Abstract
Two intrastrain variants of herpes simplex virus type 1 (HSV-1) were isolated from a newborn with fatal disseminated infection. A small-plaque-producing variant (SP7) was the predominant virus (>99%) in the brain, and a large-plaque-producing variant (LP5) was the predominant virus (>99%) in the lung and gastrointestinal tract. EcoRI and BamHI restriction fragment patterns indicated that SP7 and LP5 are related strains. The large-plaque variants produced plaques similar in size to those produced by HSV-1 KOS. Unlike LP5 or KOS, SP7 was highly cell associated and processing of glycoprotein C and glycoprotein D was limited to precursor forms in infected Vero cells. The large-plaque phenotype from KOS could be transferred into SP7 by cotransfection of plasmids containing the EK or JK EcoRI fragment or a 3-kb plasmid with the UL34.5 gene of HSV-1 KOS together with SP7 DNA. PCR analysis using primers from within the ICP34.5 gene indicated differences for SP7, LP5, and KOS. Sequencing data indicated two sets of deletions in the UL34.5 gene that distinguish SP7 from LP5. Both SP7 and LP5 variants were neurovirulent (lethal following intracranial inoculation of young BALB/c mice); however, the LP5 variant was much less able to cause lethal neuroinvasive disease (footpad inoculation) whereas KOS caused no disease. Passage of SP7 selected for viruses (SLP-5 and SLP-10) which were attenuated for lethal neuroinvasive disease, were not cell-associated, and differed in the UL34.5 gene. UL34.5 from SLP-5 or SLP-10 resembled that of KOS. These findings support a role for UL34.5 in promoting virus egress and for neuroinvasive disease.
Herpes simplex virus (HSV) is a neurotropic virus responsible for a variety of diseases in humans, including localized mucocutaneous infection, encephalitis, and disseminated disease (16, 29, 39, 43). Human neonates are particularly susceptible to HSV infection (8, 43). Neonatal HSV infections may remain localized to the site of inoculation (e.g., skin, eye, or mouth), extend to the central nervous system (CNS), or disseminate to multiple organ sites (8). Infection of the CNS causes significant morbidity and mortality. Factors influencing the extent of HSV disease in the neonate include maternal and host immunity, viral load, site of inoculation, and virulence properties of the virus strain (41, 43).
Differences in the virulence of HSV have been observed between types (intertypic) (16, 32), between strains within a type (interstrain) (16, 20), and within a single strain (intrastrain) (16, 29). Comparison of the properties of intrastrain variants can help identify the relevant genetic differences.
In studies using animal models, progress has been made toward identifying the viral genes which are required for neurovirulence (ability to replicate and cause clinical disease upon direct injection into the mouse brain) (reviewed in reference 39). Many of these genes encode activities which are important for replication in neuronal cells, e.g., thymidine kinase, ribonucleotide reductase, the US3 protein kinase (41), and ICP34.5 (12). Much less is known about viral genes which are important for establishing a primary encephalitis following inoculation of peripheral body sites (e.g., mouse footpad) (lethal neuroinvasiveness). Factors influencing lethal neuroinvasiveness include ability to replicate in neuronal and other tissues, ability to escape immune detection, ability to infect the ennervating neuron and travel to the brain, and ability to initiate tissue damage prior to immune control. Izumi and Stevens compared the lethal neuroinvasive ANG-PATH and the closely related, attenuated ANG strain and mapped the attenuation to a mutation in glycoprotein D (gD) (22). Similarly, Yuhasz and Stevens (44) mapped an attenuating mutation to gB for HSV type 1 (HSV-1) KOS (42), indicating that there is more than one genetic approach to attenuation of neuroinvasiveness. Unlike ANG-PATH, the attenuated ANG and KOS strains induced more active immune responses, suggesting this as the reason for the lack of lethal neuroinvasiveness. Both ANG (29) and KOS exhibit lethal neuroinvasiveness in cyclophosphamide-treated mice (2, 29).
In this study, we isolated and characterized the properties of two intrastrain variants obtained from a neonate with disseminated, fatal HSV-1 infection. The presence of multiple HSV-1 strain variants in human disseminated HSV-1 infections (1, 20) but not neonatal infection has been observed. One isolate predominated in samples from neuronal tissues (SP7), whereas the other isolate was predominant in nonneuronal tissues. The neonatal intrastrain variants also differed in the potential for lethal neuroinvasive disease and in tissue culture behavior. The linkage of these characteristics suggests a genetic relationship between the tissue culture phenotype and a viral gene(s) which influences the disease potential of the virus.
Previous studies from this laboratory identified a tissue culture phenotype which distinguished clinical isolates from the KOS laboratory strain (15). The clinical isolates produce small plaques on Vero cells and are cell associated, and viral glycoprotein processing appears to be restricted to the endoplasmic reticulum (small-plaque phenotype). We report herein that the strain isolated from the brain of the neonate (SP7) possesses a small-plaque phenotype as well as an enhanced ability to induce lethal neuroinvasive disease. The small-plaque phenotype mapped to sequences containing the ICP34.5 gene. Sequence differences in this gene distinguished the SP7, LP5, and KOS viruses. As for ANG and ANG-PATH (22, 29, 39, 44), comparison of SP7 and LP5 may give insight into the mechanisms of HSV virulence. This is also the first report of intrastrain variants of HSV in neonates or newborns.
(Portions of this study were presented at the 22nd and 23rd International Herpesvirus Workshops, 2–8 August 1997, La Jolla, Calif., and 1–7 August 1998, York, United Kingdom, respectively.)
MATERIALS AND METHODS
Viruses and tissue culture.
Virus was obtained postmortem from the brain, lung, kidney, gastrointestinal (GI) tract, and cerebrospinal fluid (CSF) of a 10-day-old infant with disseminated HSV-1 disease. The baby had been in good health until the previous day. The baby presented to the emergency room in shock and died 6 h later. No maternal problems were noted. All HSV isolates were typed by monoclonal antibody, using an indirect fluorescent antibody method. HSV-1 strain KOS 321 (provided by Tom Holland, Wayne State University School of Medicine), the source of the EcoRI genomic library used in this study (18), was used for comparisons. Viral isolates were grown in Vero (African green monkey kidney) cells in growth medium (medium 199 supplemented with 5% fetal calf serum and 2.25 mM NaHCO3) at 37°C in an atmosphere of 5% CO2.
Plaque size and morphology were examined for each tissue isolate under normal liquid medium growth conditions and with 0.5% methylcellulose overlay. Individual plaque stocks were prepared from each tissue isolate by twice plaque purifying representative small plaques (SP7) and/or large plaques (LP5) from tissue culture plates containing fewer than 10 plaques.
Efficiency of viral egress (inverse of cell association) was determined at 20 h postinfection (p.i.). A half volume of spent medium was removed, and the cells were scraped into the remaining media. Both aliquots were twice frozen and thawed and then evaluated by plaque assay. The amount of infectious virus in the cellular fraction was corrected for cell-free virus, and a ratio was calculated.
Plaque-purified SP7 virus was serially passaged in Vero cells at low multiplicity of infection (MOI) (<0.01) on two separate occasions. A small-to-large-plaque convertant was observed after five (SLP-5) or nine (SLP-9) passages and then passed an additional time (SLP-10). Care was taken to ensure that the SP7 virus was not contaminated with other virus strains during the passage procedure.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Whole-cell extracts of HSV-1-infected Vero cells (MOI of approximately 1) were prepared in Tris (pH 7.4)–1% Triton X-100–1% sodium deoxycholate–0.1% SDS–0.01% aprotinin–10 U of bovine pancreatic DNase I (Sigma), electroblotted onto polyvinylidene difluoride membranes (Schleicher & Schuell, Keene, N.H.), and analyzed with either rabbit polyclonal anti-gC or rabbit polyclonal anti-gD antibody (kindly provided by G. Cohen and R. Eisenberg) by Western blotting. The glycoproteins were visualized by enhanced chemiluminescence (Amersham, Arlington Heights, Ill.). Images were scanned into the computer and prepared for publication by using Corel Draw.
Restriction enzyme analysis.
DNA was isolated from nucleocapsids as described by Robbins et al. (33). Restriction digests were performed with the restriction enzymes BamHI and EcoRI as recommended by the manufacturer (Promega, Madison, Wis.). DNA digests were electrophoresed in 0.8% agarose gels containing ethidium bromide.
Marker transfer studies.
An EcoRI library of the HSV-1 KOS genome cloned into PBR325 (18) (graciously provided by Joe Glorioso) was used in the marker transfer studies. Minipreparations of purified PBR325-EcoRI plasmid DNA were prepared by standard techniques (18, 35). Viral genomic DNA was obtained as described by Robbins et al. (33) from purified nucleocapsids from roller bottle cultures of Vero cells infected at an MOI of greater than 1.
Genomic DNA and plasmid DNA containing the appropriate EcoRI fragment (fivefold molar excess) were cotransfected into subconfluent monolayers of Vero cells by lipofection. Lipofection was performed as instructed by the manufacturer of Lipofectamine (BRL-Gibco) under conditions which generate 50 to 100 plaques per μg of viral DNA. Successful recombination was indicated by transfer of the large-plaque characteristic (marker) and subsequently the extent of processing of gC. Extracellular virus was harvested, tested for tissue culture properties, and plaque purified at least three times prior to analysis by PCR.
PCR.
HSV-1 DNA was isolated (Wizard genomic purification kit; Promega) from Vero cells grown in six-well tissue culture dishes and infected with the appropriate virus strain. Primers (5′-CTG-CAC-GCA-CAT-GCT-TGC-CT-3′ and 5′-CTC-GGG-TGT-AAC-GTT-AGA-CC-3′; Research Genetics, Inc.) were chosen based on their unique sequences and to bracket the ICP34.5 gene (8). Due to the high GC content of the sequence and the failure of several other approaches, PCR was performed with a MasterAmp PCR optimization kit (Epicentre) and Tfl polymerase (Epicentre). The program for amplification consisted of 30 cycles of 5 min at 96°C, 1 min at 60°C, 1 min at 68°C, and 1 min at 95°C, with an additional incubation of 10 min at 68°C. Resultant DNA was analyzed by electrophoresis on 1.5% ethidium bromide-containing agarose gels.
DNA sequencing and analysis.
DNA sequences were analyzed by using programs accessible over the Internet as described in reference 37. They included MAP multiple alignment (19) and 6 Frame translation conversion (37) programs.
Animal studies.
Four-week-old male BALB/c mice (Charles Rivers Breeding Laboratories, Wilmington, Mass.), were used, and all animal studies were performed with Institutional Animal Care and Use Committee approval. Intracerebral inoculation was performed by introducing virus (in 15 μl) directly into the right cerebral hemisphere. Peripheral inoculation was accomplished by subcutaneous injection into the ventral surface of the right hind foot with a virus inoculum containing approximately 104 to 107 PFU in a volume of 40 μl. All animals were examined daily for the progression of symptoms from paralysis to encephalitis and death.
In vivo viral growth curves were prepared as described by Izumi and Stevens (22). Groups of three animals were inoculated in the right hind foot with the virus. The groups were euthanized on day 0, 1, 3, 5, 7, or 9 postchallenge, and the foot, sciatic nerve, dorsal root ganglia (DRGs), spinal cord, and brain were removed. Tissues were homogenized and clarified by centrifugation. The supernatants were titrated for virus by plaque assay. Titers are representative of the amount of virus in the total tissue sample.
RESULTS
Characterization of viral isolates obtained from different tissues.
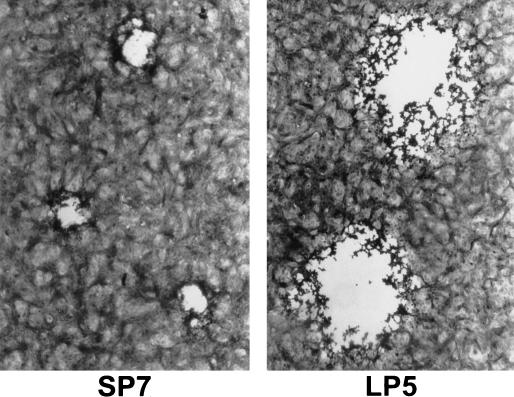
HSV-1 isolates obtained postmortem from the brain, CSF, lung, kidney and GI tract contained two plaque size variants (Fig. 1). The plaques for the small-plaque variant measured 0.51 ± 0.09 mm (72 h p.i.), and the plaques for the large-plaque variant measured 1.03 ± 0.14 mm in Vero cells. All isolates formed plaques with rounded cells, and no syncytial plaques were observed. For comparison, the laboratory strain, KOS, had a plaque size of 1.0 ± 0.16 mm and the cytopathic effect appeared similar to that for the large-plaque strain. The plaque morphology for SP7 and LP5 on HEp-2 cells was similar to that for Vero cells (data not shown). Unlike the large-plaque variants, the small-plaque variants did not spread through the monolayer even in the absence of methylcellulose.
FIG. 1.
Plaque morphology of the small-plaque (SP7) and large-plaque (LP5) variants grown in Vero cells. The cells were fixed and stained with crystal violet in ethanol.
The small-plaque variant exhibited a preferential distribution to neuronal tissue (Table 1). The small-plaque variant was the predominant virus (>99%) in the brain isolate. The lung and GI tract isolates yielded primarily a large-plaque variant (>99%). The CSF and kidney isolates contained a mixture of both the small- and large-plaque variants (Table 1). The kidney sample included adrenal tissue which shares characteristics with neuronal tissue (31).
TABLE 1.
Tissue distribution of plaque variants
| Tissue source of isolate | % of isolatea
|
|
|---|---|---|
| Large-plaque virus | Small-plaque virus | |
| Brain | 0.5 | 99.5 |
| GI tract | 99.5 | 0.5 |
| Lung | 99 | 1 |
| CSF | 47 | 53 |
| Kidney | 22 | 78 |
Plaques were examined 72 h p.i. Sample size was 200 plaques.
Representative large- and small-plaque variants from each tissue were picked and grown for stocks. The SP7 small-plaque virus was developed from a triple-plaque-purified brain isolate, and the LP5 large-plaque virus was developed from a triple-plaque-purified lung isolate.
Viral growth and egress.
Differences in viral growth and egress from tissue culture cells were compared for the plaque-purified small- and large-plaque variants. Total virus production (72 h in Vero cells) by the large-plaque isolates from lung (2.2 × 108 PFU/ml) and CSF (2.4 × 108 PFU/ml) was greater than for the small-plaque variants from the brain (3.3 × 107 PFU/ml) and CSF (3.6 × 107 PFU/ml). The ratios of extracellular to cell-associated virus, determined at 20 h p.i., were 0.9 for the LP5 variant, 0.46 for KOS, and 0.06 for the SP7 variant. These results indicate that the SP7 variant was competent for replication but more cell associated than the LP5 variant or KOS.
Glycoprotein analysis.
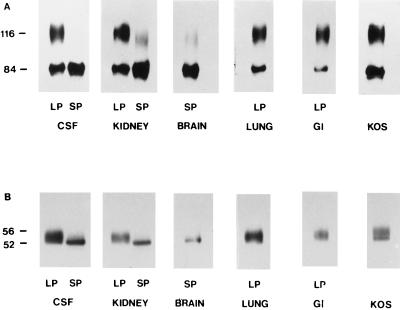
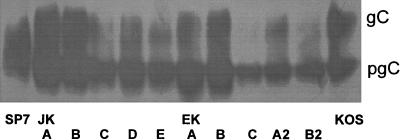
gC and gD were examined by SDS-PAGE and immunoblotting to determine if the SP7 virus expresses characteristics consistent with defects in virion maturation and egress observed in other small-plaque-producing clinical isolates (12, 13, 15). Whole-cell detergent extracts were prepared from Vero cells infected with plaque-picked small- and large-plaque virus obtained from each of the neonatal tissues. An extract prepared in this manner would be expected to contain the precursor and mature glycoforms of viral glycoproteins expressed on intracellular and plasma membranes as well as on virions. Figure 2A shows that the small-plaque variant present in brain, CSF, and kidney yielded almost exclusively a glycoform of gC with an apparent molecular mass of 84,000 Da. The large-plaque variant from the lung, kidney, CSF, and GI tract yielded two species of gC with apparent molecular masses of 84,000 and 116,000 Da. The 84,000- and 116,000-dalton species correspond to the precursor and mature forms of gC, respectively (28). The gC profile for KOS was similar to that for the LP5 variant.
FIG. 2.
HSV-1 glycoprotein differences associated with large-plaque and small-plaque variants. Whole-cell detergent extracts from Vero cells infected with large- or small-plaque variants from the CSF, kidney, brain, lung, or GI tract were examined by SDS-PAGE and Western blot analysis using either polyclonal anti-gC (A) or anti-gD (B). The laboratory strain KOS was included as a control, and molecular weights of the major glycoforms (kilodaltons) are indicated.
gD was similarly examined (Fig. 2B). The small-plaque variant yielded a single glycopeptide for gD of 52,000 Da. The large-plaque variant and KOS, however, yielded at least two gD glycopeptides of 52,000 to 55,000 Da, which agree with the expected sizes for precursor gD and mature gD, respectively (28).
Little or no mature gC was present in extracts of cells infected with the SP7 variant even 34 h postinfection (data not shown). This suggests that viral glycoprotein processing is blocked at the precursor level rather than slowed.
Observation of limited glycoprotein maturation for two viral glycoproteins, gC and gD, and the limited viral release of SP7 is consistent with a block in viral maturation and egress rather than mutation in the structural genes for the glycoproteins. This correlation between small-plaque production and restriction in glycoprotein processing is similar to our previous findings for two other clinical isolates (15).
Restriction enzyme analysis.
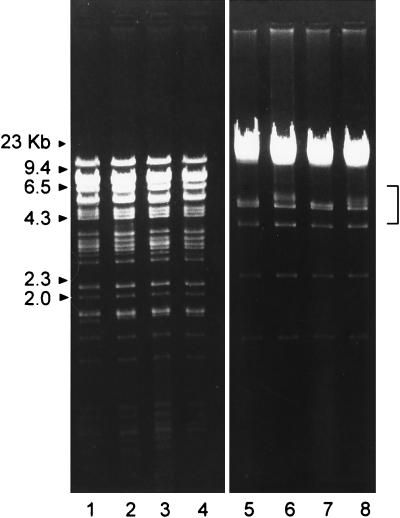
Restriction fragment length polymorphism (RFLP) analysis of small-plaque and large-plaque variants was performed to determine whether the variants were derived from a single strain or represented distinct infecting strains. Genomic DNA was digested with BamHI and EcoRI and examined by agarose gel electrophoresis (Fig. 3). The restriction enzyme cleavage patterns following reaction with either the EcoRI or BamHI were very similar for the two viruses. In the EcoRI digestion pattern of the small-plaque (SP7) variant (brain and CSF), an additional fragment with a slower electrophoretic mobility was present that was absent in the large-plaque variants (kidney, lung, and GI tract). The mobility of this fragment corresponds to the K or L EcoRI fragment of HSV-1 KOS (18, 25). The BamHI cleavage patterns for LP5 and SP7 were very similar except for small differences in the mobility of fragments of approximately 5 kb and approximately 1 to 2 kb. These DNA fragments appear to correspond to the K and the X, Y, and Z fragments, respectively (25, 26). The K fragment contains sequences from the terminal repeats at the ends of the L and S regions of the HSV genome and are associated with intrastrain variation due to insertion or deletion of discrete DNA sequences (26). These results suggest that the small- and large-plaque isolates are closely related viruses and appear to be intrastrain variants derived from the same parental strain.
FIG. 3.
RFLP of large- and small-plaque variants isolated from different tissues. DNA was digested with BamHI (lanes 1 to 4) or EcoRI (lanes 5 to 8) and electrophoresed in 0.8% agarose gels containing ethidium bromide. Lanes 1 and 5, large-plaque variant from the kidney; lanes 2 and 6, small-plaque variant from the kidney; lanes 3 and 7, large-plaque variant from the GI tract; lanes 4 and 8, small-plaque variant from the brain. Bracket indicates fragments exhibiting RFLP.
Marker transfer studies.
Marker transfer was attempted by cotransfection of genomic SP7 DNA and plasmid DNA containing EcoRI sequences from HSV-1 KOS into Vero cells. KOS was used for these studies because it expresses the large-plaque phenotype, and in an earlier study (15) we showed that coinfection of KOS with either of two small-plaque clinical strains could complement the small-plaque phenotype and promote efficient glycoprotein processing. In addition, the KOS genome is well characterized, and cloned subgenomic sequences are available. Plaque morphology was used as an initial screening procedure to identify relevant DNA sequences from KOS. Cell association and the extent of glycoprotein processing of gC were also analyzed to confirm whether a KOS sequence converted SP7 from the small-plaque phenotype to the large-plaque phenotype. Unlike conversion of a large-plaque- to a small-plaque-producing strain (which could represent a virus weakened in many different ways), conversion of the small-plaque producer to a large-plaque producer represents enhancement of growth in tissue culture, a significant event which should require a unique genetic change. Plaque size has been used as a selective marker in other marker transfer studies, e.g., association of the UL11 gene with its phenotype (3).
Plaques generally developed by day 3 or 4 after cotransfection. Virus from small plaques did not spread through the monolayer. In contrast, virus from large plaques spread readily and extensively through the monolayer. This facilitated detection of a successful marker transfer.
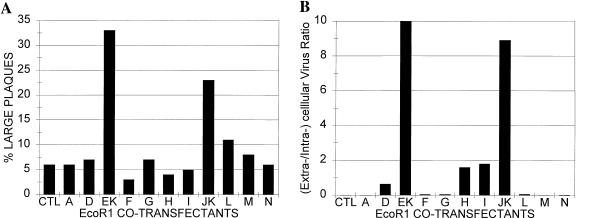
As shown in Fig. 4A, the highest percentage of large-plaque production was obtained following cotransfection of SP7 genomic DNA with plasmid DNA containing EcoRI fragment EK (SP7EK) or JK (SP7JK) but not other sequences from HSV-1 KOS. In several subsequent experiments, cotransfection of SP7 DNA with plasmids containing the EK and JK fragments of HSV-1 KOS would consistently yield large plaques and at relatively high frequency. Following each of the cotransfections, crude virus stocks were obtained by freeze-thaw lysis of the infected cells in the spent culture medium.
FIG. 4.
Evaluation of initial isolates from cotransfection of genomic DNA from HSV-1 SP7 and plasmid DNA containing EcoRI fragments of HSV-1 KOS. (A) Plaque size. Vero cells were cotransfected with HSV-1 and plasmid DNA by using lipofection. The cells were maintained in growth medium for 4 days, and large and small plaques were counted. (B) Extracellular-to-intracellular virus ratios. Virus obtained from cell cultures described in the legend to panel A was applied to Vero cells and allowed to infect cells for 20 h. Virus obtained from the medium (0.5 volumes) and virus from the cells and the remaining medium were quantitated by plaque assay.
The crude virus stocks obtained from the cotransfections were evaluated for other large-plaque-associated characteristics of KOS such as efficient viral release and glycoprotein processing (Fig. 4B). Consistent with the plaque size results, the ratios of extracellular to intracellular virus were higher for SP7 virus containing an EK or JK sequence than for the parental SP7 virus or other cotransfectants. In addition, the mature, fully processed form of gC was consistently observed in Vero cells infected with the SP7EK or SP7JK virus. The EK and JK fragments, but not other sequences, appeared to carry all three parameters of the large-plaque phenotype into SP7.
The correlation between plaque size and extent of glycoprotein processing was also observed for the plaque-purified isolates (Fig. 5). For example, isolate SP7EK-C or SP7JK-C produced smaller plaques and also showed little processing of gC, in contrast to SP7EK-A and SP7JK-A, which produced large plaques and showed levels of gC processing similar to those for KOS. Several of the SP7JK-A isolates were triple plaque purified and analyzed by PCR (described below).
FIG. 5.
Comparison of glycoforms of gC from cotransfectants of HSV-1 SP7 and HSV-1 KOS EcoRI JK and EK fragments. Virus obtained from the cotransfection experiment described in the legend to Fig. 4 was triple plaque purified. Infected Vero cell detergent extracts were prepared and analyzed by SDS-PAGE and Western blotting. Images were scanned into the computer.
The EK and JK regions of the HSV-1 genome contain sequences from the UL inverted repeat regions and had approximately 11-kb sequences in common. Subsequent cotransfections with plasmid pL/ST-Nx from HSV-1 KOS (provided by Lily Yeh Lee and Priscilla Schaeffer) (24) and SP7 DNA promoted the generation of 15% (versus 4.5% for controls) large plaques. A similar but greater extent of conversion was obtained upon pL/ST-Nx cotransfection with DNA from another small-plaque clinical isolate (HSV-1 490) (data not shown). Plasmid pL/ST-Nx contains 3.5 kb from the terminal portion of the UL of the inverted repeat region and contains the UL34.5 gene, ORF-P, ORF-O, and portions of latency-associated transcript (LAT) and L/ST sequences. These studies indicate that an activity encoded within the EK and JK EcoRI sequences and the pL/ST-Nx sequences transfers the large-plaque tissue culture phenotype from KOS into the SP7 virus.
PCR amplification of ICP34.5 coding sequences.
PCR was attempted to take a closer look at the gene for ICP34.5 since it is a major portion of plasmid pL/ST-Nx and the protein has been associated with neurovirulence (11, 12) and with virion maturation and egress (6). The UL34.5 gene is rich in GC and required special procedures for PCR amplification. DNA primers were chosen to have unique sequences and to bracket the ICP34.5 coding sequences (13, 24).
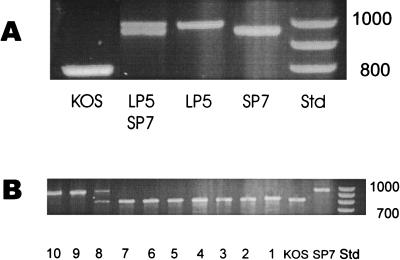
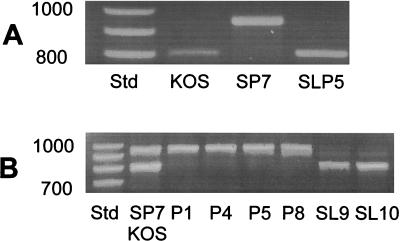
Unexpected large differences in size of the PCR products were obtained for the different strains (Fig. 6). The SP7 product electrophoresed as an approximately 900-bp fragment, whereas the LP5 product was slightly larger. Confirmation of the larger size of the SP5 product was obtained by mixing the SP7 and LP5 products prior to electrophoresis. The KOS product electrophoresed as a much smaller fragment of approximately 800 bp.
FIG. 6.
PCR amplification products of UL34.5 sequences. DNA purified from Vero cells infected with the indicated virus was amplified by PCR using unique primers from the termini of the UL34.5 gene and evaluated by electrophoresis on 1.5% agarose gels. (A) Individual PCR products from HSV-1 KOS, LP5, and SP7 DNA and a mixture of the PCR product of LP5 and SP7 to illustrate differences in mobility. (B) Lanes 1 to 8, PCR products from different plaque-picked isolates from suspected recombinants of SP7/JK; lane 9, SP7/EK plaque isolate; lane 10, is another SP7/JK isolate. Std, size standards, positions of which are indicated in base pairs.
PCR analysis of individual triple-plaque-purified SP7JK and SP7EK isolates was also performed (Fig. 6B). Analysis of plaque isolates 1 to 7 showed the KOS-like PCR product, whereas isolates 9 and 10 showed the parental SP7-like PCR product. Isolate 8 was most likely a mixture of SP7 and a recombinant. The plaque isolates which contained the KOS-like product yielded gC glycoforms similar to KOS, whereas the plaque isolates which contained the SP7-like PCR product yielded only the precursor gC similar to SP7 (data not shown). These results confirm that the phenotypic change in SP7JK-A was due to a recombination event between SP7 and KOS sequences.
Sequence analysis of the PCR-amplified UL34.5 genes.
The products obtained by PCR amplification of the UL34.5 sequences were submitted for DNA sequence analysis. The forward and reverse PCR primers and an intermediate primer were used in the automated DNA analysis. DNA sequences were assembled, aligned, and converted to peptide sequences by using the MAP multiple alignment (21) and 6 Frame translation (37) programs.
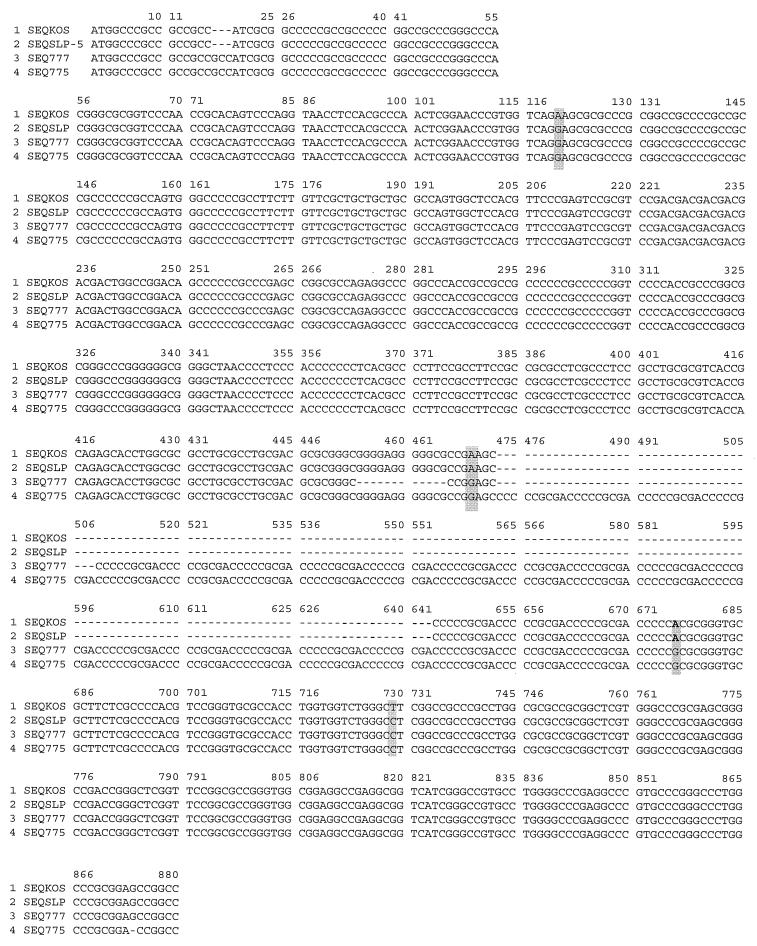
Analysis of the DNA sequences obtained by PCR indicated extensive homology within the UL34.5 gene for the SP7, LP5, and KOS strains (Fig. 7). The major sequence difference between the three strains was in the number of repeats of CCCGCGACC, encoding proline-alanine-threonine (PAT). The amino acid differences between LP5, SP7, and KOS are summarized in Table 2. The LP5 strain had 22 repeats, SP7 had 18, while KOS had only 3 repeats of the PAT unit in the predicted peptide sequence (Fig. 8). The number of PAT repeats in LP5 and SP7 is larger than has been reported for other HSV-1 strains (13). The only other difference between LP5 and SP7 is a deletion of a sequence preceding the nine nucleotide repeats, causing the loss of glycine-glutamic acid-glycine-alanine (GEGA). There were other differences between SP7, LP5, and KOS, including a deletion of arginine near the N terminus, two conservative mutations, and two nonconservative mutations.
FIG. 7.
Comparisons of UL34.5 DNA sequences. DNA obtained by PCR amplification of the UL34.5 gene, as described for Fig. 6, were submitted for sequencing. The sequences were aligned by using the MAP alignment program. Differences as well as deletions in the sequences are indicated.
TABLE 2.
Genetic differences in ICP34.5 among HSV strains
| Strain | No. of PAT repeats | Other differences in ICP34.5 from LP5a |
|---|---|---|
| LP5 | 22 | |
| SP7 | 18 | Deletion of GEGA (153–156) |
| KOS 70 or 321 (KOS 63) | 3 | Deletion of R (6) |
| K instead of E (158) | ||
| T instead of A (227) | ||
| SLP5 | 3 | Deletion of R (6) |
| K instead of E (158) | ||
| T instead of A (227) | ||
| SLP10 | 3 | Deletion of RH (6, 8) |
| K instead of E (158) | ||
| T instead of A (227) |
Amino acids are designated in single-letter code. Numbers in parentheses refer to amino acid positions in Fig. 8.
FIG. 8.
Comparisons of the predicted peptide sequences of UL34.5 from KOS, SP7, and LP5. Dashes denote lack of corresponding sequence; shading denotes differences in sequence; hatched boxes enclose HSV-1 conserved sequence with similarity to the murine MyD116 myeloid differentiation primary response protein. KOS 70 and KOS 321 (marked with asterisks) have the same sequence.
Comparison of neuroinvasive disease production in the mouse model.
Differences in virulence between SP7 and LP5 variants were examined by using the mouse model. This model allows evaluation of encephalitis and lethality following intracranial inoculation (neurovirulence) and of ascending myelitis, encephalitis, and lethality following peripheral footpad inoculation (neuroinvasiveness) (16). The results are summarized in Table 3. Intracranial inoculation of LP5 with 100 PFU killed five of six and six of eight mice in two separate experiments, whereas intracranial inoculation of 100 PFU of SP7 killed three of six and three of eight mice. These results show that both LP5 and SP7 viruses can replicate and kill upon direct injection into the brain. In contrast, SP7 was significantly more virulent upon footpad inoculation than LP5 (P < 0.005). Similar to SP7, the related small-plaque virus obtained from the kidney of the baby was capable of causing lethal neuroinvasive disease, whereas like LP5, the large-plaque virus from the kidney could not. These results indicate that the SP7 virus is more lethal for neuroinvasive disease upon peripheral inoculation than LP5 despite the similar abilities of the two viruses to kill upon direct intracerebral inoculation. KOS showed no killing upon peripheral inoculation at 92 × 105 PFU.
TABLE 3.
Comparison of virulence properties
| Expt | Intracerebral
|
Footpada
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SP7
|
LP5
|
SP7
|
LP5
|
SLP5
|
KOS
|
|||||||
| PFU | FKb | PFU | FK | PFU | FK | PFU | FK | PFU | FK | PFU | FK | |
| 1 | 1 | 0/6 | 1 | 3/6 | ||||||||
| 100 | 3/6 | 100 | 5/6 | |||||||||
| 1,000 | 5/6 | |||||||||||
| 10,000 | 6/6 | |||||||||||
| 2 | 100 | 3/8 | 100 | 6/8 | ||||||||
| 3 | 7.2 × 105 | 3/5* | ||||||||||
| 7.2 × 106 | 5/5* | 1.6 × 106 | 0/5# | |||||||||
| 1.6 × 107 | 2/5# | 9.2 × 106 | 0/5 | |||||||||
| 4 | 1.5 × 106 | 7/8* | 4.5 × 106 | 1/8# | ||||||||
| 5 | 2.1 × 107 | 0/6 | ||||||||||
Highly significant difference between values noted by * and # (P = 0.005) per logistic regression analysis.
FK, fraction killed.
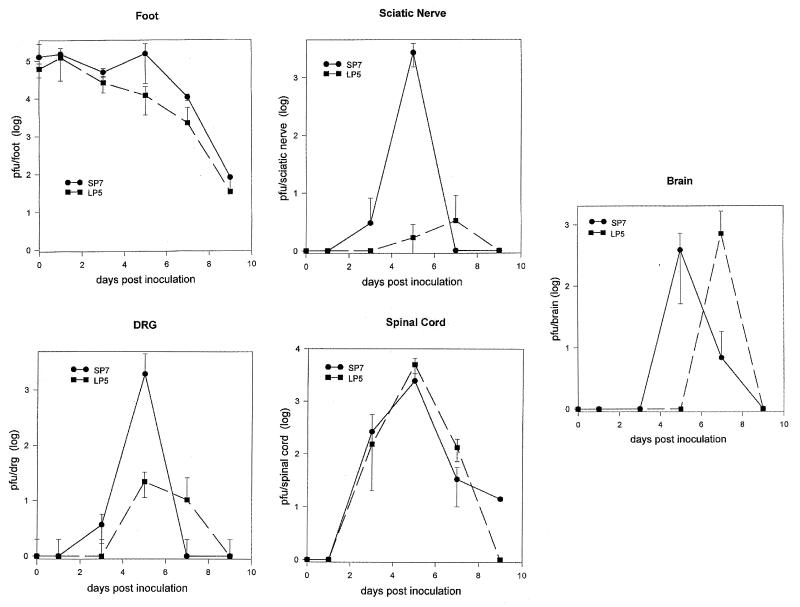
In vivo growth curves of the SP7 and LP5 variants were performed to analyze the progression of virus through the CNS following peripheral inoculation (Fig. 9). BALB/c mice were infected in the right rear footpad with either the LP5 or SP7 variant, and viral titers in the foot, sciatic nerve, sacral DRG, spinal cord, and brain were determined for each day. Viral titers for the SP7 and LP5 variants differed in the foot by only two- to threefold except on day 5, when a small peak in the SP7 titer was observed. This difference in virus titer on day 5 was also observed in the sciatic nerve and the DRG. The virus titer in the DRG for one mouse on day 5 was experimentally low, diminishing the difference between the other two mice infected with SP7 and those infected with LP5. Stanberry et al. attribute such a peak in virus titer to anteriograde transport of virus from the DRG (38). The SP7 variant appeared to reach the DRG and the sciatic nerve sooner and yield 100- and 1,000-fold-greater titers than the LP5. The two viruses were detected in the spinal cord at the same time, and equivalent titers were obtained. Interestingly, the SP7 variant was detected in the brain 2 days prior to the LP5 virus, yet the two viruses attained similar titers in the brain. The data for SP7 on day 9 are from the one surviving mouse. Consistent with other experiments, only SP7 caused lethal disease.
FIG. 9.
Neuroinvasive progression of SP7 and LP5 variants in the mouse. BALB/c mice were inoculated in the right rear footpad with 5 × 105 PFU of either the SP7 or LP5 variant. Mice were euthanized (three mice per time point) on 0, 1, 3, 5, 7, and 9 days p.i. The foot, sciatic nerve, DRG, spinal cord, and brain were removed, virus was extracted into 1 ml of medium, and viral titers were determined. Virus titers are representative of the amount of virus in the total tissue sample.
Passage-related changes in SP7.
SP7 was serially passed in Vero cells at low MOI to determine whether a large-plaque-producing virus that was also attenuated for neuroinvasive disease would be selected. Infections were performed at low MOI to facilitate selection of a virus that can be released and will spread more efficiently through the cell culture than SP7.
Conversion of SP7 to a large-plaque-producing virus occurred after five (SLP-5) passages in the first experiment and nine (SLP-9 and SLP-10 [an additional passage of SLP-9]) passages during the second experiment. The change was noted within one passage, suggesting that the large-plaque substrain has the dominant phenotype. Change in tissue culture behavior with passage of HSV-1 has been noted for other small-plaque-producing clinical strains (17, 27).
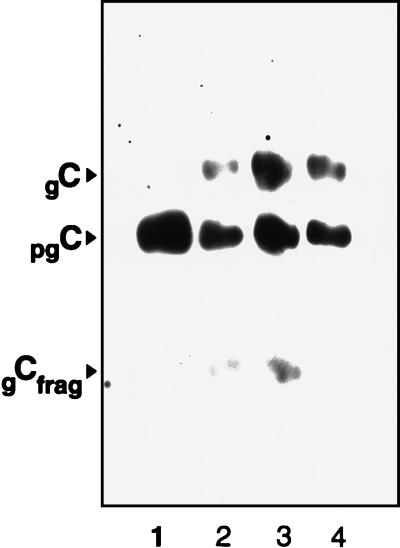
Analysis of SLP-5 showed that the EcoRI cleavage pattern of SLP-5 DNA was very similar but could be distinguished from that of SP7 (data not shown). Western blot analysis of gC produced upon SLP-5 infection of Vero cells showed both precursor and mature forms, resembling the glycoform profiles of LP5 and KOS, in contrast to only the precursor gC from the parental SP7 (Fig. 10).
FIG. 10.
HSV-1 gC profiles from SP7, SLP7, LP5, and KOS. Whole-cell detergent extracts from infected Vero cells were examined by SDS-PAGE and Western blot analysis using polyclonal anti-gC. Lanes: 1, SP7; 2, SLP5; 3, LP5; 4, KOS.
Unlike the SP7 parent, the SLP-5 virus was attenuated for lethal neuroinvasive disease upon peripheral inoculation (Table 3). The SLP-5 virus caused no symptoms and no deaths following footpad inoculation with doses as high as 3.2 × 107 PFU. This is in contrast to a dose of 1.5 × 106 PFU of SP7 virus, which killed six of eight mice. The virulence of the SP7 virus resembles that reported for other CNS clinical isolates (3), and the lack of virulence of SLP-5 resembles that reported for other highly passaged laboratory strains, such as KOS 321. These results suggest that tissue culture passage of SP7 in Vero cells selects for a spontaneous mutant, SLP, which has the same tissue culture and virulence characteristics as the highly passaged KOS used in these studies. In addition, the change in tissue culture characteristics correlates with attenuation of the virus.
Surprisingly, PCR amplification of the UL34.5 gene from SLP-5 produced a product with the same agarose gel electrophoretic mobility as KOS (Fig. 11). Analysis of the product indicated a sequence that resembled KOS, with only three repeats of the PAT-encoding nucleotide sequence (Fig. 7 and 8 and Table 2). Other changes can also be noted. The similarity to KOS, and a fear of contamination, prompted the second passage experiment of SP7 to produce the SLP-9 and SLP-10 viruses. The PCR product of SLP-10 had the same mobility as SLP-5 and KOS and also had only three repeats of PAT. The ICP34.5 sequences for both SLP-5 and SLP-10 resemble that of KOS more than that of the SP7 parent. The coincidental passage related conversion of SP7 to a KOS-like virus with regard to plaque size, efficiency of virus release, sequence of the UL34.5 gene, and attenuation of lethal neuroinvasive disease supports the interrelationship of these parameters.
FIG. 11.
Electrophoretic mobilities of UL34.5 PCR products from SLP, LP5, SP7, and KOS strains. (A) PCR products from KOS 321, SP7, and SLP5. (B) PCR products from SP7 and KOS 321 (mixed to show differences) and from SP7 virus obtained after passages 1, 4, 5, 8, 9, and 10. Virus from passages 9 and 10 showed phenotypic differences from the parent and are therefore designated SL (small to large plaque) and SLP9 and SLP10 in the text. Positions of size standards (Std) are indicated in base pairs.
DISCUSSION
A human neonate with disseminated HSV-1 infection yielded two intrastrain variants (SP7 and LP5) which differed in tissue distribution, tissue culture and biochemical behavior, and ability to cause lethal neuroinvasive infection of the mouse. The close relationship between these strains was confirmed by the overall similarity of the restriction endonuclease fragmentation pattern. This is the first description of two genetically distinct but related strains within an infant, and these intrastrain variants differ in virulence and related properties.
The small-plaque-producing variant (SP7) was obtained from neuronal sites and was the overwhelmingly predominant strain isolated from the brain. The large-plaque-producing virus (LP5) was isolated from the lungs and the GI tract. The reason for the difference in the tissue distribution is not known. Although the initial virus dissemination in the neonate was likely to be by the hematogenous route, the small-plaque-producing virus may have found a preferential route to the brain, or different tissue environments may have selected or restricted the growth of one virus or the other. The immaturity of the immune response of the neonate (9) may also be a factor in allowing both virus substrains to coexist in the baby and may have influenced their tissue distribution.
Small-plaque production by clinical isolates (14, 15, 17, 27) and its correlation with limited virion release and a block in processing of multiple glycoproteins have been described for other low-passage HSV-1 isolates (15). HSV glycoproteins are incorporated into the envelope within the endoplasmic reticulum and are processed from the high-mannose precursor form to the sialylated mature form as the virion progresses through the Golgi apparatus, and then the virus is released by exocytosis or cell lysis (36). Individual HSV glycoproteins are also processed in this manner. For SP7, the lack of processing of multiple viral glycoproteins and the inhibition of virus release is consistent with a block in the progression of virus through the cell. A similar block in the processing of viral glycoproteins and HSV egress occurs upon disruption of vesicular transport or Golgi apparatus function with monensin (23) or brefeldin A (10). Inhibition of virus release would cause production of a small plaque in tissue culture.
The identity of the gene(s) involved in limiting the SP7 virus to a small-plaque phenotype was indicated by the marker transfer experiments. DNA containing the UL34.5 sequence from KOS transferred its large-plaque phenotype into the SP7 background. The transfer included a concomitant change in plaque size, efficiency of virus release, and glycoprotein processing. PCR confirmed the transfer of a KOS-like UL34.5 gene into SP7. In support of the association between the small-plaque phenotype and ICP34.5, marker transfer experiments with the same sequences from KOS were able to convert an unrelated small-plaque-producing clinical isolate, 490, to the large-plaque phenotype (data not shown). The linkage between UL34.5 sequences and this small/large-plaque phenotype is strengthened by our findings that passage of SP7 in Vero cells selects for large-plaque-producing virus (SLP-5 and SLP-10). The SLP-5 and SLP-10 viruses produce mature glycoproteins, are attenuated for neuroinvasive disease, and also have UL34.5 gene sequences resembling those for the highly passaged, attenuated KOS strains used herein. Our results are in agreement with those of Brown and coworkers, who demonstrated by using the 1716 deletion mutant that virions lacking UL34.5 are defective in virion maturation and egress, but the deletion mutant is unable to grow in neuronal cells (6). SP7, like strain 1716, produces small plaques and is defective in virion maturation and egress; however, unlike strain 1716, SP7 contains the UL34.5 gene in proper reading frame and maintains its ability to cause lethal infection upon intracerebral inoculation (neurovirulence).
Analysis of the sequence for ICP34.5 indicates that the protein has three distinct portions: an N-terminal unit of approximately 155 amino acids; a bridge unit with a strain-dependent number of PAT repeats; and a C-terminal unit of approximately 65 amino acids. As expected, the nucleotide and protein sequences for UL34.5 and ICP34.5 were more similar between SP7 and LP5 than with KOS (Table 2). Unexpectedly, the SLP-5 and SLP-10 sequences resembled KOS more than the parental strain but are consistent with their phenotype.
The C-terminal portion of ICP34.5 is important for growth in neurons (34), and the sequences were in proper frame for the SP7, LP5, the SLP viruses and were identical to KOS and other HSV strains (13). This portion of the protein is interchangeable with a peptide from the mammalian GADD34 (growth arrest and DNA damage) genes (19). The peptide binds to PCNA, a cell cycle regulatory protein (7), and can also activate protein phosphatase 1α to reverse the double-stranded RNA-dependent protein kinase-induced inhibition of protein synthesis in HSV-infected cells (13).
The most obvious sequence difference between the viruses was within the PAT repeat portion. The LP5 strain has the largest number of PAT repeats reported in the literature, with 22, while the SP7 strain has 18 PAT repeats. In comparison, SLP-5, SLP-10, and KOS have only three repeats. Passage of SP7 appears to generate deletion events which reduced the number of PATs to three, the same as for the highly passaged KOS. The nucleotide sequence for this region is highly repetitive and GC rich, and apparently it is susceptible to deletion. This may be the minimal number of PAT repeats necessary to retain basic function of the ICP34.5 protein. Alternatively, the number of nine nucleotide repeats encoding PAT may affect the ORF-O, ORF-P, LATs, or L/STs, which share sequences with UL34.5 (24).
Only two differences in the predicted protein sequences of ICP34.5 distinguish the SP7 strains from LP5 strains: SP7 virus has fewer PAT repeats and a GEGA deletion that precedes the PAT repeats. It is tempting to suggest that either or both of these differences are responsible for the small-plaque phenotype and the greater ability to cause lethal neuroinvasive disease of the SP7 virus, but this must be tested.
In addition to the number of PAT repeats, the passaged/attenuated viruses differ from both the SP7 and LP5 viruses in that they have a deletion of an arginine near the N terminus, a substitution of a glutamic acid for lysine prior to the PAT repeats, and an alanine-to-threonine change following the PAT repeats but preceding the GADD34 sequence. Although the N-terminal units of ICP34.5 are very similar between the different strains, the point mutations in this and other parts of the protein may also be significant.
Although the small-plaque-producing SP7 virus may be at a disadvantage for routine growth in tissue culture due to a limited ability to spread through the media, it has greater potential for causing lethal disease upon peripheral inoculation (SP7 > LP5 >>> SLP-5 = KOS). The order of virulence by this route of infection reflects the similarities and the differences within ICP34.5 between the strains. Since SP7 and LP5 have similar capacities to kill upon intracerebral inoculation of the mouse, the significant difference between these strains must be in their ability to progress from the local site lesion through the CNS to the brain. As shown in Fig. 9, SP7 reached and replicated to higher levels in the DRG than LP5, and the SP7 virus reached the brain before LP5.
Studies by Stevens (39) and Alexander and Rosenthal (2) indicate the importance of the immune response for limiting the neuroinvasiveness of certain HSV strains (2, 39). In immunocompromised mice, the absence of host immune responses allows the normally attenuated KOS (2, 29) and ANG strains to cause lethal disease upon peripheral infection (22, 29). In addition, the attenuated KOS and F strains induce a greater mononuclear cell infiltration into the CNS following infection than does the virulent ANG-PATH strain (29, 39). Mutations in gB and gD were implicated as important for lethal neuroinvasive disease upon comparison of the ANG-PATH strain to the attenuated ANG (22) and KOS (29, 44) strains.
In a primary infection, host responses take time to mature, and Stevens (39) and Mitchell and Stevens (29) suggest that HSV competes in a race between the development of host defenses and the progression of infection in order to cause lethal disease. This may be relevant to SP7 and LP5 since SP7 progresses to the brain from the periphery faster than does LP5 and is more lethal. For SP7, the reduced extent of virus egress and hence release of viral antigen may reduce its visibility to host protective responses. This may allow the disease to progress further prior to initiation of host control mechanisms. In contrast, viruses which release more virus, like LP5 and KOS, are likely to initiate host protective responses sooner to limit progression of the virus and disease. Further studies will be necessary to address the pathological mechanism of virulence of SP7.
Many studies have shown the importance of ICP34.5 for neurovirulence (10, 24; reviewed in reference 34). Further studies will be necessary to confirm the association of ICP34.5 and the specific differences in ICP34.5 with neuroinvasiveness. The gene encoding ICP34.5 shares sequences with LATs, ORF-O and ORF-P, and L/STs (24), and differences in these activities may also influence the character of the virus.
ACKNOWLEDGMENTS
We thank Gary Cohen and Roselyn Eisenberg for providing the rabbit anti-gC and gD. We thank John Docherty, Ming Ming Fu, and Josephine Dick for valuable help and discussions.
This study was supported by a Pediatric Scientist Training Program Fellowship, grant NICHD-22297, funds from the Department of Pediatrics, Children’s Hospital Medical Center of Akron, and other funds. M. Detweiller was supported in part by Cancer Center Core Grant CA16056.
REFERENCES
- 1.Alam T M, Joncas J H, Ozanne G. DNA polymorphism among isolates from multiple sites of a patient with chronic herpes simplex virus, type 1 infection. J Med Virol. 1989;29:186–191. doi: 10.1002/jmv.1890290308. [DOI] [PubMed] [Google Scholar]
- 2.Alexander T S, Rosenthal K S. Autologous antibody is protective against HSV-1 infection of the immunocompromised mouse. J Lab Clin Med. 1990;116:400–407. [PubMed] [Google Scholar]
- 3.Baines J D, Roizman B. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol. 1992;66:5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackshear P J. Systems for polyacrylamide gel electrophoresis. Methods Enzymol. 1984;104:237–255. doi: 10.1016/s0076-6879(84)04093-3. [DOI] [PubMed] [Google Scholar]
- 5.Bloom D C, Stevens J G. Neuron-specific restriction of a herpes simplex virus recombinant maps to the UL5 gene. J Virol. 1994;68:3761–3772. doi: 10.1128/jvi.68.6.3761-3772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown S M, MacLean A R, Aitken J D, Harland J. ICP 34.5 influences herpes simplex virus type 1 maturation and egress from infected cells in vitro. J Gen Virol. 1994;75:3679–3686. doi: 10.1099/0022-1317-75-12-3679. [DOI] [PubMed] [Google Scholar]
- 7.Brown S M, MacLean A R, McKie E A, Harland J. The herpes simplex virus virulence factor ICP34.5 and the cellular protein MyD116 complex with proliferating cell nuclear antigen through the 63-amino-acid domain conserved in ICP34.5, MyD116, and GADD34. J Virol. 1997;71:9442–9449. doi: 10.1128/jvi.71.12.9442-9449.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown Z A, Vontver L A, Benedetti J, Critchlow C W, Sells C J, Berry S, Corey L. Effects on infants of a first episode of genital herpes during pregnancy. N Engl J Med. 1987;317:1246–1251. doi: 10.1056/NEJM198711123172002. [DOI] [PubMed] [Google Scholar]
- 9.Burchett S K, Corey L, Mohan K M, Westall J, Ashley R, Wilson C B. Diminished interferon-gamma and lymphocyte proliferation in neonatal and postpartum primary herpes simplex virus infection. J Infect Dis. 1992;165:813–818. doi: 10.1093/infdis/165.5.813. [DOI] [PubMed] [Google Scholar]
- 10.Cheung P, Banfield B W, Tufaro F. Brefeldin A arrests the maturation and egress of herpes simplex virus particles during infection. J Virol. 1991;65:1893–1904. doi: 10.1128/jvi.65.4.1893-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou J, Chen J-J, Gross M, Roizman B. Association of a novel M1 90,000 phosphoprotein with PKR kinase in cells exhibiting enhanced phosphorylation of eIF-2α and premature shutoff of protein synthesis after infection with γ134.5− mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou J, Kern E R, Whitley R J, Roizman B. Mapping of herpes simplex virus 1 neurovirulence to γ134.5, a gene nonessential for growth in cell culture. Science. 1990;252:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 13.Chou J, Roizman B. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+ J Virol. 1990;64:1014–1020. doi: 10.1128/jvi.64.3.1014-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costanzo F, Borgatti M, Bartoletti A M, Foa-Tomasi L, Cassai E, Mannini-Palenzona A. Further characterization of virus obtained from herpes simplex virus type 1 recurrences and primary infection. Influence of the temperature of incubation upon glycoprotein synthesis and virus release. Arch Virol. 1986;88:293–299. doi: 10.1007/BF01310883. [DOI] [PubMed] [Google Scholar]
- 15.Dick J W, Rosenthal K S. A block in glycoprotein processing correlates with small plaque morphology and virion targetting to cell-cell junctions for an oral and an anal strain of herpes simplex virus type-1. Arch Virol. 1995;140:2163–2181. doi: 10.1007/BF01323238. [DOI] [PubMed] [Google Scholar]
- 16.Dix R D, McKendall R R, Baringer J R. Comparative neurovirulence of herpes simplex virus type 1 strains after peripheral or intracerebral inoculation of BALB/c mice. Infect Immun. 1983;40:103–112. doi: 10.1128/iai.40.1.103-112.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epstein A L, Lyon M, Michal Y, Jacquemont B. In vitro divergence of HSV-1 populations propagated in different cell lines. Arch Virol. 1990;111:133–140. doi: 10.1007/BF01310511. [DOI] [PubMed] [Google Scholar]
- 18.Goldin A L, Sandri-Goldin R M, Levine M, Glorioso J C. Cloning of herpes simplex virus type 1 sequences representing the whole genome. J Virol. 1981;38:50–58. doi: 10.1128/jvi.38.1.50-58.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He B, Chou J, Liebermann D A, Hoffman B, Roizman B. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the γ134.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J Virol. 1996;70:84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heller M, Dix R D, Baringer J R, Schachter J, Conte J E., Jr Herpetic proctitis and meningitis: recovery of two strains of herpes simplex virus type 1 from cerebrospinal fluid. J Infect Dis. 1982;146:584–588. doi: 10.1093/infdis/146.5.584. [DOI] [PubMed] [Google Scholar]
- 21.Huang X. On global sequence alignment. Comp Appl Biosci. 1994;10:227–235. doi: 10.1093/bioinformatics/10.3.227. [DOI] [PubMed] [Google Scholar]
- 22.Izumi K M, Stevens J G. Molecular and biological characterization of a herpes simplex virus type 1 (HSV-1) neuroinvasiveness gene. J Exp Med. 1990;172:487–496. doi: 10.1084/jem.172.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson D C, Spear P G. Monensin inhibits the processing of HSV glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y L, Schaffer P A. A virus with a mutation in the ICP4-binding site in the L/ST promoter of herpes simplex virus type 1, but not a virus with a mutation in open reading frame P, exhibits cell-type-specific expression of γ134.5 transcripts and latency-associated transcripts. J Virol. 1998;72:4250–4264. doi: 10.1128/jvi.72.5.4250-4264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locker H, Frenkel N. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: occurrence of heterogeneities in defined regions of the viral DNA. J Virol. 1979;32:429–441. doi: 10.1128/jvi.32.2.429-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lonsdale D M, Brown S M, Lang J, Subak-Sharpe J H, Koprowski H, Warren K G. Variations in herpes simplex virus isolated from human ganglia and a study of clonal variation in HSV-1. Ann N Y Acad Sci. 1980;354:291–308. doi: 10.1111/j.1749-6632.1980.tb27973.x. [DOI] [PubMed] [Google Scholar]
- 27.Mannini-Palenzona A, Bartoletti A M, Foa-Tomasi L, Costanzo F, Borgatti M, Tognon M, Cassai E. Study of herpes simplex virus type 1 populations obtained from recurrences and primary infections. J Med Virol. 1985;15:17–28. doi: 10.1002/jmv.1890150104. [DOI] [PubMed] [Google Scholar]
- 28.Marsden H. Herpes simplex virus glycoproteins and pathogenesis. Symp Soc Gen Microbiol. 1987;40:259–288. [Google Scholar]
- 29.Mitchell B M, Stevens J G. Neuroinvasive properties of herpes simplex virus type I glycoprotein variants are controlled by the immune response. J Immunol. 1996;156:246–255. [PubMed] [Google Scholar]
- 30.Montgomery A, Centifanto Y. Heterogeneity within an HSV-1 wild type strain and its importance in pathogenesis. Proc Soc Exp Biol Med. 1989;191:362–369. doi: 10.3181/00379727-191-42934. [DOI] [PubMed] [Google Scholar]
- 31.Podlech J, Hengerer F, Fleck M, Walev I, Falke D. Replication of herpes simplex virus type 1 and 2 in medulla of the adrenal gland after vaginal infection of mice. Arch Virol. 1996;141:1999–2008. doi: 10.1007/BF01718210. [DOI] [PubMed] [Google Scholar]
- 32.Richards J T, Kern E R, Overall J C, Glasgow L A. Differences in neurovirulence among isolates of herpes simplex virus type 1 and 2 in mice using four routes of infection. J Infect Dis. 1981;144:464–471. doi: 10.1093/infdis/144.5.464. [DOI] [PubMed] [Google Scholar]
- 33.Robbins A K, Whealy M E, Enquist L W. Centrifugation procedures for studying herpes viruses using the Sorvall RC-28 SUPRAspeed centrifuge. Biotechnology Update. 1988;3:1–3. [Google Scholar]
- 34.Roizman B, Markovitz N. Herpes simplex virus virulence: the functions of the γ134.5 gene. J NeuroVirol. 1997;3(Supp. 1):S1–S2. [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sommer M, Courtney R J. Differential rates of processing and transport of herpes simplex virus type 1 glycoproteins gB and gC. J Virol. 1991;65:520–525. doi: 10.1128/jvi.65.1.520-525.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith R F, Wiese B A, Wojzynski M K, Davison D B, Worley K C. BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- 38.Stanberry L R, Bourne N, Bravo F J, Bernstein D I. Capsaicin-sensitive peptidergic neurons are involved in the zosteriform spread of herpes simplex virus infection. J Med Virol. 1992;38:142–146. doi: 10.1002/jmv.1890380213. [DOI] [PubMed] [Google Scholar]
- 39.Stevens J G. HSV-1 neuroinvasiveness. Intervirology. 1993;35:152–163. doi: 10.1159/000150306. [DOI] [PubMed] [Google Scholar]
- 40.Thompson R L, Cook M L, Devi-Rap G B, Wagner E K, Stevens J G. Functional and molecular analyses of the avirulent wild-type herpes simplex virus type 1 strain KOS. J Virol. 1986;58:203–211. doi: 10.1128/jvi.58.1.203-211.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward P L, Roizman B. Herpes simplex genes: the blueprint of a successful human pathogen. Trends Genet. 1994;10:267–274. doi: 10.1016/0168-9525(90)90009-u. [DOI] [PubMed] [Google Scholar]
- 42.Weise K, Kaerner H C, Glorioso J, Schroder C H. Replacement of gB gene sequences in HSV-1 strain ANG by corresponding sequences of the strain KOS causes changes of plaque morphology and neuropathogenicity. J Gen Virol. 1987;68:1909–1919. doi: 10.1099/0022-1317-68-7-1909. [DOI] [PubMed] [Google Scholar]
- 43.Whitley R J, Kimberlin D W, Roizman B. Herpes simplex viruses. Clin Infect Dis. 1998;26:541–555. doi: 10.1086/514600. [DOI] [PubMed] [Google Scholar]
- 44.Yuhasz S A, Stevens J G. Glycoprotein B is a specific determinant of herpes simplex virus type 1 neuroinvasiveness. J Virol. 1993;67:5948–5954. doi: 10.1128/jvi.67.10.5948-5954.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zemanick M C, Strick P L, Dix R D. Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc Natl Acad Sci USA. 1991;88:8048–8051. doi: 10.1073/pnas.88.18.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]