Abstract
Objective
Both obesity and exposure to chemicals may induce non-alcoholic fatty liver disease (NAFLD). Pregnane X Receptor (PXR) is a central target of metabolism disrupting chemicals and disturbs hepatic glucose and lipid metabolism. We hypothesized that the metabolic consequences of PXR activation may be modified by existing obesity and associated metabolic dysfunction.
Methods
Wildtype and PXR knockout male mice were fed high-fat diet to induce obesity and metabolic dysfunction. PXR was activated with pregnenolone-16α-carbonitrile. Glucose metabolism, hepatosteatosis, insulin signaling, glucose uptake, liver glycogen, plasma and liver metabolomics, and liver, white adipose tissue, and muscle transcriptomics were investigated.
Results
PXR activation aggravated obesity-induced liver steatosis by promoting lipogenesis and inhibiting fatty acid disposal. Accordingly, hepatic insulin sensitivity was impaired and circulating alanine aminotransferase level increased. Lipid synthesis was facilitated by increased liver glucose uptake and utilization of glycogen reserves resulting in dissociation of hepatosteatosis and hepatic insulin resistance from the systemic glucose tolerance and insulin sensitivity. Furthermore, glucagon-induced hepatic glucose production was impaired. PXR deficiency did not protect from the metabolic manifestations of obesity, but the liver transcriptomics and metabolomics profiling suggest diminished activation of inflammation and less prominent changes in the overall metabolite profile.
Conclusions
Obesity and PXR activation by chemical exposure have a synergistic effect on NAFLD development. To support liver fat accumulation the PXR activation reorganizes glucose metabolism that seemingly improves systemic glucose metabolism. This implies that obese individuals, already predisposed to metabolic diseases, may be more susceptible to harmful metabolic effects of PXR-activating drugs and environmental chemicals.
Keywords: PXR, Steatosis, Insulin resistance, Endocrine disrupting chemicals, Glycogen
Highlights
-
•
PXR is a powerful metabolic regulator in obese male mice.
-
•
PXR activation and obesity have synergistic effect on NAFLD.
-
•
PXR-induced hepatosteatosis is supported by increased glucose uptake and glycogenolysis.
-
•
PXR activation dissociates NAFLD from systemic glucose metabolism.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is associated with metabolic syndrome in bidirectional manner. While metabolic syndrome, especially obesity, is a major risk factor for NAFLD, the NAFLD may also predispose to components and complications of metabolic syndrome, especially type 2 diabetes, and cardiovascular diseases [1]. Indeed, NAFLD is tightly linked to insulin resistance and glucose intolerance [1,2].
While NAFLD is most commonly triggered by obesity, other factors including exposure to chemicals affecting metabolic functions may also contribute to the rapidly rising prevalence of NAFLD [3]. These chemicals are often called endocrine disrupting chemicals (EDCs) or, more specifically, metabolism disrupting chemicals [3]. Potentially, obesity and the EDCs may have synergistic effects on the development of NAFLD.
Pregnane X Receptor (PXR, NR1I2) is a nuclear receptor activated by numerous, structurally variable chemicals, including multiple drugs and environmental chemicals, or mixtures of them, making it a central sensor of chemical exposure [4]. Originally, PXR was mainly studied as a regulator of drug metabolism [5]. More recently, its function has been extended to cover the regulation of lipid and glucose metabolism, and PXR is considered an important mediator of metabolic effects of EDCs [6,7]. Activation of PXR induces steatosis in mouse liver and in human hepatic cell models [8,9]. Furthermore, PXR activation impairs glucose tolerance both in animal models and humans [[10], [11], [12]]. PXR knockout has been shown to protect mice from diet-induced obesity and metabolic dysfunction by increasing energy consumption and mitochondrial β-oxidation [[13], [14], [15]]. However, in our recent study using a different sub-strain of C57BL/6 mice, we did not observe any effect of PXR deficiency on high-fat diet (HFD) induced weight gain [16].
In the current study, we investigated the metabolic consequences of PXR activation or PXR deficiency in a mouse model of diet-induced obesity. We have already previously described that PXR activation in these mice induces hepatic cholesterol level due to increase in cholesterol synthesis [16]. We now show that PXR activation aggravates liver steatosis and that this is supported by remodulation of glucose metabolism. Consequently, despite the intensified hepatic insulin resistance, this emerges as improvement of systemic glucose metabolism.
2. Materials and methods
2.1. Animal experiments
All experiments on animals were approved by the National Animal Experiment Board of Finland (license numbers ESAVI/6357/04.10.07/2014, ESAVI/8240/04.10.07/2017 and ESAVI/23252/2020 according to the EU directive 2010/63/EU). Male C57BL/6N mice were housed individually with 12-hour light cycle in Oulu Laboratory Animal Centre, University of Oulu, Oulu, Finland. Mice had ad libitum access to water and food.
PXR knockout mouse line (PXR-KO) were kindly provided by Professor Wen Xie, University of Pittsburgh School of Pharmacy Center for Pharmacogenetics, PA, USA [17]. Mice were backcrossed from C57BL/6J to C56BL/6N strain for six generations before they were utilized.
To evaluate the effect of PCN treatment on the glucose tolerance of obese mice, WT C57BL/6N and PXR-KO male mice were fed high-fat diet (HFD) (60% of calories from fat, Envigo td. 06414) for 15 and 18 weeks, respectively. One group in each mouse line remained on standard chow (2018 Teklad Global 18% Protein Rodent Diet (Envigo)). HFD-fed mice were randomly allocated to experimental groups (HFD, HFD Vehicle and HFD PCN) so that there was no significant weight difference between the groups. WT chow group contained 10 mice, HFD group 9 mice and treatment groups 8 mice. PXR-KO groups contained six mice. Vehicle group was treated with 30% DMSO in corn oil and PCN group by 50 mg/kg PCN in 30% DMSO-corn oil by intraperitoneal injections once a day for four days. To assay oral glucose tolerance, WT mice were fasted for 12 h overnight and anesthetized with midazolam/fentanyl-fluanisone and orally gavaged 2 g/kg glucose. PXR-KO mice were fasted 12 h overnight, but they were not anesthetized during the glucose tolerance test. Blood glucose was monitored from hind leg veins from the WT mice and from the tail tip from the PXR-KO mice with Freestyle Precision Neo glucometer (Abbott Laboratories) at 0, 15, 30, 60, 90 and 120 min timepoints. After the last time point, mice were sacrificed with carbon dioxide inhalation and cervical dislocation. Blood was collected from vena cava with EDTA-primed syringe and plasma was separated by centrifugation 3500 rpm for 10 min at room temperature. Tissues were collected and snap-frozen in liquid nitrogen or fixed with formalin and embedded in paraffin for further analyses.
To investigate the ability of HFD itself, without metabolic dysfunction, to sensitize mice for PXR-induced liver steatosis, 7-week-old male C57BL/6N were fed with HFD for 15 days, while the control group remained on standard chow. Both groups contained four mice. All mice were treated with daily intraperitoneal PCN (50 mg/kg) injections once a day for four days before the sacrifice. Mice were sacrificed with carbon dioxide and cervical dislocation and tissues were collected to liquid nitrogen or fixed with formalin and embedded in paraffin.
To investigate PCN-mediated liver steatosis in lean mice, nine-week-old male C57BL/6N mice were treated with daily intraperitoneal PCN (50 mg/kg) or vehicle (30% DMSO in corn oil) injections once a day for four days. Both groups contained seven mice. Before the sacrifice, the mice were fasted for 12 h and orally gavaged 2 g/kg glucose. One hour after glucose dosing, mice were sacrificed with carbon dioxide inhalation and cervical dislocation. Tissues were collected to liquid nitrogen or fixed with formalin and embedded in paraffin.
Pyruvate and insulin tolerance tests were performed for a different set of male C57BL/6N mice. Obesity was induced and group allocation done as described before. Chow group contained 10 mice, HFD group 10 mice, and treatment groups nine mice. After 14 weeks of the HFD, PCN and vehicle treatments were performed. For pyruvate tolerance test, mice were fasted for 12 h overnight, and 2 g/kg pyruvate was administered intraperitoneally. Blood glucose was monitored from the tail tip at 0, 15, 30, 60, 90 and 120 min timepoints. After the test, diets were continued for two weeks. Then, PCN and vehicle treatments were performed again to the same animals. To assay insulin tolerance, mice were fasted for 6 h, and were then given 1 U/kg short-acting insulin (Actrapid, Novo Nordisk). Blood glucose was monitored from tail tip at 0, 15, 30, 60, 90 and 120 min timepoints. To collect insulin-stressed tissue samples, mice were left to recover for 2 h, and were then given 1 U/kg insulin intraperitoneally. 15 min after the insulin dose, the mice were sacrificed with carbon dioxide inhalation and cervical dislocation, and blood and tissues were collected and processed as describe above.
To measure glucose uptake, male C57BL/6N mice were fed HFD for 13 weeks, after which the mice were allocated to HFD vehicle and HFD PCN groups randomly. Mice were then treated with vehicle or PCN (50 mg/kg) for four days. After a 12-hour fast, mice were orally gavaged glucose (2 g/kg) spiked with 3H-labeled 2-deoxyglucose (0.7 μCi/g; Perkin Elmer). Mice were sacrificed 1 h after the glucose dose with carbon dioxide inhalation and cervical dislocation and tissues were snap frozen in liquid nitrogen. Both groups contained 10 mice, but due to technical errors two PCN-treated mice were excluded from the final analyses.
To assay glucagon response, male C57BL/6N mice were fed HFD for 14 weeks before allocation to HFD vehicle and HFD PCN groups, which both contained 10 mice. The mice were treated with vehicle or PCN for four days prior to the glucagon challenge. The mice were fasted for 5 h, and were then given somatostatin (Sigma Aldrich) (7 mg/kg) intraperitoneally 15 min before subcutaneous glucagon administration (20 μg/kg). Somatostatin-priming was done to inhibit glucagon-induced insulin secretion. Blood glucose was measured from tail tip at 0, 15, 30, 60, 90 and 120 min timepoints.
2.2. RNA extraction, reverse transcription, and quantitative PCR
Total tissue RNA was extracted with RNAzol RT reagent (Sigma Aldrich) except RNeasy Lipid Tissue Mini kit (QIAGEN) was used for adipose tissue. RNA was reverse transcribed to complementary DNA with RevertAid Reverse Transcriptase (Thermo Scientific) with random hexamers per manufacturer's protocol. Quantitative PCR (qPCR) was performed either in ABI 7300 thermal cycler (Applied Biosystems) or in QuantStudio 5 (Applied Biosystems) either with FastStart Universal SYBR Green Master Mix (Roche) or with PowerUp SYBR Green Master Mix (Thermo Scientific), respectively. Fold changes were determined with 2−ΔCT method, where Fold change = 2−ΔCt sample/2−ΔCt control sample. Primers are listed in Suppl. table 1. The qPCR methods comply with minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines.
2.3. RNA sequencing and pathway analyses
Liver RNA sequencing of the WT mice has been described before [16]. PXR-KO liver and WT adipose tissue and muscle RNA sequencing protocols were essentially the same. Shortly, RNA samples were DNase treated with RNase-Free DNase Set (QIAGEN) coupled with RNeasy MinElute Cleanup Kit (QIAGEN). Concentration and quality of the RNA were determined with Qubit RNA BR Assay kit (Thermo Fisher Scientific) and Agilent RNA 6000 Nano Kit, respectively. TruSeq Stranded Total RNA with Ribo-Zero Gold kit (Illumina) was used for cDNA library preparations. Sequencing was performed with NextSeq550 (Illumina) in the Biocenter Oulu Sequencing Center, University of Oulu, Oulu, Finland. Data was analysed with Chipster software [18]. 75 bp paired-end reads were aligned to mouse Ensembl reference genome GRCm38.95 with HISAT2 and the reads were counted with HTSeq. DESeq2 was used to determine differentially expressed genes. A gene was considered differentially expressed when Benjamini-Hochberg adjusted p < 0.05 and log2fold change ≥0.6 or ≤ -0.6. Ingenuity Pathway Analysis (IPA) software [19] (QIAGEN) was used for pathway analyses of differentially expressed genes. IPA uses Fisher's exact test to evaluate p-values of overlap for pathway enrichment and upstream regulator analyses. The PXR-KO liver and the WT liver, muscle and adipose tissue complete data sets are available in Gene Expression Omnibus (GEO) database with accession numbers GSE162196, GSE136667, GSE168196 and GSE168497, respectively.
2.4. Histology
HE staining: 5 μm thick liver paraffin sections were subjected to hematoxylin-eosin (HE) staining with Mayer's hematoxylin. Oil Red O: To stain neutral lipids, 10 μm liver cryosections were subjected to Oil Red O staining and counterstained with Cole's hematoxylin. PAS staining: 5 μm thick liver paraffin sections were subjected to Periodic Acid Schiff (PAS) staining to stain glycogen.
2.5. Steatosis quantitation
To quantify steatosis from HE-stained liver sections, slides were scanned with Nanozoomer S60 (Hamamatsu). Steatosis application of Visiopharm software (Visiopharm) was used to quantify lipid droplet areas. In the quantification of steatosis with ORO staining, the slides were digitized with a Leica Aperio AT2 slide scanner with a 20× objective. Representative areas of 1.5–2 mm2 in the achieved virtual slides were analyzed using the Qupath program [20] by counting areas that were stained red. Microvacuolar steatosis was defined as droplets with an area under 176.625 μm2 (corresponding to a diameter of 15 μm) and macrovacuolar steatosis as droplets larger than that [21]. Droplets with an area less than 1 μm2 were regarded as nonspecific staining reaction and thus not counted.
2.6. Adipocyte size determination
To determine adipose tissue cell size, gonadal fat tissue samples were fixed with formalin and embedded in paraffin. 5 μm thick tissue sections were subjected to hematoxylin-eosin (HE) staining with Mayer's hematoxylin. Representative pictures from tissue sections were taken with 10× magnification and cell sizes of 50 biggest looking cells per mouse were determined with Adobe Photoshop. Average cell size was calculated per mouse.
2.7. Transmission electron microscopy
Tissues were prepared, and transmission electron microscopy (TEM) was performed in Biocenter Oulu Tissue Imaging Center, University of Oulu, Oulu, Finland. Pieces of liver tissue were fixated in 1% glutaraldehyde, 4% paraformaldehyde, 0.1M phosphate buffer for 12 h. Tissues were stained with potassium ferrocyanide in the presence of osmium tetroxide to visualize glycogen. To enhance contrast, tissues were further stained with uranyl acetate. After staining, tissues were dehydrated with acetone and embedded in plastic resin. Thin sections were imaged with Tecnai G2 Spirit 120 kV with Veleta and Quemesa CCD cameras.
2.8. ALT activity determination
Alanine aminotransferase (ALT) levels were determined from EDTA-plasma with Alt Activity Assay kit (Sigma) according to manufacturer's protocol.
2.9. 2-Deoxyglucose quantification
Tissue pieces were homogenized to phenol:chloroform (2:1) with TissueLyser II (Qiagen). Homogenates were shaken for 20 min, saline was added and shaking was continued for 20 min. Then, the samples were centrifuged and water-phase scintillated for 3H activity with Optiphase Hisafe 2 scintillation solution (Perkin Elmer) and MicroBeta Trilux (Perkin Elmer) counter. 2-deoxyglucose-6-phosphate was quantified with a method taking advantage of differential solubility of 2-deoxyglucose and 2-deoxyglucose-6-phosphate to HClO4 and Ba(OH)2/ZnSO4. The method has been described in detail before [22]. In brief, the polar phase after phenol:chloroform extraction was dried with Savant Speed Vac (Thermo Fisher), pellet resuspended to water and equal volumes of the obtained solution was either subjected to Ba(OH)2/ZnSO4 precipitation or added to HClO4 solution. The difference in the scintillation of these two solutions represented the amount of 2-deoxyglucose 6-phosphate in the tissue.
2.10. Liver glycogen quantitation
To isolate glycogen, liver pieces were dissolved in 30% KOH solution by boiling. Glycogen was precipitated with ethanol and pelleted. Pellet was dissolved in water and precipitated again. Finally, pellet was dissolved in water and glycogen was assayed with Glycogen Assay Kit (Sigma Aldrich).
2.11. Immunoblotting
Liver and muscle tissues were homogenized in lysis buffer (20 mM Tris-Cl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton-X100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, pH 7.5) with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific), 1 mM DTT and 50 mM NAF with Tissuelyser II for one and 2 min, respectively. Homogenate was cleared by centrifugation at 12,500 rpm for 20 min at +4 °C. Protein content was determined with Bio-Rad Protein Assay. 30 μg protein was separated on polyacrylamide gels. and transferred to nitrocellulose membranes with TransBlot Turbo (Bio-Rad). Membranes were blocked with ECL Prime Blocking Agent (GE Healthcare) and primary antibody incubations were performed overnight in tris-buffered saline with 0.1% Tween20 at +4 °C. Primary antibodies were rabbit polyclonal AKT antibody (Cell Signaling Technology; #9272; 1:1000), rabbit polyclonal phospho-AKT antibody targeted to serine 473 phosphorylation (Cell Signaling Technology; #9271; 1:1000), mouse monoclonal beta-actin antibody (Sigma Aldrich; #A3854; 1:25000), rabbit polyclonal IRS1 antibody (Thermo Fisher Scientific; #PA1-1059; 1:900), rabbit polyclonal pIRS1 antibody targeted to serine 307 phosphorylation (Thermo Fisher Scientific; #PA1-1054; 1:500), mouse monoclonal S6 antibody (Cell Signaling Technology; #2317, 1:1000) and rabbit polyclonal S6 antibody targeted to serine 235/236 phosphorylation (Cell Signaling Technology; #2211S, 1:1000).
2.12. Homeostatic model assessment of insulin resistance (HOMA-IR)
HOMA-IR index was calculated using fasting values of glucose and insulin. The formula was HOMA-IR = ([Glucose mmol/L]x [Insulin mU/L])/14,1 [23]
2.13. Plasma enzyme-linked immunosorbent analysis and plasma glycerol measurement
Plasma adiponectin, leptin and resistin levels were determined with Novex mouse ELISA kits KMP0041, KMC2281 and EMRETN (Invitrogen), respectively, according to manufacturer's protocol. Fasting insulin from plasma samples was determined with Insulin Mouse Serum Assay (Cisbio). Plasma glycerol levels were measured using Glycerol Assay Kit (Sigma MAK117) according to instructions.
2.14. Liver and plasma triglyceride measurements
Liver and plasma total triglyceride levels were measured with Trigyceride Quantification Colorimetric/Fluorometric Kit (Sigma MAK266) with the colorimetric protocol.
2.15. Relative triglyceride quantification in mouse liver samples
Frozen mouse liver samples (−80 °C) were put into a liquid nitrogen-cooled mortar and gently crushed with a pestle to smaller pieces, which were quickly put to 2 mL tubes, weighed, and refrozen at −80 °C until further use. Liver portions (20–50 mg) were thawed on ice, complemented with 1.0 (20–40 mg) or 1.5 mL (>40–50 mg) buffer (RLT lysis buffer, RNeasy kit, Qiagen, Hilden, Germany), and lysed with an ultrasound head (Sonopuls UW 2070, Bandelin electronic, Berlin, Germany: six times: cycle 6, 10 s, 60–65% power). Lysed samples were stored at −80 °C until extraction.
Triglyceride extraction was performed at room temperature using glassware and in three technical replicates, which were processed on different days. Ten-mL tubes were filled with 0.25 mL lysed liver sample and 5 mL extraction solvent (iso-octane:ethyl acetate 3:1) containing 4 mg/L internal standard (glyceryl tritridecanoate, C39). Air was replaced by nitrogen, the tube sealed, and the mixture vortexed, sonicated for 1–3 min, vortexed again, and rotated overhead for 15 min. Afterwards, 1 mL 0.9% (w/v) aqueous NaCl was added, air was replaced by nitrogen, and the mixture vortexed and centrifuged (10 min, 13083 g). Four mL organic phase was removed, the extraction procedure repeated with addition of 2 mL extraction solvent without internal standard and final removal of 2 mL organic phase. A 0.65 mL aliquot of the combined extracts was dried under a nitrogen stream, solubilized in 0.1 mL iso-octane, transferred to a vial, and stored at −20 °C until analysis.
A 7890A gas chromatograph (Agilent Technologies, Böblingen, Germany) equipped with a splitless liner (2879305-U, Supelco/Sigma-Aldrich, Munich, Germany), a Zebron ZB-5HT Inferno column (0.25 mm∗30 m, 0.1 μm film, Phenomenex, Aschaffenburg, Germany), and connected to a MPS2 autosampler (Gerstel, Mülheim, Germany) was used. He (1.5 mL/min) served as carrier gas and the instrument was operated in FID mode (350 °C, 45 mL/min H2, 450 mL/min air, 20 mL/min He as make-up gas). Two μL sample were injected (splitless, 350 °C, 3 mL/min He septum purge flow) and analyzed with the following oven program: initial 90 °C (1 min) followed by linear ramps to 230 °C (60 °C/min) and 380 °C (10 °C/min) and a final hold at 380 °C for 8 min. Liver samples were run in randomized order separated by a solvent blank and a quality control sample.
2.16. Relative triglyceride quantification
Triglycerides containing 44, 46, 48, 50, 52, and 54 fatty acid carbon atoms in total (C42, C44, C46, C48, C52, C54) were identified by retention time comparison with standards (C39, C42, C45, C48, C50, C52, C54) contained in the quality control sample (level). Peak integration was performed with enhanced data analysis using the area sum function at the appropriate retention time windows, peak areas were normalized to the internal standard, then averaged over the three technical replicates. Linearity was proven within suitable ranges (9–150 ng for C39 and 9–35 to 800 (C42, C45, C48, C50, C54) or 2500 ng (C52), respectively) using serial 1:3 dilution series (n = 3) of the standards prior and after sample measurement. Furthermore, two-level standard addition to liver extracts proved linearity also in matrix-containing material (data not shown). Intra- and interday precision (n = 5) were below 20 and 25% in most cases, respectively.
2.17. Statistical analysis
Student's unpaired t-test was used to evaluate the difference between chow-fed and HFD-fed mice, and vehicle-treated and PCN-treated mice. Two-way ANOVA coupled with Tukey's post hoc test was used in Figure 3D to evaluate the difference the diets caused in WT and PXR-KO mice. Differences were considered significant when p < 0.05. Statistical analyses of the RNA sequencing data and the mass spectrometry have been described as part of the method description.
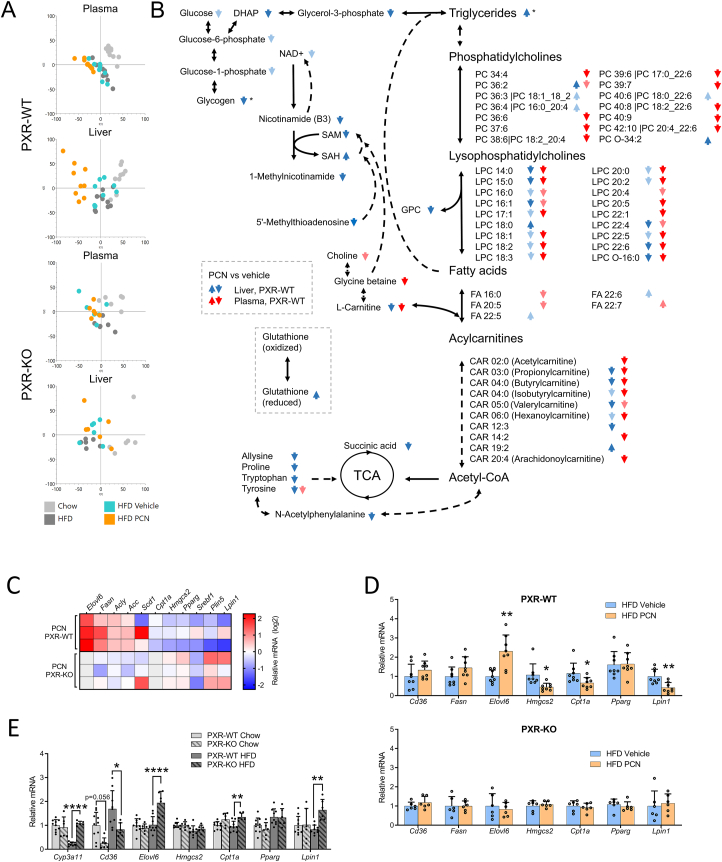
Figure 3.
The effect of high-fat diet and PXR activation on plasma and liver metabolome and on liver lipogenetic gene responses. A) The principal component analyses (PCA) of WT and PXR-KO mouse plasma and liver samples in metabolomic analyses. B) PXR activation by PCN treatment reduced plasma and liver phospholipids and acylcarnitines in WT plasma samples and altered liver energy metabolism pathways. These effects were not seen in the PXR-KO mice. Dark blue/red arrow = p-value <0.001; light blue/red = p-value between 0.05 and 0.001. ∗ Triglycerides and glycogen were not among the metabolites measured in the metabolomics and these data come from separate measurements. C) Expression of liver lipid metabolism related genes in HFD-fed, wildtype and PXR-KO mice after PCN treatment in RNA sequencing data set. The data indicates relative response to the vehicle treated groups. D) Relative mRNA expression of lipid metabolism related genes in the HFD-fed and PCN or vehicle treated WT or PXR-KO mouse livers (QPCR measurement). E) Relative mRNA expression of Cyp3a11 and lipogenic genes regulated by PXR in the WT and PXR-KO mice on chow and HFD. Statistical analyses in B were done by Welch's one-way ANOVA followed by Bonferroni correction for multiple testing with α of 0.0010, in D by unpaired Student's t-test and in E with Two-way ANOVA coupled with Tukey's post hoc test. CAR; acylcarnitine; DHAP, dihydroxyacetone phosphate; LPC, lysophosphatidylcholine; NAD+, nicotinamide adenine dinucleotide; PC, phosphatidylcholine; SAH, S-Adenosyl-homocysteine; SAM, S-Adenosyl-methionine; TCA, the tricarboxylic acid cycle (the Krebs cycle, the citric acid cycle). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
2.18. Non-targeted liquid chromatography mass spectrometry metabolomics
Non-targeted metabolite profiling analysis of liver and plasma was carried out at the LC-MS metabolomics center (Biocenter Kuopio, University of Eastern Finland). The workflow of the metabolomics analysis has been described in detail elsewhere [24]. The analysis order of the samples was randomized.
Plasma samples were thawed on ice and then 100 μL of each sample was mixed by pipette with 400 μL of acetonitrile (LC-MS grade, VWR International) in the well of a Captiva ND filter plate (Agilent Technologies). The filter plate was centrifuged at 700 rcf at 4 °C for 5 min. Then the protein-free filtrate was collected on a 96-well polypropylene plate. A pooled QC sample was prepared for quality control and injected after every 12 analytical samples.
Liver samples were cryo-weighed and 80% methanol was added (v/v H2O,LC–MS Ultra CHROMASOLV®, Fluka), at a ratio of 300 μL solvent/100 mg tissue. Samples were then homogenized using Omni 2 mL tubes with 1.4 mm ceramic beads in Omni Bead Ruptor Elite with a cryo cooling unit (1 cycle, 30 s, speed 5; Omni International). The samples were shaken for 5 min, centrifuged for 10 min at 4 °C (14,000 rpm), and the supernatants were filtered using 0.45 μm Acrodisc® Syringe Filters, with a PTFE membrane (PALL Corporation). Samples were stored in freezer overnight. Because there was sediment in the samples, the samples were centrifuged for 5 min at 4 °C (10,000 rpm), and an aliquot of 50 μL was taken and mixed with 50 μL of 80% MeOH. Further aliquots of 2 μL were taken from each tissue sample to make QC sample, and injected after every 12 analytical samples.
For mass spectrometry analysis, we utilized an ultra-high performance liquid chromatography (Vanquich Flex UHPLC system, Thermo Scientific, Bremen, Germany) coupled online to a high-resolution mass spectrometry (HRMS, Q Exactive Classic, Thermo Scientific, Bremen, Germany). For more hydrophilic compounds, we used hydrophilic interaction (HILIC) chromatography (Acquity UPLC® BEH Amide 1.7 μm, 2.1 × 100 mm, Waters Corporation, Milford, MA, USA) and both positive and negative ionization. For amphiphilic compounds, we used reversed-phase chromatography (RP) column (Zorbax Eclipse XDBC18, 2.1 × 100 mm, 1.8 μm, Agilent Technologies, Palo Alto, CA, USA) and both positive and negative ionization.
2.19. Metabolomics data preprocessing and statistical analysis
We used an open-source software MS-DIAL version 4.36 [25] for peak picking and molecular feature alignment. The used parameters for peak picking were: mass range 40–1600 Da, MS tolerance 0.008 Da, MSMS tolerance 0.015 Da, minimum peak width 8, minimum peak height 700000, mass slice width 0.1. The used parameter for feature alignment were: retention time tolerance 0.1 min, MS tolerance 0.015, detected in at least 50% of the samples in one group, and gap filling by compulsion.
The preprocessing procedure has described in more detail in [24]. In brief, the molecular features were corrected for the drift pattern caused by the LC-HRMS procedures using regularized cubic spline regression, fit separately for each feature on the QC samples. The smoothing parameter was chosen from an interval between 0.5 and 1.5 using leave-one-out cross validation to prevent overfitting. After the drift correction, feature quality was assessed, and low-quality features were flagged. Features were kept if their RSD∗ was below 20% and their D-ratio below 40%. In addition, features with classic RSD, RSD∗ and basic D-ratio all below 10% were kept. This additional condition prevents the flagging of features with very low values in all but a few samples. These features tend to have a very high value of D-ratio∗, since the median absolute deviation of the biological samples is not affected by the large concentration in a handful of samples, causing the D-ratio∗ to overestimate the significance of random errors in measurements of QC samples. Thus, other quality metrics were applied with conservative limit of 0.1 to ensure that only good quality features were kept this way. Missing values were imputed using random forest imputation. The imputation was done in two phases: first, only the good quality features were imputed, to prevent the flagged features from affecting the imputation. Next, flagged features were imputed. QC samples were removed prior to imputation to prevent them from affecting imputation.
For multivariate analysis, we used principal component analysis (PCA). For univariate analysis, we first used Welch's one-way ANOVA to identify significantly altered metabolites between the study groups. Group-to-group comparisons were done using Welch's t-test between the PCN and vehicle treated groups, to observe effects associated with the PXR agonism treatment, and between the control chow diet and the HFD group to observe effects associated with the HFD.
To account for multiple testing, we used Bonferroni's method to adjust the α level by the number of principal components needed to explain 95% of the variation in the metabolomics data. Here 51 and 48 principal components were needed to explain 95% variance in the metabolomics data from the plasma and liver samples, respectively. Therefore, we adjusted the α level to 0.0010 to account for multiple testing. Differences between the study groups with p-values between 0.05 and 0.001 were considered to be trends.
2.20. Metabolite identification
Metabolite identification was focused to molecular features with p-value below 0.05 in either of the experiments. Metabolite identification was done using an open-source software MS-DIAL version 4.36 [25]. Metabolite identifications were ranked according to the guidelines from Sumner et al. [26]. Metabolites in level 1 were matched against accurate mass, retention time, and product ion (MSMS) spectra of fragmented ions from the in-house library of chemical standards built using the same instrument and experimental conditions. Level 2 includes metabolites with matching exact mass and MSMS spectra from public libraries (METLIN, Lipidmaps and Human Metabolome DataBase were used) or in the case of lipids, the built-in MS-DIAL library version 4.00. In level 3, only the chemical group of the compound (but not the exact compound) could be identified.
3. Results
3.1. PXR activation aggravates obesity-induced hepatosteatosis
Male mice underwent 15-week HFD-feeding and for the last four days the mice were treated with vehicle or pregnenolone-16α-carbonitrile (PCN). To control the metabolic effects of the diet, one HFD group was left untreated and was compared to a chow-fed group. The experiment was replicated with PXR knockout mice (PXR-KO) to proof that PXR mediates the effect of PCN. The treatment of the wildtype (WT) mice was repeated multiple times with small modifications to perform all the needed metabolic tests as described in the methods section.
The HFD-fed mice were visibly obese and heavier than the chow-fed controls (Figure 1A,B.). Vehicle or PCN did not affect the weight (Figure 1B.). HFD alone increased liver size, but only in the PXR-KO mice (Figure 1C.). PCN treatment doubled the liver size in the WT mice but not in the PXR-KO mice (Figure 1C.).
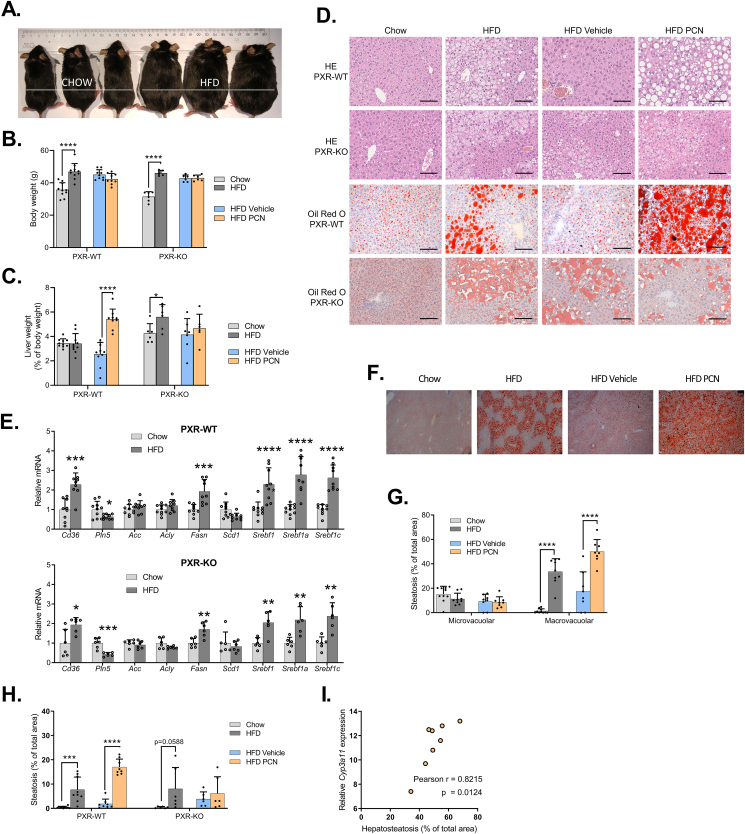
Figure 1.
The effect of high-fat diet and PXR activation on hepatic steatosis. WT and PXR-KO mice were fed regular chow or high-fat diet (HFD) for 15 or 18 weeks, respectively. In the end of the experiment subset of the HFD-fed mice were treated with PCN or vehicle for 4 days. A) Pictures of wildtype mice fed with either normal chow diet or HFD for 16 weeks. B) Body weights at the time of sacrifice. C) Liver weights as percentage of body weight. D) Hematoxylin-eosin (HE) and Oil Red O-stained liver sections of WT and PXR-KO mouse livers. Scale bar = 100 μm. E) Relative expression of liver lipogenic PXR-regulated genes in the WT and PXR-KO mice on chow and HFD. F) Oil Red O-stained liver sections of WT mice with 4× magnification. G) Quantification of Oil Red O-stained microvacuolar and macrovacuolar liver lipid droplets in the WT mice. H) Quantification of lipid droplet areas from Hematoxylin-eosin (HE)-stained liver sections in the WT and PXR-KO mice. I) Correlation of Cyp3a11 mRNA expression and Oil Red O-stained liver lipid droplet areas of the wildtype mice on HFD and treated with PCN. Difference between chow and HFD or HFD vehicle and HFD PCN were evaluated with unpaired Student's t-test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Hematoxylin-eosin (HE) and Oil Red O-stained liver sections indicated similar HFD-induced fat accumulation in both genotypes (Figure 1D.). Accordingly, the liver mRNA expression of central lipid metabolism-related genes was similarly affected in both genotypes (Figure 1E.). 4-day vehicle treatment reduced liver fat, however, PCN treatment dramatically aggravated the steatosis even if compared to the non-treated HFD group (Figure 1D.). No signs of steatohepatitis were observed. HFD-induced steatosis was focused to the pericentral region of a liver lobule, and additional PCN treatment displayed steatosis throughout the lobule (Figure 1D. & 1F.). Quantitative analysis of the lipid droplet areas confirmed the visual observations and showed that macrovacuolar steatosis was induced by both HFD and PCN-treatment (Figure 1G. & 1H.). The expression of liver Cyp3a11, a marker of PXR activation, correlated strongly with the lipid droplet areas in the PCN-treated mice (Figure 1I.).
HFD increased liver triglycerides in both genotypes but had no effect on plasma triglycerides in the wildtypes (Figure 2A. & 2B.). PCN increased hepatic triglycerides in the wildtypes but not in the PXR-KO mice (Figure 2A.). HFD raised the amount of hepatic long-chain fatty acids (triglyceride chains containing 44–54 carbons) (Figure 2C.) in both the WT and PXR-KO mice. PCN increased triglycerides with 44, 46, 52 and 54 fatty acid carbons in the WT mice (Figure 2C.). HFD increased plasma alanine aminotransferase (ALT) level irrespective of the PXR genotype, and PCN further elevated ALT in the WT mice (Figure 2D.).
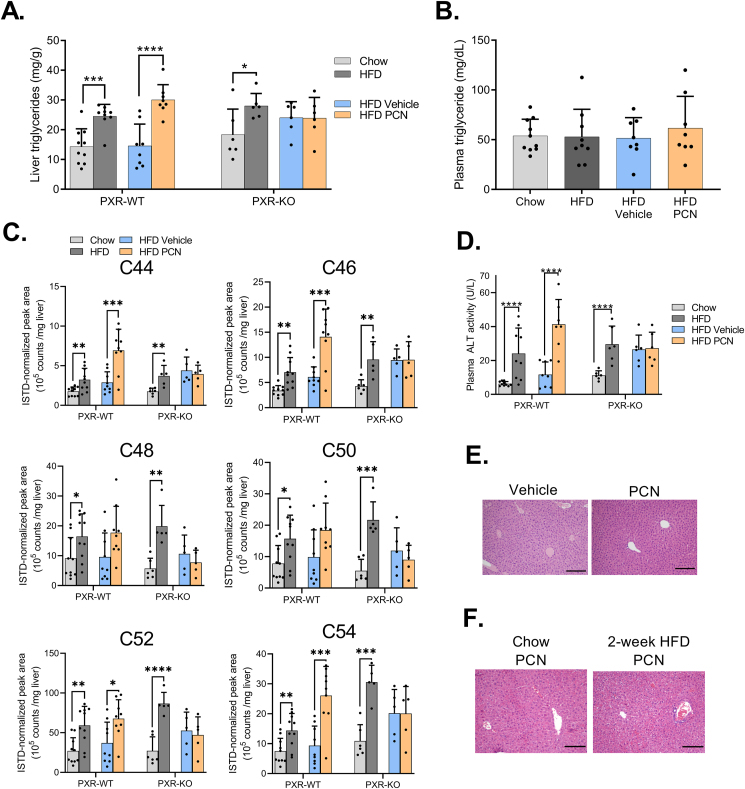
Figure 2.
PCN increases liver triglycerides and plasma ALT in HFD-fed WT mice. A) Liver triglycerides in the WT and PXR-KO mice. B) Plasma triglyceride levels in the WT mice. C) GC-FID quantification of liver triglycerides with different length fatty acid chains (e.g. C44 refers to 44 carbons in triglyceride fatty acids in total) in WT and PXR-KO mice. D) Plasma ALT activities. E) HE-stained liver sections of lean mice treated with PCN or vehicle for four days. 10× magnification, scale bar = 200 μm. F) HE-stained liver sections of WT mice treated with PCN after two-week regular chow or high-fat diet. 10× magnification, scale bar = 200 μm. Difference between chow and HFD or HFD vehicle and HFD PCN were evaluated with unpaired Student's t-test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
4-day PCN treatment did not induce visible steatosis in chow-fed lean mice (Figure 2E.) or mice fed HFD for only two weeks (Figure 2F.) indicating that the HFD-induced metabolic state and not the diet itself predisposes to PXR-induced liver steatosis. This also indicates that the liver triglycerides do not originate from the HFD per se.
3.2. PXR-induced steatosis stems from multiple modulations of hepatic lipid metabolism
Triglycerides accumulate in the liver through increased lipid acquisition (dietary absorption, synthesis) or decreased lipid disposal (very low-density lipoprotein synthesis, β-oxidation) [1]. To elucidate the mechanisms of PXR-induced liver fat accumulation, we performed non-targeted metabolomics analyses for liver and plasma samples.
All significantly altered metabolites are shown in Supplementary table 2. (plasma samples) and in Supplementary table 3 (liver samples). In principal component analysis, chow-fed mice liver and plasma samples were separated from the HFD-fed groups in both genotypes, but to a lesser degree in the PXR-KO mice (Figure 3A.). PCN-treated WT mice were separated further from the other HFD groups (Figure 3A.). In the WT mice, PCN reduced plasma and liver phospholipids and acylcarnitines (Figure 3B.). Importantly, reductions were observed in all the pathways involved in triglyceride synthesis, including anaplerotic amino acids, which replenish Krebs cycle, and dihydroxyacetone phosphate and glycerol-3-phosphate derived from glucose. These findings suggests that the liver metabolism is adjusted to support triglyceride synthesis. Furthermore, S-adenosyl-methionine (SAM)/S-adenosyl-homocysteine (SAH)-ratio and reduced phosphatidylcholine levels (Figure 3B.) are known to associate with reduced VLDL secretion [27] that also may contribute to increased liver triglyceride levels and explain the lack of increase in the plasma triglyceride (Figure 2B).
In agreement with the lipid acquisition by de novo synthesis and increase in long-chain triglycerides, RNA-sequencing of the liver samples suggested upregulation of several fatty acid synthesis genes by PCN treatment; of these fatty acid elongase 6 (Elovl6) and fatty acid synthase (Fasn) were induced statistically significantly (Figure 3C). Further confirmation in the whole sample set with QPCR indicated 2.5-fold induction of Elovl6, while the Fasn-induction was not significant. Furthermore, PCN repressed carnitine palmitoyltransferase (Cpt1a) and 3-hydroxy-3-methylglutaryl-CoA synthase 2 (Hmgcs2) involved in β-oxidation and ketogenesis, while the expression of fatty acid uptake transporter Cd36 was not affected (Figure 3D.). LPIN1 is a bifunctional protein, which catalyzes the penultimate step in the triglyceride synthesis [28], but also functions as a transcriptional cofactor promoting fatty acid oxidation and repressing sterol regulatory element-binding protein 1 (SREBP1), a key regulator of lipogenesis. Lpin1 gene expression was repressed by PCN that fits to the concept that downregulation of LPIN1 level removes a transcriptional brake from lipogenesis. All these effects of PCN on lipid metabolism genes were abolished in the PXR-KO mice (Figure 3D.).
PXR deficiency selectively affected the expression of lipid metabolism genes. Cd36 was the only gene which expression was downregulated by PXR deficiency in both chow and HFD-fed mice (Figure 3E.). Elovl6, Cpt1a and Lpin1 had higher liver expression in the PXR-KO mice than in the WT mice but only after HFD feeding (Figure 3E.). Interestingly, the prototypic PXR target gene, Cyp3a11, was repressed by the HFD only in the WT mice (Figure 3E.).
3.3. PXR does not affect white adipose tissue lipid storage
While adipose tissue (WAT) functioning as an energy storage and an endocrine organ is greatly affected by obesity and inflammation. Indeed, the HFD doubled the sizes of adipocytes (Suppl. figure 1A.) and altered the WAT gene expression profile by inducing the expression of proinflammatory genes and modulating adipokine (adiponectin, leptin, resistin) expression and secretion (Suppl. figures 1B.-1F.). PCN did not affect adipocyte sizes but induced WAT Tnfa and Ccl3 expression in the WT mice, however, the adipokine expression or secretion were not affected by PCN (Suppl. figures 1A.-1F.). Plasma glycerol levels, indicative of adipose tissue lipolysis, were not affected by the HFD (Suppl. figure 1G.), however, the oleic acid level was increased (Suppl. figure 1H.). PCN had a small increasing effect on plasma glycerol in the wildtype mice (Figure 1G.). In contrast, plasma palmitic acid was slightly reduced, and the plasma oleic acid was not affected by PCN (Suppl. figure 1H.). These results do not suggest any major effect of PCN on adipose tissue or on modulation of lipolysis.
3.4. Hepatosteatosis dissociates from glucose tolerance impairment and whole-body insulin resistance after PXR activation
NAFLD is tightly linked to insulin resistance and glucose homeostasis disturbances [2]. Expectedly, the HFD-fed mice displayed impaired glucose tolerance in an oral glucose tolerance test (OGTT) irrespective of PXR genotype (Figure 4A. and 4C). While the PCN treatment was found to aggravate liver steatosis in the WT mice (Figure 1D.), it tended to improve glucose tolerance as PCN lowered blood glucose 15 min after glucose ingestion (Figure 4B.). However, the total and incremental AUCs were not affected (Figure 4C.). PCN did not affect glucose tolerance in the PXR-KO mice.
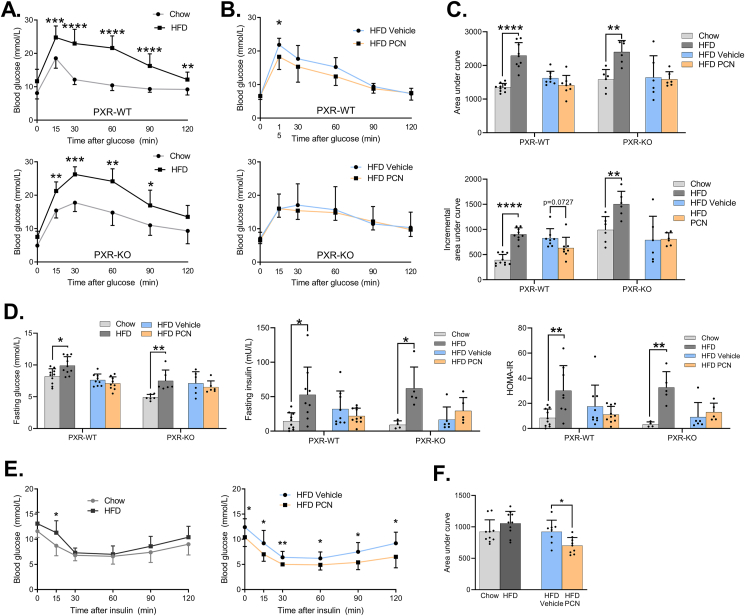
Figure 4.
Glucose tolerance and systemic insulin sensitivity after HFD feeding and PXR activation by PCN in the WT and PXR-KO mice. A) Oral glucose tolerance test in WT and PXR-KO mice fed chow or HFD. Mice were fasted 12 h prior to the test. B) Oral glucose tolerance test in HFD-fed WT and PXR-KO mice treated either with vehicle or PCN. C) Area under curve and incremental area under curve analyses of oral glucose tolerance tests. D) Fasting glucose, fasting insulin and HOMA-IR index in study groups after a 12-hour fast. E) Insulin tolerance tests of WT mice. Mice were fasted 6 h prior to the test. F) Area under curve values of insulin tolerance test. Difference between chow and HFD or HFD vehicle and HFD PCN were evaluated with unpaired Student's t-test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
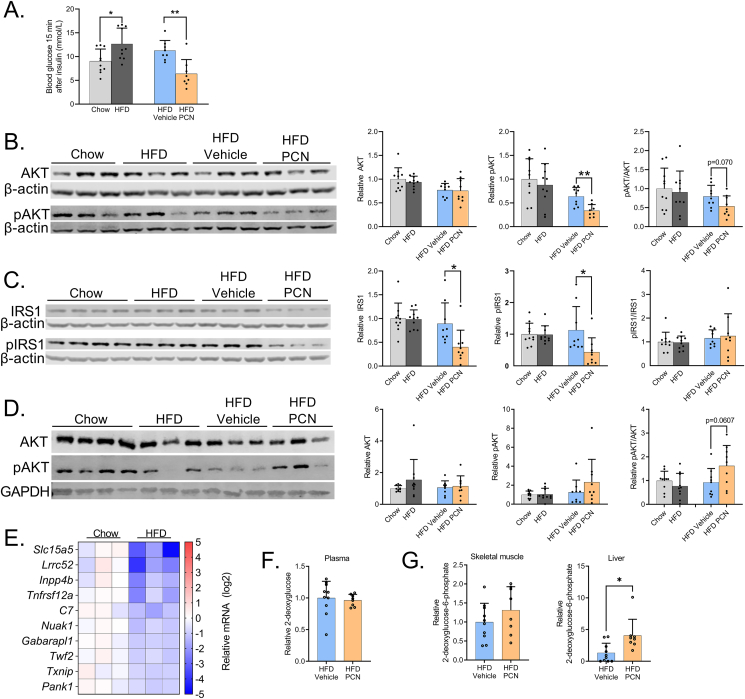
12-hour fasting blood glucose, insulin, and homeostatic model assessment for insulin resistance (HOMA-IR) were all increased by the HFD both in the WT and PXR-KO mice. PCN treatment did not affect these values (Figure 4D.). Interestingly, the PCN-treated mice demonstrated lower blood glucose after a 6-h fast, (Figure 4E.). In insulin tolerance test (ITT), the HFD-fed WT mice had higher blood glucose 15 min after insulin (Figure 4E.). PCN decreased blood glucose at all timepoints and decreased the ITT AUC value (Figure 4E,F).
In summary, the PXR-mediated exacerbation of hepatosteatosis did not promote whole body insulin resistance. In contrast, the PCN treatment appeared to slightly improve glucose metabolism.
3.5. PXR activation impairs hepatic insulin signaling but increases glucose uptake
Insulin increases glucose uptake in muscle, adipose tissue and liver, of which PXR is only expressed in the liver. To investigate tissue-specific insulin sensitivity chow- and HFD-fed WT mice were sacrificed 15 min after insulin bolus. HFD increased and PCN treatment decreased blood glucose at the time of sacrifice (Figure 5A.). Liver insulin signaling was studied by measuring expression of two key proteins in the process: AKT kinase (AKT) and insulin receptor substrate 1 (IRS1) and their Ser473 and Ser307 phosphoproteins, respectively [29,30]. HFD feeding did not affect expression or phosphorylation either of the proteins compared to lean chow-fed mice despite obvious systemic insulin resistance (Figure 5B. & 5C.). This is probably explained by calculation of the insulin dose based on weight; the higher insulin dose of the obese mice may have compensated for the insulin resistance. In contrast, the PCN treatment reduced hepatic pAKT levels and tended to reduce pAKT/AKT ratio (Figure 5B.). Furthermore, the PCN treatment decreased both total IRS1 and phosphorylated IRS1 protein in the liver, without affecting the pIRS/IRS1 ratio Figure 5C.). Reduced AKT phosphorylation and lower IRS1 level in liver suggest impaired insulin signaling as a sign of insulin resistance in response to PCN treatment.
Figure 5.
PCN treatment impairs hepatic insulin signaling but increases hepatic glucose uptake in wildtype mice. A) Blood glucose at the time of sacrifice, 15 min after insulin administration. B) Liver AKT and p-AKT immunoblotting and their quantification. β-actin was used as a reference protein for normalization. C) Liver IRS1 and p-IRS1 immunoblotting and their quantification. β-actin was used as a reference protein for normalization. D) Muscle AKT and pAKT immunoblotting and their quantification. GAPDH was used as a reference protein for normalization. E) Differentially regulated genes in the skeletal muscle in response to HFD. 10 genes with the lowest p-values are presented. F) Plasma relative 3H activity, indicative of 2-deoxyglucose level. G) Tissue 2-deoxyglucose-6-phosphate content. Difference between chow and HFD or HFD vehicle and HFD PCN were evaluated with unpaired Student's t-test. ∗p < 0.05; ∗∗p < 0.01.
Next, we estimated the insulin signaling in skeletal muscle, where total AKT or pAKT were not affected by the HFD (Figure 5D.). Furthermore, PCN did not have a significant effect on the total AKT level or AKT phosphorylation (Figure 5D.). In transcriptomic analyses of muscle samples, the HFD modulated transcriptomic profile (Figure 5E.) and differentially regulated (adjusted p < 0.5, log2fold change <-0.6 or >0.6; data not shown) 26 genes, while PCN treatment did not lead to differential gene regulation.
To further understand the tissue contribution to glucose tolerance, glucose uptake was determined by giving the WT mice 3H-labeled 2-deoxyglucose along with normal glucose after a 12-hour fast. After cellular uptake, 2-deoxyglucose is phosphorylated but is unable to undergo glycolysis. Plasma 2-deoxyglucose (Figure 5F.) and skeletal muscle 2-deoxyglucose-6-phosphate (Figure 5G.) were not affected by PCN. In contrast, PCN treatment induced three-fold increase in liver 2-deoxyglucose-6-phosphate indicating increased hepatic glucose uptake (Figure 5G.).
3.6. PCN-aggravated hepatic insulin resistance does not induce gluconeogenesis
Liver insulin resistance impairs normal repression of gluconeogenesis by insulin promoting hyperglycemia [31]. On the other hand, previous studies have shown that PXR activation represses the key gluconeogenic genes, glucose-6-phosphatase catalytic subunit (G6pc) and phosphoenol pyruvate carboxykinase 1 (Pck1) in mice [32].
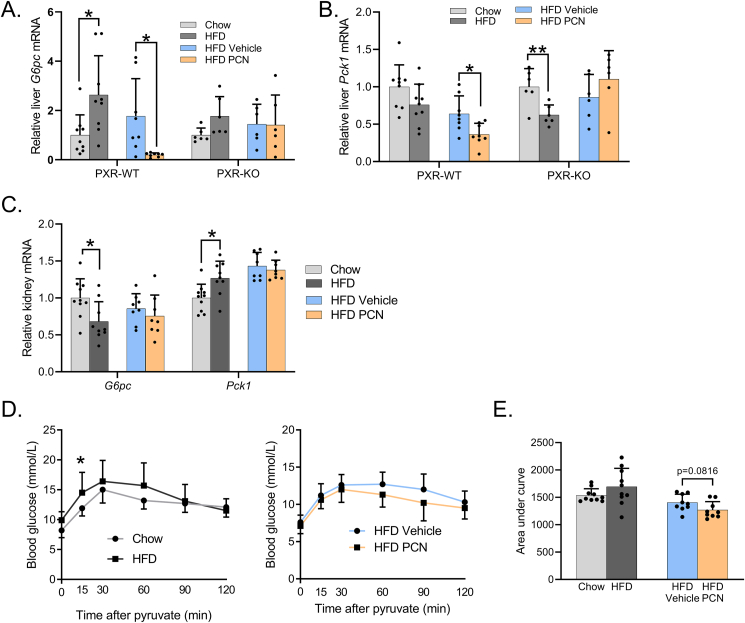
The HFD induced hepatic G6pc mRNA in the WT mice; in the PXR-KOs G6pc expression was slightly but not significantly induced (Figure 6A.). Pck1 was repressed by the HFD, but statistical significance was reached only in the PXR-KO mice (Figure 6B.). PCN repressed expression of both genes in the WT mice (Figure 6A,B.). Since renal gluconeogenesis may sometimes compensate for the reduced hepatic gluconeogenesis [33], we measured expression of the gluconeogenic genes also in the kidney. The HFD repressed renal G6pc and induced Pck1, while PCN did not have any effect (Figure 6C.). In pyruvate tolerance test, which measures pyruvate-to-glucose conversion (i.e. gluconeogenesis), the HFD increased blood glucose at 15-minute timepoint in the WT mice (Figure 6D.) but did not affect the AUC values (Figure 6E). While PCN impaired liver insulin signaling (see Figure 5B,C), and, on the other hand, downregulated G6pc and Pck1 expression (Figure 6A,B), PCN treatment did not affect pyruvate-to-glucose conversion (Figure 6D,E).
Figure 6.
Gluconeogenesis is not affected by PXR activation despite the NAFLD progression. A) Relative hepatic G6pc expression. B) Relative hepatic Pck1 expression. C) Relative Expression of gluconeogenic genes Pck1 and G6pc in the kidney. D) Pyruvate tolerance test in the WT mice fed with chow or HFD-fed mice and in the WT mice fed with HFD and treated with vehicle or PCN. E) Area under curve analysis of pyruvate tolerance test. Difference between chow and HFD or HFD vehicle and HFD PCN were evaluated with unpaired Student's t-test. ∗p < 0.05.
3.7. PCN depletes hepatic glycogen and impairs glucagon-mediated hepatic glucose production
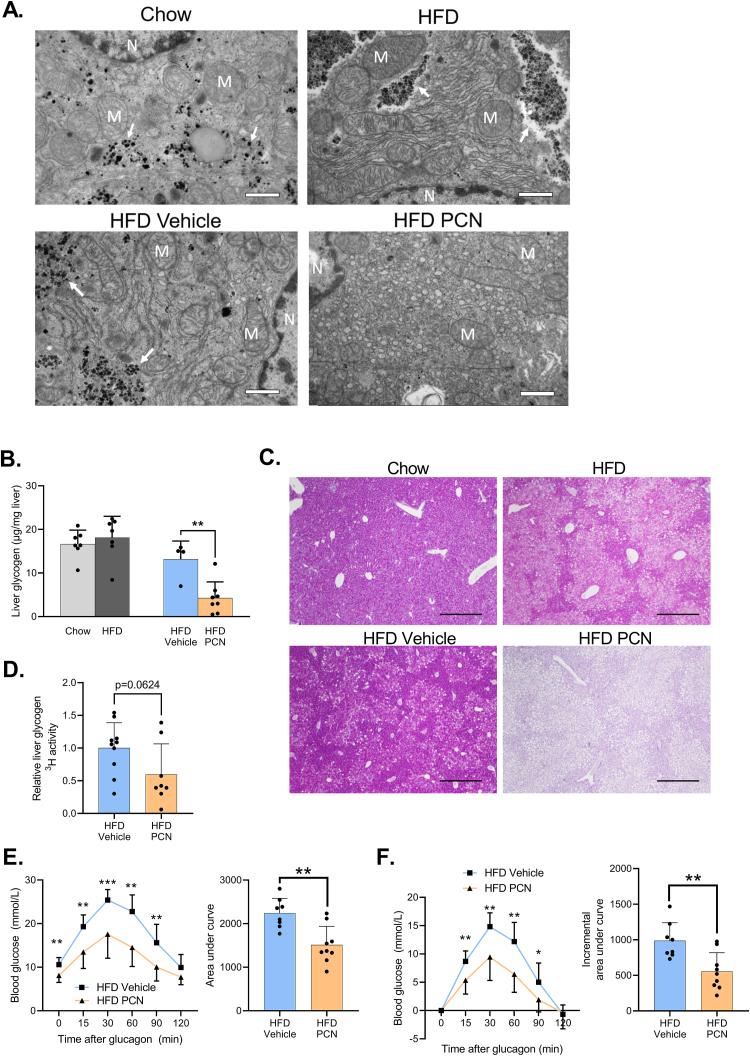
The periportal regions of livers of the WT mice were imaged with transmission electron microscopy (TEM). The HFD-fed mice demonstrated more pronounced rough endoplasmic reticulum (ER) and differently organized glycogen (Figure 7A.). PCN triggered a striking proliferation of smooth ER and abolished glycogen storage (Figure 7A.). In agreement with the TEM imaging, PCN decreased hepatic glycogen content by 45% (Figure 7B.), which was also supported by PAS staining of the liver sections (Figure 7C.).
Figure 7.
PCN treatment depletes hepatic glycogen, inhibits glycogen synthesis, and impairs glucagon response. A) Transmission electron microscopy images of the livers of WT mice on regular chow or HFD for 15 weeks, and treated with vehicle or PXR. 6800× magnification. Scale bar = 1 μm. N = nucleus, M = mitochondria. Arrows point to glycogen. B) Liver glycogen content in the WT study groups. C) PAS staining of the WT liver sections. 4× magnification, scale bar = 500 μm. D) Liver glycogen 3H activity, indicative of 3H-labeled 2-deoxyglucose incorporation into glycogen after glucose bolus in the WT mice. E) Glucagon challenge of WT mice plotted as blood glucose values and area under the curve values. F) Glucagon challenge plotted as incremental change from the 0 timepoint and area under the curve values. Difference between chow and HFD or HFD vehicle and HFD PCN were evaluated with unpaired Student's t-test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To determine if glycogen synthesis was affected by PCN treatment, glycogen 3H activity was measured from the livers collected 1 h after the administration of 3H-2-deoxyglucose, which may be incorporated into glycogen [34]. PCN decreased liver glycogen 3H activity, however, the difference was not statistically significant (Figure 7D.)
In physiological fasting response, an increase in plasma glucagon triggers glycogen mobilization to maintain blood glucose levels. In HFD-fed WT mice, PCN decreased the ability of glucagon to increase blood glucose (Figure 7E. & 7F.), implicating impaired hepatic glucose production. This is probably mainly due to the depleted glycogen reserves.
3.8. PXR-KO protects from activation of inflammatory hepatic gene response under HFD
To further understand the PXR-mediated molecular mechanisms in obesity-related liver disease, we studied the hepatic transcriptome by RNA sequencing. Liver RNA sequencing dataset of the WT mice has been published previously and we reported that PCN treatment caused widespread induction of the cholesterol synthesis genes [16].
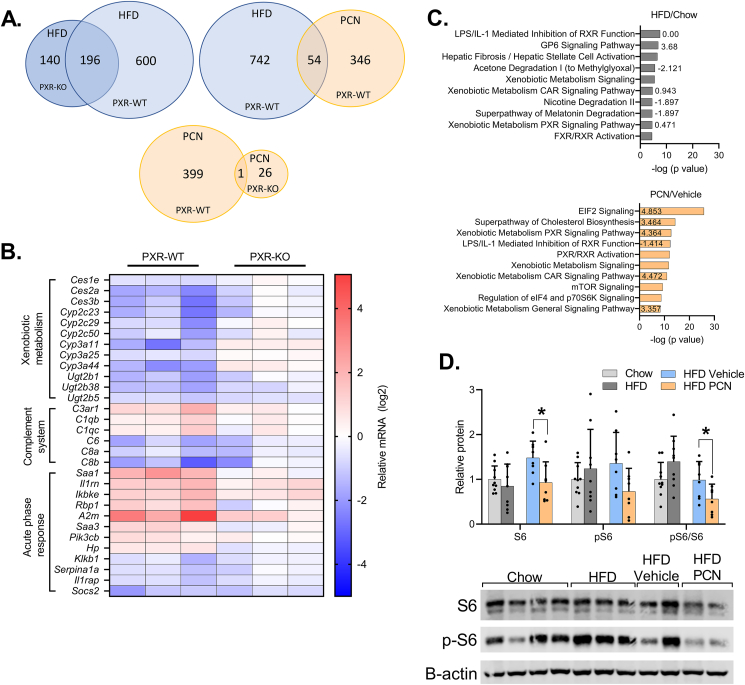
In the WT mice, the HFD differentially regulated 796 genes compared to mice on regular chow (Figure 8A.). In PXR-KO mice, the HFD differentially regulated 336 genes, of which 196 (58%) were shared with the WT mice (Figure 8A.). The direction of gene regulation, i.e. induction or repression, was also similar (Figure 8A.). In the WT mice, of the 400 genes that were regulated by PCN treatment, 54 were also regulated by the HFD (Figure 8A.). Among the PCN-regulated genes only one was significantly regulated in both WT and the PXR-KO mice.
Figure 8.
The effect of HFD and PCN on the liver transcriptome of WT and PXR deficient mice. A) Venn diagram of differentially regulated genes in RNA Sequencing. B) Heatmap of selected differentially expressed genes in response to HFD in the WT and PXR PXR-KO mice livers. These genes were differentially expressed only in the WT mice. C) Genes differentially regulated by the HFD vs. chow or by the PCN treatment vs. vehicle in the WT mice were analyzed with Ingenuity Pathway Analysis for pathway enrichment. Top ten most enriched pathways and their activation z-scores are presented. D) Immunoblotting of liver S6 and pS6. Difference between chow and HFD or HFD vehicle and HFD PCN were evaluated with unpaired Student's t-test. ∗p < 0.05.
The genes that were regulated similarly by the HFD in the WT and the PXR-KO included genes related to lipid synthesis, such as Srebf1 and Fasn, several collagen genes and cytochrome P450 (CYP) 2A and 4A gene family members (Suppl. table 4.). The pathway Hepatic Fibrosis/Hepatic Stellate Cell Activation was predicted to be activated regardless of the PXR genotype, however, in the PXR-KO the pathway was restricted to collagens, while in the WT mice additional factors, such as transforming growth factor beta receptor 2 (Tgfbr2), was upregulated (Suppl. Tables 4 and 5). TGF-β, an agonist for TGFBR2, is one of the key pathways promoting fibrogenic response through hepatic stellate cell (HSC) activation in NAFLD.
As a sign of inflammatory activation, the Acute Phase Response Signaling pathway, with positive Z-score, was enriched in the WT but not in the PXR-KO mice in response to HFD (Figure 8B, Suppl. Tables 5 and 6) The most highly induced gene in this pathway was serum amyloid A 1 (Saa1) with a recently described mechanistic role in the progression of NAFLD [35]. The genes of the Complement System were affected by the HFD only in the WT mice (Figure 8B, Suppl. table 5) in a bidirectional manner: whereas the expression of C1qb, C1qc and C3ar1 was upregulated, C6, C8a and C8b belonging to the membrane attack complex were repressed.
Furthermore, multiple pathways containing different CYP enzymes, such as Superpathway of Melatonin Degradation and Xenobiotic Metabolism PXR signaling pathway were enriched among the HFD-regulated genes only in the WT mice (Suppl. table 5.). Notably, the repressive effect of HFD on Ces, Cyp2c, Cyp3a, and Ugt2b gene families was absent in the PXR-KO mice (Figure 8B.). In the PXR-KO mice, the HFD uniquely affected several cholesterol synthesis pathways (Suppl. table 6.), as reported in our previous publication [16].
In the WT mice, HFD and PCN treatments enriched several shared pathways among the differentially induced genes (Figure 8C.), including LPS/IL-1 Mediated Inhibition of RXR Function and Xenobiotic Metabolism Signaling. Common for these pathways is the involvement of xenobiotic-metabolizing enzymes, including CYPs, which were widely repressed by the HFD and induced by the PCN treatment.
Some of the top PCN enriched pathways were involved in translation and mTOR signaling, such as EIF2 Signaling, mTOR signaling, and Regulation of eIF4 and p706S6K Signaling (Figure 8C. and Suppl. table 7). This was mainly due to the wide induction of ribosomal proteins by PCN. To gain a deeper insight into function of mTOR, which is a major regulator of SREBP1 and lipogenesis, we measured the phosphorylation status of ribosomal protein S6, an mTOR activation marker [36]. Unexpectedly, PCN decreased total S6 and the pS6/S6 ratio (Figure 8D.) and it remains possible that some compensatory mechanisms have already started to inhibit the pathway in the studied time point.
4. Discussion
In the present study we evaluated the metabolic role of xenobiotic-sensing nuclear receptor PXR in obese mice. We show for the first time that in the mice with existing metabolic disorder PXR activation results in dichotomous metabolic response: aggravated liver steatosis but improved systemic glucose tolerance.
PXR activation by a ligand dramatically aggravated liver steatosis and impaired liver insulin signaling. PXR activation in lean mice for a similar time did not induce steatosis, showing that HFD-induced obesity and associated metabolic disturbances sensitized mice to PXR-induced hepatosteatosis. Liver fat accumulation was, at least partly, due to increased lipid synthesis, which was facilitated by increased liver glucose uptake and utilization of glycogen reserves. Systemically, these rearrangements of liver metabolism led to apparent dissociation of systemic insulin sensitivity and glucose tolerance from the liver fat accumulation. We and others have previously shown that short-term PXR activation impairs glucose tolerance in healthy humans, mice and rats [[10], [11], [12]]. Therefore, it was unexpected to find that PXR activation in obese mice did not impair glucose tolerance but contrastingly had some tendency to improve it.
In agreement with increased liver fat and plasma ALT activity, liver insulin sensitivity was impaired in the PCN-treated mice indicated by decreased liver AKT phosphorylation and lower IRS1 protein level [29]. PCN did not affect skeletal muscle insulin sensitivity or glucose uptake. Furthermore, PCN did not affect the muscle transcriptome and only very few changes in gene expression could be observed in WAT suggesting that the systemic metabolic effects of PCN treatment are mediated through liver that agrees with the expression of PXR in the liver and absence in the other major metabolic tissues. This agrees with a recent study reporting no effect of adipocyte-specific PXR deficiency on metabolic parameters under HFD [37].
A central feature of hepatic insulin resistance is the inability of insulin to suppress gluconeogenesis through AKT-directed phosphorylation of the transcription factor forkhead box O1 (FOXO1), which in turn regulates the expression of gluconeogenic G6pc and Pck1 [38]. On the other hand, PXR has been shown to bind to FOXO1 to downregulate G6pc and Pck1 [32]. In agreement, we observed that also in the obese mice PXR activation suppressed hepatic G6pc and Pck1 expression, despite the aggravated insulin resistance. Gluconeogenesis was assayed by a pyruvate tolerance test. PCN-treated mice did not differ from the vehicle-treated mice, and thus, the pyruvate tolerance test results do not suggest any PCN-elicited changes in gluconeogenesis. Therefore, despite the downregulation of the genes coding for the gluconeogenic enzymes, the liver was able to release glucose normally.
Besides gluconeogenesis, glycogenolysis is another key process in maintaining a stable blood glucose level during fasting periods. Interestingly, PCN treatment depleted hepatic glycogen. This agrees with an earlier study done in female rats [39]. Hepatic glycogen reserves are a dynamic buffer increasing and decreasing in response to energy surplus or demand. In agreement with the glycogen depletion, glucagon administration resulted in diminished blood glucose increase in the PCN-treated mice.
We utilized labeled 2-deoxyglucose to assess glycogen synthesis. Although 2-deoxyglucose is considered metabolically inactive, it may be incorporated into glycogen [34]. Glycogen 3H activity tended to be lower, but the result was not statistically significant. Glycogen synthase, the enzyme responsible for glycogenesis, is primarily activated by glucose-6-phosphate [40]. The PCN-treated mice displayed increased glucose uptake and had more hepatic 2-deoxyglucose-6-phosphate, but less of it was incorporated to glycogen. This may suggest that glycogenesis was indeed suppressed, and glucose-6-phosphate directed to other metabolic pathways. It is tempting to speculate that PXR activation redirected the glucose-6-phosphate flux to supply the needs of the increased de novo lipogenesis. Alternatively, it remains possible that the increase in glucose utilization was compensatory to the reduced fatty acid oxidation.
Interestingly, the PCN-treated mice displayed lower fasting glucose than the controls after a 6 h fast but not after a 12-hour fast. Glycogenolysis plays a bigger role in maintenance of blood glucose during the early stages of fast while the role of gluconeogenesis increases in the prolonged fasting [41]. Thus, it is likely that impaired glycogenolysis, due to the low glycogen reserves, explains the reduced blood glucose in the PCN-treated mice after a short fast. In conclusion, both increased hepatic glucose uptake and reduced glucose production play a role in the dissociation of hepatic insulin resistance and systemic glucose tolerance in the PCN-treated, obese mice.
Although PXR activation in obese mice induced hepatosteatosis, the gene expression profiling and subsequent pathway enrichment analysis did not identify lipogenic pathways among the major pathways activated. Previously, PXR activation has been shown to associate with activation of SREBP1, the main regulator of de novo lipogenesis, in ob/ob mice and in human hepatocytes [8,13]. Further, the induction of ribosomal proteins pointed at activation of mTOR, a central regulator of SREBP1 [42]. However, in our previous study [16] we described that PXR activation in these mice resulted in SREBP2 activation but did not affect SREBP1 activity or expression of SREBP1 target genes. Further, the current study shows PXR activation to inhibit hepatic mTOR activity, a central determinant of SREBP1 activity [43]. In conclusion, PXR activation did not activate SREBP1 in the obese mice. PXR has been shown to regulate lipid metabolism genes, including the fatty acid synthesis gene Elovl6, the β-oxidation genes Hmgcs2 and Cpt1a, and multifunctional Lpin1 independent of SREBP1 [9,13,44]. In line with the previous reports, we observed induced Elovl6 and repressed Hmgcs2 and Cpt1a in response to PCN. However, PXR activation repressed Lpin1. Altogether, the gene expression changes suggest PXR to increase lipogenesis and suppress β-oxidation independently of SREBP1. Another lipogenic pathway suggested to be induced by PXR is PPARγ, however, we did not detect activation of that either [45]. It should, however, be kept in mind that the RNA-Seq and protein results represent a snapshot in one time point and therefore all the mechanisms involved may not have been detected. In addition to direct effects of PXR to liver, additional mechanisms may play some role in the PXR-induced metabolic alterations. For example, the metabolomics analysis indicated that the liver corticosterone level was increased by PCN treatment in the WT but not in the PXR-KO mice (Supplementary table 2.) in agreement with a previous report [46]. Intestine is another major site of PXR expression and may in certain conditions play role in liver steatosis, however, this was described to require several months of PXR activation [47].
Some earlier studies have shown PXR deficiency to protect mice from diet-induced obesity and metabolic dysfunction [[13], [14], [15]]. In the current study, with a different mouse substrain, PXR-KO mice had similar-to-WT increases in bodyweight, liver steatosis, glucose tolerance AUC values and plasma ALT activities in response to HFD-feeding. Also, some gene responses were different; while He et al. reported downregulation of Lpin1 in the HFD-fed PXR-KO mouse, in our study Lpin1 was upregulated in similar conditions [13]. The difference in weight gain may potentially be the primary reason for all these differences but this requires further clarification in future studies. Although the metabolic phenotype did not suggest any clear protective effect of PXR deficiency, the “omics” analyses still suggest that PXR-KO has some protective effect against NAFLD progression. In the metabolomics analysis the HFD-feeding induced clear distinction to the chow-feeding in the overall metabolomics profile only in the WT mice and not in the PXR-KO mice. RNA sequencing of the livers of the WT and PXR-KO mice indicated that the HFD regulates fewer hepatic genes in the PXR-KO mice. Some of the genes altered by HFD only in the WT mice, such as genes related to inflammation, can be assumed to play a mechanistic role in the progression of simple steatosis to more severe forms of NAFLD.
Interestingly, the HFD repressed expression of multiple CYP enzymes but only in WT and not in PXR-KO mice. Furthermore, basal Cyp3a11 expression was not affected by PXR-KO, but it was only repressed by the HFD in the WT mice. These observations suggest PXR to be indispensable for HFD-induced CYP repression. NAFLD has been shown to suppress CYP3A4 expression and function in humans [48,49]. Hence, it is possible that metabolic dysfunction and NAFLD could suppress drug metabolism through PXR-mediated mechanism in humans, too.
In conclusion, this study shows that PXR is a powerful regulator of hepatic lipid and glucose metabolism in obesity. While PXR activation resulted in aggravation of liver steatosis this was not reflected to glucose tolerance. We propose this to be due to remodeling of the glucose metabolism to enable the maximal lipid synthesis, which may be considered unbeneficial for the metabolic health. Thus, we call the phenomenon pseudo-improvement in glucose tolerance. The prominent effect of PXR activation in obese mice raises the concern that also obese humans may be more susceptible to harmful metabolic effects of PXR ligands.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Olli Kärkkäinen is a cofounder of Afekta Technologies Ltd., a company providing metabolomics analysis services. The other authors declare no conflict of interest.
Acknowledgements
This study was funded by a grant from the Academy of Finland (323706). This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 825762 (EDCMET). We thank Ritva Tauriainen, Miia Reponen and Marlies Sagmeister for technical assistance. Biocenter Oulu (University of Oulu) Tissue Imaging Center and Sequencing Center are acknowledged for the help with TEM imaging and RNA sequencing. The animal work was carried out with the support of the Oulu University Laboratory Animal Centre Research Infrastructure. We appreciate Biocenter Finland and Biocenter Kuopio for supporting the LC-MS laboratory facility.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101779.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meex R.C.R., Watt M.J. Hepatokines: linking nonalcoholic fatty liver disease and insulin resistance. Nat Rev Endocrinol. 2017;13(9):509–520. doi: 10.1038/nrendo.2017.56. [DOI] [PubMed] [Google Scholar]
- 3.Foulds C.E., Treviño L.S., York B., Walker C.L. Endocrine-disrupting chemicals and fatty liver disease. Nat Rev Endocrinol. 2017;13(8):445–457. doi: 10.1038/NRENDO.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delfosse V., Dendele B., Huet T., Grimaldi M., Boulahtouf A., Gerbal-Chaloin S., et al. Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds. Nat Commun. 2015;6 doi: 10.1038/NCOMMS9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kliewer S.A., Moore J.T., Wade L., Staudinger J.L., Watson M.A., Jones S.A., et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92(1):73–82. doi: 10.1016/S0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 6.Hakkola J., Rysä J., Hukkanen J. Regulation of hepatic energy metabolism by the nuclear receptor PXR. Biochimica et Biophysica Acta - Gene Regulatory Mechanisms. 2016;1859(9):1072–1082. doi: 10.1016/j.bbagrm.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Toporova L., Balaguer P. Nuclear receptors are the major targets of endocrine disrupting chemicals. Mol Cell Endocrinol. 2020;502 doi: 10.1016/J.MCE.2019.110665. [DOI] [PubMed] [Google Scholar]
- 8.Bitter A., Rümmele P., Klein K., Kandel B.A., Rieger J.K., Nüssler A.K., et al. Pregnane X receptor activation and silencing promote steatosis of human hepatic cells by distinct lipogenic mechanisms. Arch Toxicol. 2015;89(11):2089–2103. doi: 10.1007/s00204-014-1348-x. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J., Zhai Y., Mu Y., Gong H., Uppal H., Toma D., et al. A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem. 2006;281(21):15013–15020. doi: 10.1074/jbc.M511116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassani-Nezhad-Gashti F., Rysä J., Kummu O., Näpänkangas J., Buler M., Karpale M., et al. Activation of nuclear receptor PXR impairs glucose tolerance and dysregulates GLUT2 expression and subcellular localization in liver. Biochem Pharmacol. 2018;148:253–264. doi: 10.1016/J.BCP.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Ling Z., Shu N., Xu P., Wang F., Zhong Z., Sun B., et al. Involvement of pregnane X receptor in the impaired glucose utilization induced by atorvastatin in hepatocytes. Biochem Pharmacol. 2016;100:98–111. doi: 10.1016/j.bcp.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 12.Rysä J., Buler M., Savolainen M.J., Ruskoaho H., Hakkola J., Hukkanen J. Pregnane X receptor agonists impair postprandial glucose tolerance. Clin Pharmacol Ther. 2013;93(6):556–563. doi: 10.1038/clpt.2013.48. [DOI] [PubMed] [Google Scholar]
- 13.He J., Gao J., Xu M., Ren S., Stefanovic-Racic M., O'Doherty R.M., et al. PXR ablation alleviates diet-induced and genetic obesity and insulin resistance in mice. Diabetes. 2013;62(6):1876–1887. doi: 10.2337/db12-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spruiell K., Richardson R.M., Cullen J.M., Awumey E.M., Gonzalez F.J., Gyamfi M.A. Role of pregnane X receptor in obesity and glucose homeostasis in male mice. J Biol Chem. 2014;289(6):3244–3261. doi: 10.1074/jbc.M113.494575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao L.Y., Xu J.Y., Shi Z., Englert N.A., Zhang S.Y. Pregnane X receptor (PXR) deficiency improves high fat diet-induced obesity via induction of fibroblast growth factor 15 (FGF15) expression. Biochem Pharmacol. 2017;142:194–203. doi: 10.1016/j.bcp.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 16.Karpale M., Käräjämäki A.J., Kummu O., Gylling H., Hyötyläinen T., Orešič M., et al. Activation of nuclear receptor PXR induces atherogenic lipids and PCSK9 through SREBP2-mediated mechanism. Br J Pharmacol. 2021 doi: 10.1111/bph.15433. [DOI] [PubMed] [Google Scholar]
- 17.Xie W., Barwick J.L., Downes M., Blumberg B., Simon C.M., Nelson M.C., et al. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406(6794):435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- 18.Kallio M.A., Tuimala J.T., Hupponen T., Klemelä P., Gentile M., Scheinin I., et al. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genom. 2011;12(1):507. doi: 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krämer A., Green J., Pollard J., Tugendreich S., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., et al. QuPath: open source software for digital pathology image analysis. bioRxiv. 2017 doi: 10.1101/099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaitoun A.M., Al Mardini H., Awad S., Ukabam S., Makadisi S., Record C.O. Quantitative assessment of fibrosis and steatosis in liver biopsies from patients with chronic hepatitis C. J Clin Pathol. 2001;54:461–465. doi: 10.1136/jcp.54.6.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferre P., Leturque A., Burnol A.F., Penicaud L., Girard J. A method to quantify glucose utilization in vivo in skeletal muscle and white adipose tissue of the anaesthetized rat. Biochem J. 1985;228(1):103–110. doi: 10.1042/bj2280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Dijk T.H., Laskewitz A.J., Grefhorst A., Boer T.S., Bloks V.W., Kuipers F., et al. A novel approach to monitor glucose metabolism using stable isotopically labelled glucose in longitudinal studies in mice. Lab Anim. 2013;47(2):79–88. doi: 10.1177/0023677212473714. [DOI] [PubMed] [Google Scholar]
- 24.Klåvus A., Kokla M., Noerman S., Koistinen V.M., Tuomainen M., Zarei I., et al. “Notame”: workflow for non-targeted LC–MS metabolic profiling. Metabolites. 2020;10(4):135. doi: 10.3390/METABO10040135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12(6):523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sumner L.W., Alexander A.E., Ae A., Ae D.B., Ae M.H.B., Beger R., et al. Proposed minimum reporting standards for chemical analysis. Metabolomics. 2007;3(3):211–221. doi: 10.1007/S11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs R.L., van der Veen J.N., Vance D.E. Finding the balance: the role of S-adenosylmethionine and phosphatidylcholine metabolism in development of nonalcoholic fatty liver disease. Hepatology (Baltimore, Md. 2013;58(4):1207–1209. doi: 10.1002/HEP.26499. [DOI] [PubMed] [Google Scholar]
- 28.Schweitzer G.G., Chen Z., Gan C., McCommis K.S., Soufi N., Chrast R., et al. Liver-specific loss of lipin-1-mediated phosphatidic acid phosphatase activity does not mitigate intrahepatic TG accumulation in mice. JLR (J Lipid Res) 2015;56(4):848–858. doi: 10.1194/JLR.M055962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeFronzo R.A., Ferrannini E., Groop L., Henry R.R., Herman W.H., Holst J.J., et al. Type 2 diabetes mellitus. Nat Rev Dis Prim. 2015;1(1):1–22. doi: 10.1038/nrdp.2015.19. [DOI] [PubMed] [Google Scholar]
- 30.Hiratani K., Haruta T., Tani A., Kawahara J., Usui I., Kobayashi M. Roles of mTOR and JNK in serine phosphorylation, translocation, and degradation of IRS-1. Biochem Biophys Res Commun. 2005;335(3):836–842. doi: 10.1016/j.bbrc.2005.07.152. [DOI] [PubMed] [Google Scholar]
- 31.Jitrapakdee S. Transcription factors and coactivators controlling nutrient and hormonal regulation of hepatic gluconeogenesis. Int J Biochem Cell Biol. 2012:33–45. doi: 10.1016/j.biocel.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Kodama S., Koike C., Negishi M., Yamamoto Y. Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol. 2004;24(18):7931–7940. doi: 10.1128/mcb.24.18.7931-7940.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cappel D.A., Deja S., Duarte J.A.G., Kucejova B., Iñigo M., Fletcher J.A., et al. Pyruvate-carboxylase-mediated anaplerosis promotes antioxidant capacity by sustaining TCA cycle and redox metabolism in liver. Cell Metabol. 2019;29(6):1291–1305.e8. doi: 10.1016/j.cmet.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colwell D.R., Higgins J.A., Denyer G.S. Incorporation of 2-deoxy-D-glucose into glycogen. Implications for measurement of tissue-specific glucose uptake and utilisation. Int J Biochem Cell Biol. 1996;28(1):115–121. doi: 10.1016/1357-2725(95)00110-7. [DOI] [PubMed] [Google Scholar]
- 35.Jiang B., Wang D., Hu Y., Li W., Liu F., Zhu X., et al. Serum amyloid A1 exacerbates hepatic steatosis via TLR4-mediated NF-κB signaling pathway. Mol Metabol. 2022;59 doi: 10.1016/J.MOLMET.2022.101462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H., Liu Y., Wang D., Xu Y., Dong R., Yang Y., et al. The upstream pathway of mTOR-mediated autophagy in liver diseases. Cells. 2019;8(12):1597. doi: 10.3390/cells8121597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F., Liu J., Hernandez R., Park S.-H., Lai Y.-J., Wang S., et al. 2023. Adipocyte-derived PXR signaling is dispensable for diet-induced obesity and metabolic disorders in mice. Drug Metabolism and Disposition: the Biological Fate of Chemicals: DMD-AR-2023-001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farmer S.R. The forkhead transcription factor Foxo1: a possible link between obesity and insulin resistance. Mol Cell. 2003:6–8. doi: 10.1016/S1097-2765(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 39.Tuchweber B., Solymoss B., Khandekar J.D., Kovacs K., Garg B.D., Zsigmond G., et al. Effect of pregnenolone-16α-carbonitrile on the hepatic ultrastructure, glycogen content and ethylmorphine N-demethylase activity in pregnant, fetal, and newborn rats. Exp Mol Pathol. 1972;17(3):281–295. doi: 10.1016/0014-4800(72)90041-X. [DOI] [PubMed] [Google Scholar]
- 40.Roach P. Glycogen and its metabolism. Curr Mol Med. 2005;2(2):101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- 41.Sharabi K., Tavares C.D.J., Rines A.K., Puigserver P. Molecular pathophysiology of hepatic glucose production. Mol Aspect Med. 2015;46:21–33. doi: 10.1016/J.MAM.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S., Brown M.S., Goldstein J.L. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci U S A. 2010;107(8):3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakan I., Laplante M. Connecting mTORC1 signaling to SREBP-1 activation. Curr Opin Lipidol. 2012;23(3):226–234. doi: 10.1097/MOL.0b013e328352dd03. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura K., Moore R., Negishi M., Sueyoshi T. Nuclear pregnane X receptor cross-talk with FoxA2 to mediate drug-induced regulation of lipid metabolism in fasting mouse liver. J Biol Chem. 2007;282(13):9768–9776. doi: 10.1074/jbc.M610072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoekstra M., Lammers B., Out R., Li Z., Van Eck M., Van Berkel T.J.C. Activation of the nuclear receptor PXR decreases plasma LDL-cholesterol levels and induces hepatic steatosis in LDL receptor knockout mice. Mol Pharm. 2009;6(1):182–189. doi: 10.1021/MP800131D. [DOI] [PubMed] [Google Scholar]
- 46.Zhai Y., Pai H.V., Zhou J., Amico J.A., Vollmer R.R., Xie W. Activation of pregnane X receptor disrupts glucocorticoid and mineralocorticoid homeostasis. Molecular Endocrinology (Baltimore, Md. 2007;21(1):138–147. doi: 10.1210/ME.2006-0291. [DOI] [PubMed] [Google Scholar]
- 47.Cheng J., Krausz K.W., Tanaka N., Gonzalez F.J. Chronic exposure to rifaximin causes hepatic steatosis in pregnane X receptor-humanized mice. Toxicol Sci : An Official Journal of the Society of Toxicology. 2012;129(2):456–468. doi: 10.1093/TOXSCI/KFS211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jamwal R., De La Monte S.M., Ogasawara K., Adusumalli S., Barlock B.B., Akhlaghi F. Nonalcoholic fatty liver disease and diabetes are associated with decreased CYP3A4 protein expression and activity in human liver. Mol Pharm. 2018;15(7):2621–2632. doi: 10.1021/acs.molpharmaceut.8b00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woolsey S.J., Mansell S.E., Kim R.B., Tirona R.G., Beaton M.D. CYP3A activity and expression in nonalcoholic fatty liver disease. Drug Metabol Dispos. 2015;43(10):1484–1490. doi: 10.1124/dmd.115.065979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.