Abstract
Typical defective interfering (DI) RNAs are more successful in the competition for viral polymerase than the parental (helper) virus, which is mostly due to an altered DI promoter composition. Rabies virus (RV) internal deletion RNAs which possess the authentic RV terminal promoters, and which therefore are transcriptionally active and can be used as vectors for foreign gene expression, are poorly propagated in RV-infected cells and do not interfere with RV replication. To allow DI-like amplification and high-level gene expression from such mini-RNA vectors, we have used an engineered 3′ copy-back (ambisense) helper RV in which the strong replication promoter of the antigenome was replaced with the 50-fold-weaker genome promoter. In cells coinfected with ambisense helper virus and mini-RNAs encoding chloramphenicol acetyltransferase (CAT) and luciferase, mini-RNAs were amplified to high levels. This was correlated with interference with helper virus replication, finally resulting in a clear predominance of mini-RNAs over helper virus. However, efficient successive passaging of mini-RNAs and high-level reporter gene activity could be achieved without adding exogenous helper virus, revealing a rather moderate degree of interference not precluding substantial HV propagation. Compared to infections with recombinant RV vectors expressing CAT, the availability of abundant mini-RNA templates led to increased levels of CAT mRNA such that CAT activities were augmented up to 250-fold, while virus gene transcription was kept to a minimum. We have also exploited the finding that internal deletion model RNAs behave like DI RNAs and are selectively amplified in the presence of ambisense helper virus to demonstrate for the first time RV-supported rescue of cDNA after transfection of mini-RNA cDNAs in ambisense RV-infected cells expressing T7 RNA polymerase.
The genetic information of nonsegmented negative-strand RNA viruses (order Mononegavirales) is contained in a ribonucleoprotein complex (RNP). Both the negative-strand genome RNA, from which all virus genes are expressed, and the complementary positive-sense antigenome RNA, which represents the replicative intermediate, are tightly encapsidated by nucleoprotein (N) and associated with the viral RNA polymerase (L) and a polymerase cofactor (P). Only RNPs are templates for all viral RNA synthesis. Genome RNPs may act as templates for transcription of a short leader RNA and monocistronic mRNAs, as well as for replication of full-length RNPs which involves cotranscriptional encapsidation. As a corollary, infection of cells leads to the synthesis of huge amounts of genome strand RNPs, while antigenome RNPs are found at low levels. In the case of paramyxoviruses such as Sendai virus, the bias is moderate, with a 4- to 10-fold preponderance of genome RNAs. Very different values have been reported for members of the Rhabdoviridae family. Whereas a modest three- to fourfold predominance of genome RNPs was observed in vesicular stomatitis virus (VSV)-infected cells, the ratios between genome and antigenome RNAs may reach 50:1 in rabies virus (RV)-infected cells (genus Lyssavirus of the Rhabdoviridae family) (15).
The observed biases have been attributed to different activities of the cis-acting sequences present at the 3′ end of the genome and antigenome RNAs, respectively, which are required to direct replication and encapsidation (5, 6, 9, 12, 28, 30, 31, 34, 45, 46). Since the sequences engaged in the different functions, such as polymerase binding, replication initiation and enhancement, encapsidation, and elongation, are poorly defined, these RNA ends are so far regarded as the genome promoter (GP) and the antigenome promoter (AGP), respectively (5). Accordingly, the AGP should represent a strong replication promoter, whereas the GP should function as a weak replication promoter. This view was first supported by the structure of naturally occurring defective interfering (DI) RNAs of VSV and paramyxoviruses (36, 37). Most of the DI RNAs which replicated efficiently while heavily interfering with helper virus (HV) replication were of the so-called 5′ copy-back type, containing a copy of the strong AGP at both RNA ends. In contrast, RV internal deletion RNAs containing the promoter sequences of the original virus, i.e., one copy of the GP and one copy of the AGP, replicate less efficiently, and interference with virus replication is not observed (9, 41).
Since the GP is active not only in replication but also in the synthesis of a leader RNA and transcription of subgenomic RNAs, a competition between replication and transcription might contribute to the apparently weak replication capacity of the GP. Although this cannot be totally ruled out, recent studies support the assumption that the two promoter sequences indeed differ in the ability to serve as an initiation site for RNA replication (5, 14, 20, 23, 45). However, the factors that determine their activity remain incompletely understood.
We previously addressed the function of RV promoters by constructing a recombinant 3′ copy-back virus in which the AGP was replaced with a copy of the transcriptionally active GP. This virus (SAD Ambi-CAT) was able to express genes from both RNA strands, thus exhibiting ambisense gene expression. In SAD Ambi-CAT-infected cells, genome and antigenome RNAs were present in equal amounts, revealing that the striking 50:1 bias in RV-infected cells is exclusively due to the competition between the GP and AGP. Most remarkably, the total rate of SAD Ambi-CAT replication was not much lower than that of RV. Thus, the replication capacity of the RV GP is not low per se; rather, the GP is competed out by the stronger AGP in the engagement of a limiting amount of polymerase (15).
In this study, we examined whether the presence of the parental AGP in transcriptionally active internal deletion RNAs would allow successful competition with the replication of an HV of the ambisense type. We analyzed the propagation and gene expression of bicistronic model RNAs encoding chloramphenicol acetyltransferase (CAT) and luciferase reporter genes in cells infected with standard RV or with ambisense RV as the helper. Whereas the mini-RNAs were poorly replicated and expressed in RV-infected cells, they behaved as true DI RNAs in ambisense virus-infected cells. Preferential amplification of the model RNAs was correlated with interference with the replication of the ambisense HV. The degree of interference, however, was moderate enough to allow high-level HV-supported reporter gene expression from the model RNAs throughout successive passages.
In contrast to paramyxoviruses, for which HV-mediated recovery of artificial RNAs into RNPs is easily achieved, rescue of rhabdovirus-like RNAs by HVs has not been demonstrated. The finding that standard RV RNAs can be rendered the phenotype of DI RNAs by selecting an appropriate HV led us to reason that ambisense RVs might not only represent valuable tools for efficient expression of foreign genes from transcriptionally active mini-RNAs but also allow recovery of cDNA-derived RNA transcripts. Indeed, after transfection of plasmids encoding the mini-RNA in T7 RNA polymerase-expressing cells previously infected with ambisense RV, mini-RNAs were rescued reproducibly.
MATERIALS AND METHODS
Construction of cDNA clones.
As a basis for the construction of the bicistronic cDNA plasmid pSDI-CL(NP), we used the previously described pSDI-CAT, which contains the CAT gene between the cDNA sequences of the terminal regions of RV (9). For integration of additional restriction sites, two synthetic oligonucleotides (AdapI [5′-ctagtgttaacaggcctgcgcgcagatctggctagct-3′] and AdapII [5′-ctagagctagccagatctgcgcgcaggcctgttaaca-3′]) were annealed and inserted as a 37-bp DNA fragment in an XbaI site downstream of the CAT gene. The downstream XbaI site was restored and was used after Klenow fill-in for the insertion of the firefly luciferase gene, which was excised from pT3/T7-luc (Clontech) as a BsmI/SmaI DNA fragment. In a further step, the two reporter gene sequences were separated by the insertion of an 86-bp MaeIII/AsuII/Klenow DNA fragment (positions 1412 to 1498 of RV SAD B19 [7]) from the RV full-length cDNA clone pSAD L16 (42), which comprises the cis-acting transcription signals for N mRNA stop/polyadenylation and P mRNA start.
For the construction of pSAD Ambi, a DNA fragment comprising the complete leader RNA sequence and the N start signal of RV SAD B19 (position 1 to 67) was PCR amplified from pSDI-1 (9) with primer 49M (5′-cgcgcggttaacaggggtgttacatttttgc-3′) and reversed primer (5′-ggaaacagctatgaccatg-3′). The PCR product was subcloned as a NotI/HpaI DNA fragment in pSKADAP− (NotI/HpaI). pSKADAP− was generated by insertion of the synthetic 37-bp DNA fragment described above into the XbaI site of pBluescript SKII− (Stratagene). The resulting plasmid pleaHpa contained the RV leader sequence and the N start sequence, followed by an HpaI restriction site. By replacement of the 1.1-kb HpaI/NotI fragment of pSAD Ambi-CAT (15), which comprises the CAT reporter gene sequence and the leader sequence with the 0.3-kb HpaI/NotI fragment of pleaHpa, the cDNA full-length clone pSAD Ambi (organization, T7 promoter-leader-RV genes-leader-hepatitis delta virus-T7 terminator) was generated.
For expression of RV proteins N, P, and L in T7 polymerase-expressing cells, the coding sequences of the RV genes were inserted in the vector pTIT, which comprises the internal ribosome entry site (IRES) of the encephalomyocarditis virus (organization, T7 promoter-IRES-multiple cloning site-T7 terminator [4]). pTIT-N was constructed by insertion of a 150-bp NcoI/EcoRV fragment that was PCR amplified from pT7T-N (9) with the primers N-ATG (5′-aataccatggatgccgacaagattg-3′) and N2M (5′-cccatatagcatcctac-3′) in pTIT (NcoI/EcoRV) and subsequent integration of a 1.3-kb EcoNI/PstI fragment of pT7T-N. pTIT-P was generated after insertion of a 180-bp BspHI/NcoI PCR fragment (primers P-ATG [5′aatatcatgagcaagatctttgtca-3′] and NS3M [5′-tccactgatagatcatcc-3′]) from pT7T-P (9) in pTIT (BspHI/NcoI) and subsequent integration of an NcoI/ HindIII fragment of pT7T-P. pTIT-L was constructed by insertion of a 480-bp SphI/NsiI-digested PCR fragment (primers L-ATG [5′-agcaggcatgctcgatcctgg-3′] and 6410M [5′-aagttgactaactttgtctttt-3′]) of pT7T-L (9) in pTIT (NcoI/PstI) and subsequent insertion of a 6.4-kb BsgI/SpeI fragment of pTIT-L.
Cells, viruses, and cDNA rescue experiments.
RV SAD L16 (42) and SAD Ambi-CAT (15) were grown on BSR (a BHK-21 clone) cell monolayers. For vaccinia virus-free recovery of RV or HV-driven recovery of mini-RNAs, we used the BSR T7/5 clone, which stably expresses T7 RNA polymerase (4).
Infectious SDI-CL(NP) particles were recovered from pSDI-CL(NP) as described previously for pSDI-CAT (9). Four micrograms of pSDI-CL(NP) was cotransfected with T7T plasmids encoding the RV proteins N, P, M, G, and L in BSR cells expressing T7 RNA polymerase from recombinant vTF7-3. Three days after transfection, cell culture supernatants were harvested, partially cleared of vaccinia virus by centrifugation, and transferred on fresh BSR cells. The cells were superinfected with helper virus at a multiplicity of infection (MOI) of 1 after 1 h.
The recombinant RV SAD Ambi was recovered in a new, vaccinia virus-free recovery system, using BSR T7/5 cells that constitutively express T7 RNA polymerase (4). For virus recovery, 10 μg of full-length cDNA and plasmids pTIT-N (5 μg), pTIT-P (2.5 μg), and pTIT-L (2.5 μg) were cotransfected in BSR T7/5 cells. After 3 days, fresh cell culture medium was added; after a further 3 days, cell culture supernatants were harvested and transferred on BSR cells. Two days after passage, infectious virus was detected by immunostaining against RV N protein (42). DNA transfections were performed after CaPO4 precipitation (Stratagene) in 8-cm2 culture dishes containing ∼106 cells.
RNA analysis.
Total RNA from cells or from virions pelleted from supernatants by ultracentrifugation (Beckman TLA45 rotor; 45,000 rpm, 1 h, 4°C) was isolated 2 days after infection with an RNeasy Mini kit (Qiagen) according to the supplier’s instructions. Agarose gel electrophoresis and Northern blotting were performed as described previously (8). RV N and CAT DNA fragments were labeled with [α-32P]dCTP (3,000 Ci/mmol; ICN) by nick translation (nick translation kit; Amersham). After hybridization, RNA amounts were measured in a phosphorimager (Fuji-BAS).
Luciferase assay.
Two days after infection, the cell monolayers were lysed with 1 ml of luciferase lysis buffer, and enzyme activities in cell extracts were determined as described previously (41).
RESULTS
Previous studies on synthetic RV model RNAs have shown that transcribing internal deletion-type RNAs possessing the authentic parental 3′ and 5′ ends are replicated inefficiently by wild-type (wt) helper RV and do not interfere with HV replication (9, 41). Since in RV-infected cells the AGP directs the synthesis of an approximately 50-fold-higher amount of RNPs than the GP (15), we reasoned that internal deletion RNAs might be able to successfully compete with an HV containing only GPs, such as the previously described SAD Ambi-CAT (15).
To study the competition between the two different promoters, we analyzed the replication activities and gene expression of a bicistronic model RNA, SDI-CL(NP), in cells coinfected with SAD Ambi-CAT or with standard RV SAD L16. As for the previously described monocistronic mini-RNAs SDI-CAT (9) or SDI-flash (41), SDI-CL(NP) contained the authentic RV 3′ and 5′ ends (68 and 164 residues, respectively [Fig. 1]) but encoded two nonviral reporter gene products, CAT and firefly luciferase. The reporter genes were separated by the N/P gene border sequence of RV strain SAD B19 which directs transcription termination/polyadenylation of the upstream CAT mRNA and restart of luciferase mRNA transcription. SDI-CL(NP) was initially recovered from cDNA in a standard RV recovery system (9). After infection of BSR cells with recombinant vaccinia virus vTF7-3 (16), providing T7 RNA polymerase and transfection of T7 promoter-controlled plasmids encoding RV proteins N, P, M, G, and L and of pSDI-CL(NP), the mini-RNAs were rescued by encapsidation into RNPs. Supernatants containing SDI-CL(NP) particles were harvested 3 days after transfection, pooled, and used as a stock for coinfection experiments.
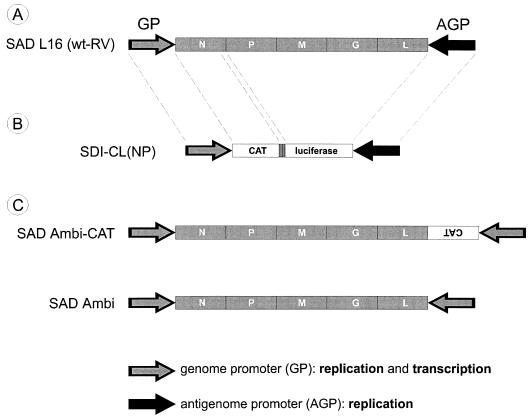
FIG. 1.
(A) Organization of the RV genome. The RV genes (marked by filled boxes) are flanked by noncoding sequences, which contain the terminal promoters for replication and transcription. The 3′-terminal sequence of the positive-sense RNA comprises the AGP, which is inactive in transcription but functional in replication as a strong promoter. The 3′ end of the negative-sense genome RNA comprises the transcriptionally active GP, which is a weak promoter in the replication mode. (B) Organization of the bicistronic model RNA SDI-CL(NP). The reporter genes are separated by the N/P gene border sequence of wt RV, which comprises cis-acting signals for transcription stop/polyadenylation and restart. As in wt RV, the terminal RNA sequences consist of the GP and AGP (positions 1 to 68 and 11763 to 11928 of RV SAD B19). (C) Organization of recombinant ambisense RVs. In SAD Ambi-CAT, the AGP sequence is exchanged for a copy of the GP sequence and the CAT gene sequence is coded on the positive-sense RNA strand in orientation opposite that of the RV genes (15). SAD Ambi lacks the CAT reporter gene.
Ambisense RV supports high-level gene expression from mini-RNAs.
In the first DI passage experiment, 106 BSR cells were infected with identical aliquots of the SDI-CL(NP) particle stock resulting from rescue experiments. One hour postinfection, the cells were superinfected with SAD Ambi-CAT or with RV SAD L16 at an MOI of 1. After 48 h of coinfection, cell culture supernatants were harvested and 1 ml of supernatant of a total of 2 ml was transferred on fresh BSR cells, without additional HV. Five further passages were performed according to the same protocol.
Mini-RNA gene expression was first analyzed by monitoring the activity of the luciferase reporter gene product (Fig. 2). Two days after the initial coinfection (passage 1), luciferase activities were about 100-fold higher in cells infected with SAD Ambi-CAT than in cells infected with SAD L16 HV. Thus, SAD Ambi-CAT supported gene expression from the mini-RNA much more efficiently than wt RV. During the further passages, the luciferase activity was greatly increased in SAD Ambi-CAT-infected cells, whereas SAD L16 allowed only low-level luciferase expression. From the second passage on, SAD Ambi-CAT yielded 104-fold-higher luciferase expression than SAD L16. Infection of cells with SAD Ambi-CAT, SAD L16, or SDI-CL(NP) alone did not result in detectable luciferase activity.
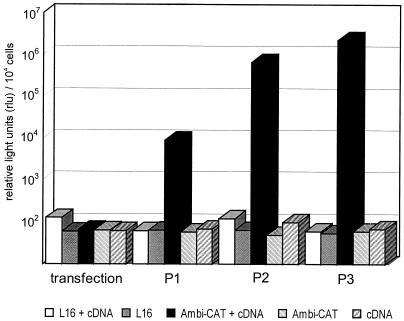
FIG. 2.
Mini-RNA-derived luciferase expression is enhanced in SAD Ambi-CAT-infected cells. BSR cells were infected with a SDI-CL(NP) stock and were superinfected with SAD Ambi-CAT or SAD L16 (MOI of 1). Successive passages were performed by transferring supernatants on fresh cells every 48 h without adding exogenous HV. Luciferase activities in cell extracts prepared from each passage experiment are shown as relative light units per 104 infected cells.
RNA synthesis of mini-RNAs interferes with HV replication.
The augmented expression of mini-RNA-encoded luciferase in ambisense virus-infected cells and the efficient passaging of luciferase activity indicated that SDI-CL(NP) was used preferentially as a template for RNA synthesis. To analyze RNA synthesis directly, RNA was isolated from cells in parallel experiments. Northern hybridization with a luciferase-specific DNA probe showed that in SAD Ambi-CAT-infected cells the mini-RNA-encoded luciferase gene was transcribed in large amounts as a monocistronic mRNA (Fig. 3A). As generally found for RV mRNAs, a minor fraction of luciferase-specific mRNA was transcribed as a bicistronic CAT/luciferase mRNA by partial readthrough at the N/P gene border separating the two reporter genes. Since the bicistronic mRNA [2.8 kb + poly(A)] and the mini-RNA template (3.1 kb) have nearly the same length, discrimination of the two RNAs was not possible in this experiment. In SAD Ambi-CAT-infected cells luciferase mRNA was readily detectable after the first passage. The level of luciferase mRNA strongly increased after the second passage and remained at a high level after the third passage, reflecting the observed reporter gene activities. Although identical aliquots of SDI-CL(NP) were used for the initial coinfections (first passage), luciferase-specific RNA was not detectable in SAD L16-infected cells after the first passage. Even after loading of 10 times more RNA, only a slight hybridization signal was visible (not shown). In contrast to SAD Ambi-CAT, SAD L16 HV was not able to augment the level of mini-RNA-derived transcripts during the following passage experiments (not shown).
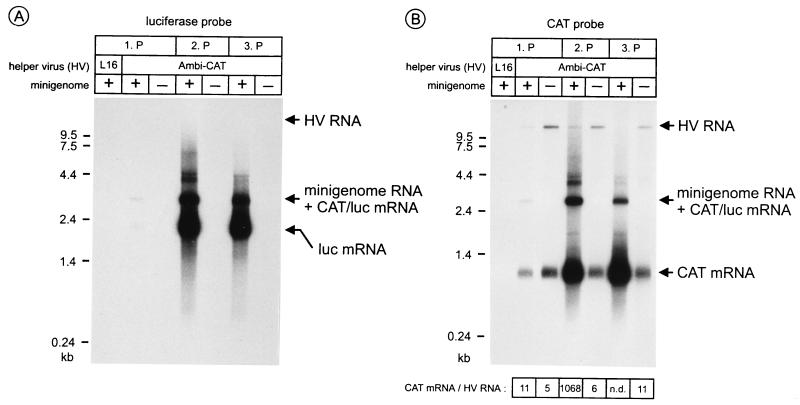
FIG. 3.
SDI-CL(NP) is selectively amplified in SAD Ambi-CAT-infected cells and interferes with HV replication. BSR cells were infected with SDI-CL(NP) stock and helper HVs, and passages (1.P, 2.P, and 3.P) were performed as described for Fig. 2. Cell RNA was analyzed with luciferase (A) and CAT (B) gene-specific DNA probes recognizing both plus- and minus-strand RNAs. Mini-RNA-derived luciferase mRNA was detectable in SAD Ambi-CAT-infected cells after the first passage, in contrast to SAD L16-infected cells. In SAD Ambi-CAT-infected cells, CAT RNAs are represented by a 1-kb monocistronic CAT mRNA, derived either from SDI-CL(NP) or from HV, a 3.1-kb RNA band composed of full-length mini-RNA and CAT/luciferase (luc) readthrough transcripts, and HV full-length RNA. The 3.1-kb hybridization signal was detectable after the first passage and much stronger after the second passage. At the bottom, the ratios of CAT mRNA and HV RNA are given.
Due to the presence of the CAT gene in both SAD Ambi-CAT HV and the mini-RNA, hybridization with a CAT gene-specific DNA probe allowed us to directly compare HV full-length RNA, full-length SDI-CL(NP) RNA, and CAT mRNAs (Fig. 3B). An increase in the level of SDI-CL(NP) RNA-derived RNA was correlated with a striking decrease of HV RNA. Thus, SDI-CL(NP) interfered heavily with HV replication. The total amount of monocistronic CAT mRNAs which could be transcribed from both SAD Ambi-CAT and SDI-CL(NP) was considerably greater in coinfected cells than in cells infected only with SAD Ambi-CAT. This obviously is due to the increasing amount of SDI-CL(NP) CAT templates in coinfected cells. The amount of HV template, which has been shown to represent 50% of the total full-length RNA of SAD Ambi-CAT (15) and which must provide all essential RV proteins, was reduced to a strikingly low level after passage 2 and was not detectable under these hybridization conditions after passage 3. However, HV levels were maintained high enough to allow efficient replication of mini-RNAs and highly efficient passaging of the defective RNAs.
Quantitation of the hybridization signals by phosphorimaging showed that after the second passage the ratio of monocistronic CAT mRNA to SAD Ambi-CAT full-length RNA was nearly 1,000:1 in SAD Ambi-CAT–SDI-CL(NP)-infected cells, whereas it was only 5:1 when cells were infected with SAD Ambi-CAT alone. Thus, CAT mRNA synthesis relative to HV full-length RNA was increased approximately 200-fold in the presence of SDI-CL(NP) compared to SAD Ambi-CAT alone. As for the luciferase probe, direct quantitation of SDI-CL(NP) RNA was not possible due to the comigrating bicistronic CAT/luciferase transcript. However, from other studies (to be published elsewhere) it is known that in SAD L16-infected cells readthrough at the N/P gene border in SDI-CL(NP) yields up to 7% of CAT mRNAs. Since the 3.1-kb RNA hybridization signal reached 15% of the monocistronic CAT mRNA signal, at least half of the 3.1-kb band represents full-length mini-RNA. This finding further confirmed that mini-RNA was much more abundant than SAD Ambi-CAT HV RNA in coinfected cells.
To allow direct comparison, RNA was prepared from supernatant virions, which do not contain mRNAs. Northern hybridizations reflected the findings obtained with cell extracts: mini-RNA was detectable after the first passage and was strongly amplified in the further passages, while the level of HV RNA was greatly decreased (Fig. 4). In supernatants from the second passage, the amount of mini-RNA exceeded that of HV by a factor of 240. Due to hardly detectable amounts of HV RNA in the following passages, reliable ratios could not be determined; however, mini-RNAs were kept at levels comparable to those after passage 2. In contrast to supernatants from SDI-CL(NP)-infected cells, supernatants from cells infected only with ambisense HV contained quite stable amounts of HV RNA throughout the passages. Mini-RNA was not detectable in supernatants from SAD L16–SDI-CL(NP)-infected cells. Taken together, these results demonstrate that the internal deletion RNA SDI-CL(NP) behaves like a DI RNA when SAD Ambi-CAT is used as an HV, whereas it is not amplified in standard RV-infected cells.
FIG. 4.
Formation of supernatant virions. RNA was isolated from virion pellets after ultracentrifugation of cell culture supernatants from passage experiments (Fig. 3) and was analyzed by Northern hybridization with a CAT gene-specific DNA probe. In all passages with SAD Ambi-CAT HV, SDI-CL(NP) virion RNA was detectable. With increasing amounts of mini-RNA, the amount of SAD Ambi-CAT HV decreased after the passage 2 (2.P) and was not detectable after the passage 3 (3.P). In supernatants from SAD L16/SDI-CL(NP)-infected cells or in supernatants from cells infected with SDI-CL(NP) alone, mini-RNAs were not detectable. n.d., not detectable.
Interference with ambisense HV is not due to the presence of the additional CAT gene.
After having confirmed the DI phenotype of SDI-CL(NP) for SAD Ambi-CAT, we wished to exclude the possibility that the expression of the additional CAT gene from the HV antigenome somehow contributed to the degree of interference. To this end, we constructed an ambisense RV from cDNA in which the CAT reporter gene was deleted (SAD Ambi [Fig. 1]).
SAD Ambi was recovered in a new vaccinia virus-free recovery system that makes use of T7 RNA polymerase-expressing cells (BSR T7/5 [4]). These cells were transfected with T7 promoter-controlled plasmids encoding RV proteins N, P, and L downstream of the encephalomyocarditis virus IRES and with pSAD Ambi. Six days after transfections, the supernatants were transferred onto fresh BSR cell monolayers (for details, see Materials and Methods). Virus recovery was monitored 2 days after passage by immunofluorescence, and the recombinant virus was grown for two further passages on BSR cells.
Coinfections of SDI-CL(NP) and SAD Ambi were performed as described above, and cell RNA was prepared after three passages. Hybridizations with the CAT probe verified that CAT gene-specific RNAs were synthesized in abundant amounts when cells were additionally infected with SDI-CL(NP) (Fig. 5). Full-length SAD Ambi RNA was readily detected with an N probe in cells infected with SAD Ambi alone, but in coinfections with SDI-CL(NP), full-length RNA was reduced to levels not detectable with the N probe. However, HV-derived N transcripts could be demonstrated and were found to be reduced fourfold in comparison with cells infected with SAD Ambi or SAD Ambi-CAT alone. Thus, SDI-CL(NP) interfered with SAD Ambi as with SAD Ambi-CAT, confirming that the interference is due exclusively to the difference in promoter composition of ambisense HVs and mini-RNA.
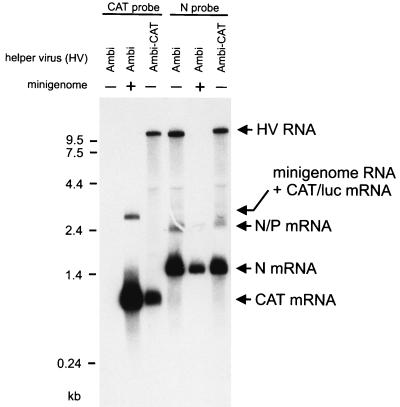
FIG. 5.
Model RNA-derived gene expression exceeds HV gene expression. SDI-CL(NP) was passaged with SAD Ambi as described above for SAD Ambi-CAT. For the third passage, BSR cells were infected with the HV-DI stock (Ambi; +) at an HV MOI of 1. Cells were also infected with SAD Ambi alone (Ambi; −) or with SAD Ambi-CAT (Ambi-CAT) at an MOI of 1. Cell RNA was analyzed by Northern blotting 2 days postinfection by CAT- and RV N gene-specific DNA probes. In SAD Ambi–SDI-CL(NP)-coinfected cells, the level of monocistronic CAT mRNA was 11-fold higher than in cells infected with SAD Ambi-CAT. The level of N mRNA was fourfold lower in SAD Ambi–SDI-CL(NP)-coinfected cells than in SAD Ambi- or SAD Ambi-CAT-infected cells. luc, luciferase.
In contrast to SAD Ambi-CAT, SAD Ambi HV does not produce CAT mRNA. However, virtually the same amounts of CAT mRNAs were observed in cells coinfected with SDI-CL(NP) or with either SAD Ambi or SAD Ambi-CAT, showing that the contribution of SAD Ambi-CAT to CAT transcription was negligible. Compared to cells infected with SAD Ambi-CAT alone, an 11-fold amount of CAT mRNA was determined. Further experiments in which different HV MOIs and incubation times were used (not shown) revealed that CAT activities may be 40- to 250-fold higher in cells infected with SDI-CL(NP) and SAD-Ambi than in cells infected with SAD Ambi-CAT alone. We have previously shown that recombinant RVs expressing a CAT gene from the genome RNA (SAD XCAT and SAD VCAT [24]) and SAD Ambi-CAT produced comparable amounts of CAT protein (15). Thus, the described binary system involving a transcriptionally active DI RNA in combination with an ambisense HV provides the possibility of expressing heterologous genes much more efficiently than is possible with standard recombinant virus vectors. Moreover, while mini-RNA-encoded genes are expressed preferentially, the expression of HV genes is reduced to a minimum.
Ambisense helper RV support recovery of mini-RNAs from cDNA.
In striking contrast to paramyxoviruses and orthomyxoviruses, HV-driven rescue of artificial RNAs into RNPs has not been described for rhabdoviruses, and own attempts to recover mini-RNAs in RV-infected cells were unsuccessful (unpublished results). The highly preferential amplification of RV mini-RNAs by ambisense HVs prompted us to investigate whether it is possible to demonstrate rescue of mini-RNAs in cells infected with ambisense viruses.
To this end, 106 T7 RNA polymerase-expressing BSR T7/5 cells were infected with SAD Ambi-CAT at an MOI of 1. Twenty hours postinfection, the cDNA-plasmid pSDI-CL(NP), which should direct transcription of a genomic-sense SDI-CL(NP) RNA, was transfected into the cells. After 3 days, cell culture supernatants were harvested and 1 ml was transferred on fresh BSR cells without additional HV. Further passages were performed every 48 h, and mini-RNA-derived gene expression was monitored by detection of luciferase activity in cell extracts. As shown in Fig. 6, rescue of mini-RNA was successful in SAD Ambi-CAT-infected cells. Significant luciferase activity was not detectable in the transfected cells but appeared after passaging, demonstrating encapsidation of T7 RNA polymerase transcripts into RNPs and DI particle formation. As predicted from earlier experiments using a variety of monocistronic mini-RNA analogs, no significant luciferase activity was detectable in SAD L16-infected cells.
FIG. 6.
Mini-RNA rescue from cDNA by ambisense HV. T7 RNA polymerase-expressing cells were infected with SAD Ambi-CAT or SAD L16 at an MOI of 1; 20 h postinfection pSDI-CL(NP), from which negative-strand model RNA is transcribed by T7 RNA polymerase, was transfected into the cells. After 3 days supernatants were transferred on fresh BSR cells. Two further passage experiments were performed. SDI-CL(NP)-derived gene expression was monitored by determining the luciferase enzyme activities (relative light units) in cell extracts prepared from transfected-infected cells. P1, P2, and P3, passages 1, 2, and 3.
DISCUSSION
Most DI RNAs interfere with the growth of parental viruses because they are more successful in the competition for trans-acting factors required for replication, presumably the viral polymerase (17). This can be achieved, for example, by point mutations in promoter sequences (8) or the acquisition of new promoters allowing more efficient polymerase binding and/or faster initiation and elongation. As we have previously shown for RV, competition between promoters of different strengths indeed represents the physiological mechanism to maintain exact levels of protein-encoding genome and replicative intermediate and to ensure coordinated, optimal virus gene expression and virus propagation (15). Such functional constraints do not apply to DI RNAs, which are therefore rather free in promoter use. The overwhelming amount of naturally occurring VSV and paramyxovirus DI particles are 5′ copy-back DI RNAs which contain two copies of the strong parental AGP promoter (21, 22, 35, 37, 40). While the gain of a second AGP provides a selective advantage in replication over the parental virus, and also over internal deletion DI RNAs in mixed infections (36), it is coupled with the loss of the GP and the ability to transcribe mRNAs. Several internal deletion DIs RNAs which have promoters derived from the parental GP and AGP and which transcribe to various extents have also been found in nature (1, 8, 13, 38). The more recent possibility of constructing defective RNAs with exact copies of parental promoter sequences, however, has confirmed that RV internal deletion constructs (mini-RNAs) do not interfere with HV replication (9).
Rather than modifying the promoters of a mini-RNA, we have here used another strategy to render it the phenotype of a DI RNA while maintaining its capability for effective transcription and foreign gene expression, namely, by providing an appropriate HV. The clue for this was the availability of a nondeficient virus in which the highly competitive AGP was replaced with the sequence of the GP (15). Interestingly, this virus replicated efficiently, showing that it is the competition with the AGP that makes the GP weak. We have shown here that the principle of competition between promoters can be used for preferential amplification and gene expression of a standard model RNA in an HV-dependent system.
Indeed, compared to wt RV, SAD Ambi-CAT and SAD Ambi supported up to 104-fold-higher luciferase expression from SDI-CL(NP). A low level of luciferase expression in wt RV-infected cells, and scarcely detectable Northern hybridization signals, showed that the mini-RNA was also propagated by wt RV through the passages but was not amplified. As was previously observed for other internal deletion mini-RNAs (9), SDI-CL(NP) was completely lost after slight dilution of the supernatants during passages (not shown). In contrast, as indicated by high-level luciferase expression and confirmed by Northern analyses, there was considerable interference of SDI-CL(NP) with the replication of SAD Ambi-CAT.
Preferential amplification of DI RNAs is sometimes accompanied by drastic interference with HV replication, leading to an depletion of the virus protein supply. This might also shut down DI RNA replication, especially when the initial amount of DI RNAs in infections is high (3, 18, 29). In most cases, steeply oscillating titer curves are observed over time, reflecting the interaction between HV and DI RNA. The passage of SDI CL(NP), as monitored by luciferase activities, occurred also in a somehow oscillatory manner but remained at a high level throughout the six passages. Thus, the interference of DI RNA with HV replication appeared moderate and more regulated, leading to rather stable infections and constant levels of gene expression throughout the passages.
The extent of interference with ambisense HV RNA replication was analyzed by Northern hybridizations with RNA from cells and from virions that were released into the cell culture supernatants. Since the GP and AGP promoters of RV direct a ratio of genome and antigenome RNA of 50:1, we concluded that the proportion of mini-RNA and HV RNAs should at least not be less. Already in the second passage the ratio was found to be greater than 50:1. Because the hybridization signal of full-length SDI-CL(NP) in cell RNA is composed of viral RNA and bicistronic readthrough transcripts and gave a 150-fold-higher value than that of full-length HV RNA, the actual ratio was less than 150:1. This ratio was further increased in the third passage. The total amount of both mini-RNA and HV was found to be decreased, such that HV RNA was hardly detectable and a reasonable direct determination of ratios was not possible.
Remarkably, the ratio of mini-RNA and HV particles present in cell culture supernatants was higher than that observed in cells. After the second passage, an approximately 240-fold dominance of DI particles was observed, compared to the maximal 150-fold dominance in cells. Since RV genome and antigenome RNAs are incorporated equally well into virus envelopes (15), this indicated that formation of SDI-CL(NP) particles occurs a bit more efficiently than HV formation. This is consistent with previous work indicating that budding of internally deleted Sendai virus RNPs was positively influenced by decreasing RNP size (39). However, increasing budding efficiencies with increasing RNP size were also reported for Sendai virus internal deletion DI RNAs (27). For Sendai virus 5′ copy-back DI RNAs, it was consistently observed that budding efficiency was negatively influenced by small RNP sizes (27, 39). Thus, it remains unclear how budding efficiency is correlated with the size of RNPs and whether other factors may contribute to the ability of RNPs to be released into the supernatant.
In view of the magnitude of interference, it was astonishing that the low ambisense HV production was sufficient for allowing continuous passaging of DI RNAs and high-level DI RNA-derived reporter gene expression without additional HV. One factor contributing to this is probably a high intracellular stability of viral RNPs. For VSV, it was previously shown that the biological half-life of intracellular DI RNPs was between 6 and 12.5 h (11). It was also shown for measles virus and Sendai virus that defective RNPs remain quite stable after infection of cells in the absence of HV (26). In cells coinfected with few RV ambisense HV and many interfering mini-RNAs, a high stability of RNPs might become an important factor allowing long-term gene expression and for providing sufficient virus proteins for RNA replication and for release of virus particles. Another factor that might contribute is a delayed accumulation of intracellular N protein in ambisense RV-DI RNA-infected cells. According to the widely accepted view that the N protein concentration is involved in determining the mode of RNA synthesis of negative-strand RNA viruses, high amounts of soluble N protein should direct the viral polymerase into the replication mode (2, 10, 33, 49). We hypothesize that in SAD Ambi–SDI-CL(NP)-infected cells a relative low level of N protein is available due to the competition between DI RNA and HV for N protein and that this may be responsible for the relatively high level transcription from both HV and mini-RNAs. The transcription mode, which is not dependent on N protein supply, might be favored or elongated at the expense of replication, which is N dependent. Indeed, in cells coinfected with VSV and 5′ copy-back DI RNAs we observed a prolonged synthesis of viral proteins that resulted in slower accumulation of virus proteins and delayed release of both virus and DI particles (47). Approaches can now be developed to investigate whether transcription and replication rates can be influenced by a transcriptionally active DI RNA.
Finally, the most important factor responsible for maintaining high-level gene expression in coinfected cells is the promoter composition of HV and DI RNAs. In RV-infected cells, the 50:1 bias of genome and antigenome RNA is extreme in comparison with other negative-strand RNA viruses. Even in the closely related rhabdovirus VSV, only an approximately fourfold dominance of genome RNA over antigenome RNA is observed in infected cells (44, 48), and in the more distantly related Sendai virus 13 to 28% of cellular virus full-length RNA is antigenome RNA (19). The relative difference in promoter strength, which obviously must influence the degree of interference of DI RNAs with HV, should be reflected by the DI biology in the different virus systems. The most atypical behavior in this respect is indeed exhibited by RV. In striking contrast to VSV and paramyxovirus infections, where 5′ copy-back DI RNAs are readily amplified after undiluted passaging, this is not observed for RV. Indeed, all natural RV DI RNAs that we have obtained and analyzed so far are of the internal deletion type (reference 8 and unpublished results). This observation introduces the idea that the interference of putative RV 5′ copy-back DI RNAs which have two strong AGPs with HV replication may be so pronounced that HV infection is completely shut down. As verified by hybridization experiments with strand-specific probes, SDI-CL(NP) virions contain abundant amounts of negative-strand RNA (not shown). Newly coinfected cells should thus contain equal levels of HV genome and antigenome RNPs and high amounts of negative-strand DI RNA. Only upon the first steps of replication does the strong AGP of the DI antigenome become available and able to enter in the competition for polymerase. Thus, a moderate degree of interference especially in the early stage of infection may allow HV the opportunity for propagation. In contrast, in the case of 5′ copy-back DI RNAs, the strongly interfering AGP is available from the initiation of infection. Moreover, the hot start of 5′ copy-back DI RNAs immediately initiates the exponential phase of amplification, since both strands possess the competitive AGP promoters, so that HV replication may be rapidly suppressed to insufficient levels.
We have also exploited the features of ambisense RV to provide evidence that recovery of a rhabdovirus genome analog from cDNA is possible in an HV-driven support system. So far, rescue has been demonstrated after coexpression of support proteins N, P, and L from transfected DNA, mostly by using the transient vaccinia virus T7 RNA polymerase expression system (9, 32), but not in rhabdovirus-infected cells. This is in striking contrast to the situation in paramyxoviruses, where expression of genome-analogous RNAs in virus-infected cells faithfully resulted in rescue into RNPs, mini-RNA gene expression, and formation of passageable particles (6, 12, 30, 43). Encapsidation of naked SDI-CL(NP) RNA in ambisense HV-infected cells was possible but was very inefficient, as no specific reporter gene activity was found in the transfected cells. In the case of paramyxovirus helper-driven rescue, this is readily observed in most cases. Enzyme activity was detectable only after passaging and rose rapidly, owing to the competition of SDI-CL(NP) RNPs with ambisense HV, while no detectable activity was found with SAD L16 virus. A few SDI-CL(NP) RNPs might also be formed after transfection of SAD L16-infected cells, as the levels of N protein are comparable in SAD Ambi- and SAD L16-infected cells. Since the newly formed RNPs are not selectively amplified by SAD L16, HV rescue appears to fail. However, it cannot be ruled out that efficiencies of encapsidation of T7 RNA polymerase transcripts are different in SAD Ambi- and in SAD L16-infected cells. Competition between promoter sequences may occur at the encapsidation step. This may be caused by a high N binding activity of the standard virus’s AGP, such that in SAD L16-infected cells a lower amount of N protein is available for illegitimate encapsidation of T7 transcripts.
The described infection system making use of a nondefective ambisense virus and a transcriptionally active mini-RNA combines the principles of DI interference and effective gene expression, as described previously for a wt Sendai virus that also amplified transcriptionally active mini-RNAs (25). The rather moderate degree of interference allows fairly stable infection conditions. Preferential amplification to a certain degree of SDI-CL(NP) RNPs led to the accumulation of high amounts of the desired templates. These RNP templates competed with the HV RNPs also in transcription, leading to high-level expression of the mini-RNA-encoded genes. This combination is superior to nondeficient RV vectors. Besides offering the possibility of efficient foreign gene expression, the system also may considerably enhance the total capacity of RV for additional genes. Although a size limit for integration of additional sequences is not yet defined for any nonsegmented negative-strand RNA virus, one can expect that greatly increasing the length of at least 12-kb full-length constructs reduces the recovery rate. Thus, defective RNAs that contain only 237 nucleotides of the RV terminal sequences seem to be the choice for integration of large foreign sequences. What makes this approach even more attractive and powerful is the possibility that the respective cDNA constructs are reproducibly rescued by the ambisense HV after transfection into the newly established BSR T7/5 cells expressing T7 RNA polymerase.
ACKNOWLEDGMENT
This work was supported by grant BEO 031171 from BMBF.
REFERENCES
- 1.Amesse L S, Pridgen C L, Kingsbury D W. Sendai virus DI RNA species with conserved virus genome termini and extensive internal deletions. Virology. 1982;118:17–27. doi: 10.1016/0042-6822(82)90315-4. [DOI] [PubMed] [Google Scholar]
- 2.Arnheiter H, Davis N L, Wertz G, Schubert M, Lazzarini R A. Role of the nucleocapsid protein in regulating vesicular stomatitis virus RNA synthesis. Cell. 1985;41:259–267. doi: 10.1016/0092-8674(85)90079-0. [DOI] [PubMed] [Google Scholar]
- 3.Bangham C R, Kirkwood T B. Defective interfering particles: effects in modulating virus growth and persistence. Virology. 1990;179:821–826. doi: 10.1016/0042-6822(90)90150-p. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz U, Finke S, Conzelmann K-K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in vitro and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calain P, Roux L. Functional characterisation of the genomic and antigenomic promoters of Sendai virus. Virology. 1995;212:163–173. doi: 10.1006/viro.1995.1464. [DOI] [PubMed] [Google Scholar]
- 6.Collins P L, Mink M A, Stec D S. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc Natl Acad Sci USA. 1991;88:9663–9667. doi: 10.1073/pnas.88.21.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conzelmann K K, Cox J H, Schneider L G, Thiel H J. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. [DOI] [PubMed] [Google Scholar]
- 8.Conzelmann K K, Cox J H, Thiel H J. An L (polymerase)-deficient rabies virus defective interfering particle RNA is replicated and transcribed by heterologous helper virus L proteins. Virology. 1991;184:655–663. doi: 10.1016/0042-6822(91)90435-e. [DOI] [PubMed] [Google Scholar]
- 9.Conzelmann K K, Schnell M. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J Virol. 1994;68:713–719. doi: 10.1128/jvi.68.2.713-719.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis N L, Wertz G W. Synthesis of vesicular stomatitis virus negative-strand RNA in vitro: dependence on viral protein synthesis. J Virol. 1982;41:821–832. doi: 10.1128/jvi.41.3.821-832.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePolo N J, Holland J J. The intracellular half-lives of nonreplicating nucleocapsids of DI particles of wild type and mutant strains of vesicular stomatitis virus. Virology. 1986;151:371–378. doi: 10.1016/0042-6822(86)90057-7. [DOI] [PubMed] [Google Scholar]
- 12.Dimock K, Collins P L. Rescue of synthetic analogs of genomic RNA and replicative-intermediate RNA of human parainfluenza virus type 3. J Virol. 1993;67:2772–2778. doi: 10.1128/jvi.67.5.2772-2778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelhorn M, Stricker R, Roux L. Molecular cloning and characterization of a Sendai virus internal deletion defective RNA. J Gen Virol. 1993;74:137–141. doi: 10.1099/0022-1317-74-1-137. [DOI] [PubMed] [Google Scholar]
- 14.Fearns R, Peeples M E, Collins P L. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 15.Finke S, Conzelmann K K. Ambisense gene expression from recombinant rabies virus: random packaging of positive- and negative-strand ribonucleoprotein complexes into rabies virions. J Virol. 1997;71:7281–7288. doi: 10.1128/jvi.71.10.7281-7288.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giachetti C, Holland J J. Vesicular stomatitis virus and its defective interfering particles exhibit in vitro transcriptional and replicative competition for purified L-NS polymerase molecules. Virology. 1989;170:264–267. doi: 10.1016/0042-6822(89)90375-9. [DOI] [PubMed] [Google Scholar]
- 18.Grabau E A, Holland J J. Analysis of viral and defective-interfering nucleocapsids in acute and persistent infection by rhabdoviruses. J Gen Virol. 1982;60:87–97. doi: 10.1099/0022-1317-60-1-87. [DOI] [PubMed] [Google Scholar]
- 19.Kolakofsky D, Bruschi A. Antigenomes in Sendai virions and Sendai virus-infected cells. Virology. 1975;66:185–191. doi: 10.1016/0042-6822(75)90189-0. [DOI] [PubMed] [Google Scholar]
- 20.Kuo L, Grosfeld H, Cristina J, Hill M G, Collins P L. Effects of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J Virol. 1996;70:6892–6901. doi: 10.1128/jvi.70.10.6892-6901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazzarini R A, Keene J D, Schubert M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell. 1981;26:145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- 22.Leppert M, Kort L, Kolakofsky D. Further characterization of Sendai virus DI-RNAs: a model for their generation. Cell. 1977;12:539–552. doi: 10.1016/0092-8674(77)90130-1. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Pattnaik A K. Replication signals in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication. Virology. 1997;232:248–259. doi: 10.1006/viro.1997.8571. [DOI] [PubMed] [Google Scholar]
- 24.Mebatsion T, Schnell M J, Cox J H, Finke S, Conzelmann K K. Highly stable expression of a foreign gene from rabies virus vectors. Proc Natl Acad Sci USA. 1996;93:7310–7314. doi: 10.1073/pnas.93.14.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mottet G, Muhlemann A, Tapparel C, Hoffmann F, Roux L. A Sendai virus vector leading to the efficient expression of mutant M proteins interfering with virus particle budding. Virology. 1996;221:159–171. doi: 10.1006/viro.1996.0362. [DOI] [PubMed] [Google Scholar]
- 26.Mottet G, Curran J, Roux L. Intracellular stability of nonreplicating paramyxovirus nucleocapsids. Virology. 1990;176:1–7. doi: 10.1016/0042-6822(90)90223-e. [DOI] [PubMed] [Google Scholar]
- 27.Mottet G, Roux L. Budding efficiency of Sendai virus nucleocapsids: influence of size and ends of the RNA. Virus Res. 1989;14:175–187. doi: 10.1016/0168-1702(89)90037-3. [DOI] [PubMed] [Google Scholar]
- 28.Murphy S K, Ito Y, Parks G D. A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3′ terminus. J Virol. 1998;72:10–19. doi: 10.1128/jvi.72.1.10-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palma E L, Huang A. Cyclic production of vesicular stomatitis virus caused by defective interfering particles. J Infect Dis. 1974;129:402–410. doi: 10.1093/infdis/129.4.402. [DOI] [PubMed] [Google Scholar]
- 30.Park K H, Huang T, Correia F F, Krystal M. Rescue of a foreign gene by Sendai virus. Proc Natl Acad Sci USA. 1991;88:5537–5541. doi: 10.1073/pnas.88.13.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattnaik A K, Ball L A, LeGrone A, Wertz G W. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology. 1995;206:760–764. doi: 10.1016/S0042-6822(95)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 33.Patton J T, Davis N L, Wertz G W. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J Virol. 1984;49:303–309. doi: 10.1128/jvi.49.2.303-309.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224:405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- 35.Perrault J. Origin and replication of defective interfering particles. Curr Top Microbiol Immunol. 1981;93:151–207. doi: 10.1007/978-3-642-68123-3_7. [DOI] [PubMed] [Google Scholar]
- 36.Perrault J, Semler B L. Internal genome deletions in two distinct classes of defective interfering particles of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1979;76:6191–6195. doi: 10.1073/pnas.76.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao D D, Huang A S. Interference among defective interfering particles of vesicular stomatitis virus. J Virol. 1982;41:210–221. doi: 10.1128/jvi.41.1.210-221.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Re G G, Gupta K C, Kingsbury D W. Genomic and copy-back 3′ termini in Sendai virus defective interfering RNA species. J Virol. 1983;45:659–664. doi: 10.1128/jvi.45.2.659-664.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Re G G, Kingsbury D W. Paradoxical effects of Sendai virus DI RNA size on survival: inefficient envelopment of small nucleocapsids. Virology. 1988;165:331–337. doi: 10.1016/0042-6822(88)90577-6. [DOI] [PubMed] [Google Scholar]
- 40.Schlesinger S. The generation and amplification of defective interfering RNAs. In: Domingo E, Holland J J, Ahlquist P, editors. RNA genetics. Boca Raton, Fla: CRC Press Inc.; 1988. pp. 167–184. [Google Scholar]
- 41.Schnell M J, Conzelmann K K. Polymerase activity of in vitro mutated rabies virus L protein. Virology. 1995;214:522–530. doi: 10.1006/viro.1995.0063. [DOI] [PubMed] [Google Scholar]
- 42.Schnell M J, Mebatsion T, Conzelmann K K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sidhu M S, Chan J, Kaelin K, Spielhofer P, Radecke F, Schneider H, Masurekar M, Dowling P C, Billeter M A, Udem S A. Rescue of synthetic measles virus minireplicons: measles genomic termini direct efficient expression and propagation of a reporter gene. Virology. 1995;208:800–807. doi: 10.1006/viro.1995.1215. [DOI] [PubMed] [Google Scholar]
- 44.Soria M, Little S P, Huang A S. Characterization of vesicular stomatitis virus nucleocapsids. I. Complementary 40 S RNA molecules in nucleocapsids. Virology. 1974;61:270–280. doi: 10.1016/0042-6822(74)90261-x. [DOI] [PubMed] [Google Scholar]
- 45.Tapparel C, Maurice D, Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tapparel C, Roux L. The efficiency of Sendai virus genome replication: the importance of the RNA primary sequence independent of terminal complementarity. Virology. 1996;225:163–171. doi: 10.1006/viro.1996.0584. [DOI] [PubMed] [Google Scholar]
- 47.Von Laer D M, Mack D, Kruppa J. Delayed formation of defective interfering particles in vesicular stomatitis virus-infected cells: kinetic studies of viral protein and RNA synthesis during autointerference. J Virol. 1988;62:1323–1329. doi: 10.1128/jvi.62.4.1323-1329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wertz G W. Isolation of possible replicative intermediate structures from vesicular stomatitis virus-infected cells. Virology. 1978;85:271–285. doi: 10.1016/0042-6822(78)90431-2. [DOI] [PubMed] [Google Scholar]
- 49.Wertz G W, Levine M. RNA synthesis by vesicular stomatitis virus and a small plaque mutant: effects of cycloheximide. J Virol. 1973;12:253–264. doi: 10.1128/jvi.12.2.253-264.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]