Abstract
Aleutian mink disease parvovirus (ADV) is the etiological agent of Aleutian disease of mink. The acute disease caused by ADV consists of permissive infection of alveolar type II cells that results in interstitial pneumonitis. The permissive infection is experimentally modeled in vitro by infecting Crandell feline kidney (CrFK) cells with a tissue culture-adapted isolate of ADV, ADV-G. ADV-G VP2 empty virions expressed in a recombinant baculovirus system were analyzed for the ability to bind to the surface of CrFK cells. Radiolabeled VP2 virions bound CrFK cells specifically, while they did not bind either Mus dunni or Spodoptera frugiperda cells, cells which are resistant to ADV infection. The binding to CrFK cells was competitively inhibited by VP2 virions but not by virions of cowpea chlorotic mottle virus (CCMV), another unenveloped virus similar in size to ADV. Furthermore, preincubation of CrFK cells with the VP2 virions blocked infection by ADV-G. The VP2 virions were used in a virus overlay protein binding assay to identify a single protein of approximately 67 kDa, named ABP (for ADV binding protein), that demonstrates specific binding of VP2 virions. Exogenously added VP2 virions were able to competitively inhibit the binding of labeled VP2 virions to ABP, while CCMV virions had no effect. Polyclonal antibodies raised against ABP reacted with ABP on the outer surface of CrFK cells and blocked infection of CrFK cells by ADV-G. In addition, VP2 virion attachment to CrFK cells was blocked when the VP2 virions were preincubated with partially purified ABP. Taken together, these results indicate that ABP is a cellular receptor for ADV.
Aleutian mink disease parvovirus (ADV) causes both chronic and acute disease in mink. The chronic disease, termed Aleutian disease, is associated with a persistent infection of adult mink and is characterized by viral persistence, hypergammaglobulinemia, plasmacytosis, increased CD8+ lymphocytes, and immune complex disorder (reviewed in reference 12). Macrophages have been identified as sites of restricted virus replication, and infection of these cells is thought to lead to the immune disturbance (3, 36, 37). ADV gains entry into macrophages by Fc-mediated uptake of antibody-virus complexes, a process called antibody-dependent enhancement of infection (22, 28, 31).
The acute disease, which occurs in newborn mink, is a fulminant, fatal interstitial pneumonitis. A permissive ADV infection occurs in alveolar type II cells, which leads to disturbances in surfactant secretion within the lung (12). The mechanism by which ADV attaches to and gains entry into these cells is not understood. Unlike macrophages, type II cells are not phagocytic and do not bear Fc receptors. The permissive infection is mimicked by infecting Crandell feline kidney (CrFK) cells with a tissue culture isolate of ADV named ADV-G (9, 13, 41). It is believed that ADV entry into both type II cells and CrFK cells is receptor mediated. ADV is capable of infecting many different mustelid hosts, including mink, ferrets, weasels, fishers, marten, skunks, otters, raccoons, and foxes (5, 24, 27). The broad host range exhibited by ADV suggests that it may utilize a cellular receptor that is widely distributed among the different mustelids.
The initial event in a virus infection is the attachment of viruses to the surface of the cell. Virus attachment is usually dependent on a specific virus receptor on the cell surface, and the presence of the receptor can be a major factor in determining viral tissue tropism and host range. A large variety of molecules have been identified as virus receptors. Virus receptors range from ubiquitous cell surface moieties, such as carbohydrates, to cell-specific membrane proteins with various functions (8, 20, 38, 47). Attachment to the surface of cells by some viruses requires only the presence of specific carbohydrates, as in the case of adeno-associated virus (44). Other viruses, such as echoviruses, utilize specific glycoproteins as receptors, where both the protein and carbohydrate moiety are necessary for receptor function (32). In addition, some viruses (e.g., dengue virus) utilize receptors consisting only of protein without a carbohydrate moiety (42). Regardless of which molecule or combination of molecules is utilized as the virus receptor, the net effect is the same: virus entry into the cell to establish infection. Characterization of virus receptors can provide insight into the basis of virus host range and disease pathogenesis. In addition, defining the specific chemical interactions that occur between a virus and its receptor may enable the design of chemical therapies that can perturb this interaction and prevent virus infection.
ADV, as well as many of the parvoviruses, exhibits many advantages for examining virus-receptor interactions. The parvoviruses are small, unenveloped viruses with single-stranded, negative-sense DNA genomes of about 5,000 nucleotides (10). The T=1 ADV virion is comprised of two virion structural proteins designated VP1 and VP2 (2). ADV virions are extremely stable and are virtually impossible to dissociate in vitro without using reagents that unfold or hydrolyze the individual proteins. Capsid proteins from several parvoviruses, including ADV, have been shown to self-assemble into empty capsids when expressed in insect cells by using recombinant baculoviruses (16, 17, 26, 43, 49). The ADV VP2 empty virion (baculovirus-expressed) structure has recently been solved to 22-Å resolution, and studies to solve the structure at a higher resolution are under way (33). The baculovirus-expressed VP2 virions provide an efficient, relatively easy system to purify large quantities of virions for use in structural and receptor studies.
The cellular receptor has been characterized for 2 of the 31 described eukaryotic parvoviruses. The human parvovirus B19 uses the erythrocyte P antigen, a glycosophingolipid, as its cellular receptor (15). The parvovirus adeno-associated virus type 2 was recently reported to employ membrane-associated heparan sulfate proteoglycans as cellular receptors (44). A requirement for sialic acid has been demonstrated for hemagglutination by canine parvovirus (CPV) and feline panleukopenia virus and for infection by minute virus of mice (6, 7, 18, 25, 35, 46). However, their specific cellular receptors have not been identified.
Because ADV permissively infects alveolar type II cells as well as CrFK cells, it is possible that these cells have similar receptors for ADV. The low number of alveolar type II cells in the lung combined with the lack of an efficient method for isolating these cells technically hinders the ability to identify the ADV cellular receptor in type II cells. However, the advantages of working with the CrFK-ADV system permitted us to initiate experiments to isolate the CrFK cell receptor. In this study we have identified a single protein, denoted ADV binding protein (ABP), with a molecular mass of 67 kDa that specifically binds ADV-G VP2 virions.
MATERIALS AND METHODS
Viruses, cells, and plasmids.
The ADV-G strain used in this study was derived from a molecularly cloned virus stock (XXI-Q-3-15) (11). ADV-G was propagated in CrFK cells. CrFK and Mus dunni cells were cultured at 37°C in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (Gibco-BRL, Gaithersburg, Md.). Spodoptera frugiperda (SF9) cells, used for baculovirus expression, were cultured in Grace’s insect medium (Gibco-BRL) supplemented with 10% fetal calf serum (Gibco-BRL).
Baculovirus expression of VP2 empty virions and iodination.
The VP2 virions were expressed, as previously described, in a recombinant baculovirus, Autographa californica nuclear polyhedrosis virus, encoding the ADV VP2 protein (17). One modification to the previous protocol included maintaining the SF9 cells as suspension cultures by using spinner flasks. The Grace’s insect medium was supplemented with 0.05% Pluronic Polyol F-68 (Gibco-BRL). The SF9 cells were infected at a multiplicity of infection (MOI) of 1 and cultured for 5 days postinfection. VP2 virions were purified by collecting the infected SF9 cells, resuspending them in lysis buffer (0.05 M Tris-HCl [pH 8.0], 0.15 M NaCl, 0.2% Triton X-100, protease inhibitor cocktail [Pefabloc, 1 μg/ml; pepstatin, 1 μg/ml; aprotinin, 1 μg/ml; leupeptin, 5 μg/ml]), and freeze-thawing four times. The suspension was sonicated on ice for three cycles of 15 s each, chloroform extracted, sonicated for two cycles of 15 s each, and centrifuged for 30 min at 10,000 rpm in a JA-14 rotor (Beckman, Piscataway, N.J.). The aqueous phase was subjected to a second chloroform extraction. The VP2 virions were further purified by making the suspension 10% with polyethylene glycol 8000, stirring overnight at 4°C, and collecting the precipitated virions by centrifugation at 10,000 rpm for 15 min in a JA-14 rotor. The virion pellet was resuspended in 1/10 volume (of that of original lysis buffer suspension) of storage buffer (0.05 M Tris-HCl [pH 8.0], 0.05 M NaCl) with protease inhibitor cocktail, dialyzed against storage buffer, and stored at 4°C.
VP2 virions were labeled with 125I by incubating 100 μg of VP2 virions with 1 mCi of Na125I (Amersham, Arlington Heights, Ill.) in the presence of IODO-BEADS (Pierce, Rockford, Ill.) as specified by the manufacturer. The labeled virions were purified from free Na125I by using D-Salt desalting columns (Pierce) and stored in Tris-EDTA buffer containing 50 mM NaCl. The specific activity of the labeled virions was determined by protein quantitation and radioactivity quantitation with a gamma counter.
Virion-cell binding assay.
CrFK cells were plated (5 × 106 cells/well) in a 12-well tissue culture plate and incubated overnight at 37°C. The plate was incubated for 2 h at 4°C to chill the cells. The cells were incubated at 4°C for 1 h with 100,000 cpm of radiolabeled ADV VP2 virions (50 μg) in DMEM, washed four times with 0.5 ml of ice-cold DMEM, and then lysed with a buffer containing 0.01 M phosphate-buffered saline (PBS) (pH 7.2), 0.15 M CaCl2, 1% Triton X-100, and 0.1% sodium dodecyl sulfate (SDS). The radioactivity was quantitated with a gamma counter. Competition assays were performed by preincubating the cells with 900 μg of unlabeled virions for 15 min at 4°C in DMEM, followed by addition of 100,000 cpm of radiolabeled ADV VP2 virions (as described above). In blocking experiments, 125I-ADV VP2 empty virions (100,000 cpm) were preincubated for 15 min with either 500 μg of total CrFK cell membrane proteins, 200 μg of partially purified ABP (approximately 70% pure as estimated by Coomassie blue staining of an SDS-polyacrylamide gel), or 500 μg of SF9 cell membrane proteins and then added to the cells and processed as described above. In each experiment the assays were performed in triplicate, and the average values from the three assays along with the standard errors are reported.
Virus overlay protein binding assay (VOPBA).
CrFK membrane proteins were purified from confluent cultures in T-150 tissue culture flasks by washing three times with buffer A (0.02 M sodium phosphate [pH 8.0], 0.005 M EDTA, 0.002 M N-ethylmaleimide, protease inhibitor cocktail) at room temperature and scraping the cells into 5 ml of buffer A. The suspended cells were collected by centrifugation at 1,500 rpm for 5 min in a Beckman J-6 swinging-bucket centrifuge. Pellets were resuspended in 2.5 ml of buffer A containing 0.5% deoxycholate and 1% Triton X-100, incubated on ice for 10 min, and centrifuged at 3,000 rpm for 15 min in a Beckman J-6 swinging-bucket centrifuge. The supernatant, containing the membrane proteins, was collected, supplemented with protease inhibitor cocktail, and stored at −20°C.
For further analysis, the membrane proteins (50 to 100 μg) were separated in a nonreducing SDS–8% polyacrylamide gel, transferred to a polyvinylidene difluoride membrane (Sigma, St. Louis, Mo.), and refolded by incubating the blot in refolding buffer (0.01 M HEPES [pH 7.4], 0.01 M MgCl2, 0.05 M NaCl, 0.001 M dithiothreitol, 10% glycerol, protease inhibitor cocktail) for 12 h at 4°C. The blot was blocked in PBS containing 5% nonfat dry milk for 5 h at 4°C and then reacted with 100,000 cpm of 125I-ADV VP2 empty virions (50 μg) in hybridization buffer (DMEM containing 10% fetal calf serum and 0.25 M NaCl) for 5 h at room temperature. Finally, the blots were washed four times with hybridization buffer and exposed to film. Competition experiments were performed by adding unlabeled virions (amounts are indicated in the figure legends) to the hybridization buffer just prior to addition of the iodinated ADV VP2 virions.
Production of polyclonal antibody to ABP.
Comparison of VOPBA blots with a parallel Coomassie blue-stained gel identified a single protein band with an approximate molecular mass of 67 kDa as the ABP, which was excised from the gel. The protein was electroeluted by using a protein elution apparatus from Bio-Rad (Hercules Calif.) and quantified by rerunning on an SDS-polyacrylamide gel, staining, and comparing the band intensity with those of known standards. To confirm that the eluted protein was a single protein rather than multiple proteins with similar electrophoretic mobilities, a two-dimensional gel was run. The first dimension was near-equilibrium pH gel electrophoresis with ampholines with a pH range of 3.5 to 9.5, and the second dimension was SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Silver staining of the gel revealed a single protein. Rabbits were injected subcutaneously with approximately 100 μg of purified ABP emulsified in Freund complete adjuvant. At 3, 6, and 9 weeks postinoculation, the rabbits were boosted with 100 μg of purified ABP emulsified in Freund incomplete adjuvant. At 12 weeks postinoculation, the rabbits were exsanguinated under anesthesia and sera were collected.
Western blotting.
Purified CrFK membrane proteins (500 ng) were separated in a nonreducing SDS–10% polyacrylamide gel, transferred to a Hybond-C extra-nitrocellulose membrane (Amersham), and blocked for 2 h in PBS containing 15% nonfat dry milk. The blots were probed with anti-ABP serum diluted in PBS–0.05% Tween 20 (PBST) containing 15% nonfat dry milk at room temperature for 1 h, washed three times with PBST, and probed with horseradish peroxidase-conjugated goat anti-rabbit antibody (ICN, Aurora Ohio) diluted 1:3,000 in PBST containing 15% nonfat dry milk for 30 min at room temperature. The blots were washed four times in PBST with a final wash in PBS, developed by using the ECL Western blotting system (Amersham) according to the manufacturer’s specifications, and exposed to film.
Blocking of ADV infection with ABP polyclonal antibody.
CrFK cells (105 cells/well) were seeded in four-well Chamber-Slides (Nunc Inc., Naperville, Ill.) and incubated overnight at 37°C. The slides were incubated at 4°C for 2 h, at which time the medium was removed and the cells were flooded with 0.25 ml of ice-cold DMEM containing various dilutions of anti-ABP polyclonal serum or normal rabbit serum (collected as prebleeds from rabbits used to raise anti-ABP antibodies). The cell-antibody suspension was incubated for 30 min at 4°C, after which ADV-G was added at an MOI of 1 focus-forming unit/cell. Incubation was continued at 4°C for 30 min, and the cultures were then washed four times with 1.0 ml of ice-cold DMEM. The cells were flooded with 1.0 ml of DMEM and incubated at 32°C for 3 days. Infectivity was analyzed by immunofluorescence microscopy as described below.
Immunofluorescence microscopy.
In experiments examining ADV-G infectivity, a polyclonal antibody to the ADV nonstructural 1 protein (NS1) diluted 1:20 in PBST was utilized to stain infected cultures of CrFK cells (39). The primary NS1 antibody was incubated with the cells for 30 min at room temperature, followed by washing. A secondary fluorescein isothiocyanate (FITC)-conjugated goat antirabbit antibody, diluted 1:1,000 in PBST, was incubated with the cells for 30 min, followed by several washes and viewing with a Nikon Microphot-SA microscope. The number of infected cells was determined by averaging the counts from 20 fields of view in areas of equal cell density.
In examining ABP reactivity with CrFK cells, cells (105/well) were seeded in four-well Chamber-Slides and incubated overnight at 37°C. The medium was removed from the cells, the slides were briefly washed in PBS, and the cells were fixed by incubation in 3.7% formaldehyde diluted in PBS for 1 h at room temperature. Blocking was accomplished by addition of PBS containing 5% nonfat dry milk, 4% bovine serum albumin fraction V, and 5% bovine fetal calf serum and incubation for 1 h at room temperature. The blocking buffer was removed by aspiration, and PBS-diluted polyclonal ABP antisera were added and left for 30 min at room temperature. The slides were washed three times with PBS, and then FITC-conjugated goat antirabbit antibody (diluted 1/200) was added and left for 30 min at room temperature, followed by additional washing in PBS. A final rinse in 95% ethanol for 5 min was performed before mounting. The slides were dried and viewed with a Nikon Microphot-SA microscope. Photographs were taken with a Nikon 35DX camera.
RESULTS
Specific attachment of baculovirus-expressed VP2 virions to CrFK cells.
In order to identify the cellular receptor from CrFK cells, it was necessary to obtain large quantities of virions to conduct the experiments described below. Due to the difficulty in obtaining the large quantities of purified native ADV virions needed, we used ADV VP2 empty virions (hereafter referred to as VP2 virions) expressed in a recombinant baculovirus system to experimentally identify the ADV cellular receptor. Initially we sought to determine if the VP2 virions attached to cells in a similar fashion as native ADV-G virions. To address this question, the VP2 virions were radioactively labeled with 125I and assayed for the ability to bind CrFK cells. The iodinated VP2 virions bound to CrFK cells but did not bind to M dunni (mouse) cells or SF9 (insect) cells, cell lines that are not permissive for ADV replication (Fig. 1). Additionally, the VP2 virion binding to the CrFK cell surface could be blocked by adding exogenous unlabeled VP2 virions (96% reduction) but not by adding virions of cowpea chlorotic mottle virus (CCMV), an unenveloped plant virus similar in size to ADV (Fig. 1). This indicated that the VP2 virions specifically bound to the surface of CrFK cells.
FIG. 1.
Binding of baculovirus-expressed VP2 virions to CrFK cells. 125I-VP2 virions were incubated with CrFK cells and washed, and cell associated radioactivity was quantitated. VP2 virion binding to the indicated cells was determined. Competition experiments examining the ability of iodinated VP2 virions to bind CrFK cells in the presence of an 18-fold excess of unlabeled VP2 virions or unlabeled CCMV virions demonstrated that only the VP2 virions could inhibit the cell binding of radiolabeled VP2 virions. All values are represented as a percentage of the CrFK-VP2 virion binding. Each experiment was performed in triplicate, and standard errors are shown.
The demonstration that the VP2 virions bound to the CrFK cell surface in a specific manner suggested that they behaved like native ADV virions but did not prove that the two types of virions bound to CrFK cells by using the same receptor. Competitive binding experiments examining the ability of ADV-G virions and VP2 virions to bind CrFK cells were conducted to address this concern. CrFK cells were incubated with VP2 virions at low temperature, a condition that allows attachment but not entry of virions into the cell. Following preincubation with VP2 virions, native ADV-G virions were added and incubated for 1 h. The unbound virus was washed away, and the cells were incubated for 3 days at an optimal temperature for ADV replication. The number of infected cells was determined by immunofluorescence microscopy with an antibody to NS1, a nonstructural protein that is produced only during active ADV replication. VP2 virions inhibited CrFK infection by ADV-G by nearly 95%, indicating that prior binding of VP2 virions to the CrFK cell surface prevented ADV-G binding and entry (Fig. 2). In contrast, CCMV virions did not inhibit ADV-G infection. Similar results were obtained with several preparations of labeled VP2 virions. Taken together, these results indicate that VP2 virions bind CrFK cells in a functional manner similar to native ADV-G virions.
FIG. 2.
Blocking of ADV-G infection of CrFK cells with VP2 virions. VP2 or CCMV virions were preincubated with chilled CrFK cells, after which ADV-G was added. The cells were then washed and incubated for 3 days. Infectivity was measured by immunofluorescence microscopy with ADV NS1 antibody. Each value is represented as a percentage of that for ADV-G infection of CrFK cells alone, and the standard error is shown.
Identification of an ABP by VOPBA analysis.
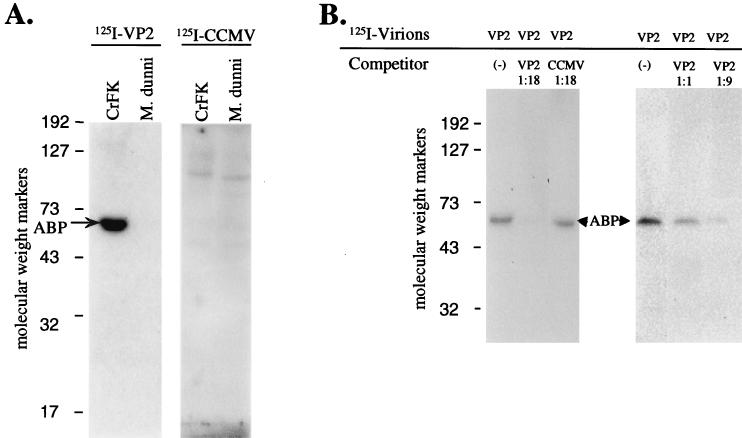
In order to identify membrane proteins from CrFK cells that could bind VP2 virions, we chose to use VOPBA, a biochemical approach proven to be useful in the identification of several virus receptors (14, 19, 30, 34, 42). The CrFK cell membrane proteins were isolated and used in the VOPBA to identify proteins that specifically bound radiolabeled VP2 virions. VP2 virions specifically bound a CrFK membrane protein (with an approximate molecular mass of 67 kDa) that we named ABP (Fig. 3A). No binding of VP2 virions was observed with membrane proteins from M. dunni cells, indicating that the binding was cell specific. In addition, labeled CCMV virions did not bind ABP but interacted nonspecifically with several other proteins from both CrFK and M. dunni cells. This indicated that VP2 virion binding by ABP was both cell and virus specific.
FIG. 3.
(A) Identification of a single VP2 virion binding protein from CrFK cells by VOPBA. One hundred micrograms of either CrFK or M. dunni membrane proteins was separated on a nonreducing SDS–8% polyacrylamide gel and processed for VOPBA. 125I-VP2 virions (100,000 cpm/50 μg) were used to probe the blot on the left, while 125I-CCMV virions (100,000 cpm/55 μg) were used with the blot on the right. (B) Inhibition of 125I-VP2 virion binding to ABP by addition of unlabeled VP2 virions. One hundred micrograms of CrFK membrane proteins was separated on a nonreducing SDS–8% polyacrylamide gel and processed for VOPBA. Competing virions were added to the blots just prior to addition of the 125I-VP2 virions (100,000 cpm/50 μg). Left blot, 125I-VP2 virions alone (left lane), addition of an 18-fold excess (900 μg) of unlabeled VP2 virions (middle lane), and addition of an 18-fold excess (900 μg) of unlabeled CCMV virions (right lane). Right blot, 125I-VP2 virions alone (left lane), addition of an equivalent amount (50 μg) of unlabeled VP2 virions as labeled VP2 virions (middle lane), and addition of a ninefold excess (450 μg) of unlabeled VP2 virions (right lane). Sizes of molecular weight markers are in thousands.
The specificity of the ABP-VP2 virion binding was further analyzed in competitive binding experiments. Radiolabeled virions were analyzed for the ability to bind ABP in the VOPBA when other unlabeled virions were present (Fig. 3B). The radiolabeled VP2 virion binding to ABP was inhibited in a dose-dependent manner by addition of unlabeled VP2 virions (Fig. 3B). Addition of 18-fold more unlabeled VP2 virions (900 μg) than radiolabeled VP2 virions resulted in complete inhibition of radiolabeled VP2 virion binding (100% competition). Addition of an 18-fold excess of unlabeled CCMV virions was unable to competitively inhibit radiolabeled VP2 virion binding, providing additional evidence that the ABP-VP2 virion binding was a specific interaction.
A final experiment was conducted to confirm the specificity of the ABP-VP2 virion interaction. A virion-cell binding assay was employed to determine if preincubation of ABP with VP2 virions could inhibit VP2 virion binding to CrFK cells. Partially purified ABP was mixed with radiolabeled VP2 virions for 15 min and tested for the ability to bind to CrFK cells. The binding of VP2-ABP to CrFK cells was reduced by 75% compared to that of unbound VP2 virions (Fig. 4). This was in comparison to a relative 27% reduction in CrFK binding observed when VP2 virions were preincubated with total CrFK membrane proteins. Incubation of SF9 cell membrane proteins with the VP2 virions had no effect on VP2 virion binding to CrFK cells. Thus, the binding of ABP to the surface of VP2 virions prevented the virions from attaching to the CrFK cells, presumably by competition for receptor binding sites on the virion surface.
FIG. 4.
Reduced CrFK binding by VP2 virions when complexed with ABP. ABP was preincubated with 125I-VP2 virions for 15 min prior to addition of CrFK cells. The ability of the 125I-virions to bind CrFK cells was then examined. VP2 virions were preincubated either alone or with 500 μg of total CrFK membrane proteins, 200 μg of partially purified ABP, 500 μg of total SF9 membrane proteins. Each value is represented as a percentage of that for VP2 alone, and standard errors from triplicate experiments are shown.
Production and characterization of anti-ABP antibodies.
The previous experiments demonstrated that VP2 virions specifically bound ABP but did not address the relevance of the binding in the biology of virus infection. To determine if the interaction between ABP and ADV is important in virus binding and subsequent entry into the cell, we raised antibodies against ABP and tested their ability to block ADV infection of CrFK cells. If native ADV-G attached to CrFK cells through interactions with ABP, antibody against ABP could potentially interfere with binding of ADV-G to ABP on the cell surface.
Two polyclonal anti-ABP rabbit antisera specifically reacted with ABP in a Western blot of CrFK membrane proteins (Fig. 5A). The antisera reacted with a single protein migrating at the ABP molecular mass of 67 kDa. The prebleed serum control did not show any such specific reactivity. To ensure that the antibodies raised against ABP were specific for only the ABP protein, 100 μg of total CrFK membrane proteins was separated by two-dimensional gel electrophoresis, Western blotted, and probed with the ABP antisera. The results of such analysis are shown for the R2 anti-ABP serum (Fig. 5B). The R2 ABP antiserum reacted strongly and specifically with only one protein having an approximate molecular mass of 67 kDa. Similar results were found with the R1 ABP antiserum (data not shown). This analysis also demonstrated that the ABP protein has a pI in the range of 7.0 to 8.0. Further analysis will be required to accurately determine the exact pI.
FIG. 5.
(A) Western blot of CrFK membrane proteins with ABP polyclonal antisera. Five hundred nanograms of CrFK membrane proteins was separated by SDS-PAGE, transferred, and incubated with either normal rabbit serum (prebleed), rabbit R1 anti-ABP polyclonal antiserum, or rabbit R2 anti-ABP polyclonal antiserum. Each serum was diluted 1:200. (B) Western blot of CrFK membrane proteins separated by using a two-dimensional gel where the first dimension was near-equilibrium pH gel electrophoresis (with ampholines with a pH range of 3.5 to 9.5) and the second dimension was SDS-PAGE. The blot was probed with rabbit R2 anti-ABP polyclonal antiserum diluted 1:200. Sizes of molecular weight markers are in thousands.
One of the anti-ABP antisera, the R2 anti-ABP antiserum, was chosen to test the ability of the sera to bind ABP on the surface of CrFK cells. Immunofluorescence microscopy of CrFK cells incubated with the R2 anti-ABP antibodies showed selective staining of the CrFK cell surfaces, whereas that with the prebleed serum did not (Fig. 6A). The anti-ABP sera did not react with either M. dunni or SF9 cells (data not shown). ADV-G infection of CrFK cells was blocked when the cells were preincubated with these antibodies (Fig. 6B). Both of the anti-ABP polyclonal antisera tested blocked ADV-G infection of CrFK cells (73 and 67% blocking, respectively), with an optimal dilution of 1:10. The prebleed serum had no effect on ADV-G infectivity. The blocking of ADV-G infection indicated that ADV binding to ABP was required for binding and entry into the cell to establish infection.
FIG. 6.
(A) Immunofluorescence microscopy of CrFK cells with rabbit R2 anti-ABP polyclonal antiserum. CrFK cells, fixed with formaldehyde, were stained with with a 1:10 dilution of either rabbit prebleed serum or rabbit R2 anti-ABP polyclonal antiserum. An FITC-conjugated goat antirabbit polyclonal antibody was used as the secondary stain. The two fields contained similar numbers of cells. The photograph for the prebleed serum was taken with an exposure four times longer than that for the R2 anti-ABP serum. (B) Blocking of ADV-G infection in CrFK cells with anti-ABP polyclonal antisera. Rabbit sera, diluted 1:10 in medium, were preincubated with CrFK cells for 30 min at 4°C. ADV-G was added at an MOI of 1 and incubated for 30 min. The cells were washed and incubated for 3 days, after which they were analyzed by immunofluorescence microscopy with ADV NS1 antibody. NRS, normal rabbit prebleed serum. The results are graphed as relative percentages of that for the ADV-G–normal rabbit serum experiment, and standard errors are shown.
DISCUSSION
In this study we have identified a putative cellular receptor for ADV, a single protein with a molecular mass of 67 kDa that specifically binds ADV VP2 virions. This protein, which we have named ABP, has many characteristics expected of a cellular receptor: (i) ABP is expressed on the surface of CrFK cells and specifically binds VP2 virions, (ii) ABP binding of VP2 virions prevents virion binding to CrFK cells, (iii) VP2 virion binding to ABP on the CrFK cell surface prevents native ADV-G virions from infecting the cells, and (iv) antibodies directed against ABP bind to ABP on the CrFK cell surface and block infection by ADV-G.
In our identification of ABP, we used baculovirus-expressed VP2 virions. Although such artificially expressed virions potentially differ from native ADV virions in terms of cell binding and entry, we addressed this concern by demonstrating that VP2 virions compete with native ADV-G virions for cell binding. The baculovirus-expressed VP2 empty virions are morphologically and antigenically indistinguishable from ADV-G (8a). For other parvovirus systems, determination of the atomic structures of the native virions and empty VP2 empty virions has shown very few differences between the two virion types (1). In addition, it was demonstrated that the baculovirus-expressed B19 VP2 virions are antigenically and immunologically similar to the native virions (26). Thus, there is no reason to believe that the ADV VP2 empty virions behave differently from ADV-G in cell binding.
By using immunofluorescence microscopy, antibodies raised against ABP were reacted specifically with the surfaces of live and fixed CrFK cells. The ABP antisera, when preincubated with CrFK cells, blocked infection of the cells by ADV-G. The simplest explanation for this result is that the antibodies bind ABP and in doing so prevent ADV-G virions from binding to the cell surface. Precedence for this exists in a number of virus systems. Antibodies raised against two VOPBA-identified surface proteins from C6/36 cells specifically blocked dengue virus type 4 infection (42). In addition, antibodies raised against biochemically identified cell surface proteins from echovirus, Norwalk virus, Venezuelan equine encephalitis virus, papillomavirus, and porcine reproductive respiratory syndrome virus systems have been shown to specifically block attachment to and thus infection of susceptible cells (21, 23, 30, 32, 48). The possibility does exist that anti-ABP antibodies are nonspecifically blocking ADV-G attachment by steric hindrance to another, yet-unidentified, protein that is the real receptor for ADV. However, the additional data showing specific binding of ADV to ABP argues against this interpretation. Complete blocking of ADV-G infection was not obtained. Reductions of 73 and 67% were observed with the two antisera. This is most likely due to the technical inability to block every single ABP molecule on every cell. A single unbound ABP molecule could result in virus binding, and the cell would score positive for infection in our assay.
Experiments demonstrating the blocking of CrFK cell binding by ABP-complexed VP2 virions further support the hypothesis that ABP is the ADV cellular receptor. This, along with the antibody evidence discussed above, suggests that ABP is necessary for ADV binding to CrFK cells. The partially purified ABP used in the blocking experiments still contains other proteins (the ABP preparation was estimated to be approximately 70% pure) which could be playing a role in binding inhibition. However, the significant difference seen in binding inhibition between total CrFK membrane proteins and partially purified ABP suggests that ABP is the cause of the dramatic decrease in virion binding. We did not observe a significant increase in concentration of any other proteins in the partially purified ABP that could contribute to the dramatic decrease in virus binding observed. Experiments to purify ABP to homogeneity for more detailed analyses are under way.
During optimization of the VOPBA, we found that the inclusion of detergents in VOPBA buffers decreased the specificity of binding to CrFK membrane proteins by VP2 virions. Omitting detergents from the buffers resulted in the specific binding of ABP by VP2 virions. One reason that the detergents may promote nonspecific binding is that the detergents are interacting with hydrophobic, transmembrane regions of the membrane proteins on the VOPBA blot. The virions may be nonspecifically interacting with the membrane protein-associated detergents. Work by Alexandersen et al. demonstrated that ADV has a propensity to interact with hydrophobic cell membrane components due to the amphiphilic nature of the virion exterior (4). Thus, when detergents are present, the VP2 virions are likely interacting with the immobilized protein-detergent complexes in a nonspecific manner.
Previous work by two different laboratories with another parvovirus, CPV, attempted to use a similar approach in identifying its cellular receptor (6, 46). Both of those reports identified multiple proteins that bound CPV. The proteins identified were different in each report, most likely because the cell lines used differed. The report by Barbis et al. (6) demonstrated that binding of some of the proteins from rhesus monkey erythrocytes, required for hemagglutination, was dependent on protein-associated sialic acid moieties, while others were not. While both ADV and CPV are parvoviruses, the differ greatly and are only distantly related among the parvoviruses. ADV has approximately 40 more amino acid residues in VP2 than does CPV. A significant proportion of these amino acid residues are positioned in the loop 3-4 region of VP2, a region of the protein that is positioned on the virion surface at, or near, the virion threefold axis of symmetry (33). This region is hypothesized to be important for receptor binding and has been demonstrated to contain amino acid residues that affect host range in the CPV and feline panleukopenia virus systems as well as in ADV (23a, 40, 45).
Native ADV virions contain 60 protein molecules of which about 10% are VP1. The VP1 protein contains all of the amino acids encoded by the VP2-coding region with approximately 40 additional amino acid residues on the N terminus. Our results indicate that the receptor binding component of ADV virions is contained within the VP2-coding region, since the baculovirus-expressed VP2 empty virions do not contain VP1. VP1 empty virions can be expressed in baculovirus and contain approximately 90% VP1 and 10% VP2 (the opposite of what is found in native virions). The VP1 virions were analyzed for their ability to bind ABP in the VOPBA and were found to behave similarly to VP2 virions (data not shown). These results indicate that the VP1 unique region does not significantly influence receptor binding.
The identification of the ADV receptor utilized in permissive infection will undoubtedly contribute to future studies aimed at understanding the biology of ADV pathogenesis. An important but unanswered question in ADV pathogenesis is which cells in the mink are capable of being infected. Experiments identifying infected cells have been limited by the sensitivity of assays detecting virus replication. Using the antibodies to ABP, we plan to screen various mink cell populations for the cell surface expression of an ABP homologue. If these antibodies are capable of cross-reacting with the mink equivalent of ABP, we will be able to identify which cell populations can potentially be infected. In addition, defining the ADV-receptor complex in chemical terms will offer the possibility of designing therapies that could potentially interfere with this interaction and block virus infection. This would be highly desirable for the treatment of mink kits that have contracted ADV, because the mortality rate is often high and there is not currently a method to either prevent or cure the infection in such kits.
ACKNOWLEDGMENTS
We thank Klaus Jensen, Mavis Agbandge-McKenna, and Mary Ann Stevenson for helpful discussions. We also thank Kim Hasenkrug, John Portis, and Sue Priola for critical reading of the manuscript.
REFERENCES
- 1.Agbandje M, Parrish C R, Rossmann M G. The structure of parvoviruses. Semin Virol. 1995;6:299–309. [Google Scholar]
- 2.Alexandersen S, Bloom M E, Perryman S. Detailed transcription map of Aleutian mink disease parvovirus. J Virol. 1988;62:3684–3694. doi: 10.1128/jvi.62.10.3684-3694.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandersen S, Bloom M E, Wolfinbarger J. Evidence of restricted viral replication in adult mink infected with Aleutian disease of mink parvovirus. J Virol. 1988;62:1495–1507. doi: 10.1128/jvi.62.5.1495-1507.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandersen S, Hau J, Larsen S. Examination of Aleutian disease virus in charge-shift crossed immunoelectrophoresis. Acta Pathol Microbiol Immunol Scand Sect B. 1984;92:331–334. doi: 10.1111/j.1699-0463.1984.tb02842.x. [DOI] [PubMed] [Google Scholar]
- 5.Alexandersen S, Uttenthal-Jensen A, Hansen M, Aasted B. Experimental transmission of Aleutian disease virus (ADV) to different animal species. Acta Pathol Microbiol Immunol Scand Sect B. 1985;93:195–200. doi: 10.1111/j.1699-0463.1985.tb02876.x. [DOI] [PubMed] [Google Scholar]
- 6.Barbis D P, Chang S-F, Parrish C R. Mutations adjacent to the dimple of the canine parvovirus capsid structure affect sialic acid binding. Virology. 1992;191:301–308. doi: 10.1016/0042-6822(92)90192-r. [DOI] [PubMed] [Google Scholar]
- 7.Barbis D P, Parrish C R. Characterization of canine parvovirus (CPV) interactions with 3201 T cells: involvement of GPI-anchored protein(s) in binding and infection. Braz J Med Biol Res. 1994;27:401–407. [PubMed] [Google Scholar]
- 8.Bass D M, Greenberg H B. Strategies for the identification of icosahedral virus receptors. J Clin Invest. 1992;89:3–9. doi: 10.1172/JCI115575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Bloom, M. E. Unpublished data.
- 9.Bloom M E, Alexandersen S, Mori S, Wolfinbarger J B. Analysis of parvovirus infections using strand-specific hybridization probes. Virus Res. 1989;14:1–26. doi: 10.1016/0168-1702(89)90066-x. [DOI] [PubMed] [Google Scholar]
- 10.Bloom M E, Alexandersen S, Perryman S, Lechner D, Wolfinbarger J B. Nucleotide sequence and genomic organization of Aleutian mink disease parvovirus (ADV): sequence comparisons between a nonpathogenic and a pathogenic strain of ADV. J Virol. 1988;62:2903–2915. doi: 10.1128/jvi.62.8.2903-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bloom M E, Berry B D, Wei W, Perryman S, Wolfinbarger J B. Characterization of chimeric full-length molecular clones of Aleutian mink disease parvovirus (ADV): identification of a determinant governing replication of ADV in cell culture. J Virol. 1993;67:5976–5988. doi: 10.1128/jvi.67.10.5976-5988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloom M E, Kanno H, Mori S, Wolfinbarger J B. Aleutian mink disease: puzzles and paradigms. Infect Agents Dis. 1994;3:279–301. [PubMed] [Google Scholar]
- 13.Bloom M E, Race R E, Wolfinbarger J B. Characterization of Aleutian disease virus as a parvovirus. J Virol. 1980;35:836–843. doi: 10.1128/jvi.35.3.836-843.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrow P, Oldstone M B. Characterization of lymphocytic choriomeningitis virus-binding protein(s): a candidate cellular receptor for the virus. J Virol. 1992;66:7270–7281. doi: 10.1128/jvi.66.12.7270-7281.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown K E, Anderson S M, Young N S. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science. 1993;262:114–117. doi: 10.1126/science.8211117. [DOI] [PubMed] [Google Scholar]
- 16.Christensen J, Alexandersen S, Bloch B, Aasted B, Uttenthal A. Production of mink enteritis parvovirus empty capsids by expression in a baculovirus vector system: a recombinant vaccine for mink enteritis parvovirus in mink. J Gen Virol. 1994;75:149–155. doi: 10.1099/0022-1317-75-1-149. [DOI] [PubMed] [Google Scholar]
- 17.Christensen J, Storgaard T, Bloch B, Alexandersen S, Aasted B. Expression of Aleutian mink disease parvovirus proteins in a baculovirus vector system. J Virol. 1993;67:229–238. doi: 10.1128/jvi.67.1.229-238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotmore S F, Tattersall P. A genome-linked copy of the NS-1 polypeptide is located on the outside of infectious parvovirus particles. J Virol. 1989;63:3902–3911. doi: 10.1128/jvi.63.9.3902-3911.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Verdugu V R, Selinka H C, Huber M, Kramer B, Kellermann J, Hofschneider P H, Kandolf R. Characterization of a 100-kilodalton binding protein for the six serotypes of coxsackie B viruses. J Virol. 1995;69:6751–6757. doi: 10.1128/jvi.69.11.6751-6757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dimmock N J. Initial stages in infection with animal viruses. J Gen Virol. 1982;59:1–22. doi: 10.1099/0022-1317-59-1-1. [DOI] [PubMed] [Google Scholar]
- 21.Duan X, Nauwynck H J, Favoreel H W, Pensaert M B. Identification of a putative receptor for porcine reproductive and respiratory syndrome virus on porcine alveolar macrophages. J Virol. 1998;72:4520–4523. doi: 10.1128/jvi.72.5.4520-4523.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dworak L J, Wolfinbarger J B, Bloom M E. Aleutian mink disease parvovirus infection of K562 cells is antibody-dependent and is mediated via an Fc(gamma)RII receptor. Arch Virol. 1997;142:363–373. doi: 10.1007/s007050050082. [DOI] [PubMed] [Google Scholar]
- 23.Evander M, Frazer I H, Payne E, Qi Y M, Hengst K, McMillan N A. Identification of the alpha6 integrin as a candidate receptor for papillomaviruses. J Virol. 1997;71:2449–2456. doi: 10.1128/jvi.71.3.2449-2456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Fox, J. M., and M. E. Bloom. Unpublished data.
- 24.Gorham J R, Henson J B, Crawford T B, Padgett G A. The epizootiology of Aleutian disease. In: Kimberlin R H, editor. Slow virus diseases of animals and man. Amsterdam, The Netherlands: North-Holland Publishing Co.; 1976. pp. 135–158. [PubMed] [Google Scholar]
- 25.Goto H. Feline panleukopenia in Japan. II. Hemagglutinability of the isolated virus. Nippon Juigaku Zasshi. 1975;37:239–245. doi: 10.1292/jvms1939.37.5_239. [DOI] [PubMed] [Google Scholar]
- 26.Kajigaya S, Fujii A, Field A M, Anderson S, Shimada T, Young N S. B19 parvovirus capsids produced in a baculovirus system are antigenically and immunologically similar to native virions. Proc Natl Acad Sci USA. 1991;88:4646–4650. doi: 10.1073/pnas.88.11.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenyon A J, Kenyon B J, Hahn E C. Protides of the Mustelidae: immunoresponse of mustelids to Aleutian mink disease virus. Am J Vet Res. 1978;39:1011–1015. [PubMed] [Google Scholar]
- 28.Krilov L R, Anderson L J, Marcoux L, Bonagura V R, Wedgwood J F. Antibody-mediated enhancement of respiratory syncytial virus infection in two monocyte/macrophage cell lines. J Infect Dis. 1989;160:777–782. doi: 10.1093/infdis/160.5.777. [DOI] [PubMed] [Google Scholar]
- 29.Linser P, Bruning H, Armentrout R W. Specific binding sites for a parvovirus, minute virus of mice, on cultured mouse cells. J Virol. 1977;24:211–221. doi: 10.1128/jvi.24.1.211-221.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig G V, Kondig J P, Smith J F. A putative receptor for Venezuelan equine encephalitis virus from mosquito cells. J Virol. 1996;70:5592–5599. doi: 10.1128/jvi.70.8.5592-5599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mady B J, Erbe D V, Kurane I, FAnger M W, Ennis F A. Antibody-dependent enhancement of dengue virus infection mediated by bispecific antibodies against surface molecules other than Fcgamma receptors. J Immunol. 1991;147:3139–3144. [PubMed] [Google Scholar]
- 32.Mbida A D, Pozzetto B, Gaudin O G, Grattard F, Le B J, Akono Y, Ros A. A 44,000 glycoprotein is involved in the attachment of echovirus-11 onto susceptible cells. Virology. 1992;189:350–353. doi: 10.1016/0042-6822(92)90714-z. [DOI] [PubMed] [Google Scholar]
- 33.McKenna, R., N. H. Olson, P. R. Chipman, T. S. Baker, T. F. Booth, J. Christensen, B. Aasted, J. M. Fox, M. E. Bloom, and M. Agbandje-McKenna. The three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 34.Mizukami H, Young N S, Brown K E. Adeno-associated virus type 2 binds to a 150-kilodalton cell membrane glycoprotein. Virology. 1996;217:124–130. doi: 10.1006/viro.1996.0099. [DOI] [PubMed] [Google Scholar]
- 35.Mochizuki M, Konishi S, Ogata M. Studies on feline panleukopenia. II. Antigenicities of the virus. Nippon Juigaku Zasshi. 1978;40:375–383. doi: 10.1292/jvms1939.40.375. [DOI] [PubMed] [Google Scholar]
- 36.Mori S, Wolfinbarger J B, Dowling N, Wei W, Bloom M E. Simultaneous identification of viral proteins and nucleic acids in cells infected with Aleutian mink disease parvovirus. Microb Pathog. 1991;9:243–253. doi: 10.1016/0882-4010(90)90013-g. [DOI] [PubMed] [Google Scholar]
- 37.Mori S, Wolfinbarger J B, Miyazawa M, Bloom M E. Replication of Aleutian mink disease parvovirus in lymphoid tissues of adult mink: involvement of follicular dendritic cells and macrophages. J Virol. 1991;65:952–956. doi: 10.1128/jvi.65.2.952-956.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norkin L C. Virus receptors: implications for pathogenesis and the design of antiviral agents. Clin Microbiol Rev. 1995;8:293–315. doi: 10.1128/cmr.8.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oleksiewicz M B, Costello F, Huhtanen M, Wolfinbarger J B, Alexandersen S, Bloom M E. Subcellular localization of Aleutian mink disease parvovirus proteins and DNA during permissive infection of Crandell feline kidney cells. J Virol. 1996;70:3242–3247. doi: 10.1128/jvi.70.5.3242-3247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker J S L, Parrish C R. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J Virol. 1998;71:9214–9222. doi: 10.1128/jvi.71.12.9214-9222.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porter D D, Larsen A E, Cook N A, Porter H G, Suffin S L. Isolation of Aleutian disease virus of mink in cell culture. Intervirology. 1977;8:129–144. doi: 10.1159/000148888. [DOI] [PubMed] [Google Scholar]
- 42.Salas-Benito J S, del Angel R M. Identification of two surface proteins from C6/C36 cells that bind dengue type 4 virus. J Virol. 1997;71:7246–7252. doi: 10.1128/jvi.71.10.7246-7252.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saliki J T, Mizak B, Flore H P, Gettig R R, Burand J P, Carmichael L E, Wood H A, Parrish C R. Canine parvovirus empty capsids produced by expression in a baculovirus vector: use in analysis of viral properties and immunization of dogs. J Gen Virol. 1992;73:369–374. doi: 10.1099/0022-1317-73-2-369. [DOI] [PubMed] [Google Scholar]
- 44.Summerford C, Samulski R J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truyen U, Parrish C R. Canine and feline host ranges of canine parvovirus and feline panleukopenia virus: distinct host cell tropisms of each virus in vitro and in vivo. J Virol. 1992;66:5399–5408. doi: 10.1128/jvi.66.9.5399-5408.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uwatoko K, Kano R, Sunairi M, Nakajima M, Yamaura K. Canine parvovirus binds to multiple cellular membrane proteins from both permissive and nonpermissive cell lines. Vet Microbiol. 1996;51:267–273. doi: 10.1016/0378-1135(96)00049-1. [DOI] [PubMed] [Google Scholar]
- 47.Weir D M. Carbohydrates as recognition molecules in infection and immunity. FEMS Microbiol Immunol. 1989;1:331–340. doi: 10.1111/j.1574-6968.1989.tb02418.x. [DOI] [PubMed] [Google Scholar]
- 48.White L J, Ball J M, Hardy M E, Tanaka T N, Kitamoto N, Estes M K. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J Virol. 1996;70:6589–6597. doi: 10.1128/jvi.70.10.6589-6597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu W-H, Bloom M E, Berry B D, McGinley M J, Platt K B. Expression of Aleutian mink disease parvovirus capsid proteins in a baculovirus expression system for potential diagnostic use. J Vet Diagn Invest. 1994;6:23–29. doi: 10.1177/104063879400600105. [DOI] [PubMed] [Google Scholar]