (100), the alignments of our example were filtered with a mapping quality threshold of 20 (Supplementary Figure S6). Further filtering criteria in FFPE-DNA analyses include thresholds for minimal coverage and minimal base quality (80).
Probabilistic variant callers use statistical models that assess multiple characteristics of observed variants and compute their respective probabilities of being artefacts. However, the underlying models used for determining probabilities can be radically different, and the validity of their results may be limited to their specific application area only. For the challenging task of true somatic mutation calling at low allelic frequency, some model strategies incorporate criteria to detect alignment artefacts, strand and orientation bias artefacts, polymerase slippage artefacts, and contamination. Other models assess observed variants based on global nucleotide or local mismatch rates (80) or call variants above a model-determined sample-specific noise threshold (101).
More recently, machine learning techniques have been leveraged for a broader feature set to classify variants (102). Just as for probabilistic models, machine learning models are restricted to their specific use case. The dependency is even restricted to their training data set: when this dataset is not comprehensive enough and contains a variety of different samples and preparation strategies they cannot easily be applied to other data.
Eventually, a combination of predictions from different models may increase both the precision and sensitivity of variant calling (103,104). With tools such as GenSearchNGS (105), variant-lists from different programs can be imported and combined, and deterministic filters (e.g. VAF, strand and position balance) can be applied in real-time, after which the variants of interest can be manually assessed.
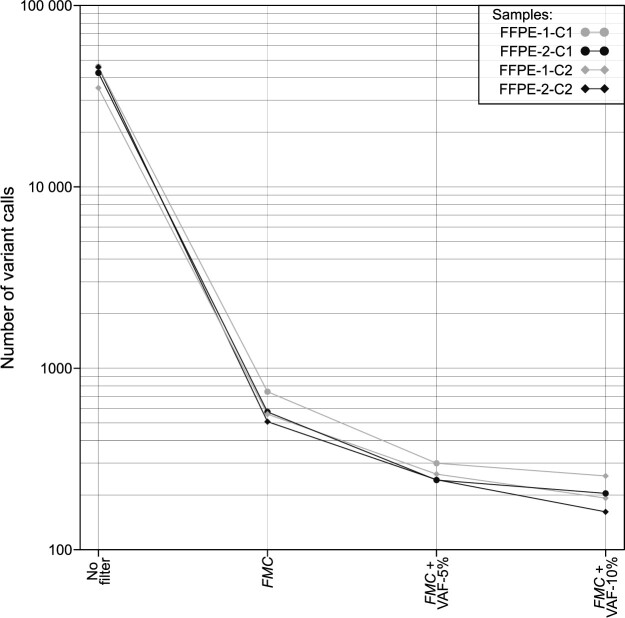
To demonstrate the effectiveness of suppressing false positive variant calls (artefacts), Figure 8 shows the number of false positives for a probabilistic bioinformatic filter alone and in combination with deterministic VAF filtering. Supplementary Table S1 lists all settings used in this figure, from raw sequencing files to analysis. The probabilistic variant caller GATK Mutect2 (10,106) reduces false positive variant calls based on the combined likelihood of diverse parameters, and even more when the FilterMutectCalls (FMC) postprocessing program is applied. FMC alone removed approximately 98% of initial unfiltered false positive variant calls, achieving a reduction by a factor of 58. The application of FMC and additionally a VAF filter of 5% or 10% decreased false positives 250-fold or 400-fold compared to unfiltered data. However, such additional VAF filters limit the sensitivity of variant calling.
Figure 8.
Probabilistic bioinformatic filters consistently reduce artefacts. This figure shows the number of false-positive variant calls (y-axis) in four untreated FFPE-DNA replicates processed in two different sequencing centres (C1, C2). ‘No filter’ refers to the total number of false-positive variant calls prior to filtering. The number of false positives was reduced using the probabilistic filter FMC (FilterMutectCalls) of GATK Mutect2 variant calling alone, or in combination with VAF-based filtering (VAF threshold 5% or 10%). All variant calls in this figure are false positives resulting from FFPE-DNA damage or other causes (e.g. sequencing error). Over 100 false positives remain even after combined FMC and 10% VAF-filtering.
Generally, we recommend adjusting the bioinformatic settings according to the sample quality. While removing most FFPE artefacts, weak true signals in low-covered regions may potentially be lost if filters are set with too strict thresholds. Therefore, we suggest the careful application of deterministic and probabilistic filters when the purity of a sample is high (e.g. a tumour cell content ≥ 50% as has been described (10)) and a more conservative, cautious approach when sample purity or DNA yield is lower.
Bioinformatic UMI filtering
The effects of the three different bioinformatic UMI-filtering approaches that were summarised in Figure 7 were put to the test with the 13-year-old FFPE-DNA vs. FF-DNA sample pair obtained from the same surgical resection specimen. Figure 9 summarises the effects of the different bioinformatic approaches that were applied to the same down-sampled datasets. The data were processed using (i) a simple deduplication approach based on the start/stop coordinates of the reads, (ii) a deduplication approach additionally involving the UMI information, (iii) a more sophisticated approach generating an error-corrected molecular consensus by single read families on the single strand level and (iv) generating a duplex consensus by combined read families representing the double stranded source molecules.
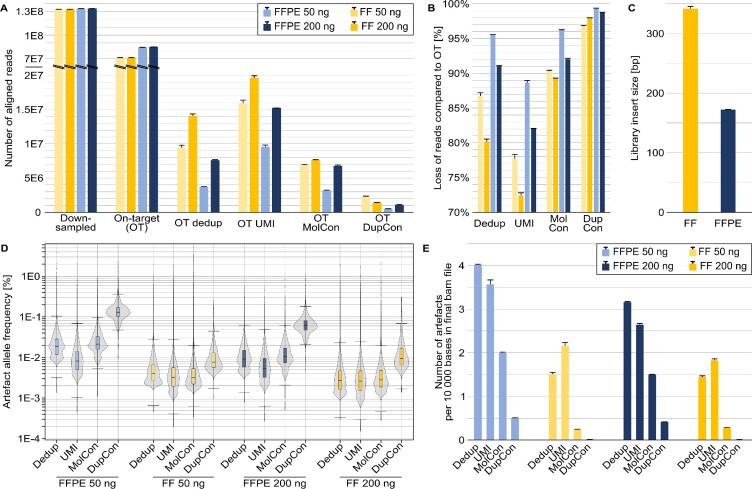
Figure 9.
Effect of four bioinformatic read filtering methods on library sequences with dual UMIs. Data are shown for the 13-year-old FFPE and FF sample pair, with library preparation in replicates for each input amount of 50 and 200 ng (light and dark colours, respectively) of FFPE-DNA (blue) and FF-DNA (orange). The eight libraries were target-enriched and deep sequenced. The sequence data were bioinformatically down-sampled to the identical number of 1.3E8 raw sequencing reads per library and aligned to the human reference genome, referred to as on-target (OT) reads and off-target reads. Four different bioinformatic filtering approaches are shown: standard deduplication by the read start-stop positions (dedup), deduplication by additionally using the UMI information (UMI), molecular consensus (MolCon) error correction by collapsing single read families, and duplex consensus (DupCon) where error correction was done by collapsing combined read families. (A) Number of reads per experiment. Note the y-axis scale break. (B) Percentual loss of reads following the recommended data processing compared to the raw OT data. (C) Differences in median insert size for the FF and FFPE libraries. (D) Artefact allele frequencies for the different approaches used. (E) Number of artefacts observed per 10 000 bases in the final alignment file.
Figure 9A shows how the number of reads is reduced depending on the filtering approach. In most cases the loss of reads is greater for FFPE-DNA than for FF-DNA. The loss of reads in comparison to the raw on-target reads (Figure 9B) ranged from 72% to 99%. UMI filtering (ii) gives a representative picture of unique sequencing reads, whereas simple deduplication (i) results in extensive loss of reads. Simple deduplication potentially underestimates the library complexity (107), which can lead to the removal of weak but true signals.
Significant read loss occurs when UMI-based error correction through read family consensus computation (iii, iv) is performed, resulting in severely decreased coverage. The true coverage is further impaired if the library insert sizes are as short as 120 bp for FFPE-DNA (Figure 9C), resulting in overlap of the forward and reverse reads. These two coverage reduction effects have the consequence that some AAFs can become unexpectedly high: in the 13-year-old test sample, despite very deep sequencing of a relatively small genomic target, the outliers almost reached 5% AAF (Figure 9D). The higher input amount of FFPE-DNA (200 ng versus 50 ng) led to higher library complexity and reduced the AAF.
Significant read loss can be a fair price to pay if artefacts are reliably removed by the UMI-based consensus sequence approaches. As an example, the number of artefacts per 10 000 bases in the final bam file of consensus sequences was comprehensively reduced (Figure 9E) in FFPE-DNA and almost completely eliminated in FF-DNA. However, due to the extensive read loss, the remaining artefacts after the consensus-based filter approaches reached relatively high AAFs, especially in FFPE-DNA after the DupCon approach. In summary, the dual UMI-based error correction may not always be the optimal approach to analyse severely damaged FFPE-DNA. If the target region size is large (as in whole exome sequencing), a filtering approach leveraging UMI deduplication in combination with an optimally applied VAF-threshold for variant calling may be more economic than consensus error correction that requires very deep sequencing.
ASSESSING THE RIGOR OF PUBLISHED RESEARCH
Without complete information on study design, methodological details and exact parameters that were applied, it is difficult to assess the quality of results described in publications. For example, based on a meta-analysis of mutations predominantly derived from FFPE tissue samples, Murray et al. (108) reported that 62.2% of all discovered mutations were only reported once. These singleton mutations have been challenged by others (66) as they may (all or in part) represent artefacts. Whether singletons are true findings or artefacts is hard to interpret unless detailed technical information is available. Therefore, it is of utmost importance that studies using FFPE samples adhere to minimal scientific standards of technical information and that these standards are precisely formulated and commonly agreed upon. Here, we describe a number of criteria that we consider to be essential information for reviewers to assess the validity of reported FFPE-DNA variant calls.
Study design
As reviewed in previous sections, FFPE samples are distinctly different from FF samples. A study design should therefore be adapted accordingly. Coverage non-uniformity and high duplication ratios can result in inflated coverage of specific genomic regions. After bioinformatic duplicate removal, their actual coverage can be significantly lower than the expected or overall coverage. Key considerations in a study design include: exclusion of low quality samples (e.g. high degree of DNA fragmentation), maximisation of DNA input amounts where possible, inclusion of DNA repair when the sample quality is low, adapted sample preparation strategies as needed (e.g. use of UMIs or replicates), and appropriate bioinformatic data processing.
In general, every study involving FFPE-DNA should employ a transparent sample processing strategy, that, based on sample quality, allocates samples to respective processing arms containing measures for artefact mitigation. The simplest and most cost-effective processing arm constitutes the exclusion of samples that do not meet minimal quality criteria, which must be defined beforehand. The sequencing depth should be generously increased compared to FF-DNA, before which a pilot experiment can be performed to obtain guidance data.
If amplicon-based targeting of a genomic region is used, UMIs are encouraged, as they allow deduplication to be performed. Alternatively, if the library was prepared for capture-based targeting, deduplication algorithms using the start and end points of the reads perform satisfactorily without UMIs. However, UMIs can also be combined with capture-based library approaches, to more accurately identify the read families originating from the same source molecule.
The data analysis methods should be shown to be appropriate, for example by inclusion of formalin-compromised reference samples matching the lowest-quality study sample. If this is not possible, alternative verification of the suitability of the analysis method should be demonstrated, e.g. by using replicate strategies or FF tissue-matching samples.
Minimal information in publications
Reviewers and editors have the task to check that sufficient information is available for the readers of a scientific article. For studies with FFPE samples, the following information must be included: the type of tissue fixation (buffered or unbuffered formalin); for cancer tissue the tumour cell content; the method used for DNA quantification; the DNA fragment size range; the amount of DNA used for library preparation; the library kit; the number of PCR cycles performed in library preparation and targeted enrichment; a statement whether sequencing was done with single or paired-end reads; the read length; the sequencing equipment used; the method of targeted enrichment (amplicon vs. capture); and bioinformatic thresholds for variant calling (variant sequencing depth, variant reads and coverage at variant position). Reported results should include coverage statistics and duplicate ratios.
To aid reviewers, authors, and study designers, based on similar checklists (109,110), we provide the ‘Essential Recommendations for Reporting On Results from FFPE-DNA (ERROR-FFPE-DNA)’ checklist as a supplemental file to this article.
Misleading focus on artefact count
Many studies postulating FFPE artefact repair strategies have focused on decreasing the absolute count of artefacts, while few have considered AAFs. However, artefact count is a one-sided criterion to benchmark against, as it does not include a quantification of the observed AAF. AAF is more important than the artefact count per se, as low-intensity artefacts can easily be filtered out by VAF filters. To give an extreme example, FFPE-DNA treatment with DNAse would reduce the number of artefacts (appearing to be a good treatment from a one-sided viewpoint), but it would also digest a considerable amount of DNA and significantly decrease the number of useable reads and hence reduce the amount of derivable information. In reality, a higher number of unique reads improves data quality, despite increasing the overall artefact count. The information derived from every additional sequenced DNA molecule improves the identification and discrimination of artefacts and can potentially increase the number of identified true variants, at all genomic coordinates spanned by the read from the recovered DNA molecule.
SUMMARY AND PERSPECTIVES
Biomedical biobanks across the world harbour an immense collection of FFPE tissue specimens, which are generated as part of a diagnostic or treatment approach, e.g. surgical resection of diseased tissue. Linked with relevant patient demographics and clinical data these biorepositories captured the interest of researchers of various disciplines. However, they come with a number of technical challenges, which may have influenced the decision of the 100 000 Genomes Project to collect and use only FF samples for NGS and FFPE samples for surgical-pathological diagnostics. These challenges and consequences of formalin fixation have been summarised by Xiaoet al. (10), however they did not provide FFPE-specific recommendations.
Biobanking of FF samples and alternative fixation methods circumvent the hurdles of formalin fixation (111). In practice, a fresh tissue specimen is split into a sample for FFPE tissue pathological diagnostics and FFPE biobanking, and if sufficient tissue is available, a sample for freezing or non-formalin fixation. This concept has already been adopted by some pathology departments, but it is certainly not yet in universal practice. Therefore, many, and especially small specimens such as fine needle biopsies are usually exclusively available as FFPE specimens. The most important considerations for sequencing these and other FFPE samples from the existing collections are summarised below.
For specimens stored a couple of years, FFPE-DNA sequencing can be carried out reliably and relatively easily for germline and other studies where variants with allele frequencies below 50% are of minor importance. Here, DNA fragmentation criteria can be used to select suitable samples and exclude unsuitable ones for sequencing. For mixed cell populations or somatic mutations, where low-frequency alleles can be important, it is crucial that the study designers consider DNA repair, the most suitable library preparation and appropriate bioinformatic analysis.
It is recommended to use all available FFPE-DNA for sequencing library preparation, rather than a standardised aliquot amount as is common when sequencing fresh, unfixed DNA. FFPE-DNA is fragile and must be handled with care. Library protocols optimised specifically for FFPE-DNA are required to ensure DNA-to-library conversion success. If ultrasonication fragmentation is needed, it should be done in a buffered solution that minimises DNA oxidation. Good alternatives are tagmentase- and enzyme-based protocols. These provide a gentle alternative to ultrasonication and are suitable for larger target region sizes than PCR-amplicon based protocols.
The main challenge in analysing FFPE-DNA sequence data is non-uniform coverage despite a satisfactory total sequencing read output. Drop-outs during library preparation lead to these critical low-covered regions. Consequently, without a sufficient sequencing depth of unique reads in the regions of interest, artefacts cannot be distinguished from true variants. Deeper sequencing to compensate for the drop-outs may recover more unique reads.
The number of unique reads may also be increased by enzymatic repair of damaged DNA using a BER-based protocol, thereby increasing the amount of usable DNA. However, in our experience, BER-based repair protocols (NEBRepair, IQBErepair) lead to a reduction of the measured DNA amounts by 10–40% after repair, in particular in low concentrated samples. Hence, BER-based protocols are only beneficial if the increased amount of sequenced unique bases outweighs the enzymatic repair DNA losses. In our hands, NEBrepair did not improve coverage uniformity but IQBErepair is able to increase unique bases by between 53–80% (Figure 5A).
As a perspective of what may come in future years, we predict that the scientific evaluation of tens of millions of FFPE samples could be performed using the methods available today. Projects such as the International Cancer Genome Consortium (ICGC), The Cancer Genome Atlas (TCGA) and 100 000 Genomes have only begun to scratch the surface of the diversity of cancers. For example, in May 2023, the NIH’s GDC Cancer Portal contained only 1420 sequenced oesophageal cancer entries, a common cancer with a dismal prognosis and limited treatment options. Clearly, extending this repository by using FFPE sequencing would be desirable and beneficial to patients. In this decade of ambitious scientific initiatives, the vast collection of available FFPE cancer tissue samples has still not been systematically investigated. Rare tumour entities and soft tissue tumours might be particular targets for sequencing of FFPE-DNA. Studying this collection will enable cancer entities to be analysed and stratified into specific cancer sub-entities with distinct mutation profiles, to provide a basis for a better understanding of prognosis and treatment failures or improve survival and treatments.
Further research is necessary to address the challenges of limited DNA amounts (e.g. needle biopsies) and poor quality in many FFPE samples - including improvements to DNA extraction, DNA repair and DNA-to-library conversion rates. New library conversion protocols for FFPE ssDNA have recently raised hopes of significantly increasing the amount of usable DNA. Restoration of a broader spectrum of formalin-induced DNA alterations by further refined repair techniques may also help to improve coverage uniformity. Finally, the specific bioinformatic demands related to FFPE-DNA analyses will require new expertise of molecular biologists, mathematicians, statisticians, and bioinformaticians. Currently, the evaluation of FFPE sequence datasets can be difficult and time-consuming highlighting the need for new innovative statistical algorithms that could extract the best possible data from FFPE sequencing replicates. The new algorithms would need to include easily interpretable graphics that summarise the results reliably and comprehensively to maximise the effectiveness and deployment of these tools.
CONCLUSION
FFPE tissue is both a boon and bane for nucleic acid researchers. Tens of millions of well-documented FFPE tissue specimens are immediately available worldwide and suitable for molecular and biomedical research. However, FFPE-DNA sequencing studies are more complex, laborious, and costly, compared to the sequencing of FF-DNA. The necessary considerations and key conclusions of this critical review with respect to sequencing study design are listed in Table 1. For reviewers dealing with the peer review of such studies, we provide a checklist called “ERROR-FFPE-DNA” that summarises recommendations for the minimal, essential technical information that should be provided in a scientific manuscript.
Table 1.
Considerations and key conclusions of this article
| Considerations | Conclusions |
|---|---|
| Formalin-induced alterations to DNA | Modifications in FFPE-DNA mostly occur pseudorandomly, but more frequently in AT-rich genomic regions, leading to a higher prevalence of GC-rich sequences than in FF-DNA. Base modifications, inter-strand cross-linking, base excision, polydeoxyribose fragmentation and cytosine deamination, among others, constitute to the artefact repertoire of FFPE-DNA. |
| Consequences of formalin fixation | Formalin modifications are complex and still not completely understood. Artefacts can be mistaken as true variants, especially if their allelic frequency exceeds filter thresholds, which arises in regions of locally low coverage, which in turn is caused by reduced library complexity and non-uniform coverage. |
| Pre-analytical sample quality and its specifications (Parameter I) | Specimens of a decade or older can be considered if fixed in buffered formalin. Target tissue (e.g. tumour area) should be optimally enriched. FFPE-DNA extraction is critical and should be performed with caution. The average fragment length is an easy-to-determine metric that correlates with coverage uniformity, one of the most important quality criteria in FFPE-DNA sequencing. |
| Optional application of DNA repair treatment (Parameter II) | Repair should be considered especially for severely impaired specimens in smaller hypothesis-driven studies. Simple repair (e.g. UDG treatment) should be avoided in favour of BER-based enzymatic repair protocols. We recommend a BER-based repair protocol that restores fragments to increase coverage evenness and decrease artefact allele frequency. |
| Analytical sample preparation (Parameter III) | When FFPE-DNA is prepared for sequencing, individualised workflow adaptations can improve the outcome. Useful adaptations include higher DNA input amounts, mild shearing conditions or tagmentase, FFPE-specific kits, the use of UMIs, and replicate strategies. |
| Sequencing | In general, up to four-fold deeper sequencing is necessary than for undamaged FF-DNA. Very deep sequencing (e.g. 5000–7500×) is necessary if dual-UMI strand-specific error correction is the aim. |
| Bioinformatic analysis (Parameter IV) | Bioinformatic filters can facilitate the discrimination between true variants and artefacts. Different UMI filter and error correction strategies can drastically reduce artefacts but at the cost of coverage. This might impair the sensitivity in detecting true variants with low VAFs. |
| Assessing the rigor of published research | Not providing enough technical and methodological details limits the scientific quality and integrity of FFPE-studies. An adapted study design and providing a minimal set of information can improve the situation in the future. When developing mitigation strategies, the sole focus on artefact count reduction is too simplistic as the restoration of additional fragments might be more effective. |
By shedding light on the paradigms of FFPE-DNA sequencing, our goal was to suggest standards that can help to make research more comparable and reproducible in this challenging field. Achieving these objectives will help to leverage the power of FFPE-DNA sequencing and provide reliable datasets for their algorithmic exploitation. Ultimately, these steps are a necessary prerequisite for bringing precision medicine another step closer towards its ambitious promises.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yewgenia Dolshanskaya, Regina Fredrik, Dorina Oelsner, Sören Franzenburg, Janina Fuß (all CCGA Kiel) for support on laboratory methods, and Marc Höppner, Georg Hemmrich-Stanisak and Teide Boysen for support on bioinformatics. We gratefully acknowledge Penelope Kungl (Diagnostic & Research Center for Molecular Biomedicine, Graz, Austria) and Steven McGinn (Centre National de Recherche en Génomique Humaine, Évry, France) for their careful proofreading. Furthermore, we thank Krzysztof Grzyb and the Department of Pathology, Oslo University Hospital, Oslo, Norway for their contribution.
Contributor Information
Tim A Steiert, Institute of Clinical Molecular Biology, Christian-Albrechts-University and University Medical Center Schleswig-Holstein, Kiel 24105, Germany.
Genís Parra, Center for Genomic Regulation, Centro Nacional de Análisis Genómico, Barcelona 08028, Spain.
Marta Gut, Center for Genomic Regulation, Centro Nacional de Análisis Genómico, Barcelona 08028, Spain.
Norbert Arnold, Department of Gynaecology and Obstetrics, University Medical Center Schleswig-Holstein, Campus Kiel, Kiel 24105, Germany.
Jean-Rémi Trotta, Center for Genomic Regulation, Centro Nacional de Análisis Genómico, Barcelona 08028, Spain.
Raúl Tonda, Center for Genomic Regulation, Centro Nacional de Análisis Genómico, Barcelona 08028, Spain.
Alice Moussy, Le Centre de référence, d’innovation, d’expertise et de transfert (CRefIX), PFMG 2025, Évry 91057, France.
Zuzana Gerber, Centre National de Recherche en Génomique Humaine (CNRGH), Institut de Biologie François Jacob, CEA, Université Paris-Saclay, Évry 91057, France.
Peter M Abuja, Diagnostic & Research Center for Molecular Biomedicine, Diagnostic & Research Institute of Pathology, Medical University of Graz, Graz 8010, Austria.
Kurt Zatloukal, Diagnostic & Research Center for Molecular Biomedicine, Diagnostic & Research Institute of Pathology, Medical University of Graz, Graz 8010, Austria.
Christoph Röcken, Department of Pathology, University Medical Center Schleswig-Holstein, Campus Kiel, Kiel 24105, Germany.
Trine Folseraas, Norwegian PSC Research Center Department of Transplantation Medicine, Division of Surgery, Inflammatory Medicine and Transplantation, Oslo University Hospital Rikshospitalet, Oslo 0372, Norway; Section of Gastroenterology, Department of Transplantation Medicine, Division of Surgery, Inflammatory Diseases and Transplantation, Oslo University Hospital Rikshospitalet, Oslo 0372, Norway.
Marit M Grimsrud, Norwegian PSC Research Center Department of Transplantation Medicine, Division of Surgery, Inflammatory Medicine and Transplantation, Oslo University Hospital Rikshospitalet, Oslo 0372, Norway; Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo 0372, Norway.
Arndt Vogel, Department of Gastroenterology, Hepatology and Endocrinology, Hannover Medical School, Hanover 30625, Germany.
Benjamin Goeppert, Institute of Pathology, University Hospital Heidelberg, Heidelberg 69120, Germany; Institute of Pathology and Neuropathology, RKH Klinikum Ludwigsburg, Ludwigsburg 71640, Germany.
Stephanie Roessler, Institute of Pathology, University Hospital Heidelberg, Heidelberg 69120, Germany.
Sebastian Hinz, Department of General Surgery, University Medicine Rostock, Rostock 18057, Germany.
Clemens Schafmayer, Department of General Surgery, University Medicine Rostock, Rostock 18057, Germany.
Philip Rosenstiel, Institute of Clinical Molecular Biology, Christian-Albrechts-University and University Medical Center Schleswig-Holstein, Kiel 24105, Germany.
Jean-François Deleuze, Le Centre de référence, d’innovation, d’expertise et de transfert (CRefIX), PFMG 2025, Évry 91057, France; Centre National de Recherche en Génomique Humaine (CNRGH), Institut de Biologie François Jacob, CEA, Université Paris-Saclay, Évry 91057, France.
Ivo G Gut, Center for Genomic Regulation, Centro Nacional de Análisis Genómico, Barcelona 08028, Spain.
Andre Franke, Institute of Clinical Molecular Biology, Christian-Albrechts-University and University Medical Center Schleswig-Holstein, Kiel 24105, Germany.
Michael Forster, Institute of Clinical Molecular Biology, Christian-Albrechts-University and University Medical Center Schleswig-Holstein, Kiel 24105, Germany.
Data Availability
The datasets generated and analysed during the current study are available in the European Genome-phenome Archive (EGA) repository, EGAS00001005757. The EGA is a controlled-access human data repository subject to European data protection laws. Therefore, data access is subject to an application, ethics approval by the applicant's ethics board and a data access agreement.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union's Horizon 2020 research and innovation program European Advanced infraStructure for Innovative Genomics, EASI-Genomics [824110] (NGS analyses were carried out at the EASI-Genomics infrastructure in Spain, France, and Germany); Integrated and standardised NGS workflows for Personalised therapy, Instand-NGS4P [874719]; German Research Foundation (DFG) Research Infrastructure NGS_CC and CRC SFB/TR 209 Liver Cancer [407495230, 314905040, 497786653, 493697503 to S.R. and B.G.]; as part of the Next Generation Sequencing Competence Network, Competence Center for Genomic Analysis (CCGA) Kiel [423957469]; German Cancer Aid [70113922]; Spanish Instituto de Salud Carlos III, Fondo de Investigaciones Sanitarias and cofounded with ERDF funds [PI19/01772]; Spanish Ministry of Science and Innovation through the Instituto de Salud Carlos III and the 2014–2020 Smart Growth Operating Program, to the EMBL partnership and co-financing with the European Regional Development Fund [MINECO/FEDER, BIO2015-71792-P]; Centro de Excelencia Severo Ochoa, and the Generalitat de Catalunya through the Departament de Salut, Departament d’Empresa i Coneixement and the CERCA Programme. Funding for open access charge: EU funds (EASI-Genomics).
Conflict of interest statement. The authors have no relevant affiliations or financial involvement with any organisation or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
REFERENCES
- 1. Blum F. Notiz über die Anwendung des Formaldehyds (Formol) als Härtungs-und Konservierungsmittel. Anat. Anz. 1894; 9:229–231. [Google Scholar]
- 2. Seiler C., Sharpe A., Barrett J.C., Harrington E.A., Jones E.V., Marshall G.B.. Nucleic acid extraction from formalin-fixed paraffin-embedded cancer cell line samples: a trade off between quantity and quality. BMC Clin. Pathol. 2016; 16:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lewis F., Maughan N., Smith V., Hillan K., Quirke P.. Unlocking the archive–gene expression in paraffin-embedded tissue. J. Pathol. 2001; 195:66–71. [DOI] [PubMed] [Google Scholar]
- 4. Arreaza G., Qiu P., Pang L., Albright A., Hong L.Z., Marton M.J., Levitan D. Pre-analytical considerations for successful next-generation sequencing (NGS): challenges and opportunities for formalin-fixed and paraffin-embedded tumor tissue (FFPE) samples. Int. J. Mol. Sci. 2016; 17:1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Pineros M., Znaor A., Bray F.. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019; 144:1941–1953. [DOI] [PubMed] [Google Scholar]
- 6. Asslaber M., Zatloukal K.. Biobanks: transnational, European and global networks. Brief. Funct. Genomics Proteomics. 2007; 6:193–201. [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y., Blomquist T.M., Kusko R., Stetson D., Zhang Z., Yin L., Sebra R., Gong B., Lococo J.S., Mittal V.K.. Deep oncopanel sequencing reveals within block position-dependent quality degradation in FFPE processed samples. Genome Biol. 2022; 23:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo Q., Lakatos E., Bakir I.A., Curtius K., Graham T.A., Mustonen V.. The mutational signatures of formalin fixation on the human genome. Nat. Commun. 2022; 13:4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thapa M.J., Fabros R.M., Alasmar S., Chan K.. Analyses of mutational patterns induced by formaldehyde and acetaldehyde reveal similarity to a common mutational signature. G3 - Genes Genomes Genet. 2022; 12:jkac238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao W., Ren L., Chen Z., Fang L.T., Zhao Y., Lack J., Guan M., Zhu B., Jaeger E., Kerrigan L.. Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat. Biotechnol. 2021; 39:1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Srinivasan M., Sedmak D., Jewell S.. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 2002; 161:1961–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beland F.A., Fullerton N.F., Heflich R.H.. Rapid isolation, hydrolysis and chromatography of formaldehyde-modified DNA. J. Chromatogr. B Biomed. Appl. 1984; 308:121–131. [DOI] [PubMed] [Google Scholar]
- 13. McGhee J.D., Von Hippel P.H. Formaldehyde as a probe of DNA structure. 3. Equilibrium denaturation of DNA and synthetic polynucleotides. Biochemistry. 1977; 16:3267–3276. [DOI] [PubMed] [Google Scholar]
- 14. Gilbert M.T.P., Haselkorn T., Bunce M., Sanchez J.J., Lucas S.B., Jewell L.D., Van Marck E., Worobey M.. The isolation of nucleic acids from fixed, paraffin-embedded tissues–which methods are useful when?. PLoS One. 2007; 2:e537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Douglas M.P., Rogers S.O.. DNA damage caused by common cytological fixatives. Mutat. Res. - Fundam. Mol. Mech. 1998; 401:77–88. [DOI] [PubMed] [Google Scholar]
- 16. Lindahl T., Andersson A.. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972; 11:3618–3623. [DOI] [PubMed] [Google Scholar]
- 17. An R., Jia Y., Wan B., Zhang Y., Dong P., Li J., Liang X.. Non-enzymatic depurination of nucleic acids: factors and mechanisms. PLoS One. 2014; 9:e115950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sagher D., Strauss B.. Insertion of nucleotides opposite apurinic apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983; 22:4518–4526. [DOI] [PubMed] [Google Scholar]
- 19. Sikorsky J.A., Primerano D.A., Fenger T.W., Denvir J.. DNA damage reduces Taq DNA polymerase fidelity and PCR amplification efficiency. Biochem. Biophys. Res. Commun. 2007; 355:431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Craig J.M., Vena N., Ramkissoon S., Idbaih A., Fouse S.D., Ozek M., Sav A., Hill D.A., Margraf L.R., Eberhart C.G.. DNA fragmentation simulation method (FSM) and fragment size matching improve aCGH performance of FFPE tissues. PLoS One. 2012; 7:e38881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bonin S., Petrera F., Niccolini B., Stanta G.. PCR analysis in archival postmortem tissues. Mol. Pathol. 2003; 56:184–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen G., Mosier S., Gocke C.D., Lin M.-T., Eshleman J.R.. Cytosine deamination is a major cause of baseline noise in next-generation sequencing. Mol. Diagn. Ther. 2014; 18:587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Do H., Wong S.Q., Li J., Dobrovic A.. Reducing sequence artifacts in amplicon-based massively parallel sequencing of formalin-fixed paraffin-embedded DNA by enzymatic depletion of uracil-containing templates. Clin. Chem. 2013; 59:1376–1383. [DOI] [PubMed] [Google Scholar]
- 24. Do H., Dobrovic A.. Limited copy number-high resolution melting (LCN-HRM) enables the detection and identification by sequencing of low level mutations in cancer biopsies. Mol. Cancer. 2009; 8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lamy A., Blanchard F., Le Pessot F., Sesboüé R., Di Fiore F., Bossut J., Fiant E., Frébourg T., Sabourin J.-C.. Metastatic colorectal cancer KRAS genotyping in routine practice: results and pitfalls. Mod. Pathol. 2011; 24:1090–1100. [DOI] [PubMed] [Google Scholar]
- 26. Wong C., DiCioccio R.A., Allen H.J., Werness B.A., Piver M.S.. Mutations in BRCA1 from fixed, paraffin-embedded tissue can be artifacts of preservation. Cancer Genet. Cytogenet. 1998; 107:21–27. [DOI] [PubMed] [Google Scholar]
- 27. Goelz S.E., Hamilton S.R., Vogelstein B.. Purification of DNA from formaldehyde fixed and paraffin embedded human tissue. Biochem. Biophys. Res. Commun. 1985; 130:118–126. [DOI] [PubMed] [Google Scholar]
- 28. Rogers B.B., Alpert L., Hine E., Buffone G.. Analysis of DNA in fresh and fixed tissue by the polymerase chain reaction. Am. J. Pathol. 1990; 136:541–548. [PMC free article] [PubMed] [Google Scholar]
- 29. Ben-Ezra J., Johnson D.A., Rossi J., Cook N., Wu A.. Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by the polymerase chain reaction. J. Histochem. Cytochem. 1991; 39:351–354. [DOI] [PubMed] [Google Scholar]
- 30. Millán-Esteban D., Reyes-García D., García-Casado Z., Bañuls J., López-Guerrero J.A., Requena C., Rodríguez-Hernández A., Traves V., Nagore E.. Suitability of melanoma FFPE samples for NGS libraries: time and quality thresholds for downstream molecular tests. Biotechniques. 2018; 65:79–85. [DOI] [PubMed] [Google Scholar]
- 31. Sie D., Snijders P.J., Meijer G.A., Doeleman M.W., van Moorsel M.I., van Essen H.F., Eijk P.P., Grünberg K., van Grieken N.C., Thunnissen E.. Performance of amplicon-based next generation DNA sequencing for diagnostic gene mutation profiling in oncopathology. Cell. Oncol. 2014; 37:353–361. [DOI] [PubMed] [Google Scholar]
- 32. Filia A., Droop A., Harland M., Thygesen H., Randerson-Moor J., Snowden H., Taylor C., Diaz J.M.S., Pozniak J., Nsengimana J.. High-resolution copy number patterns from clinically relevant FFPE material. Sci. Rep. 2019; 9:8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hedegaard J., Thorsen K., Lund M.K., Hein A.-M.K., Hamilton-Dutoit S.J., Vang S., Nordentoft I., Birkenkamp-Demtröder K., Kruhøffer M., Hager H.. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One. 2014; 9:e98187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shibutani S., Takeshita M., Grollman A.P.. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991; 349:431–434. [DOI] [PubMed] [Google Scholar]
- 35. Wong S.Q., Li J., Tan A.Y., Vedururu R., Pang J.-M.B., Do H., Ellul J., Doig K., Bell A., McArthur G.A.. Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med. Genom. 2014; 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robbe P., Popitsch N., Knight S.J., Antoniou P., Becq J., He M., Kanapin A., Samsonova A., Vavoulis D.V., Ross M.T.. Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 Genomes Project. Genet. Med. 2018; 20:1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carrick D.M., Mehaffey M.G., Sachs M.C., Altekruse S., Camalier C., Chuaqui R., Cozen W., Das B., Hernandez B.Y., Lih C.J.et al.. Robustness of next generation sequencing on older formalin-fixed paraffin-embedded tissue. PLoS One. 2015; 10:e0127353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bonnet E., Moutet M.-L., Baulard C., Bacq-Daian D., Sandron F., Mesrob L., Fin B., Delépine M., Palomares M.-A., Jubin C.. Performance comparison of three DNA extraction kits on human whole-exome data from formalin-fixed paraffin-embedded normal and tumor samples. PloS One. 2018; 13:e0195471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oh E., Choi Y.-L., Kwon M.J., Kim R.N., Kim Y.J., Song J.-Y., Jung K.S., Shin Y.K.. Comparison of accuracy of whole-exome sequencing with formalin-fixed paraffin-embedded and fresh frozen tissue samples. PLoS One. 2015; 10:e0144162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kerick M., Isau M., Timmermann B., Sültmann H., Herwig R., Krobitsch S., Schaefer G., Verdorfer I., Bartsch G., Klocker H.. Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med. Genom. 2011; 4:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cho M., Ahn S., Hong M., Bang H., Van Vrancken M., Kim S., Lee J., Park S.H., Park J.O., Park Y.S.. Tissue recommendations for precision cancer therapy using next generation sequencing: a comprehensive single cancer center's experiences. Oncotarget. 2017; 8:42478–42486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abuja P.M., Pabst D., Bourgeois B., Loibner M., Ulz C., Kufferath I., Fackelmann U., Stumptner C., Kraemer R., Madl T.et al.. Residual humidity in paraffin-embedded tissue reduces nucleic acid stability. Int. J. Mol. Sci. 2023; 24:8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Werner M., Chott A., Fabiano A., Battifora H.. Effect of formalin tissue fixation and processing on immunohistochemistry. Am. J. Surg. Pathol. 2000; 24:1016–1019. [DOI] [PubMed] [Google Scholar]
- 44. Cross S., Start R., Smith J.. Does delay in fixation affect the number of mitotic figures in processed tissue?. J. Clin. Pathol. 1990; 43:597–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kingsbury A.E., Foster O.J., Nisbet A.P., Cairns N., Bray L., Eve D.J., Lees A.J., Marsden C.D.. Tissue pH as an indicator of mRNA preservation in human post-mortem brain. Mol. Brain Res. 1995; 28:311–318. [DOI] [PubMed] [Google Scholar]
- 46. Kofanova O., Bellora C., Frasquilho S.G., Antunes L., Hamot G., Mathay C., Mommaerts K., Muller A., DeWitt B., Betsou F.. Standardization of the preanalytical phase of DNA extraction from fixed tissue for next-generation sequencing analyses. N. Biotechnol. 2020; 54:52–61. [DOI] [PubMed] [Google Scholar]
- 47. Groelz D., Viertler C., Pabst D., Dettmann N., Zatloukal K.. Impact of storage conditions on the quality of nucleic acids in paraffin embedded tissues. PLoS One. 2018; 13:e0203608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Patel P.G., Selvarajah S., Guérard K.-P., Bartlett J.M., Lapointe J., Berman D.M., Okello J.B., Park P.C.. Reliability and performance of commercial RNA and DNA extraction kits for FFPE tissue cores. PloS One. 2017; 12:e0179732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. ISO/TC 212 ICS: 11.100.10 In vitro diagnostic test systems, Molecular in vitro diagnostic examinations - specifications for pre-examination processes for formalin-fixed and paraffin-embedded (FFPE) tissue, Part 3: isolated DNA. Int. Org. Standard. 2018; 12:17. [Google Scholar]
- 50. Amemiya K., Hirotsu Y., Oyama T., Omata M.. Relationship between formalin reagent and success rate of targeted sequencing analysis using formalin fixed paraffin embedded tissues. Clin. Chim. Acta. 2019; 488:129–134. [DOI] [PubMed] [Google Scholar]
- 51. Nagahashi M., Shimada Y., Ichikawa H., Nakagawa S., Sato N., Kaneko K., Homma K., Kawasaki T., Kodama K., Lyle S.. Formalin-fixed paraffin-embedded sample conditions for deep next generation sequencing. J. Surg. Res. 2017; 220:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Watanabe M., Hashida S., Yamamoto H., Matsubara T., Ohtsuka T., Suzawa K., Maki Y., Soh J., Asano H., Tsukuda K.. Estimation of age-related DNA degradation from formalin-fixed and paraffin-embedded tissue according to the extraction methods. Exp. Ther. Med. 2017; 14:2683–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. So A.P., Vilborg A., Bouhlal Y., Koehler R.T., Grimes S.M., Pouliot Y., Mendoza D., Ziegle J., Stein J., Goodsaid F.. A robust targeted sequencing approach for low input and variable quality DNA from clinical samples. NPJ Genom. Med. 2018; 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grossman L., Braun A., Feldberg R., Mahler I.. Enzymatic repair of DNA. Annu. Rev. Biochem. 1975; 44:19–43. [DOI] [PubMed] [Google Scholar]
- 55. Dedhia P., Tarale S., Dhongde G., Khadapkar R., Das B.. Evaluation of DNA extraction methods and real time PCR optimization on formalin-fixed paraffin-embedded tissues. Asian Pac. J. Cancer Prev. 2007; 8:55–59. [PubMed] [Google Scholar]
- 56. Einaga N., Yoshida A., Noda H., Suemitsu M., Nakayama Y., Sakurada A., Kawaji Y., Yamaguchi H., Sasaki Y., Tokino T.. Assessment of the quality of DNA from various formalin-fixed paraffin-embedded (FFPE) tissues and the use of this DNA for next-generation sequencing (NGS) with no artifactual mutation. PLoS One. 2017; 12:e0176280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Flores Bueso Y., Walker S.P., Tangney M.. Characterization of FFPE-induced bacterial DNA damage and development of a repair method. Biol. Methods Protoc. 2020; 5:bpaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Do H., Dobrovic A.. Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil-DNA glycosylase. Oncotarget. 2012; 3:546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tchou J., Kasai H., Shibutani S., Chung M., Laval J., Grollman A., Nishimura S.. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:4690–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen L., Liu P., Evans T.C., Ettwiller L.M.. DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification. Science. 2017; 355:752–756. [DOI] [PubMed] [Google Scholar]
- 61. Mitra S., Hazra T.K., Roy R., Ikeda S., Biswas T., Lock J., Boldogh I., Izumi T.. Complexities of DNA base excision repair in mammalian cells. Mol. Cells. 1997; 7:305–312. [PubMed] [Google Scholar]
- 62. Do H., Molania R., Mitchell P.L., Vaiskunaite R., Murdoch J.D., Dobrovic A.. Reducing artifactual EGFR T790M mutations in DNA from formalin-fixed paraffin-embedded tissue by use of thymine-DNA glycosylase. Clin. Chem. 2017; 63:1506–1514. [DOI] [PubMed] [Google Scholar]
- 63. Bessho T., Roy R., Yamamoto K., Kasai H., Nishimura S., Tano K., Mitra S.. Repair of 8-hydroxyguanine in DNA by mammalian N-methylpurine-DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:8901–8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Haile S., Corbett R.D., Bilobram S., Bye M.H., Kirk H., Pandoh P., Trinh E., MacLeod T., McDonald H., Bala M.. Sources of erroneous sequences and artifact chimeric reads in next generation sequencing of genomic DNA from formalin-fixed paraffin-embedded samples. Nucleic Acids Res. 2019; 47:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Picelli S., Björklund Å.K., Reinius B., Sagasser S., Winberg G., Sandberg R.. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014; 24:2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Do H., Dobrovic A.. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin. Chem. 2015; 61:64–71. [DOI] [PubMed] [Google Scholar]
- 67. Serizawa M., Yokota T., Hosokawa A., Kusafuka K., Sugiyama T., Tsubosa Y., Yasui H., Nakajima T., Koh Y.. The efficacy of uracil DNA glycosylase pretreatment in amplicon-based massively parallel sequencing with DNA extracted from archived formalin-fixed paraffin-embedded esophageal cancer tissues. Cancer Genet. 2015; 208:415–427. [DOI] [PubMed] [Google Scholar]
- 68. Chun S.-M., Sung C.O., Jeon H., Kim T.-I., Lee J.-Y., Park H., Kim Y., Kim D., Jang S.J.. Next-generation sequencing using s1 nuclease for poor-quality formalin-fixed, paraffin-embedded tumor specimens. J. Mol. Diagn. 2018; 20:802–811. [DOI] [PubMed] [Google Scholar]
- 69. Costello M., Pugh T.J., Fennell T.J., Stewart C., Lichtenstein L., Meldrim J.C., Fostel J.L., Friedrich D.C., Perrin D., Dionne D. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013; 41:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xiong K., Shea D., Rhoades J., Blewett T., Liu R., Bae J.H., Nguyen E., Makrigiorgos G.M., Golub T.R., Adalsteinsson V.A. Duplex-Repair enables highly accurate sequencing, despite DNA damage. Nucleic Acids Res. 2021; 50:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bruinsma S., Burgess J., Schlingman D., Czyz A., Morrell N., Ballenger C., Meinholz H., Brady L., Khanna A., Freeberg L.. Bead-linked transposomes enable a normalization-free workflow for NGS library preparation. BMC Genom. 2018; 19:722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Astolfi A., Urbini M., Indio V., Nannini M., Genovese C.G., Santini D., Saponara M., Mandrioli A., Ercolani G., Brandi G.. Whole exome sequencing (WES) on formalin-fixed, paraffin-embedded (FFPE) tumor tissue in gastrointestinal stromal tumors (GIST). BMC Genom. 2015; 16:892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kim J., Kim D., Lim J.S., Maeng J.H., Son H., Kang H.-C., Nam H., Lee J.H., Kim S. The use of technical replication for detection of low-level somatic mutations in next-generation sequencing. Nat. Commun. 2019; 10:1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kwok C.K., Ding Y., Sherlock M.E., Assmann S.M., Bevilacqua P.C.. A hybridization-based approach for quantitative and low-bias single-stranded DNA ligation. Anal. Biochem. 2013; 435:181–186. [DOI] [PubMed] [Google Scholar]
- 75. Chung J., Son D.-S., Jeon H.-J., Kim K.-M., Park G., Ryu G.H., Park W.-Y., Park D. The minimal amount of starting DNA for Agilent's hybrid capture-based targeted massively parallel sequencing. Sci. Rep. 2016; 6:26732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Stiller M., Sucker A., Griewank K., Aust D., Baretton G.B., Schadendorf D., Horn S.. Single-strand DNA library preparation improves sequencing of formalin-fixed and paraffin-embedded (FFPE) cancer DNA. Oncotarget. 2016; 7:59115–59128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gansauge M.-T., Gerber T., Glocke I., Korlević P., Lippik L., Nagel S., Riehl L.M., Schmidt A., Meyer M.. Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase. Nucleic Acids Res. 2017; 45:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Saunderson E.A., Baker A.-M., Williams M., Curtius K., Jones J.L., Graham T.A., Ficz G.. A novel use of random priming-based single-strand library preparation for whole genome sequencing of formalin-fixed paraffin-embedded tissue samples. NAR Genom. Bioinform. 2020; 2:lqz017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Meléndez B., Van Campenhout C., Rorive S., Remmelink M., Salmon I., D’Haene N. Methods of measurement for tumor mutational burden in tumor tissue. Transl. Lung Cancer Res. 2018; 7:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yost S.E., Smith E.N., Schwab R.B., Bao L., Jung H., Wang X., Voest E., Pierce J.P., Messer K., Parker B.A.. Identification of high-confidence somatic mutations in whole genome sequence of formalin-fixed breast cancer specimens. Nucleic Acids Res. 2012; 40:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bhagwate A.V., Liu Y., Winham S.J., McDonough S.J., Stallings-Mann M.L., Heinzen E.P., Davila J.I., Vierkant R.A., Hoskin T.L., Frost M.. Bioinformatics and DNA-extraction strategies to reliably detect genetic variants from FFPE breast tissue samples. BMC Genom. 2019; 20:689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang M., Escudero-Ibarz L., Moody S., Zeng N., Clipson A., Huang Y., Xue X., Grigoropoulos N.F., Barrans S., Worrillow L.. Somatic mutation screening using archival formalin-fixed, paraffin-embedded tissues by fluidigm multiplex PCR and Illumina sequencing. J. Mol. Diagn. 2015; 17:521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Spencer D.H., Sehn J.K., Abel H.J., Watson M.A., Pfeifer J.D., Duncavage E.J.. Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. J. Mol. Diagn. 2013; 15:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Islam S., Zeisel A., Joost S., La Manno G., Zajac P., Kasper M., Lönnerberg P., Linnarsson S.. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014; 11:163–166. [DOI] [PubMed] [Google Scholar]
- 85. Kivioja T., Vähärautio A., Karlsson K., Bonke M., Enge M., Linnarsson S., Taipale J.. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods. 2012; 9:72–74. [DOI] [PubMed] [Google Scholar]
- 86. Shiroguchi K., Jia T.Z., Sims P.A., Xie X.S.. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:1347–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Peng Q., Vijaya Satya R., Lewis M., Randad P., Wang Y.. Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genom. 2015; 16:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B.. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A.. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Waalkes A., Penewit K., Wood B.L., Wu D., Salipante S.J.. Ultrasensitive detection of acute myeloid leukemia minimal residual disease using single molecule molecular inversion probes. Haematologica. 2017; 102:1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gregory M.T., Bertout J.A., Ericson N.G., Taylor S.D., Mukherjee R., Robins H.S., Drescher C.W., Bielas J.H.. Targeted single molecule mutation detection with massively parallel sequencing. Nucleic Acids Res. 2015; 44:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Newman A.M., Lovejoy A.F., Klass D.M., Kurtz D.M., Chabon J.J., Scherer F., Stehr H., Liu C.L., Bratman S.V., Say C.. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016; 34:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ghannam J., Dillon L.W., Hourigan C.S.. Next-generation sequencing for measurable residual disease detection in acute myeloid leukaemia. Br. J. Haematol. 2020; 188:77–85. [DOI] [PubMed] [Google Scholar]
- 94. Cohen J.D., Douville C., Dudley J.C., Mog B.J., Popoli M., Ptak J., Dobbyn L., Silliman N., Schaefer J., Tie J.. Detection of low-frequency DNA variants by targeted sequencing of the Watson and Crick strands. Nat. Biotechnol. 2021; 39:1220–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kennedy S.R., Schmitt M.W., Fox E.J., Kohrn B.F., Salk J.J., Ahn E.H., Prindle M.J., Kuong K.J., Shen J.-C., Risques R.-A.. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat. Protoc. 2014; 9:2586–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Frank M.S., Fuß J., Steiert T.A., Streleckiene G., Gehl J., Forster M.. Quantifying sequencing error and effective sequencing depth of liquid biopsy NGS with UMI error correction. Biotechniques. 2021; 70:226–232. [DOI] [PubMed] [Google Scholar]
- 97. Pel J., Choi W.W., Leung A.O., Shibahara G., Gelinas L., Despotovic M., Ung W.L., Marziali A.. Duplex Proximity Sequencing (Pro-Seq): a. [DOI] [PMC free article] [PubMed]
- 98. Woerner A.E., Mandape S., King J.L., Muenzler M., Crysup B., Budowle B.. Reducing noise and stutter in short tandem repeat loci with unique molecular identifiers. Forensic Sci. Int. Genet. 2021; 51:102459. [DOI] [PubMed] [Google Scholar]
- 99. Smith T., Heger A., Sudbery I.. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017; 27:491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Li H., Durbin R.. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Muller E., Goardon N., Brault B., Rousselin A., Paimparay G., Legros A., Fouillet R., Bruet O., Tranchant A., Domin F.. OutLyzer: software for extracting low-allele-frequency tumor mutations from sequencing background noise in clinical practice. Oncotarget. 2016; 7:79485–79493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kalatskaya I., Trinh Q.M., Spears M., McPherson J.D., Bartlett J.M., Stein L.. ISOWN: accurate somatic mutation identification in the absence of normal tissue controls. Genome Med. 2017; 9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Alioto T.S., Buchhalter I., Derdak S., Hutter B., Eldridge M.D., Hovig E., Heisler L.E., Beck T.A., Simpson J.T., Tonon L.. A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat. Commun. 2015; 6:10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. de Schaetzen van Brienen L., Larmuseau M., Van der Eecken K., De Ryck F., Robbe P., Schuh A., Fostier J., Ost P., Marchal K.. Comparative analysis of somatic variant calling on matched FF and FFPE WGS samples. BMC Med. Genom. 2020; 13:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wolf B., Kuonen P., Dandekar T., Atlan D. DNAseq workflow in a diagnostic context and an example of a user friendly implementation. Biomed Res. Int. 2015; 2015:403497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G.. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013; 31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mansukhani S., Barber L.J., Kleftogiannis D., Moorcraft S.Y., Davidson M., Woolston A., Proszek P.Z., Griffiths B., Fenwick K., Herman B.. Ultra-sensitive mutation detection and genome-wide DNA copy number reconstruction by error-corrected circulating tumor DNA sequencing. Clini. Chem. 2018; 64:1626–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Murray S., Dahabreh I.J., Linardou H., Manoloukos M., Bafaloukos D., Kosmidis P.. Somatic mutations of the tyrosine kinase domain of epidermal growth factor receptor and tyrosine kinase inhibitor response to TKIs in non-small cell lung cancer: an analytical database. J. Thorac. Oncol. 2008; 3:832–839. [DOI] [PubMed] [Google Scholar]
- 109. Mirzayi C., Renson A., Furlanello C., Sansone S.-A., Zohra F., Elsafoury S., Geistlinger L., Kasselman L.J., Eckenrode K., van de Wijgert J.et al.. Reporting guidelines for human microbiome research: the STORMS checklist. Nat. Med. 2021; 27:1885–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Knottnerus A., Tugwell P.. STROBE—a checklist to Strengthen the Reporting of Observational Studies in Epidemiology. J. Clin. Epidemiol. 2008; 61:323. [DOI] [PubMed] [Google Scholar]
- 111. Gündisch S., Slotta-Huspenina J., Verderio P., Ciniselli C.M., Pizzamiglio S., Schott C., Drecoll E., Viertler C., Zatloukal K., Kap M.et al.. Evaluation of colon cancer histomorphology: a comparison between formalin and PAXgene tissue fixation by an international ring trial. Virchows Arch. 2014; 465:509–519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available in the European Genome-phenome Archive (EGA) repository, EGAS00001005757. The EGA is a controlled-access human data repository subject to European data protection laws. Therefore, data access is subject to an application, ethics approval by the applicant's ethics board and a data access agreement.