Abstract
Herpes simplex virus 1 encodes two multifunctional regulatory proteins, infected-cell proteins 22 and 0 (ICP22 and ICP0). ICP0 is a promiscuous transactivator, whereas ICP22 is required in vivo and for efficient replication and expression of a subset of late (γ2) genes in rodent or rabbit cell lines and in primary human cell strains (restrictive cells) but not in HEp-2 or Vero (permissive) cells. We report the identification in the yeast two-hybrid system of a cellular protein designated p60 that interacts with ICP22. This protein (apparent Mr of 60,000) has not been previously described and has no known motifs. Analyses of p60 revealed the following. (i) p60 bound fast-migrating, underprocessed wild-type ICP22 and ICP22 lacking the carboxyl-terminal 24 amino acids but not ICP22 lacking the carboxyl-terminal 40 amino acids, whereas the previously identified cellular protein p78 (R. Bruni and B. Roizman, J. Virol. 72:8525–8531, 1998) bound all forms of ICP22. The interaction of p60 with only one isoform of ICP22 supports that hypothesis that each isoform of herpes simplex virus proteins performs a specific function that may be different from that of other isoforms. (ii) p60 also bound ICP0; the binding of ICP0 was independent of that of ICP22. (iii) p60 localized in uninfected rabbit skin cells in both nuclei and cytoplasm. In rabbit skin cells infected with wild-type virus, p60 was posttranslationally processed to a higher apparent Mr but was not redistributed. Posttranslational processing required the presence of the genes encoding ICP22 and UL13 protein kinase. (iv) In uninfected HEp-2 cells, p60 localized primarily in nuclei. Soon after infection with wild-type virus, the p60 localized in discrete small nuclear structures with ICP0. Late in infection, both ICP0 and p60 tended to disperse but p60 did not change in apparent Mr. The localization of p60 was independent of ICP22, but p60 tended to be more localized in small nuclear structures and less dispersed in cells infected with mutants lacking the genes encoding the UL13 or US3 protein kinases. The results suggest that posttranslational modification of p60 is mediated either by ICP0 (permissive cells) or by ICP22 and UL13 protein kinase (restrictive rabbit skin cells) and that the restrictive phenotype of rabbit skin cells may be related to the failure to process p60 by mutants lacking the genes encoding UL13 or ICP22.
The herpes simplex virus 1 (HSV-1) genome contains at least 84 genes, whose expression is coordinately regulated and sequentially ordered in a cascade fashion (16, 17, 36). The first genes to be expressed are the α genes, whose products enable the expression of the β and γ genes. With the exception of infected-cell protein 47 (ICP47) (43), all products of the α genes are regulatory proteins (36). ICP4, encoded by the α4 gene, is an essential viral regulatory protein, which binds DNA at specific sites and can both transactivate and repress the expression of viral genes (36). ICP27, the product of the α27 gene, mediates the processing and translocation of mRNA (14, 15, 39). The two remaining α proteins, ICP22 and ICP0, are the subjects of the present study.
ICP22 is a 420-amino-acid protein encoded by the α22 gene. A smaller transcript promoted from the amino-terminal domain of the α22 gene encodes a polypeptide colinear with the carboxyl-terminal 60% of ICP22 and designated US1.5 (7). The α22 gene is dispensable in Vero and HEp-2 (permissive) cell lines, but α22− mutant viruses replicate poorly in rodent or rabbit skin (restrictive) cell lines and in confluent primary human fibroblasts (23, 31, 40). One of the viruses tested in these studies, R325, lacked the carboxyl-terminal 220 codons of the α22 gene (31). In the restrictive cells, the mutant virus exhibited a reduction of α0 and of US11 mRNA and proteins (33). Early in infection, ICP22 localizes in punctuate nuclear structures. At the time of onset of viral DNA synthesis, ICP22 colocalizes in infected nuclei with ICP4, viral DNA, RNA polymerase II, and a small cellular protein (EAP) named on the basis of its association with Epstein-Barr virus small nuclear RNAs. This aggregation requires the presence of a functional protein kinase encoded by UL13 and is necessary for optimal late-gene expression (21, 33). ICP22 is extensively modified in infected cells. These modifications include phosphorylation by the viral protein kinases US3 and UL13 (31, 33) and nucleotidylylation by casein kinase II (4, 24, 25). A recombinant virus carrying a deletion in the UL13 gene was found to be similar to R325 with respect to several properties. Studies of the UL13− virus in restricted cells led to the conclusion that the phosphorylation of ICP22 is necessary for the functions described above. At least one of the sequences required for posttranslational modification of ICP22 maps in the carboxyl-terminal domain of ICP22. Thus, the ICP22 encoded by two mutants used in this study, R7820 and R7810, lacking the carboxyl-terminal 24 and 40 amino acids, respectively, is not posttranslationally processed (27). These mutants have properties similar to those of R325 (27).
In other recent studies, it was shown that ICP22 is required for the alternative splicing of the α0 gene and for the accumulation of the viral host shutoff protein in infected cells (8, 26). ICP22 has also been reported to be responsible for the aberrant phosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II (34, 35). More recently, this laboratory reported that ICP22 interacts with a previously unknown cell cycle-regulated cellular protein, p78 (6). Interestingly, ICP22 also accumulated in a cell cycle-specific fashion and novel forms of ICP22 could be detected during the cell cycle. These forms differ from the previously published isoforms with respect to their electrophoretic mobility. These results suggest that ICP22 is able to interact with cell cycle-regulated proteins (6).
ICP0, a protein of 775 amino acids, is encoded by the α0 gene and is best described as a promiscuous transactivator inasmuch as it transactivates both viral and cellular genes (11). HSV-1 strains lacking the α0 gene replicate less efficiently than the wild-type parent (37, 41). Recent studies indicate that ICP0 is a multifunctional protein inasmuch as it interacts with several cellular proteins. For example, the protein has been shown to bind and colocalize with the cell cycle regulator cyclin D3 (19). ICP0 also colocalizes with a ubiquitin-specific protease in nuclear dense bodies known as PML or ND10s (12, 22). Later in infection, ICP0 is translocated into the cytoplasm and interacts with the elongation factor EF-1δ (18). The possible role of ICP0 in the cytoplasm and especially its interactions with EF-1δ are reflected in the finding that EF-1δ is phosphorylated by the viral protein kinase UL13 (20). ICP0 is also phosphorylated by the UL13 protein kinase (29).
The interaction of viral regulatory proteins among themselves has been the subject of many studies. Thus, ICP4 interacts with ICP0 (42) and, as noted above, ICP4 and ICP22 colocalize late in infection in structures containing viral DNA and RNA polymerase (21). In this report, we describe the identification of a hitherto unknown cellular protein, p60, that binds independently both to ICP0 and ICP22. In rabbit skin cells infected with wild-type virus, the p60 protein was processed to a form with a higher apparent molecular weight and the processing required ICP22 and UL13 protein. In HEp-2 cells, processing to a higher apparent Mr did not take place, but p60 was translocated to small nuclear structures containing ICP0. In this instance the translocation was independent of ICP22 and of the UL13 or US3 protein kinases. Our data suggest a correlation between the sequestering or posttranslational modification of p60 and the permissivity of cells for optimal HSV gene expression.
MATERIALS AND METHODS
Cell lines and viruses.
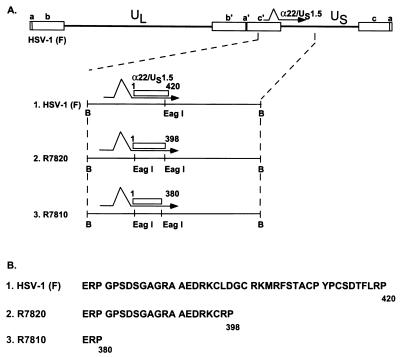
HeLa and HEp-2 cell lines were obtained from the American Type Culture Collection. Rabbit skin cells were originally obtained from J. McClaren. HSV-1(F) is the prototype HSV-1 strain used in this laboratory (10). R7810 and R7820 (Fig. 1) are recombinant viruses from which the carboxyl-terminal 40 and 24 codons of α22, respectively, have been deleted (27). Recombinant virus R325 has been described elsewhere (31). Recombinants lacking the genes encoding the protein kinase US3 (R7041) or UL13 (R7356) were described elsewhere (32, 33).
FIG. 1.
(A) Schematic representation of the DNA sequence arrangements of the HSV-1 genome of new recombinant viruses used in this study. Top line, linear representation of the HSV-1 genome. The open rectangles represent the terminal repeats flanking the unique long (UL) and unique short (US) sequences. The location of the α22/US1.5 gene is shown. Line 1, representation of the BamHI N fragment which contains the α22/US1.5 gene. The transcript is represented by the arrow, and the coding domain of the α22/US1.5 genes is represented by the open rectangle. Line 2, recombinant R7820, in which the C-terminal 22 amino acids encoded by the α22 open reading frame have been deleted. Line 3, recombinant R7810, in which the C-terminal 40 amino acids encoded by the α22 open reading frame have been deleted. B, BamHI. (B) Polypeptide sequence of the carboxyl terminus of ICP22. Line 1, the 43 amino acids of the carboxyl-terminal domain of ICP22. Line 2, sequence of the 21 amino acids remaining within the carboxyl-terminal domain of R7820. Line 3, the 3 amino acids remaining within the carboxyl-terminal domain of R7810.
Plasmids.
pBH1025 contained a 1-kb cDNA insert encoding 300 amino acids of p60 in the yeast two-hybrid system vector pACT (Clontech). pBH1026 was constructed by ligating 550 bp of the 5′ end of the cDNA insert of pBH1025 in frame with the glutathione S-transferase (GST) gene in pGEX4T-3 (Pharmacia). pRB4965 contained the carboxyl-terminal 370 codons of p78 in pGEX4T-3 and has been described elsewhere (6). pRB5113 contained the entire coding sequence of α22/US1.5 genes in pGBT9 (6).
Yeast two-hybrid system and isolation of cDNA clones.
The yeast strain HF7c (Clontech) was transformed with pRB5113, grown on a large scale, and transformed with a cDNA library derived from an Epstein-Barr virus-immortalized human peripheral-blood B-lymphocyte cell line (Clontech) cloned in pACT. Positive clones were selected and isolated as described elsewhere (5). cDNA clones containing larger p60 inserts were also isolated as described elsewhere (5), using the 1-kb insert of pBH1025 as a probe.
GST pull-down experiments.
Subconfluent HeLa cells grown in a 150-cm2 flask were exposed to 10 PFU of HSV-1(F), R7810, or R7820 per cell. After 18 h at 37°C, the infected cells were scraped into phosphate-buffered saline (PBS), rinsed once with PBS, and resuspended in 1 ml of PBS* (PBS containing 1% deoxycholate, 1% Nonidet P-40, 100 μg of phenylmethylsulfonyl fluoride per ml, 50 μg of α-tosyl-l-lysine chloromethyl ketone per ml, and 100 μg of tolyl-l-phenylanalyl chloromethyl ketone per ml). Recombinant pGEX vectors were grown in BL21, and fusion proteins were isolated as recommended by the manufacturer (Pharmacia). Binding-reaction mixtures containing 300 μl of cell extract mixed with 3 to 5 μg of fusion protein were incubated at 4°C for 4 to 5 h. Beads were collected by centrifugation, rinsed three times with PBS* (1 ml), and resuspended with 100 μl of 2× disruption buffer (100 mM Tris-Cl [pH 6.8], 200 mM dithiothreitol, 4% sodium dodecyl sulfate, 0.2% bromophenol blue, 20% glycerol).
Electrophoretic separation of proteins.
Subconfluent rabbit skin cells or HEp-2 cells grown in 25cm2-flasks were infected with 10 PFU of virus per cell. After 18 h at 37°C, the cells were scraped in PBS, rinsed once with PBS, and resuspended with 130 μl of 2× disruption buffer (38). They were then sonicated for 10 s, and 40 to 60 μl was subjected to electrophoretic separation.
Antibodies.
Monoclonal antibody to ICP0, purchased from the Goodwin Institute (Plantation, Fla.), and polyclonal antiserum R77 to ICP22 are described elsewhere (1, 2). The polyclonal antiserum to p60 was generated as follows. pBH1026 was grown in BL21, and the fusion protein was purified from large-scale culture as recommended by the manufacturer (Pharmacia). Two rabbits were injected subcutaneously at Josman Laboratories (Napa, Calif.) with 1 mg of fusion protein per injection at 14-day intervals. The serum used in this study was collected 1 week after the fifth immunization.
Immunoblots.
Cell extracts were separated in sodium dodecyl sulfate–7% denaturing polyacrylamide gels as previously described (31). Proteins were electrically transferred to nitrocellulose sheets, blocked for 1 h at room temperature in 5% milk (in PBS), and then reacted for 2 to 4 h at room temperature with the primary antibody diluted in 1% bovine serum albumin (BSA) in PBS. The monoclonal antibody to ICP0 and polyclonal antiserum to ICP22 were diluted 1:1,000, and the polyclonal antiserum to p60 was diluted 1:6,000. The nitrocellulose sheets were rinsed four times in 5% milk (in PBS) and reacted for 1 h at room temperature with the secondary antibody conjugated to either alkaline phosphatase (Bio-Rad) or horseradish peroxidase (Amersham). The sheets were then rinsed four times in PBS, and enzymatic reactions were done as recommended by the manufacturer.
Immunofluorescence.
HEp-2 or rabbit skin cells were grown in wells on 1 by 3-in. slides, exposed to 10 PFU of HSV-1(F) per cell, maintained for 18 h at 37°C, and then fixed in methanol at −20°C for 20 min. The cells were blocked at room temperature for 60 min in 1% BSA in PBS containing 20% normal human serum, rinsed once with PBS, and then reacted for 18 to 24 h at 4°C with the primary antibodies diluted in 1% BSA in PBS containing 10% normal human serum. Dilutions were 1:2,000 for the polyclonal serum to p60 and 1:300 for the monoclonal antibody to ICP0. The cells were rinsed three times with PBS and then reacted for 1 h at room temperature with goat anti-rabbit immunoglobulin G (IgG) conjugated to Texas red (Molecular Probes) and goat anti-mouse IgG conjugated to fluorescein isothiocyanate (FITC; Sigma). The cells were rinsed again with PBS and mounted in PBS containing 90% glycerol and 1 mg of p-phenylenediamine per ml of solution. The slides were examined with a Zeiss confocal fluorescence microscope, and digitized images were acquired with software provided by the manufacturer and printed with a Tektronix 440 phaser printer. Single-color images were acquired by excitation with an argon-krypton laser at 488 nm (FITC) or 568 nm (Texas red). Double-stained images were acquired by using a split image of both fluorochromes filtered by 515- to 540-nm band-pass (FITC) and 590-nm long-pass (Texas red) filters. Overlays were acquired with the software provided by the manufacturer. Each set of images was acquired with the same settings and the images were not modified.
RESULTS
ICP22 and ICP0 interact with a novel cellular protein.
In the first series of experiments, we screened an Epstein-Barr virus-immortalized human peripheral blood B-lymphocyte cell line cDNA cloned in the yeast two-hybrid system vector pACT with the full-length α22 gene as a bait. Several positive clones were identified and further analyzed for specificity (see Materials and Methods). One of the clones, designated 22.6, interacted with full-length ICP22 and, in a subsequent experiment, with ICP0 amino acids 111 to 241 but not with a truncated ICP22 protein extending from amino acids 1 to 267 or with an irrelevant protein (e.g., ORF P or ICP0 amino acids 543 to 775).
Clone 22.6 contained a cDNA insert of approximately 1 kb. The sequence of this clone revealed an open reading frame encoding 300 amino acids (data not shown). A larger 1.5-kb cDNA clone was isolated from a HeLa cell library and on sequencing was found to contain an open reading frame encoding 441 amino acids (Fig. 2). The protein encoded by this open reading frame was designated p60 on the basis of its apparent molecular weight, as described below. Analyses of data banks with either BLAST or FastA software (3, 30) failed to reveal known homologs. This clone does not contain an initiator methionine residue. Although the sequence shows two leucine residues at positions 15 and 17, which could potentially act as initiator amino acids (Fig. 2), it is likely that the 1.5-kb clone does not encode a full-length p60 and that a portion of the amino terminus is missing.
FIG. 2.
Nucleotide and amino acid sequences of p60. The top line represents the nucleotide sequence, and the bottom line shows the amino acid sequence. Numbers on the right refer to the nucleotide sequence. The underlined sequences indicate the domain containing leucine repeats.
p60 interacts in vitro with ICP22 and ICP0.
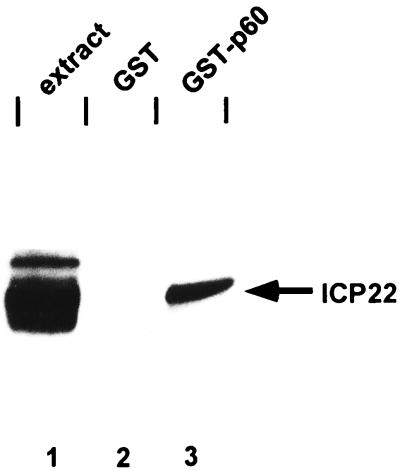
To characterize the interactions between p60 and ICP22 or ICP0, a GST-p60 fusion protein was reacted with an HSV-1(F)-infected HeLa cell extract. The proteins pulled down by the GST-p60 protein were solubilized, electrophoretically separated on a denaturing gel, transferred to a nitrocellulose sheet, and reacted with antibody to ICP22 as described in Materials and Methods. In a first experiment, GST-p60 was reacted with HSV-1(F)-infected extract. As can be seen in Fig. 3, GST-p60 bound ICP22, whereas GST alone did not (compare lanes 2 and 3). Interestingly, only the fastest-migrating, underprocessed form of ICP22 was able to bind to GST-p60 (compare lanes 1 and 3). This is in contrast to another ICP22-binding cellular protein, p78, which we found to bind all forms of ICP22 (6) (see below).
FIG. 3.
Photographic image of cell proteins bound to GST fusion protein, electrophoretically separated in a denaturing gel, and reacted with antiserum to ICP22. Fusion proteins were grown in BL21 cells and purified as recommended by the manufacturer (Pharmacia). GST and GST-p60 were mixed with HSV-1(F)-infected HeLa extract, and beads were collected and washed as described in Materials and Methods. Proteins were separated in 7% denaturing polyacrylamide gel, transferred to nitrocellulose, blocked, reacted with R77 and then with a goat anti-rabbit antibody conjugated to horseradish peroxidase, and processed as described by the manufacturer (Amersham). Lanes: 1, cell extract from HSV-1(F)-infected HeLa cells; 2 and 3, infected HeLa cell extract bound to GST and GST-p60, respectively.
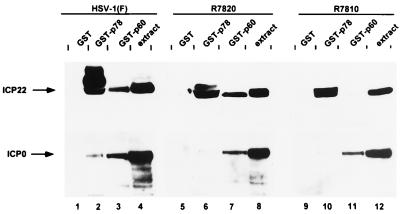
In the next series of experiments, we reacted several different GST fusion proteins with extracts from HeLa cells infected with HSV-1(F), with R7820 (a recombinant virus lacking the carboxyl-terminal 24 amino acids of ICP22), or with 7810 (a recombinant virus lacking the carboxyl-terminal 40 amino acids of ICP22) (Fig. 1). The proteins bound to the fusion proteins were processed as above and reacted with antibodies to ICP22 and ICP0. The results (Fig. 4) indicate the following. (i) GST-p60 bound the full-length ICP22 and the ICP22 lacking the carboxyl-terminal 24 amino acids but not the protein lacking the terminal 40 amino acids (Fig. 4, top, lanes 3, 7, and 11). (ii) In contrast to GST-p60, GST-p78 bound all three forms of the ICP22 protein (Fig. 4, top, lanes 2, 6, and 10). (iii) GST-p60 pulled down ICP0 from all the three cell extracts, whereas GST-p78 bound ICP0 at best in trace amounts (Fig. 4, bottom, lanes 2, 3, 6, 7, 10, and 11). These results indicate that we have verified the physical interaction of p60 with ICP22 demonstrated in the yeast two-hybrid system; p60 interacts specifically with the fast-migrating, minimally processed forms of ICP22; the binding site on ICP22 for p60 maps in the carboxyl-terminal 40 amino acids, approximately between amino acids 24 and 40 from the carboxyl terminus of ICP22; and p60 interacts with ICP0 independently of ICP22.
FIG. 4.
Photographic image of cell proteins bound to GST fusion proteins, electrophoretically separated in denaturing gels and reacted with a serum to ICP22 (top) or ICP0 (bottom). GST, GST-p78 and GST-p60 were mixed with HSV-1(F)-infected HeLa cell extract and processed as described in Materials and Methods. Proteins were separated in 7% denaturing polyacrylamide gels, transferred to nitrocellulose, blocked, reacted with R77 (top) or with a monoclonal antibody to ICP0 (bottom) and then with a goat anti-rabbit antibody (ICP22) or a goat anti-mouse antibody (ICP0) conjugated to horseradish peroxidase, and processed as described by the manufacturer (Amersham). Lanes: 1 to 3; HSV-1(F)-infected HeLa cell extract bound to GST, GST-p78, and GST-p60, respectively; 5 to 7; R7820-infected HeLa cell extract bound to GST, GST-p78, and GST-p60, respectively; 9 to 11, R7810-infected HeLa cell extract bound to GST, GST-p78, and GST-p60, respectively; 4, 8, and 12, cell extract from HSV-1(F)-, R7820-, and R7810-infected HeLa cells, respectively. Note that ICP22 made in cells infected with R7820 (lane 6) or R7810 (lane 10) is not posttranslationally processed by UL13 and hence only the underprocessed forms are pulled down by GST-p78 (27).
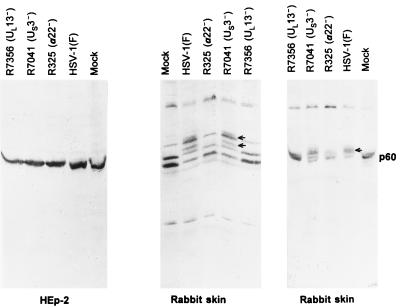
p60 is modified in a cell-type-specific fashion upon infection, and this modification is mediated by the products of the α22/US1.5 and UL13 genes.
To further investigate the relationship between ICP22, ICP0, and p60, we raised a rabbit polyclonal antiserum to GST-p60 as described in Materials and Methods. This antiserum reproducibly detected a closely migrating doublet bands with an apparent Mr of 60,000 in electrophoretically separated lysates of mock-infected HEp-2 cells (Fig. 5, left). On one occasion (Fig. 5, middle), the antibody reacted with well-resolved doublet of bands in lysates of mock-infected rabbit skin cells. These bands migrated with a mobility similar to that of bands from mock-infected HEp-2 cells. The pattern shown in Fig. 5 (right) was more reproducible. In addition, the antibody reacted with a higher-molecular-weight band in lysates of rabbit skin cells. In preliminary experiments, we noted that the electrophoretic mobility of p60 in lysates of rabbit skin cells infected with the R325 (α22−/US1.5−) mutant p60 was processed to a higher apparent molecular weight. The experiments in Fig. 5 were designed to investigate this observation further. The results shown in Fig. 5 may be summarized as follows. (i) The electrophoretic mobility of p60 from HEp-2 cells infected with wild-type virus could not be differentiated from that of p60 from mock-infected cells or cells infected with R325, R7041, or R7356. These findings were reproducible in several experiments. (ii) Electrophoretically separated p60 from lysates of rabbit skin cells infected with wild-type virus formed several additional bands (Fig. 5, middle and right). These bands were also formed by p60 from lysates of cells infected with the US3− virus (R7041)-infected cells but were not formed by the p60 present in lysates of mock-infected cells or cells infected with UL13− (R7356) or α22−/US1.5− (R325) viruses.
FIG. 5.
Photographic image of uninfected or infected rabbit skin cells and HEp-2 cells electrophoretically separated in denaturing gels and reacted with a serum to p60. HEp-2 cell extracts (left) and rabbit skin cell extracts (middle and right) were separated on 10% denaturing polyacrylamide gels, transferred to nitrocellulose, blocked, and reacted with a serum to p60 and then with a goat anti-rabbit antibody conjugated to alkaline phosphatase. The immunoblots were developed as described by the manufacturer (Bio-Rad). The identity of the upper band reacting with the polyclonal anti-p60 rabbit antibody in rabbit skin cells is unknown.
The results shown in Fig. 5 indicate that in wild-type virus-infected rabbit skin cells, p60 is posttranslationally modified to a higher apparent molecular weight and that this posttranslational modification requires the UL13 protein kinase and ICP22 and or US1.5 protein.
p60 is translocated to the nucleus and colocalizes with ICP0 in infected HEp-2 cells but not in infected rabbit skin cells.
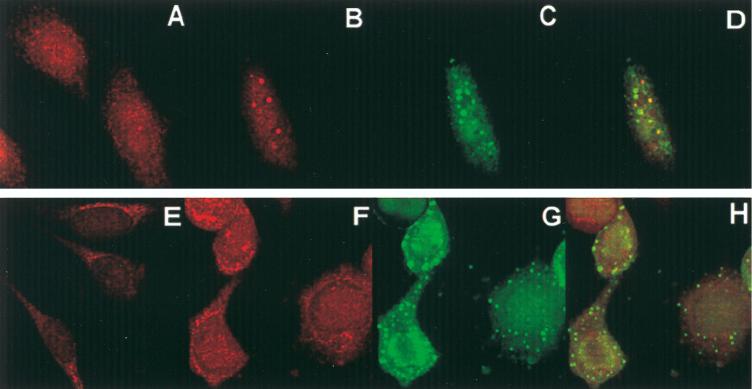
In these experiments, uninfected or HSV-1(F)-infected HEp-2 or rabbit skin cells were prepared for immunofluorescence assays as described in Materials and Methods. The results (Fig. 6) show the following. (i) In uninfected HEp-2 cells, p60 was present predominantly in the nucleus (Fig. 6A). After infection with HSV-1(F), p60 was detected diffused throughout the nucleus along with diffused ICP0 and in small dense nuclear structures in the nucleus colocalized with ICP0 (Fig. 6B to D). Note that ICP0 colocalized with p60 in some but not all dense nuclear structures. At this time after infection, ICP0 accumulated in the cytoplasm in many cells (see Fig. 7). (ii) In uninfected rabbit skin cells, p60 was localized mainly in the cytoplasm, with some perinuclear presence (Fig. 6E). In infected rabbit skin cells, p60 seemed to be redistributed throughout the cell (Fig. 6F) but mainly in the cytoplasm (Fig. 6G). Unlike the pattern observed in infected HEp-2 cells, p60 did not colocalize with ICP0 in rabbit skin cells (Fig. 6F to H). ICP0 was localized mainly in the cytoplasm in these cells (Fig. 6G).
FIG. 6.
Photomicrographs of uninfected (A and E) and HSV-1(F)-infected (B to D and F to H) HEp-2 cells (A to D) and rabbit skin cells (E to H) reacted with antibodies to p60 and ICP0. Cells were fixed 18 h after infection and processed for immunofluorescence as described in Materials and Methods. (A, B, E, and F) Texas red-labeled anti-rabbit IgG to p60 serum; (C and G) FITC-labeled anti-mouse IgG to the ICP0 antibody; (D and H) overlays of panels B and C and panels F and G, respectively.
FIG. 7.
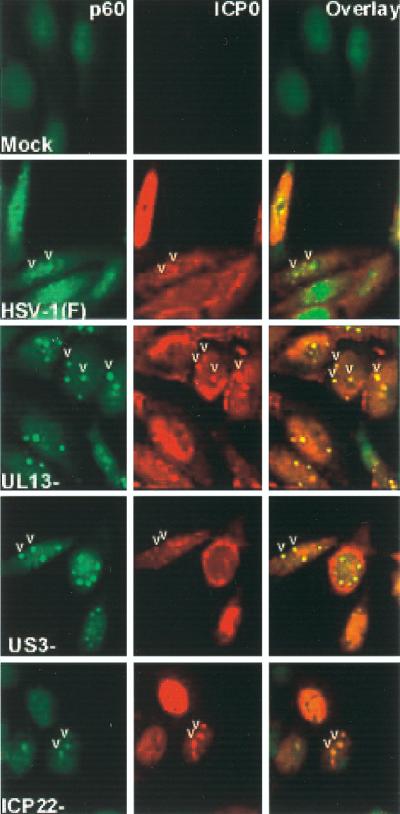
Photographs of uninfected and infected cells fixed at 16 h after mock infection or infection with HSV-1(F) parent virus, UL13−, US3−, or the recombinant R325 (ICP22−/US1.5−) and reacted with polyclonal antibody to p60 (left) and monoclonal antibody to ICP0 (center). The right column shows an overlay of the FITC-conjugated anti rabbit IgG and the Texas red-conjugated anti-mouse IgG. The images were acquired as described in Materials and Methods. Note that in this figure, the colors are reversed with respect to those in Fig. 6 FITC (green) identifies p60, whereas Texas red (red) identifies ICP0. The arrowheads point to representative small nuclear structures present in all photographs except those of mock-infected cells.
The colocalization of p60 with ICP0 in HEp-2 cells does not require the α22 US1.5, US3, or UL13 genes.
The purpose of this series of experiments was to determine the requirement for change in the distribution of p60 in infected cells. In the experiment in Fig. 7, HEp-2 cells grown as described in Materials and Methods were exposed to 10 PFU of HSV-1(F), R7356 (UL13−), R7041, or R325 per cell. The cultures were fixed and reacted with antibody to p60 (FITC) or ICP0 (Texas red). The results (Fig. 7) were as follows. (i) In uninfected HEp-2 cells, p60 was distributed mainly in the nucleus. (ii) In cells infected with wild-type virus, p60 was distributed in some cells in small nuclear bodies [Fig. 7, HSV-1(F), arrowheads] and in others throughout the nucleus. p60 colocalized with ICP0 in the small nuclear bodies but not in cells in which p60 was dispersed throughout the nucleus. The distribution of p60 in cells infected with the R325 (α22−/US1.5−) mutant was similar to that observed in wild-type-infected cells. As shown in Fig. 7, ICP0 also localized in the cytoplasm of infected cells. (iii) In cells infected with the UL13− mutant, p60 was localized almost exclusively in the small nuclear bodies with ICP0 (Fig. 7, UL13−, arrowheads). The infected cells exhibited additional ICP0 that was dispersed throughout the nucleus and in the cytoplasm. (iv) The cells infected with the US3− mutant exhibited a pattern closer to that of wild-type virus. p60 was readily found in the small nuclear bodies. In this instance, a small fraction of the small nuclear dense bodies also contained ICP0 (Fig. 7, US3−, arrowheads). ICP0 was also diffused throughout the nucleus and in the cytoplasm.
In other experiments (results not shown), we found that in cells exposed to phosphonoacetate (300 μg/ml of medium) 1 h before, during, and for 16 h after infection, the pattern of localization of p60 could not be differentiated from that of wild-type infected cells.
We conclude that the redistribution of p60 from a diffuse pattern into small nuclear structures is independent of ICP22, US1.5, UL13, or US3 proteins. Late in infection, p60 redistributed again in a diffuse pattern. This redistribution requires the presence of UL13 and, to a lesser extent, of US3 protein kinase.
DISCUSSION
We report the identification of a novel cellular protein, designated p60, that interacts with two HSV-1 regulatory proteins, ICP22 and ICP0, in the yeast two-hybrid system and in in vitro biochemical assays. The salient features of results, summarized in part in Fig. 8, are as follows.
FIG. 8.
Schematic representation of the interaction of p60 with ICP0 and ICP22 (A) and the distribution of p60 in HEP-2 and rabbit skin cells (B). (A) Although p60 interacts with ICP0 and ICP22, there is currently no evidence that it can interact with both proteins simultaneously. (B) The schematic diagram shows only the distribution of p60 and not those of ICP0 or ICP22. Note that at 16 h after infection, the small structures containing p60 decrease in number in cells infected with wild-type or ICP22−/US1.5− virus but are present in increasing numbers in cells infected with UL13− or US3− viruses. PAA, phosphonoacetic acid.
(i) p60 binds only one of the multiple electrophoretic isoforms of ICP22. Inasmuch as phosphorylation and concurrent alteration in the electrophoretic mobility of ICP22 are necessary for the up-regulation of selected HSV-1 genes (33), this interaction suggests that each ICP22 isoform performs a specific function. The p60-ICP22 interaction is different from the p78-ICP22 interaction in that in the latter case all isoforms of ICP22 interact with p78. The significance of the results presented in this report stems from two considerations. First, HSV proteins frequently accumulate in several isoforms differing in electrophoretic mobility. To our knowledge, this is the first evidence that an HSV protein isoform expresses a function distinct from that of other isoforms of the same protein. Second, all ICP22 isoforms are present in infected cells throughout the viral replicative cycle. This observation raises the possibility that the processing is at least in part mutually exclusive rather than cumulative and that the underprocessed or unprocessed isoforms are available to perform their functions throughout infection rather than transiently at specified times during the viral replicative cycle.
(ii) The p60-binding domain was mapped to a stretch of approximately 16 amino acids located close to the carboxyl terminus of ICP22. This domain is essential for the processing of ICP22 (27, 28). It is conceivable that phosphorylation of ICP22 could interfere with binding to p60.
(iii) p60 binds to ICP0, and the domain responsible for binding has been mapped to the carboxyl-terminal 130 amino acids encoded by exon 2 of ICP0. The observation that in the course of infection p60 colocalizes with ICP0 in infected HEp-2 cells lends credence to and supports the conclusions based on genetic and physical interactions of ICP0 and p60. Although the data indicate that p60 can bind to both proteins, as illustrated in Fig. 8A, we have not rigorously proven that p60 can simultaneously bind to both proteins. However, similarities in the functions of ICP22 and ICP0 suggest that this is likely to occur at specific stages of viral replicative cycle. Thus, ICP0 and ICP22 are both multifunctional proteins, and recent studies indicate that both interact with cell cycle-dependent cellular proteins. ICP0 binds and stabilizes cyclin D3, whereas ICP22 interacts with p78, a protein detected only briefly early in the S phase (19). The accumulation of ICP22 itself was also found to be cell cycle dependent.
(iv) Both rabbit skin and HEp-2 cell lines are highly permissive to wild-type virus. They differ in two respects. First, rabbit skin cells restrict the replication of α22− mutants whereas HEp-2 cells are equally permissive to both wild-type and α22− viruses. The evidence presented in this report shows that p60 is similarly dispersed in uninfected rabbit skin and HEp-2 cells but that the localization differs quite significantly after infection. In infected HEp-2 cells, p60 is translocated into dense nuclear bodies containing ICP0. At late times after infection, p60 is also found dispersed throughout the nucleus of infected cells (Fig. 8B). Translocation of p60 to the dense nuclear bodies does not require ICP22/US1.5 protein since p60 is translocated to these structures in cells infected with virus mutants lacking the carboxyl-terminal domain of ICP22 (13). Dispersal of the p60 throughout the nucleus requires the presence of UL13 protein kinase and, to a slightly lesser extent, of US3 protein kinase. In rabbit skin cells, p60 is processed to isoforms of higher apparent molecular weight and is not translocated into the dense nuclear structures. Moreover, the modification of p60 to isoforms of higher apparent molecular weight requires the participation of ICP22/US1.5 and of UL13 protein kinase. The simple sum of the data is that in the permissive HEp-2 cells p60 most probably interacts with ICP0 since it colocalizes with it. The roles of α22/US1.5, UL13, and US3 proteins are less clear, although we have observed that late in infection p60 is dispersed in cells infected with wild-type virus or with the α22− UL1.5− mutant but tend to remain aggregated in cells infected with UL13− or US3− mutants. p60 is not redistributed in rabbit skin cells infected with wild-type virus. In this instance, p60 is processed to a higher apparent Mr and the posttranslational processing requires ICP22/US1.5 protein and UL13 protein kinase.
The exact role of p60 in the life cycle of the virus is not known. Does p60 support or inhibit viral replication? It is customary to look at virus-cell protein interactions as leading to recruitment of cellular proteins necessary for viral replication. There are, however, lines of evidence that indicate that viral proteins also bind cellular proteins capable of blocking viral replication. In this category are the studies showing that ICP47 binds TAP1/TAP2 and blocks the presentation of antigenic peptides to the cell surface (43) and the evidence that US11 protein can bind protein kinase R to block phosphorylation of the α subunit of the translation initiation factor 2 (9). For heuristic reasons, it seems useful to consider p60 a possible antagonist rather than agonist of HSV replication. One scenario is that HSV evolved functions embodied in two different viral proteins, ICP0 and ICP22/US1.5, to neutralize p60. In permissive cells (e.g., HEp-2), ICP0 causes p60 to aggregate into discrete, small nuclear structures. Deletion of either UL13 or ICP22/US1.5 protein has little effect, since p60 is effectively neutralized, although p60 undergoes further relocation in the nucleus in the presence of these proteins. In restrictive cells (e.g., the rabbit skin cells) only one pathway dependent on ICP22/US1.4 protein and UL13 protein kinase is operative in neutralization of p60 by posttranslational processing enabling wild-type virus to replicate. The problem arises in rabbit skin cells infected with mutants lacking the genes encoding the α22/US1.5 or UL13 proteins; in these cells, viral gene expression could be affected by an unfettered p60. Further exploration of this scenario requires a better understanding of the normal functions of p60. These studies are in progress.
ACKNOWLEDGMENTS
These studies were supported by grants from the National Cancer Institute (CA47451, CA71933, and CA78766), U.S. Public Health Service. B.F. is a postdoctoral trainee (5-32-CA-09273-21).
We thank Alice P. W. Poon for careful reading of the manuscript.
REFERENCES
- 1.Ackermann M, Braun D K, Pereira L, Roizman B. Characterization of HSV-1 α proteins 0, 4 and 27 with monoclonal antibodies. J Virol. 1984;52:108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann M, Sarmiento M, Roizman B. Application of antibody to synthetic peptides for characterization of the intact and truncated α22 protein specified by herpes simplex virus 1 and the R325 α22− deletion mutant. J Virol. 1985;56:207–215. doi: 10.1128/jvi.56.1.207-215.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Blaho J A, Mitchell C, Roizman B. Guanylylation and adenylylation of the α regulatory proteins of the herpes simplex virus require a viral β or γ function. J Virol. 1993;67:3891–3900. doi: 10.1128/jvi.67.7.3891-3900.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruni R, Roizman B. Open reading frame P—a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc Natl Acad Sci USA. 1996;93:10423–10427. doi: 10.1073/pnas.93.19.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruni R, Roizman B. Herpes simplex virus 1 regulatory protein ICP22 interacts with a new cell cycle-regulated factor and accumulates in a cell cycle-dependent fashion in infected cells. J Virol. 1998;72:8525–8531. doi: 10.1128/jvi.72.11.8525-8531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter K L, Roizman B. The promoter and transcriptional unit of a novel herpes simplex virus 1 α gene are contained in, and encode a protein in frame with, the open reading frame of the α22 gene. J Virol. 1996;70:172–178. doi: 10.1128/jvi.70.1.172-178.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter K L, Roizman B. Alternatively spliced mRNAs predicted to yield frame-shift proteins and stable intron 1 RNAs of the herpes simplex virus 1 regulatory gene α0 accumulate in the cytoplasm of infected cells. Proc Natl Acad Sci USA. 1996;93:12535–12540. doi: 10.1073/pnas.93.22.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassady K A, Gross M, Roizman B. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the α subunit of eukaryotic translation initiation factor 2. J Virol. 1998;72:8620–8626. doi: 10.1128/jvi.72.11.8620-8626.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 11.Everett R D, Preston C M, Stow N D. Functional and genetic analysis of the role of Vmw 110 in herpes simplex virus replication. In: Wagner E K, editor. Herpesvirus transcription and its regulation. Boston, Mass: CRC Press, Inc.; 1991. pp. 50–76. [Google Scholar]
- 12.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fineschi, B., and B. Roizman. Unpublished data.
- 14.Hardwicke M A, Sandri-Goldin R M. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honess R W, Roizman B. Regulation of herpes simplex virus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honess R W, Roizman B. Regulation of herpes simplex virus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1Δ: ICP0 affects translational machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulatory cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaguchi Y, Van Sant C, Roizman B. Eukaryotic elongation factor 1Δ is hyperphosphorylated by the protein kinase encoded by the UL13 gene of herpes simplex virus 1. J Virol. 1998;72:1731–1736. doi: 10.1128/jvi.72.3.1731-1736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leopardi R, Ward P L, Ogle W O, Roizman B. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J Virol. 1997;71:1133–1139. doi: 10.1128/jvi.71.2.1133-1139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 23.Meignier B, Longnecker R, Mavromara-Nazos P, Sears A E, Roizman B. Virulence of and establishment of latency by genetically engineered mutants of herpes simplex virus 1. Virology. 1988;162:251–254. doi: 10.1016/0042-6822(88)90417-5. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell C, Blaho J A, Roizman B. Casein kinase II specifically nucleotidylylates in vitro the amino acid sequence of the protein encoded by the α22 gene of herpes simplex virus 1. Proc Natl Acad Sci USA. 1994;91:11864–11868. doi: 10.1073/pnas.91.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell C, Blaho J A, McCormick A L, Roizman B. The nucleotidylylation of herpes simplex virus 1 regulatory protein α22 by human casein kinase II. J Biol Chem. 1997;272:25394–25400. doi: 10.1074/jbc.272.40.25394. [DOI] [PubMed] [Google Scholar]
- 26.Ng T I, Chang Y E, Roizman B. Infected cell protein 22 of herpes simplex virus 1 regulates the expression of virion host shutoff gene UL41. Virology. 1997;234:226–234. doi: 10.1006/viro.1997.8659. [DOI] [PubMed] [Google Scholar]
- 27.Ogle W O, Roizman B. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J Virol. 1999;73:4305–4315. doi: 10.1128/jvi.73.5.4305-4315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogle, W. O., and B. Roizman. Unpublished data.
- 29.Ogle W O, Ng T I, Carter K L, Roizman B. The UL13 protein kinase and the infected cell type are determinants of posttranslational modification of ICP0. Virology. 1997;235:406–413. doi: 10.1006/viro.1997.8710. [DOI] [PubMed] [Google Scholar]
- 30.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: α gene 22 of herpes simplex virus is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 32.Purves F C, Longnecker R M, Leader D P, Roizman B. The herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J Virol. 1987;61:2896–2901. doi: 10.1128/jvi.61.9.2896-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purves F C, Ogle W O, Roizman B. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of α and γ mRNAs and proteins in infected cells. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice S A, Long M C, Lam V, Spencer C A. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J Virol. 1994;68:988–1001. doi: 10.1128/jvi.68.2.988-1001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice S A, Long M C, Lam V, Schaffer P A, Spencer C A. Herpes simplex virus immediate early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J Virol. 1995;69:5550–5559. doi: 10.1128/jvi.69.9.5550-5559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Field’s virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 2231–2295. [Google Scholar]
- 37.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sandri-Goldin R M, Mendoza G E. A herpes virus regulatory protein appears to act posttranscriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 40.Sears A E, Halliburton I W, Meignier B, Silver S, Roizman B. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J Virol. 1985;55:338–346. doi: 10.1128/jvi.55.2.338-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 42.Yao F, Schaffer P A. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J Virol. 1994;68:8158–8168. doi: 10.1128/jvi.68.12.8158-8168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.York I A, Roop C, Andrews D W, Riddell S R, Graham F L, Johnson D C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]