Abstract
The study of genes and their products is an essential prerequisite for fundamental research. Characterization can be achieved by analyzing mutants or overexpression lines or by studying the localization and substrate specificities of the resulting proteins. However, functional analysis of specific proteins in complex eukaryotic organisms can be challenging. To overcome this, the use of heterologous systems to express genes and analyze the resulting proteins can save time and effort. Yeast is a preferred heterologous model organism: it is easy to transform, and tools for genomics, engineering, and metabolomics are already available. Here, we describe a well-established and simple method to analyze the activity of plant monosaccharide transporters in the baker’s yeast, Saccharomyces cerevisiae, using a simple growth complementation assay. We used the famous hexose-transport-deficient yeast strain EBY.VW4000 to express candidate plant monosaccharide transporters and analyzed their transport activity. This assay does not require any radioactive labeling of substrates and can be easily extended for quantitative analysis using growth curves or by analyzing the transport rates of fluorescent substrates like the glucose analog 2-NBDG. Finally, to further simplify the cloning of potential candidate transporters, we provide level 0 modular cloning (MoClo) modules for efficient and simple Golden Gate cloning. This approach provides a convenient tool for the functional analysis of plant monosaccharide transporters in yeast.
Key features
Comprehensive, simple protocol for analysis of plant monosaccharide transporters in yeast
Includes optional MoClo parts for cloning with Golden Gate method
Includes protocol for the production and transformation of competent yeast cells
Does not require hazardous solutions, radiolabeled substrates, or specialized equipment
Keywords: Yeast, Heterologous expression, Plant monosaccharide transporters, EBY.VW4000, Drop-out assay
Background
In the past decade, the availability of full genome sequences, powerful genome engineering tools, and innovations in transcriptomics and metabolomics have revolutionized plant research (Rai et al., 2019; Purugganan and Jackson, 2021; Wang and Doudna, 2023). Plant sugar transporters are an important area of research, as they play a crucial role in delivering sugars as energy sources for developmental processes, contribute to disease resistance mechanisms, and are key determinants of yield (Julius et al., 2017; Bezrutczyk et al., 2018; Breia et al., 2021). However, the analysis of individual transporter candidates or transporter mutants is challenging because plant genomes are complex and sugar transporters are often organized in large gene families. To solve this bottleneck, heterologous systems, such as the baker’s yeast Saccharomyces cerevisiae, are powerful tools for analyzing plant sugar transporter functions on a single scale (Boles, 2003). Yeast is easy to transform, and many different strains with various phenotypes exist, which are suitable for analyzing specific sugar-transport abilities. In this protocol, we used the well-known hexose-transport-deficient strain EBY.VW4000 to analyze the activity of plant monosaccharide transporters and their mutants (Wieczorke et al., 1999). Instead of measuring the uptake of radioactively labeled substrates, as described in other protocols (Milne et al., 2017), this protocol describes a simple and cost-effective drop-out assay. Therefore, the hexose-transport-deficient strain EBY.VW4000 is transformed with expression constructs for the monosaccharide transporter of interest. A dilution series of the complemented EBY.VW4000 strains is spotted on selective plates to determine their growth on different carbon sources like maltose or glucose. The described protocol has already been extensively used in many studies dealing with sugar transporters (e.g., Chen et al., 2010; Moore et al., 2015; Skoppek et al., 2022; Tamayo et al., 2022; Yue et al., 2023). To provide a comprehensive workflow, we also implemented a protocol for the production of competent yeast cells and for transformation. This approach is based on the protocols described by Gietz and Schiestl (2007a and 2007b) and has been optimized for EBY.VW4000. Finally, we provide optional level 0 modular cloning (MoClo) modules for the yeast PMA1 promoter (plasma membrane H+ ATPase 1) and the ADH2 (alcohol dehydrogenase 2) terminator, which are commonly used for the expression of transporters in yeast (Meyer et al., 2006; Skoppek et al., 2022). These modules allow an easy and cost-effective cloning of new constructs using the Golden Gate cloning method (Engler et al., 2009; Werner et al., 2012).
The described drop-out assay is a simple method to rapidly screen for the transport activities of transporter candidates. However, if quantitative analysis is needed, we suggest extending this analysis to a growth curve or by analyzing the transport rate of fluorescent sugar analogs [for example esculin (sucrose analog) or 2-NBDG (glucose analog)] (Gora et al., 2012; Roy et al., 2015).
Material and reagents
Biological materials
-
Saccharomyces cerevisiae strain EBY.VW4000 (hexose transport-deficient)
Genotype: CEN.PK2-1C hxt13Δ::loxP hxt15Δ::loxP hxt16Δ::loxP hxt14Δ::loxP hxt12Δ::loxP hxt9Δ::loxP hxt11Δ::loxP hxt10Δ::loxP hxt8Δ::loxP hxt4-1-5Δ::loxP hxt2Δ::loxP hxt3-6-7Δ::loxP gal2Δ::(ura3/FOA) stl1Δ::loxP agt1Δ::loxP mph2(ydl247w)Δ::loxP mph3(yjr160c)Δ::loxP
For more information about the choice of suitable yeast strains see General Note 1.
Reagents
Agar-agar, Kobe I (Carl Roth, catalog number: 5210.5)
Peptone from meat (Carl Roth, catalog number: 2366.1)
Yeast synthetic drop-out medium supplements, without uracil (amino acid mix -Ura) Merck KGaA, catalog number: Y1501-20G)
Yeast extract (Carl Roth, catalog number: 2363.4)
Yeast nitrogen base without amino acids (YNB) (Merck KGaA, catalog number: Y0626-250G)
D-Glucose monohydrate (Duchefa B.V., catalog number: G0802.1000)
D(+)-maltose monohydrate (Carl Roth, catalog number: 8951.4)
Salmon sperm DNA sodium salt (Carl Roth, catalog number: 5434.1)
Lithium acetate dihydrate (LiAc) (LiCH3COO·2H2O) (Carl Roth, catalog number: 6713.1)
Poly(ethylene glycol) 3350, PEG3350, H(OCH2CH2)nOH (Merck KGaA, catalog number: P4338)
Dimethyl sulfoxide (DMSO), (CH3)2SO (Merck KGaA, catalog number: 276855-1L)
Glycerol (C3H8O3) 86% (Carl Roth, catalog number: 4043.3)
Sodium chloride (NaCl) (Carl Roth, catalog number: 3957.2)
Potassium chloride (KCl) (AppliChem GmbH, catalog number: A1039.1000)
Tris (C4H11NO3) (Carl Roth, catalog number: AE15.3)
Ethylenediamine tetra acetic acid disodium salt dihydrate (EDTA), C10H14N2Na2O8·2H2O (Carl Roth, catalog number: 8043.2)
1 N hydrochloric acid (HCl) or any other non-fuming HCl (Carl Roth, catalog number: 6792.1)
1 N sodium hydroxide (NaOH) or similar (Carl Roth, catalog number: 6785.1)
ddH2O (sterile)
Optional: MoClo plasmids level 0 and ready-to-use level 1
pJS506 [PMA1 promoter (Pro5U module), pICH41295, GGAG-AATG (BsaI)] (Addgene, #200712)
pJS237 [ADH2 terminator (3′U+Ter module), pUCGent, GCTT-CGCT (BsaI)] (Addgene, #200713)
pJS288 [Hxt1 transcription unit (Level 1 TU, P1), pAGT572, TGCC-GCAA (BpiI)] (Addgene, #200714)
Acceptor vectors: pAGT572 (selection marker for uracil) was a kind gift from Sylvestre Marillonnet and Alain Tissier (Scheler et al., 2016). Alternative yeast-compatible vectors is, for example, pJOG417 (Addgene #105341; selection marker for leucine) (Gantner et al., 2018)
Solutions
20% glucose solution (sterile filtered)
20% maltose solution (sterile filtered)
14% glycerol solution
YPM medium (see Recipe 1)
Frozen competent cell solution (FCC) (see Recipe 2)
10× yeast synthetic drop-out medium supplements, without uracil, (amino acid mix -Ura) (see Recipe 3)
YNB medium with amino acid mix -Ura and carbon source (see Recipe 4)
50% PEG 3350 (autoclaved) (see Recipe 5)
1 M LiAc (sterile filtered) (see Recipe 6)
Salmon sperm carrier DNA (2 mg/mL) (see Recipe 7)
1 M Tris-HCl (pH 8.0) (see Recipe 8)
0.2 M EDTA (pH 8.0) (see Recipe 9)
10 mM TE buffer (pH 8.0) (see Recipe 10)
Recipes
-
YPM medium (full medium with maltose)
Reagent Final concentration Amount Yeast extract 1% (w/v) 5 g Peptone 2% (w/v) 10 g Maltose 2% (w/v) 10 g Agar-agar (optional for plates) 2% (w/v) 10 g ddH2O n/a Ad 500 mL Autoclave. Pour plates with solid medium.
Note: If needed for a certain yeast strain, the carbon source can be replaced by glucose, galactose, or glycerol. Maltose was most suitable to grow EBY.VW4000.
-
Frozen competent cell solution (FCC)
Reagent Final concentration Amount Glycerol 5% (v/v) 5 mL DMSO 10% (v/v) 10 mL Total n/a Ad 100 mL Store FCC at room temperature (RT).
-
10× yeast synthetic drop-out medium supplements, without uracil (amino acid mix -Ura)
Reagent Final concentration Amount Yeast synthetic drop-out medium supplements, without uracil (amino acid mix -Ura) 10× 1.92 g ddH2O n/a Ad 100 mL Filter sterilize. Store at 4 °C and protect from light.
Note: To dissolve the supplements in water, stir the solution on a magnetic stirrer. Carefully warm the solution (maximum 50 °C) if the supplements do not dissolve well. Always tightly close the stock bottle with the supplements after usage, to prevent the powder from absorbing water. For additional information on how to choose the yeast synthetic drop-out medium, see General Note 2.
-
YNB medium with amino acid mix -Ura and carbon source
To prepare 500 mL of YNB medium with amino acid mix -Ura and carbon source, first weigh the required amount of YNB and dissolve in 400 mL of water. For solid plates, add 2% agar-agar. Autoclave and store until usage.
Reagent Final concentration Amount YNB n/a 3.35 g Agar-agar (optional for plates) 2% (w/v) 10 g ddH2O n/a Ad 400 mL For solid medium: Carefully boil up the medium in a microwave. Secondly, add the following supplements before use:
Reagent Final concentration Amount 20% carbon source1 2% (v/v) 50 mL 10× amino acid mix -Ura (Recipe 3) 1% (v/v) 50 mL For solid medium: pour plates.
1The carbon source is chosen according to the intended use of the medium. For the required carbon source, e.g., glucose, maltose, or similar, a 20% stock solution is prepared in water, filter sterilized, and stored at RT until use. For selection and cultivation of transformed EBY.VW4000 strains, add maltose as carbon source. For performing the drop-out assay on different carbon sources, add the required sugar, e.g., glucose.
-
50% PEG 3350
Reagent Final concentration Amount PEG 3350 50 % 50 g ddH2O n/a Ad 100 mL Autoclave, store at RT.
-
1 M LiAc
Reagent Final concentration Amount PEG 3350 1 M 10.2 g ddH2O n/a Ad 100 mL Autoclave, store at RT.
-
Salmon sperm carrier DNA (2 mg/mL)
Reagent Final concentration Amount Salmon sperm carrier DNA 2 mg /mL (w/v) 8 g 10 mM TE buffer (Recipe 8) 10 mM Ad 4 mL Dissolve the salmon sperm carrier DNA in 10 mM TE buffer on ice for 3–4 h with gentle shaking. Prepare aliquots, for example with 500 μL each. Incubate the aliquots for 10 min at 95 °C and immediately transfer to ice. Store at -20 °C.
-
1 M Tris-HCl (pH 8.0)
Reagent Final concentration Amount Tris 1 M 12.114 g ddH2O n/a Ad 100 mL Dissolve Tris in 80 mL of ddH2O and adjust the pH to 8.0 by carefully adding drops of HCl (1 N or any other non-fuming HCl). Fill up the volume to 100 mL.
-
0.2 M EDTA
Reagent Final concentration Amount EDTA 200 mM 7.44 g ddH2O n/a Add 100 mL Dissolve EDTA in 80 mL of ddH2O and adjust the pH to 8.0 by carefully adding NaOH (1 N NaOH solution or pellets). Fill up the volume to 100 mL.
-
10 mM TE-buffer (pH 8.0)
Reagent Final concentration Amount Tris-HCl pH 8.0 10 mM 1 mL EDTA pH 8.0 1 mM 0.5 mL ddH2O n/a 98.5 mL
Laboratory supplies
Squared Petri dishes for drop-out (Carl Roth, catalog number: EL50.1)
Petri dish (Sarstedt, catalog number: 82.1473.001)
2 mL reaction tube (Sarstedt, catalog number: 72.695.500)
Parafilm
Disposable gloves
50 mL reaction tubes (Sarstedt, catalog number: 62.547.254)
Equipment
Biosafety cabinet or bench with flame
Pipettes
Photometer (e.g., IMPLEN, OD600 DiluPhotometer; or similar)
Tabletop centrifuge (e.g., Eppendorf Micro Centrifuge 5425, maximum 21,300 rcf or similar)
Centrifuge for 50 mL reaction tubes (e.g., Eppendorf 5810R, maximum 20,913 rcf or similar)
Mobile phone or any other camera device
Optional: INTEGRA multichannel pipet Voyager (INTEGRA Biosciences GmbH, catalog number: 4721)
Optional: INTEGRA tips (INTEGRA Biosciences GmbH, e.g., as box 3416 or as bag 4411)
Procedure
-
Optional: available modular cloning (MoClo) modules for construct design
A helpful protocol on how to design and clone constructs by using the Golden Gate cloning and the MoClo system can be found in Engler et al. (2014).
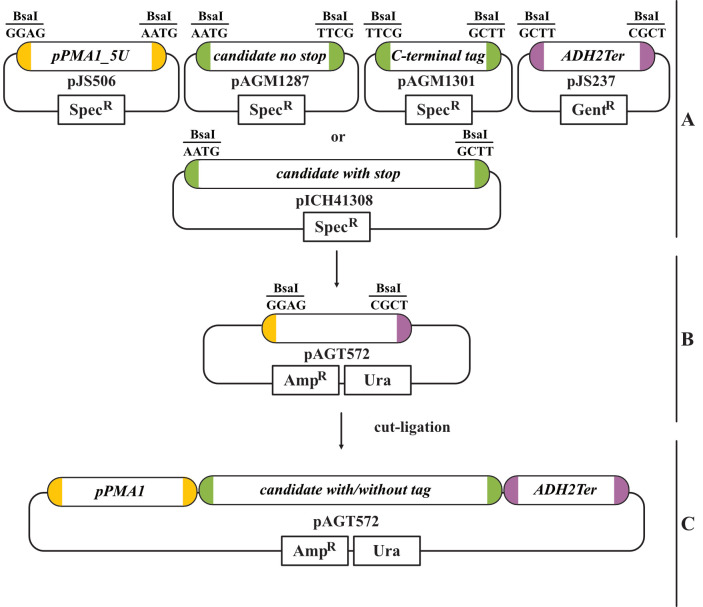
If cloning is planned with this method, the provided level 0 PMA1 promoter and ADH2 terminator module can be used (Figure 1A).
As level 1 acceptor vector, choose a yeast-compatible one. Here, the MoClo level 1 acceptor vector pAGT572 (uracil selection) was used (Figure 1B and 1C).
Remember to clone suitable negative and positive controls for the drop-out assay. We usually use an empty plasmid or GFP on a plasmid as negative control and the yeast monosaccharide transporter Hxt1 (NCBI 856494) as positive control. The transcription unit (pJS288) for the positive control used in this protocol (Hxt1 under control of the PMA1 promoter and ADH2 terminator cloned in pAGT572) is available via Addgene #200714.
For more background information, see General Note 3.
-
Preparation of competent EBY.VW4000 cells
A helpful protocol on how to prepare competent yeast cells can be found in Gietz and Schiestl (2007a).
For EBY.VW4000, use the following modifications of this protocol:
Streak out a stock of EBY.VW4000 cells on a plate with solid YPM medium (Recipe 1) (single colony needed). Incubate at 28 °C for two days.
Pick a single yeast colony and inoculate a 50 mL preculture by using liquid YPM medium. Incubate the culture overnight at 28 °C on a shaker at 140 rpm.
Take a 1 mL sample and dilute 1:10 with liquid YPM. Mix the sample before diluting and before measuring. Measure the OD600 of the 1:10 dilution.
Calculate the volume of preculture needed to inoculate a 200 mL main culture with an OD600 of 0.1 in YPM medium. Do not forget to include the dilution factor in your calculation.
-
Incubate the yeast cells at 28 °C on a shaker at 140 rpm until an OD600 between 0.6 and 0.8 is reached.
Note: Regularly check the OD600. Do not overgrow the culture. In our hands, it took approximately 5 h to reach an OD600 between 0.6 and 0.8.
Perform the next steps at RT.
-
Split the culture equally into six 50 mL reaction tubes (approximately 33.33 mL for each tube).
Note: For centrifugation, we never fill the tubes with the maximum possible volume. We usually centrifuge a maximum of 2/3 of the total tube volume. This prevents spilling during centrifugation, as tubes are not tightly closed.
Centrifuge at 3,000× g for 5 min.
Discard the supernatant.
Add 16.6 mL (0.5 V of the starting culture, divided by six tubes: 0.5 × 200 mL/6) of sterile ddH2O to each tube and resuspend the pellet.
Combine two resuspended pellets in one tube. This results in three tubes with approximately 33.2 mL each.
Centrifuge at 3,000× g for 5 min.
Discard the supernatant.
Resuspend the three pellets by adding 0.666 mL (0.01 V of the starting culture, divided by three tubes: 0.01 × 200 mL/3) of sterile ddH2O to each tube. Combine the resuspended pellets of all three tubes into one tube (total volume approximately 1.99 mL).
Centrifuge at 3,000× g for 5 min.
Discard the supernatant.
Resuspend the pellet by adding 2 mL (0.01 V of the starting volume: 0.01 × 200 mL) of FCC solution (Recipe 2) to each tube.
Prepare aliquots of 50 μL competent cells in 1.5 mL tubes for subsequent transformation.
-
Store at -20 °C for at least 3 h (overnight is also possible).
Note: Slowly freezing the cells is essential for survival.
Store at -80 °C for long-term storage.
Optional: To check the general viability of the competent cells, plate a 1:100 dilution (in ddH2O) of one aliquot on YPM medium and incubate overnight at 28 °C. The plate should be nicely overgrown with yeast.
-
Yeast EBY.VW4000 transformation
This method is based on the protocol for LiAc/SS carrier DNA/PEG transformation by Gietz and Schiestl (2007b), with the following modifications:
Thaw competent EBY.VW4000 cells on ice (50 μL per transformation).
For each plasmid, one transformation mix is prepared:
Mix the plasmid with ddH2O for a total volume of 14 μL and 600 ng plasmid in a 1.5 mL reaction tube. Vortex. Prepare one negative control with 14 μL of ddH2O only.
Add 260 μL of 50% PEG3350 (Recipe 5), 36 μL of 1 M LiAc (Recipe 6), and 50 μL of salmon sperm carrier DNA (2 mg/mL) (Recipe 7).
-
Mix well.
Note: PEG3350 is very viscous. Pipette slowly and ensure that the transformation mix is well mixed.
Store transformation mix at RT until use.
Centrifuge competent yeast cells at 13,000× g for 2 min at RT.
Discard the supernatant.
Add the transformation mix, including the plasmid.
Vortex or mix by pipetting up and down until the pellet is resuspended.
-
Incubate for 60 min at 42 °C (water bath or heat block)
Note: If not in stock, pour YNB plates with selective amino acid mix during the incubation time (Recipe 3 and Recipe 4).
Centrifuge at 13,000× g for 30 s.
Discard the supernatant.
Add 1 mL of sterile ddH2O.
Resuspend the pellet by pipetting up and down.
-
Plate 50–200 μL on YNB medium (with selective amino acid mix, in our setup -Ura, and maltose).
Note: We usually prepare two plates: one with 50 μL and a backup with 200 μL of transformed cells. The plasmids we are using contain a 2 μ origin of replication and typically yield between 50 and 100 clones when we plate 50 μL. The transformed cells can be stored at 4 °C and plated again one day later if needed.
Incubate the cells for 2–3 days at 28 °C.
Transfer 2–4 single colonies per transformation to a new YNB selection plate and grow overnight at 28 °C.
To generate long-term stocks: scrape off one of the clones, resuspend in sterile 14% glycerol solution, and store at -80 °C. Usually, we prepare stocks of 2–3 individual clones.
-
Yeast sugar uptake analysis via drop-out assay
-
Prepare YNB plates with selective amino acid mix and the required carbon source in squared Petri dishes for the drop-out assay. Squared Petri dishes offer more space to analyze several strains and dilutions on one plate. In our experiment, we used YNB -Ura + maltose and YNB -Ura + glucose.
Note: For yeast sugar uptake analysis, we usually include a negative control strain with an empty plasmid or GFP on a plasmid. As positive control, we use the yeast monosaccharide transporter Hxt1. We usually analyze at least two independent clones of the transporter of interest.
Plate two clones per strain (fresh transformed or from stock) onto fresh YNB selective medium with maltose two days prior to the drop-out assay.
The following steps are prepared at RT with sterile conditions, for example by working in a biosafety cabinet or next to a flame. Performing the assay under sterile conditions is recommended to avoid contamination of the yeast strains or plates.
Prepare a 1.5 mL reaction tube with 1 mL of ddH2O for each clone.
Scrape the colonies from the plates and resuspend in the tubes.
Prepare a 1:10 dilution with ddH2O and measure the OD600.
Calculate the volume needed for 1.5 mL of cell suspension with an OD600 of 0.4. Do not forget to include the dilution factor.
Prepare 1.5 mL of cell suspension with an OD600 of 0.4 in a new 2 mL reaction tube. Vortex.
Take 1 mL to check the OD600. Adjust again if needed.
Prepare a serial dilution (500 μL volume) for each transformant in a new sterile 1.5 mL tube.
Always vortex the solutions before preparing the next dilution.
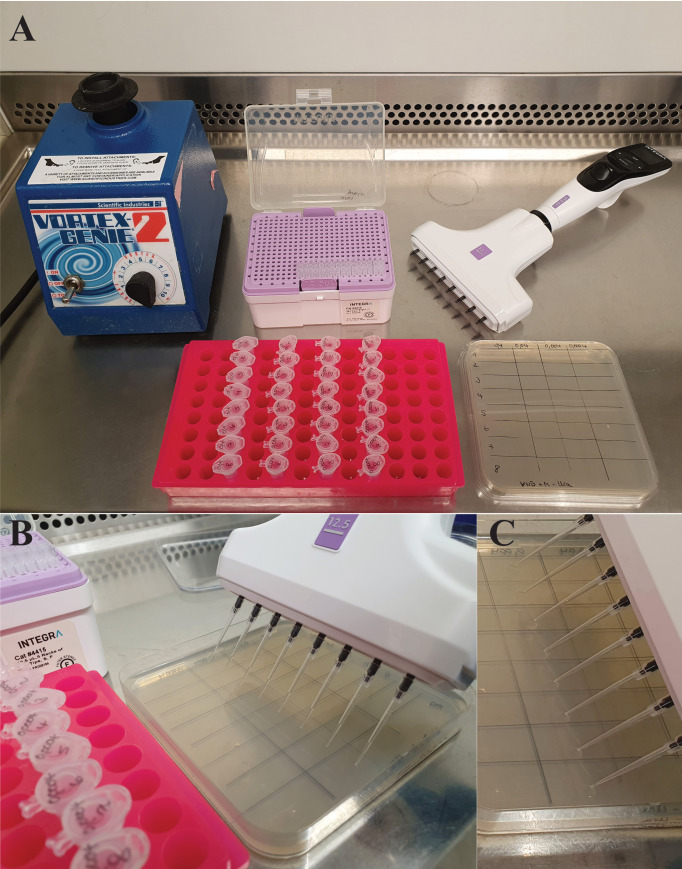
Label the squared plates for the drop-out assay. We usually use grid lines to generate nicely distributed drops (Table 1 and Figure 2A).
Figure 2A shows an example of a possible labeling of the plates; the individual yeast transformants are ordered vertically and the corresponding serial dilution horizontally. At least two types of plates are required for the assay: one with YNB -Ura + maltose and one with YNB -Ura + glucose.
Organize your tubes in a rack according to the droplet scheme on the plates (e.g., yeast transformants vertically, serial dilutions horizontally).
Always vortex each tube immediately before pipetting.
-
We prefer using a multichannel pipette for the drops as it is faster. This might require a little bit of training to place the drops correctly. You can also use a normal single-channel pipette.
Note: The serial dilution and the spotting of the droplets can also be performed using a sterile 96-well plate and a microplate replicator (e.g., Millipore Sigma, Replica plater for 96-well plate, R2508 or similar).
Drop 3 μL of each sample on the two plates. Always ensure that the pipette tips in the multichannel pipette are loaded equally. Avoid pricking the tips into the agar (Figure 2B). After pipetting, the drops should be visible on the plate (Figure 2C).
Dry the plates (opened lid) under sterile conditions until no liquid drops are visible.
Incubate the plates for 48 h at 28 °C. If the colonies are not clearly visible, the incubation time can be prolonged.
Document the growth of the colonies by using a normal mobile phone camera or any other device. We typically take pictures with the Bio-Rad Chemidoc using the colorimetric mode (Figure 3).
-
Figure 1. Golden Gate cloning strategy with the provided modular cloning (MoClo) modules.
A. We provide the level 0 MoClo modules for the PMA1 promoter (Pro+5U module) and the ADH2 terminator (3U+Ter module). The candidate transporter gene has to be amplified with CDS1 stop or without stop (CDS1 no stop) according to the MoClo guidelines. The latter can be used to fuse a C-terminal tag (CT). The required BsaI overhangs for cloning are indicated for each module. B. In this example, the yeast-compatible level 1 acceptor vector pAGT572 with uracil selection was used. Cloning of the level 0 modules into the level 1 acceptor vector is done in a cut-ligation with BsaI. C. The final level 1 transcription unit contains all parts required for yeast transformation and expression of the candidate transporter.
Table 1. Example layout for drop-out grid lines.
| OD600 | 0.4 | 0.04 | 0.004 | 0.0004 |
|---|---|---|---|---|
| Strain 1 | ||||
| Strain 2 | ||||
| Strain 3 | ||||
| Strain 4 | ||||
| Strain 5 | ||||
| Strain 6 |
Figure 2. Preparation and procedure of the yeast drop-out assay.
A. Material needed. Prepare a vortex, pipette, and pipette tips. Prepare the plates needed with a pipetting scheme. In this case, a grid line was used, with the yeast strains ordered vertically and serial dilutions horizontally. Prepare a rack with the tubes ordered in the same scheme as the plate. B. A multichannel pipette is used to place the drops on the plate. C. Liquid drops visible on the plate after pipetting.
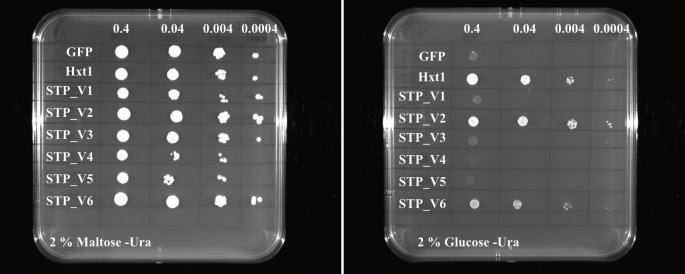
Figure 3. Example pictures of a drop-out assay result for different sugar transport protein (STP) variants (STP_V1 to STP_V6) expressed in yeast.
The STP variants (STP_V1 to STP_V6) were transformed to the hexose-transport-deficient yeast strain EBY.VW4000. A serial dilution of the respective strains was dropped on selective medium either containing 2% maltose or 2% glucose as carbon source. Selective medium lacking uracil was used (-Ura). Plates were grown for two days at 28 °C and growth was documented using the Chemidoc from Bio-Rad. GFP was used as negative control and the yeast hexose transporter Hxt1 served as positive control.
Data analysis
The colonies growing on YNB plates with maltose should all grow in a comparable range. The colonies on YNB plates containing glucose generally grow a little bit slower compared to maltose. If the expressed plant sugar transporter complements the growth defect of EBY.VW4000 on hexose-containing medium, these colonies should grow. The negative control (empty vector or other protein cloned, e.g., GFP) should not grow on glucose-containing medium. As positive control, we usually use Hxt1, a sugar transporter from yeast.
General notes and troubleshooting
Yeast is a highly suitable heterologous system to express and analyze the functions of certain proteins on an individual scale. Besides the described plant monosaccharide transporters (sugar transport protein family), other transporters, for other substrates or from other organisms, can be analyzed using the described drop-out assay and growth complementation. Therefore, a suitable yeast strain has to be chosen with a deficiency in transporting a specific substrate. This deficiency can be complemented by the expression of the respective transporter, thus conferring growth on selective medium or transport of fluorescent substrates. For example, members of the barley and potato SWEET sugar transporter family were also analyzed using yeast strain EBY.VW4000 and the substrates glucose, fructose, galactose, or mannose (Tamayo et al., 2022; Yue et al., 2023). The type I sucrose transporters StSUT1 from potato and AtSUC2 from Arabidopsis were analyzed using the yeast strain SEY6210 and BY4742 and the fluorescent sucrose analog esculin (Gora et al., 2012). The Arabidopsis γ-aminobutyric acid (GABA) transporter AtGAT1 was analyzed using the yeast strain 22574d and selective medium with different nitrogen sources (citrulline, proline, or GABA) (Meyer et al., 2006).
Different yeast synthetic drop-out medium supplements are available. Always choose the required supplements fitting to your plasmid and the genotype of the used yeast strain. In this protocol, we used the yeast strain EBY.VW4000, that is auxotrophic for uracil, leucine, histidine, and tryptophan. The plasmid pAGT572 encodes the URA3 selection marker to identify transformed EBY.VW4000 clones on selective medium lacking uracil.
For our protocol, we provide plasmids that allow the Golden Gate–based cloning of yeast expression constructs. To clone the transporter candidates, a commonly used vector is pDR196 (Meyer et al., 2006; Chen et al., 2010; Gora et al., 2012; Roy et al., 2015; Tamayo et al., 2022; Yue et al., 2023). Here, the candidate transporter sequence is inserted between the PMA1 promoter (plasma membrane H+ ATPase 1) and the ADH2 (alcohol dehydrogenase 2) terminator by using a multiple cloning site and type IIP restriction enzyme (e.g., EcoRI). In our laboratory, we use the simple and efficient Golden Gate cloning with the standardized modular cloning (MoClo) syntax to flexibly generate single or multigene constructs. The Golden Gate cloning method is based on the use of type IIS restriction enzymes that cut outside of their recognition sequence and generate user-defined overhangs. These overhangs allow the pre-defined, overhang-dependent, and scarless fusion of modules by using a one-pot restriction and ligation reaction (Engler et al., 2009; Weber et al., 2011; Werner et al., 2012). In addition, the standardized MoClo syntax allows an easy and flexible exchange of modules according to the requirement of the user. To our knowledge, no modules were available for the standard MoClo syntax to flexibly clone yeast expression constructs comparable to the pDR196 setup. Therefore, we provide publicly available level 0 MoClo modules for the yeast PMA1 promoter and the ADH2 terminator, to clone a transcription unit for the candidate transporter by using Golden Gate. In addition, we also provide a ready-to-use transcription unit to express the positive control Hxt1 (Skoppek et al., 2022).
Acknowledgments
This work was funded by university core funding only. We thank Jens Boch for general support. The protocol was used in Skoppek et al. (2022).
Competing interests
The authors declare to have no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1.Bezrutczyk M., Yang J., Eom J. S., Prior M., David. Sosso, Hartwig T., Boris. Szurek, Oliva R., Vera-Cruz C., White F. F., et al.(2018). Sugar flux and signaling in plant-microbe interactions. Plant J 93(4): 675-685. [DOI] [PubMed] [Google Scholar]
- 2.Boles E.(2003). Yeast as a Model System for Studying Glucose Transport. In: Sibley, D. R. and Quick, M. W.(Eds.). Transmembrane Transporters. Wiley‐Liss, Inc. [Google Scholar]
- 3.Breia R., Conde A., Badim H., Fortes A. M., Gerós H. and Granell A.(2021). Plant SWEETs: from sugar transport to plant–pathogen interaction and more unexpected physiological roles. Plant Physiol 186(2): 836-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L. Q., Hou B. H., Lalonde S., Takanaga H., Hartung M. L., Qu X. Q., Guo W. J., Kim J. G., Underwood W., Chaudhuri B., et al.(2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468(7323): 527-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engler C., Gruetzner R., Kandzia R. and Marillonnet S.(2009). Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4(5): e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engler C., Youles M., Gruetzner R., Ehnert T. M., Werner S., Jones J. D., Patron N. J. and Marillonnet S.(2014). A golden gate modular cloning toolbox for plants. ACS Synth Biol 3(11): 839-843. [DOI] [PubMed] [Google Scholar]
- 7.Gantner J., Ordon J., Ilse T., Kretschmer C., Gruetzner R., Lofke C., Dagdas Y., Burstenbinder K., Marillonnet S. and Stuttmann J.(2018). Peripheral infrastructure vectors and an extended set of plant parts for the Modular Cloning system. PLoS One 13(5): e0197185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gietz R. D. and Schiestl R. H.(2007). Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2(1): 1-4. [DOI] [PubMed] [Google Scholar]
- 9.Gietz R. D. and Schiestl R. H.(2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 31-34. [DOI] [PubMed] [Google Scholar]
- 10.Gora P. J., Reinders A. and Ward J. M.(2012). A novel fluorescent assay for sucrose transporters. Plant Methods 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julius B. T., Leach K. A., Tran T. M., Mertz R. A. and Braun D. M.(2017). Sugar Transporters in Plants: New Insights and Discoveries. Plant Cell Physiol 58(9): 1442-1460. [DOI] [PubMed] [Google Scholar]
- 12.Meyer A., Eskandari S., Grallath S. and Rentsch D.(2006). AtGAT1, a high affinity transporter for γ-aminobutyric acid in Arabidopsis thaliana. J Biol Chem 281(11): 7197-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milne R. J., Dibley K. E. and Lagudah E. S.(2017). Yeast as a heterologous system to functionally characterize a multiple rust resistance gene that encodes a hexose transporter. In: Periyannan, S.(Ed.). Wheat Rust Diseases. Methods in Molecular Biology, vol 1659. Humana Press, New York, NY. [DOI] [PubMed]
- 14.Milne R. J., Dibley K. E. and Lagudah E. S.(2017). Yeast as a heterologous system to functionally characterize a multiple rust resistance gene that encodes a hexose transporter. In: Periyannan, S.(Ed.). Wheat Rust Diseases. Methods in Molecular Biology(pp. 265-274). Humana Press. [DOI] [PubMed] [Google Scholar]
- 15.Moore J. W., Herrera-Foessel S., Lan C., Schnippenkoetter W., Ayliffe M., Huerta-Espino J., Lillemo M., Viccars L., Milne R., Periyannan S., et al.(2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat Genet 47(12): 1494-1498. [DOI] [PubMed] [Google Scholar]
- 16.Purugganan M. D. and Jackson S. A.(2021). Advancing crop genomics from lab to field. Nat Genet 53: 595-601. [DOI] [PubMed] [Google Scholar]
- 17.Rai A., Yamazaki M. and Saito K.(2019). A new era in plant functional genomics. Curr Opin Syst Biol 15: 58-67. [Google Scholar]
- 18.Roy A., Dement A. D., Cho K. H. and Kim J. H.(2015). Assessing glucose uptake through the yeast hexose transporter 1(Hxt1). PLoS One 10(3): e0121985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheler U., Brandt W., Porzel A., Rothe K., Manzano D., Božić D., Papaefthimiou D., Balcke G. U., Henning A., Lohse S., et al.(2016). Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast. Nat Commun 7: 12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skoppek C. I., Punt W., Heinrichs M., Ordon F., Wehner G., Boch J. and Streubel J.(2022). The barley HvSTP13GR mutant triggers resistance against biotrophic fungi. Mol Plant Pathol 23(2): 278-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamayo E., Figueira-Galan D., Manck-Gotzenberger J. and Requena N.(2022). Overexpression of the Potato Monosaccharide Transporter StSWEET7a Promotes Root Colonization by Symbiotic and Pathogenic Fungi by Increasing Root Sink Strength. Front Plant Sci 13: 837231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J. Y. and Doudna J. A.(2023). CRISPR technology: A decade of genome editing is only the beginning. Science. 379(6629): eadd8643. [DOI] [PubMed] [Google Scholar]
- 23.Weber E., Engler C., Gruetzner R., Werner S. and Marillonnet S.(2011). A modular cloning system for standardized assembly of multigene constructs. PLoS One 6(2): e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werner S., Engler C., Weber E., Gruetzner R. and Marillonnet S.(2012). Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng Bugs 3(1): 38-43. [DOI] [PubMed] [Google Scholar]
- 25.Wieczorke R., Krampe S., Weierstall T., Freidel K., Hollenberg C. P. and Boles E.(1999). Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett 464(3): 123-128. [DOI] [PubMed] [Google Scholar]
- 26.Yue W., Cai K., Xia X., Liu L. and Wang J.(2023). Genome-wide identification, expression pattern and genetic variation analysis of SWEET gene family in barley reveal the artificial selection of HvSWEET1a during domestication and improvement. Front Plant Sci 14: 1137434. [DOI] [PMC free article] [PubMed] [Google Scholar]