Abstract
Reprogramming cellular behaviour is one of the hallmarks of synthetic biology. To this end, prokaryotic allosteric transcription factors (aTF) have been repurposed as versatile tools for processing small molecule signals into cellular responses. Expanding the toolbox of aTFs that recognize new inducer molecules is of considerable interest in many applications. Here, we first establish a resorcinol responsive aTF-based biosensor in Escherichia coli using the TetR-family repressor RolR from Corynebacterium glutamicum. We then perform an iterative walk along the fitness landscape of RolR to identify new inducer specificities, namely catechol, methyl catechol, caffeic acid, protocatechuate, L-DOPA, and the tumour biomarker homovanillic acid. Finally, we demonstrate the versatility of these engineered aTFs by transplanting them into the model eukaryote Saccharomyces cerevisiae. This work provides a framework for efficient aTF engineering to expand ligand specificity towards novel molecules on laboratory timescales, which, more broadly, is invaluable across a wide range of applications such as protein and metabolic engineering, as well as point-of-care diagnostics.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Bacteria have evolved complex networks for transcriptional regulation of gene expression to respond to environmental and chemical cues (1–4). Allosteric transcription factors (aTFs) are regulatory proteins that conditionally bind DNA promoters to regulate gene expression in response to small molecules (5,6). Bacterial aTFs are frequently employed as tools in synthetic biology for processing biochemical signals to cellular responses. This is in part due to the ease of their heterologous expression as relatively small soluble cytoplasmic proteins, in addition to the wealth of knowledge of aTF structures, DNA recognition sequences (operators), and small molecule inducers (7–9). As a result, aTF-based biosensors are used in a multitude of applications including but not limited to: screening for environmental contaminants (10,11), screening metagenomic libraries to identify genes encoding functional enzymes (12), point-of-care disease diagnostics (13), as well as controlling synthetic bacterial consortia (14) and bacterial behaviour in situ (15). As well, aTFs are used in metabolic engineering, which aims to precisely engineer native and heterologous metabolism to produce value-added chemicals (16,17).
Another attractive aspect of aTF-based biosensors is their tunability. Well-established methods exist for tuning the response curves of genetic circuits such as optimizing promoter and ribosomal binding site (RBS) strength as well as implementing amplifier circuits (18–21). Tuning the ligand specificity of aTFs however remains a challenging undertaking. aTFs have often evolved to respond to a limited set of inducers, which restricts their use in many applications (22). The existing repertoire of aTFs could be expanded by continuous bioprospecting (23,24), creating synthetic chimeras (25,26), or by directed evolution to alter their ligand specificity (8). Directed evolution is widely used to introduce beneficial properties to proteins, where rounds of genetic sequence diversification (either random or structure-guided) are followed by variant library screening or selection (27,28). One of the challenges of aTF directed evolution is maintaining allosteric interdomain signalling for proper protein function (29,30), with the possible localized unfolding and intrinsic disorder of the DNA-binding domain further complicating this task (31,32).
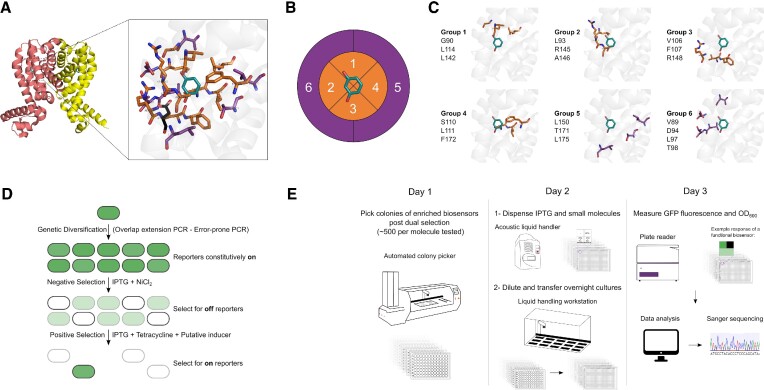
In the present work, we report a broadly applicable automated framework for engineering aTFs through the semi-rational directed evolution of the TetR-family member RolR from Corynebacterium glutamicum (33,34). We used combinatorial active site saturation test (CAST) (35) and iterative saturation mutagenesis (ISM) (36) methods for the grouping of proximal residues in the ligand-binding pocket when viewed as a Cartesian 3-dimensional space. This allowed us to target different regions of the binding pocket separately and sequentially to generate variants of RolR responsive to aromatic small molecules (Figure 1A–C). We first target 19 residues in the ligand-binding pocket for mutagenesis, coupled with under-sampling for further variant library size reduction (35). We then employ the TetA antiporter (36,37) for dual selection over several variant generations to identify activity towards other aromatic molecules, coupled with an automated screening strategy (Figure 1D, E). RolR-regulated promoters are induced by resorcinol, a molecule with various industrial applications (38). We start our engineering efforts by targeting catechol biosensing, which is a lignin degradation by-product (39), and a precursor to the bioproduction of value-added chemicals including muconic acid (40), as well as the chemical synthesis of fragrances, dyes, and pesticides (41). We then use the identified catechol-responsive variants as a template for further evolution towards substituted catechol moieties such as lignin degradation by-products (methyl catechol) (39), pathway intermediates to value-added chemicals (protocatechuate (40), 2,3-dihydroxybenzoate (42), L-DOPA (43), caffeic acid (44)), catecholamine neurotransmitters (epinephrine, dopamine) and 3-O-methylated catecholamines that act as tumour biomarkers (homovanillic acid and methoxytyramine (45)). We perform an in-depth characterization of different RolR variant attributes such as response, crosstalk, dynamic range, and leakiness, followed by transplantation of the engineered aTFs from E. coli to the model eukaryotic system Saccharomyces cerevisiae. This work provides a framework to perform a gradual walk across the fitness landscape of the RolR scaffold in a high-throughput manner to generate variants with altered ligand-binding properties that could function as biosensors towards sensing molecules previously undetectable by the wild-type (WT), and is broadly applicable to other aTFs and ligand-binding proteins.
Figure 1.
RolR mutagenesis, selection, and semi-automated high-throughput screening workflows. (A) RolR dimer in the holo conformation (PDB:3AQT), and the structure of the ligand-binding pocket with residue D149 in black, resorcinol in teal, and the 19 residues chosen for mutagenesis within 5 Å in orange, and those between 5 and 8 Å in purple. (B) Cartesian binding pocket map for combinatorial active site saturation test (CAST). (C) The six amino acid groups comprising the 19 residues to be targeted for mutagenesis. (D) Principle of biosensor TetA dual selection using NiCl2 for negative selection towards transcriptional repression competency, and tetracycline for positive selection towards the induction by ligands of interest. (E) Semi-automated high-throughput screen. On day 1, ∼500 colonies were picked for each candidate molecule. On day 2, IPTG and small molecules were dispensed using an acoustic liquid handler into 384 well plates. Growing colonies were diluted and dispensed into 384 plates testing the different states of the sensor using a liquid handling workstation. On day 3, fluorescence was measured using a plate reader. Data was then analyzed, and positive clones were genotyped by Sanger sequencing.
MATERIALS AND METHODS
Strains and growth media
E. coli DH5α was used for all molecular cloning, plasmid propagation, genetic circuit characterization, and protein engineering and directed evolution experiments. Unless otherwise stated, E. coli cells were cultured at 37°C for ∼16 h in liquid or solid LB medium (10 g l−1 NaCl, 10 g l−1 tryptone, 5 g l−1 yeast extract). Biosensor selection and response characterization was performed in M9 minimal medium (6.78 g l−1 Na2HPO4, 3 g l−1 KH2PO4, 1 g l−1 NH4Cl, 0.5 g l−1 NaCl, 4 g l−1 Dextrose, 2 mM MgSO4, and 0.1 mM CaCl2) supplemented with 0.34 g l−1 thiamine hydrochloride, 2 g l−1 casamino acids, and 100 mM ascorbic acid as an antioxidant. Kanamycin (50 μg ml−1) was added for plasmid selection as needed.
S. cerevisiae manipulations were conducted in the prototroph CEN.PK113-7D, which was grown at 30°C in liquid or solid Yeast Peptone Dextrose medium (YPD; 10 g l−1 yeast extract, 20 g l−1 peptone, 20 g l−1 dextrose) for ∼16 h or 48 h, respectively. Biosensor characterization was performed in Yeast Nitrogen Base medium (YNB; 6.7 g l−1 yeast nitrogen base, 20 g l−1 dextrose) and Synthetic Complete medium (SC; 6.7 g l−1 yeast nitrogen base, 20 g l−1 dextrose, 1.92 g l−1 yeast synthetic drop-out medium supplement without histidine, 0.08 g l−1 histidine). Hygromycin (200 μg ml−1) and G418 (200 μg ml−1) were added for plasmid selection as needed.
Chemicals
Chemicals for biosensor testing were purchased from Sigma Aldrich and dissolved in water, unless otherwise specified, to obtain the stock concentrations of 100 mM resorcinol, 100 mM catechol, 50 mM protocatechuate, 100 mM dopamine, 5 mM L-DOPA (dissolved directly in media), 100 mM epinephrine, 100 mM methyl catechol, 100 mM caffeic acid (dissolved in 99% ethanol), 50 mM 2,3-dihydroxybenzoate, 50 mM homovanillic acid, 50 mM methoxytyramine. IPTG 1 M stock solutions were prepared by dissolving in water. Toxicity of the chemicals used to E. coli and S. cerevisiae was tested by measuring OD600 values in the presence of each molecule (Supplementary Figure S6).
Synthetic genes, oligonucleotides and plasmids
The rolR (cgl1157) gene from Corynebacterium glutamicum was codon-optimized for yeast expression using the IDT tool and was purchased from Thermo Scientific. Oligonucleotides were purchased from Thermo Scientific and suspended in nuclease-free water to a concentration of 100 μM. Sequences of oligonucleotides used in this study are listed in Supplementary data S1. Plasmids and sequences of parts used in this work are listed in Supplementary Table S1 and Supplementary data S2, respectively.
RolR genetic circuit construction
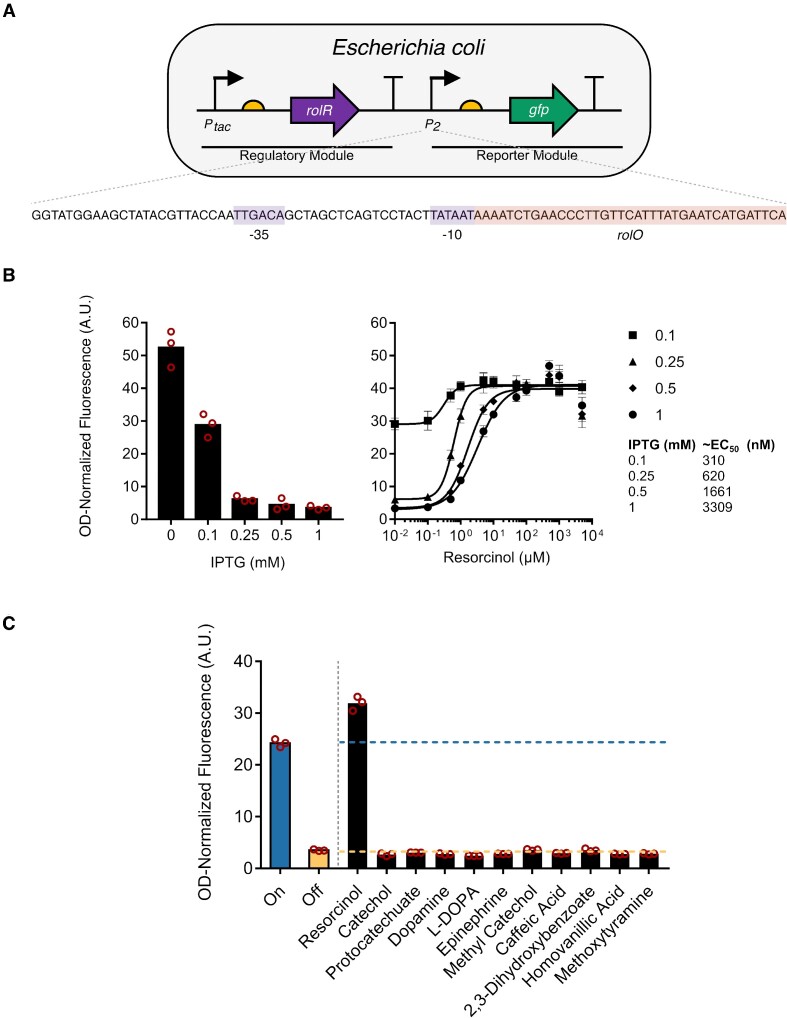
Genetic circuits were constructed using the template plasmid pQacR-Q2 (Addgene plasmid # 74690) (46). The qacR gene was replaced with the codon-optimized rolR gene using primer sets 1–4 by polymerase incomplete primer extension (PIPE) cloning (47) by mixing equal volumes of PCR products of rolR and the vector backbone each amplified without a final extension step, and each containing a 20 bp homology arm to the other part. Promoter PQacR was modified by replacing qacO with rolO, and optimizing its –10 sequence to the consensus TATAAT, creating promoter ‘P2’. As well, eyfp was mutated to egfp. Finally, a second ribosomal binding site followed by TetA (amplified from pBBR1MCS3) were inserted downstream of eGFP.
RolR variant library construction
Our variant library construction strategy exploits the combinatorial active-site saturation test (CAST) (48) and iterative saturation mutagenesis (ISM) (49) methodologies, combined with library under-sampling (35) in order to build focused RolR variant libraries. For mutagenesis, we selected all residues within 5 Å of the inducer molecule in addition to some residues between 5 and 8 Å that we posited to have an influence on binding pocket structure as well as interactions with the inducer molecule, for a total of 19 amino acids. We chose not to mutate residue D149, as it has been established that it is essential for RolR ligand recognition through direct interaction with one of the hydroxyl groups of resorcinol, and mutating it was shown to abolish induction (34) (Figure 1A). We then used site saturation mutagenesis to mutate the selected positions in the ligand-binding pocket by splitting them into six groups of spatially proximal residues, which were built as separate variant libraries targeting 3–4 amino acids each (Figure 1B, C). Then we subjected these libraries to dual selection (Figure 1D), followed by an automated high-throughput screen to interrogate the post-selection enriched variants (Figure 1E).
Amino acid-randomized libraries of RolR were generated using overlap extension PCR (50). The residues of interest were randomized by inserting the NNK degenerate codon using the primer sets listed in Supplementary data S1. Overlap extension PCR products were gel-purified, PCR-amplified, and assembled with the linear parent PCR-amplified backbone using either PIPE or LIC cloning (51), the latter of which incorporates an additional T4 polymerase digest of the insert and backbone mix prior to transformation. Plasmid libraries were electroporated into E. coli DH5α to reach complete coverage (100 000–150 000 clones depending on library). To confirm substitutions, five clones from each library were subjected to Sanger sequencing. Plasmid library colonies were scraped off of agar plates into LB medium containing 50 μg ml−1 kanamycin and normalized by colony number and OD to remove any biases that could skew the selection results using the following equation:
 |
After combining, libraries were stored both as glycerol stocks and as plasmid preparations. RolR random variant libraries were constructed by error-prone PCR (epPCR) of the ligand-binding domain (residues 73–183). To generate epPCR libraries, a 50 μl PCR reaction was prepared by mixing the following: Taq buffer, 4 mM MgCl2, 2 mM freshly prepared MnCl2, 1 μM each oligo, 1 μM CTP, 1 μM TTP, 0.2 μM ATP, 0.2 μM GTP, 20 ng template DNA, nuclease-free water and Taq polymerase. For this library, we aimed for 1–2 amino acid substitutions per gene. To confirm the number of mutated residues, three clones from each library were subjected to Sanger sequencing. PCR products were gel-purified, PCR-amplified, and assembled with the linear parent PCR-amplified backbone using either PIPE or LIC cloning. Plasmid libraries were electroporated into E. coli Dh5α to reach a coverage of ∼700 000 clones. Transformations were recovered in 1 ml LB medium for 1 h. After recovery, 50 ul were plated to identify the number of clones, and the remaining cells were pelleted and suspended in 5 ml LB medium containing 50 μg ml−1 kanamycin followed by incubating for 16 h at 37°C. Cultures were normalized by colony number and OD using the same equation described above, combined, and stored both as glycerol stocks and as a plasmid preparation.
Variant library selection
Dual selection was employed to simultaneously select for functional repressors that are capable of repressing DNA expression (negative selection), and for repressors that are induced by a ligand of interest (positive selection). To start dual selection, E. coli DH5α glycerol stocks containing the plasmid variant libraries were thawed, and 5 ml cultures were inoculated at a final OD600 of 0.05 into M9 medium containing 50 μg ml−1 kanamycin, and incubated for 16 h at 37°C. For dual selection on solid media, which was implemented for the first-generation variant library, cells were plated on M9 agar plates containing 50 μg ml−1 kanamycin, 1 mM IPTG, and 0.6 mM NiCl2, and incubated at 37°C for 48 h. For positive selection, colonies were scraped off, washed twice with M9 medium, plated on M9 agar plates containing 50 μg ml−1 kanamycin, 1 mM IPTG, 7.5 μg ml−1 tetracycline and 0.5 mM catechol, and incubated for 16 h at 37°C. This was followed by a final negative selection step for a total of three selection rounds (negative–positive–negative). The negative selection plates from the final round were subjected to an activity screening step described in the next section. Importantly, negative selection media components were mixed in the following order to avoid salt precipitation: NiCl2, CaCl2, MgSO4, followed by M9 salts. The rest of the media components (glucose, thiamine hydrochloride, casamino acids, kanamycin) were added in any order.
Alternatively, for liquid dual selection, which was implemented for the second and third variant library generations as they incorporated a larger number of small molecules, we followed the same procedure except for the addition of more selection rounds. Briefly, cells from overnight cultures were inoculated at a final OD600 of 0.05 into M9 medium containing 50 μg ml−1 kanamycin and 1 mM IPTG, and incubated for 16 h at 37°C. For negative selection, cells were inoculated at a final OD600 of 0.005 into M9 medium containing 50 μg ml−1 kanamycin, 1 mM IPTG and 0.4 mM NiCl2, and incubated at 37°C for 16 h. Cells were then inoculated at a final OD600 of 0.05 into M9 medium containing 50 μg ml−1 kanamycin, 1 mM IPTG, and 0.5 mM of the inducer of interest (catechol, protocatechuate, dopamine, L-DOPA, epinephrine, methyl catechol, caffeic acid, 2,3-dihydroxybenzoate, homovanillic acid, methoxytyramine), and incubated at 37°C for 12 hours. Cells were then inoculated at a final OD600 of 0.005 into M9 medium containing 50 μg ml−1 kanamycin, 1 mM IPTG, and 7.5 μg ml−1 tetracycline, and 0.5 mM of the inducer of interest, and incubated at 37°C for 12 h. This process was repeated one more time ensuring that the last round is negative selection, for a total of five selection rounds (negative–positive–negative–positive–negative). Cells from the final liquid negative selection round were plated on LB plates containing 50 μg ml−1 kanamycin to obtain isolated colonies that were subjected to activity screening. Importantly, our selection scheme also included the addition of 50 mM of each molecule apart from the target for evolution in the negative selection step in an effort to minimize crosstalk, as higher concentrations were not possible due to the toxicity of these molecules.
Biosensor activity screening
For variant characterization, ∼480 colonies were picked from agar plates using a colony picker (Qpix 460) into five 96 well-plates with LB medium containing 50 μg ml−1 kanamycin, and were incubated for 16 h at 37°C. The next day, 384-well plates were prepared for screening by dispensing 1 mM IPTG and 0.5 mM small molecules using an acoustic liquid handler (Labcyte Echo 550). This scheme will generate four data points per variant: an on state (no IPTG), an off state (IPTG), and 2 data points for testing inducibility by a small molecule of interest. Growing cells were diluted 1:100 into M9 medium containing 50 μg ml−1 kanamycin and transferred using a liquid handler (Biomek), and in some cases a 96-channel pipette (Gilson), into the 384 well-plates containing IPTG and small molecules, and plates were incubated for 20 h at 30°C. The following day, eGFP fluorescence (excitation: 475–10 nm, emission: 510–10 nm) and OD600 of cultures were measured using a microplate reader (CLARIOstar, BMG Labtech, Germany). Data was processed using Excel, and positive hits that exhibited on, off, and small molecule-induced states were then subjected to two rounds of colony purification by streaking on LB plates containing 50 μg ml−1 kanamycin. Single colonies underwent a second activity screen, and positive hits were then subjected to Sanger sequencing.
Biosensor characterization and data analyses
For biosensor response characterization, E. coli cells carrying genetic circuit plasmids were grown in LB medium containing 50 μg ml−1 kanamycin at 37°C for 16 h. The following day, different volumes of IPTG and small molecules were dispensed into 96-well flat or deep-well using an acoustic liquid handler (Labcyte Echo 550) or an electronic multi-channel pipette (Thermo Scientific Finipipette), respectively. E. coli cells were diluted 1:100 in M9 medium containing 50 μg ml−1 kanamycin and 1 mM IPTG, incubated at 37°C for 4 h, and then diluted 1:100 in fresh M9 medium with the addition of small molecule ligands as needed, and incubated at 30°C for 20 h. The following day, eGFP and OD600 of cultures were measured using a microplate reader (CLARIOstar, BMG Labtech, Germany) for first generation responses as well as dynamic range measurements for all variants, or using an Accuri C6 flow cytometer (BD Biosciences) for the second and third generation responses and crosstalk analyses. Cells were diluted 1:5 in deionized water to obtain an average rate of ∼1000 events/second for a total of 10 000 single-cell events. The mean fluorescence of the total ungated population was plotted for each molecule and/or concentration tested. Dose-response data points are averages of technical triplicates, with error bars representing standard deviations. When applicable, curves were fitted using GraphPad Prism by modelling them to a four-parameter non-linear regression equation, and EC50 values were determined as a result. Dynamic range was determined by measuring fluorescence response in the absence and presence of IPTG (on and off states, respectively).
Homology models and protein docking
When needed, homology models for variants were generated with Swiss-prot (https://swissmodel. expasy.org/) using RolR either in its apo (RCSB: 3AQS) or holo (RCSB: 3AQT) forms as a template. Models were then energy-minimized using the GROMOS96 force field tool of Swiss-PDB Viewer. Small molecule structures downloaded from PubChem or RCSB were docked into RolR variants using Autodock Vina (52), and visualized using PyMol (DeLano Scientific). The lowest energy conformation with the ligand positioned in the designated binding pocket between helices 4, 5, 6 and 7 was used for further analyses.
Saccharomyces cerevisiae strain construction
Biosensors were integrated in the genome of S. cerevisiae using CRISPR-Cas9-mediated genomic integration and in vivo DNA homologous recombination (53,54). Briefly, yeast cells were transformed with a DNA pool comprising the following parts: PCR-amplified promoters, aTF, Envy GFP, and terminators with ∼40 bp homology to the preceding and/or following part, mixed in equimolar ratios; two ∼500 bp homology arms to the genomic locus targeted for integration (200 ng each); linearized pCas-G418 and pCas-Hyg vectors encoding Cas9 (55) (100 ng each) (Supplementary table S1); PCR-amplified gRNAs targeting the chromosomal locus Flagfeldt site 16 (56) and with homology to the Cas9 plasmid for assembly of the plasmid by in vivo homologous recombination (200 ng per transformation). The DNA pool was transformed into yeast using a 25 μl lithium acetate/PEG heat shock transformation (57). Briefly, cells were heat-shocked at 42°C for 30 min, recovered for 16 hours in YPD, and plated on YPD-Hyg/G418 dual selection plates. Successful genomic integrations were confirmed by colony PCR and Sanger sequencing. Finally, the Cas9 plasmid was cured by streaking colonies two times on YPD plates with no selection. Sequences of promoters, gene expression cassettes, terminators, and gRNAs are found in Supplementary data S2.
S. cerevisiae biosensor characterization
Cells were grown overnight at 30°C and 300 rpm in 96 deep-well plates in an orbital shaker. The following day, yeast cells were diluted 1:500 into 500 μl SC or YNB medium to an OD600 of ∼0.01 in 96 deep-well plates with or without different concentrations of small molecules from the low μM range up to to 5–10 mM (depending on the toxicity of the molecule), and 100 mM in the case of protocatechuate. The plates were then incubated in an orbital shaker at 30°C and 300 rpm for 20 h, after which fluorescence was measured using an Accuri C6 flow cytometer. Cells were diluted 1:5 in deionized water to obtain an average rate of ∼1000 events/second for a total of 10 000 single-cell events. The mean fluorescence of the total ungated population was plotted for each molecule and/or concentration tested. Unless otherwise stated, dose-response data points are averages of technical triplicates, shown either as three data points, or a single data point with an error bar representing standard deviation. When applicable, curves were fitted using GraphPad Prism by modelling them to a four-parameter nonlinear regression equation.
RESULTS
Construction and inducer-characterization of resorcinol biosensors
RolR acts by negatively regulating the rolHMD cluster involved in resorcinol catabolism in C. glutamicum (33,34). We have introduced the rolR-rolO repressor-operator pair into a previously described genetic circuit (46), whereby a regulatory module comprising rolR is expressed from the IPTG-inducible tac promoter, and a reporter module is expressed from a second, RolR-regulated promoter (Figure 2A). The engineered circuit comprises the modified promoter P2, which contains the consensus promoter elements (–35: TTGACA, –10: TATAAT) and 38 bp that include the rolO operator element immediately downstream of the –10 sequence (Figure 2A). This circuit design allows the reporter module to be expressed in the absence of regulator binding, and to be repressed when RolR is produced upon induction of the regulatory module by IPTG supplementation, as seen in the gradual decrease in GFP fluorescence on increasing IPTG concentration (Figure 2B). Adding the RolR inducer resorcinol, in the presence of IPTG, causes the gradual induction of the reporter module and increasing GFP fluorescence. This circuit setup enables tuning of EC50 values for resorcinol from the mid nM to low μM range (Figure 2B).
Figure 2.
Description and functional characterization of RolR resorcinol biosensors in Escherichia coli. (A) Structure of the one-plasmid RolR biosensor genetic circuit. RolR expression is driven by the IPTG-inducible tac promoter (regulatory module), and GFP expression is driven by a second engineered resorcinol-responsive promoter (reporter module), which contains one copy of the rolO operator sequence inserted immediately downstream of the –10 promoter element. (B) Left panel: gradually increasing IPTG concentrations reduces whole-cell OD-normalized fluorescence levels. Right panel: Gradually increasing resorcinol concentrations in fixed IPTG concentrations results in an increase in OD-normalized fluorescence levels, and resorcinol EC50 values in different IPTG concentrations were measured. (C) Ligand-specificity of RolR towards all molecules in this study in the presence of 1 mM IPTG. Data points are mean ± SD, n = 3. A.U., arbitrary units.
Most aTFs have evolved towards specific ligands, yet some exhibit promiscuity towards multiple small molecules, and are sometimes called ‘generalists’ (24,58–60). Thus, we sought to determine if expression from P2 could be induced by other molecules of interest. We have tested the RolR circuit by introducing a range of aromatic small molecules containing catechol moieties and did not observe induction by any molecules other than resorcinol (Figure 2C).
A selection-screening framework to identify RolR variants that respond to new ligands
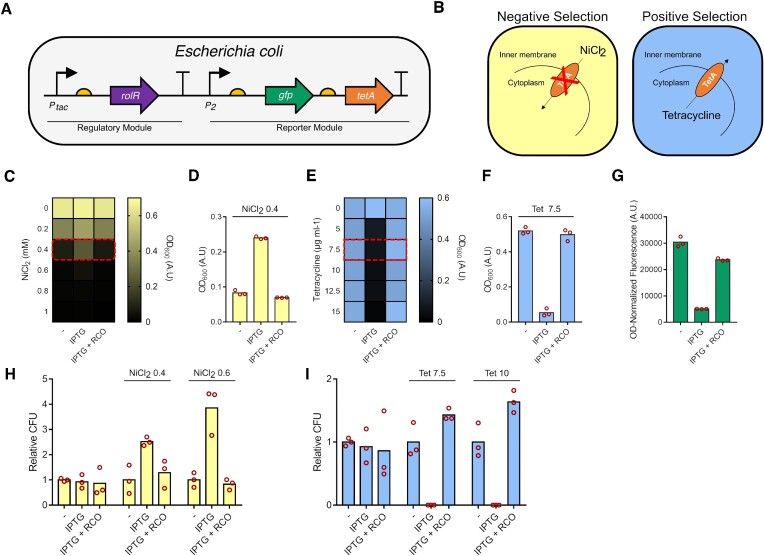
Many successful aTF directed evolution efforts have resulted in expanding ligand specificity towards new molecules (61–63). This motivated us to create high quality variant libraries that sample the protein sequence landscape more effectively, in an attempt to perform a gradual walk across the fitness landscape towards molecules with divergent structures (see ‘Materials and Methods’). Some amino acid substitutions in aTFs can affect their DNA binding affinity, resulting in altered transcriptional regulation (61,64). Dual selection schemes are essential for aTF directed evolution to select for and maintain DNA-binding competency (transcriptional repression), and induction by the ligand of interest, the former which could be easily perturbed by substitutions due to the nature of allostery (31). Fluorescence activated cell-sorting has been used extensively for dual selection (62,65), and genes that encode multiple phenotypes such as TolC (66) and thymidine kinases (67) have been used as dual selection markers. To this end, we employed the E. coli TetA antiporter as a dual selection marker to couple aTF activity to cell growth (36,37,68,69), which requires tetracycline and NiCl2 for positive and negative selection, respectively (70). We extended the reporter module by inserting TetA downstream of GFP for polycistronic expression (Figure 3A), and characterized the system in both liquid and solid media. In this selection scheme, negative selection is first employed to select for DNA-binding-competent variants of RolR that repress TetA expression, allowing the cells to survive in media supplemented with NiCl2 (71). This is then followed by positive selection to identify variants that are induced by a ligand of interest, where induction of TetA expression causes a tetracycline resistant phenotype (Figure 3B). In a liquid medium, we identified the concentrations for selection with the most growth difference between repressed and induced states to be 0.4 mM NiCl2 (Figure 3C, D) and 7.5 μg ml−1 or higher for tetracycline (Figure 3E, F). As well, given that TetA and GFP are expressed from the same promoter, we observed a correlation between the expected NiCl2 and tetracycline resistance or susceptibility phenotypes and cell population fluorescence levels (Figure 3G). Furthermore, on solid medium, negative selection performed better at a higher concentration of NiCl2 (0.6 mM) compared to liquid medium, whereas positive selection on tetracycline performed equally on liquid and solid medium (Figure 3H, I).
Figure 3.
Characterization of the TetA dual selection system. (A) The reporter module was extended by introducing TetA downstream of GFP. (B) Mechanism of TetA dual selection. Negative selection selects for the ability to repress expression from promoter P2 which would not allow for toxic NiCl2 to enter the cell. Positive selection selects for induction of expression from P2 by a ligand of interest, which would allow for tetracycline resistance. (C) Characterization of negative selection in liquid media in different NiCl2 concentrations, and (D) in 0.4 mM NiCl2 (RCO = resorcinol). (E) Characterization of positive selection in liquid media at different tetracycline concentrations, and (F) in 7.5 μg ml−1 tetracycline. (G) GFP fluorescence levels correlate with expected phenotypes associated with TetA expression. (H) Negative selection in solid media in 0.4 and 0.6 mM NiCl2. (I) Positive selection in solid media in 7.5 and 10 μg ml−1 tetracycline. Data points are mean ± SD, n= 3. A.U., arbitrary units. CFU, colony forming units.
First generation variants towards catechol
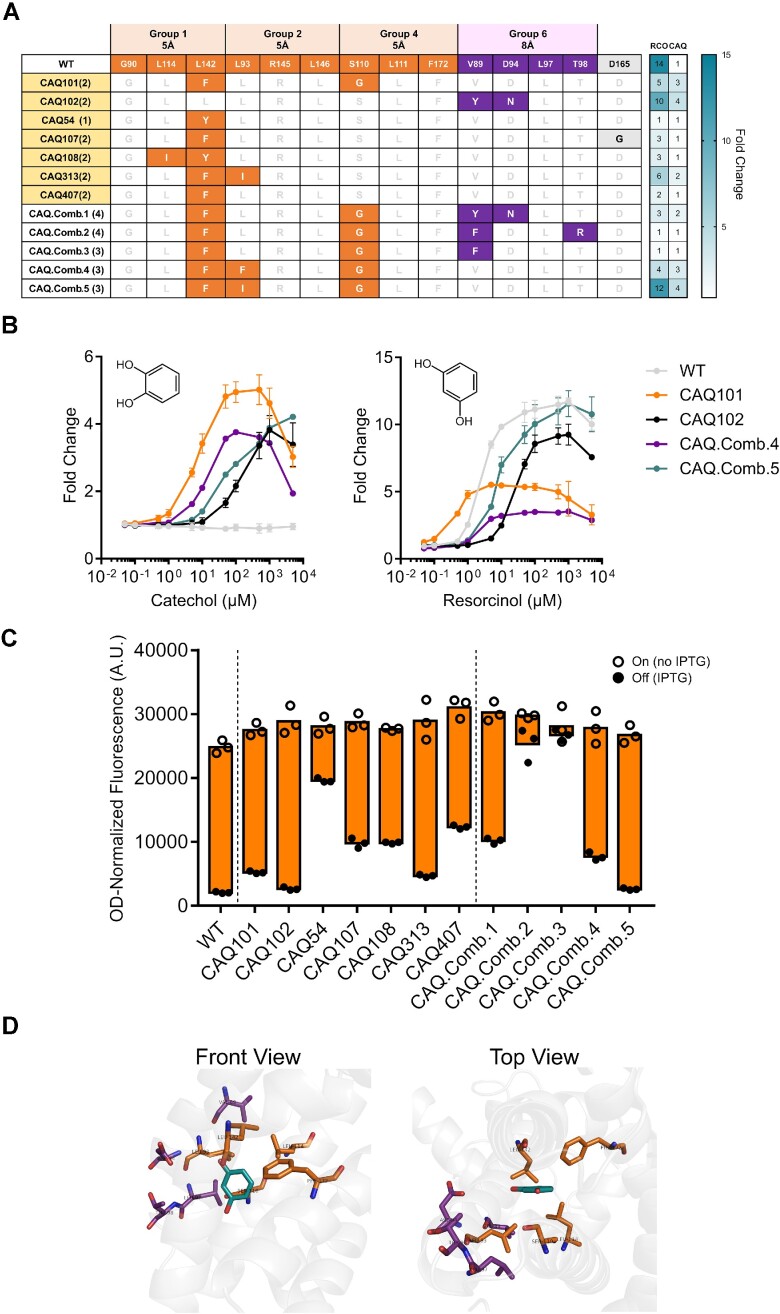
In the first generation of variant library hits, we identified 19 unique variants that are induced by catechol to varying degrees (Figure 4A, Supplementary Figure S1a). Variants induced by catechol show an overrepresentation of substitutions in amino acid positions within groups 1 and 6, which are spatially close to the OH groups of resorcinol (Figure 4A, Supplementary Figure S1a). Our top-performing catechol-responsive variants, named CAQ101, CAQ102, CAQ.Comb.4 and CAQ.Comb.5, had maximum induction values of ∼4–5 fold in catechol, while retaining responsiveness to resorcinol (Figure 4B, Supplementary Figure S1b). We found variant CAQ101 to have a bell-shaped response, seen in the increase in fluorescence followed by a subsequent decrease in increasing catechol concentrations. This type of response was previously described, yet the precise mechanism behind it remains unknown (61,72). Variants from this library also exhibited varying dynamic ranges defined here as the difference in signal between on and off states in the absence and presence of IPTG, respectively (Figure 4C, Supplementary Figure S1d). We have also observed that all 19 identified variants with increased response to catechol always have substitutions in at least one of positions V89, L93, or L142, with substitutions to the aromatic hydrophobic residues phenylalanine, tyrosine, or in only one case to hydrophobic aliphatic isoleucine. These residues cluster close to the top OH group of resorcinol, which would suggest that the substitutions contribute by slightly reshaping the pocket to accommodate the hydroxyl group of a benzenediol isomer of resorcinol at an ortho position relative to the bottom OH group, and as a result being able to accommodate catechol. Furthermore, when combining these substitutions, only variants CAQ.Comb.4 and CAQ.Comb.5 had a significant induction fold change in their response to catechol, as well as dynamic ranges comparable to that of the wild type in response to resorcinol (Figure 4B, C, Supplementary Figure S1c). CAQ.Comb.5 does not exhibit the bell-shaped response that is seen in its parent variant CAQ101, and the loss of that effect was achieved by the single additional substitution L93I (Figure 4A–C), unlike CAQ.Comb.4, which contains a phenylalanine residue at position 93 and retains the bell-shaped response. Lastly, some of the variants were found to have a D165G substitution, which was due to a gene synthesis error that appeared to slightly reduce leakiness (off state), and was observed when comparing the responses of variants CAQ107 and CAQ407 (Figure 4A, C).
Figure 4.
First generation variant hits towards catechol. (A) Table depicting a subset of identified variants responsive to catechol, their substitutions coloured according to group, and their response towards resorcinol (RCO) and catechol (CAQ). Variants highlighted in yellow were used as a template for further mutagenesis. (B) Dose response curves of the top-performing variants compared to WT RolR with catechol (left panel) and resorcinol (right panel). (C) Dynamic range of WT RolR and identified variants. (D) Position of residues mutated in the RolR binding pocket around resorcinol using PDB:3AQT. Data points are mean ± SD, n = 3. A.U., arbitrary units.
Second and third generation variants towards substituted catechol moieties
In addition to catechol, we sought to add diversity to the range of recognized inducer molecules by identifying variants that respond to catechol derivatives with substitutions in positions 3 and 4 of the benzene ring. For the second generation library, we used seven variants from the first library as templates (Figure 4A). We then created variant libraries targeting groups 3, 4, and 5, which align with the hydrophobic side of the ring where the required substitutions occur. Furthermore, we revisited the structure of RolR and identified the additional and adjacent residues of interest L142, G143, and A146, as well as targeted a modified group 1 that included residues G90, L114, and F172. Following dual selection and screening, we identified variants that were enriched from pools where methyl catechol, caffeic acid, or L-DOPA were used for positive selection, and that displayed activity towards these molecules. We also identified variants enriched in the epinephrine-, dopamine-, and protocatechuate-selected pools that displayed no change in GFP expression in the presence of these molecules (Supplementary Figure S2). We used a subset of these variants, in addition to the catechol-responsive variant CAQ101, as a template for a final round of random mutagenesis and selection targeting the entire ligand-binding domain of RolR, in a strategy similar to that used to improve the response of weak biosensors (Supplementary Table S2) (64). We then characterized the identified variants in terms of response, crosstalk, dynamic range, and we also investigated the position of substitutions in the binding pocket.
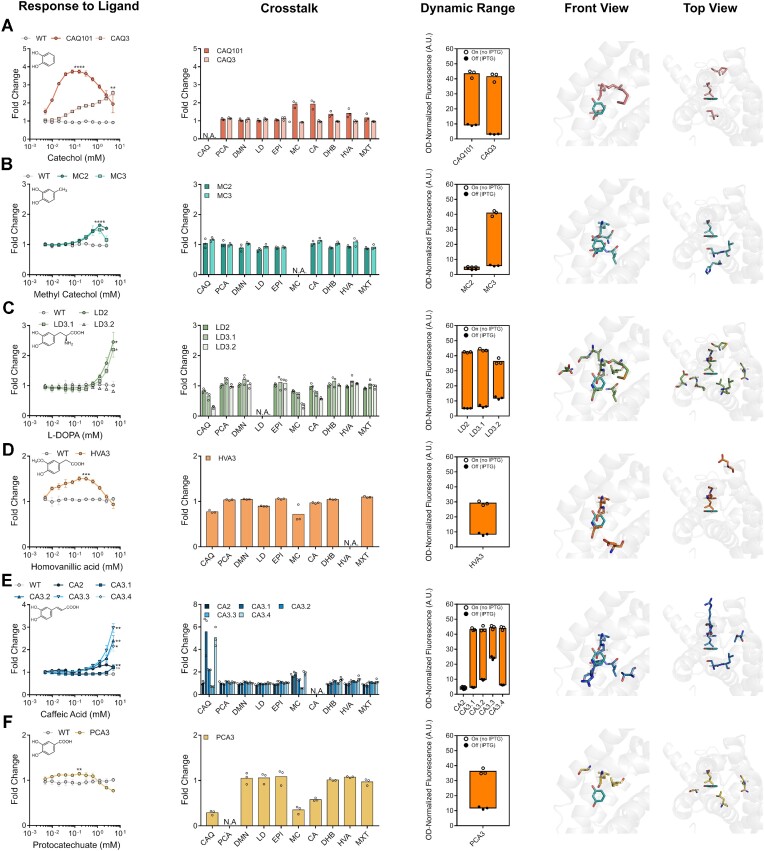
For variants selected for response to catechol, the third generation variant CAQ3 notably lacked the bell-shaped response of CAQ101 (Figure 5A). Furthermore, CAQ3 behaved favourably by having significantly reduced the crosstalk with methyl catechol and caffeic acid that was seen with CAQ101, as well as by having reduced leakiness (Figure 5A). This was all achieved by a single distal M139L substitution, wherein methionine is replaced with another hydrophobic residue, leucine (Figure 5A, Supplementary Table S2). In the case of methyl catechol, the second and third generation variants MC2 and MC3 had a similar response, although it was not possible to test higher concentrations of methyl catechol due to its toxicity to E. coli (Figure 5B). Both variants exhibited minimal crosstalk with other molecules, particularly catechol, but MC3 had a significantly improved dynamic range, which is a desirable attribute for aTF-based biosensors (Figure 5B). This was achieved also by the single substitution H113L, rendering this position hydrophobic (Figure 5B, Supplementary Table S2). For L-DOPA, the second and third generation variants LD2 and LD3.1, respectively, both exhibited ∼2-fold increase in fluorescence in the presence of L-DOPA, with limited crosstalk, near wild-type dynamic range levels, and with most substitutions clustering around the hydrophobic side of the ring (Figure 5C, Supplementary Table S2). In the case of the tumour marker homovanillic acid, which differs from the rest of the molecules in that the phenol has an ortho-methoxy substitution in addition to a para-carboxymethyl substitution, we observed a minimal bell-shaped response that reached a maximum induction of ∼1.5-fold at 0.1 mM homovanillic acid, as well as minimal crosstalk and a dynamic range that is approximately that of the wild-type (Figure 5D). Substitutions in variant HVA3 to accommodate this molecule were mainly in the newly substituted adjacent residues 142, 143 and 146 (Figure 5D, Supplementary Table S2). For caffeic acid, the second generation variant CA2 performed poorly, with better variants identified in the third generation (Figure 5E). All caffeic acid-responsive variants identified in the third generation exhibited significant crosstalk with catechol and less so with methyl catechol, except for variant CA3.3, which, as a tradeoff, exhibited a poorer dynamic range relative to other third generation variants (Figure 5E). Substitutions were once again found to cluster around the hydrophobic side of the ring where the substitution occurs (Figure 5E, Supplementary Table S2). Lastly, of all identified aTF variants, the protocatechuate-responsive variant PCA3 appeared to be the weakest of all, as GFP expression is induced by approximately 1.2 fold, which is further weakened by its bell-shaped response (Figure 5F). PCA3 exhibited limited crosstalk, an acceptable dynamic range, with substitutions clustering around the hydrophobic side (Figure 5F), and with the same substitutions seen in variant LD3.2 (Supplementary Table S2). Lastly, we identified variants that exhibited an abolished response to the cognate inducer, resorcinol, such as LD2 (Supplementary Figures S3 and S4).
Figure 5.
Second and third generation variant hits. Characterizing variant biosensor attributes of response to ligand, crosstalk (CAQ = catechol, PCA = protocatechuate, DMN = dopamine, LD = L-DOPA, EPI = epinephrine, MC = methyl catechol, CA = caffeic acid, DHB = 2,3-dihydroxybenzoate, HVA = homovanillic acid, MXT = methoxytyramine), dynamic range, and the positions of the identified substitutions in the binding pocket, respectively, for variants identified for catechol (A), methyl catechol (B), L-DOPA (C), homovanillic acid (D), caffeic acid (E) and protocatechuate (F). Data points are mean ± SD, n = 3. Statistical significance was determined by a two-tailed Student's t-test for the data point with the highest fluorescence. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; n.s., P > 0.05. A.U., arbitrary units.
Transplantation of biosensor in saccharomyces cerevisiae
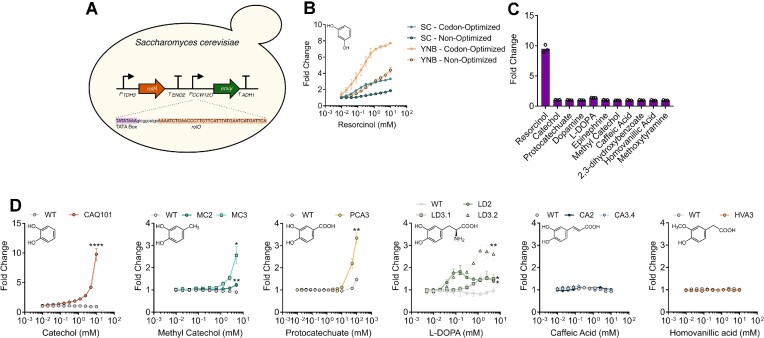
Bacterial aTFs have been successfully transplanted into eukaryotes, such as the yeasts S. cerevisiae and Yarrowia lipolytica, in addition to mammalian cell lines (73–78). Here, to demonstrate the portability of RolR and its variants, we transplanted a subset of RolR variants into genetic circuits in S. cerevisiae. We integrated a single copy of our genetic circuit, which consists of a regulatory module comprising RolR expressed from the strong constitutive yeast promoter PTDH3 (Figure 6a). For the reporter module, we employed the PCCW12 promoter to drive expression of the Envy fluorescent protein (79). We engineered the promoter for RolR regulation by inserting a single copy of the rolO operator 11 bp downstream of the terminal consensus TATA box sequence of the promoter, TATAWAWR (W = A/T R = A/G), as previously described (80). The WT RolR biosensor was functional in S. cerevisiae, in which we identified additional methods for tuning the response curve of genetic circuits, namely codon optimization and the type of media used. This gave rise to a maximal induction of ∼8-fold change in the minimal YNB medium with the codon-optimized RolR variant (Figure 6B). As seen when using E. coli as host, WT RolR is specific to resorcinol in the S. cerevisiae system, with no observable response towards any of the other molecules in this work at concentrations up to 5 mM (Figure 6C). Of all our generated variants, the catechol-responsive variant CAQ101 exhibited the strongest response to its cognate inducer (∼10-fold change), but without the bell-shaped response phenotype that was seen in E. coli (Figure 6D). Methyl catechol-responsive variants MC2 and MC3 are both functional, with the ∼3-fold induction of the third generation variant MC3 significantly better than the response of MC2 (Figure 6D). PCA3 exhibited a modest 1.2-fold change at 10 mM protocatechuate but when the concentration was increased to 100 mM, which is possible due to the tolerance of S. cerevisiae to this molecule, a ∼3-fold induction was observed relative to ∼1.5-fold from the WT, and, similar to CAQ101 without the bell-shaped response observed in E. coli (Figure 6D). Furthermore, L-DOPA-responsive variants also exhibited a ∼1.5–3 fold induction in 5 mM L-DOPA (Figure 6d). However, variant LD3.2, which exhibited the strongest response in S. cerevisiae to L-DOPA lacked an observable response in E. coli, which highlights possible variations in biosensor response in different microbial chassis. Lastly, we could see no response with the generated caffeic acid- and homovanillic acid-responsive variants when transplanted to S. cerevisiae, further emphasizing these differences (Figure 6D).
Figure 6.
Construction and characterization of wild-type and variant RolR biosensors in Saccharomyces cerevisiae. (A) Design of chromosomally integrated RolR genetic circuit in S. cerevisiae. RolR is expressed from the strong constitutive promoter PTDH3, and Envy GFP is expressed from the PCCW12promoter, which was engineered for resorcinol-responsiveness by inserting 1 copy of rolO downstream of the TATA box. (B) Response of the resorcinol biosensor in S. cerevisiae and tuning of the circuit response by altering codon-optimization status and inoculation media. The sensor exhibited the best induction and EC50 when codon-optimized for yeast expression and when minimal YNB media was used. (C) Response of the WT RolR with all the small molecules included in this study. (D) Dose-response curves of representative variant RolR biosensors compared to WT RolR with catechol, methyl catechol, protocatechuate, L-DOPA, caffeic acid, and homovanillic acid. Data points are mean ± SD, n = 3. Statistical significance was determined by a two-tailed Student's t-test for the data point with the highest fluorescence. ****P < 0.0001; **P < 0.01; *P < 0.05.
It has been shown that introducing substitutions in aTF repressors can reverse the activities towards their ligands, causing them to exhibit a stronger affinity to DNA in the presence of their ligand instead of the loss of affinity to DNA that occurs under normal induction conditions. This was seen with the introduction of two substitutions in TetR to generate revTetR, which is used in the Tet-OFF system for regulating gene expression in response to tetracycline and its analogues (81). Furthermore, some aTFs exhibit a dual function with different molecules such as TyrR, which has its regulated promoter activated by phenylalanine and repressed by tyrosine (82). Here, on characterizing variant PCA3 in E. coli, in addition to the weak, 1.2-fold bell-shaped induction response to protocatechuate, we noticed a significant reduction in fluorescence in response to catechol (Figure 5, Supplementary Figure S5a). When the PCA3 circuit was introduced to S. cerevisiae, we observed a robust and stronger response, where we identified what strongly suggests three stable and distinct expression states from that promoter: an intermediate state in the absence of any effector molecule, an induced state in presence of protocatechuate, and a repressed state in the presence of catechol (Supplementary Figure S5b). Titrating these molecules yielded a significant response in S. cerevisiae to both molecules, but not in E. coli (Supplementary Figure S5c, d).
DISCUSSION
In the present work, we engineered a diverse collection of aTF-based biosensors with expanded specificity and minimal crosstalk towards aromatic molecules by combining a structure-guided approach with a dual selection scheme and automated screening. We generated and screened small variant libraries that sampled a fraction of the aTF’s sequence space with mutations focused around the resorcinol-binding region of RolR followed by randomized libraries targeting the ligand-binding domain. This protein engineering/evolution approach resulted in biosensors that respond to catechol, protocatechuate, and caffeic acid, in addition to the molecules homovanillic acid, L-DOPA, and methyl catechol for which whole-cell aTF biosensors had not been reported to date.
Directed evolution campaigns to evolve generalist aTFs to be more specific towards a single target molecule show a high rate of success (59,65,83,84). Evolving narrow or ligand-specific aTFs for induction by new molecules was also recently reported using one or a combination of random mutagenesis, computational protein design, and site saturation mutagenesis (61,62,85–87). Similar to other proteins, evolving new activities in ligand-specific aTFs is traditionally more challenging, and is facilitated by both structural knowledge and chemical intuition (88,89). Here, we demonstrate the evolvability of the resorcinol-specific aTF RolR (Figure 2C) by engineering biosensor variants that respond to six molecules that are not recognized by the parent protein and with minimal crosstalk between ligands (Figure 5). This demonstrates the benefit of structure-guided site-saturation mutagenesis of the ligand-binding pocket coupled with random mutagenesis of the entire ligand-binding domain to simultaneously evolve a single ligand-specific aTF to respond to multiple molecules. Similar methods were used for example to improve response and eliminate crosstalk of the promiscuous aTFs RamR (83) and LysG (65), or to expand substrate specificity of AraC (85), PcaV (86) and VanR (87) and over a single generation of mutagenesis.
Of the six molecules RolR variants were evolved to respond to in this study, five had variants that were identified intentionally (e.g. a catechol responsive variant from the catechol-selected pool) and one molecule, protocatechuate, had the variant PCA3/LD3.2 identified incidentally as responsive to a molecule that was not used for its selection. Variant PCA3/LD3.2 responds to both protocatechuate and L-DOPA, but was identified from a pool that underwent selection using methoxytyramine. This particular variant emphasizes a general issue with aTF engineering that could be regarded as a double-edged sword. PCA3/LD3.2 likely escaped our dual selection due to its leaky off state (or intermediate state considering its tri-stability) (Figure 5F), which would have positioned this state in a sweet spot that is able to survive both negative and positive selection without entirely repressing transcription nor being induced by the inducer of interest. As a comparison, this variant would have been missed in the extensively used fluorescence-activated cell sorting methodologies, where the highest and lowest signallers are usually selected for, with intermediate variants entirely eliminated (62,65). This overlooks the inherent complexity of allostery, and leads to losing significant diversity from the variant library that could be seen in leaky variants as well as intermediate signallers. This bifunctional/tri-stable biosensor PCA3/LD3.2 was identified as an unintentional outcome of our selection method, which, in an effort to eliminate crosstalk was modified in the third generation to include a negative selection step including all molecules from our panel other than our desired target for evolution. Adding catechol to our negative selection pools led to the identification of this biosensor that repressed expression in the presence of catechol, and induced it in the presence of protocatechuate, with an intermediate state of promoter expression in the absence of either molecule. These findings suggest a new and tantalizing methodology for aTF engineering, wherein instead of evolving aTFs for activity towards a single molecule, one could evolve them directly for induction by one molecule and repression by another. This would ultimately add to the in vivo pathway dynamic control biosensor toolbox in the field of metabolic engineering (90), and, more importantly, is truly inspired by nature, which has already evolved bi-functional aTFs (82).
In addition to resorcinol, our engineered biosensor variants were responsive to molecules that are either intermediates or final products in several metabolic engineering pathways, and to concentrations that are relevant in these contexts. These molecules include catechol (40), protocatechuate (40,91), L-DOPA (92) and caffeic acid (93,94). Moreover, we identified variants that are responsive to methyl catechol, a lignin degradation byproduct (39) and a metabolic engineering pathway intermediate (95). As well, we were able to observe a response of our evolved RolR towards the tumour marker homovanillic acid (Figure 5D). This molecule along with methoxytyramine could be used for screening of catecholamine-secreting tumours such as paraganglioma, phaeochromocytoma, among others, as well as for monitoring disease progression following treatment (45). We observed maximum fluorescence induction at 0.1 mM homovanillic acid, or ∼18 mg l−1, which is higher than the limit of detection of current diagnostic methods (0.03–0.3 mg l−1) (96). Additional protein engineering or the use of cascaded amplification circuits (21) will be needed to enhance the sensitivity and response to homovanillic acid to enable clinical applications of this sensor (96,97).
Transplanting aTFs into S. cerevisiae has been accomplished using repressors (73), aporepressors (98), and activators (74). Engineering promoters for regulation by repressors and aporepressors have the advantage of being more straightforward than activators, as they require the operator to be placed downstream of the terminal TATA box of the promoter, compared to a variable and unique location for every activator (80,98,99). Here, to demonstrate the versatility of RolR and the generated variants, we transplanted them as components of engineered genetic circuits into S. cerevisiae, and observed responses that were generally stronger than those observed from E. coli (Figure 6). This observation could be a result of differences in small molecule membrane transport between these organisms or differences between the biosensor contexts in both organisms, which exist as a multi-copy plasmid in E. coli and a single-copy genomic integration in S. cerevisiae. This was observed specifically with variant LD3.2, which exhibited undetectable activity for L-DOPA in E. coli but was functional in S. cerevisiae, as well as with the tri-stable switch PCA3. Conversely, sensors for homovanillic acid and caffeic acid showed no activity in S. cerevisiae compared to E. coli. In particular, we have previously seen caffeic acid-responsive biosensors in E. coli that did not function in S. cerevisiae, further emphasizing these differences (24). The highest fold induction we achieved however was ∼10-fold in the case of wildtype RolR with resorcinol and the CAQ3 variant with catechol. Prokaryotic transcription activator-based biosensors in S. cerevisiae have achieved higher fold inductions of up to 26-fold with several rounds of promoter optimization (74,99). This suggests potential room for improvement of aTF repressor biosensor dynamic range in S. cerevisiae.
We were not able to engineer RolR to respond to the catecholamines dopamine, epinephrine, and the tumour marker methoxytyramine. The only catecholamine that gave a response was L-DOPA, which has a terminal carboxylic acid and an internal amine group, as opposed to the terminal amine group seen in the other targeted catecholamines. We hypothesize that this terminal amine could be the contributor behind the failure to identify variants that respond to them, and that further structure-guided engineering will be needed to engineer RolR response to catecholamines. We also were not able to identify variants responsive to 2,3-dihydoxybenzoate, which is the only benzene derivative in this work with substitutions on positions 1, 2 and 3, as opposed to 1-, 2- and 4-substitutions for the other trisubstituted benzenes, suggesting that different residues will have to be targeted to evolve response towards substitutions in these positions.
Our methodical approach for engineering aTFs provides a process for iteratively and systemically traversing their fitness landscapes to acquire novel ligand specificities. This approach has the potential for dramatically increasing the repertoire of inducer molecules for aTFs that are functional in broad host backgrounds, subsequently expanding opportunities for small molecule-directed programming of cellular activity in many metabolic engineering, cell, and synthetic biology applications. Additional directions for RolR engineering could focus on expanding its biosensor capabilities towards further diverse structures, such as the resorcinol-containing stilbene moiety seen in cannabinoid precursors (100), or the catechol-containing tetrahydroisoquinoline alkaloids (43), as well as other tumour biomarkers such as metanephrines.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Logan Timmins for contributing towards preliminary experiments. We thank Dr Roberto Chica, Olivier Gagnon, and Dr Antony St.-Jacques for their help in studying the structure of RolR, and with the choice of residues to target for mutagenesis. We thank the Concordia Genome Foundry for use of automation equipment, as well as James Bagley, Nicholas Gold and Dr Smita Amaranth for assistance with implementing and troubleshooting automation workflows.
Author contributions: M.A.N., D.H.K. and V.J.J.M. designed the research. M.A.N. performed experiments and analyzed data. D.H.K. and V.J.J.M. supervised the research. M.A.N. wrote the manuscript with editing help from D.H.K. and V.J.J.M.
Contributor Information
Mohamed A Nasr, Centre for Applied Synthetic Biology, Concordia University, Montréal, Québec, Canada; Department of Biology, Concordia University, Montréal, Québec, Canada; PROTEO, Québec Network for Research on Protein Function, Structure, and Engineering, Québec City, Québec, Canada.
Vincent J J Martin, Centre for Applied Synthetic Biology, Concordia University, Montréal, Québec, Canada; Department of Biology, Concordia University, Montréal, Québec, Canada.
David H Kwan, Centre for Applied Synthetic Biology, Concordia University, Montréal, Québec, Canada; Department of Biology, Concordia University, Montréal, Québec, Canada; Department of Chemistry and Biochemistry, Concordia University, Montréal, Québec, Canada; PROTEO, Québec Network for Research on Protein Function, Structure, and Engineering, Québec City, Québec, Canada.
Data Availability
The data underlying this article are available in the article and in its online supplementary material.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NSERC Discovery Grants [RGPIN-2016–05464 to D.H.K., RGPIN-2017-06703 to V.J.J.M.]; M.A.N. was supported by a Concordia International Tuition Award of Excellence, and an FRQNT B2X Doctoral Research Award; V.J.J.M. was supported by a Concordia University Research Chair. Funding for open access charge: Natural Sciences and Engineering Research Council of Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1. Balleza E., López-Bojorquez L.N., Martínez-Antonio A., Resendis-Antonio O., Lozada-Chávez I., Balderas-Martínez Y.I., Encarnación S., Collado-Vides J.. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol. Rev. 2009; 33:133–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lozada-Chávez I., Janga S.C., Collado-Vides J.. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 2006; 34:3434–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seshasayee A.S., Bertone P., Fraser G.M., Luscombe N.M.. Transcriptional regulatory networks in bacteria: from input signals to output responses. Curr. Opin. Microbiol. 2006; 9:511–519. [DOI] [PubMed] [Google Scholar]
- 4. Browning D.F., Busby S.J.W.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 5. Lutz R., Bujard H.. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997; 25:1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ulrich L.E., Koonin E.V., Zhulin I.B.. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005; 13:52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mahr R., Frunzke J.. Transcription factor-based biosensors in biotechnology: current state and future prospects. Appl. Microbiol. Biotechnol. 2016; 100:79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Paepe B., Peters G., Coussement P., Maertens J., De Mey M.. Tailor-made transcriptional biosensors for optimizing microbial cell factories. J. Ind. Microbiol. Biotechnol. 2017; 44:623–645. [DOI] [PubMed] [Google Scholar]
- 9. Matilla M.A., Velando F., Martín-Mora D., Monteagudo-Cascales E., Krell T.. A catalogue of signal molecules that interact with sensor kinases, chemoreceptors and transcriptional regulators. FEMS Microbiol. Rev. 2022; 46:fuab043. [DOI] [PubMed] [Google Scholar]
- 10. Wegner S.V., Boyaci H., Chen H., Jensen M.P., He C.. Engineering A uranyl-specific binding protein from NikR. Angew. Chem. Int. Ed. 2009; 48:2339–2341. [DOI] [PubMed] [Google Scholar]
- 11. Belkin S., Yagur-Kroll S., Kabessa Y., Korouma V., Septon T., Anati Y., Zohar-Perez C., Rabinovitz Z., Nussinovitch A., Agranat A.J.. Remote detection of buried landmines using a bacterial sensor. Nat. Biotechnol. 2017; 35:308–310. [DOI] [PubMed] [Google Scholar]
- 12. Ho J.C.H., Pawar S.V., Hallam S.J., Yadav V.G.. An improved whole-cell biosensor for the discovery of lignin-transforming enzymes in functional metagenomic screens. ACS Synth Biol. 2018; 7:392–398. [DOI] [PubMed] [Google Scholar]
- 13. Wen K.Y., Cameron L., Chappell J., Jensen K., Bell D.J., Kelwick R., Kopniczky M., Davies J.C., Filloux A., Freemont P.S.. A cell-free biosensor for detecting quorum sensing molecules in P. aeruginosa -infected Respiratory samples. ACS Synth. Biol. 2017; 6:2293–2301. [DOI] [PubMed] [Google Scholar]
- 14. Scott S.R., Din M.O., Bittihn P., Xiong L., Tsimring L.S., Hasty J.. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis. Nat. Microbiol. 2017; 2:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim S., Kerns S.J., Ziesack M., Bry L., Gerber G.K., Way J.C., Silver P.A.. Quorum sensing can Be repurposed to promote information transfer between bacteria in the mammalian gut. ACS Synth. Biol. 2018; 7:2270–2281. [DOI] [PubMed] [Google Scholar]
- 16. Mitchler M.M., Garcia J.M., Montero N.E., Williams G.J.. Transcription factor-based biosensors: a molecular-guided approach for natural product engineering. Curr. Opin. Biotechnol. 2021; 69:172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dinh C.V., Prather K.L.. Layered and multi-input autonomous dynamic control strategies for metabolic engineering. Curr. Opin. Biotechnol. 2020; 65:156–162. [DOI] [PubMed] [Google Scholar]
- 18. Chen Y., Ho J.M.L., Shis D.L., Gupta C., Long J., Wagner D.S., Ott W., Josić K., Bennett M.R.. Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 2018; 9:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brophy J.A.N., Voigt C.A.. Principles of genetic circuit design. Nat. Methods. 2014; 11:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ang J., Harris E., Hussey B.J., Kil R., McMillen D.R.. Tuning response curves for synthetic biology. ACS Synth. Biol. 2013; 2:547–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wan X., Volpetti F., Petrova E., French C., Maerkl S.J., Wang B.. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nat. Chem. Biol. 2019; 15:540–548. [DOI] [PubMed] [Google Scholar]
- 22. Fernandez-López R., Ruiz R., de la Cruz F., Moncalián G.. Transcription factor-based biosensors enlightened by the analyte. Front. Microbiol. 2015; 6:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grazon C., Baer R.C., Kuzmanović U., Nguyen T., Chen M., Zamani M., Chern M., Aquino P., Zhang X., Lecommandoux S.et al.. A progesterone biosensor derived from microbial screening. Nat. Commun. 2020; 11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nasr M.A., Timmins L.R., Martin V.J.J., Kwan D.H.. A versatile transcription factor biosensor system responsive to multiple aromatic and indole inducers. ACS Synth Biol. 2022; 11:1692–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Feng J., Jester B.W., Tinberg C.E., Mandell D.J., Antunes M.S., Chari R., Morey K.J., Rios X., Medford J.I., Church G.M.et al.. A general strategy to construct small molecule biosensors in eukaryotes. Elife. 2015; 4:e10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Juárez J.F., Lecube-Azpeitia B., Brown S.L., Johnston C.D., Church G.M.. Biosensor libraries harness large classes of binding domains for construction of allosteric transcriptional regulators. Nat. Commun. 2018; 9:3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Packer M.S., Liu D.R.. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015; 16:379–394. [DOI] [PubMed] [Google Scholar]
- 28. Arnold F.H. Design by directed evolution. Acc. Chem. Res. 1998; 31:125–131. [Google Scholar]
- 29. Saenger W., Hinrichs W., Orth P., Schnappinger D., Hillen W.. Structural basis of gene regulation by the tetracycline inducible Tet repressor–operator system. Nat. Struct. Biol. 2000; 7:215–219. [DOI] [PubMed] [Google Scholar]
- 30. Zaim J., Kierzek A.M.. The structure of full-length LysR-type transcriptional regulators. Modeling of the full-length OxyR transcription factor dimer. Nucleic Acids Res. 2003; 31:1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Motlagh H.N., Wrabl J.O., Li J., Hilser V.J.. The ensemble nature of allostery. Nature. 2014; 508:331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reichheld S.E., Yu Z., Davidson A.R.. The induction of folding cooperativity by ligand binding drives the allosteric response of tetracycline repressor. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:22263–22268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li T., Zhao K., Huang Y., Li D., Jiang C.Y., Zhou N., Fan Z., Liu S.J.. The TetR-type transcriptional repressor RolR from Corynebacterium glutamicum regulates resorcinol catabolism by binding to a unique operator, rolO. Appl. Environ. Microbiol. 2012; 78:6009–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li D.F., Zhang N., Hou Y.J., Huang Y., Hu Y., Zhang Y., Liu S.J., Wang D.C.. Crystal structures of the transcriptional repressor rolr reveals a novel recognition mechanism between inducer and regulator. PLoS One. 2011; 6:e19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nov Y. When second best is good enough: another probabilistic look at saturation mutagenesis. Appl. Environ. Microbiol. 2012; 78:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nomura Y., Yokobayashi Y.. Dual selection of a genetic switch by a single selection marker. Biosystems. 2007; 90:115–120. [DOI] [PubMed] [Google Scholar]
- 37. Yang J., Seo S.W., Jang S., Shin S.I., Lim C.H., Roh T.Y., Jung G.Y.. Synthetic RNA devices to expedite the evolution of metabolite-producing microbes. Nat. Commun. 2013; 4:1–7. [DOI] [PubMed] [Google Scholar]
- 38. Gioia C., Banella M.B., Vannini M., Celli A., Colonna M., Caretti D.. Resorcinol: a potentially bio-based building block for the preparation of sustainable polyesters. Eur. Polym. J. 2015; 73:38–49. [Google Scholar]
- 39. Du L., Wang Z., Li S., Song W., Lin W.. A comparison of monomeric phenols produced from lignin by fast pyrolysis and hydrothermal conversions. Int. J. Chem. React. Eng. 2013; 11:135–145. [Google Scholar]
- 40. Pyne M.E., Narcross L., Melgar M., Kevvai K., Mookerjee S., Leite G.B., Martin V.J.J.. An engineered Aro1 protein degradation approach for increased cis,cis-muconic acid biosynthesis in saccharomyces cerevisiae. Appl. Environ. Microbiol. 2018; 84:e01095-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fiege H., Voges H.-W., Hamamoto T., Umemura S., Iwata T., Miki H., Fujita Y., Buysch H.-J., Garbe D., Paulus W.. Phenol derivatives. Ullmann's Encyclopedia of Industrial Chemistry. 2000; John Wiley & Sons Ltd. [Google Scholar]
- 42. Thompson B., Pugh S., Machas M., Nielsen D.R.. Muconic acid production via alternative pathways and a synthetic “metabolic funnel”. ACS Synth. Biol. 2018; 7:565–575. [DOI] [PubMed] [Google Scholar]
- 43. Pyne M.E., Kevvai K., Grewal P.S., Narcross L., Choi B., Bourgeois L., Dueber J.E., Martin V.J.J.. A yeast platform for high-level synthesis of tetrahydroisoquinoline alkaloids. Nat. Commun. 2020; 11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang J., Mahajani M., Jackson S.L., Yang Y., Chen M., Ferreira E.M., Lin Y., Yan Y.. Engineering a bacterial platform for total biosynthesis of caffeic acid derived phenethyl esters and amides. Metab. Eng. 2017; 44:89–99. [DOI] [PubMed] [Google Scholar]
- 45. Lam L., Woollard G.A., Teague L., Davidson J.S.. Clinical validation of urine 3-methoxytyramine as a biomarker of neuroblastoma and comparison with other catecholamine-related biomarkers. Ann. Clin. Biochem. 2017; 54:264–272. [DOI] [PubMed] [Google Scholar]
- 46. Nielsen A.A.K., Der B.S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E.A., Ross D., Densmore D., Voigt C.A.. Genetic circuit design automation. Science. 2016; 352:aac7341. [DOI] [PubMed] [Google Scholar]
- 47. Klock H.E., Lesley S.A.. The polymerase incomplete primer extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol. Biol. 2009; 498:91–103. [DOI] [PubMed] [Google Scholar]
- 48. Reetz M.T., Bocola M., Carballeira J.D., Zha D., Vogel A.. Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test. Angew. Chem. Int. Ed. 2005; 44:4192–4196. [DOI] [PubMed] [Google Scholar]
- 49. Reetz M.T., Carballeira J.D.. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat. Protoc. 2007; 2:891–903. [DOI] [PubMed] [Google Scholar]
- 50. Bryksin A.V., Matsumura I.. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. BioTechniques. 2010; 48:463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stevenson J., Krycer J.R., Phan L., Brown A.J.. A practical comparison of ligation-independent cloning techniques. PLoS One. 2013; 8:8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trott O., Olson A.J.. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010; 31:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Horwitz A.A., Walter J.M., Schubert M.G., Kung S.H., Hawkins K., Platt D.M., Hernday A.D., Mahatdejkul-Meadows T., Szeto W., Chandran S.S.et al.. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst. 2015; 1:88–96. [DOI] [PubMed] [Google Scholar]
- 54. Shao Z., Zhao H., Zhao H.. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009; 37:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bean B.D.M., Whiteway M., Martin V.J.J.. The MyLO CRISPR-Cas9 toolkit: a markerless yeast localization and overexpression CRISPR-Cas9 toolkit. G3: Genes, Genomes, Genet. 2022; 12:jkac154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Flagfeldt D.B., Siewers V., Huang L., Nielsen J.. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast. 2009; 26:545–551. [DOI] [PubMed] [Google Scholar]
- 57. Gietz R.D., Schiestl R.H.. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007; 2:35–37. [DOI] [PubMed] [Google Scholar]
- 58. Ruegg T.L., Pereira J.H., Chen J.C., DeGiovanni A., Novichkov P., Mutalik V.K., Tomaleri G.P., Singer S.W., Hillson N.J., Simmons B.A.et al.. Jungle Express is a versatile repressor system for tight transcriptional control. Nat. Commun. 2018; 9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ellefson J.W., Ledbetter M.P., Ellington A.D.. Directed evolution of a synthetic phylogeny of programmable Trp repressors. Nat. Chem. Biol. 2018; 14:361–367. [DOI] [PubMed] [Google Scholar]
- 60. D’oelsnitz S., Nguyen V., Alper H.S., Ellington A.D.. Evolving a generalist biosensor for bicyclic monoterpenes. ACS Synth. Biol. 2022; 2022:272. [DOI] [PubMed] [Google Scholar]
- 61. Taylor N.D., Garruss A.S., Moretti R., Chan S., Arbing M.A., Cascio D., Rogers J.K., Isaacs F.J., Kosuri S., Baker D.et al.. Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods. 2016; 13:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Snoek T., Chaberski E.K., Ambri F., Kol S., Bjørn S.P., Pang B., Barajas J.F., Welner D.H., Jensen M.K., Keasling J.D.. Evolution-guided engineering of small-molecule biosensors. Nucleic Acids Res. 2020; 48:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ray S., Panjikar S., Anand R.. Structure guided design of protein biosensors for phenolic pollutants. ACS Sens. 2017; 2:411–418. [DOI] [PubMed] [Google Scholar]
- 64. Meyer A.J., Segall-Shapiro T.H., Glassey E., Zhang J., Voigt C.A.. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nat. Chem. Biol. 2019; 15:196–204. [DOI] [PubMed] [Google Scholar]
- 65. Della Corte D., van Beek H.L., Syberg F., Schallmey M., Tobola F., Cormann K.U., Schlicker C., Baumann P.T., Krumbach K., Sokolowsky S.et al.. Engineering and application of a biosensor with focused ligand specificity. Nat. Commun. 2020; 11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Raman S., Rogers J.K., Taylor N.D., Church G.M.. Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:17803–17808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tashiro Y., Fukutomi H., Terakubo K., Saito K., Umeno d.. A nucleoside kinase as a dual selector for genetic switches and circuits. Nucleic Acids Res. 2011; 39:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Griffith J.K., Cuellar D.H., Fordyce C.A., Hutchings K.G., Mondragon A.A.. Structure and function of the class C tetracycline/H + antiporter. Mol. Membr. Biol. 1994; 11:271–277. [DOI] [PubMed] [Google Scholar]
- 69. Podolsky T., Fong S.-T., Lee B.T.O.. Direct selection of tetracycline-SensitiveEscherichia coliCells using nickel salts. Plasmid. 1996; 36:112–115. [DOI] [PubMed] [Google Scholar]
- 70. Hinrichs W., Kisker C., Düvel M., Müller A., Tovar K., Hillen W., Saenger W.. Structure of the tet repressor-tetracycline complex and regulation of antibiotic resistance. Science. 1994; 264:418–420. [DOI] [PubMed] [Google Scholar]
- 71. Stavropoulos T.A., Strathdee C.A.. Expression of the tetA(C) tetracycline efflux pump in Escherichia coli confers osmotic sensitivity. FEMS Microbiol. Lett. 2000; 190:147–150. [DOI] [PubMed] [Google Scholar]
- 72. Tack D.S., Tonner P.D., Pressman A., Olson N.D., Levy S.F., Romantseva E.F., Alperovich N., Vasilyeva O., Ross D.. The genotype-phenotype landscape of an allosteric protein. Mol. Syst. Biol. 2021; 17:e10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ikushima S., Zhao Y., Boeke J.D.. Development of a tightly controlled off switch for saccharomyces cerevisiae regulated by camphor, a low-cost natural product. G3: Genes, Genomes, Genet. 2015; 5:1983–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Skjoedt M.L., Snoek T., Kildegaard K.R., Arsovska D., Eichenberger M., Goedecke T.J., Rajkumar A.S., Zhang J., Kristensen M., Lehka B.J.et al.. Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. Nat. Chem. Biol. 2016; 12:951–958. [DOI] [PubMed] [Google Scholar]
- 75. Xie M., Fussenegger M.. Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol. 2018; 19:507–525. [DOI] [PubMed] [Google Scholar]
- 76. Wang R., Cress B.F., Yang Z., Hordines J.C., Zhao S., Jung G.Y., Wang Z., Koffas M.A.G.. Design and characterization of biosensors for the screening of modular assembled Naringenin Biosynthetic library in Saccharomyces cerevisiae. ACS Synth. Biol. 2019; 8:2121–2130. [DOI] [PubMed] [Google Scholar]
- 77. Dabirian Y., Li X., Chen Y., David F., Nielsen J., Siewers V.. Expanding the dynamic range of a transcription factor-based biosensor in saccharomyces cerevisiae. ACS Synth. Biol. 2019; 8:1968–1975. [DOI] [PubMed] [Google Scholar]
- 78. Lv Y., Gu Y., Xu J., Zhou J., Xu P.. Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield. Metab. Eng. 2020; 61:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Slubowski C.J., Funk A.D., Roesner J.M., Paulissen S.M., Huang L.S.. Plasmids for C-terminal tagging in saccharomyces cerevisiae that contain improved GFP proteins, Envy and Ivy. Yeast. 2015; 32:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Murphy K.F., Balázsi G., Collins J.J.. Combinatorial promoter design for engineering noisy gene expression. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:12726–12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kamionka A., Bogdanska-Urbaniak J., Scholz O., Hillen W.. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 2004; 32:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yang J.I., Hwang J.S., Camakaris H., Irawaty W., Ishihama A., Pittard J.. Mode of action of the TyrR protein: repression and activation of the tyrP promoter of Escherichia coli. Mol. Microbiol. 2004; 52:243–256. [DOI] [PubMed] [Google Scholar]
- 83. Oelsnitz S., Kim W., Burkholder N.T., Javanmardi K., Thyer R., Zhang Y., Alper H.S., Ellington A.D.. Using fungible biosensors to evolve improved alkaloid biosyntheses. Nat. Chem. Biol. 2022; 18:981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Flachbart L.K., Gertzen C.G.W., Gohlke H., Marienhagen J.. Development of a biosensor platform for phenolic compounds using a transition Ligand strategy. ACS Synth Biol. 2021; 10:2002–2014. [DOI] [PubMed] [Google Scholar]
- 85. Frei C.S., Qian S., Cirino P.C.. New engineered phenolic biosensors based on the AraC regulatory protein. Protein Eng. Des. Sel. 2018; 31:213–220. [DOI] [PubMed] [Google Scholar]
- 86. F.M. L., Currin A., Dixon N.. Directed evolution of the PcaV allosteric transcription factor to generate a biosensor for aromatic aldehydes. J. Biol. Eng. 2019; 13:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yao J., He Y., Su N., Bharath S.R., Tao Y., Jin J.M., Chen W., Song H., Tang S.Y.. Developing a highly efficient hydroxytyrosol whole-cell catalyst by de-bottlenecking rate-limiting steps. Nat. Commun. 2020; 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Romero P.A., Arnold F.H.. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009; 10:866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Trudeau D.L., Tawfik D.S.. Protein engineers turned evolutionists-the quest for the optimal starting point. Curr. Opin. Biotechnol. 2019; 60:46–52. [DOI] [PubMed] [Google Scholar]
- 90. Gupta A., Reizman I.M.B., Reisch C.R., Prather K.L.J.. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 2017; 35:273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang G., Øzmerih S., Guerreiro R., Meireles A.C., Carolas A., Milne N., Jensen M.K., Ferreira B.S., Borodina I.. Improvement of cis, cis-muconic acid production in saccharomyces cerevisiae through biosensor-aided genome engineering. ACS Synth. Biol. 2020; 9:634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fordjour E., Adipah F.K., Zhou S., Du G., Zhou J.. Metabolic engineering of Escherichia coli BL21 (DE3) for de novo production of l-DOPA from d-glucose. Microb. Cell Fact. 2019; 18:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Huang Q., Lin Y., Yan Y.. Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol. Bioeng. 2013; 110:3188–3196. [DOI] [PubMed] [Google Scholar]
- 94. Zhou P., Yue C., Shen B., Du Y., Xu N., Ye L.. Metabolic engineering of saccharomyces cerevisiae for enhanced production of caffeic acid. Appl. Microbiol. Biotechnol. 2021; 105:5809–5819. [DOI] [PubMed] [Google Scholar]
- 95. Kruyer N.S., Wauldron N., Bommarius A.S., Peralta-Yahya P.. Fully biological production of adipic acid analogs from branched catechols. Sci. Rep. 2020; 10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hwang N., Chong E., Oh H., Cho H.W., Lee J.W., Sung K.W., Lee S.Y.. Application of an LC–MS/MS method for the simultaneous quantification of homovanillic acid and vanillylmandelic acid for the diagnosis and follow-up of neuroblastoma in 357 patients. Mol. 2021. 2021; 26:3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Grouzmann E., Centeno C., Eugster P.J.. Quantification of vanillylmandelic acid, homovanillic acid and 5-hydroxyindoleacetic acid in urine using a dilute-and-shoot and ultra-high pressure liquid chromatography tandem mass spectrometry method. Clin. Chem. Lab. Med. 2018; 56:1533–1541. [DOI] [PubMed] [Google Scholar]
- 98. D’Ambrosio V., Jensen M.K.. Lighting up yeast cell factories by transcription factor-based biosensors. FEMS Yeast Res. 2017; 17:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ambri F., D’ambrosio V., Di Blasi R., Maury J., Jacobsen S.A.B., Mccloskey D., Jensen M.K., Keasling J.D. High-resolution scanning of optimal biosensor reporter promoters in yeast. ACS Synth. Biol. 2020; 9:218–226. [DOI] [PubMed] [Google Scholar]
- 100. Luo X., Reiter M.A., D’Espaux L., Wong J., Denby C.M., Lechner A., Zhang Y., Grzybowski A.T., Harth S., Lin W.et al.. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature. 2019; 567:123–126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.