Abstract
Problem
Human papillomavirus infection is integral to developing invasive cervical cancer in the majority of patients. In a recent genome-wide association study, rs9357152 and rs4243652 have been associated with seropositivity for HPV16 or HPV18, respectively. It is unknown whether these variants also associate with cervical cancer triggered by either HPV16 or HPV18.
Methods
We investigate whether the two HPV susceptibility variants show association with type-specific cervical cancer in a genetic case-control study with cases stratified by HPV16 or HPV18, respectively. We further tested whether rs9357152 modulates gene expression of any of 36 genes at the human leukocyte antigen locus in 256 cervical tissues.
Results
rs9357152 was associated with invasive HPV16-positive cervical cancer (OR 1.33, 95%CI 1.03–1.70, p = 0.03), and rs4243652 was associated with HPV18-positive adenocarcinomas (OR 2.96, 95%CI 1.18–7.41, p = 0.02). These associations remained borderline significant after testing against different sets of controls. rs9357152 was found to be an eQTL for HLA-DRB1 in HPV-positive cervical tissues (pANOVA = 0.0009), with the risk allele lowering mRNA levels.
Conclusions
We find evidence that HPV seropositivity variants at chromosome 6 and 14 may modulate type-specific cervical cancer risk. rs9357152 may exert its effect through regulating HLA-DRB1 induction in the presence of HPV. In regard of multiple testing, these results need to be confirmed in larger studies.
Keywords: Human leukocyte antigen, Single nucleotide polymorphism, Association study, HPV infection, eQTL, VASH1
Highlights
-
•
Recent GWASs identified genetic risk factors associated with HPV infection.
-
•
These variants replicate in a German case-control series for cervical cancer and dysplasia.
-
•
The risk variant on chromosome 6 associates with decreased HLA-DRB1 expression levels in cervical tissue samples.
-
•
Unexplained genetic risk for cervical cancer may be informed by large scale studies of HPV seropositivity.
List of abbreviations used
- cDNA
Complementary DNA
- CI
Confidence interval
- CIN
Cervical intraepithelial neoplasia
- CIS
Carcinoma in situ
- EDTA
Ethylenediaminetetraacetic Acid
- eQTL
Expression quantitative trait loci
- GWAS
Genome wide association study
- HLA
Human Leukocyte Antigen
- HPV
Human papillomavirus
- HWE
Hardy Weinberg Equilibrium
- ICC
Invasive cervical cancer
- MAF
Minor Allele frequency
- OR
Odds ratio
- qRT-PCR
Quantitative real time PCR
- SNP
Single nucleotide polymorphism
1. Introduction
Cervical cancer is the fourth most prevalent cancer in women worldwide [1]. A persistent infection with high-risk human papillomavirus types (HPV 16, 18, 31, 33, 52, 58, among others) is seen in most invasive cervical cancers [2]. Not every infected woman develops invasive cancer, pointing to underlying environmental or lifestyle factors or genetic risk factors involved in persistent infection, failure to clear the viral load and induction of tumor formation [2]. Among various genome-wide association studies (GWASs) published for cervical cancer so far [[3], [4], [5], [6], [7], [8], [9], [10]], the majority of signals arise at the Human Leukocyte Antigen locus (HLA) at 6p21.32–33. Other GWASs and follow up studies for human papillomavirus seropositivity have identified type-specific signals at the same chromosomal region [5,11,12]. Chen et al. reported a variant, rs9357152 at 6p21.32, to be associated with HPV8 seropositivity [11], which was replicated in a recent HPV16 seropositivity GWAS from the UK biobank [12]. The latter study also identified a novel locus potentially associated with HPV18 seropositivity at 14q24.3 (rs4243652) [12]. Here, we test the association of these two variants with cervical cancer and dysplasia in HPV16- and HPV18- positive samples, respectively, from the German Cervigen cohort in a case-control study. We additionally test the impact of the chromosome 6 variant on cervical mRNA transcript levels of 36 genes at the HLA locus using eQTL analyses.
2. Materials and methods
2.1. Patient material
We investigated a total of 395 cancer cases from the German Cervigen consortium [[13], [19]], including samples that had been collected in nine German hospitals in Hannover, Wolfsburg, Jena, Erlangen, Dresden, Halle, Munich, Berlin and Bad Münder. The HPV status of the patients was taken as an inclusion criterion: only cases with HPV16-positive status were tested for the HPV16 seropositivity variant rs9357152, and only cases with HPV18-positive status were taken for genotyping the HPV18 seropositivity variant rs4243652. This stratification served to avoid dilution of effects and thereby increase the power of the study to detect potentially specific associations. 26 samples were excluded because they had double infections with HPV16 and HPV18 so that the viral driver could not be unambiguously defined. We finally included 307 patients with cancer or dysplasia having a HPV16-positive test as cases for testing rs9357152. Similarly, for testing rs4243652, a total of 88 HPV18-positive tested cervical cancer or dysplasia patients were taken.
The main comparison control series consisted of 569 healthy females and was the same for both case groups. These samples were randomly drawn from the Cervigen study [19] and were genotyped by PCR-RFLP in the same way as the cases. Ten women with non-European ancestry were excluded from statistical analysis (5 cases and 5 controls), leaving 564 cancer-free healthy controls as “control group 1”. We also defined a second control series, “control group 2”, of 379 independent samples that had been genotyped on the OncoArray during a previous GWAS [21]. In that GWAS, rs9357152 had been directly genotyped and rs4243652 had been imputed. Finally, we defined a third control series, “control group 3”, of known HPV-negative status from a cervical tissue collective derived from women undergoing routine colposcopy (see below). After exclusion of women with a previous cervical dysplasia or cancer, we remained with 129 samples in the third control group. Age characteristics differed between cases with dysplasia or invasive cancer as well as within the three control series and are summarized in Supplementary Table 1.
From our cervical tissue collective (n = 256), RNA, genomic DNA and HPV status information was available, as described previously [[13], [19]]. From total RNA, cDNA was synthesized for gene expression analysis. The samples were stratified into HPV-negative or HPV-positive groups (16+, 18+, or other high risk HPV type+), along with lesion negative or positive subgroups (where the lesion status PAPIII and above was considered positive). These cDNA were taken for gene expression analyses.
2.2. Methods
2.2.1. SNP genotyping
Genomic DNA from peripheral white blood cells was isolated from 5 ml venous EDTA blood from each patient using standard phenol-chloroform extraction and subjected to SNP genotyping via restriction fragment length polymorphism (RFLP) analysis. Primer pairs were designed for both rs4243652(A/G) and rs9357152(C/T) as follows: for rs4243652, a 168bp fragment was PCR amplified with 5′-GAG TAG AAG CAG CTG ATT CCT TC-3′ and 5′-CCT TCC TTC AAG CAC AGA CAC-3′, and PCR products were digested with the enzyme Hpy188III (TC^NNGA, New England Biolabs). For rs9357152, a 162bp fragment was amplified with 5′-GAT GAA TTT TAT GTC CAA ATC TAG GC-3′ and 5′-GGA GAA ATT GAA GAT CCC TGA TTG C-3′ and digested with the enzyme Sau96I (G^GNCC, New England Biolabs). The digested products were separated on a 2% agarose gel containing GelRed, and visualized with a UV transilluminator (Intas) (Supplementary Fig. 1). For rs9357152, 560 healthy controls and 333 HPV16-positive cancer/dysplasia samples were successfully genotyped (call rate 98.9%), and for rs4243652, 558 healthy controls and 114 HPV18-positive cancer/dysplasia cases were successfully genotyped (call rate 98.8%). Twenty-six samples were both HPV16- and HPV18-positive and have been genotyped for both variants but were excluded from statistical analysis later.
2.2.2. Sanger sequencing
Some samples having common homozygous, rare homozygous and heterozygous genotypes for each of the two variants were randomly selected for validation sequencing. PCR-products were sequenced using BigDye™ Terminator chemistry on a SeqStudio Genetic Analyzer (Applied Biosystems/Thermo Fisher Scientific), and sequencing results were analysed using Sequencing Analysis 5.1.1 software (Applied Biosystems) and FinchTV v1.4.0 (Geospiza). All sequencing results were in concordance with RFLP genotyping results (Supplementary Fig. 2).
2.2.3. Statistical analysis of genetic data
We performed logistic regression analyses using STATA v17 with case-control status as the outcome and genotype as predictor variable. The median age at diagnosis for cases overall was 38 years (range 17–78 years). A stratified analysis was performed based on the disease severity. For this, samples were grouped into LSIL/low-grade dysplasia (CIN1 together with CIN2 patients at age <30 years (CIN2<30)), HSIL/high-grade dysplasia (CIN2 cases at age ≥30 years (CIN2≥30) together with CIN3 patients), and invasive cervical cancer. Invasive carcinomas were further grouped histologically into squamous cell carcinoma or adenocarcinoma. Odds ratios were calculated under an allelic model and reported with 95% confidence intervals. In regard of the prior identification of the two variants under study as seropositivity variants, two-sided P-values below 0.05 were regarded as confirmatory evidence. All stratified analyses were performed with control group 1. Results at p < 0.05 were then tested for robustness against control bias in additional logistic regression analyses using the control groups 2 and 3 for separate and combined comparisons with cases in the stratum of interest.
2.2.4. Transcript expression and eQTL analysis
For the 256 cervical tissue samples where the genotype of rs9357152 was available, gene expression analysis was performed using cDNA for genes of interest at chromosome 6 (Supplementary Table 2). Data was generated after real time qPCR on the Fluidigm Biomark HD system using DeltaGene assays (listed in Supplementary Table 2) via 48.48 gene expression arrays. Normalization was performed using qBASE + software [14] with B2M and RPL13A as housekeeping genes. After removing outliers (ROUT method, GraphPad Prism v9.5.1), log10 normalized expression values were tested for association with the SNP genotype in eQTL analysis. For comparison of two groups, student's t-test was employed, whereas for comparing three groups, one way ANOVA was used, together with a linear test for trend. P-values less than 0.05 were reported. Since we tested 36 genes in three groups, multiple testing penalty was applied and only p-values below 0.0005 were considered to be statistically significant associations after Bonferroni correction. In case of some genes that were detected at very low levels in samples, an additional chi-square test was performed to test whether the SNP genotype predicts detection of gene transcripts.
3. Results
A total of 894 samples (333 HPV16-positive cases and 561 controls) were successfully genotyped for rs9357152, and 672 samples (114 HPV18-positive cases and 558 controls) were genotyped successfully for rs4243652 (Table 1). From these, 26 samples were both HPV16- and HPV18-positive, from which 13 samples harboured the rare allele of rs9357152 (10 heterozygotes, 3 rare homozygotes) and 2 samples harboured the rare allele of rs4243652 (1 heterozygote, 1 rare homozygote). These double positive patients were excluded from the study to increase the stringency of subtype analysis. Around 10% of all samples were repeated and showed 100% concordance in their genotype results. Both variants were in Hardy-Weinberg equilibrium (p > 0.05) in the tested cohort.
Table 1.
Association of rs9357152 and rs4243652 with case-control status.
| Stratum |
rs9357152 (HPV16+ve) |
rs4243652 (HPV18+ve) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ncases | Ncontrols | NTotal | OR | p | Ncases | Ncontrols | NTotal | OR | p | |
| Overall | 307 | 561 | 868 | 1.16 (0.93–1.43) | 0.19 | 88 | 558 | 646 | 1.56 (0.79–3.09) | 0.20 |
| LSIL (CIN1 + CIN2< 30y) | 19 | 561 | 580 | 0.52 (0.21–1.24) | 0.14 | 4 | 558 | 562 | n.a. | |
| HSIL (CIN2≥30y + CIN3) | 89 | 561 | 650 | 1.01 (0.71–1.43) | 0.97 | 11 | 558 | 569 | 1.11 (0.15–8.40) | 0.92 |
| Invasive | 197 | 561 | 758 | 1.33 (1.03–1.70) | 0.03 | 73 | 558 | 631 | 1.72 (0.84–3.52) | 0.14 |
| - Adenocarcinoma | 19 | 561 | 580 | 1.58 (0.81–3.07) | 0.18 | 27 | 558 | 585 | 2.96 (1.18–7.41) | 0.02 |
| - Squamous carcinoma | 165 | 561 | 726 | 1.25 (0.96–1.64) | 0.10 | 43 | 558 | 601 | 1.13 (0.40–3.23) | 0.81 |
Stratified logistic regression analyses restricted to the disease type. Cervical intraepithelial neoplasia was differentiated into LSIL/low-risk (CIN1 + CIN2<30) and HSIL/high-risk (CIN2≥30 + CIN3) groups. Invasive cervical cancer was stratified into squamous epithelial cell carcinoma or adenocarcinoma. High-risk dysplasia (CIN2≥30 + CIN3) and invasive cancer were also combined together. For the LSIL analysis for rs4243652, numbers of HPV 18 positive samples were too small for statistical analysis. CI, confidence interval; OR, odds ratio for minor allele; P, P value from logistic regression analysis.
We did not observe an overall association with cervical disease for either of the two variants. However, in stratified case-control comparisons of HPV16-positive patients and healthy controls using logistic regression, rs9357152 showed evidence of association with invasive cervical cancer (rs9357152: OR 1.33, 95% CI 1.03–1.70, p = 0.03) (Table 1). The effect of rs9357152 tended to increase with severity of disease (p = 0.02, linear-by-linear test for trend). For the less common variant rs4243652, we found evidence of a specific association with HPV18-positive adenocarcinomas (OR 2.96, 95%CI 1.18–7.41, p = 0.02) (Table 1). This association was stronger in women who were diagnosed with adenocarcinoma before the median age of 40 years (OR 5.31, 95%CI 1.83–15.39, p = 0.002).
We tested whether the possible associations above remained stable when using different sets of controls and compared the genotype distribution in HPV16-positive and HPV18-positive patients to 379 independent control females that had previously been genotyped on a GWAS array [21]. HPV16 seropositivity variant rs9357152 showed an effect in the same direction but the association with invasive cervical cancer was not significant when using this second control group (OR 1.16, 95%CI 0.89–1.59; p = 0.27). HPV18 seropositivity variant rs4243652 remained associated with adenocarcinoma (OR 2.72, 95%CI 1.03–7.18; p = 0.04), in particular with a diagnosis before age 40 (OR 5.29, 95%CI 1.68–16.65; p = 0.004). No significant associations were detected when cases were compared to a third control group consisting of PAP smear samples derived from HPV-negative women but the numbers were small (Table 2). Logistic regression analysis with all controls combined still revealed evidence for association of HPV16 seropositivity variant rs9357152 with invasive cervical cancer (OR 1.26, 95%CI 1.00–1.58, p = 0.049) and HPV18 seropositivity variant rs4243652 with adenocarcinoma (OR 2.49, 95%CI 1.02–6.06, p = 0.044) (Table 2). When analyses were adjusted for age, the association was not significant for rs9357152 but remained nominally significant for rs4243652 (OR 1.19, 95%CI 0.94–1.52; p = 0.19 for rs9357152 with invasive cervical cancer, OR 2.72, 95%CI 1.11–6.67; p = 0.03 for rs4243652 with adenocarcinoma).
Table 2.
Association results for rs9357152 and rs4243652 in distinct control groups.
| Control series |
rs9357152 (HPV16+ve invasive cancer) |
rs4243652 (HPV18+ve adenocarcinoma) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ncases | Ncontrols | NTotal | OR | p | Ncases | Ncontrols | NTotal | OR | p | |
| Control group 1 | 197 | 561 | 758 | 1.33 (1.03–1.70) | 0.03 | 27 | 558 | 585 | 2.96 (1.18–7.41) | 0.02 |
| Control group 2 | 197 | 379 | 576 | 1.16 (0.89–1.50) | 0.27 | 27 | 379 | 406 | 2.72 (1.02–7.18) | 0.04 |
| Control group 3 | 197 | 129 | 326 | 1.32 (0.94–1.86) | 0.11 | 27 | 119 | 146 | 1.30 (0.49–3.44) | 0.60 |
| Controls combined | 197 | 1069 | 1266 | 1.26 (1.00–1.58) | 0.05* | 27 | 1056 | 1083 | 2.49 (1.02–6.06) | 0.04 |
Logistic regression analyses restricted to either HPV16-positive invasive cancer (for rs9357152) or HPV18-positive adenocarcinoma (for rs4243652) and three distinct control groups. Control group 1: random controls from Cervigen, RFLP-genotyped; Control group 2: additional controls from Cervigen, array-genotyped; Control group 3: HPV-negative controls derived from cervical tissue cohort, RFLP-genotyped. CI, confidence interval; OR, odds ratio for minor allele; P, P value from logistic regression analysis. *Exact P value 0.049.
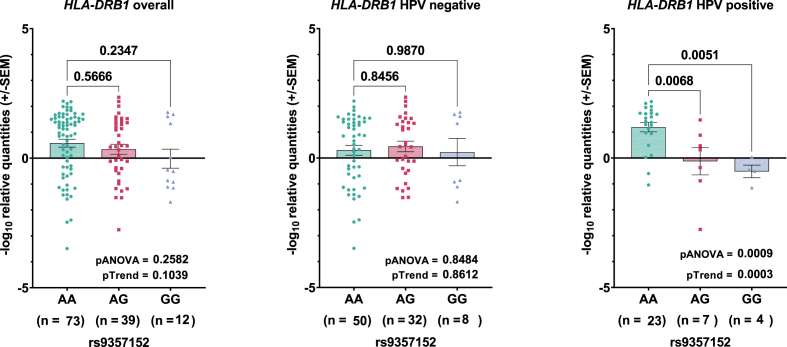
To understand how the HPV16 infection susceptibility variant rs9357152 works at the molecular level in cervical tissues, we performed eQTL analyses on the expression of 36 genes within the HLA locus on chromosome 6 (Supplementary Fig. 3). We identified potential eQTL effects on seven of the 36 targets, with the most significant association detected for transcript levels of HLA-DRB1 with rs9357152 (Fig. 1, Supplementary Fig. 4). Notably, rs9357152 was found to be a strong eQTL for HLA-DRB1 selectively in HPV-positive samples (pANOVA = 0.0009, pTrend = 0.0003) (Fig. 1).
Fig. 1.
Correlation of rs9357152 genotype with HLA-DRB1 levels in cervical tissue specimens. Association of transcript levels was tested overall and in HPV positive and negative tissues. P values after student's t-test between two groups or ANOVA between all groups is indicated.
4. Discussion
Genomic factors underlying susceptibility to cervical cancer have been sought in multiple candidate gene based studies and GWASs so far [8]. Previously reported susceptibility loci, mainly arising from the HLA region, appeared from studies where cervical cancer was the tested outcome, and as those studies usually do not stratify by the type of HPV infection, type-specific odds ratios are rarely provided [5,15,16]. Since a consistent infection by human papillomavirus is seen in almost all invasive cancer cases, GWASs for HPV seropositivity might be informative to identify type-specific genetic risk factors for developing CC. Previously, a HPV seropositivity GWAS had identified a signal on chromosome 6 for HPV8 seropositivity [11]. While this is not a high-risk HPV type for cervical cancer, a more recent GWAS from the UK Biobank replicated the same variant (rs9357152) for HPV16 seropositivity and further identified a novel potential hit on chromosome 14 for HPV18 seropositivity (rs4243652) [12]. This GWAS also identified a variant at the HLA locus, rs601148, that is correlated with the known risk variant rs9272143 [17] and closely linked variants associated with cervical cancer in multiple studies [6,7,18,19]. The example of rs601148 illustrates how a seropositivity GWAS is consistent with and can inform association studies for cervical cancer. However, the two other new candidate variants rs9357152 (HPV16 seropositivity) and rs4243652 (HPV18 seropositivity) did not emerge from genome-wide association studies of cervical cancer risk, suggesting that they may have been missed because the cases were not stratified by HPV type. This prompted us to perform the present stratified replication study in the Cervigen cohort where HPV type information is documented in the majority of cases. rs9357152 is located between the genes HLA-DQB1 and HLA-DQA2 on chromosome 6, whereas rs4243652 resides between the genes ESRRB and VASH1 on chromosome 14 and thus constitutes an independent promising novel locus.
Consistent with our hypothesis, rs9357152 was found to be associated with invasive carcinoma in the Cervigen cohort with the rare allele increasing the risk of cancer. At further inspection of available GWAS data, this variant also associated with cervical cancer in the Finnish biobank GWAS at sub-genome-wide level (p = 2 × 10E-4, malignant neoplasm of cervix uteri, https://r8.finngen.fi/variant/6:32697183-A-G), and in a meta-analysis performed in-house including five available cervical cancer GWAS datasets so far (p = 4 × 10E-4, datasets included: UK Biobank GWAS: 20001_1041.gwas.imputed_v3.female.tsv, Finnish biobank GWAS version 4: finngen_R4_C3_CERVIX_UTERI_EXALLC, CeC Japanese Biobank: Sakaue et al., 2020 [20], Leo et al., 2017 [5], and Cervigen 2022 [21]). The GTEx database lists eQTL evidence for rs9357152 in whole blood for multiple genes related to HLA-DQ, such as HLA-DQB2 (p = 5.2 × 10E-23), HLA-DQB1 (p = 8.9 × 10E-14), HLA-DQA2 (p = 1.3 × 10E-10), HLA-DQB1-AS1 (p = 6.0 × 10E-8) and HLA-DQA1 (p = 7.9 × 10E-8). However, we find that rs9357152 is an eQTL for HLA-DRB1 specifically in HPV-positive cervical tissues with the risk allele associating with decreasing levels of HLA-DRB1, located about 100 kBp apart. HLA-DRB1 belongs to class II MHC molecules that are constitutively expressed on antigen presenting cells. However, epithelial cells are also known to express HLA-DR genes, in particular when exposed to infection or inflammatory environment [[22], [23], [24]]. Our results thus raise the interesting possibility that this induced expression detected in HPV-positive samples is being modified by the risk genotype. The HLA-DRB1 gene is highly polymorphic with more than 4395 alleles discovered so far (https://www.ebi.ac.uk/ipd/imgt/hla/) and has been investigated in detail for association with cervical cancer [25] as well as with HPV titer and viral load in the cervix uteri [15,26]. While HLA-DRB1 alleles are mostly defined by amino acid variation, it is not without precedence that mRNA levels of this gene have been linked to a cervical cancer risk variant. A previous study on cervical cancer identified another variant at chromosome 6 (rs9272143) that acts as a cis-eQTL for HLA-DRB1 [27]. As the eQTL effect on HLA-DRB1 mediated via rs9357152 was only seen in HPV positive cells, lowered levels of HLA-DRB1 in the presence of the risk allele may contribute to immune evasion and a higher risk of developing invasive cancer.
rs4243652, discovered in a GWAS for HPV18 seropositivity by Kachuri and co-workers [12], was found to be associated with adenocarcinoma in our cohort and thus may contribute to the higher risk of adenocarcinomas that is observed for HPV18-positive women [28,29]. The variant showed minor evidence for replication (p = 0.01) in our meta-analysis performed in the five available cervical cancer GWAS datasets [5,20,21] including the Finnish, Japanese and UK biobanks as mentioned above. Notably, rs4243652 has up to ten-fold different minor allele frequencies in Europeans (MAF 0.04), Africans (MAF 0.45) and Asians (MAF 0.18) and thus might impact cervical cancer risk in a population-specific manner. The low MAF of the variant in our population precluded us from eQTL analyses in our tissue samples, but it is predicted to be a strong eQTL in whole blood for the Vasohibin-1 gene (VASH1) (p = 8.7 × 10E-8) in the GTEx browser, with the risk allele increasing VASH1 levels. Vasohibin-1 is an inhibitor of angiogenesis under the control of VEGF and FGF2 [30,31]. Levels of VASH1 were reported to be increased in cervical cancers [32], and high levels of VASH1 have also been observed and correlated with poor prognosis in head and neck squamous carcinomas, another HPV-associated tumor entity [32,33]. VASH1 is a predicted target for a HPV16 miRNA, HPV16-miR-H2-1 [34]. No such miRNA is known for HPV18, so that this HPV type may be more dependent on genomic host factors that regulate VASH1. Further studies will be needed to elucidate the specific interaction of rs4243652 with HPV18 at the molecular level.
Although the present study did not uncover an increased overall risk for cervical disease associated with the two tested variants, their potential associations with cervical cancer subgroups deserve attention. Limitations of our study included the relatively small sample size and the unavailability of HPV status in most of our population controls which did not permit a selection for HPV-negativity (“super-controls”) and could have obscured a stronger effect. While the fact that we were able to find evidence for both seropositivity variants as likely risk variants for cervical cancer is encouraging, the associations are marginally significant and do not survive correction for multiple testing of subgroups so that there is clearly a need for additional case-control studies on these variants in cohorts stratified by HPV type.
5. Conclusion
Cervical cancer is the paradigm for a virally induced cancer, with disease severity and progression attributed to the viral load, host immune response, lesion persistence and infection recurrence. In the present study, we link HPV16- and HPV18-specific seropositivity loci at 6p21.32 and 14q24.3, respectively, with disease risk of cervical cancer and provide some evidence for the HPV16-associated variant to act through HLA-DRB1. Our data indicate the existence of novel cervical cancer susceptibility loci that can be specific for HPV16 or HPV18 and warrant further investigation in cohorts with HPV-induced cancer.
Funding statement
F.S. received a stipend from the StrucMed program of the Hannover Biomedical Research School. D.R. received funding from the Tumorstiftung at Hannover Medical School. T.D. received funding from the Bruno and Helene Jöster foundation.
Ethics approval statement
This study was approved by the Ethics committee of Hannover Medical School (Votes 441 and 10737) and the samples as well as data used were in accordance with German medical council regulations.
Patient consent statement
Written informed consent was collected from each patient.
Author contributions
Finja Seifert, Rieke Eisenblätter, Julia Beckmann, Peter Schürmann: Formal analysis, Investigation, Visualization, Writing - original draft; Patricia Hanel: Data curation, Writing - original draft; Matthias Jentschke, Gerd Böhmer, Hans-Georg Strauβ, Christine Hirchenhain, Monika Schmidmayr, Florian Müller, Peter Fasching, Alexander Luyten, Norman Häfner, Matthias Dürst, Ingo B. Runnebaum, Peter Hillemanns: Resources, Data curation, Writing - original draft; Thilo Dörk: Conceptualization, Data curation, Funding acquisition, Resources, Supervision, Writing - original draft, Writing - review & editing; Dhanya Ramachandran: Conceptualization, Formal analysis, Funding acquisition, Investigation, Resources, Supervision, Visualization, Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Peter A. Fasching received grants from BioNTech and Cepheid, and personal fees from Novartis, Roche, Pfizer, Daiichi-Sankyo, Eisai, AstraZeneca, Merck Sharp & Dohme, Pierre Fabre, SeaGen, Hexal, Agenida and Lilly during the conduct of our study. This funding had no connection to this study and did not influence our study design and results. All other authors declare no conflicts of interest in the writing or preparation of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tvr.2023.200269.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Arbyn M., Weiderpass E., Bruni L., de Sanjosé S., Saraiya M., Ferlay J., et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. The Lancet Global Health. 2020;8(2):e191–e203. doi: 10.1016/S2214-109X(19)30482-6. https://linkinghub.elsevier.com/retrieve/pii/S2214109X19304826 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crosbie E.J., Einstein M.H., Franceschi S., Kitchener H.C. Human papillomavirus and cervical cancer. Lancet. 2013;382(9895):889–899. doi: 10.1016/S0140-6736(13)60022-7. [DOI] [PubMed] [Google Scholar]
- 3.Chen D., Juko-Pecirep I., Hammer J., Ivansson E., Enroth S., Gustavsson I., et al. Genome-wide association study of susceptibility loci for cervical cancer. J Natl Cancer Inst. 2013;105(9):624–633. doi: 10.1093/jnci/djt051. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y., Li L., Hu Z., Li S., Wang S.S., Liu J.J., et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat. Genet. 2013;45(8):918–922. doi: 10.1038/ng.2687. [DOI] [PubMed] [Google Scholar]
- 5.Leo P.J., Madeleine M.M., Wang S., Schwartz S.M., Newell F., Pettersson-Kymmer U., et al. Defining the genetic susceptibility to cervical neoplasia—a genome-wide association study. PLoS Genet. 2017;13(8):1–20. doi: 10.1371/journal.pgen.1006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rashkin S.R., Graff R.E., Kachuri L., Thai K.K., Alexeeff S.E., Blatchins M.A., et al. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-18246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowden S.J., Bodinier B., Kalliala I., Zuber V., Vuckovic D., Doulgeraki T., et al. Genetic variation in cervical preinvasive and invasive disease: a genome-wide association study. Lancet Oncol. 2021 Apr;22(4):548–557. doi: 10.1016/S1470-2045(21)00028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramachandran D., Dörk T. Genomic risk factors for cervical cancer. Cancers. 2021 Oct 13;13(20):5137. doi: 10.3390/cancers13205137. https://www.mdpi.com/2072-6694/13/20/5137 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miura Kiyonori, Mishima H., Kinoshita A., Hayashida C., Abe S., Tokunaga K., et al. Genome-wide association study of HPV-associated cervical cancer in Japanese women. J. Med. Virol. 2014;86:1153–1158. doi: 10.1002/jmv.23943. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi F., Kukimoto I., Li Z., Li S., Li N., Hu Z., et al. Genome-wide association study of cervical cancer suggests a role for ARRDC3 gene in human papillomavirus infection. Hum. Mol. Genet. 2019;28(2):341–348. doi: 10.1093/hmg/ddy390. [DOI] [PubMed] [Google Scholar]
- 11.Chen D., McKay J.D., Clifford G., Gaborieau V., Chabrier A., Waterboer T., et al. Genome-wide association study of HPV seropositivity. Hum. Mol. Genet. 2011;20(23):4714–4723. doi: 10.1093/hmg/ddr383. [DOI] [PubMed] [Google Scholar]
- 12.Kachuri L., Francis S.S., Morrison M.L., Wendt G.A., Bossé Y., Cavazos T.B., et al. The landscape of host genetic factors involved in immune response to common viral infections. Genome Med. 2020 Oct;12(1):93. doi: 10.1186/s13073-020-00790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramachandran D., Wang Y., Schürmann P., Hülse F., Mao Q., Jentschke M., et al. Association of genomic variants at PAX8 and PBX2 with cervical cancer risk. Int. J. Cancer. 2021;149(January):893–900. doi: 10.1002/ijc.33614. [DOI] [PubMed] [Google Scholar]
- 14.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2008;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beskow A.H., Moberg M., Gyllensten U.B. HLA class II allele control of HPV load in carcinoma in situ of the cervix uteri. Int. J. Cancer. 2005;117(3):510–514. doi: 10.1002/ijc.21204. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y., Zhang Q., Liu B., Yu G. The analysis of the entire HLA, partial non-HLA and HPV for Chinese women with cervical cancer. J. Med. Virol. 2008 Oct;80(10):1808. doi: 10.1002/jmv.21251. https://onlinelibrary.wiley.com/doi/10.1002/jmv.21251 13. Available from: [DOI] [PubMed] [Google Scholar]
- 17.Chen D., Juko-Pecirep I., Hammer J., Ivansson E., Enroth S., Gustavsson I., et al. Genome-wide association study of susceptibility loci for cervical cancer. J Natl Cancer Inst. 2013;105(9):624–633. doi: 10.1093/jnci/djt051. [DOI] [PubMed] [Google Scholar]
- 18.Chen D., Enroth S., Liu H., Sun Y., Wang H., Yu M., et al. Pooled analysis of genome-wide association studies of cervical intraepithelial neoplasia 3 (CIN3) identifies a new susceptibility locus. Oncotarget. 2016;7(27):42216–42224. doi: 10.18632/oncotarget.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramachandran D., Schürmann P., Mao Q., Wang Y., Bretschneider L.M., Speith L.M., et al. Association of genomic variants at the human leukocyte antigen locus with cervical cancer risk, HPV status and gene expression levels. Int. J. Cancer. 2020;147(9):2458–2468. doi: 10.1002/ijc.33171. [DOI] [PubMed] [Google Scholar]
- 20.Sakaue S., Hirata J., Kanai M., Suzuki K., Akiyama M., Lai Too C., et al. Dimensionality reduction reveals fine-scale structure in the Japanese population with consequences for polygenic risk prediction. Nat. Commun. 2020;11(1):1–11. doi: 10.1038/s41467-020-15194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramachandran D., Dennis J., Fachal L., Schürmann P., Bousset K., Hülse F., et al. Genome-wide association study and functional follow-up identify 14q12 as a candidate risk locus for cervical cancer. Hum. Mol. Genet. 2022 Aug 17;31(15):2483–2497. doi: 10.1093/hmg/ddac031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Axelrod M.L., Cook R.S., Johnson D.B., Balko J.M. Biological consequences of MHC-II expression by tumor cells in cancer. Clin. Cancer Res. 2019;25(8):2392–2402. doi: 10.1158/1078-0432.CCR-18-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wosen J.E., Mukhopadhyay D., Macaubas C., Mellins E.D. Epithelial MHC class II expression and its role in antigen presentation in the gastrointestinal and respiratory tracts. Front. Immunol. 2018 Sep 25;9:2144. doi: 10.3389/fimmu.2018.02144. https://www.frontiersin.org/article/10.3389/fimmu.2018.02144/full Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabibzadeh S. Induction of HLA-DR expression in endometrial epithelial cells by endometrial T-cells: potential regulatory role of endometrial T-cells in vivo. J. Clin. Endocrinol. Metab. 1991 Dec;73(6):1352. doi: 10.1210/jcem-73-6-1352. https://academic.oup.com/jcem/article-lookup/doi/10.1210/jcem-73-6-1352 9. Available from: [DOI] [PubMed] [Google Scholar]
- 25.Kamiza A.B., Kamiza S., Mathew C.G. HLA-DRB1 alleles and cervical cancer: a meta-analysis of 36 case-control studies. Cancer Epidemiol. 2020;67(March) doi: 10.1016/j.canep.2020.101748. [DOI] [PubMed] [Google Scholar]
- 26.Beskow A.H., Gyllensten U.B. Host genetic control of HPV 16 titer in carcinomain situ of the cervix uteri. Int. J. Cancer. 2002 Oct 20;101(6):526–531. doi: 10.1002/ijc.90010. https://onlinelibrary.wiley.com/doi/10.1002/ijc.90010 Available from: [DOI] [PubMed] [Google Scholar]
- 27.Chen D., Hammer J., Lindquist D., Idahl A., Gyllensten U. A variant upstream of HLA-DRB1 and multiple variants in MICA influence susceptibility to cervical cancer in a Swedish population. Cancer Med. 2014;3(1):190–198. doi: 10.1002/cam4.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodman C.B., Collins S., Rollason T.P., Winter H., Bailey A., Yates M., et al. Human papillomavirus type 18 and rapidly progressing cervical intraepithelial neoplasia. Lancet. 2003 Jan;361(9351):40. doi: 10.1016/S0140-6736(03)12120-4. https://www.sciencedirect.com/science/article/pii/S0140673603121204 3. Available from: [DOI] [PubMed] [Google Scholar]
- 29.Bulk S., Berkhof J., Bulkmans N.W.J., Zielinski G.D., Rozendaal L., van Kemenade F.J., et al. Preferential risk of HPV16 for squamous cell carcinoma and of HPV18 for adenocarcinoma of the cervix compared to women with normal cytology in The Netherlands. Br. J. Cancer. 2006 Jan 13;94(1):171. doi: 10.1038/sj.bjc.6602915. http://www.nature.com/articles/6602915 5. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato Y. The vasohibin family: a novel family for angiogenesis regulation. J. Biochem. 2013 Jan 1;153(1):5–11. doi: 10.1093/jb/mvs128. https://academic.oup.com/jb/article-lookup/doi/10.1093/jb/mvs128 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du H., Zhao J., Hai L., Wu J., Yi H., Shi Y. The roles of vasohibin and its family members: beyond angiogenesis modulators. Cancer Biol. Ther. 2017;18(11):827–832. doi: 10.1080/15384047.2017.1373217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshinaga K., Ito K., Moriya T., Nagase S., Takano T., Niikura H., et al. Roles of intrinsic angiogenesis inhibitor, vasohibin, in cervical carcinomas. Cancer Sci. 2011;102(2):446–451. doi: 10.1111/j.1349-7006.2010.01812.x. [DOI] [PubMed] [Google Scholar]
- 33.Torii C., Hida Y., Shindoh M., Akiyama K., Ohga N., Maishi N., et al. Vasohibin-1 as a novel prognostic factor for head and neck squamous cell carcinoma. Anticancer Res. 2017 Mar 17;37(3):1219–1226. doi: 10.21873/anticanres.11437. http://ar.iiarjournals.org/content/37/3/1219.abstract Available from: [DOI] [PubMed] [Google Scholar]
- 34.Qian K., Pietilä T., Rönty M., Michon F., Frilander M.J., Ritari J., et al. vol. 8. 2013 Jul 30. Identification and validation of human papillomavirus encoded microRNAs.https://dx.plos.org/10.1371/journal.pone.0070202 (PLoS One). 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.