Abstract
Based on previous structural and functional studies, a potential receptor-binding site composed of residues that form a pocket at one end of the two long antiparallel helices in the receptor-binding domain of Friend 57 murine leukemia virus envelope protein (RBD) has been proposed. To test this hypothesis, directed substitutions for residues in the pocket were introduced and consequences for infection and for receptor binding were measured. Receptor binding was measured initially by a sensitive assay based on coexpression of receptor and RBD in Xenopus oocytes, and the findings were confirmed by using purified proteins. Three residues that are critical for both binding and infection (S84, D86, and W102), with side chains that extend into the pocket, were identified. Moreover, when mCAT-1 was overexpressed, the infectivity of Fr57-MLV carrying pocket substitutions was partially restored. Substitutions for 18 adjacent residues and 11 other previously unexamined surface-exposed residues outside of the RBD pocket had no detectable effect on function. Taken together, these findings support a model in which the RBD pocket interacts directly with mCAT-1 (likely residues, Y235 and E237) and multiple receptor-envelope complexes are required to form the fusion pore.
For infection by enveloped viruses to occur, a pore that allows the virion core to enter the cytoplasm must form between the virus and the host cell membranes (10). Pore formation is a consequence of localized mixing of the viral and cell membrane lipids at the site of virus attachment. Previous studies of influenza virus infection suggest that formation of the fusion pore is initiated by insertion of the amino terminus of the envelope protein, HA2, into the target cell membrane (6, 10). On the virion surface, trimeric HA2 is surrounded by three overlying HA1 subunits which make extensive contact with each other and with HA2. Upon endocytosis, there is pH-dependent weakening of the HA1-HA1 and HA1-HA2 contacts along the threefold axis (6) and large conformational changes which expose the amino terminus of HA2 in a way that is conducive to the mixing of lipids between adjacent membranes (10, 12).
Recent studies have revealed striking structural and functional similarities between the envelope proteins of influenza virus and two retroviruses: type C murine leukemia virus (type C MLV) (14) and human immunodeficiency virus (HIV) (7). Unlike influenza virus HA2, exposure of the amino-terminal domain of retroviral membrane (TM) proteins is coupled to receptor binding and does not require endosomal acidification. At present, a direct interaction between cell receptors and TM proteins has not been identified; rather, TM-mediated fusion is activated by binding of a receptor(s) to the envelope surface protein (SU; analogous to HA1), which destabilizes the envelope trimer, exposing TM. The distinct tropism of each class of MLVs is determined by a specific receptor-SU interaction (4, 5).
Although the receptors for type C MLVs (2, 31, 33) and HIV (15) have been identified, as yet these studies have provided little insight into how binding to SU results in fusion. Toward this end, the receptor-binding domain in the SU protein of Friend 57 MLV (Fr57-MLV) has been identified (residues 1 to 236 of SU; Fr57-RBD), the stoichiometry and binding affinity have been determined by using purified proteins (11), and the structure has been resolved to 2.0-Å resolution by X-ray crystallography (13). Residues conserved in the SU proteins of ecotropic MLV (5), which utilize mCAT-1 as a receptor, were found among intertwined loops and helices (variable region A [VRA] domain) which abut a “stalk”-like β-barrel (see Fig. 1B). Based on the results of previous studies of mutant SU proteins (16, 20, 29), an mCAT-1 binding site composed of a pocket formed by residues in the VRA of Fr57-RBD was proposed (13). To test this hypothesis, substitutions for residues that form the pocket and other residues exposed on the surface of Fr57-RBD that have not been previously studied have been introduced and receptor binding and infection have been measured. These experiments demonstrate that the three residues with side chains which project into the pocket strongly influence mCAT-1 binding and infection, suggesting that the pocket is the critical site of receptor contact.
FIG. 1.
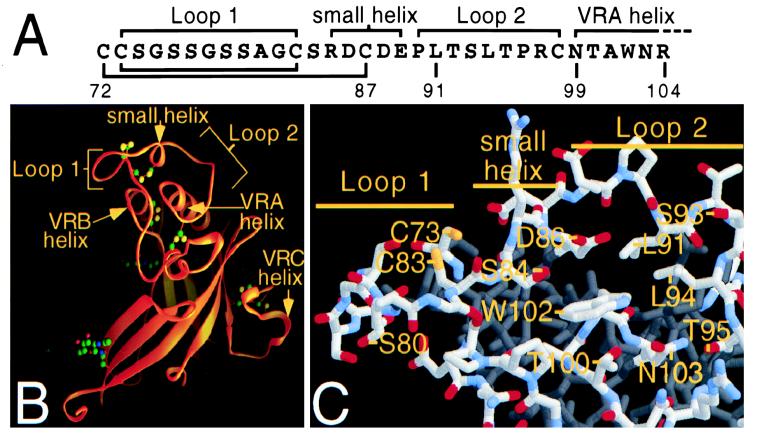
Models of the Fr57-RBD based on coordinates supplied from Fass et al. (13). (A) Primary sequence of a portion of Fr57-RBD showing the small and VRA helices and loops 1 and 2. (B) Ribbon representation of the Fr57-RBD structure. Indicated with arrows are the VRA, VRB, and VRC helices and loops 1 and 2 from VRA which are bridged by the small helix. Together with residues at the proximal end of the juxtaposed VRA and VRB helices, the loops and small helix form a pocket (13). (C) A stick model of residues within and lining the pocket. Oxygen, nitrogen, sulfur, and carbon atoms are colored red, blue, yellow, and gray, respectively.
MATERIALS AND METHODS
Cells and viruses.
Mouse-derived fibroblast cell lines, NIH 3T3, NFM, and mCAT-1−/− (27), were grown in Dulbecco minimal essential medium (DMEM) and fetal calf serum (FCS; 10%). NFM was created by clonal selection after transfection with an expression plasmid (pCDNA3; Invitrogen) encoding mCAT-1 bearing an influenza virus HA epitope tag at the carboxyl terminus (11). The packaging cell line Anjou, which expresses MLV Gag and Pol proteins, was obtained from Warren Pear (26) and also grown in DMEM and fetal calf serum (10%).
Computer analysis.
The Fr57-RBD structure was displayed by using Rasmol 2.6-UCB version 1.0 (28) on a Macintosh PowerPC computer. Fr57-RBDs bearing amino acid substitutions were modeled by using the program Swiss-PdbViewer version 2.11 (17).
Mutagenesis.
To facilitate mutagenesis, the cDNA encoding the Fr57-MLV envelope protein gp85 (19) (GenBank accession no. J02192) was initially altered by oligonucleotide-directed mutagenesis, using PCR to add a XhoI site at nucleotides 573 to 577 and a KpnI site at nucleotides 806 to 811 (numbered from the first nucleotide of the initiation codon). These changes did not affect the amino acid sequence. The resulting construct was inserted into pCDNA3 (Invitrogen) by using a 5′ HindIII site (present in the native DNA at bp −540) and a 3′ EcoRI site (added 10 nucleotides after the termination codon by PCR), and the sequence was confirmed. The XhoI and KpnI restriction endonuclease sites were used to insert fragments of the SU gene further altered by oligonucleotide-directed mutagenesis using PCR. The sequences of the subcloned regions were verified.
Production and characterization of viruses bearing recombinant envelope proteins.
To obtain recombinant Fr57-MLV bearing altered envelope proteins, a plasmid encoding Fr57-MLV envelope protein was introduced into the Anjou packaging line (26) by transfection after calcium phosphate precipitation. As a marker of infection, DNA from pBABE-β-gal, a plasmid containing a provirus encoding β-galactosidase (23), was included in the transfection. Typically, 10 μg of each plasmid was used per 10-cm-diameter tissue culture plate containing 2 × 105 to 3 × 105 Anjou cells. Twenty-four hours after transfection, the cells were refed, and virus-containing supernatant was collected 24 h later. After filtering the supernatant through a 0.45-μm-pore-size cellulose acetate filter (Corning), the titer of infectious virus was determined on murine NIH 3T3 fibroblasts. These cells were incubated overnight in virus-containing supernatant and Polybrene (8 μg/ml), then refed on the following day, and stained for acquired β-galactosidase activity by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), as described previously (1). The transfected Anjou cells were also stained with X-Gal to determine transfection efficiency.
Incorporation of recombinant SU proteins into virions was analyzed by immunoblotting. Virions were purified from filtered supernatant by density gradient centrifugation (20% sucrose in 100 mM NaCl–10 mM Tris-HCl [pH 7.4]; 14,000 × g for 1 h at 4°C). Pelleted material was resuspended in gel loading buffer (1% sodium dodecyl sulfate [SDS], 20% glycerol, 25 mM Tris-HCl, 0.5% [vol/vol] β-mercaptoethanol [pH 6.8]) and boiled for 5 min, and proteins were separated on an SDS–8% polyacrylamide gel electrophoresis (PAGE) gel and immunoblotted by using goat anti-Rauscher MLV gp69/71 antibody (Quality Biotech Inc., catalog no. 80S000019). The secondary antibody was a mouse anti-goat antibody–horseradish peroxidase conjugate (Pierce), and blots were developed with chemiluminescent substrate (NEN, DuPont).
Receptor binding studies in oocytes.
Relative binding of mutant Fr57-RBD proteins to receptor was determined by using an assay based on expression in Xenopus oocytes that was described previously (18). Briefly, Xenopus oocytes were injected with 10 ng of cRNA encoding mCAT-1 (receptor; 10 ng) alone or with 25 ng of cRNA encoding wild-type or mutant Fr57-RBD. Two days later, binding of purified 125I-MLV SU protein (104 cpm/ng) to injected oocytes was determined. Previous experiments demonstrated that mCAT-1-dependent binding of 125I-MLV SU is inversely correlated with the expression of Fr57-RBD (18).
Purification of recombinant Fr57-RBD proteins.
cDNAs encoding Fr57-RBDs bearing S84I and S84A or D86A mutations were subcloned into the pFASTBAC vector (Gibco-BRL) modified to add six histidine residues in frame at the carboxyl terminus as previously described (11). Baculoviruses expressing each of these proteins were obtained by following the manufacturer’s instructions. Recombinant Fr57-RBD proteins were obtained from the medium of Hi five cells (BTI-TN-5B1-4; Invitrogen) 2 days postinfection and purified by nickel chelation chromatography as previously described (11). No differences were observed in the recovery or stability of the wild-type and mutant Fr57-RBD proteins.
Fr57-RBD binding to cells.
NIH 3T3, NFM, or mCAT-1−/− fibroblasts were harvested from culture plates by scraping and resuspended in 120 mM NaCl–5 mM KCl–12 mM glucose–20 mM HEPES (pH 7.4) (assay buffer) to give 107 cells/ml. Aliquots of cells (106 in 0.1 ml) were mixed with an equal volume of 2% bovine serum albumin in assay buffer with 2 mM CaCl2, 1 mM MgCl2, and 50 ng of 125I-labeled Fr57-RBD. After incubation for 1 h at 20°C, the cells were washed five times with 1.5 ml of ice-cold assay buffer and solubilized, and bound 125I-SU was determined with a gamma counter.
RESULTS
Contribution of residues in the proposed receptor-binding pocket to binding and infection.
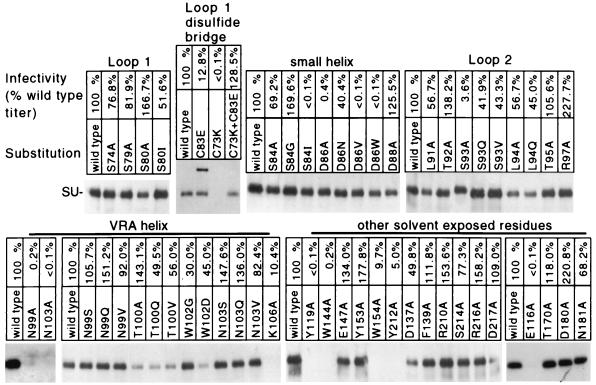
MacKrell et al. previously reported that substitutions for residue D84 in the SU protein reduced the infectivity of Moloney MLV without affecting the incorporation of SU into the virus membrane (20). The residue analogous to D84 in Fr57-MLV SU is D86, which is located on one side of a small pocket (Fig. 1B and C) created by the confluence of loops and helices at one end of the Fr57-RBD (13). To determine if this pocket forms a critical portion of the receptor binding site, D86 and adjacent residues within the Fr57-RBD pocket (Fig. 1A) were changed and receptor binding and virus infectivity were measured. PCR was used to introduce mutations into an expression plasmid encoding Fr57-MLV gp85 (SU and TM proteins), and recombinant Fr57-MLVs were obtained by transient transfection of the altered plasmids into a human 293-derived cell line, Anjou, that constitutively expresses MLV Gag-Pol proteins (26). An equal amount of pBABE-β-gal, a plasmid containing proviral DNA encoding β-galactosidase (23), was included in each transfection to provide a marker of infection. By using this protocol, more than 1,000 β-galactosidase-positive cells were typically observed when the supernatant from 106 Anjou cells transfected with the plasmid encoding wild-type Fr57-MLV envelope protein was applied to a 10-cm-diameter culture plate containing 5 × 105 permissive mouse NIH 3T3 fibroblasts. Two days after transfection of the Anjou cells, recombinant virions were purified from the culture medium by density gradient centrifugation and lysates were examined by immunoblotting to determine incorporation of altered SU protein. When 293 cells were transfected with Fr57-MLV envelope protein-encoding plasmid, the envelope protein released from cells into the supernatant did not penetrate the sucrose cushion, indicating a likely requirement for association with gag-pol products. Using this protocol with Anjou cells, we observed that replacement of the aspartic acid 86 (D86) with alanine reduced infectivity of Fr57-MLV to 0.4% of that of the wild type and replacement by valine or tryptophan completely abrogated detectable infectivity (<0.1%), without reducing the incorporation of SU into virions (Fig. 2). The negatively charged carboxylate group on the side chain of D86 is not critical, since replacement by an amide group through the substitution, D86N, had only a small effect on infection (40.4% of control).
FIG. 2.
Effect of substitutions for residues in the Fr57-RBD on SU incorporation and Fr57-MLV infection (titer is percent of wild-type virus infectivity, combined from data from at least two separate experiments). Immunoblots of pelleted material from culture supernatants of Anjou cells transfected with DNA encoding mutant SUs are shown. Blots were probed with goat anti-Rauscher MLV SU antibody which recognizes Fr57-MLV SU.
D86 is located on the small helix where its side chain projects into the center of the pocket in close proximity to the side chains of S84 and W102 (Fig. 1C). Indeed, substitution of isoleucine for S84 reduced infectivity by more than 1,000-fold. However, the substitutions S84A or S84G as well as W102G had no detectable effect on Fr57-MLV infection, suggesting that the role of the S84 and D86 side chains is not simply to tether the small loop to the proximal end of the VRA helix (Fig. 1C) by interacting with the side chain of W102.
Single substitutions for the remainder of the residues at the amino-terminal end of the VRA helix that face the pocket opposite D86, including N99Q/S/V, T100A/V/Q, and N103Q/S/V, also had minimal effects (<10-fold) on envelope processing or infectivity (Fig. 2). Also, single substitutions in the remainder of the small helix and in loop 2, including D88A, L91A, T92A, S93V/Q, L94A/Q, and T95A, did not noticeably alter function. One exception, S93A, reduced infectivity to 3% of that of the control, likely the result of altered posttranslational processing, since SU bearing this change demonstrated slightly slower mobility on an SDS-PAGE gel (Fig. 2). R97A was consistently associated with enhanced infection (more than twofold), despite reduced incorporation of SU into virions. Single substitutions for 11 other surface-exposed residues on Fr57-RBD that had not been examined by others (3, 16, 20, 29) and that did not impair incorporation into virions were also studied (Fig. 2).
The functional importance of residues adjacent to the pocket in the large loop (loop 1) formed by the disulfide bond between C73 and C83 (Fig. 1) was also examined. Not surprisingly, if either one of these cysteine residues was replaced, SU processing was altered (Fig. 2). SU carrying the C73K mutation was not incorporated into virions, and no infection was detected. SU carrying the C83E mutation likely formed intersubunit dimers, since on SDS-PAGE, an additional protein with a mobility corresponding to a 140-kDa protein was observed in unreduced lysates (Fig. 2); this protein disappeared in the presence of β-mercaptoethanol (data not shown). Despite the loss of the disulfide bond that forms the loop, virions bearing SU containing C83E remained infectious (13% of wild-type infectivity). Virions with SU proteins containing both C73K and C83E mutations, predicted by computer-based modeling (Swiss-PdbViewer) (17) to form a salt bridge, infected NIH 3T3 fibroblasts normally. No deleterious effects of substitutions for other residues in the loop (S74A, S79A, S80A/I) were observed (Fig. 2). Therefore, a strict requirement for specific residues and/or for intact loop structure was not observed.
Envelope binding studies.
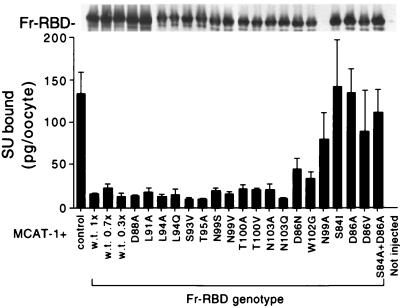
Previously, we observed that specific binding of 125I-labeled Moloney MLV SU (125I-MLV-SU) to the plasma membranes of oocytes injected with cRNA encoding its receptor, mCAT-1, was specifically blocked by coexpression of ecotropic MLV SU proteins, including Fr57-RBD (11, 18). The capacity of altered Fr57-RBD to block 125I-MLV-SU binding was measured by using this assay and correlated with effects on infectivity. When wild-type Fr57-RBD, but not control (kinesin light chain), protein was coexpressed, 125I-MLV-SU binding was reduced from 130 ± 20 to 20 ± 10 pg/oocyte (mean ± standard deviation) (Fig. 3, lower panel). Expression of the Fr57-RBD proteins carrying the substitutions D88A, L91A, L94A/Q, S93V, T95A, N99S/V, T100A/V, and N103A/Q also reduced 125I-MLV-SU binding to <20 ± 10 pg/oocyte. In contrast, 125I-MLV-SU binding to oocytes that expressed mCAT-1 was not significantly reduced by coexpression of Fr57-RBD proteins bearing D86A, D86V, or S84I substitutions, despite comparable steady-state levels of expression as determined by immunoblotting (Fig. 3, upper panel). Fr57-RBD bearing the substitution N99A was not expressed and failed to block 125I-MLV-SU binding. Expression of Fr57-RBD carrying W102G partially blocked 125I-MLV-SU binding (40 pg/oocyte). The behavior of Fr57-RBDs bearing the double substitutions S84A plus D86A or D86A plus W102G (data not shown) was not significantly different from the behavior of Fr57-RBDs carrying D86A alone. These observations are unlikely to be explained by small differences in expression, since injection of only 5 ng (30% of control) of mRNA encoding the wild-type Fr57-RBD was sufficient to completely block 125I-MLV-SU binding (Fig. 3, lower panel). Therefore, the failure of the SU proteins carrying changes in S84, D86, and/or W102 to block 125I-MLV-SU binding is likely the result of reduced binding affinity for mCAT-1.
FIG. 3.
Indirect measure of relative affinity of Fr57-RBD–receptor interaction. (Lower panel) Fr57-RBD proteins bearing substitutions for residues within and lining the pocket were analyzed for their ability to inhibit the binding of 125I-labeled SU to receptor expressed on Xenopus oocytes. (Upper panel) Extracts were made from the oocytes and analyzed by immunoblotting. Since the mutant Fr57-RBDs were tagged with a hemagglutinin epitope, the blot was probed with the monoclonal antibody 12CA5.
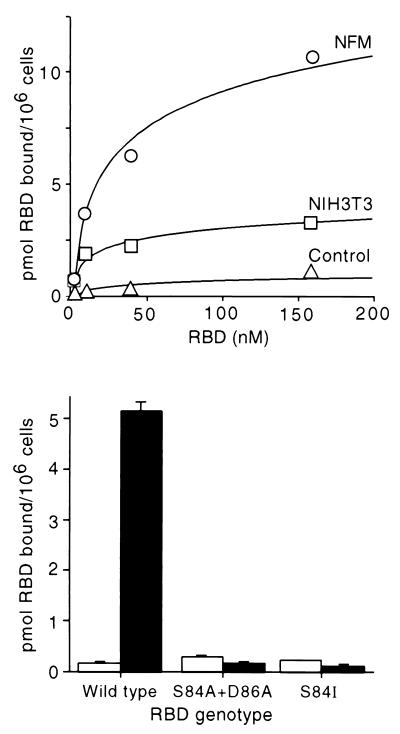
To verify this conclusion, Fr57-RBD proteins bearing S84I and S84A plus D86A were purified and binding to mouse fibroblasts was measured directly. To enhance the sensitivity of these studies, a clonal cell line (NFM) that binds 4.7-fold more 125I-Fr57-RBD than that bound by parental NIH 3T3 fibroblasts was utilized (Fig. 4, upper panel). NFM was created by forced expression of mCAT-1 from an expression plasmid with a cytomegalovirus early region promoter. NFM cells bound 10 pmol of 125I-Fr57-RBD/106 cells, 4.7-fold more than the parent NIH 3T3 fibroblasts and 22-fold more than a mouse fibroblast cell line that lacks receptor (Fig. 4, upper panel) because of targeted deletion of the mCAT-1 gene (27). However, binding of 125I-Fr57-RBDs bearing S84I and S84A plus D86A to NFM was not significantly different than the binding to the fibroblast cell line that lacks mCAT-1 (Fig. 4, lower panel), indicating that S84 and D86 have critical roles in receptor binding.
FIG. 4.
Comparison of surface expression of the ecotropic MLV receptor (mCAT-1) on NFM cells, NIH 3T3 fibroblasts, and a fibroblast cell line derived from the mCAT-1 knockout mouse. (Upper panel) Saturation binding curve for 125I-labeled Fr57-RBD. Symbols: ○, NFM cells; □, NIH 3T3 fibroblasts; ▵, nonpermissive human 293 cells. (Lower panel) Comparison of binding of 125I-labeled Fr57-RBDs, carrying the substitutions indicated, to fibroblasts from the mCAT-1 knockout mouse (open bars) and NFM cells (solid bars). Each Fr57-RBD was used at 50 nM.
Overexpression of receptor abrogates the deleterious effects of mutations in the Fr57-RBD pocket.
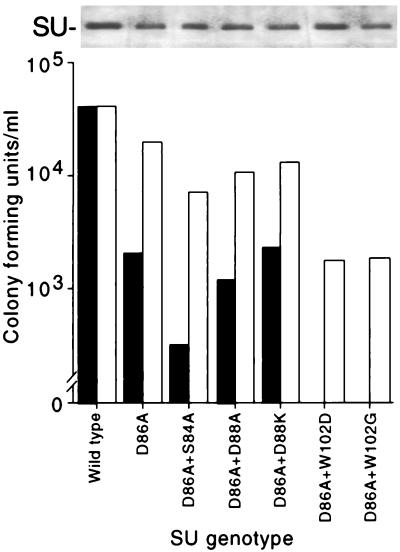
Substitution of glycine for W102 partially inhibited Fr57-RBD binding to mCAT-1 expressed on oocyte membranes (Fig. 3), consistent with the close contact between its side chain and the side chains of S84 and D86 in the putative binding pocket. Unlike substitutions for S84 and D86, however, replacement of W102 had no apparent effect on Fr57-MLV infection (Fig. 2). The effects of these substitutions on infection were reexamined in the presence of D86A with NIH 3T3 and NFM fibroblasts as indicators. These cell lines were equally susceptible to wild-type Fr57-MLV infection (determined by end point dilution; 4.0 × 104 and 4.2 × 104 CFU/ml, respectively) (Fig. 5). When D86A was introduced into SU, the titer of Fr57-MLV decreased to 2 × 103 CFU/ml on NIH 3T3 fibroblasts but was unchanged (1.9 × 104 CFU/ml) on NFM cells. The addition of S84A, but not D88A or D88K (located outside the pocket), further reduced the apparent titer of Fr57-MLV bearing D86A to 3 × 102 CFU/ml on NIH 3T3 fibroblasts and to 7 × 103 on NFM cells. Introduction of W102G/D also impaired infectivity of Fr57-MLV bearing D86A. Indeed, no infectious virions bearing D86A plus W102D or D86A plus W102G were detected with NIH 3T3 cells; however, the titer of both recombinant viruses was 2 × 103 CFU/ml on NFM cells. Therefore, like the substitution S84A, impairment of Fr57-MLV infection by the substitution W102G/D was detected only in the presence of D86A, consistent with a direct interaction of the side chains of these residues with each other and with mCAT-1. In addition, the deleterious effect of these substitutions on infectivity was significantly less when assayed on NFM cells that express higher levels of receptor than that expressed by NIH 3T3 cells. This finding suggests that Fr57-MLV infection requires formation of multiple functional receptor-envelope complexes.
FIG. 5.
(Lower panel) Titers of recombinant Fr57-MLVs on NIH 3T3 (solid bars) and NFM (open bars) fibroblasts; (Upper panel) immunoblot of SU recovered from virus pellets.
DISCUSSION
Previously, we proposed that a pocket created by the apposition of two helices and two loops at the top of Fr57-RBD formed the critical portion of the receptor binding site (13). In this report, we provide evidence for this hypothesis by demonstrating that three residues which form the pocket strongly influence Fr57-MLV infectivity. Moreover, when the remainder of the binding pocket and adjacent loops or helices were examined, additional residues that are crucial to binding and/or to infection were not identified. Since, in this study, the identification of residues involved in receptor interaction was limited to single amino acid substitutions that did not affect SU processing or incorporation into virions, residues with smaller contributions to receptor binding may have been overlooked.
The observation that a small number of adjacent residues are critical for binding of two proteins has also been observed in the analysis of growth hormone binding to its receptor (8). In crystallographic studies, a large binding interface (1,300 Å2) composed of at least 30 residues in growth hormone was identified (8). However, systematic studies of each residue revealed that only two tryptophans (W104 and W169) contributed >75% of the free energy of binding (8). Similarly, the residues in the Fr57-RBD pocket may be the critical portion of a larger interface formed during binding of SU to receptor. If true, additional contacts between SU and mCAT-1 will only be elucidated after multiple amino acid substitutions and/or structural studies of Fr57-RBD bound to receptor. Indeed, Bae et al. recently observed that substitutions for either R85 on the small helix or R97 on loop 2 (Fig. 1) were each innocuous but, when combined, reduced infectivity by more than 100-fold (3).
Substitution of valine and alanine, but not asparagine, for D86 in Fr57-MLV SU (in this study), and not serine or glutamic acid (20) for the analogous residue, D84, in the SU protein of Moloney MLV, are deleterious to binding or infection, demonstrating the critical importance of the side chain. This preliminary analysis suggests that only one of the two oxygen atoms on the side chain is required. At the 2-Å resolution of the current Fr57-RBD structure, oxygen atoms on the side chain of D86 and also S84 are adjacent to the aromatic ring of W102 (Fig. 1C), perhaps attracted to the relative positive charge manifested at the edge of the ring dipole (9). However, only replacement of S84 by isoleucine was inhibitory and the participation of W102 was revealed only in the presence of substitutions for D86. Therefore, it is unlikely that the role of S84 and/or D86 is simple tethering of the small helix to the proximal end of the VRA helix through an interaction with the aromatic ring on the side chain of W102. Additional studies, likely requiring resolution beyond that achievable through the study of simple amino acid substitutions, will be required to determine the chemical basis for the role of the pocket in receptor binding and infection.
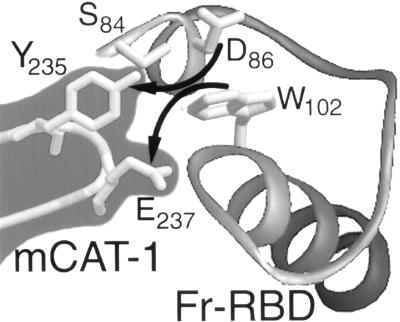
Adjacent tryptophan and glutamic acid (235-YGE-237) residues that are required for SU binding and ecotropic MLV infection have been identified in an extracellular loop of mCAT-1 (1). Further analysis demonstrated that an aromatic side chain at position 235 is required and a potential electron donor on the side chain of residue 237 is optimal for infection (21). Moreover, exhaustive mutagenesis of extracellular domains of mCAT-1 failed to identify additional critical residues (30). Together, these findings suggest that Y235 and E237 interact directly with the Fr57-RBD pocket, perhaps by providing alternative binding partners for D86 and W102 (Fig. 6). If true, peptides corresponding to the extracellular domain of mCAT-1 carrying YGE should competitively inhibit SU binding. Moreover, it may be possible to identify SU proteins with changes in pocket residues that can complement substitutions for Y235 or E237.
FIG. 6.
Proposed model of mCAT-1 binding to the pocket of Fr57-RBD. Residues Y235 and E237 in mCAT-1 may disrupt the interaction of S84 and/or D86 with W102 in the Fr57-RBD pocket.
An ecotropic MLV, TR1.3, causes hind-limb paralysis in mice by inducing syncytia in midbrain endothelial cells, resulting in local thrombosis and neuronal injury (24). The virus also induces widespread syncytia of SC-1 fibroblasts in tissue culture (25). Studies of chimeric viruses using nonpathogenic Friend 29 MLV (Fr29-MLV) revealed that the W102G change in SU is sufficient to explain the pathogenic properties of TR1.3 (25). In our experiments using Fr57-MLV, the presence of W102G in SU did not induce syncytium formation; however, Fr57-MLV SU differs from Fr29-MLV SU at eight additional residues that are candidates for key amino acids that modulate infection, possibly by affecting receptor binding. Fr29-MLV SU contains an alanine instead of a serine at position 84 and an asparagine instead of an aspartic acid at residue 88. In the Fr57-RBD structure derived by X-ray crystallography, D88 forms a salt bridge with K179 on the VRB helix (Fig. 1B and C), possibly increasing the relative stability of the Fr57-RBD to that of Fr29-RBD. Further studies of SU proteins carrying the W102G substitution coupled with these and the other changes in Fr29-MLV may reveal additional details of receptor binding and/or postbinding conformational change.
Forced overexpression of mCAT-1 did not increase Fr57-MLV infection, demonstrating that, normally, receptor density is not limiting on NIH 3T3 cells. However, since substitutions in the pocket reduced infectivity, the dependence of infectivity on receptor density was increasingly evident. This strongly suggests that Fr57-RBD interacts directly with mCAT-1 and that more than one interaction occurs during infection. These findings suggest that, to obtain a fusion pore, the formation of more than one receptor-envelope complex is required, likely restricted in time and space. A similar model of influenza virus fusion pore formation has been proposed previously (32). This model suggests that clustering of mCAT-1, recently observed in studies of non-clathrin-coated pits (22), may facilitate infection.
ACKNOWLEDGMENTS
This work was supported by funding from the Howard Hughes Medical Institute and a National Institutes of Health grant (2R01CA/AI61246-06).
REFERENCES
- 1.Albritton L M, Kim J W, Tseng L, Cunningham J M. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 3.Bae Y, Kingsman S M, Kingsman A J. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J Virol. 1997;71:2092–2099. doi: 10.1128/jvi.71.3.2092-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battini J L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battini J L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 7.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 8.Clackson T, Wells J A. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 9.Creighton T E. Proteins: structures and molecular properties. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1993. pp. 1–22. [Google Scholar]
- 10.Danieli T, Pelletier S L, Henis Y I, White J M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey R A, Hamson C A, Healey J J, Cunningham J M. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J Virol. 1997;71:8096–8102. doi: 10.1128/jvi.71.11.8096-8102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doms R W, Gething M J, Henneberry J, White J, Helenius A. Variant influenza virus hemagglutinin that induces fusion at elevated pH. J Virol. 1986;57:603–613. doi: 10.1128/jvi.57.2.603-613.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 14.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guex N, Peitsch M C. Swiss-PdbViewer: a fast and easy-to-use PDB viewer for the Macintosh and PC. Protein Data Bank Q Newsl. 1996;77:7. [Google Scholar]
- 18.Kim J W, Cunningham J M. N-linked glycosylation of the receptor for murine ecotropic retroviruses is altered in virus-infected cells. J Biol Chem. 1993;268:16316–16320. [PubMed] [Google Scholar]
- 19.Koch W, Hunsmann G, Friedrich R. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J Virol. 1983;45:1–9. doi: 10.1128/jvi.45.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacKrell A J, Soong N W, Curtis C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malhotra S, Scott A G, Zavorotinskaya T, Albritton L M. Analysis of the murine ecotropic leukemia virus receptor reveals a common biochemical determinant on diverse cell surface receptors that is essential to retrovirus entry. J Virol. 1996;70:321–326. doi: 10.1128/jvi.70.1.321-326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald K K, Zharikov S, Block E R, Kilberg M S. A caveolar complex between the cationic amino acid transporter 1 and endothelial nitric-oxide synthase may explain the “arginine paradox.”. J Biol Chem. 1997;272:31213–31216. doi: 10.1074/jbc.272.50.31213. [DOI] [PubMed] [Google Scholar]
- 23.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park B H, Lavi E, Blank K J, Gaulton G N. Intracerebral hemorrhages and syncytium formation induced by endothelial cell infection with a murine leukemia virus. J Virol. 1993;67:6015–6024. doi: 10.1128/jvi.67.10.6015-6024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park B H, Matuschke B, Lavi E, Gaulton G N. A point mutation in the env gene of a murine leukemia virus induces syncytium formation and neurologic disease. J Virol. 1994;68:7516–7524. doi: 10.1128/jvi.68.11.7516-7524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkins C P, Mar V, Shutter J R, del Castillo J, Danilenko D M, Medlock E S, Ponting I L, Graham M, Stark K L, Zuo Y, Cunningham J M, Bosselman R A. Anemia and perinatal death result from loss of the murine ecotropic retrovirus receptor mCAT-1. Genes Dev. 1997;11:914–925. doi: 10.1101/gad.11.7.914. [DOI] [PubMed] [Google Scholar]
- 28.Roger A, Milner-White E J. RasMol: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 29.Skov H, Andersen K B. Mutational analysis of Moloney murine leukaemia virus surface protein gp70. J Gen Virol. 1993;74:707–714. doi: 10.1099/0022-1317-74-4-707. [DOI] [PubMed] [Google Scholar]
- 30.Tang, H., and J. M. Cunningham. Unpublished data.
- 31.van Zeijl M, Johann S V, Closs E, Cunningham J M, Eddy R, Shows T B, O’Hara B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White J M, Danieli T, Henis Y I, Melikyan G, Cohen F S. Membrane fusion by the influenza hemagglutinin: the fusion pore. Soc Gen Physiol Ser. 1996;51:223–229. [PubMed] [Google Scholar]
- 33.Yang Y-L, Lei G, Holland C, Kitamura T, Hunter K, Cunningham J M. Receptors for polytropic and xenotropic mouse leukemia viruses encoded by a single gene at Rmc1. Nat Genet. 1999;21:216–219. doi: 10.1038/6005. [DOI] [PubMed] [Google Scholar]