Abstract
The natural life cycle of alphaviruses, a group of plus-strand RNA viruses, involves transmission to vertebrate hosts via mosquitoes. Chronic infections are established in mosquitoes (and usually in mosquito cell cultures), but infection of susceptible vertebrate cells typically results in rapid shutoff of host mRNA translation and cell death. Using engineered Sindbis virus RNA replicons expressing puromycin acetyltransferase as a dominant selectable marker, we identified mutations allowing persistent, noncytopathic replication in BHK-21 cells. Two of these adaptive mutations involved single-amino-acid substitutions in the C-terminal portion of nsP2, the viral helicase-protease. At one of these loci, nsP2 position 726, numerous substitution mutations were created and characterized in the context of RNA replicons and infectious virus. Our results suggest a direct correlation between the level of viral RNA replication and cytopathogenicity. This work also provides a series of alphavirus replicons for noncytopathic gene expression studies (E. V. Agapov, I. Frolov, B. D. Lindenbach, B. M. Prágai, S. Schlesinger, and C. M. Rice, Proc. Natl. Acad. Sci. USA 95:12989–12994, 1998) and a general strategy for selecting RNA viral mutants adapted to different cellular environments.
Alphaviruses are a globally distributed group of important human and animal pathogens that often cause transient febrile illness or more severe disease such as encephalitis (18). Representative and well-studied members of this positive-strand RNA virus genus include Sindbis virus (SIN), Semliki Forest virus, Ross River virus, and Venezuelan equine encephalitis virus. These viruses are transmitted to susceptible vertebrate hosts by mosquitoes. Insect vectors remain chronically infected for life and shed high levels of virus from the salivary glands. In contrast, vertebrate infections are typically self-limited. In the vertebrate host, rapid replication and high levels of viremia must be achieved to facilitate transmission prior to clearance by virus-specific immune responses. Perhaps as a consequence of this life cycle, remarkable differences in alphavirus infection of mosquito versus vertebrate cells in culture are often observed. Persistently infected mosquito cell cultures can be readily established (47), whereas infection of vertebrate cells usually results in suppression of host macromolecular synthesis, cytopathic effects (CPE), and cell death within a few days (19, 50). Although many aspects of alphavirus RNA replication, virion assembly, and pathogenesis have been studied, the virus-host interactions leading to the dramatically different biological outcomes of mosquito versus vertebrate cell infection are poorly understood.
In this study, we employed a genetic approach to define virus-specific determinants responsible for the deleterious effects of SIN infection on baby hamster kidney (BHK) cells. BHK cells are highly permissive for SIN replication. Essentially 100% of these cells can be productively infected with virus or transfected with infectious RNA transcribed from full-length SIN cDNA clones (29, 35). Like for other alphaviruses, early stages in SIN infection involve binding to a specific receptor, endocytosis, and a fusion between the virion envelope and the endosomal membrane at acidic pH (for a review, see reference 50). After uncoating, the 5′ two-thirds of the 11.7-kb SIN genome RNA is translated into two RNA replicase polyproteins, P123 and P1234. P1234 is produced in smaller quantities via readthrough of an opal termination codon. RNA replication proceeds through a full-length minus-strand intermediate which serves as the template for synthesis of both genome-length RNA and a 3′-coterminal 4.1-kb subgenomic RNA, via an internal promoter element. Subgenomic RNA accumulates to high levels and encodes the SIN structural proteins. Synthesis of all three SIN RNA species is highly regulated, with shutoff of minus-strand RNA synthesis occurring by 3 to 4 h postinfection (41, 42). This temporal regulation appears to occur via a proteolytic processing cascade orchestrated by a papain-like protease domain residing in the nsP2 region of P123/P1234 (23, 27, 45). This proteolytic activity is responsible for processing of P123 and P1234 by cis cleavage at the 3/4 site and trans cleavage at the 1/2 and 2/3 sites to yield four nonstructural proteins: nsP1, nsP2, nsP3, and nsP4 (reviewed in reference 50). Replicases that contain uncleaved nsP2-containing polyproteins (either P123 or P23) are required for efficient minus-strand initiation, whereas complexes containing the four mature nsPs are defective for synthesis of minus-strand RNAs but efficient for synthesis of genomic and subgenomic plus-strand RNAs. Release of progeny virus, which occurs by budding from the plasma membrane, begins a few hours after infection, with peak yields approaching 10,000 particles per cell achieved by 12 to 24 h.
Along with these replicative events, alphavirus infection results in marked effects on vertebrate cell metabolism (19). One of the most dramatic virus-induced changes is the rapid shift from host to viral mRNA translation. Based on a number of studies, multiple mechanisms have been proposed to explain this phenomenon (see reference 50 for a review). These include (i) competition between viral and host mRNAs for limiting translational components, (ii) changes in the intracellular Na+/K+ concentrations favoring translation of viral mRNAs, and (iii) interaction of the viral capsid protein with ribosomes, resulting in selective inhibition of host mRNA translation initiation. However, neither expression of alphavirus structural proteins nor high levels of subgenomic mRNA are required, since engineered replicons lacking the structural protein-coding region or containing a cis-acting mutation that severely depresses subgenomic mRNA synthesis still shut off host mRNA translation with kinetics indistinguishable from those of unmodified parental virus (12). Later virus-induced changes include cell rounding, loss of adherence, and eventually cell death. Expression of the SIN structural proteins clearly accelerates the onset of such CPE (12). In several cell types, death appears to occur via apoptosis (14). Although some virus-specific determinants which influence apoptosis or cell survival have been mapped to the viral glycoproteins (17, 55), little is known about the mechanism(s) by which they trigger cellular apoptotic pathways.
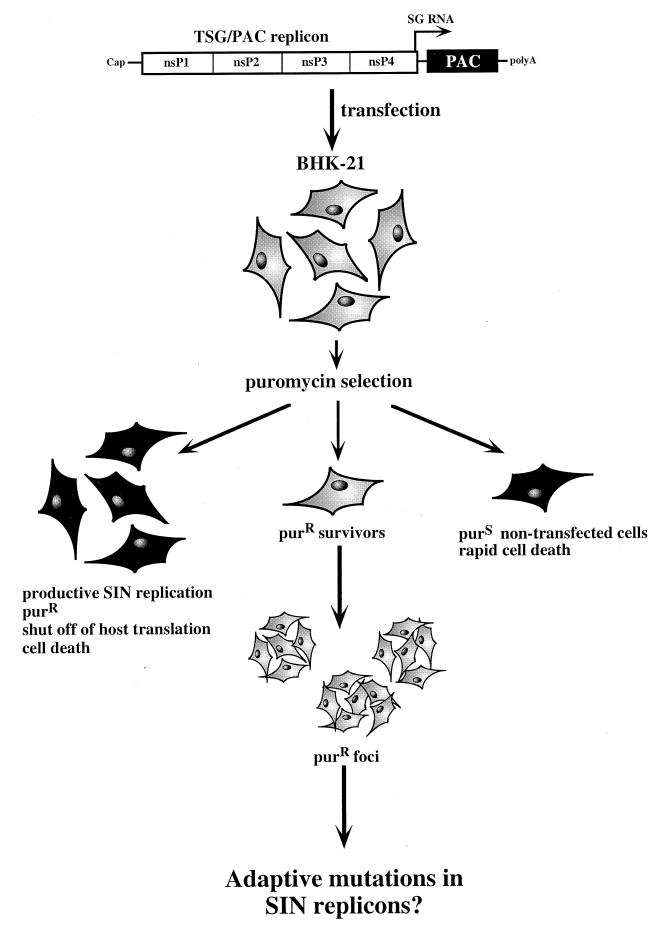
Although the deleterious effects of SIN on vertebrate cell biology undoubtedly result from multiple factors, one approach to understanding this virus-host interaction is to select for and study SIN mutants capable of noncytopathic replication in vertebrate cells. Mapping of such adaptive mutations should help to identify virus-specific functions which modulate host translation and cell death. To pursue this strategy, we placed the dominant selectable marker puromycin N-acetyltransferase (PAC) (8, 57) under the control of the SIN replication machinery (Fig. 1). BHK cells were transfected with these engineered PAC-expressing RNA replicons and allowed to recover and initiate SIN RNA replication, and then puromycin selection was imposed. At low frequency, stable puromycin-resistant (Purr) BHK foci grew out under selection. Characterization of adaptive mutations in SIN replicons recovered from these cell lines has allowed us to identify single-substitution mutations in the SIN nonstructural region that lead to noncytopathic replication in BHK cells. Furthermore, phenotypic characterization of these and other site-directed mutants suggests a strong correlation between the level of SIN replication and cytopathogenicity.
FIG. 1.
Strategy. A flow chart of the method used to select for adaptive SIN mutations is shown. At the top is a diagram of a SIN RNA replicon (TSG/PAC) in which the structural genes have been replaced with the pac gene (black box). The nonstructural region (open box), the four SIN nonstructural proteins (nsPs), and the start site for subgenomic (SG) mRNA transcription on the minus-strand genome-length RNA template (arrow) are indicated. After transfection, sequential steps in the selection scheme are shown. Viable and nonviable BHK cells are depicted in gray and black, respectively. See the text for further explanation.
MATERIALS AND METHODS
Cell cultures and virus plaque assays.
Chicken embryo fibroblasts (CEF) were grown in minimal essential medium (MEM) supplemented with 3% fetal bovine serum. BHK-21 cells (American Type Culture Collection) were grown in alpha MEM containing 10% fetal bovine serum.
SIN plaque formation was assayed by using BHK or CEF monolayers as previously described (24, 33).
Plasmid constructions.
Plasmids were assembled and propagated by standard methods (38). All regions amplified by PCR were verified by sequence analysis. Brief descriptions of the constructs are presented below; maps and sequences are available from the authors upon request. The cassette encoding PAC was obtained from pBABE Puro (31). This version of the pac gene differs from the originally reported sequence (GenBank accession no. M25346) (21) at the following positions in the PAC-coding region: 52 (A instead of C), 114 (T instead of C), and 550 to 567 (5′-GTGCCCGAAGGACCGCGC-3′ instead of 5′-TGCCCGAAGGACCGCGCG-3′).
(i) Double subgenomic SIN constructs.
Double subgenomic SIN vectors expressing chloramphenicol acetyltransferase (CAT) have been described previously (15). pTE/5′2J/CAT contains the cat gene following the first SIN subgenomic mRNA promoter followed by a second promoter driving expression of the subgenomic mRNA encoding the SIN structural proteins (15a). pTE/5′2J/PAC was constructed by subcloning the HindIII-ClaI fragment from pBABE Puro, containing the pac gene, into the XbaI site of pTE/5′2J (15a). Prior to ligation, 5′ overhangs were filled in by using Klenow fragment and deoxynucleoside triphosphates.
(ii) SIN replicons.
Four SIN RNA replicons expressing PAC but lacking the SIN structural genes were constructed. pTSG/PAC, which expresses a single subgenomic mRNA encoding PAC, was constructed by subcloning the ApaI-XhoI fragment from pTR2003 [containing the SIN 3′ nontranslated region and poly(A) tract] (29a) into the corresponding sites of pTE/5′2J/PAC. pT*SG/PAC contains Val substitutions at the P2 positions of the nsP1/nsP2 and nsP2/nsP3 cleavage sites (44). pT*SG/PAC was constructed by subcloning the Eco47III-SpeI fragment of pTM3/SIN1*2*3V (26) into the corresponding sites of pTSG/PAC. pT4.1/EMCPAC contains a 3-base insertion in the subgenomic mRNA promoter (13) followed by an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES)/PAC cassette. pT4.1/EMCPAC was constructed by ligating the ApaI-XhoI fragment from pT4.1/EMC-S (33a), the ApaI-BglII fragment from p1568 (30a), and the XbaI-XhoI fragment from pToto/3′2J/PAC (33b). Prior to ligation, the BglII and XbaI sites were filled in by using Klenow fragment and deoxynucleoside triphosphates. pT*4.1/EMCPAC was constructed by subcloning the Eco47III-SpeI fragment of pTM3/SIN1*2*3V (26) into the corresponding sites of pT4.1/EMCPAC. For all four constructs, productive SIN RNA replication and PAC gene expression were verified in first-cycle analyses after replicon RNA transfection of BHK-21 cells (data not shown).
(iii) SIN replicons with substitutions at nsP2 residue 726.
pTSG/PAC derivatives containing various substitutions at nsP2 residue 726 were constructed by PCR mutagenesis (38). For each mutant, an NheI (3437)-PstI (3949) fragment containing the mutagenized region was subcloned into pRS2 (10a), sequenced, and used to replace the corresponding region of pTSG/nsP2-726L/PAC (see below). pTSG/nsP2-726L/PAC was used as a cloning vector because the C3856T mutation leading to the nsP2-726 Leu substitution creates an AvrII site which can be used as a convenient marker to distinguish between vector background and mutant subclones. Substitutions relative to the parental nsP2-726 Pro (CCA codon in the SIN cDNA) include pTSG/nsP2-726F/PAC (TTT), pTSG/nsP2-726V/PAC (GTA), pTSG/nsP2-726A/PAC (GCA), pTSG/nsP2-726Q/PAC (CAA), pTSG/nsP2-726Y/PAC (TAC), pTSG/nsP2-726T/PAC (ACA), pTSG/nsP2-726G/PAC (GGA), and pTSG/nsP2-726R/PAC (AGA). pTSG/nsP2-726S/PAC was constructed by replacement of the BglII-AvrII fragment of pTSG/nsP2-726L/PAC with the corresponding fragment of VnsP2 (9).
(iv) Full-length SIN derivatives with substitutions at nsP2 residue 726.
Full-length SIN cDNA clones with mutations at nsP2 residue 726 were made by using the pToto1101 background (35). As described above, the NheI-PstI fragment containing each substitution was used to replace the corresponding region of pToto/nsP2-726L to create pToto/nsP2-726F, pToto/nsP2-726V, pToto/nsP2-726A, pToto/nsP2-726Q, pToto/nsP2-726Y, pToto/nsP2-726T, pToto/nsP2-726G, and pToto/nsP2-726R. The Toto1101 derivative containing the SIN-1 mutation (pToto/nsP2-726S) (9) was constructed by replacing the BamHI-XhoI fragment of pTSG/nsP2-726S/PAC with the corresponding fragment from Toto1101.
(v) Other plasmids.
A pTSG/PAC derivative with a hyperprocessing mutation in nsP2 was constructed for cell-free processing analyses (614Asp) (48). pTSG/nsP2-614D/PAC was derived by subcloning the ClaI-SpeI fragment of pTM3/SIN123N>D (27) into the corresponding sites of pTSG/nsP2-726L/PAC.
RNA transcription and transfection.
Purified plasmid DNAs were linearized by digestion with XhoI and transcribed in the presence of cap analog by using SP6 RNA polymerase (35). The yield and integrity of RNA transcripts were monitored by incorporation of [3H]UTP and gel electrophoresis. Transcription reactions were aliquoted, stored at −70°C, thawed, and used for transfection of BHK-21 cells by electroporation (29) or with Lipofectin (34).
Selection, passaging, and storage of puromycin-resistant cell lines.
Puromycin stocks (1 mg/ml) were made up in MEM, sterile filtered, and stored at −20°C. Following transfection, cells were allowed to recover for 4 to 12 h before addition of puromycin (Sigma) to 5 to 10 μg/ml. To select for puromycin-resistant foci, the medium was replaced every 2 to 3 days with fresh puromycin-containing medium. Individual foci were cloned, expanded, and stored frozen in liquid nitrogen. After trypsinization, cells were seeded in medium lacking puromycin and allowed to recover for 12 h before addition of puromycin to 5 μg/ml. For routine passaging, this recovery period in the absence of the drug was later found to be unnecessary. At 1 to 2 h prior to freezing of cell lines, the medium was replaced with fresh medium lacking puromycin. Cells were trypsinized and frozen in alpha MEM containing 10% dimethyl sulfoxide and 40% fetal calf serum. Cells thawed after liquid nitrogen storage were allowed to recover for 12 h prior to addition of puromycin.
Mapping of adaptive mutations.
Total cellular RNA from Purr cell lines was isolated by using RNAzol B as described by the manufacturer (Tel-Test, Inc., Friendswood, Tex.). For some experiments, poly(A)-containing RNA was purified from total cellular RNA by oligo(dT) cellulose chromatography (38). Total or poly(A)+ RNA from Purr BHK cell lines was assayed for its ability to confer puromycin resistance by transfection of naive BHK cells, as described above.
For the S1 and S24 Purr cell lines, cDNAs were synthesized by using poly(A+) RNA from approximately 107 cells. A negative-sense oligonucleotide complementary to SIN nucleotides 4629 to 4644 served as the primer for cDNA synthesis with avian myeloblastosis virus reverse transcriptase (Promega). Following reverse transcription, cDNAs were amplified by 15 cycles of PCR with Vent polymerase (New England Biolabs), the same negative-sense primer, and a positive-sense oligonucleotide (SIN nucleotides 2282 to 2300). The resulting 2,291-bp fragment was digested with BglII (2288) and AflII (4572) and cloned into the corresponding sites of pTSG/PAC. Plasmid DNAs from individual transformants were linearized and transcribed, and the resulting RNA transcripts were assayed for their ability to confer puromycin resistance by transfection of BHK-21 cells, as described above. This subregion allowed cell survival and division in the presence of puromycin and was sequenced by using the Sequenase DNA sequencing kit (Amersham Life Sciences). Single-base changes leading to substitutions in nsP2 were identified. These correspond to nsP2 Pro726Leu (S1; C3856T) and Asn779Lys (S24; C4017A). pTSG/PAC derivatives containing these causal mutations are designated pTSG/nsP2-726L/PAC and pTSG/nsP2-779K/PAC.
Packaging of replicons.
For pTSG/PAC, stocks of packaged replicons were produced by cotransfection of TSG/PAC and DH-BB(5′SIN) helper RNAs as previously described (3).
RNA analysis.
SIN-specific RNAs were labeled with [3H]uridine (30 μCi/ml) in the presence of dactinomycin (1 μg/ml). Specific labeling periods for different experiments are given in the figure legends. Total cellular RNA was isolated by the RNAzol B procedure, denatured by treatment with glyoxal and dimethyl sulfoxide, and separated by electrophoresis in 1% agarose gels containing 10 mM phosphate buffer (2). Gels were fixed in methanol, impregnated with 3% PPO (2,5-diphenyloxazole) in methanol, equilibrated in water, dried, and exposed to X-ray film at −70°C. In some experiments, bands were localized by fluorography and excised, and the radioactivity was determined by liquid scintillation counting.
Protein analysis.
Cell-free translation mixtures, with nuclease-treated rabbit reticulocyte lysates (Promega) and [35S]methionine (Amersham), were incubated for 90 min at 30°C according to the manufacturer’s instructions. Final reaction volumes were 10 μl and contained 10 ng of capped transcript RNA. Translation reactions were terminated by the addition of Laemmli sample buffer (4 volumes), and the products were heated for 10 min at 70°C and separated by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis (22). Gels were fixed in 10% acetic acid–30% methanol, dried, and exposed to X-ray film without fluorographic enhancement. Relative levels of incorporation into P123 and nsP2 were determined by using a Bio-Rad phosphorimaging system. 14C-methylated molecular weight marker proteins were purchased from Amersham.
RESULTS
PAC as a dominant selectable marker.
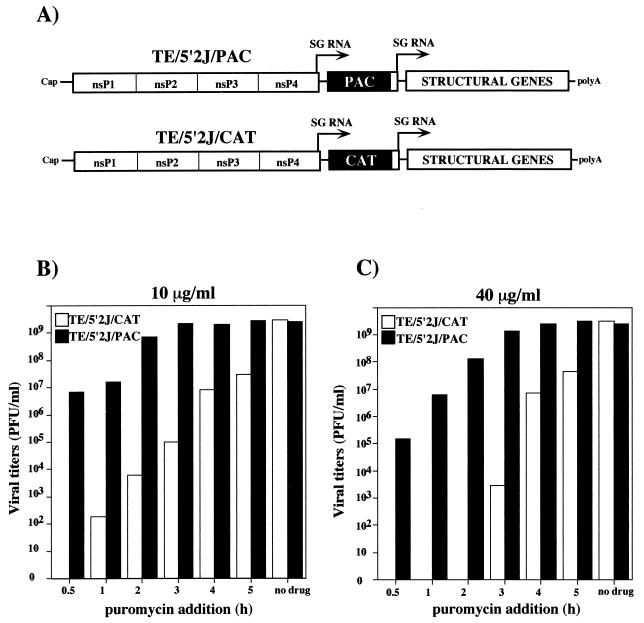
PAC from Streptomyces alboniger is a single polypeptide of 199 amino acids that inactivates the translational inhibitor puromycin by acetylation of its tyrosinyl moiety (21). PAC has been successfully used as a rapidly selectable marker in several systems, including mammalian cells (8, 57). To determine whether this marker could be used in the context of SIN replication, we constructed double subgenomic promoter vectors (4, 15) designed to express either PAC or CAT, as a negative control (Fig. 2A) (15). BHK-21 cells were transfected with capped RNAs transcribed from either pTE/5′2J/PAC or pTE/5′J/CAT, and puromycin was added to 10 (Fig. 2B) or 40 (Fig. 2C) μg/ml at different times posttransfection. Without added puromycin, both constructs yielded similar titers of infectious virus, in excess of 109 PFU/ml at 18 h posttransfection. When puromycin was added at 0.5 h posttransfection, no virus was detected in the supernatant of TE5′2J/CAT-transfected cells, whereas ∼107 or 105 PFU/ml was found for cells transfected with TE5′2J/PAC RNA and incubated with 10 or 40 μg of puromycin per ml, respectively. If the drug was added at 3 to 4 h posttransfection, the yield of TE5′2J/PAC virus was essentially the same as that found when cells were not treated with puromycin. Puromycin selection allowed for as much as 108-fold discrimination between virus expressing PAC and the CAT-expressing negative control (addition of 40 μg/ml at 2 h posttransfection [Fig. 2C]), and only 3 to 4 h of viral gene expression postelectroporation was needed for complete resistance to be achieved. Although a short incubation period in the absence of the drug was required to establish SIN replication, subgenomic RNA transcription, and PAC production, these experiments demonstrated that SIN-expressed PAC conferred efficient resistance to even high concentrations of puromycin.
FIG. 2.
Puromycin as a dominant selectable marker. (A) Double subgenomic (SG) promoter constructs expressing either CAT (pTE/5′2J/CAT) or PAC (pTE/5′2J/PAC). In addition to the features described in Fig. 1, these engineered RNAs contain a second subgenomic promoter driving expression of the SIN structural protein-coding region (STRUCTURAL) and are therefore capable of making infectious virus. (B and C) Effect of the time of puromycin addition on yield of infectious virus. Transcripts (2.5 μg) from linearized pTE/5′2J/CAT (open bars) or pTE/5′2J/PAC (solid bars) templates were used to electroporate BHK-21 cells, as described in Materials and Methods. At the indicated times postelectroporation, puromycin was added to 10 (B) or 40 (C) μg/ml and incubation was continued. At 18 h postelectroporation, viral titers in the culture supernatants were determined by plaque assay on CEF monolayers. Control samples without added drug were also assayed.
Derivation of Purr cell lines.
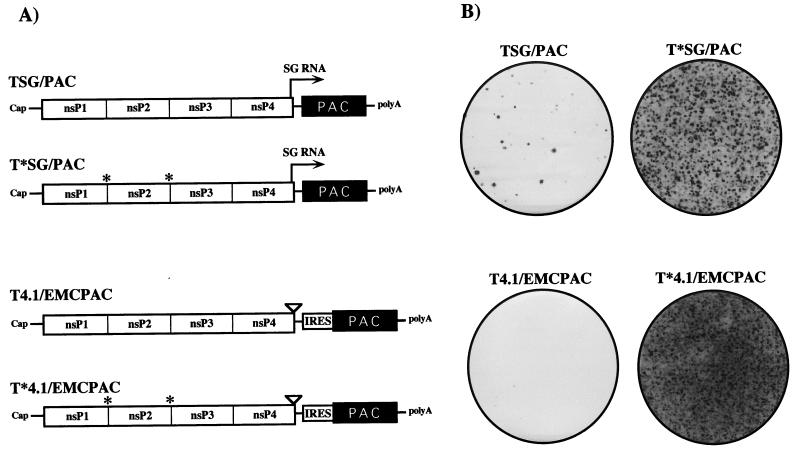
Although expression of the SIN structural proteins contributes to cytopathogenicity, BHK cells transfected or infected with SIN RNA replicons lacking the structural protein-coding region still shut off host translation with kinetics similar to those of the parental virus, and the cells eventually die (12). In our attempt to select for mutations in SIN nonstructural proteins or cis RNA elements that would allow noncytopathic replication in BHK cells, we tested four different replicons (Fig. 3A). TSG/PAC is essentially an unmodified SIN replicon in which the structural protein genes have been replaced by the pac gene. This replicon expresses high levels of a subgenomic mRNA encoding PAC. A second derivative, T*SG/PAC, was constructed with substitutions at the nsP1/nsP2 and nsP2/nsP3 cleavage sites to block processing by the SIN nsP2 papain-like protease (7, 44). According to the current model for regulation of SIN RNA replication, this replicon should be defective in minus-strand shutoff and impaired in the synthesis of genomic and subgenomic plus strands (27, 45). Since high levels of viral subgenomic mRNA accumulation might be toxic for cells, two additional derivatives were made with a 3-base insertion in the subgenomic promoter region (called the 4.1 mutation). This mutation has been shown to decrease subgenomic RNA accumulation approximately 100-fold (13). In order to ensure synthesis of PAC, translation of the pac gene in the 4.1 constructs was driven by the EMCV IRES, which can function in the context of replicon genomic RNA (33a). Preliminary transfection experiments confirmed that all four constructs could replicate and express detectable levels of PAC (data not shown).
FIG. 3.
Replicon constructs. (A) Schematic of SIN replicon RNAs expressing PAC. PAC is expressed either by synthesis of a subgenomic (SG) mRNA (arrow) or by internal translation initiation mediated by an IRES derived from EMCV. The 4.1 derivatives contain the 3-base CR4.1 insertion, which virtually inactivates the subgenomic mRNA promoter (13). Constructs with blocked nsP1/nsP2 and nsP2/nsP3 cleavage sites (indicated by asterisks) should produce only uncleaved P123 and nsP4. For other features, see the legend to Fig. 1. (B) Puromycin-resistant foci produced after RNA transfection. BHK-21 cells (107) were electroporated with 5 μg of each replicon RNA, plated in 100-mm-diameter dishes, and allowed to recover for 12 h before addition of puromycin to 10 μg/ml. The medium was changed every 2 to 3 days, and Purr foci were stained with crystal violet after 8 days.
To compare the abilities of these replicons to confer puromycin resistance and establish noncytopathic replication, BHK cells were transfected by electroporation (which allows essentially 100% transfection efficiency [29]) with each construct, including TSG/CAT and no-RNA negative controls. Cells were allowed to recover for 12 h, and then medium containing puromycin at 10 μg/ml was added. Relative to the TSG/CAT- or mock-transfected cells, which died and detached from the plates within 12 h following addition of the drug, three of the four PAC-expressing replicons (the exception being T4.1/EMCPAC) allowed prolonged cell survival. However, the majority of the transfected cells died after 3 to 4 days, presumably due to SIN-induced cytotoxic effects or insufficient PAC expression. Incubation was continued in the presence of puromycin, and after 1 week Purr foci were detected at various frequencies for each of the four replicons (Fig. 3B). No foci were detected for TSG/CAT- or mock-transfected cells (data not shown). Numerous foci were observed for the replicons with blocked 1/2 and 2/3 cleavage sites (T*SG/PAC and T*4.1/EMCPAC), many fewer were observed for the parental TSG/PAC replicon, and only two or three were observed for the T4.1/EMCPAC replicon. Individual Purr foci from each of the transfections were cloned, expanded, and frozen. In an attempt to identify adaptive mutations which could function in the context of an unmodified SIN replicon, subsequent experiments focused on characterizing Purr cell lines derived by transfection with the TSG/PAC replicon.
SIN RNA phenotype in Purr cell lines.
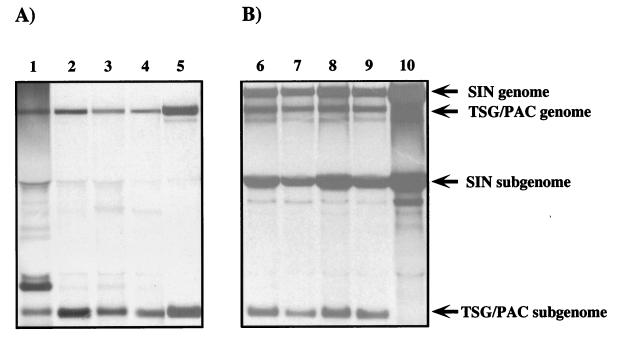
Given the different focus-forming frequencies observed for the four replicons, it seemed likely that puromycin resistance in these cell lines was mediated by SIN-expressed PAC and not by nuclear integration of a functional pac cassette (template DNA was degraded prior to transfection) or acquisition of spontaneous cellular resistance mutations. To examine this directly, SIN-specific RNAs were selectively labeled with [3H]uridine in the presence of dactinomycin to inhibit cellular RNA synthesis, isolated, denatured, and analyzed by agarose gel electrophoresis (Fig. 4A). As shown for four of the TSG/PAC-derived cell lines (S22, S1, S24, and S25 [Fig. 4A, lanes 1 to 4, respectively]), replicon genomic and subgenomic RNAs which comigrated with the corresponding RNAs produced by the parental TSG/PAC replicon (Fig. 4A, lane 5) were found. For the S22 line, an additional dactinomycin-resistant RNA species was present but has not been further characterized. In all four Purr cell lines, the levels of SIN-specific RNAs were lower (approximately 100-fold) than the level observed for the parental TSG/PAC replicon (the signal shown in lane 5 is from one-fifth the number of cells and was exposed for 6 h, as opposed to 60 h for lanes 1 to 4). The Purr cell lines could be successfully infected with SIN, which resulted in a dramatic increase in levels of replicon genomic and subgenomic RNAs (Fig. 4B). Although a slight delay in SIN replication and virus production was observed in these cells (data not shown), cellular translation was shut off and the Purr cells showed all the hallmarks of SIN-induced CPE by 18 to 24 h after infection. Thus, the cellular environment of the Purr cell lines was permissive for high-level SIN replication, and the cells were still susceptible to SIN-induced cytopathogenicity. This is somewhat surprising given that SIN superinfection exclusion is established rapidly and requires synthesis of nsPs (5). However, we believe that the levels of nsPs or RNA replication complexes in the Purr cells may be too low to establish this state (1). In any case, SIN appeared to be able to trans activate replication of the replicons, suggesting that downregulation of replicon RNA synthesis did not involve mutation of cis RNA elements required for efficient replication and transcription.
FIG. 4.
Analysis of SIN RNAs in Purr cell lines. (A) Purr cell lines were labeled for 18 h with [3H]uridine (30 μCi/ml) in the presence of dactinomycin (1 μg/ml). To provide a control for replicon genomic and subgenomic RNAs, BHK-21 cells were infected at a high multiplicity (10 infectious units/cell) with packaged TSG/PAC replicons (lane 5). (B) Cells were superinfected or infected with Toto1101 (10 PFU/cell). Infected cells were also labeled as described above for 18 h beginning 1 h postinfection. The Purr cell lines analyzed were S22 (lanes 1 and 6), S1 (lanes 2 and 7), S24 (lanes 3 and 8), S25 (lanes 4 and 9), and BHK-21 cells (lane 10). RNAs were extracted and analyzed by agarose gel electrophoresis as described in Materials and Methods. Lanes 1 to 4 contain RNAs from 5 × 105 cells, and the gel was fluorographed and exposed for 60 h. Lanes 5 to 10 contain RNAs isolated from 105 cells, and the exposure time for these samples was only 6 h. The positions of replicon and SIN genomic and subgenomic RNAs are indicated on the right of panel B.
Puromycin resistance can be conferred by RNA.
The presence of SIN replicons in the Purr cells suggested that they, as opposed to adaptive changes in the BHK host cells, might be responsible for drug resistance. Further support for this idea was obtained from RNA transfection experiments. Total or poly(A)+ RNA from the Purr cell lines was isolated and used to transfect naive BHK cells. For each cell line tested (data for S1 is shown in Fig. 5), RNA transfection conferred puromycin resistance with high efficiency compared to the original TSG/PAC transfection results (Fig. 3B). No Purr cell or focus was obtained after transfection with poly(A)+ RNA isolated from control BHK cells. Furthermore, RNase treatment of RNA isolated from Purr cell lines abolished the recovery of Purr cells after transfection (data not shown). In contrast to the original Purr BHK cell lines, which had varied morphologies and growth rates, naive cells transduced with these RNAs and selected with puromycin had normal appearance and doubling times (data not shown). These experiments demonstrate that RNA from Purr cells is sufficient to confer long-term resistance to puromycin in naive BHK cells, a result which is consistent with the notion that PAC expression is mediated by adapted SIN replicons capable of persistent noncytopathic replication in this cellular environment.
FIG. 5.
Transfection of naive BHK cells with RNA from the Purr S1 cell line. BHK-21 cells were electroporated with poly(A)+ RNA isolated from 2 × 106 cells of the S1 Purr cell line (dish 1), total RNA isolated from 5 × 105 cells of the S1 line (dish 2), or poly(A)+ RNA isolated from 1 × 106 BHK-21 cells (dish 3). Puromycin (10 μg/ml) was added to the medium at 16 h post-transfection. Purr foci were stained with crystal violet after 5 days (dish 1) or 7 days (dishes 2 and 3).
Mapping of adaptive mutations.
We mapped the adaptive mutations in replicons derived from two of the cell lines, S1 and S24. Poly(A)+ RNAs from these cell lines were used as templates for cDNA synthesis and amplification by high-fidelity PCR with a limited number of cycles. Fragments were digested with convenient restriction endonucleases and subcloned into the parental TSG/PAC plasmid, and multiple independent clones were assayed for their ability to confer puromycin resistance by transfection of BHK cells and puromycin selection. Nucleotide sequences were determined for minimal subregions allowing efficient cell survival in the presence of puromycin. By this approach, adaptive mutations for S1 and S24 were mapped to single-amino-acid substitutions in the C-terminal region of SIN nonstructural protein nsP2 (Fig. 6). S1 contained a Leu substitution for Pro at nsP2 residue 726. S24 contained a Lys substitution for Asn at residue 779. Either of these substitutions on the TSG/PAC background allowed survival of naive BHK cells with essentially 100% efficiency after electroporation and puromycin selection. Thus, two different single-amino-acid substitutions in nsP2, located in a region immediately downstream of the papain-like protease domain, could convert a cytopathic replicon into self-replicating RNAs that could persist in BHK cells without apparent deleterious effects on these host cells.
FIG. 6.
Sequence alignment and adaptive mutations of S1 and S24. An alignment of nsP2 residues 720 to 790 (SIN numbering [49]) for several alphaviruses is shown. EEE, Eastern equine encephalitis virus (58); VEE, Venezuelan equine encephalitis virus (20); SFV, Semliki Forest virus (52); RRV, Ross River virus (10); ONN, O’nyong nyong virus (28); OCK, Ockelbo virus (43). Residues identical to those in the SIN sequence are indicated by dashes. The adaptive SIN-1 (726S) (9), S1 (726L) (this study), and S24 (779K) (this study) mutations and the mutation in ts24 responsible for temperature sensitivity and impaired subgenomic RNA synthesis (16) are highlighted.
Mutagenesis of nsP2 residue 726.
To our surprise, a different substitution at nsP2 residue 726 (Ser) was previously shown to be partially responsible for the phenotype of SIN-1 (9), a virus which was isolated from persistently infected BHK cells (59). This substitution downregulates RNA replication, has minimal effects on host mRNA translation and virus yields, and facilitates establishment of persistent viral infections in BHK cells (9, 59). However, replicons with the Ser substitution are significantly more cytopathic than those with the S1 Leu substitution. Given the remarkable convergence of the S1 and SIN-1 mutations at position 726, yet their dramatically different phenotypes with respect to RNA replication and cytopathogenicity, we created and studied several additional mutations at this locus in the context of the TSG/PAC replicon (Fig. 7). Substitutions included amino acids with small side chains (Ala and Gly), as well as polar (Thr and Gln), bulkier hydrophobic (Val, Phe, Tyr, and Leu), and charged (Arg) residues. To assay for differences in RNA replication efficiency, we transfected BHK cells with the mutant or parent replicons and labeled SIN-specific RNAs with [3H]uridine for 18 h beginning at 5 h posttransfection (Fig. 7A). A spectrum of RNA replication efficiencies was observed, ranging from 100% (wild-type Pro and the Ala substitution) to less than 1% (Gln) (Fig. 7B; Table 1). As observed previously for other nsP2 mutants, differences in the ratio of genomic to subgenomic RNAs were noted (Fig. 7B) (16, 39). The ability of the wild-type or mutant replicons to establish noncytopathic replication was assayed by transfection of BHK cells and puromycin selection. In the case of Phe, Tyr, Arg, and Gln substitutions, Purr cell populations were readily established, with efficiencies similar to that seen for the S1 Leu substitution (essentially 100% survival following puromycin selection). For the wild-type TSG/PAC replicon, as seen before, the majority of the cells transiently survived but eventually died, with only a few Purr foci developing. A picture similar to that for the wild type was obtained for the Ala and Val substitutions. For the Gly, Ser, and Thr substitutions, an intermediate phenotype was observed. Although most of the initially transfected cells died, increasing numbers of foci were recovered for these mutants with prolonged incubation (Thr > Ser > Gly) (Fig. 7C). Although it is unverified, we believe that these foci probably result from additional adaptive mutations that arose during in vitro transcription or, more likely, during SIN replication. Overall, there was a striking correlation between cytopathogenicity and the level of RNA replication. Replicons with high levels of RNA replication (>5 to 100% of the wild-type level) were cytopathic, whereas replicons with lower levels (<2% of the wild-type level) were able to establish noncytopathic replication in BHK cells.
FIG. 7.
Mutagenesis of nsP2 residue 726. (A) First-cycle RNA and replicon phenotypes. BHK-21 cells were transfected with the parental replicon or the various substitution mutants. At 5 h posttransfection, RNAs were labeled for 18 h with [3H]uridine (30 μCi/ml) in the presence of dactinomycin (2 μg/ml). Total RNA was isolated, and equivalent portions were analyzed by agarose gel electrophoresis. The RNAs analyzed were the parent 726P (lane P), 726A (lane A), 726V (lane V), 726G (lane G), 726S (lane S), 726T (lane T), 726F (lane F), 726Y (lane Y), 726L (lane L), 726R (lane R), 726Q (lane Q), and no RNA (lane Mock). Below the gel, the abilities of replicons to confer Purr and maintain cell viability are summarized. At 48 h postelectroporation and after 40 h of puromycin selection, cells were observed and stained with crystal violet. Healthy monolayers were observed for replicons containing the Phe, Tyr, Leu, Arg, and Gln substitutions (no CPE [−]). Few viable cells remained for the parent and the Ala, Val, Gly, Ser, and Thr substitution mutants (CPE [+]). No viable cells were present in mock-transfected BHK-21 cells. (B) Levels of SIN-specific RNA accumulation relative to that for the parent were determined by excision of radiolabeled bands and liquid scintillation counting. Values (with background [mock] subtracted) are expressed as percentages of the wild-type (wt) parental (Pro) level (set at 100%). Molar ratios of subgenomic (SG) to genomic (G) RNA were also determined for each sample. (C) Low-frequency focus formation by the parental TSG/PAC replicon (Pro) and the Ala, Val, Gly, Ser, and Thr substitution mutants. Purr foci present at 8 days posttransfection were visualized by crystal violet staining. (D) Polyprotein-processing phenotypes. Replicon RNAs were translated in a reticulocyte cell-free system and analyzed by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis as described in Materials and Methods. Samples included the parental replicon 726P (P) and the following nsP2 substitution mutants: 726A (A), 726V (V), 726G (G), 726S (S), 726T (T), 726F (F), 726Y (Y), 726L (L), 726R (R), 726Q (Q), and 779K (779N>K). pTSG/nsP2-614D/PAC transcripts were also analyzed as an example of a mutation that confers a hyperprocessing phenotype (123N>D). A measure of processing efficiency was obtained by determining the molar ratio of P123 to nsP2 and was normalized to that observed for the parental replicon (set at 100%). The graph shows the results of three independent experiments (means and standard errors of the means).
TABLE 1.
Replicon and virus phenotypes of nsP2-726 mutants
| Mutationa | RNA level (%)b | Processing (mean ± SEM)c | CPE
|
|
|---|---|---|---|---|
| Replicond | Viruse | |||
| 726Pro (wild type) | 100 | 1 | Yes | Yes |
| 726Ala | 101 | 0.77 ± 0.13 | Yes | Yes |
| 726Val | 30 | 0.32 ± 0.06 | Yes | Yes |
| 726Gly | 17 | 0.28 ± 0.06 | Yes | Nof |
| 726Ser | 9.9 | 0.16 ± 0.04 | Yes | Nof |
| 726Thr | 5.1 | 0.12 ± 0.03 | Yes | Nof |
| 726Phe | 1.6 | 0.09 ± 0.01 | No | —g |
| 726Tyr | 1.4 | 0.09 ± 0.01 | No | — |
| 726Leu | 1.1 | 0.10 ± 0.01 | No | — |
| 726Arg | 1.2 | 0.10 ± 0.03 | No | — |
| 726Gln | 0.8 | 0.08 ± 0.03 | No | — |
| 779Lys | NDh | 0.10 ± 0.01 | No | ND |
| 614Asp | ND | 0.20 ± 0.03 | ND | Lethal |
Amino acid substitutions at nsP2 residue 726, residue 779 (S24), or residue 614 (48).
Genomic RNA accumulation for replicon derivatives relative to that for the parent as determined from the data presented in Fig. 7A. Bands were localized by fluorography and excised, and the level of incorporation was determined by liquid scintillation counting.
Results of processing-phenotype assays by cell-free translation, compared by analyzing the ratio of uncleaved P123 to nsP2 (Fig. 7D).
BHK cells were transfected with the various replicons, puromycin was added after 5 h, and the remaining cells were stained with crystal violet after 48 h. Replicons with Phe, Tyr, Leu, Arg, or Gln at position 726 were noncytopathic and produced healthy monolayers in the presence of puromycin. For the other replicons, including the parent (726Pro), the vast majority of the cells were dead by 48 h. As shown in Fig. 7C, at various frequencies, Purr foci were observed for these replicons.
Virus-induced CPE was observed after transfection with Toto1101 derivatives containing the nsP2-726 substitutions. By 24 h posttransfection, complete CPE was observed for the Toto1101 parent and the Ala and Val substitution mutants. The nsP2 hyperprocessing mutation at residue 614 is lethal for virus replication (48).
Transient CPE, recovery, and persistently infected cultures were produced after transfection with RNAs containing Ser, Gly, or Thr.
—, for the Phe, Tyr, Leu, Arg, and Gln mutants, the viral cytopathic phenotype was difficult to assay due to the rapid appearance of cytopathic revertants or pseudorevertants.
ND, not determined.
Protein-processing phenotypes.
As mentioned earlier, the nsP2 papain-like protease appears to regulate SIN RNA synthesis, including minus-strand shutoff, and alterations in P123 processing could affect the ability of SIN replicons to establish and maintain persistent replication in dividing cells. To examine the effect of the S1 and S24 adaptive mutations and the engineered nsP2 residue 726 substitutions on P123 processing, TSG/PAC RNA and RNAs containing each mutation were translated in rabbit reticulocyte lysates. These processing patterns were also compared to that for a TSG/PAC derivative containing an Asp substitution for Asn-614 (123N>D), which leads to hyperprocessing and is lethal for SIN replication (48). Figure 7D shows a summary of the values obtained from three independent experiments. Remarkably, we observed a general correlation between P123 hyperprocessing (Fig. 7D), decreased RNA accumulation (Fig. 7A and B), and the ability to establish long-term noncytopathic replication.
Viral phenotypes of nsP2-726 and nsP2-779K substitution mutants.
To study the effect of the nsP2 substitutions in the context of virus, each substitution was incorporated into an infectious SIN cDNA clone (Toto1101) (35). Electroporated BHK cells were observed for signs of CPE, viability by trypan blue exclusion, and production of plaque-forming virus. The parental virus, Toto1101, replicated to high titers and caused dramatic CPE by 24 h posttransfection. No viable cells remained by 60 h (Fig. 8A and data not shown). Similar results were obtained for the nsP2-726 Ala and Val substitutions, which had exhibited 100 and 30% of wild-type RNA replication levels, respectively, in the context of the replicon (Fig. 7B and C). These two mutants, as seen for the wild-type parent, produced plaques on both BHK cells and CEF.
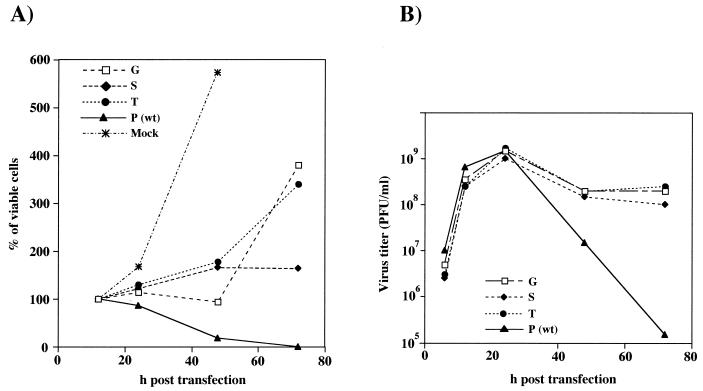
FIG. 8.
Viral phenotypes of selected nsP2-726 mutants. BHK cells were transfected with pToto1101 (wild type [wt]; 726P), pToto/nsP2-726G, pToto/nsP2-726S, or pToto/nsP2-726T transcripts or mock transfected with no RNA (Mock). At the indicated times posttransfection, the numbers of viable adherent cells (A) and titers of infectious virus in the culture supernatants (B) were determined. Viable cells were determined by trypan blue exclusion. Infectious virus titers were determined on CEF monolayers, where CPE is observed for all of these variants. Values at 24, 48, and 72 h are expressed as the percentage of viable cells present at 12 h posttransfection prior to the onset of SIN-induced CPE (set at 100%). At 12 h, the number of viable cells for the five transfected samples was 3.2 × 106 ± 0.7 × 106.
For the nsP2-726 Gly, Ser (SIN-1), and Thr substitutions, partial survival of the transfected BHK cells was observed, with viable cell counts maintained and even increasing over 48 to 72 h (Fig. 8A). As assayed by plaque formation on CEF, wild-type levels of virus were produced by these mutants (Fig. 8B); however, less-distinct small plaques were formed on BHK monolayers. As seen previously for the SIN-1 Ser substitution (9), the Gly and Thr substitution mutants readily established persistently infected BHK cell cultures, although viral titers declined after 1 to 2 weeks (data not shown). These three mutations had all exhibited lower replicon RNA replication levels (17, 10, and 5% of the wild-type level, respectively).
For the remaining group of mutants with significantly impaired RNA replication (<2% of wild type; nsP2-726 Phe, Tyr, Leu, Arg, and Gln and nsP2-779 Lys), transfected cells survived somewhat longer but eventually succumbed to complete CPE. Plaque-forming virus was recovered from transfection supernatants, suggesting that cytopathic revertants or pseudorevertants had arisen. In the case of the Leu substitution, this was characterized further. Immunostaining for SIN structural proteins early after transfection demonstrated that productive replication initiated in a majority (70 to 80%) of transfected BHK cells. In a parallel infectious-center assay, the Leu mutant produced plaques but at a frequency that was 2 orders of magnitude lower than that of parental transcripts (2 × 104 versus 3 × 106 PFU/μg). Viruses from 12 independent plaques exhibited differences in growth rate, plaque size, cytopathogenicity, and host range (data not shown). Sequence analysis revealed that 11 of these isolates were pseudorevertants and retained the Leu substitution at position 726; 1 isolate was a true revertant with the wild-type Pro residue at this position.
DISCUSSION
In theory, a spectrum of possible mutations in a cytopathic SIN replicon could lead to persistent noncytopathic replication in vertebrate cells. For instance, mutations which result in diminished accumulation of toxic viral components might allow survival of the host cell and replicon. Such mutations might exert their effect by decreasing the synthesis and/or increasing the turnover rates of toxic viral components. Mutations which generally downregulate the efficiency of RNA replication (this could be at the translational level or via direct effects of mutations on replicase function) would be expected to fall in the former class. Alternatively, mutations may occur in the toxic components themselves such that high levels of SIN-specific RNA synthesis and accumulation can occur without toxic effects on the vertebrate host cell.
In this study, expression of a selectable drug resistance marker (PAC) by a normally cytopathic RNA replicon has allowed us to identify mutations which enable persistent noncytopathic RNA replication in BHK cells. Presumably, adaptive mutations were selected from a population of variants generated either by in vitro transcription with SP6 RNA polymerase or very early during SIN RNA amplification. Two independent adaptive mutations mapped to nonstructural protein nsP2, which is a bifunctional replicase component consisting of a predicted N-terminal RNA helicase domain and a C-terminal papain-like protease domain (reviewed in reference 50). Previously characterized mutations in nsP2 have implicated this protein as a critical RNA replicase component (16) which can modulate shutoff of negative-strand RNA synthesis (6, 40) and transcription of subgenomic mRNA (16, 39, 51). A substantial fraction of alphavirus nsP2 also translocates to the nucleus and nucleolus (32) and may play a role in inhibiting host DNA replication (36).
Both of the adaptive mutations, 726Leu (S1) and 779Lys (S24), are located downstream of the canonical nsP2 papain-like protease domain, in a region of unknown function. Although nuclear localization of these mutant nsP2s has not been examined, neither mutation disrupts the SIN sequence (PRKRI, beginning at position 656) homologous to the nuclear localization signal determined for Semliki Forest virus (37). A common phenotype shared by replicons containing either of these two mutations was the dramatic downregulation of SIN RNA accumulation relative to that for the parental replicon. In the case of the nsP2-726 Leu substitution, genomic RNA accumulated to only about 1% of the parental level. Further mutagenesis of nsP2 residue 726 revealed a strong correlation between diminished SIN RNA replication and loss of cytopathogenicity. Amino acid substitutions allowing replication to levels greater than 5% of the parental level gave rise to replicons that were cytotoxic; those with replication levels below 2% were able to establish noncytopathic replication in BHK cells and stable Purr cell populations. The simplest explanation for this correlation is that depressed SIN RNA replication leads to sublethal concentrations of toxic viral effectors responsible for translational shutoff and cell death. Multiple and redundant viral effectors are likely responsible for triggering these deleterious effects in vertebrate cells. Previous studies have implicated the alphavirus structural proteins in shutoff of host mRNA translation (56), induction of apoptosis, and acceleration of CPE (12, 17, 55). However, both translational shutoff and cell death (albeit delayed) occur in the context of SIN RNA replicons lacking the structural genes (12). Thus, the nsPs or viral RNA must also function as effectors of vertebrate cell cytotoxicity. The possible role of nsP2 in inhibition of host DNA synthesis has been mentioned. In addition, activation of the double-stranded-RNA-dependent protein kinase PKR and phosphorylation of its substrate eIF-2 alpha can mediate apoptosis (reference 46 and references therein). This raises the possibility that accumulation of high levels of SIN RNA replicative intermediates could directly trigger cell death. Thus, the most likely adaptive mutations may be those which globally downregulate SIN replication to levels which do not trigger deleterious effects on the host cell and yet still allow sufficient levels of the dominant selectable marker to be expressed.
From the standpoint of the current model for regulated synthesis of SIN-specific RNAs in vertebrate cells (27, 45, 50), maintaining persistent replication presents a dilemma. SIN-specific negative-strand RNA synthesis normally shuts off at 3 to 4 h postinfection when a plateau of about 104 molecules/cell has been reached. These negative-strand RNAs continue to serve as templates for continued genomic and subgenomic RNA production. As mentioned earlier, this shutoff is regulated by the activity of the nsP2 protease domain, which accumulates during replication to levels which rapidly process NS polyproteins to mature nsPs, which are then inactive for initiation of negative-strand RNA synthesis. An interesting correlation among mutations allowing noncytopathic replication is that all exhibited a hyperprocessing phenotype. It is remarkable that such a diverse set of substitutions in nsP2, downstream of the protease domain, should have this effect, and it seems unlikely that all directly affect the catalytic activity of the protease. Rather, these substitutions may simply alter the conformations of the P123 and P1234 polyproteins in a way that facilitates interaction of the protease domain with the 1/2 and 2/3 trans-processing sites. In any case, the regulatory model predicts that hyperprocessing mutations would shut off or downregulate negative-strand RNA synthesis earlier, leading to fewer active replication complexes per cell. This could explain the low levels of RNA replication observed for the noncytopathic mutants. However, early shutoff of negative-strand RNA synthesis would not lead to persistent replication. It is possible that diminished replication of the mutants results in low viral protease accumulation that is insufficient to completely shut off negative-strand RNA synthesis. Alternatively, dilution of the replication complexes and protease associated with cell growth and division may lead to conditions where negative-strand RNA synthesis resumes, thus allowing the SIN replicons to persist indefinitely. Finally, mutations in nsP2 (and nsP4) which allow continued negative-strand RNA synthesis by processed nsPs have been identified (6, 40), and it is possible that the adaptive nsP2 mutations function similarly. In future studies, it will be interesting to test these possibilities by using the vaccinia virus trans-complementation system for SIN replication (25–27) and by analyzing the dynamics of positive- and negative-strand RNA synthesis in synchronized cell cultures transfected with the adapted replicons.
We also studied the phenotypes of the various nsP2 mutations in the context of infectious SIN and noted some interesting differences. A previously identified SIN-1 mutation, Ser at nsP2 position 726, facilitates the establishment of persistently infected BHK cells, whereas cells transfected with a replicon containing this substitution exhibit prolonged survival but the majority eventually die (reference 9 and this study). Similar phenotypes were observed for the nsP2-726 Gly and Thr substitutions. All three of these mutations depressed SIN replication to similar levels (5 to 20% of the parental level as measured in the context of the replicon). These mutants demonstrate that the SIN structural region (at the level of either the RNA or expressed proteins) can have a positive effect on cell survival. One explanation is that productive encapsidation of genome RNA downregulates the production of toxic nsPs or sequesters viral RNA. In this model, it would be interesting to see if only the capsid protein is required for this effect. The nsP2 mutations leading to even lower levels of RNA accumulation (<2%) were unstable in the context of infectious virus. As shown for the nsP2-726 Leu substitution (and presumably the other mutants as well), cytopathic revertants and pseudorevertants quickly arose. Such variants were expected to emerge and rapidly take over the population if they produced higher levels of RNA replication and progeny virus and given the inability of the noncytopathic replicons to establish superinfection exclusion (data not shown). The fact that cytopathic variants arose for all of the highly impaired mutants reinforces the idea that cytopathogenicity is positively correlated with SIN replication efficiency, an observation which also holds for clonal mosquito cell lines which differ in their susceptibility to alphavirus-induced cytopathogenicity (47, 53).
This work also has implications for alphavirus vectors, which are beginning to be widely used for expression of heterologous RNAs and proteins in cell culture and in vivo (11, 30, 54). Although shutoff of host translation and high-level protein production can be advantageous for some applications, these properties preclude long-term studies or those requiring minimal perturbation of host cell biology. Some of the adaptive mutations identified in this report have now been incorporated into a new generation of SIN vectors useful for long-term expression in BHK, CHO, and Vero cell cultures (1a). Interestingly, the adaptive changes selected in BHK cells show a limited host range. The reasons for this are unclear, but they may relate to inherent differences in SIN RNA replication efficiency in different cellular environments or cell-specific differences in interferon production or responsiveness (which might limit or cure persistent cytoplasmic RNA replication). Nonetheless, the strategy employed here should be useful for identifying adaptive mutations in other cellular environments, including human cells, and can be extended to other viral vectors where cytopathogenicity is a concern.
In general, the selection and study of adaptive mutations should provide important mechanistic insights into the virus-host interactions that determine whether an infected cell survives or dies.
ACKNOWLEDGMENTS
I.F., E.A., T.A.H., and B.M.P. contributed equally to this work.
We thank Rebecca Moran and Carol Read for expert technical assistance and Peter Bredenbeek, Kaveh Ashrafi, and Elena Frolova for their participation in various stages of this project. We are also grateful to many colleagues for helpful discussions during the course of this work and to Brett Lindenbach for critical reading of the manuscript.
This work was supported by grants from the Public Health Service (AI24134 and AI11377).
REFERENCES
- 1.Agapov, E., and I. Frolov. Unpublished data.
- 1a.Agapov E V, Frolov I, Lindenbach B D, Praǵai B M, Schlesinger S, Rice C M. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc Natl Acad Sci USA. 1998;95:12989–12994. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1993. [Google Scholar]
- 3.Bredenbeek P J, Frolov I, Rice C M, Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bredenbeek P J, Rice C M. Animal RNA virus expression systems. Semin Virol. 1992;3:297–310. [Google Scholar]
- 5.Condreay L D, Brown D T. Exclusion of superinfecting homologous virus by Sindbis virus-infected Aedes albopictus (mosquito) cells. J Virol. 1986;58:81–86. doi: 10.1128/jvi.58.1.81-86.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dé I, Sawicki S G, Sawicki D L. Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein. J Virol. 1996;70:2706–2719. doi: 10.1128/jvi.70.5.2706-2719.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Groot R J, Hardy W R, Shirako Y, Strauss J H. Cleavage-site preferences of Sindbis virus polyproteins containing the nonstructural proteinase: evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990;9:2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Luna S, Soria I, Pulido D, Ortín J, Jiménez A. Efficient transformation of mammalian cells with constructs containing a puromycin-resistance marker. Gene. 1988;62:121–126. doi: 10.1016/0378-1119(88)90585-9. [DOI] [PubMed] [Google Scholar]
- 9.Dryga S A, Dryga O A, Schlesinger S. Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: the importance of a mutation in the nsP2 gene. Virology. 1997;228:72–83. doi: 10.1006/viro.1996.8364. [DOI] [PubMed] [Google Scholar]
- 10.Faragher S G, Meek A D, Rice C M, Dalgarno L. Genome sequences of a mouse-avirulent and a mouse-virulent strain of Ross River virus. Virology. 1988;163:509–526. doi: 10.1016/0042-6822(88)90292-9. [DOI] [PubMed] [Google Scholar]
- 10a.Frolov, I. Unpublished data.
- 11.Frolov I, Hoffman T A, Prágai B M, Dryga S A, Huang H V, Schlesinger S, Rice C M. Alphavirus-based expression systems: strategies and applications. Proc Natl Acad Sci USA. 1996;93:11371–11377. doi: 10.1073/pnas.93.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolov I, Schlesinger S. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J Virol. 1994;68:1721–1727. doi: 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grakoui A, Levis R, Raju R, Huang H V, Rice C M. A cis-acting mutation in the Sindbis virus junction region which affects subgenomic RNA synthesis. J Virol. 1989;63:5216–5227. doi: 10.1128/jvi.63.12.5216-5227.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin D E, Hardwick J M. Regulators of apoptosis on the road to persistent alphavirus infection. Annu Rev Microbiol. 1997;51:565–592. doi: 10.1146/annurev.micro.51.1.565. [DOI] [PubMed] [Google Scholar]
- 15.Hahn C S, Hahn Y S, Braciale T J, Rice C M. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc Natl Acad Sci USA. 1992;89:2679–2683. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Hahn, C. S., and C. M. Rice. Unpublished data.
- 16.Hahn Y S, Strauss E G, Strauss J H. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J Virol. 1989;63:3142–3150. doi: 10.1128/jvi.63.7.3142-3150.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joe A K, Foo H H, Kleeman L, Levine B. The transmembrane domains of Sindbis virus envelope glycoprotein induce cell death. J Virol. 1998;72:3935–3943. doi: 10.1128/jvi.72.5.3935-3943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston R E, Peters C J. Alphaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 843–898. [Google Scholar]
- 19.Kääriäinen L, Ranki M. Inhibition of cell functions by RNA virus infections. Annu Rev Microbiol. 1984;38:91–109. doi: 10.1146/annurev.mi.38.100184.000515. [DOI] [PubMed] [Google Scholar]
- 20.Kinney R M, Johnson B J B, Welch J B, Tsuchiya K R, Trent D W. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology. 1989;170:19–30. doi: 10.1016/0042-6822(89)90347-4. [DOI] [PubMed] [Google Scholar]
- 21.Lacalle R A, Pulido D, Vara J, Zalacaín M, Jimenéz A. Molecular analysis of the pac gene encoding a puromycin N-acetyl transferase from Streptomyces alboniger. Gene. 1989;79:375–380. doi: 10.1016/0378-1119(89)90220-5. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lemm J A, Bergqvist A, Read C M, Rice C M. Tempate-dependent initiation of Sindbis virus replication in vitro. J Virol. 1998;72:6546–6553. doi: 10.1128/jvi.72.8.6546-6553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemm J A, Durbin R K, Stollar V, Rice C M. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J Virol. 1990;64:3001–3011. doi: 10.1128/jvi.64.6.3001-3011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemm J A, Rice C M. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J Virol. 1993;67:1905–1915. doi: 10.1128/jvi.67.4.1905-1915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemm J A, Rice C M. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J Virol. 1993;67:1916–1926. doi: 10.1128/jvi.67.4.1916-1926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemm J A, Rümenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: A model for the temporal regulation of minus and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levinson R S, Strauss J H, Strauss E G. Complete nucleotide sequence of the genomic RNA of O’nyong-nyong and its use in the construction of alphavirus phylogenetic trees. Virology. 1990;175:110–123. doi: 10.1016/0042-6822(90)90191-s. [DOI] [PubMed] [Google Scholar]
- 29.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.London, S. D., and C. M. Rice. Unpublished data.
- 30.Lundstrom K. Alphaviruses as expression vectors. Curr Opin Biotechnol. 1997;8:578–582. doi: 10.1016/s0958-1669(97)80032-8. [DOI] [PubMed] [Google Scholar]
- 30a.Majors, J. Unpublished data.
- 31.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peränen J, Rikkonen M, Liljeström P, Käariänen L. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J Virol. 1990;64:1888–1896. doi: 10.1128/jvi.64.5.1888-1896.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce J S, Strauss E G, Strauss J H. Effect of ionic strength on the binding of Sindbis virus to chick cells. J Virol. 1974;13:1030–1036. doi: 10.1128/jvi.13.5.1030-1036.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Prágai, B. M., and C. M. Rice. Unpublished data.
- 33b.Rice, C. M. Unpublished data.
- 34.Rice C M, Grakoui A, Galler R, Chambers T J. Transcription of infectious yellow fever virus RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1989;1:285–296. [PubMed] [Google Scholar]
- 35.Rice C M, Levis R, Strauss J H, Huang H V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rikkonen M. Functional significance of the nuclear-targeting and NTP-binding motifs of Semliki Forest virus nonstructural protein nsP2. Virology. 1996;218:352–361. doi: 10.1006/viro.1996.0204. [DOI] [PubMed] [Google Scholar]
- 37.Rikkonen M, Peranen J, Kaariainen L. Nuclear and nucleolar targeting signals of Semliki Forest virus nonstructural protein nsP2. Virology. 1992;189:462–473. doi: 10.1016/0042-6822(92)90570-f. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Sawicki D L, Sawicki S G. Functional analysis of the A complementation group mutants of Sindbis HR virus. Virology. 1985;144:20–34. doi: 10.1016/0042-6822(85)90301-0. [DOI] [PubMed] [Google Scholar]
- 40.Sawicki D L, Sawicki S G. A second nonstructural protein functions in the regulation of alphavirus negative-strand RNA synthesis. J Virol. 1993;67:3605–3610. doi: 10.1128/jvi.67.6.3605-3610.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawicki D L, Sawicki S G, Keränen S, Kääriäinen L. Specific Sindbis virus coded functions for minus-strand RNA synthesis. J Virol. 1981;39:348–358. doi: 10.1128/jvi.39.2.348-358.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawicki S G, Sawicki D L, Kääriäinen L, Keränen S. A Sindbis virus mutant temperature-sensitive in the regulation of minus-strand RNA synthesis. Virology. 1981;115:161–172. doi: 10.1016/0042-6822(81)90098-2. [DOI] [PubMed] [Google Scholar]
- 43.Shirako Y, Niklasson B, Dalrymple J M, Strauss E G, Strauss J H. Structure of the Ockelbo virus genome and its relationship to other Sindbis viruses. Virology. 1991;182:753–764. doi: 10.1016/0042-6822(91)90616-j. [DOI] [PubMed] [Google Scholar]
- 44.Shirako Y, Strauss J H. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology. 1990;177:54–64. doi: 10.1016/0042-6822(90)90459-5. [DOI] [PubMed] [Google Scholar]
- 45.Shirako Y, Strauss J H. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srivastava S P, Kumar K U, Kaufman R J. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J Biol Chem. 1998;273:2416–2423. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- 47.Stollar V. Togaviruses in cultured arthropod cells. In: Schlesinger R W, editor. The togaviruses—biology, structure, replication. New York, N.Y: Academic Press, Inc.; 1980. pp. 584–621. [Google Scholar]
- 48.Strauss E G, deGroot R J, Levinson R, Strauss J H. Identification of the active site residues in the nsP2 proteinase of Sindbis virus. Virology. 1992;191:932–940. doi: 10.1016/0042-6822(92)90268-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strauss E G, Rice C M, Strauss J H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984;133:92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- 50.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suopanki J, Sawicki D L, Sawicki S G, Kääriäinen L. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J Gen Virol. 1998;79:309–319. doi: 10.1099/0022-1317-79-2-309. [DOI] [PubMed] [Google Scholar]
- 52.Takkinen K. Complete nucleotide sequence of the non-structural protein genes of Semliki Forest virus. Nucleic Acids Res. 1986;14:5667–5682. doi: 10.1093/nar/14.14.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tooker P, Kennedy S I. Semliki Forest virus multiplication in clones of Aedes albopictus cells. J Virol. 1981;37:589–600. doi: 10.1128/jvi.37.2.589-600.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tubulekas I, Berglund P, Fleeton M, Liljeström P. Alphavirus expression vectors and their use as recombinant vaccines: a minireview. Gene. 1997;190:191–195. doi: 10.1016/s0378-1119(96)00679-8. [DOI] [PubMed] [Google Scholar]
- 55.Ubol S, Tucker P C, Griffin D E, Hardwick J M. Neurovirulent strains of Alphavirus induce apoptosis in bcl-2-expressing cells: role of a single amino acid change in the E2 glycoprotein. Proc Natl Acad Sci USA. 1994;91:5202–5206. doi: 10.1073/pnas.91.11.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Steeg H, Kasperaitis M, Voorma H O, Benne R. Infection of neuroblastoma cells by Semliki Forest virus: the interference of viral capsid protein with the binding of host messenger RNAs into initiation complexes is the cause of the shut-off of host protein synthesis. Eur J Biochem. 1984;138:473–478. doi: 10.1111/j.1432-1033.1984.tb07940.x. [DOI] [PubMed] [Google Scholar]
- 57.Vara J A, Portela A, Ortín J, Jiménez A. Expression in mammalian cells of a gene from Streptomyces alboniger conferring puromycin resistance. Nucleic Acids Res. 1986;14:4617–4624. doi: 10.1093/nar/14.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volchkov V E, Volchkova V A, Netesov S V. Complete nucleotide sequence of the Eastern equine encephalomyelitis virus genome. Mol Genet Mikrobiol Virusol. 1991;5:8–15. [PubMed] [Google Scholar]
- 59.Weiss B, Rosenthal R, Schlesinger S. Establishment and maintenance of persistent infection by Sindbis virus in BHK cells. J Virol. 1980;33:463–474. doi: 10.1128/jvi.33.1.463-474.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]