SUMMARY
The claustrum (CLA) is a conspicuous subcortical structure interconnected with cortical and subcortical regions. Its regional anatomy and cell-type-specific connections in the mouse remain not fully determined. Using multimodal reference datasets, we confirmed the delineation of the mouse CLA as a single group of neurons embedded in the agranular insular cortex. We quantitatively investigated brain-wide inputs and outputs of CLA using bulk anterograde and retrograde viral tracing data and single neuron tracing data. We found that the prefrontal module has more cell types projecting to the CLA than other cortical modules, with layer 5 IT neurons predominating. We found nine morphological types of CLA principal neurons that topographically innervate functionally linked cortical targets, preferentially the midline cortical areas, secondary motor area, and entorhinal area. Together, this study provides a detailed wiring diagram of the cell-type-specific connections of the mouse CLA, laying a foundation for studying its functions at the cellular level.
Graphical Abstract
In brief
The claustrum is a subcortical structure interconnected with cortical and subcortical regions and its cellular anatomy remains not fully resolved. Wang et al. show regional and cell-type-specific inputs and outputs of the mouse claustrum. Its strong connections with the prefrontal module support the claustrum’s involvement in higher cognitive functions.
INTRODUCTION
The claustrum (CLA) is a small, elongated, and sheet-like subcortical structure located between the insular cortex and the striatum, well conserved from reptiles to mammals.1–3 It is also the most highly reciprocally connected structure with the cortex.4–7 Although its function is not understood,8 recent studies suggest that the CLA is involved in sleep,2,9,10 selective attention,11–13 engagement with the external world10 and task switching.14,15 Furthermore, top-down signals from the anterior cingulate cortex may go through the CLA to influence posterior sensory and association cortices.16 Because it is amenable to advanced transgenic tools and newly developed technologies, the laboratory mouse has become a popular animal model for studying CLA functions17–20
There are discrepancies in how the mouse CLA is defined, with a total of five parcellation schemes in existence. In one, the CLA is split into dorsal and ventral subdivisions underneath the gustatory and agranular insular areas.21 In a second scheme, the CLA is divided into core and shell.10,22–27 The third one defines CLA as a combination of the first and second schemes.28,29 In a fourth scheme, the CLA is delineated largely underneath the gustatory, agranular insular, and visceral cortical areas, but not further subdivided.1,30 A fifth scheme defines the CLA as a single, densely packed group of neurons embedded in the agranular insular area.5,31–33 Reconciling these differences is necessary for correctly quantifying and interpreting tract-tracing data, electrophysiological recording and opto/chemogenetic manipulations.
The afferent and efferent connections of CLA have been extensively studied in mammals.8,17,18,34,35 As in humans, non-human primates, and cats,4,36–41 the CLA in mice is reciprocally connected with frontal, parietal, temporal, and occipital cortices.5,6,9,11 It also receives inputs from olfactory areas, cortical subplate, striatum, pallidum, hippocampal formation, thalamus, hypothalamus, midbrain, and pons.6,9,11
The inputs to the CLA predominantly originate from cortical pyramidal neurons.5,6,11 These are classified into three major cell types: intra-telencephalic (IT) neurons in layer 2 (L2) to L6, extra-telencephalic (ET) neurons in L5, and corticothalamic (CT) neurons in L6, on the basis of their downstream targets.7,42,43 In the primary visual area (VISp), L6 IT neurons project to the CLA,44,45 while in the secondary motor area (MOs), L2/3 IT and L5 IT neurons send projections to it.46 Given that different cortical cell types convey different kinds of information,47,48 comprehensively understanding cell-type-specific connections of the mouse CLA is important.
The principal neurons within the CLA are heterogeneous in terms of their intrinsic electrical properties and projection targets. They have recently been classified into two,16 four,20,49 and five cell types25 on the basis of their intrinsic electrical properties and into four cell types on the basis of their axonal projections.7 It is not known how morphological cell types are related to electrophysiological ones.
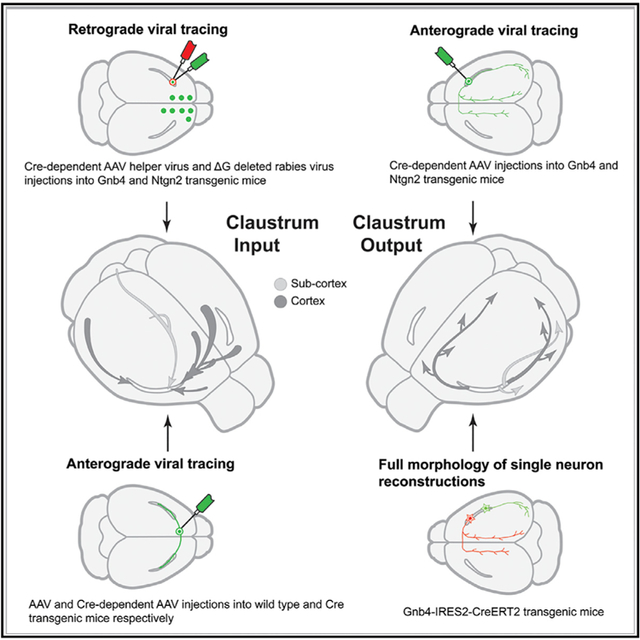
To address these issues, we characterized the location, brain-wide connectivity pattern, and full morphology of single principal neurons using systematic labeling, whole-brain imaging, and informatics approaches. Specifically, we performed new experiments by injecting monosynaptic retrograde rabies viral tracer into the CLA of transgenic mice and combined these with complementary datasets on CLA outputs and inputs mapped with anterograde Cre-dependent adeno-associated virus (AAV) tracer injections from the Allen Mouse Brain Connectivity Atlas.50,51 We reconstructed additional individual CLA principal neurons in sparsely labeled transgenic mice7 and found more diverse principal neuron types that innervate functionally linked targets. By integrating all these datasets, this study provides a detailed wiring diagram of the afferent and efferent projections of the mouse CLA at regional, laminar, and cell-type levels.
RESULTS
Delineation of the anatomical boundary of the mouse CLA
The CLA is embedded in the agranular insular area (AI) as a more densely packed group of neurons with heavy labeling in the Nissl-stained specimen compared with its adjacent structures (Figure 1A). It corresponds to a densely packed group of neurons in the transgenic mouse line Rorb-IRES2-Cre;Ai110 with red nuclear labeling (Figure 1B) and in the immunostained specimen for NeuN (nuclear protein Fox3, in red) (Figure 1C). The CLA is surrounded by densely labeled myelinated fibers revealed by immunostaining with an antibody against NF-160 (neurofilament M, in green) (Figure 1C) and an antibody against SMI-99 (basic myelin protein, in red) (Figure 1D). Similarly, the CLA is an oval shape surrounded by darkly stained AchE-positive fibers (Figure S1A). Figures 1E–1H show four Cre driver lines with enriched gene expression in the CLA and relatively weak expression in adjacent structures. Complementary to these transgenic lines, three other Cre driver lines with enriched gene expression in L6 of isocortex, tapering off from the gustatory cortical area (GU) to AI with little or no expression in CLA (Figures 1I–1K). Transgenic line Calb2-IRES2-Cre shows enriched gene expression in L2/3 of AI and GU but no expression in CLA (Figure 1L). These positive or negative gene expression patterns are consistent with the marker genes (in situ hybridization [ISH] data) (Figures S1B–S1L). Most positive marker genes are co-expressed in CLA and its adjacent structures AI, GU and the dorsal endopiriform nucleus (EPd) but with stronger expression in CLA than in its surroundings.52 Some of them express not only in L6 of lateral cortical areas, including GU and AI, but also in CLA and EPd, whereas others avoid expression in CLA but in L6 of AI, ACA and EPd, or in L6b and L6 of the cortex or in L5 and L6 of the cortex. CLA is embedded in AI, which is bordered dorsally by GU and ventrally by EPd (Figures 1 and S1). Additionally, anterograde viral tracing data show dense projections to CLA from cortical areas such as the anterior cingulate area (ACA).5 Our retrograde rabies injections into the primary motor cortex (MOp), GU and AI and the reunion nucleus (RE) of the thalamus show retrograde neurons labeled in deep layers of the GU or AI or a combination of both (Figures S2A–S2D). In our two-dimensional (2D) mouse brain atlas, CLA extends to the anterior forceps of the corpus callosum (Figure S2E), consistent with what has been shown in rats.32 Because all anatomical reference datasets show differential characteristics between CLA and its surroundings, we believe that the current and previous studies accurately delineate the boundary of CLA in CCFv3.5,33
Figure 1. Delineation of the boundary of the mouse CLA.
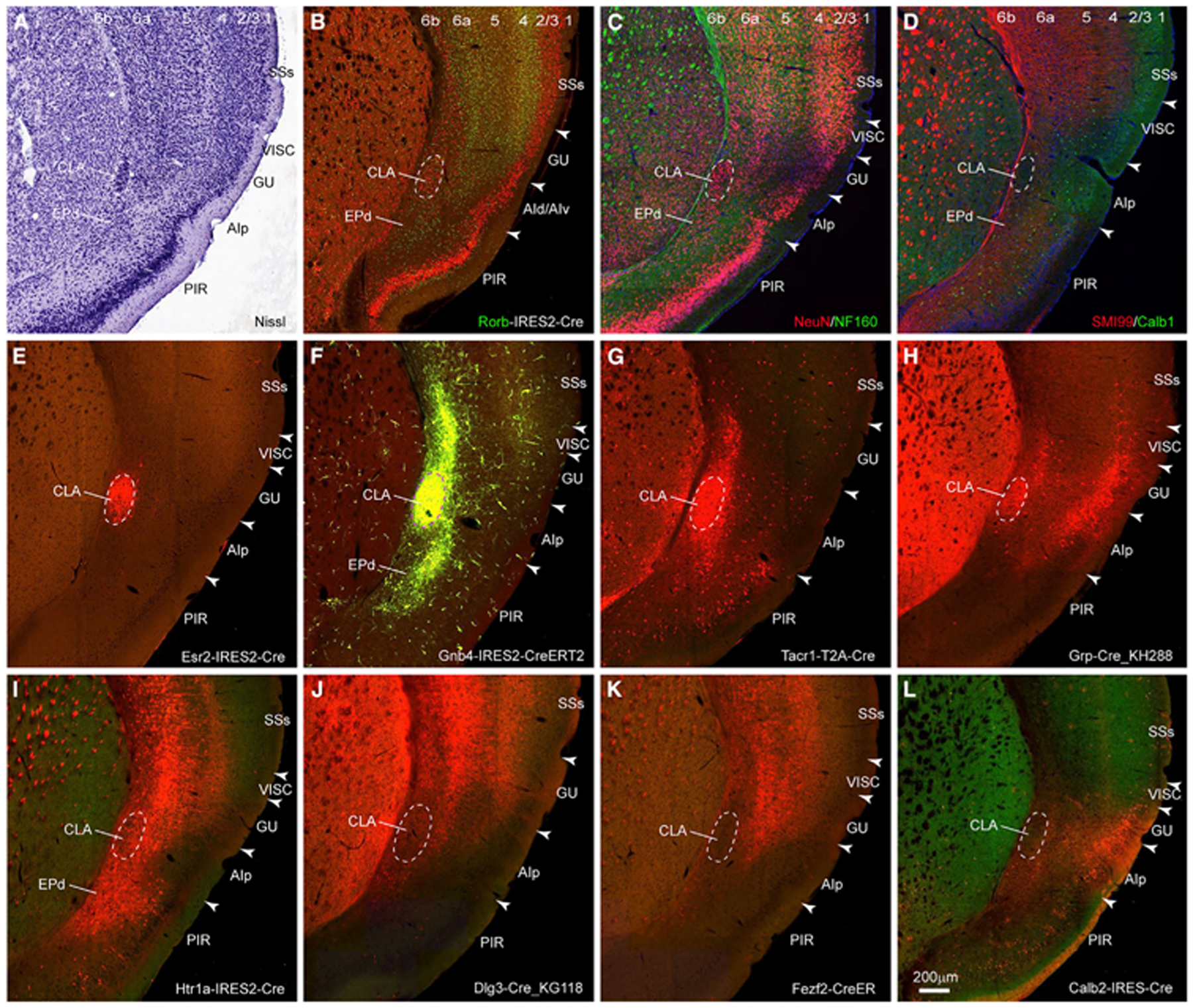
(A) The CLA is a densely packed group of neurons with heavy Nissl stain.
(B) The CLA is a densely packed group of neurons (in red) in Rorb-IRES2-Cre mouse..
(C) The CLA is a densely packed group of neurons immunostained with antibody against NeuN (in red) and is surrounded by myelin fibers immunostained with antibody against NF-160 (in green).
(D) The CLA is surrounded by myelin fibers immunostained with antibody against SMI-99 (in red). A subset of GABAergic neurons (in green) is labeled with immunostaining against calcium-binding protein calbindin (Calb1).
(E–H) Four transgenic lines, Esr2-IRES2-Cre. (E), Gnb4-IRES2-CreERT2 (F), Tacr1-T2A-Cre (G), and Grp-Cre (H), have enriched gene expression in the CLA.
(I–L) Four transgenic lines, Htr1a-IRES2-Cre (I), Dig-Cre_KG118 (J), Fezf2-CreER (K), and Calb2-IRES-Cre (L), have enriched gene expression in GU, AI, and EPd. Scale bars, 200 μm. Dashed line indicates the boundary of CLA in (A)–(L). Arrowhead indicates the border between cortical areas in (A)–(L). Numbers represent cortical layers in (A)–(D). For abbreviations, see Table S1. See also Figures S1 and S2.
Whole-brain presynaptic inputs to the CLA revealed with retrograde rabies tracing
To map whole-brain presynaptic inputs to the CLA, we used monosynaptic retrograde cell-type-specific rabies tracing method53 by injecting a Cre-dependent AAV helper virus into CLA of Gnb4-IRES2-Cre and Ntng2-IRES2-Cre mice, followed by injection of EnvA pseudotyped, glycoprotein-deleted rabies virus into the same location. Two sequential injections resulted in retrograde nuclear EGFP labeling of the presynaptic neurons that form synaptic contacts with starter cells, which were infected with both Cre-dependent AAV helper virus and rabies virus. All starter cells were manually counted in each experiment (see Method details) and were labeled more in CLA than its adjacent structures (Figure S2F). From 11 injections (Table S1), we selected 5 injections whose injection sites are mainly located in CLA and are spaced out along the anteroposterior axis of the CLA for quantitative analyses (Figure 2A, red dots) but excluded 6 injections with significant contamination, leakage along injection track, and segmentation artifacts.
Figure 2. Whole-brain presynaptic inputs to the CLA revealed with retrograde rabies tracing.
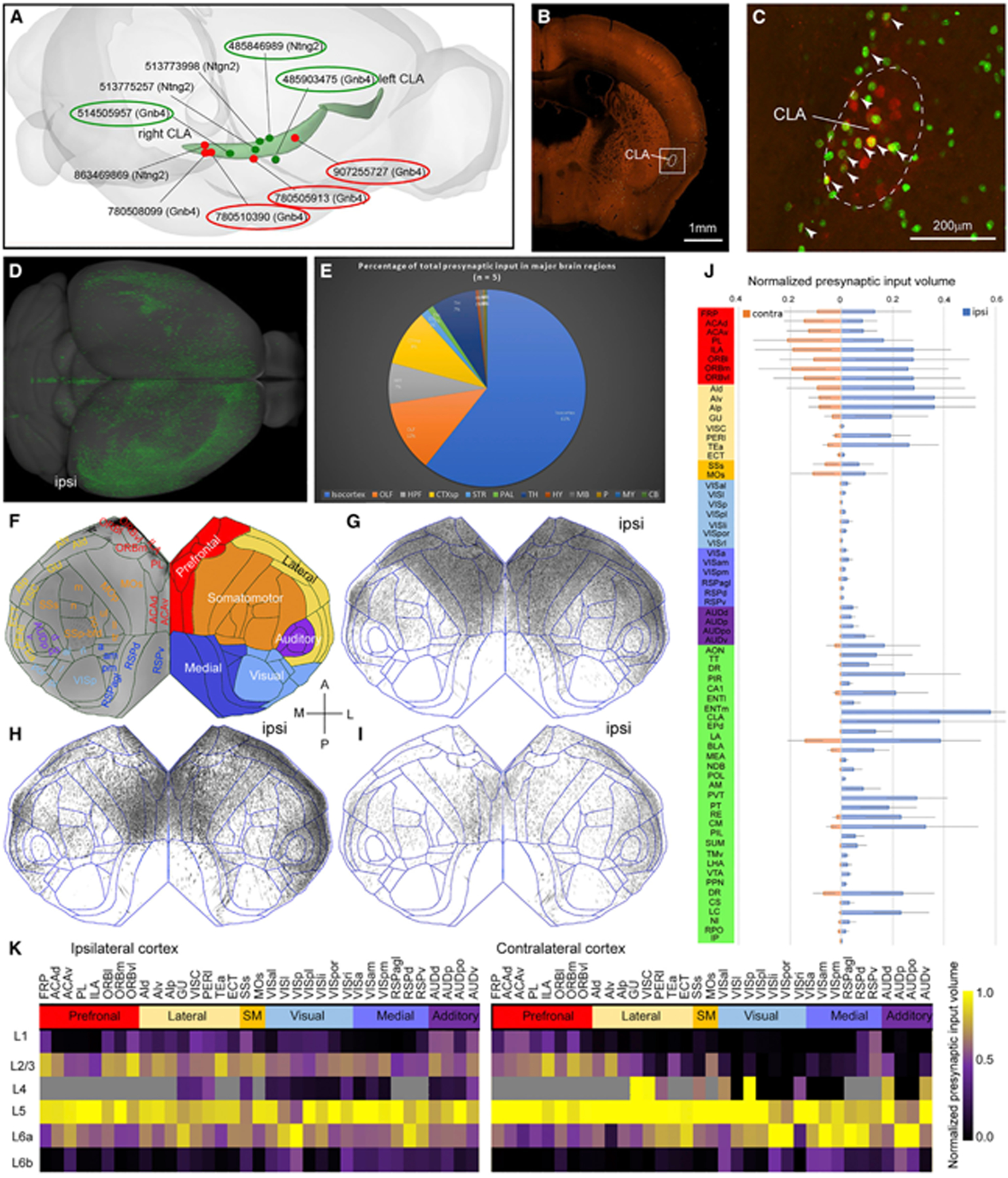
(A) Red and dark green dots indicate Cre-dependent retrograde rabies and anterograde AAV injection sites, respectively. The curved light green objects represent CLA in the left and right hemispheres. The numbers are experiment IDs. The circled injections are shown as examples in (G)–(I) and Figure 5H–5J.
(B) One representative example of retrograde rabies injection sites in coronal view at low magnification. Scale bar, 1 mm.
(C) An enlargement of the boxed area in (B). Neurons were infected with the AAV helper virus (in red) and with rabies virus (in green). Arrowheads indicate starter cells (in yellow). The dashed line indicates the border of CLA. Scale bar, 200 μm.
(D) Presynaptic neurons (in green) to the CLA in the dorsal view. “Ipsi,” ipsilateral to the injection site (same in G–I).
(E) A fraction of whole-brain presynaptic input to the CLA from 12 major brain divisions (n = 5).
(F) Flatmap showing all isocortical areas on the left side and six cortical modules color-coded differently on the right. A, anterior; P, posterior; M, medial; L, lateral.
(G–I) Flatmaps reveal similar presynaptic labeling patterns across 3 CLA injections, arranged from anterior (G), middle (H), to posterior (I). Each black dot in the flatmap represents a presynaptic neuron.
(J) Bar graph showing a fraction of total presynaptic labeling for each of the brain structures normalized by starter cells and structural volume. Blue and pink bars represent presynaptic labeling in the ipsilateral and contralateral hemispheres, respectively. Data are represented as mean ± SD (n = 5).
(K) Quantitative analysis shows the laminar origin of cortical presynaptic neurons to the CLA (n = 5). Six cortical modules are color-coded differently. Cortical areas that do not have L4 in CCFv3 are color-coded in gray in the matrix. The color scale bar presents the range between 0 and 1.
Figure 2B shows a representative example of retrograde rabies injection sites. AAV helper virus-infected neurons (in red) were labeled mostly in the CLA (Figure 2C). The starter cells (in yellow) were labeled mostly in CLA but sparsely in GU or EPd. Presynaptic neurons (in green) were labeled widely in CLA and other structures. The presynaptic neurons in the whole brain are shown in the dorsal view (Figure 2D), more frequently in the ipsilateral than the contralateral hemisphere. The fraction of total fluorescent volume detected in 5 injections is the highest in isocortex (61%), moderate in olfactory areas (OLF; 12%), hippocampal formation (HPF; 7%), cortical subplate (CTXsp; 9%) and thalamus (TH; 7%), low in striatum (STR; 1%) and pallidum (PAL; 1%), and vanishing in hypothalamus (HY), midbrain (MB), pons (P), medulla (MY), and cerebellum (CB; HY + MB + P + MY + CB < 1%) (Figure 2E; Table S2). Although the numbers of starter cells vary by more than an order of magnitude, the overall presynaptic input patterns appear similar across 5 injections.
We further analyzed presynaptic neurons at the fine structural level of the mouse ontology (314 structures in each hemisphere). As presynaptic neurons are mainly derived from the isocortex, we used the isocortical flatmap (computationally flattened 2D cortical map from CCFv3) to visualize presynaptic neuron distribution. The reference flatmap shows isocortical areas and subdivisions on the left side and the six cortical modules (prefrontal, lateral, medial, somatomotor, visual, and auditory) with different colors on the right side (Figure 2F).51 Flatmaps and quantification of the retrograde tracing data normalized by starter cells and each structural volume show that presynaptic neuron labeling is heavier in ipsilateral prefrontal and lateral modules than in the medial, visual, somatomotor, and auditory modules (Figures 2G–2J). Contralateral presynaptic labeling looks like a mirror image of ipsilateral labeling, but with fewer presynaptic neurons, except in the anterior cingulate, prelimbic (PL), and secondary motor areas whose presynaptic neurons are more numerous on the contralateral than on the ipsilateral side (Figures 2G–2J; Table S2).
To reveal the laminar distribution of presynaptic neurons, we computed averaged presynaptic input volume in each layer by dividing the sum of the total presynaptic input volume of each cortical area for 5 injections (Figure 2K; Table S2). Presynaptic neurons are denser in infragranular than in supragranular layers in both ipsilateral and contralateral cortical areas, except ventro-lateral orbital frontal area (ORBvl). Within infragranular layers, most cortical areas contain more presynaptic neurons in L5 than in L6, whereas a few cortical areas such as VISp and GU do the opposite (Figure 2K). Representative examples show the laminar distribution of presynaptic neurons in the frontal, temporal, parietal, and occipital cortical areas at high magnification (Figures 3A–3F). Presynaptic labeling shows a variety of patterns encompassing L2/3, L5, and L6 but never L4.
Figure 3. Cortical and subcortical presynaptic inputs to the CLA.
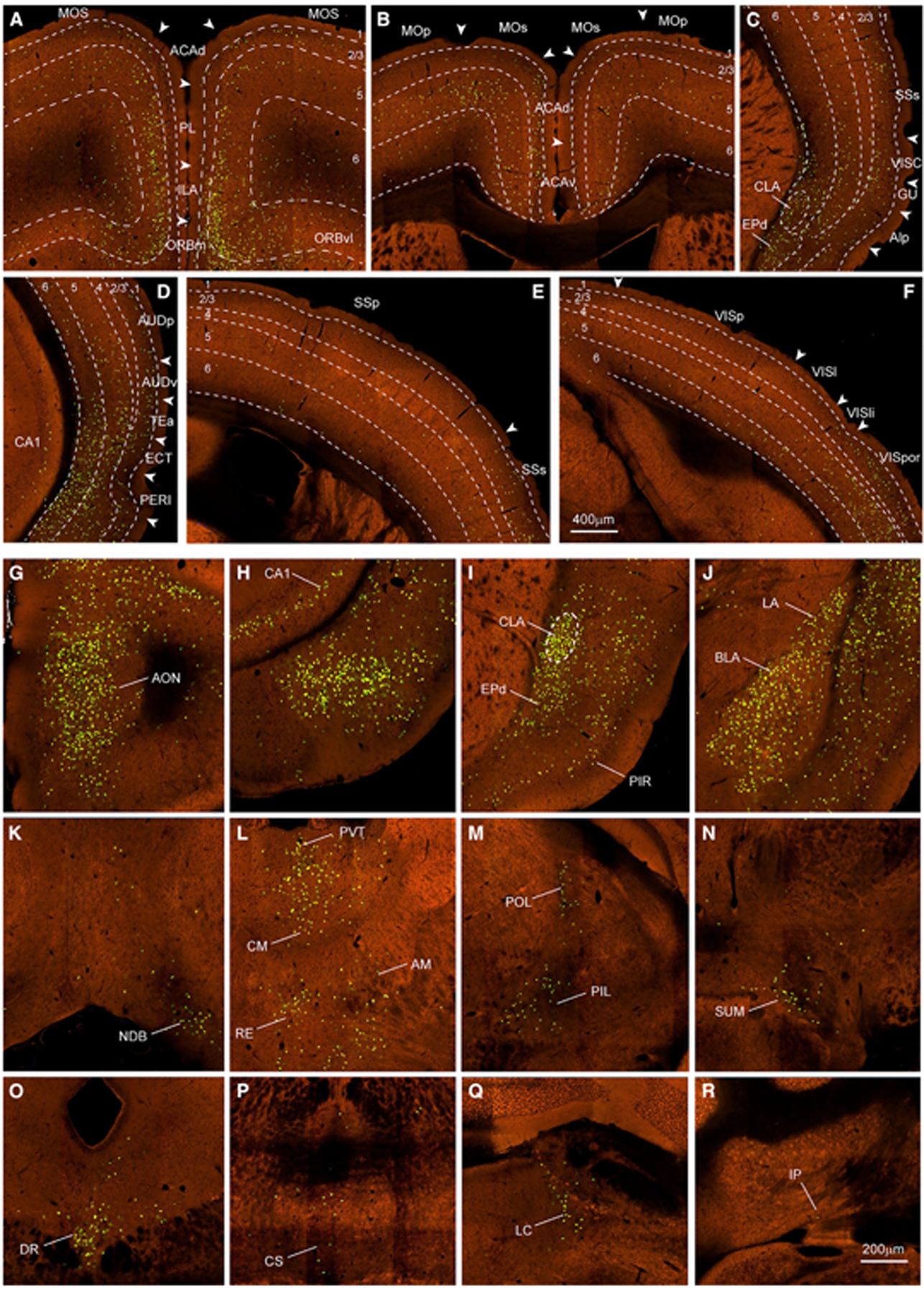
(A–F) Examples of presynaptic neurons in prefrontal (A and B), lateral (C and D), parietal (E), and occipital (F) cortices. Green dots are nuclear EGFP-labeled presynaptic neurons. Dashed lines indicate the borders of cortical layers and CLA. Arrowheads indicate the borders between cortical areas. Scale bar, 400 μm.
(G–R) Examples of presynaptic neurons labeled in other main brain regions outside isocortex are shown at high magnification, including AON (G) of the olfactory areas; ventral CA1 (H) of the hippocampal formation; CLA and EPd (I) and LA and BLA (J) of the cortical subplate; NDB (K) of the pallidum; PVT, CM, RE, and AM (L) and POL and PIL (M) of the thalamus; SUM (N) of the hypothalamus; DR (O) and CS (P) of the midbrain; LC (Q) of the pons; and IP (R) of the cerebellum. Scale bar, 200 μm.
For abbreviations, see Table S1.
Presynaptic neurons are found in many structures across major brain divisions. Thirty-one of these retrogradely labeled structures were validated with anterograde AAV injections and are listed in Figure 2J. One-third of these, especially midline structures, have bilateral presynaptic labeling with an ipsilateral predominance (Figures 3G–3R). Presynaptic neurons are more in CLA, EPd, basolateral amygdala nucleus (BLA), paraventricular nucleus (PVT), central medial nucleus (CM), and dorsal raphe nucleus (DR) than in other structures.
Inputs to CLA revealed with anterograde AAV tracing
To validate retrograde tracing results and to fully characterize cell-type-specific inputs to the CLA, we systematically reviewed experiments from the Allen Mouse Brain Connectivity Atlas.50,51 This dataset consists of 1,007 injections into isocortical areas and 1,472 injections into 175 structures across 11 other major brain regions of wild-type and Cre driver transgenic mouse lines. All injections are listed in Table S1. A total of 321 isocortical injections and 107 subcortical injections were found to send projections to CLA and these injection sites are shown (Figure S3A). Figure S3B shows examples of these injection sites in the entire cortical layers, L2/3 IT, L5 IT, L5 IT ET, L5 ET, and L6 CT of six cortical areas. Using anterograde tracing data, we confirmed that almost all cortical areas send projections to the CLA. The sole two exceptions are MOp and primary somatosensory (SSp) cortices, which do not project to the CLA but to its adjacent structures GU and AI. By quantitatively analyzing averaged axon labeling volume within CLA for each cell type of cortical pyramidal neurons, we found that the prefrontal module and MOs have more cell types projecting to the CLA than the sensory and medial modules (Figure S4). In most cortical areas, L5 IT (including L5 IT ET) neurons send stronger projections to the CLA than other cell types, whereas in VISp and VISl, L6 IT neurons predominate but with weak projections.
Figure 4A shows several examples of cell-type-specific inputs to the CLA from cortical areas PL, ACAd, ACAv, and MOs. We found two axonal projection patterns in the CLA and its surroundings from cortical areas. One pattern shows denser projections to its surroundings than to CLA itself from structures, such as PL, and another pattern shows denser projections to the CLA than to its surroundings from structures, such as ACA. L5 IT neurons in the medial entorhinal area (ENTm) send stronger projections to ipsilateral CLA than other cell types. In contrast, L2/3 IT and L5 IT neurons in the lateral entorhinal area (ENTl) project preferentially to EPd and insular cortex compared with CLA.
Figure 4. Anterograde AAV tracing reveals inputs to the CLA from cortex and subcortex.
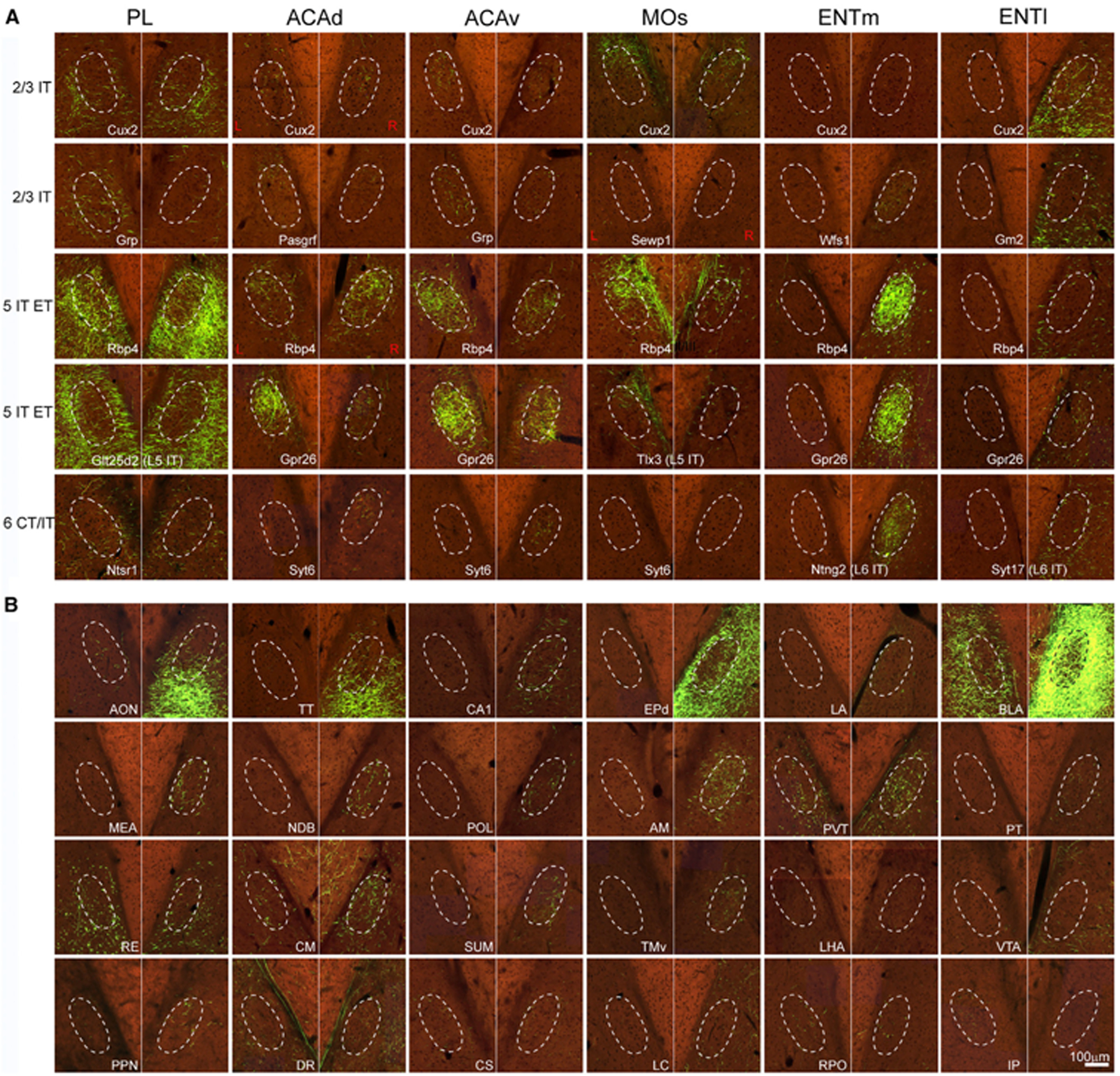
(A) Examples showing axons in the CLA and its adjacent structures from injections into cell-type-specific transgenic lines for L2/3 IT, L5 IT, L5 IT ET, L6 IT, and L6 CT of cortical areas PL, ACAd, ACAv, MOs, ENTm, and ENTl. Dashed line represents the border of the CLA (same in B). Red L (left) and R (right) indicate injection sites on the left side of the brain and the rest of these injections are on the right side. Scale bar, 100 μm.
(B) Examples showing axons in the CLA and its adjacent structures with anterograde injections into 24 subcortical structures. Scale bar, 100 μm. For abbreviations, see Table S1. See also Figures S3 and S4 and Table S3.
Presynaptic inputs to CLA from 31 subcortical structures in Figure 2J were confirmed with anterograde tracing data (Table S3). Figure 4B shows examples of axons in the CLA from 24 of these structures. Some structures such as anterior olfactory nucleus (AON), BLA, and PVT project to bilateral CLA with an ipsilateral predominance, while others such as ventral CA1 (vCA1) and anteromedial nucleus (AM) project to ipsilateral CLA. Surprisingly, the interposed nucleus (IP) in the cerebellum sends projections only to the contralateral CLA. As isocortex, these subcortical structures show two distinct projection patterns to CLA and its surroundings. One projection pattern shows preferential inputs to CLA over AI from structures, such as AM and PVT, while another pattern shows denser projections to AI over CLA from structures, such as AON and BLA (Figure 4B).
CLA outputs revealed with anterograde AAV tracing
To reveal brain-wide outputs, we re-analyzed our bulk anterograde tracing dataset by adding two newly generated CLA injections and excluding a contaminated injection from our previous study.5 Five injections into transgenic lines are shown in the rotated lateral view (Figure 2A, green dots). Figures 5A–5B shows a representative example of these bulk anterograde AAV injections. Claustro-cortical projections terminate in the lower part of L1, L2/3, L5, and L6 with different densities across isocortical areas (Figures 5C–5F). We quantified CLA output volume in each layer of each cortical area and averaged these volumes across five injections listed in Table S3. This quantitative analysis shows that axons are denser in L2/3 and L5 than in other layers in most cortical areas (Figure 5G). In a few cortical areas, such as GU and visceral area (VISC) axons are denser in L6 than in other layers. In VISp, axons are denser in L2/3 than in other layers (Figures 5F and 5G). In the retrohippocampal region, claustral outputs are much denser in the deep layers than in the superficial layers of ENTm and ENTI, while they are denser in the superficial layers than in the deep layers of the postsubiculum (POST), presubiculum (PRE) (Figure 5F) and parasubiculum (PAR).
Figure 5. Efferent projections of the CLA revealed with bulk anterograde Cre-dependent AAV tracing.
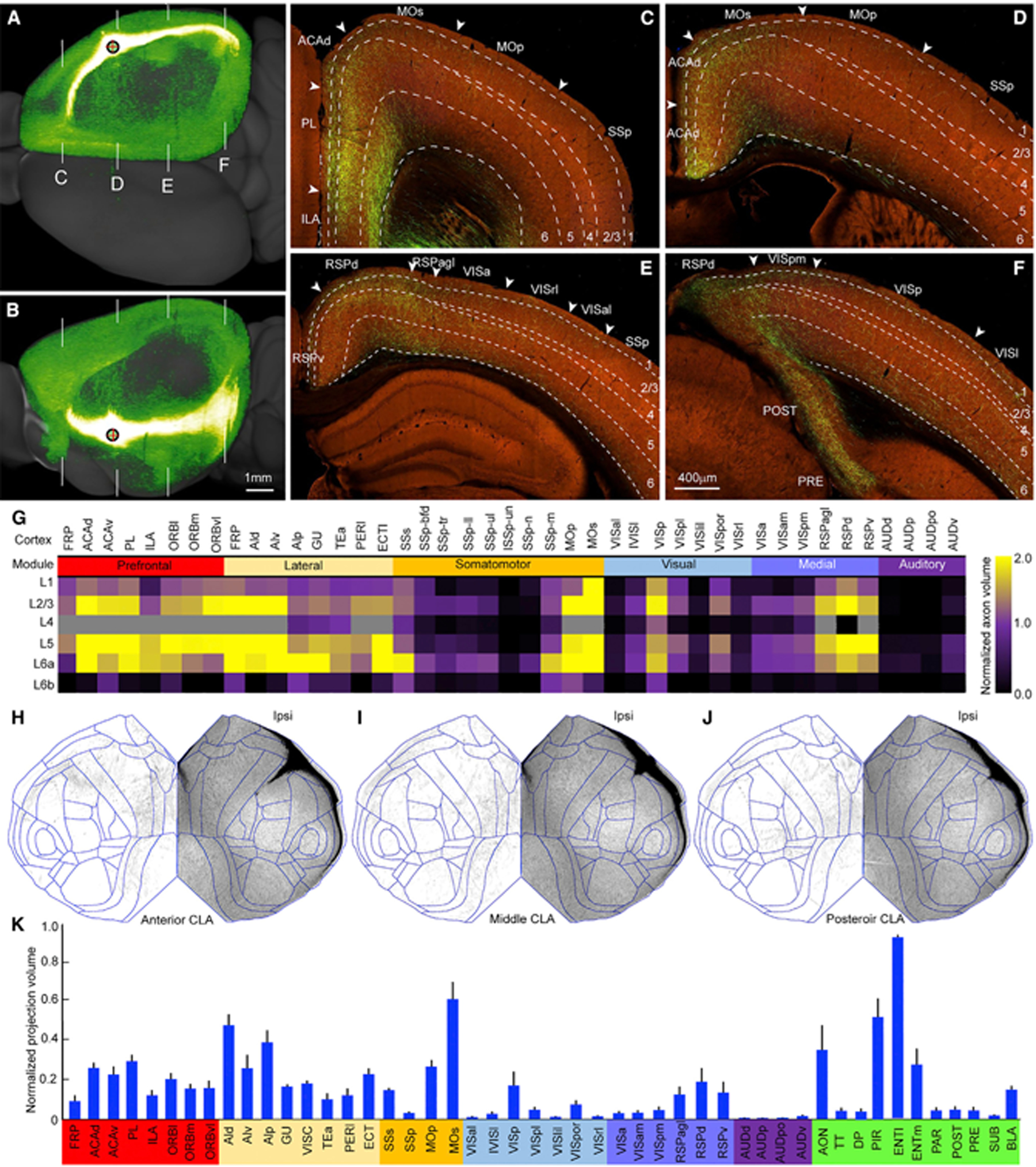
(A and B) A representative example of bulk anterograde Cre-dependent AAV injections into the CLA showing strong projections to the midline and association cortical areas in the dorsal (A) and lateral (B) views. Red cross inside a black circle represents injection site. White lines in (A) and (B) indicate coronal section levels shown in (C)–(F). Scale bar, 1 mm.
(C–F) High-power images showing axons in different cortical areas at the frontal (C), parietal (D and E), and occipital (F) levels. Arrowhead indicates the border between cortical areas. Dashed lines indicate the borders of cortical layers. For abbreviations see Table S1. Scale bar, 400 μm.
(G) Quantitative analysis of normalized axon volumes showing laminar distributions of claustro-cortical projections in different ipsilateral cortical areas on the basis of five injections. The color scale bar shows the range (0–2) of averaged axon projection volumes.
(H–J) Flatmaps of the isocortex showing similar claustro-cortical projection patterns from three anterograde Cre-dependent AAV injections. “Ipsi,” ipsilateral to the injection site.
(K) Bar graph showing the normalized axon volumes in the ipsilateral isocortical and subcortical structures by injection sites (n = 5 injections). Data are represented as mean ± SD. For abbreviations, see Table S1. See also Table S3.
To reveal axon distributions, we mapped claustro-cortical projections onto isocortical flatmap and found similar projection patterns across injections (Figures 5H–5J). Claustro-cortical projections were found in all ipsilateral isocortical areas, preferentially in the prefrontal and lateral modules and motor areas; moderately in VISp, SSs, and RSP; and sparsely in the higher visual, primary somatosensory, and auditory cortical areas (Figures 5H–5J). Sparse contralateral projections were consistently found in ACA, MOs, and RSP.
The quantitative analysis by averaging CLA output volumes in cortical areas across five injections confirmed the above qualitative observation that CLA sends stronger output to the prefrontal and lateral modules as well as motor cortices than other modules (Figure 5K). The CLA also sends projections strongly to the ENT, AON, and piriform area (PIR) and weakly to dorsal peduncular area (DP), taenia tecta (TT), subiculum complex, and BLA (Figure 5K).
Diverse cell types of CLA principal neurons
To better understand the diversity of individual neurons, we fully reconstructed 54 single CLA principal neurons using sparsely labeled transgenic Gnb4-IRES2-CreERT2 mice; 25 of these neurons were newly reconstructed in this study and 29 neurons were taken from our previous publication.7 For comparison and visualization purposes, 33 neurons reconstructed on the left side were flipped to the right side. The somata of these neurons are distributed throughout the anteroposterior extent of the CLA (Figure 6A). Examples of CLA neurons show dense axonal projections to the midline cortical targets and ENT (Figures 6B and S5A–S5C). Enlargements along axon fibers resembling synaptic boutons are barely present along the axon shafts (insets 1–3) within the CLA but are plentiful along axon branches (insets 4–6) in the neuron’s target regions. Its axon shafts travel rostrally and caudally within the CLA with almost no local collaterals.
Figure 6. Diverse cell types of CLA principal neurons revealed by single neuron reconstruction.
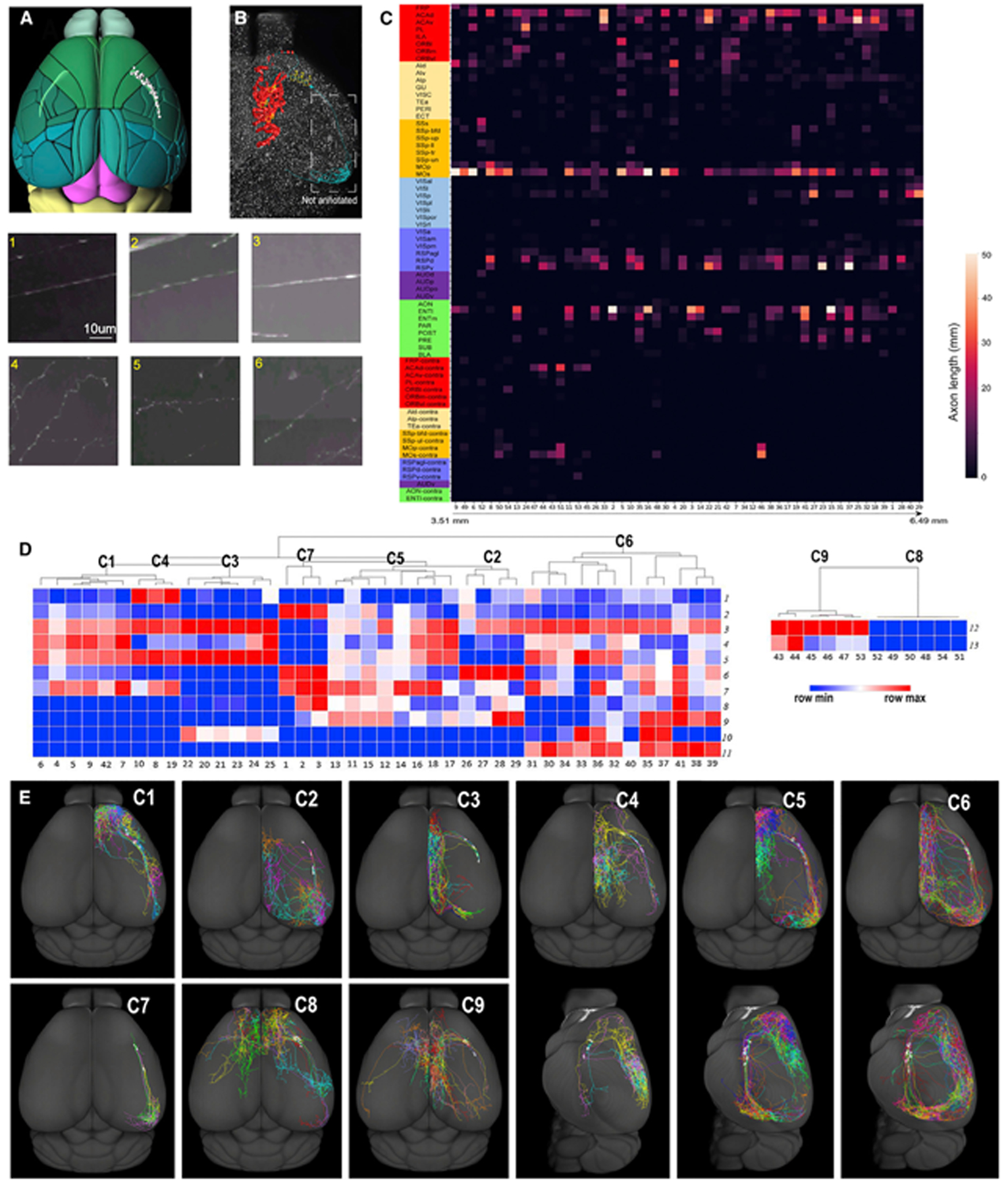
(A) Soma locations of individual principal neurons (gray balls) within the CLA (in light green) shown in the dorsal view.
(B) A representative example of synaptic bouton distribution of individual principal neurons shown in the dorsal view. Manually plotted synaptic boutons are marked (in red) in the midline cortical areas, but they are not plotted in the entorhinal cortex (in cyan). The insets (1–6) in (B) are enlarged under (A) and (B). Insets 1, 2, and 3 show axon shafts without bouton-like enlargements within the CLA, and insets 4, 5, and 6 show axonal arbors bearing synaptic bouton-like enlargements in the targets. Scale bar, 10 μm.
(C) Matrix showing the total lengths of axons of individual principal neurons in their targets. Along the x axis, the reconstructed neurons are arranged on the basis of their soma locations from rostral (3.51 mm measured from the very front of the main olfactory bulb (MOB) to caudal (6.49 mm from MOB). The y axis shows projections in target regions. For abbreviations, see Table S1.
(D) Seven clusters of ipsilaterally projecting neurons in the dendrogram on the left and 2 clusters of the bilaterally projecting neurons on the right. The numbers on the x axis are the IDs of individual neurons. The numbers on the y axis represent the morphological features.
(E) CLA neuron projections in C1–C3 and C7–C9 shown in the dorsal view and in C4–C6 shown in the dorsal and lateral views. Individual CLA principal neurons in each cluster are color-coded differently.
Individual CLA neurons densely and frequently project to PL, ACA, MOs, RSP, VISp, and ENT but sparsely and infrequently to the lateral, auditory, somatosensory, and visual modules (except VISp) (Figures S5D and S5E). Neurons project to targets topographically. Most bilaterally projecting CLA neurons (10 of 12) are distributed in the anterior part of the CLA, and a few (2 of 12) are in the middle. Anterior CLA neurons send few projections to the visual areas, POST, PRE, and PAR, while the posterior CLA neurons project substantially to these areas. Anterior and middle CLA neurons send projections frequently to ORB, PL, and infralimbic area (ILA), while posterior CLA neurons barely send projections to these areas (Figure 6C). The combined projections of all 54 neurons recapitulate our anterograde tracing results, with projections to all ipsilateral cortical areas and several contralateral ones (Figure S6A) but sparsely terminate in the ipsilateral lateral module and MOp.
To classify these cells into distinct types, we first manually separated them into ipsilaterally projecting neurons (n = 42) and bilaterally projecting neurons (n = 12). Unsupervised clustering analysis classified ipsilaterally projecting neurons into 7 clusters and bilaterally projecting neurons into 2 clusters on the basis of 11 and 2 morphological features, respectively (Figure 6D). Individual CLA neurons in the 9 clusters are visualized in dorsal and lateral views (Figure 6E) and their axonal and dendritic features are listed in Table S4.
We mapped axons of individual single neurons into isocortical flatmaps (Figures S6B–S6J). CLA neurons in C1–C7 innervate the diverse sets of target regions and in C8 and C9 project to ipsilateral ACA with or without projections to RSP.
Violin plots reveal the laminar distribution of axons by averaging total axon lengths in each cortical layer of all single CLA neurons (Figure S6K). CLA axons terminate in the lower part of L1 to L6 but with different densities, similar to our anterograde tracing results. In the frontal pole (FRP), PL, ORB, and TEa, axons are denser in L5 than in other layers. In AI, GU, and ECT, axons are denser in L6 than in other layers. In ACA, MOs, and RSP, axons are dense across all layers. In VISp, axons are denser in the lower L1 and upper L2/3 than in other layers.
Figure S7A shows fully reconstructed dendrites of 54 CLA neurons. Their dendrites arborized along the long anteroposterior axis of CLA are very narrow mediolaterally without typical apical dendrites and are restricted to the CLA boundary.54 Dendritic spines are observed on the dendritic trees of the individual CLA principal neurons (Figures S7B–S7C).
To quantitatively compare the dendritic morphology of different CLA neuron types as clustered above, we measured 11 dendritic features (Table S4). In the whole dataset of 9 clusters, significant differences were detected in 4 of the dendritic features (F < 0.05, ANOVA), including the distance of soma location measured from the very front of the CCFv3, dendritic arbor length and depth, and average bifurcate to bifurcate angle, which is an average value of the angle formed by a bifurcating node and its 2 child bifurcating nodes. For statistical significance analysis in the following, some clusters of small samples were pooled together on the basis of their similar axonal projecting directions and dominant targeting regions. The statistical difference between each pair of the groups, including data-pooled groups, was compared using a t test.
On average, the distance of bilaterally projecting neuronal soma locations is significantly shorter along the anteroposterior axis than that of ipsilaterally projecting CLA neurons (p < 0.01), of which C8 has the shortest distance. The dendrites of bilaterally projecting CLA neurons present some features differently from ipsilaterally projecting ones, including significantly thicker depth (p = 0.01) and bigger branching angles (p < 0.01). C9 has the biggest branching angle among clusters.
C2 and C7 that project only backward have significantly longer distances of soma location along the anteroposterior axis and a higher number of dendrites compared with pooled C1, C3 and C4 that project forward only or dominantly (p = 0.02, p = 0.02). In addition, C2 and C7 present significantly thinner dendritic clusters compared with pooled C1 and C5 (p = 0.04). C2 and C7 also have significantly longer distances of soma location along the anteroposterior axis (p < 0.01), a higher number of dendrites (p < 0.01) and thinner dendritic clusters (p < 0.01) compared with those of pooled C8 and C9.
DISCUSSION
The anatomical boundary of the mouse CLA
How to accurately delineate the boundary of the mouse CLA remains open to debate. As described in the Introduction, there are five distinct parcellation schemes, unlike in humans, non-human primates, and cats whose CLA is easily identifiable as a thin sheet of densely packed neurons embedded between two fiber tracts, the external and extreme capsules.55–57 Because of the lack of an obvious extreme capsule, the CLA in rodents is difficult to define. By registering multimodal anatomical reference datasets to the CCFv3, we found that the mouse CLA is restricted to the “core” or “ventral” CLA (vCLA) and that the “shell” or “dorsal” CLA (dCLA) belongs to L6 of the overlying AI and GU.5,33 The differences between CLA and AI and GU are evident in the cyto-, myelo-, geno-, chemo-architecture and connectivity. Such differences have also been reported in recent studies but are interpreted as the “core” and “shell” sending projections differentially to their targets.26,27 This interpretation is different from ours in that the “dCLA” falls into L6 of GU spatially and the “shell” is equivalent to L6 of AI. L6 neurons of GU and AI have distinct downstream targets compared with those of CLA.6,29 Our retrograde rabies injections into GU, MOp, Ald and RE clearly show retrograde neurons densely labeled in the deep layer of GU or AI or a combination of both, which is named “dCLA” or “shell” in some of the previous CLA studies.28,29 These differential connections58,59 between CLA and its surroundings could underly their physiological and functional differences.10,60 Similarly, we interpret marker genes, such as Pcp4, Nnat, and Slc30a3, as defining L6 of GU and AI, whereas others use these genes for defining the “shell” of CLA.27
Our definition of the mouse CLA meets all anatomical criteria and is consistent with a developmental study,61 a combination of tract-tracing and immunohistochemical studies62–64 and is fully compatible with projection-defined viral tracing data for those downstream targets at the network level,6 in contrast with others.20,29
The input and output connections of CLA with isocortex
The CLA has been reported to be reciprocally connected with a variety of cortical areas.5,6,9,11 However, the prior reports are inconsistent not only in the total number of efferent and afferent connections of CLA with isocortex but also in the projection strengths. By systematically analyzing retrograde and anterograde viral tract-tracing data and single neuron tracing data, we demonstrate that the CLA reciprocally connects with all cortical modules, preferentially with the prefrontal module (Figures 7A and 7B). However, we did not confirm inputs to CLA from MOp and SSp, two major nodes of the somatomotor network using anterograde tracing data. This mismatch between anterograde and retrograde results is likely due to the contamination of retrograde injections. Thus, cross-validation using multiple tracing datasets is imperative. Our finding of cortico-claustral projections is in line with electrophysiological in vivo and in vitro studies, in which CLA neurons were activated mostly by the frontal inputs but barely by single sensory or primary motor inputs.65,66 The strong and reciprocal connections with the prefrontal module, especially ACA, are consistent with other studies,6,62,65,67 supporting that CLA is involved in cognitive control.10,14,68
Figure 7. A wiring diagram of cell-type-specific input and output connections of the mouse CLA.
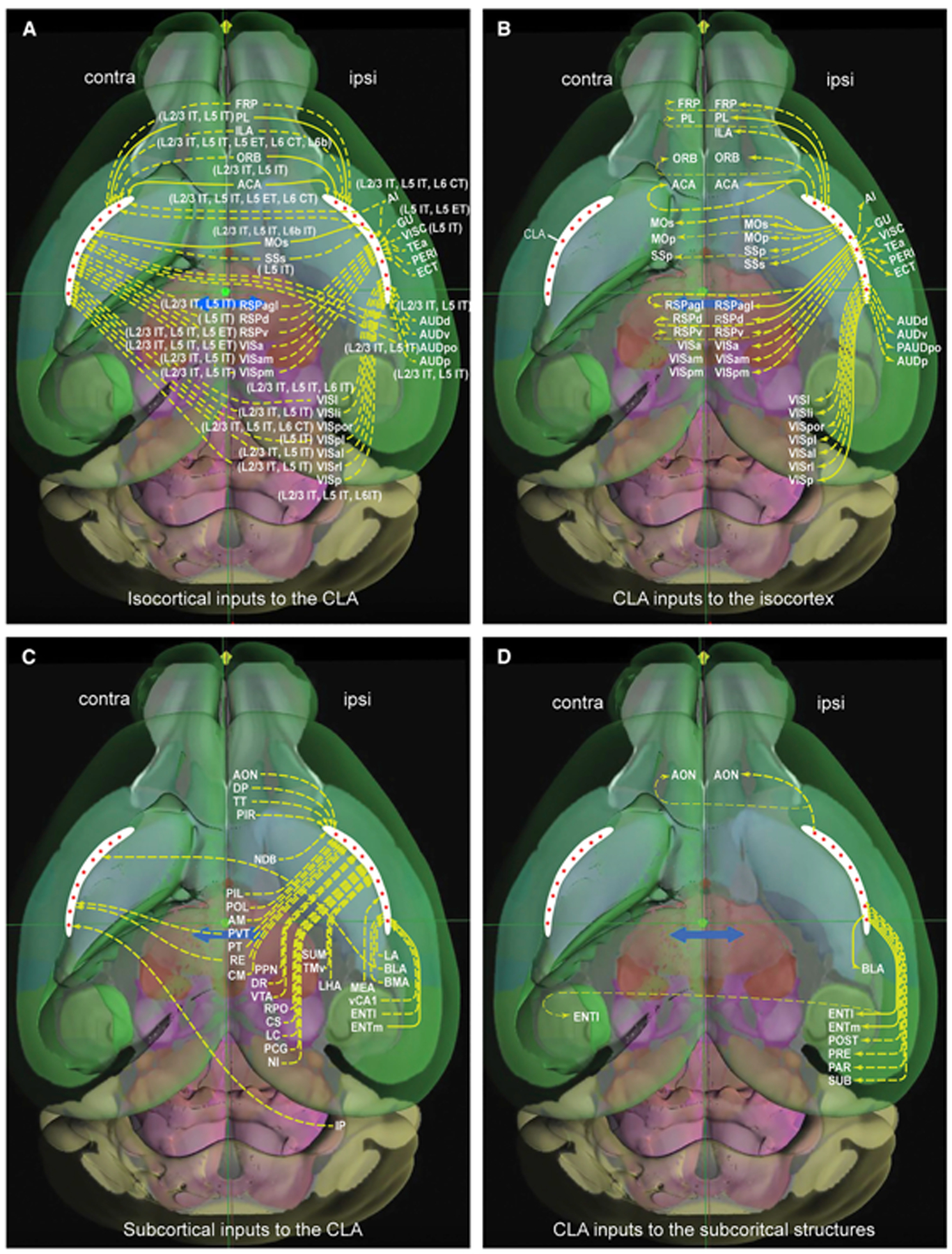
(A and B) Cortical cell-type-specific inputs to the CLA (A) and CLA outputs to isocortex (B). The CLA (in white) is an anteroposterior elongated and curved strip in each hemisphere. Red stars in the CLA represent principal neurons. These are the same in (C) and (D). Solid and dashed arrows represent strong and weak projections, respectively. Major brain divisions seen through the transparent cortex are represented by different background colors.
(C and D) Subcortical structures send inputs to bilateral, ipsilateral, or contralateral CLA (C), and the CLA sends outputs to some structures ipsilaterally or bilaterally (D).
Cell types and laminar origin of cortical pyramidal neurons projecting to the CLA
We demonstrate that cell types projecting to the CLA are determined by cortical areas and layers. Specifically, the prefrontal module has more cell types projecting to CLA than the lateral, medial, and sensory modules do. In most cortical areas, L5 IT neurons send stronger projections to the bilateral CLA than other cell types/layers, while L6 IT neurons in VISp predominate the projections. These anterograde tracing results are consistent with our retrograde tracing ones and others showing that cortical neurons projecting to CLA are denser in the infragranular layers than in the supragranular layers across cortical areas.6,69,70 Our finding of cell-type-specific inputs to CLA with different strengths could hold the key to uncovering CLA functions in the future.
Diverse morphological types of CLA principal neurons and their axonal distribution in cortical areas
The existence of diverse types of CLA principal neurons has been suggested by injecting multiple retrograde tracers into different combinations of cortical areas.6,20,26,38,71,72 The retrograde tracing method, however, has a limitation to trace many projection targets in one animal at once. Recently, principal neurons within the mouse CLA have been classified into different cell types on the basis of their intrinsic electrical properties, or their long-range axonal projections. One study using whole-cell patch-clamp recording combined with retrograde tracing classified principal neurons into 2 distinct cell types,16 while another study classified ACA-projecting principal neurons into 4 cell types.49 These 4 electrophysiological cell types may relate to our 4 morphological cell types: ipsilaterally and bilaterally ACA-projecting cell types (with or without ipsilateral RSP projections).
Our data not only reveal the topographical organization of individual CLA neurons along its anteroposterior extent but also uncover additional cell types. The nine morphological types innervate anatomically and functionally linked targets.50,51,73,74 Interestingly, all these CLA types belong to the same Car3 transcriptomic cell type.7 It will be important to establish the relationship between the morphological and electrophysiological cell types.20
The laminar distribution of claustro-cortical projections has been investigated in cats and mice.6,12,55,75–79 Our single neuron reconstruction, which does not suffer from contamination, reveals that CLA axon projections are denser in L5 and L2/3 than in L1 and L6 of the prefrontal module, denser in L6 than in other layers of the lateral cortical module (AI and GU), denser in the lower part of L1 and upper part of L2/3 than in other layers of the sensory modules but avoid branches in L4. This distribution pattern is similar to those in the previous studies6,12 but disagrees with one study showing denser axons in L6 of the frontal areas than in other layers.80 This discrepancy is most likely due to the contamination of bulk anterograde tracer injections. The laminar distribution of claustro-cortical projections can influence different neuron types in RSP compared with that of thalamo-cortical projections.81 Our findings suggest that different cell types transmit signals to different sets of anatomically and functionally linked cortical targets, rather than synchronizing the entire cortex.9
The dendritic morphology of CLA principal neurons in rats is different from that of the adjacent insular cortex.54 Our current and previous studies7 confirm such a difference in mice. Recent studies in mouse slices show two82 or four20 distinct principal neuron types based on their electrical properties and dendritic morphologies. However, slice studies cannot reveal the relationship between dendritic morphologies and axon targets of individual CLA principal neurons. In the present study, we completely reconstructed the full morphology of single CLA neurons. Our quantitative classification shows multiple types of CLA principal neurons on the basis of both axonal projections and dendritic morphologies. This finding provides the anatomical basis for further exploring their functional roles in information processing.
The connections of CLA with subcortical structures
The mouse CLA has been reported to receive inputs from subcortical structures and sends outputs back to a few of these regions.6,9,11 However, the prior reports are inconsistent in the total number of efferent and afferent connections of the claustrum with subcortical structures. In this study, we found that CLA receives inputs from 31 structures across 10 major divisions, and in turn, sends projections back to only a few of them (Figures 7C and 7D). We found 7 thalamic nuclei that project to CLA. Collectively, these thalamic nuclei have reciprocal connections with the midline and higher association cortices50,51 and have been suggested to involve arousal and higher cognitive functions.83–85 Our results clearly reveal the existence of indirect thalamo-claustro-cortical pathways.
CLA neurons receive inputs from neuromodulatory systems86–88 and have corresponding receptors to these neuromodulators.89,90 We confirmed inputs to CLA from NDB (cholinergic neurons), CS and DR (serotonergic neurons), VTA (dopaminergic neurons), and LC (noradrenergic neurons) and added inputs from PPN (cholinergic neurons) and RPO (serotonergic neurons) to the list (Figure 7C). Our finding of dopaminergic innervation of the mouse CLA is consistent with tract-tracing and immunohistochemical studies11,90 but disagrees with others.9,91 Studies of cholinergic and serotonergic inputs to CLA in acute slice recordings suggest their different influences on CLA neuron activities.90,92 Psilocybin, a partial serotonin 2A (5-HT2A) receptor agonist, has been suggested to alter the functional connectivity of CLA to cortical networks that support perception, memory, and attention.93,94
A recent study reported projections from the superior cerebellar peduncle to CLA by injecting conventional tracers into the superior cerebellar peduncle but did not specify projections from which part of the cerebellum.95 Our results showed sparse projections from IP to the contralateral CLA but not from the den-tate and fastigial nuclei. This finding suggests that the IP signal could reach the prefrontal module through CLA.
Functional implications
We here demonstrate that CLA receives inputs from more cell types in the prefrontal module than from other cortical modules, with L5 IT neurons predominating. L5 IT pyramidal neurons predominantly sending projections to CLA is consistent with what has been shown in rat prelimbic cortex with single neuron tracing.96 CLA neurons, in turn, send projections back to functionally linked cortical areas, predominantly along the midline and MOs as well as to ENT. Thus, cell-type-specific inputs and outputs of the CLA form direct, closed excitatory cortico-claustro-cortical loops, different from the indirect, inhibitory-dominant cortico-basal ganglia-thalamo-cortical loops (movement related) and the thalamo-cortico-thalamic loops (sensory relay and alertness related). Many subcortical structures, such as higher-order thalamic nuclei, send unidirectional projections to CLA, supporting the hypothesis that CLA is involved in higher cognitive processing.8,14,66 The accumulated evidence further supports such involvement of (1) CLA-ENTm pathway modulating the function of contextual memory97; (2) CLA-medial prefrontal pathway regulating impulsivity,98 attentional set-shifting,12 and salience detection11,13; and (3) CLA-ACA pathway modulating engagement with the external world.10 Recent Neuropixels recordings, combined with optogenetics, permit the investigation of multiple claustro-cortical pathways and cell types and layers simultaneously.19 Such techniques can be used to study cell-type-specific inputs and outputs of the CLA in mice that are engaged in tasks requiring competitive attentional and/or cognitive control.
Limitations of the study
Viral tracing data should be interpreted cautiously due to technical limitations.53 For retrograde rabies virus tracing, low-level leaky expression of TVA in the absence of Cre is sufficient for rabies infection. Leaky expression, in combination with retrograde neurons derived from AAV helper virus in Cre-driver expressed areas, might create undesired starter cells that complicate interpretation. Retrograde rabies tracing may not reveal all presynaptic neurons even with the higher transsynaptic efficiency of CVS-N2C used. Injection site contamination contributes to labeling for both anterograde and retrograde viral tracing, as transgene expression is not restricted to a single structure that we wanted to label. Therefore, verifying retrograde labeling with other datasets, such as anterograde tracing data, single neuron tracing data, and BARseq data, and vice versa is crucial for a better understanding of CLA circuitry.
Data segmentation and registration should also be considered. In some cases, false-positive and false-negative results were found because of uneven background signal and undetected weak fluorescent signal, respectively. The procedures for removing false-positive artifacts for both retrograde rabies tracing data and anterograde AAV tracing data are detailed in Method details. To avoid missing projection targets, false-negative results should be carefully and visually checked with high magnification. For anterograde AAV tracing data, axons of passage may not be completely excluded from terminal fields. This might cause an overestimation of axon terminal fields in some regions in some injections. As all tracing data were registered to CCFv3 for quantification, it is possible that some tracing data were not registered perfectly with CCFv3 because of different imaging modalities or other reasons.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Quanxin Wang (quanxinw@alleninstitute.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper are publicly available through the Allen institute portal (http://portal.brain-map.org/) and the archive Brain Imaging library (BIL, https://www.brainimagelibrary.org/).
No original code has been generated.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal care and use
All experimental procedures were approved by the Allen Institute Institutional Animal Care and Use Committee (IACUC) and conform to NIH guidelines. Both male and female wild-type C57BL/6J and transgenic mice ≥ P56 were used in this study listed in Table S1. All animals were housed 3–5 per cage, under constant temperature, humidity, and light conditions (14/10 h light/dark cycles) and given food and water ad libitum.
METHOD DETAILS
Histology and immunohistochemistry
The histology and double immunohistochemistry (IHC) procedures are detailed at the Allen Mouse Brain Connectivity Atlas documentation page (http://help.brain-map.org//display/mouseconnectivity/Documentation) and in our previous publications.5,33 Briefly, adult mice were anesthetized with 5% isoflurane and intracardially perfused with 10 mL saline (0.9% NaCl) followed by 50 mL freshly prepared 4% paraformaldehyde (PFA). Brains were rapidly dissected, postfixed, and transferred to a 30% sucrose solution. After sunk, brains were sectioned at 25 μm on a Leica 3050 S cryostat. A series of alternative sections from each brain were collected for Nissl-staining, or double IHC. Nissl-stained sections were delipidated with the xylene substitute Formula 83 (CBG Biotech, Columbus, OH; catalog No. CH0104) and ethanol rehydrated. The sections were stained in 0.21% thionine for 3 min and dehydrated by sequential immersion in increasing concentrations of ethanol. Differentiation and monitoring were performed at 95% ethanol before completion with pure ethanol. Dehydrated sections were subsequently incubated in Formula 83 and coverslipped with mounting medium and then scanned with the x10 objective on ScanScope (Aperio Technologies, Vista, CA).
For double IHC, sections were washed after antigen retrieval with 10 mM sodium citrate and then incubated in blocking solution (4% normal goat serum plus 0.3% Triton X-100 in PBS) for 1 h. After brief rinsing, each series of sections was incubated with a pair of the following primary antibodies: Calbindin1 (Calb1; Swant, catalog No. CB38.Rabbit final dilution 1:2,000 in blocking solution) and SMI-99 (Covance, Berkeley, CA; catalog No. SMI-99P; 1:1,000), or NeuN (Millipore, Bedford, MA; catalog No. MAB377, RRID:AB_10048713; 1:1,000) and NF-160 (Abcam, Cambridge, MA; catalog No. ab9034, RRID:AB_306956; 1:1,000) overnight and then incubated in a pair of the secondary antibodies: goat anti-rabbit-488 (final dilution 1:1,000 in blocking solution for Calb1, and 1:500 for NF160) and goat anti-mouse-594 (1:500 for SMI-99 and NeuN) overnight. After rinsing, sections were counterstained with DAPI (Invitrogen, Carlsbad, CA; catalog No. D1306) and coverslipped with Fluoromount G medium (Southern Biotechnology, Birmingham, AL; catalog No. 0100–01). Sections were scanned with VS110/120 (Olympus, Center Valley, PA) at x10 objectives.
Tracer injections
Detailed procedure for anterograde viral tracing has been described elsewhere.50,51 Surgery procedures for all experiments were the same with different tracers and mice used. A pan-neuronal AAV vector expressing EGFP under the human synapsin I promoter (AAV2/1.pSynI.EGFP.WPRE.bGH, Penn Vector Core, AV-1-PV1696, Addgene ID 105539) was as an anterograde tracer for injections into C57BL/6J mice and a Cre-dependent AAV vector expressing EGFP (AAV2/1.pCAG.FLEX.EGFP.WPRE.bGH, Penn Vector Core, AV-1-ALL854, Addgene ID 51502), or a Cre-dependent AAV virus expressing a synaptophysin-EGFP fusion protein (AAV2/1.pCAG.FLEX.sypEGFP.WPRE.bGH, Penn Vector Core) was used as an anterograde tracer for injections into Cre driver mice (Table S1). All adult mice were anesthetized with 5% isoflurane briefly and secured to a stereotaxic frame (Model# 1900, Kopf, Tujunga, CA) before surgery. During surgery, anesthesia was maintained at 2% isoflurane. After the skin incision, a small divot was made on the skull surface with a fine drill burr. To reveal the brain surface, a tiny thin layer of bone was removed with miniature forceps. For injection, a glass pipette (inner tip diameter of 10–20 μm) loaded with AAV was inserted into the desired brain depth from the pial surface. The coordinates for injections into cortical and subcortical structures were listed on our portal (connectivity.brain-map. org) based on the mouse brain atlas.21 These viral tracers were delivered by iontophoresis (current 3 μA, and 7s on/7s off duty cycle) for 5 min 2165 injections were made in the right hemisphere and 314 injections in the left hemisphere.
The procedure for monosynaptic retrograde rabies tracing has been described in detail elsewhere.53 First, Cre-dependent anterograde viral tracer AAV1-pSyn-DIO-TVA66T-dTom-CVS N2cG as helper virus was delivered to the CLA of transgenic mice (Gnb4-IRES2-Cre and Ntgn2-IRES2-Cre) by iontophoresis with the parameters as described above. Two weeks later, EnvA CVS-N2CΔg-H2B-EGFP rabies virus (500nL) was pressure-injected into the same location in the CLA. The injection coordinates for CLA were based on the mouse brain atlas21 at different anterior-posterior locations: AP −0.34 – 1.78mm from bregma, ML 1.85–3.5mm from the midline, DV 2.83–3.10mm from pia surface.
Whole-brain imaging
After the survival, mice were anesthetized with 5% isoflurane and intracardially perfused with 10 mL of saline (0.9% NaCl) followed by 50 mL of freshly prepared 4% PFA. Brains were rapidly dissected, post-fixed in 4% PFA at room temperature for 3–6 h and overnight at 4°C, then rinsed briefly with PBS and stored in PBS with 0.1% sodium azide. The brains were subsequently placed in 4.5% oxidized agarose (made by stirring 10 mM NaIO4 in agarose), transferred to a phosphate buffer solution, and put in a grid-lined embedding mold for standardized orientation in an aligned coordinate space. High-resolution 140 block-face coronal images were scanned with serial two-photon tomography (STPT) (TissueCyte 1000 system, TissueVision, Cambridge, MA) with 20× objective.
Data processing and quantification
STPT images were processed using the informatics data pipeline (IDP). The IDP contains two key algorithms: image alignment and signal detection. The global alignment process between the Cre mouse brains or AAV-injected brains and the average template consists of three steps: 1) a coarse registration, 2) a rigid registration, and 3) a 12-parameter affine registration. The signal detection algorithm was applied to each image to segment positive fluorescent signals from the background. Median filtering and large kernel low pass filter were then applied to remove noise. Two variations of the algorithm were employed, depending on the virus used for that experiment; one was tuned for EGFP, and one for SypEGFP detection. High-threshold edge information was combined with spatial distance-conditioned low-threshold edge results to form candidate signal object sets. For the SypEGFP data, filters were tuned to detect smaller objects (punctate terminal boutons vs long fibers). In addition, high-intensity pixels near the detected objects were included in the signal pixel set. Detected objects near hyper-intense artifacts occurring in multiple channels were removed. We developed an additional filtering step using a supervised decision tree classifier to filter out surface segmentation artifacts, based on morphological measurements, location context, and the normalized intensities of all three channels. The output is a full-resolution mask that classifies each 0.35 μm × 0.35 μm pixel as either signal or background. An isotropic 3-D summary of each brain is constructed by dividing each image into 10 μm × 10 μm grid voxels. Segmentation and registration results are combined to quantify the signal for each voxel in the reference space and for each structure in the reference atlas ontology by combining voxels from the same structure.
To quantify retrogradely labeled neurons in the whole brain, we modified the above algorithm to automatically detect fluorescence signal of nucleus-localized H2B-EGFP over the background and measured the pixel intensity in each anatomical structure.53 we used the automatically quantified retrograde labeling pixels as a proxy for the number of retrogradely labeled neurons in each structure.
Counting starter cells
All sections putatively containing the injection site were collected for immunostaining with an anti-RFP antibody (rabbit, Rockland Antibodies and Assays, 600-401-379) to amplify the tdTomato signal for the AAV1-pSyn-CVS-N2C-g-TVA-tdTomato virus. These immunostained sections were imaged by confocal microscopy (SP8, Leica) using 10× objective, 4 μm z-step size stacks. Maximum intensity images of each section were generated and used to manually count starter cells using ImageJ. Cells co-labeled for both tdTomato (in red) and native histone-GFP (in green) signals were identified as starter neurons (in yellow). The CLA was identified as a densely labeled cluster of neurons counterstained with DAPI.
Full morphological reconstruction of single CLA principal neurons
The full morphological reconstruction procedure has been described previously.7 Vaa3D, an open-source software was used for visualization, reconstruction, and analysis. The new modules TeraFly and TeraVR, which improved both efficiency and precision of the reconstruction, were incorporated into Vaa3D. In some cases, two or multiple similar-looking segments were intertwined or bundled together so closely that could not be separated even using Tera-VR. For those special cases, an extending segment and its branches were kept if it continued reaching all ends or given up if it was eventually connected to a main axonal stem that connect to another soma, and then continued to trace the other segment that branches further extending to normal axonal terminals. A neuron was considered fully reconstructed in the case that all dendrites and axonal arbors were traced as an example shown in Figures S5A–S5C. In order to avoid missing uncompleted ends of a complex axon, a marker was added to each trace end when reconstructing to a normal axonal terminal that typically ended with a well-labeled and enlarged bouton. Finally, a quality control (QC) procedure was performed by an experienced annotator using Tera-VR by double-checking the entire reconstruction of each reconstructed neuron. At high magnifications, special attention was paid to the proximal axonal part or a main axonal trunk of an axon cluster where axonal collaterals often emerged and branches but was more frequently missed due to the local image environment being composed of crowded high contrasting structures. The finalized reconstruction of a completed neuron had a soma node connecting multiple dendritic and axonal trees, without reconstruction errors such as breaks, loops, etc. In total, 54 single principal neurons within the CLA were reconstructed and 29 of them have been used in our previous study.7 All these single neurons were analyzed and visualized in the present study.
Quantification of axons of single CLA principal neurons
SWC files were processed and examined with Vaa3D plugins to ensure topological correctness: sorted single tree with root node as soma. SWC files were resampled with a step size of 5 microns. In this study, we used CCFv3 ontology, a manually curated set of 314 non-overlapping structures. Ipsi- and contra-lateral sides of brain regions to the somata were calculated separately. To quantitatively analyze the distribution of voxels of axons in brain-wide targets, fMOST images, and 3D reconstructed individual single CLA neurons were registered into the CCFv3. Total axon lengths in targets derived from single CLA principal neurons were measured as a proxy for projection strengths.
Unsupervised hierarchical clustering analysis of axon projections for single CLA principal neurons
Using the online software, Morpheus, (https://software.broadinstitute.org/morpheus/) to conduct unsupervised hierarchical clustering analysis, we classified single CLA neurons based on distinct axon morphological features. These morphological features are intuitive to show the similarity and dissimilarity among individual single neurons. Ipsilaterally projecting neurons have 11 axon features and bilaterally projecting neurons have 2 features. All features are listed in Table S4. Each feature was normalized to the same scale so that Euclidean distance can be used to make these clusters better discerned throughout various heights in the dendrogram.
QUANTIFICATION AND STATISTICAL ANALYSIS
Eleven dendritic features were quantified using Vaa3D. ANOVA test was performed for a parametric comparison of these dendritic features across multiple groups. For the parameters tested with a significant statistical difference in ANOVA test, t test was performed between every two individual groups.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Calbindin D-28k (Calb1) | Swant | Cat# CB38; RRID: AB_2721225 |
| Mouse monoclonal anti-NeuN | Millipore Sigma | Cat# MAB377; RRID: AB_2298772 |
| Rabbit polyclonal anti-Neurofilament, | Abcam | Cat# AB9034; RRID: AB_306956 |
| Medium, 160 kD (NF-160) | ||
| Mouse monoclonal anti-Myelin Basic Protein (SMI-99) | Covance | Cat# SMI-99P; RRID: AB_256474 |
| Alexa Fluor 488 goat anti-rabbit | Thermo Fisher Scientific | Cat# A-11008; RRID: AB_10563748 |
| Alexa Fluor 594 goat anti-mouse | Thermo Fisher Scientific | Cat# A-11005; RRID: AB_2534073 |
| anti-red fluorescent protein primary antibody | Rockland Antibodies and Assays | Cat# 600-401-379; RRID: AB_2209751 |
| Bacterial and virus strains | ||
| AAV2/1.hSynapsin.EGFP.WPRE.bGH | UPenn Vector Core | Cat# 105539-AAV1; RRID: Addgene_105539 |
| AAV2/1.CAG.FLEX.EGFP.WPRE.bGH | UPenn Vector Core | Cat# 51502-AAV1; RRID: Addgene_51502 |
| RabV-CVS-N2cdeltaG-histone-EGFP | Yao et al., 202253 | Cat# 176283-RabV; RRID: Addgene_176283 |
| pAAV-Syn-DIO-TVA66T-dTom-CVS-N2cG | Yao et al., 202253 | Cat# 176285-AAV; RRID: Addgene_176285 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Jackson Laboratory | JAX: 000664 |
| Mouse: B6.Cg-Tg(A930038C07Rik-cre) 1Aibs/J, A930038C07Rik-Tg1-Cre |
Jackson Laboratory | JAX: 017346 |
| Mouse: Adcyap1-2A-Cre | Allen Institute for Brain Science | N/A |
| Mouse: STOCK Agrptm1(cre)Lowl/J, Agrp-IRES-Cre | Jackson Laboratory | JAX:012899 |
| Mouse: B6.Cg-Avptm1.1(cre)Hze/J, Avp-IRES2-Cre | Jackson Laboratory | JAX:023530 |
| Mouse: B6.Cg-Calb1tm1.1(folA/EGFP/cre) | Jackson Laboratory | JAX:023531 |
| Hze/J, Calb1-T2A-dgCre | ||
| Mouse: B6(Cg)-Calb2tm1(cre)Zjh/J, Calb2-IRES-Cre | Jackson Laboratory | JAX: 010774 |
| Mouse: B6; 129S-Cartpttm1.1(cre)Hze/J, Cart-IRES2-Cre | Jackson Laboratory | JAX: 028533 |
| Mouse: STOCK Tg(Cartpt-cre)1Aibs/J, Cart-Tg1-Cre | Jackson Laboratory | JAX:009615 |
| Mouse: STOCK Ccktm1.1(cre)Zjh/J, Cck-IRES-Cre | Jackson Laboratory | JAX: 012706 |
| Mouse: STOCK Tg(Cdhr1-cre) KG66Gsat/Mmucd, Cdhr1-Cre_KG66 | MMRRC | MMRRC: 030952 |
| Mouse: B6; 129S6-Chattm2(cre) Lowl/J, Chat-IRES-Cre-neo | Jackson Laboratory | JAX:006410 |
| Mouse: STOCK Tg(Chrna2-cre) OE25Gsat/Mmucd, Chrna2-Cre_OE25 | MMRRC | MMRRC: 036502 |
| Mouse: STOCK Tg(Chrnb4-cre) OL57Gsat/Mmucd, Chrnb4-Cre_OL57 | MMRRC | MMRRC: 036203 |
| Mouse: STOCK Tg(Cnnm2-cre) KD18Gsat/Mmucd, Cnnm2-Cre_KD18 | MMRRC | MMRRC: 030951 |
| Mouse: Crh-IRES-Cre_BL | Bradford Lowell | N/A |
| Mouse: B6(Cg)-Crhtm1(cre)Zjh/J, Crh-IRES-Cre_ZJH | Jackson Laboratory | JAX: 012704 |
| Mouse: B6(Cg)-Cux2tm3.1(cre/ERT2) Mull/Mmmh, Cux2-CreERT2 | MMRRC | MMRRC: 032779 |
| Mouse: B6(Cg)-Cux2tm1.1(cre) Mull/Mmmh, Cux2-IRES-Cre | MMRRC | MMRRC: 031778 |
| Mouse: STOCK Tg(Dbh-cre) KH212Gsat/Mmucd, Dbh-Cre_KH212 | MMRRC | MMRRC: 032081 |
| Mouse: STOCK Tg(Dlg3-cre) KG118Gsat/Mmucd, Dlg3-Cre_KG118 | MMRRC | MMRRC: 032809 |
| Mouse: STOCK Tg(Drd1-cre) EY262Gsat/Mmucd, Drd1a-Cre_EY262 | MMRRC | MMRRC: 017264 |
| Mouse: B6.FVB(Cg)-Tg(Drd2-cre) ER44Gsat/Mmucd, Drd2-Cre_ER44 | MMRRC | MMRRC: 032108 |
| Mouse: STOCK Tg(Drd3-cre)KI196Gsat, Drd3-Cre_KI196 | MMRRC | MMRRC: 034610 |
| Mouse: STOCK Tg(Drd3-cre) KI198Gsat/Mmucd, Drd3-Cre_KI198 | MMRRC | MMRRC: 031741 |
| Mouse: STOCK Tg(Efr3a-cre) NO108Gsat/Mmucd, Efr3a-Cre_NO108 | MMRRC | MMRRC: 036660 |
| Mouse: B6.129S2-Emx1tm1(cre)Krj/J, Emx1-IRES-Cre | Jackson Laboratory | JAX: 005628 |
| Mouse: B6.Cg-Erbb4tm1.1(cre/ERT2) Aibs/J, Erbb4-T2A-CreERT2 | Jackson Laboratory | JAX: 012360 |
| Mouse: Esr1-2A-Cre | David Anderson | N/A |
| Mouse: B6; 129S-Esr2tm1.1(cre)Hze/J, Esr2-IRES2-Cre | Jackson Laboratory | JAX: 030158 |
| Mouse: B6(Cg)-Etv1tm1.1(cre/ERT2)Zjh/J, Etv1-CreERT2 | Jackson Laboratory | JAX: 013048 |
| Mouse: B6; 129S-Fezf1tm1.1(cre/folA) Hze/J, Fezf1-2A-dCre | Jackson Laboratory | JAX: 013048 |
| Mouse: B6.Cg-Foxp2tm1.1(cre/GFP) Rpa/J, Foxp2-IRES-Cre | Jackson Laboratory | JAX: 030541 |
| Mouse: B6.129P2-Gabra6tm2(cre) Wwis/Mmucd, Gabra6-IRES-Cre | MMRRC | MMRRC: 015968 |
| Mouse: STOCK Tg(Gabrr3-cre) KC112Gsat/Mmucd, Gabrr3-Cre_KC112 | MMRRC | MMRRC: 030709 |
| Mouse: STOCK GAD2tm2(cre)Zjh/J, GAD2-IRES-Cre | Jackson Laboratory | JAX: 010802 |
| Mouse: STOCK Tg(Gal-cre) KI87Gsat/Mmcd, Gal-Cre_KI87 | MMRRC | MMRRC: 031060 |
| Mouse: STOCK Tg(Colgalt2-cre) NF107Gsat/Mmucd, Glt25d2-Cre_NF107 | MMRRC | MMRRC: 036504 |
| Mouse: B6.Cg-Gnb4tm1.1(cre)Hze/J, Gnb4-IRES2-Cre | Jackson Laboratory | JAX: 029587 |
| Mouse: B6.Cg-Gnb4tm1.1(cre/ERT2) Hze/J, Gnb4-IRES2-CreERT2 | Jackson Laboratory | JAX: 030159 |
| Mouse: STOCK Tg(Gng7-cre) KH71Gsat/Mmucd, Gng7-Cre_KH71 | MMRRC | MMRRC: 031181 |
| Mouse: STOCK Tg(Gnrh1-cre)1Dlc/J, Gnrh1-Cre | Jackson Laboratory | JAX: 021207 |
| Mouse: STOCK Tg(Gpr26-cre)KO250Gsat/Mmucd, Gpr26-Cre_KO250 | MMRRC | MMRRC: 033032 |
| Mouse: C57BL/6-Tg(Grik4-cre)G32–4Stl/J, Grik4-Cre | Jackson Laboratory | JAX: 006474 |
| Mouse: STOCK Tg(Grm2-cre)MR90Gsat/Mmcd, Grm2-Cre_MR90 | MMRRC | MMRRC: 034611 |
| Mouse: STOCK Tg(Grp-cre) KH288Gsat/Mmucd, Grp-Cre_KH288 | MMRRC | MMRRC: 031183 |
| Mouse: Hcrt-Cre | Takeshi Sakurai | N/A |
| Mouse: STOCK Tg(Hdc-cre) IM1Gsat/Mmucd, Hdc-Cre_IM1 | MMRRC | MMRRC: 032079 |
| Mouse: B6; 129S-Htr1atm1.1(cre)Hze/J, Htr1a-IRES2-Cre | Jackson Laboratory | JAX: 030160 |
| Mouse: STOCK Tg(Htr2a-cre) KM207Gsat/Mmucd, Htr2a-Cre_KM207 | MMRRC | MMRRC: 031150 |
| Mouse: STOCK Tg(Htr3a-cre) NO152Gsat/Mmucd, Htr3a-Cre_NO152 | MMRRC | MMRRC: 036680 |
| Mouse: B6.Cg-Tg(Ins2-cre)25Mgn/J, Ins2-Cre_25 | Jackson Laboratory | JAX:003573 |
| Mouse: STOCK Tg(Kcnc2-Cre)K128Stl/LetJ, Kcnc2-Cre | Jackson Laboratory | JAX:008582 |
| Mouse: B6.129(SJL)-Kcng4tm1.1(cre)Jrs/J, Kcng4-Cre | Jackson Laboratory | JAX: 029414 |
| Mouse: STOCK Tg(Kiss1-cre)J2–4Cfe/J, Kiss1-Cre | Jackson Laboratory | JAX: 023426 |
| Mouse: B6.129-Leprtm2(cre)Rck/J, Lepr-IRES-Cre | Jackson Laboratory | JAX:008320 |
| Mouse: Lypd6-Cre_KL156 | Nathaniel Heintz and Charles Gerfen | N/A |
| Mouse: B6.Cg-Ndnftm1.1(folA/cre) Hze/J, Ndnf-IRES2-dgCre | Jackson Laboratory | JAX:028536 |
| Mouse: STOCK Nkx2-1tm1.1(cre/ERT2) Zjh/J, Nkx2-1-CreERT2 | Jackson Laboratory | JAX: 014552 |
| Mouse: B6; 129S-Nos1tm1.1 (cre/ERT2)Zjh/J, Nos1-CreERT2 | Jackson Laboratory | JAX: 014541 |
| Mouse: B6.Cg-Npr3tm1.1(cre)Hze/J, Npr3-IRES2-Cre | Jackson Laboratory | JAX:031333 |
| Mouse: FVB-Tg(Nr5a1-cre)2Lowl/J, Nr5a1-Cre | Jackson Laboratory | JAX:006364 |
| Mouse: B6.Cg-Ntng2tm1.1(cre)Hze/J, Ntng2-IRES2-Cre | Jackson Laboratory | JAX:029588 |
| Mouse: B6; 129S4-Ntrk1tm1(cre)Lfr/Mmucd, Ntrk1-IRES-Cre | MMRRC | MMRRC: 015500 |
| Mouse: B6.FVB(Cg)-Tg(Ntsr1-cre) GN220Gsat/Mmucd, Ntsr1-Cre_GN220 | MMRRC | MMRRC: 030648 |
| Mouse: STOCK Nkx2-1tm1.1(cre/ERT2) Zjh/J, Nxph4-T2A-CreERT2 | Jackson Laboratory | JAX:022861 |
| Mouse: B6(Cg)-Otoftm1.1(cre)Mull/Mmmh, Otof-Cre | MMRRC | MMRRC: 032781 |
| Mouse: Oxt-IRES-Cre | Bradford Lowell | N/A |
| Mouse: STOCK Tg(Oxtr-cre) ON66Gsat/Mmucd, Oxtr-Cre_ON66 | MMRRC | MMRRC: 036545 |
| Mouse: B6.Cg-Oxtrtm1.1(cre)Hze/J, Oxtr-T2A-Cre | Jackson Laboratory | JAX:031303 |
| Mouse: STOCK Tg(Pcdh9-cre) NP276Gsat/Mmucd, Pcdh9-Cre_NP276 | MMRRC | MMRRC: 036084 |
| Mouse: B6.Cg-Tg(Pcp2-cre) GN135Gsat/Mmucd, Pcp2-Cre_GN135 | MMRRC | MMRRC: 030868 |
| Mouse: B6; 129S-Pdyntm1.1(cre/ERT2) Hze/J, Pdyn-T2A-CreERT2 | Jackson Laboratory | JAX:030197 |
| Mouse: STOCK Tg(Pdzk1ip1-cre) KD31Gsat/Mmucd, Pdzk1ip1-Cre_KD31 | MMRRC | MMRRC: 030851 |
| Mouse: B6.Cg-Penktm1.1(cre/ERT2) Hze/J, Penk-2A-CreERT2 | Jackson Laboratory | JAX:022862 |
| Mouse: B6; 129S-Penktm2(cre)Hze/J, Penk-IRES2-Cre-neo | Jackson Laboratory | JAX: 025112 |
| Mouse: STOCK Tg(Plxnd1-cre) OG1Gsat/Mmucd, Plxnd1-Cre_OG1 | MMRRC | MMRRC: 036631 |
| Mouse: STOCK Tg(Pmch-cre)1Lowl/J, Pmch-Cre | Jackson Laboratory | JAX:014099 |
| Mouse: Pnmt-Cre | Steven Ebert | N/A |
| Mouse: STOCK Tg(Pomc1-cre)16Lowl/J, Pomc-Cre_BL | Jackson Laboratory | JAX:005965 |
| Mouse: B6.FVB-Tg(Pomc-cre)1Lowl/J, Pomc-Cre_ST | Jackson Laboratory | JAX: 010714 |
| Mouse: STOCK Tg(Ppp1r17-cre) NL146Gsat/Mmucd, Ppp1r17-Cre_NL146 | MMRRC | MMRRC: 036205 |
| Mouse: STOCK Tg(Prkcd-glc-1/CFP,-cre)EH124Gsat/Mmucd, Prkcd-GluCla-CFP-IRES-Cre_EH124 | MMRRC | MMRRC: 011559 |
| Mouse: B6; 129P2-Pvalbtm1(cre)Arbr/J, Pvalb-IRES-Cre | Jackson Laboratory | JAX:008069 |
| Mouse: B6.Cg-Pvalbtm1.1(cre/ERT2)Hze/J, Pvalb-T2A-CreERT2 | Jackson Laboratory | JAX: 021189 |
| Mouse: B6; 129S-Rasgrf2tm1.1 (cre/folA)Hze/J, Rasgrf2-T2A-dCre | Jackson Laboratory | JAX:022864 |
| Mouse: STOCK Tg(Rbp4-cre) KL100Gsat/Mmucd, Rbp4-Cre_KL100 | MMRRC | MMRRC: 031125 |
| Mouse: B6; 129S-Rorbtm1.1(cre)Hze/J, Rorb-IRES2-Cre | Jackson Laboratory | JAX: 023526 |
| Mouse: Rorb-IRES2-Cre-neo | Allen Institute for Brain Science | N/A |
| Mouse: STOCK Tg(Satb2-cre) MO23Gsat/Mmucd, Satb2-Cre_MO23 | MMRRC | MMRRC: 032908 |
| Mouse: B6; C3-Tg(Scnn1a-cre)2Aibs/J, Scnn1a-Tg2-Cre | Jackson Laboratory | JAX: 009112 |
| Mouse: B6; C3-Tg(Scnn1a-cre)3Aibs/J, Scnn1a-Tg3-Cre | Jackson Laboratory | JAX: 009613 |
| Mouse: STOCK Tg(Selenow-cre) NP39Gsat/Mmucd, Sepw1-Cre_NP39 | MMRRC | MMRRC: 036190 |
| Mouse: STOCK Tg(Sim1-cre) KJ18Gsat/Mmucd, Sim1-Cre_KJ18 | MMRRC | MMRRC: 031742 |
| Mouse: STOCK Slc17a6tm2(cre) Lowl/J, Slc17a6-IRES-Cre | Jackson Laboratory | JAX: 016963 |
| Mouse: B6; 129S-Slc17a7tm1.1 (cre)Hze/J, Slc17a7-IRES2-Cre | Jackson Laboratory | JAX: 023527 |
| Mouse: Tg(Slc17a8-icre)1Edw/SealJ, Slc17a8-iCre | Jackson Laboratory | JAX: 018147 |
| Mouse: B6; 129S-Slc17a8tm1.1 (cre)Hze/J, Slc17a8-IRES2-Cre | Jackson Laboratory | JAX: 028534 |
| Mouse: STOCK Tg(Slc18a2-cre) OZ14Gsat/Mmcd, Slc18a2-Cre_OZ14 | MMRRC | MMRRC: 034814 |
| Mouse: STOCK Slc32a1tm2(cre) Lowl/J, Slc32a1-IRES-Cre | Jackson Laboratory | JAX: 016962 |
| Mouse: STOCK Slc6a3tm1(cre)Xz/J, Slc6a3-Cre | Jackson Laboratory | JAX: 020080 |
| Mouse: B6.Cg-Tg(Slc6a4-cre) ET33Gsat/Mmcd, Slc6a4-Cre_ET33 | MMRRC | MMRRC: 031028 |
| Mouse: STOCK Tg(Slc6a4-cre/ERT2) EZ13Gsat/Mmcd, Slc6a4-CreERT2_EZ13 | MMRRC | MMRRC: 030071 |
| Mouse: STOCK Tg(Slc6a5-cre) KF109Gsat/Mmucd, Slc6a5-Cre_KF109 | MMRRC | MMRRC: 030730 |
| Mouse: Sst-Cre | Allen Institute for Brain Science | N/A |
| Mouse: STOCK Ssttm2.1(cre)Zjh/J, Sst-IRES-Cre | Jackson Laboratory | JAX: 013044 |
| Mouse: STOCK Tg(Syt17-cre) NO14Gsat/Mmucd, Syt17-Cre_NO14 | MMRRC | MMRRC: 034355 |
| Mouse: STOCK Tg(Syt6-cre) KI148Gsat/Mmucd, Syt6-Cre_KI148 | MMRRC | MMRRC: 032012 |
| Mouse: B6; 129S-Tac1tm1.1(cre)Hze/J, Tac1-IRES2-Cre | Jackson Laboratory | JAX: 021877 |
| Mouse: B6; 129S-Tac2tm1.1(cre)Hze/J, Tac2-IRES2-Cre | Jackson Laboratory | JAX: 021878 |
| Mouse: Tacr1-T2A-Cre | Allen Institute for Brain Science | N/A |
| Mouse: B6.FVB(Cg)-Tg(Th-cre)FI172Gsat/Mmucd, Th-Cre_FI172 | MMRRC | MMRRC: 031029 |
| Mouse: B6; 129-Thtm1(cre/Esr1)Nat/J, Th-IRES-CreER | Jackson Laboratory | JAX: 008532 |
| Mouse: STOCK Tg(Tlx3-cre)PL56Gsat/Mmucd, Tlx3-Cre_PL56 | MMRRC | MMRRC: 041158 |
| Mouse: B6.Cg-Trib2tm1.1(cre/ERT2) Hze/J, Trib2-F2A-CreERT2 | Jackson Laboratory | JAX: 022865 |
| Mouse: STOCK Tg(Ucn3-cre) KF43Gsat/Mmucd, Ucn3-Cre_KF43 | MMRRC | MMRRC: 032078 |
| Mouse: STOCK Viptm1(cre)Zjh/J, Vip-IRES-Cre | Jackson Laboratory | JAX: 010908 |
| Mouse: STOCK Tg(Vipr2-cre) KE2Gsat/Mmucd, Vipr2-Cre_KE2 | MMRRC | MMRRC: 034281 |
| Mouse: STOCK Tg(Vipr2-cre) KE2Gsat/Mmucd, Vipr2-IRES2-Cre | Jackson Laboratory | JAX: 031332 |
| Mouse: B6.Cg-Tg(Wfs1-cre/ERT2) 2Aibs/J, Wfs1-Tg2-CreERT2 | Jackson Laboratory | JAX:009614 |
| Mouse: B6; C3-Tg(Wfs1-cre/ERT2) 3Aibs/J, Wfs1-Tg3-CreERT2 | Jackson Laboratory | JAX:009103 |
| Software and algorithms | ||
| Vaa3D (version 3.604) | Vaa3D github | https://github.com/Vaa3D |
| ImageJ 2.0.02.0.0-rc-43/1.53j | NIH | https://mirror.imageJ.net |
| AllenSDK | Github | https://github.com/alleninstitute/allensdk |
| Registration code | Github | https://github.com/AllenInstitute/stpt_registration |
| Morpheus | Broad Institute | https://software.broadinstitute.org/morpheus/ |
| Other | ||
| Allen Mouse Brain Atlas | Lein et al., 200799 | http://mouse.brain-map.org |
| Allen Mouse Brain Connectivity Atlas | Oh et al., 2014; Harris et al., 201950,51 | http://connectivity.brain-map.org |
| Allen Mouse Brain Reference Atlases | Wang et al., 202033 | http://atlas.brain-map.org/ |
| Anatomic Reference Data | Allen Institute for Brain Science | http://connectivity.brain-map.org/static/referencedata |
Highlights.
The prefrontal module sends stronger input to CLA than the other 5 cortical modules
The claustrum receives stronger input from L5 IT neurons than from other cell types
The claustrum receives inputs from 31 subcortical structures
Nine types of claustrum principal neurons innervate diverse cortical targets
ACKNOWLEDGMENTS
We are grateful to the Transgenic Colony Management, Neurosurgery and Behavior, Lab Animal Services, Molecular Genetics, Imaging, Histology, Technology, and Project Management teams at the Allen Institute for technical and management support. We thank Thomas R. Reardon, Andrew J. Murray, and Ian Wickersham for providing cell lines and plasmids for the establishment of rabies virus production at the Allen Institute. This work was supported by the Allen Institute for Brain Science and by the National Institute of Mental Health (NIMH) of the National Institutes of Health (NIH) under award U19MH114830 to H.Z. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH and its subsidiary institutes. We also acknowledge funding from the Tiny Blue Dot Foundation. We thank the Allen Institute founder, Paul G. Allen, for his vision, encouragement, and support.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112118.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Bruguier H, Suarez R, Manger P, Hoerder-Suabedissen A, Shelton AM, Oliver DK, Packer AM, Ferran JL, García-Moreno F, Puelles L, and Molnár Z (2020). In search of common developmental and evolutionary origin of the claustrum and subplate. J. Comp. Neurol. 528, 2956–2977. 10.1002/cne.24922. [DOI] [PubMed] [Google Scholar]
- 2.Norimoto H, Fenk LA, Li HH, Tosches MA, Gallego-Flores T, Hain D, Reiter S, Kobayashi R, Macias A, Arends A, et al. (2020). A claustrum in reptiles and its role in slow-wave sleep. Nature 578, 413–418. 10.1038/s41586-020-1993-6. [DOI] [PubMed] [Google Scholar]
- 3.Hain D, Gallego-Flores T, Klinkmann M, Macias A, Ciirdaeva E, Arends A, Thum C, Tushev G, Kretschmer F, Tosches MA, and Laurent G (2022). Molecular diversity and evolution of neuron types in the amniote brain. Science 377, eabp8202. 10.1126/science.abp8202. [DOI] [PubMed] [Google Scholar]
- 4.Torgerson CM, Irimia A, Goh SYM, and Van Horn JD (2015). The DTI connectivity of the human claustrum. Hum. Brain Mapp. 36, 827–838. 10.1002/hbm.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Ng L, Harris JA, Feng D, Li Y, Royall JJ, Oh SW, Bernard A, Sunkin SM, Koch C, and Zeng H (2017). Organization of the connections between claustrum and cortex in the mouse. J. Comp. Neurol. 525, 1317–1346. 10.1002/cne.24047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zingg B, Dong HW, Tao HW, and Zhang LI (2018). Input-output organization of the mouse claustrum. J. Comp. Neurol. 526, 2428–2443. 10.1002/cne.245027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng H, Xie P, Liu L, Kuang X, Wang Y, Qu L, Gong H, Jiang S, Li A, Ruan Z, et al. (2021). Morphological diversity of single neurons in molecularly defined cell types. Nature 598, 174–181. 10.1038/s41586-021-03941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crick FC, and Koch C (2005). What is the function of the claustrum? Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 1271–1279. 10.1098/rstb.2005.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narikiyo K, Mizuguchi R, Ajima A, Shiozaki M, Hamanaka H, Johansen JP, Mori K, and Yoshihara Y (2020). The claustrum coordinates cortical slow-wave activity. Nat. Neurosci. 23, 741–753. 10.1038/s41593-020-0625-7. [DOI] [PubMed] [Google Scholar]
- 10.Atlan G, Matosevich N, Peretz-Rivlin N, Yvgi I, Chen E, Kleinman T, Bleistein N, Sheinbach E, Groysman M, Nir Y, et al. (2021). Claustral projections to anterior cingulate cortex modulate engagement with the external world. Preprint at bioRxiv. 10.1101/2021.06.17.448649. [DOI] [Google Scholar]
- 11.Atlan G, Terem A, Peretz-Rivlin N, Sehrawat K, Gonzales BJ, Pozner G, Tasaka GI, Goll Y, Refaeli R, Zviran O, et al. (2018). The claustrum supports resilience to distraction. Curr. Biol. 28, 2752–2762.e7. 10.1016/j.cub.2018.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodoulian L, Gschwend O, Huber C, Mutel S, Salazar RF, Leone R, Renfer JR, Ekundayo K, Rodriguez I, and Carleton A (2020). The claustrum-medial prefrontal cortex network controls attentional set-shifting. Preprint at bioRxiv. 10.1101/2020.10.14.339259. [DOI] [Google Scholar]
- 13.Terem A, Gonzales BJ, Peretz-Rivlin N, Ashwal-Fluss R, Bleistein N, Del Mar Reus-Garcia M, Mukherjee D, Groysman M, and Citri A (2020). Claustral neurons projecting to frontal cortex mediate contextual association of reward. Curr. Biol. 30, 3522–3532.e6. 10.1016/j.cub.2020.06.064. [DOI] [PubMed] [Google Scholar]
- 14.Krimmel SR, White MG, Panicker MH, Barrett FS, Mathur BN, and Seminowicz DA (2019). Resting state functional connectivity and cognitive task-related activation of the human claustrum. Neuroimage 196, 59–67. 10.1016/j.neuroimage.2019.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White MG, Mu C, Qadir H, Madden MB, Zeng H, and Mathur BN (2020). The mouse claustrum is required for optimal behavioral performance under high cognitive demand. Biol. Psychiatry 88, 719–726. 10.1016/j.biopsych.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White MG, and Mathur BN (2018). Frontal cortical control of posterior sensory and association cortices through the claustrum. Brain Struct. Funct. 223, 2999–3006. 10.1007/s00429-018-1661-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson J, Smith JB, and Lee AK (2020). The anatomy and physiology of claustrum-cortex interactions. Annu. Rev. Neurosci. 43, 231–247. 10.1146/annurev-neuro-092519-101637. [DOI] [PubMed] [Google Scholar]
- 18.Smith JB, Lee AK, and Jackson J (2020). The claustrum. Curr. Biol. 30, R1401–R1406. 10.1016/j.cub.2020.09.069. [DOI] [PubMed] [Google Scholar]
- 19.McBride EG, Gandhi SR, Kuyat JR, Ollerenshaw DR, Arkhipov A, Koch C, and Olsen SR (2023). Influence of claustrum on cortex varies by area, layer, and cell type. Neuron 111, 275–290.e5. 10.1016/j.neuron.2022.10.026. [DOI] [PubMed] [Google Scholar]
- 20.Shelton AM, Oliver DK, Grimstvedt JS, Lazarte IP, Kapoor I, Swann JA, Ashcroft CA, Williams SN, Conway N, Robinson A, et al. (2022). Single neurons and networks in the claustrum integrate input from widespread cortical sources. Preprint at bioRxiv. 10.1101/2022.05.06.490864. [DOI] [Google Scholar]
- 21.Paxinos G, and Franklin KBJ (2001). The Mouse Brain in Stereotaxic Coordinates (Elsevier Academic Press; ). [Google Scholar]
- 22.Real MA, Dávila JC, and Guirado S (2006). Immunohistochemical localization of the vesicular glutamate transporter VGLUT2 in the developing and adult mouse claustrum. J. Chem. Neuroanat. 31, 169–177. 10.1016/j.jchemneu.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Atlan G, Terem A, Peretz-Rivlin N, Groysman M, and Citri A (2017). Mapping synaptic cortico-claustral connectivity in the mouse. J. Comp. Neurol. 525, 1381–1402. 10.1002/cne.23997. [DOI] [PubMed] [Google Scholar]
- 24.Binks D, Watson C, and Puelles L (2019). A re-evaluation of the anatomy of the claustrum in rodents and primates-analyzing the effect of pallial expansion. Front. Neuroanat. 13, 34. 10.3389/fnana.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graf M, Nair A, Wong KL, Tang Y, and Augustine GJ (2020). Identification of mouse claustral neuron types based on their intrinsic electrical properties. eNeuro 7, 0216–0220.2020. 10.1523/ENEURO.0216-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marriott BA, Do AD, Zahacy R, and Jackson J (2021). Topographic gradients define the projection patterns of the claustrum core and shell in mice. J. Comp. Neurol. 529, 1607–1627. 10.1002/cne.25043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erwin SR, Bristow BN, Sullivan KE, Kendrick RM, Marriott B, Wang L, Clements J, Lemire AL, Jackson J, and Cembrowski MS (2021). Spatially patterned excitatory neuron subtypes and projections of the claustrum. Elife 10, e68967. 10.7554/eLife.68967.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimstvedt JS, Shelton AM, Hoerder-Suabedissen A, Oliver DK, Berndtsson CH, Blankvoort S, Nair RR, Packer AM, Witter MP, and Kentros CG (2022). A multifaceted architectural framework of the mouse claustrum complex. Preprint at bioRxiv. 10.1101/2022.06.02.494429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ham GX, and Augustine GJ (2022). Topographically organized networks reflect functional modularization. Front. Neuroanat. 16, 901807. 10.3389/fnana.2022.901807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong HW (2008). Allen Reference Atlas: A Digital Color Brain Atlas of the C57BL/6J Male Mouse (John Wiley & Sons Inc; ). [Google Scholar]
- 31.Real MA, Dávila JC, and Guirado S (2003). Expression of calcium-binding proteins in the mouse claustrum. J. Chem. Neuroanat. 25, 151–160. 10.1016/s0891-0618(02)00104-7. [DOI] [PubMed] [Google Scholar]
- 32.Dillingham CM, Jankowski MM, Chandra R, Frost BE, and O’Mara SM (2017). The claustrum: considerations regarding its anatomy, functions and a programme for research. Brain and Neurosci. Adv. 1, 1–9. 10.1177/2398212817718962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Ding SL, Li Y, Royall J, Feng D, Lesnar P, Graddis N, Naeemi M, Facer B, Ho A, et al. (2020). The allen mouse brain common coordinate framework: a 3D reference atlas. Cell 181, 936–953.e20. 10.1016/j.cell.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edelstein LR, and Denaro FJ (2004). The claustrum: a historical review of its anatomy, physiology, cytochemistry and functional significance. Cell. Mol. Biol. 50, 675–702. [PubMed] [Google Scholar]
- 35.Druga R (2014). The structure and connection of the claustrum. In InThe claustrum: Structural, Functional, and Clinical Neuroscience, Smythies JR, Edelstein LR, and Ramachandran VS, eds. (Elsevier Academic Press; ), pp. 29–84. [Google Scholar]
- 36.Olson CR, and Graybiel AM (1980). Sensory maps in the claustrum of the cat. Nature 288, 479–481. 10.1038/288479a0. [DOI] [PubMed] [Google Scholar]
- 37.Pearson RC, Brodal P, Gatter KC, and Powell TP (1982). The organization of the connections between the cortex and the claustrum in the monkey. Brain Res. 234, 435–441. 10.1016/0006-8993(82)90883-6. [DOI] [PubMed] [Google Scholar]
- 38.Macchi G, Bentivoglio M, Minciacchi D, and Molinari M (1983). Claus-troneocortical projections studied in the cat by means of multiple retrograde fluorescent tracing. J. Comp. Neurol. 215, 121–134. 10.1002/cne.902150202. [DOI] [PubMed] [Google Scholar]
- 39.Fernández-Miranda JC, Rhoton AL Jr., Kakizawa Y, Choi C, and Alvarez-Linera J (2008). The claustrum and its projection system in the human brain: a microsurgical and tractographic anatomical study. J. Neurosurg. 108, 764–774. 10.3171/JNS/2008/108/4/0764. [DOI] [PubMed] [Google Scholar]
- 40.Milardi D, Bramanti P, Milazzo C, Finocchio G, Arrigo A, Santoro G, Trimarchi F, Quartarone A, Anastasi G, and Gaeta M (2015). Cortical and subcortical connections of the human claustrum revealed in vivo by constrained spherical deconvolution tractography. Cereb. Cortex 25, 406–414. 10.1093/cercor/bht231. [DOI] [PubMed] [Google Scholar]
- 41.Reser DH, Majka P, Snell S, Chan JMH, Watkins K, Worthy K, Quiroga MDM, and Rosa MGP (2017). Topography of claustrum and insula projections to medial prefrontal and anterior cingulate cortices of the common marmoset (Callithrix jacchus). J. Comp. Neurol. 525, 1421–1441. 10.1002/cne.24009. [DOI] [PubMed] [Google Scholar]
- 42.Shepherd GMG (2013). Corticostriatal connectivity and its role in disease. Nat. Rev. Neurosci. 14, 278–291. 10.1038/nrn3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris KD, and Shepherd GMG (2015). The neocortical circuit: themes and variations. Nat. Neurosci. 18, 170–181. 10.1038/nn.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker A, Kalmbach B, Morishima M, Kim J, Juavinett A, Li N, and Dembrow N (2018). Specialized subpopulations of deep-layer pyramidal neurons in the neocortex: bridging cellular properties to functional consequences. J. Neurosci. 38, 5441–5455. 10.1523/JNEUROSCI.0150-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cotel F, Fletcher LN, Kalita-de Croft S, Apergis-Schoute J, and Williams SR (2018). Cell class-dependent intracortical connectivity and output dynamics of layer 6 projection neurons of the rat primary visual cortex. Cereb. Cortex 28, 2340–2350. 10.1093/cercor/bhx134. [DOI] [PubMed] [Google Scholar]
- 46.Smith JB, Klug JR, Ross DL, Howard CD, Hollon NG, Ko VI, Hoffman H, Callaway EM, Gerfen CR, and Jin X (2016). Genetic-based dissection unveils the inputs and outputs of striatal patch and matrix compartments. Neuron 91, 1069–1084. 10.1016/j.neuron.2016.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim EJ, Juavinett AL, Kyubwa EM, Jacobs MW, and Callaway EM (2015). Three types of cortical layer 5 neurons that differ in brain-wide connectivity and function. Neuron 88, 1253–1267. 10.1016/j.neuron.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Economo MN, Viswanathan S, Tasic B, Bas E, Winnubst J, Menon V, Graybuck LT, Nguyen TN, Smith KA, Yao Z, et al. (2018). Distinct descending motor cortex pathways and their roles in movement. Nature 563, 79–84. 10.1038/s41586-018-0642-9. [DOI] [PubMed] [Google Scholar]
- 49.Chia Z, Silberberg G, and Augustine GJ (2017). Functional properties, topological organization and sexual dimorphism of claustrum neurons projecting to anterior cingulate cortex. Claustrum 2, 1357412. 10.1080/20023294.2017.1357412. [DOI] [Google Scholar]
- 50.Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. (2014). A mesoscale connectome of the mouse brain. Nature 508, 207–214. 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris JA, Mihalas S, Hirokawa KE, Whitesell JD, Choi H, Bernard A, Bohn P, Caldejon S, Casal L, Cho A, et al. (2019). Hierarchical organization of cortical and thalamic connectivity. Nature 575, 195–202. 10.1038/s41586-019-1716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith JB, Alloway KD, Hof PR, Orman R, Reser DH, Watakabe A, and Watson GDR (2019). The relationship between the claustrum and endopiriform nucleus: a perspective towards consensus on cross-species homology. J. Comp. Neurol. 527, 476–499. 10.1002/cne.24537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao S, Wang Q, Hirokawa KE, Ouellette B, Ahmed R, Bomben J, Brouner K, Casal L, Caldejon S, Cho A, et al. (2022). A whole-brain monosynaptic input connectome to neuron classes in mouse visual cortex. Nat. Neurosci. 10.1038/s41593-022-01219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watakabe A, Ohsawa S, Ichinohe N, Rockland KS, and Yamamori T (2014). Characterization of claustral neurons by comparative gene expression profiling and dye-injection analyses. Front. Syst. Neurosci. 8, 98. 10.3389/fnsys.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.LeVay S, and Sherk H (1981). The visual claustrum of the cat. I. Structure and connections. J. Neurosci. 1, 956–980. 10.1523/JNEUROSCI.01-09-00956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baizer JS, Sherwood CC, Noonan M, and Hof PR (2014). Comparative organization of the claustrum: what does structure tell us about function? Front. Syst. Neurosci. 8, 117. 10.3389/fnsys.2014.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reser DH, Richardson KE, Montibeller MO, Zhao S, Chan JMH, Soares JGM, Chaplin TA, Gattass R, and Rosa MGP (2014). Claustrum projections to prefrontal cortex in the capuchin monkey (Cebus apella). Front. Syst. Neurosci. 8, 123. 10.3389/fnsys.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park S, Tyszka JM, and Allman JM (2012). The claustrum and insula in microcebus murinus: a high resolution diffusion imaging study. Front. Neuroanat. 6, 21. 10.3389/fnana.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gehrlach DA, Weiand C, Gaitanos TN, Cho E, Klein AS, Hennrich AA, Conzelmann KK, and Gogolla N (2020). A whole-brain connectivity map of mouse insular cortex. Elife 9, e55585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gogolla N (2017). The insular cortex. Curr. Biol. 27, 580–586. 10.1016/j.cub.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 61.Watson C, and Puelles L (2017). Developmental gene expression in the mouse clarifies the organization of the claustrum and related endopiriform nuclei. J. Comp. Neurol. 525, 1499–1508. 10.1002/cne.24034. [DOI] [PubMed] [Google Scholar]
- 62.Qadir H, Krimmel SR, Mu C, Poulopoulos A, Seminowicz DA, and Mathur BN (2018). Structural connectivity of the anterior cingulate cortex, claustrum, and the anterior Insula of the mouse. Front. Neuroanat. 12, 100–112. 10.3389/fnana.2018.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suárez R, Paolino A, Fenlon LR, Morcom LR, Kozulin P, Kurniawan ND, and Richards LJ (2018). A pan-mammalian map of interhemispheric brain connections predates the evolution of the corpus callosum. Proc. Natl. Acad. Sci. USA 115, 9622–9627. 10.1073/pnas.1808262115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbier M, and Risold PY (2019). The claustrum is a target for projections from the supramammillary nucleus in the rat. Neuroscience 409, 261–275. 10.1016/j.neuroscience.2019.03.045. [DOI] [PubMed] [Google Scholar]
- 65.Chia Z, Augustine GJ, and Silberberg G (2020). Synaptic connectivity between the cortex and claustrum is organized into functional modules. Curr. Biol. 30, 2777–2790.e4. 10.1016/j.cub.2020.05.031. [DOI] [PubMed] [Google Scholar]
- 66.Reus-García MM, Sánchez-Campusano R, Ledderose J, Dogbevia GK, Treviño M, Hasan MT, Gruart A, and Delgado-García JM (2021). The claustrum is involved in cognitive processes related to the classical conditioning of eyelid responses in behaving rabbits. Cereb. Cortex 31, 281–300. 10.1093/cercor/bhaa225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White MG, Cody PA, Bubser M, Wang HD, Deutch AY, and Mathur BN (2017). Cortical hierarchy governs rat claustrocortical circuit organization. J. Comp. Neurol. 525, 1347–1362. 10.1002/cne.23970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.White MG, Panicker M, Mu C, Carter AM, Roberts BM, Dharmasri PA, and Mathur BN (2018). Anterior cingulate cortex input to the claustrum is required for top-down action control. Cell Rep. 22, 84–95. 10.1016/j.celrep.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gutierrez-Ibarluzea I, Acera-Osa A, Mendizabal-Zubiaga JL, Arana-Arri E, Bueno-Lopez JL, and Reblet C (1999). Morphology and laminar distribution of cortico-claustral neurons in different areas of the rabbit cerebral cortex. Eur. J. Anat. 3, 101–109. [Google Scholar]
- 70.Zhang X, Hannesson DK, Saucier DM, Wallace AE, Howland J, and Corcoran ME (2001). Susceptibility to kindling and neuronal connections of the anterior claustrum. J. Neurosci. 21, 3674–3687. 10.1523/JNEUROSCI.21-10-03674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minciacchi D, Molinari M, Bentivoglio M, and Macchi G (1985). The organization of the ipsi- and contralateral claustrocortical system in rat with notes on the bilateral claustrocortical projections in cat. Neuroscience 16, 557–576. 10.1016/0306-4522(85)90192-7. [DOI] [PubMed] [Google Scholar]
- 72.Li ZK, Takada M, and Hattori T (1986). Topographic organization and collateralization of claustrocortical projections in the rat. Brain Res. Bull. 17, 529–532. 10.1016/0361-9230(86)90220-0. [DOI] [PubMed] [Google Scholar]
- 73.Holschneider DP, Wang Z, and Pang RD (2014). Functional connectivity-based parcellation and connectome of cortical midline structures in the mouse: a perfusion autoradiography study. Front. Neuroinform. 8, 61. 10.3389/fninf.2014.00061.eCollection2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vanni MP, Chan AW, Balbi M, Silasi G, and Murphy TH (2017). Mesoscale mapping of mouse cortex reveals frequency-dependent cycling between distinct macroscale functional modules. J. Neurosci. 37, 7513–7533. 10.1523/JNEUROSCI.3560-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carey RG, Fitzpatrick D, and Diamond IT (1979). Layer I of the striate cortex of tupaia glis and galago senegalensis: projections from thalamus and claustrum revealed by retrograde transport of horseradish peroxidase. J. Comp. Neurol. 186, 393–437. 10.1002/cne.901860306. [DOI] [PubMed] [Google Scholar]
- 76.LeVay S (1986). Synaptic organization of claustral and geniculate afferents to the visual cortex of the cat. J. Neurosci. 6, 3564–3575. 10.1523/JNEUROSCI06-12-03564.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sherk H (1986). The claustrum and the cerebral cortex. In Cerebral Cortex, Vol 5, Sensorimotor Areas and Aspects of Cortical Connectivity, Jones EG and Peters A, eds. (Plenum Publication; ), pp. 467–499. [Google Scholar]
- 78.Clascá F, Avendaño C, Román-Guindo A, Llamas A, and Reinoso-Suárez F (1992). Innervation from the claustrum of the frontal association and motor areas: axonal transport studies in the cat. J. Comp. Neurol. 326, 402–422. 10.1002/cne.903260307. [DOI] [PubMed] [Google Scholar]
- 79.da Costa NM, Fursinger €D, and Martin KAC (2010). The synaptic organization of the claustral projection to the cat’s visual cortex. J. Neurosci. 30, 13166–13170. 10.1523/JNEUROSCI.3122-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jackson J, Karnani MM, Zemelman BV, Burdakov D, and Lee AK (2018). Inhibitory control of prefrontal cortex by the claustrum. Neuron 99, 1029–1039.e4. 10.1016/j.neuron.2018.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brennan EKW, Jedrasiak-Cape L, Kailasa K, Rice SP, Sudhakar SK, and Ahmed OJ (2021). Thalamus and claustrum control parallel layer 1 circuits in the retrosplenial cortex. Elife. 10.7554/eL-ife.62207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qadir H, Stewart BW, VanRyzin JW, Wu Q, Chen S, Seminowicz DA, and Mathur BN (2022). The mouse claustrum synaptically connects cortical network motifs. Cell Rep. 41, 111860. 10.1016/j.cjca.2022.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van der Werf YD, Witter MP, and Groenewegen HJ (2002). The intra-laminar and midline nuclei of the thalamus. Anatomical and functional evidence for participation in processes of arousal and awareness. Brain Res. Brain Res. Rev. 39, 107–140. 10.1016/s0165-0173(02)00181-9. [DOI] [PubMed] [Google Scholar]
- 84.Saalmann YB (2014). Intralaminar and medial thalamic influence on cortical synchrony, information transmission and cognition. Front. Syst. Neurosci. 8, 83. 10.3389/fnsys.2014.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wolff M, and Vann SD (2019). The cognitive thalamus as a gateway to mental representations. J. Neurosci. 39, 3–14. 10.1523/JNEUROSCI.0479-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Selden NR, Gitelman DR, Salamon-Murayama N, Parrish TB, and Mesulam MM (1998). Trajectories of cholinergic pathways within the cerebral hemispheres of the human brain. Brain 121, 2249–2257. 10.1093/brain/121.12.2249. [DOI] [PubMed] [Google Scholar]
- 87.Baizer JS (2001). Serotonergic innervation of the primate claustrum. Brain Res. Bull. 55, 431–434. 10.1016/s0361-9230(01)00535-4. [DOI] [PubMed] [Google Scholar]
- 88.Pirone A, Miragliotta V, Ciregia F, Giannessi E, and Cozzi B (2018). The catecholaminergic innervation of the claustrum of the pig. J. Anat. 232, 158–166. 10.1111/joa.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sitte HH, Pifl C, Rajput AH, Hörtnagl H, Tong J, Lloyd GK, Kish SJ, and Hornykiewicz O (2017). Dopamine and noradrenaline, but not serotonin, in the human claustrum are greatly reduced in patients with Parkinson’s disease: possible functional implications. Eur. J. Neurosci. 45, 1356–2197. 10.1111/ejn.13573. [DOI] [PubMed] [Google Scholar]
- 90.Wong KLL, Nair A, and Augustine GJ (2021). Changing the cortical conductor’s tempo: neuromodulation of the claustrum. Front. Neural Circuits 15, 658228. 10.3389/fncir.2021.658228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barbier M, Houdayer C, Franchi G, Poncet F, and Risold PY (2017). Melanin-concentrating hormone axons, but not orexin or tyrosine hydroxylase axons, innervate the claustrum in the rat: an immunohistochemical study. J. Comp. Neurol. 525, 1489–1498. 10.1002/cne.24110. [DOI] [PubMed] [Google Scholar]
- 92.Nair A, Graf M, Teo YY, and Augustine GJ (2021). A functional logic for neurotransmitter co-release in the cholinergic forebrain pathway. Preprint at bioRxiv. 10.1101/2021.02.25.432623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barrett FS, Krimmel SR, Griffiths RR, Seminowicz DA, and Mathur BN (2020). Psilocybin acutely alters the functional connectivity of the claustrum with brain networks that support perception, memory, and attention. Neuroimage 218, 116980. 10.1016/j.neuro-image.2020.116980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Doss MK, Madden MB, Gaddis A, Nebel MB, Griffiths RR, Mathur BN, and Barrett FS (2022). Models of psychedelic drug action: modulation of cortical-subcortical circuits. Brain 145, 441–456. 10.1093/brain/awab406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Çavdar S,Özgur €M, Kuvvet Y, Bay H, and Aydogmus E (2018). Cortical, subcortical and brain stem connections of the cerebellum via the superior and middle cerebellar peduncle in the rat. J. Integr. Neurosci. 17, 609–618. 10.3233/JIN-180090. [DOI] [PubMed] [Google Scholar]
- 96.Lévesque M, and Parent A (1998). Axonal arborization of corticostriatal and corticothalamic fibers arising from prelimbic cortex in the rat. Cereb. Cortex 8, 602–613. 10.1093/cercor/8.7.602. [DOI] [PubMed] [Google Scholar]
- 97.Kitanishi T, and Matsuo N (2017). Organization of the claustrum-to-entorhinal cortical connection in mice. J. Neurosci. 37, 269–280. 10.1523/JNEUROSCI.1360-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu J, Wu R, Johnson B, Vu J, Bass C, and Li JX (2019). The claustrum-prefrontal cortex pathway regulates impulsive-like behavior. J. Neurosci. 39, 10071–10080. 10.1523/JNEUROSCI.1005-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. (2017). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper are publicly available through the Allen institute portal (http://portal.brain-map.org/) and the archive Brain Imaging library (BIL, https://www.brainimagelibrary.org/).
No original code has been generated.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


