Abstract
Hearing impairment (HI) is one of the most common sensory disabilities with exceptionally high genetic heterogeneity. Of genetic HI cases, 30% are syndromic and 70% are nonsyndromic. For nonsyndromic (NS) HI, 77% of the cases are due to autosomal recessive (AR) inheritance. ARNSHI is usually congenital/prelingual, severe-to-profound, affects all frequencies and is not progressive. Thus far, 73 ARNSHI genes have been identified. Populations with high rates of consanguinity have been crucial in the identification of ARNSHI genes, and 92% (67/73) of these genes were identified in consanguineous families. Recent changes in genomic technologies and analyses have allowed a shift towards ARNSHI gene discovery in outbred populations. The latter is crucial towards understanding the genetic architecture of ARNSHI in diverse and understudied populations. We present an overview of the 73 ARNSHI genes, the methods used to identify them, including next generation sequencing which revolutionized the field, and new technologies that show great promise in advancing ARNSHI discoveries.
Keywords: Hearing impairment; consanguinity; autosomal recessive, gene mapping; next generation sequencing
Introduction
Hearing impairment (HI) is a highly heterogeneous and common sensory disorder (Vona et al. 2020). The three major types of HI are conductive, sensorineural, and mixed (both conductive and sensorineural). Conductive HI is due to reduced ability of the external ear, middle ear, or both, to conduct sound, whereas sensorineural HI can be due to cochlear dysfunction, damage to stereocilia, or problems associated with vestibulocochlear nerve transmission to and from the brain and inner ear.
Congenital HI occurs in 1–2 per 1000 newborns globally (Vos et al. 2019; Vona et al. 2020). More than half of newborns who fail hearing screening have no identifiable risk factors and are presumed to have genetic HI (Vos et al. 2019). Of the genetic HI cases, 30% are syndromic and 70% are nonsyndromic (NS). For NSHI the modes of inheritance are autosomal recessive (AR) (~77%); autosomal dominant (AD) (~22%); X-linked (~1%); and mitochondrial (<1%) (Irshad et al. 2005). ARNSHI is usually sensorineural, prelingual/congenital, severe to profound, affects all frequencies, and non-progressive. While ADNSHI is usually progressive, post-lingual, mild to profound, often affecting the middle to high frequencies. To date, >120 genes have been identified for NSHI (Adadey et al. 2020) with the majority of the genes implicated in ARNSHI. The vast majority of ARNSHI genes were localized and identified through the study of consanguineous pedigrees.
Characteristics of ARNSHI in Consanguineous and Outbred Populations
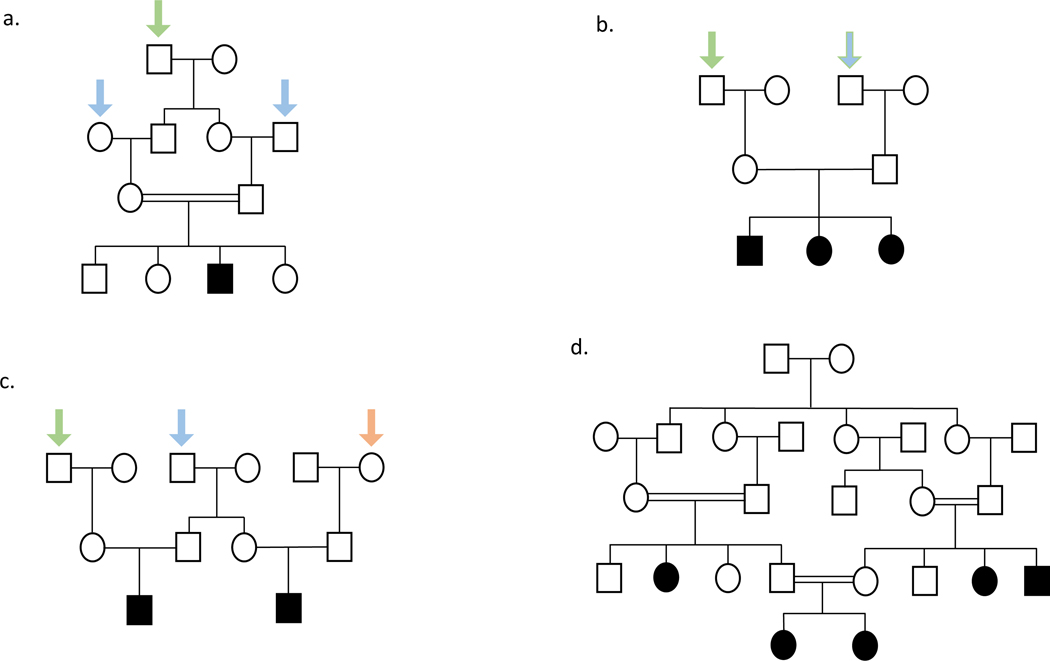
For ARNSHI, both parents are expected to be carriers of causal variants. Their hearing impaired children can either be homozygous or compound heterozygous depending on whether they inherit the same or different causal variants from each parent. When both parents are carriers of causal ARNSHI variants in the same gene, on average, ¼ of their children will be hearing impaired and ½ a causal variant carrier. Children of parents who are either carriers of causal variants or have HI due to different ARNSHI genes do not have an increased risk of being hearing impaired. Offspring of two deaf parents that carry causal variants in the same gene will all have HI. For consanguineous pedigrees, a rare causal HI variant is more likely enter the pedigree once, for example, for a pedigree segregating ARNSHI where the parents of the affected children are first cousins the causal variant is more prone to enter the pedigree through one of their two shared grandparents than twice through both the maternal and paternal lineages (Figure 1a). Therefore, for consanguineous pedigrees, it is usual to observe causal homozygous variants. In contrast, for an outbred pedigree the causal variant will enter the pedigree twice through both a maternal and paternal grandparent (Figure 1b). Although for outbred pedigrees usually compound heterozygous variants are observed, homozygous causal variants can be detected particularly those that are more frequent within a population, for example, GJB2 p.Gly12fs which has an allele frequency of 1% in non-Finnish Europeans (Genome Aggregation Database Consortium et al. 2020).
Figure 1. ARNSHI in consanguineous and outbred pedigrees.
Solid symbols represent affected individuals, clear symbols represent unaffected individuals. Squares are males and circles are females. Two parallel lines between parents indicates consanguinity. (a). Pedigree of a consanguineous family showing entrance of a causal allele once from a shared great-grandparent (indicated by a green arrow). A variant could enter through either the shared great-grandparent, but one great-grandparent was selected for the purpose of illustration, which is more likely than the variant entering twice from the maternal and paternal lineages of the carrier parents (indicated by blue arrows). (b) An outbred pedigree with a single affected child where the causal variant enters the pedigree twice through both a maternal and paternal grandparent (green arrows). The variant entering through one random grandparent is shown as an example, however entrance of the variant could be from either grandparent. (c). An outbred pedigree with two branches with affected children showing that ARNSHI variants must enter three times (blue, green, and orange arrows). The variants could have also entered through the spouses of the individuals shown with arrows. (d). Pedigree showing several consanguineous matings within a pedigree which can lead to multiple branches with affected children.
Outbred pedigrees segregating ARNSHI are usually nuclear. Since to observe more than one branch with hearing impaired members, causal variants would have to be introduced more than twice to the pedigree, for example, to observe cousins from an outbred pedigree with ARNSHI the sibling carrier parents would have to have children with spouses that are also causal variant carriers (Figure 1c). For families that hail from populations where consanguinity is a common practice it is not unusual to have several consanguineous matings within a single pedigree and multiple branches of the family with affected children (Figure 1d). Additionally, some populations where consanguinity is practiced also tend to have large families, which also impacts the ability to successfully identify the causal variant.
Early methods of ARNSHI gene identification: Positional cloning via linkage analysis and homozygosity mapping
The first methods used to identify underlying genes in Mendelian diseases included positional and functional cloning (Figure 2). Functional cloning relies on knowledge of a dysfunctional protein associated with disease, for example discovered via biochemical assays. For HI, the latter technique has only been successful in the study of deaf mice (Wang et al. 1998; Smith and Van Camp 1999). Conversely, positional cloning is a technique that focuses on the localization of the disease gene along the chromosome, without any prior knowledge needed on the gene’s product or function. Positional cloning has been the most successful technique used to identify novel HI genes in humans, including ARNSHI, especially before the availability of next generation sequencing (NGS) (Friedman et al. 1995; Chaib et al. 1996; Wang et al. 1998; Yasunaga et al. 1999). In the process of positional cloning, early studies would generate a genetic map via genotyping panels of short tandem repeat polymorphic (STRP) (aka microsatellite) markers and the application of statistical methods (Collin et al. 2008) and later, when they became available, microarray panels of single nucleotide polymorphism (SNP) markers (Basit et al. 2011). For ARNSHI, two approaches were then mainly used to map genomic loci linked to disease, i.e., linkage analysis and homozygosity mapping.
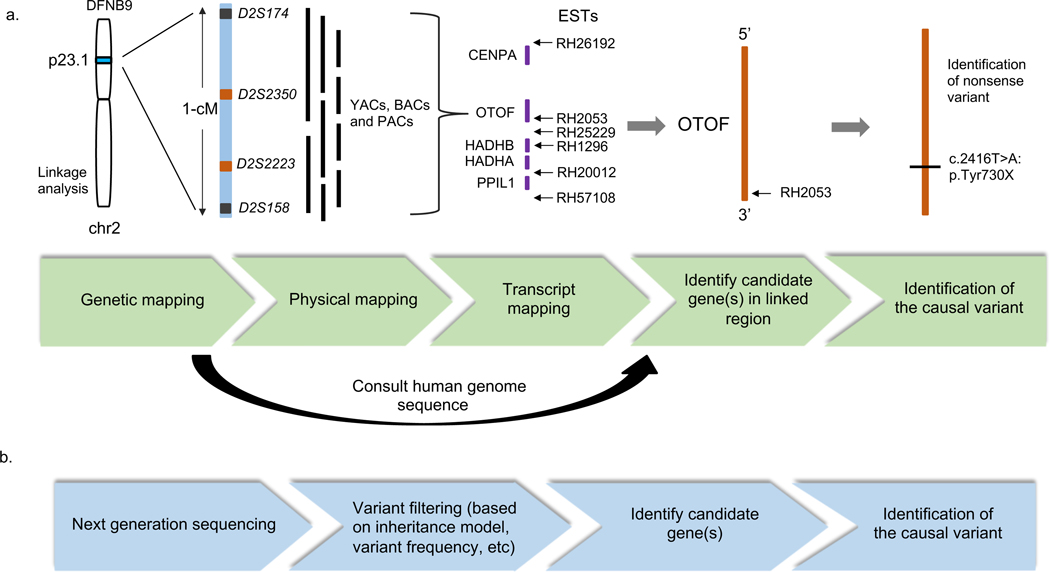
Figure 2. A comparison of traditional positional cloning and NGS approaches to ARNSHI gene identification methods.
(a). Traditional positional cloning with the identification of OTOF shown as an example. In the identification of OTOF, a small region of chromosome 2 was mapped via linkage analysis and a physical map was constructed using yeast artificial chromosomes (YACs), bacterial artificial chromosomes (BACs) and P1 phage artificial chromosomes (PACs). Following transcript mapping of all genes and expressed sequenced tags (ESTs) in the region, candidate gene OTOF was identified. Further sequencing analysis of OTOF revealed the causal variant. (b). Recent approaches evaluate next generation sequencing data via variant filtering approaches based on plausible inheritance model, variant frequency etc., to identify causal genes and variants.
Linkage analysis to identify ARNSHI loci
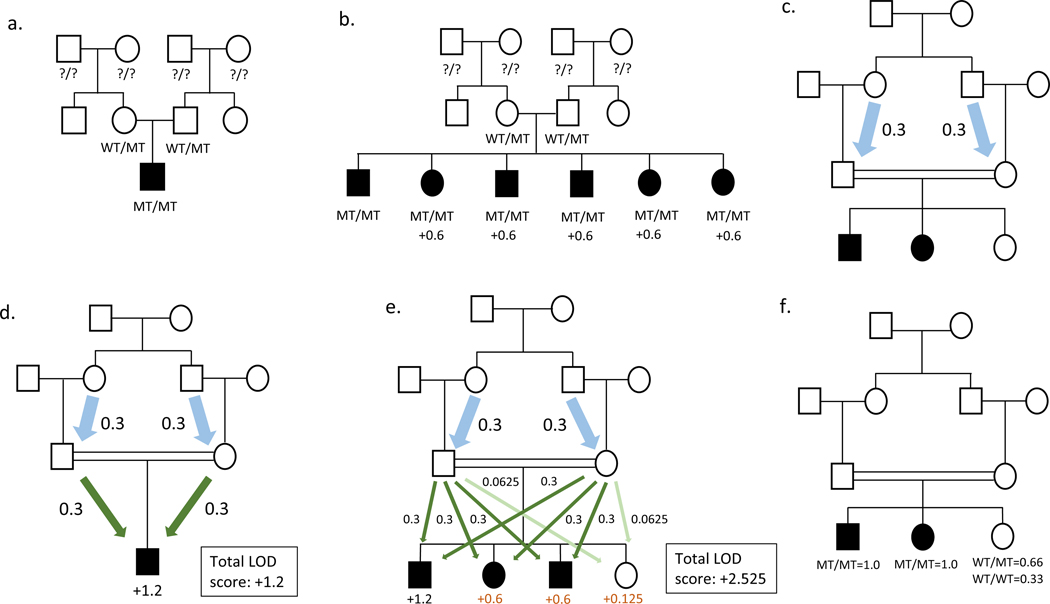
In traditional parametric linkage analysis, genetic maps of STRPs or SNPs, together with information on mode of inheritance, penetrance, and allele frequencies are used to localize Mendelian disease loci. This technique is based on the fact that genetic variants which are physically close on a chromosome segregate together during meiosis. Therefore, a genomic locus containing a causal variant can be statistically linked to disease by interrogating nearby markers, and the strength of evidence for linkage (or lack of) can be evaluated through the estimation of a logarithm of the odds (LOD) score. Due to high levels of locus heterogeneity for NSHI, the study of multiple families will not lead to significant results, even when methods that allow for heterogeneity are applied (Ott 1983). Therefore, it was important to be able to analyze families that can independently establish linkage, i.e., LOD score ≥3.0, (Morton 1955) which was later revised to a LOD score ≥3.3 (Lander and Kruglyak 1995). For outbred pedigrees it is nearly impossible to reach the significance threshold using a single pedigree. They are unphased, it is impossible to determine from which two grandparents the carrier parents received a causal variant. Therefore, the first affected child provides no linkage information and families with only a single hearing impaired child are uninformative (Figure 3a). For 1) a rare causal variant; 2) a marker in perfect linkage disequilibrium (LD) (Θ=0) with the causal variant; or 3) multiple markers that form a rare haplotype that tags the causal variant, each additional affected child adds 0.6 to the LOD score and each additional unaffected child 0.125. Therefore, for example, a nuclear outbred pedigree would need to have six affected children available for study to obtain a LOD score of 3.0 (Figure 3b), an event which is unlikely to be observed. In contrast, phase information is available for a consanguineous pedigree, since for a rare causal variant, the probability is higher that it entered the pedigree once, for example, through one of the great-grandparents for a first-cousin union (Figure 1a) or through the great-great-grandparents for a second-cousin union, then through two founder pedigree members. Since phase information is available, not only is there linkage information for the first affected child that is obtained from the meiosis received from the mother and father, but in the case of a first cousin union the meiosis from the grandparents to the parents are also informative (Figure 3c). Therefore, when analysis is performed either directly using 1) the rare causal variant; 2) a marker in perfect LD with the causal variant; or 3) multiple markers that form a rare haplotype by tagging the causal variant, a consanguineous pedigree with a single affected individual who is the offspring of a first-cousin mating can provide a LOD score of 1.2 (Figure 3d) and if the child is offspring of a second-cousin union the pedigree can provide a LOD score of 1.8. It can be observed that each informative meiosis for this situation provides a LOD score of 0.3. For either of these pedigree structures each affected child adds 0.6 and each unaffected child adds 0.125 to the LOD score (Figure 3e). The reason why an unaffected child provides so little linkage information compared to an affected child is that it cannot be determined whether or not they are a disease variant carrier; LOD score calculation must be made using 2/3 probability that they are a causal variant carrier and 1/3 probability that they are homozygous wildtype. In contrast, for an affected child the probability is 1.0 that they are carriers of two causal variants (Figure 3f) (Ott et al. 2015). Allele frequencies will also impact the LOD scores with the LOD decreasing with increasing allele frequencies, because as the causal variant allele frequency increases so too does the probability that it entered the pedigree more than once (i.e., through two founders) with more possibilities for the variant to enter a second time when consanguineous parents are more distantly related. It should be noted that consanguineous pedigrees will not provide any additional information when they segregate Mendelian traits with modes of inheritance other than AR.
Figure 3. Linkage analysis in ARNSHI pedigrees.
Solid symbols represent affected individuals, clear symbols represent unaffected individuals. Squares are males and circles are females. A double line between parents indicates consanguinity. (a). An outbred pedigree with a single affected child unphased and uninformative for linkage as it is unknown which grandparent is carrier of the variant. (b). A nuclear outbred pedigree showing affected children with carrier parents. In this case the first affected individual is uninformative, however each additional affected sibling can contribute a maximum of 0.6 to the LOD score, and 6 affected children are necessary to obtain a LOD score of 3.0. (c) Example of a pedigree providing information from meioses from the grandparents to the parents in a first-cousin union (blue arrows). Each informative meiosis provides a LOD score of 0.3 (d). Pedigree with one affected individual who is the offspring of a first-cousin mating. This pedigree will provide a LOD score of 1.2 due to the informative meioses from the grandparents to the parents (blue arrows) and the meioses from the parents to the affected child (green arrows). Each meiosis adds 0.3 to the LOD score. (e). Each affected child adds 0.6 (green arrows) to the LOD score and each unaffected child adds 0.125 (light green arrows) to the LOD score. (f). Pedigree showing the probability of the causal variant genotype of each child. The probabilities are not influenced by consanguinity.
Homozygosity mapping to identify ARNSHI loci
Another approach which was used to analyze genotype data to map ARNSHI loci is homozygosity mapping, which examines the genome for runs of homozygosity (ROH) (Lander and Botstein 1987). For this method to detect homozygous causal variants they must be surrounded by a ROH, making it an ideal approach to study consanguineous pedigrees. However, it can also be used to study outbred families. In the latter case, detection of homozygous regions has been successful when the parents are cryptically distantly related and are carriers of the same causal variant. If a causal variant is homozygous but the parents only share a very distant common ancestor, the ROH can be too small to detect. Homozygosity mapping can be performed on data obtained from a single affected individual. However, this will usually reveal many regions of homozygosity with a greater number of ROH if the parents are more closely related. The number of ROH can be reduced by analyzing data from multiple affected and unaffected family members. Unlike linkage analysis, most homozygosity mapping (Seelow et al. 2009) methods do not provide statistical evidence of a region containing a causal variant.
Candidate gene and variant identification
After an ARNSHI locus was mapped to a chromosomal location via linkage analysis or homozygosity mapping, or both, the search for the causal variant within this region began (Friedman et al. 1995; Wang et al. 1998). Before the completion or the draft of the Human Genome Project was available, a “physical map” was generated to pinpoint candidate genes within the mapped region. This physical map consists of sets of overlapping DNAs, such as yeast artificial chromosomes (YACs), that span the critical region (Figure 2 and Table 1). This process became obsolete after the completion of the Human Genome Project when genes were mapped to the full human genome. Candidate genes within the region mapped to disease were then selected to undergo sequencing step by step. This was a slow, laborious, and expensive process, since regions were almost always >1Mb and contained many genes, it would often take years to identify an ARNSHI gene.
Table 1: ARNSHI loci and genes.
Overview of all ARNSHI genes, chromosomal position, and their loci and OMIM numbers. For each gene and locus, the country of origin of the families that were used to map and identify the gene along with their consanguinity status. Information is also provided on whether linkage/homozygosity was used to map the locus and, if used, the type of next generation sequencing.
| Gene | Locus | OMIM number | Chromosome Location | Country(s) locus | Country(s) gene | Consanguineous Locus | Consanguineous Gene | L | NGS | Key reference(s)* |
|---|---|---|---|---|---|---|---|---|---|---|
| GJB2 | DFNB1A | 121011 | 13q12.11 | Tunisia | Pakistan | Yes | Yes | L |
Guilford et al. 1994a; Kelsell et al. 1997 |
|
| GJB6 a | DFNB1B | 604418 | 13q12.11 | Spain | Spain | No | No | L | del Castillo et al. 2002 | |
| MYO7A | DFNB2 | 276903 | 11q13.5 | Tunisia | Tunisia/China | Yes | Yes/ No | L | Guilford et al. 1994b; Liu et al. 1997b; Weil et al. 1997b | |
| MYO15A | DFNB3 | 602666 | 17p11.2 | Indonesia | Indonesia/ Indiac | No | No/Yes | L |
Friedman et al. 1995; Wang et al. 1998 |
|
| SLC26A4 | DFNB4 | 605646 | 7q31 | Israel | India | Yes | Yes | L | Baldwin et al. 1995; Li et al. 1998 |
|
| TMIE | DFNB6 | 607237 | 3p14-p21 | India | India/ Pakistan | Yes | Yes/Yes | L | Fukushima et al. 1995; Naz et al. 2002 |
|
| TMC1 | DFNB7 | 606706 | 9q13-q21 | India | India/ Pakistan | Yes | Yes/Yes | L | Jain et al. 1995; Kurima et al. 2002 |
|
| TMPRSS3 | DFNB8 | 605511 | 21q22 | Pakistan | Pakistan/ Palestine | Yes | Yes/Yes | L | Veske et al. 1996; Scott et al. 2001 |
|
| OTOF | DFNB9 | 603681 | 2p22-p23 | Lebanon | Lebanon | Yes | Yes | L |
Chaib et al. 1996a; Yasunaga et al. 1999 |
|
| CDH23 | DFNB12 | 605516 | 10q21-q22 | Syria | India/ Pakistan | Yes | Yes/Yes | L |
Chaib et al. 1996b; Bork et al. 2001 |
|
| GIPC3 | DFNB15 | 608792 | 19p13.3 | India | Netherlands/ India | Yes | Yes/Yes | L | Chen et al. 1997; Charizopoulou et al. 2011 | |
| STRC | DFNB16 | 606440 | 15q21-q22 | Pakistan/ Palestine/ Syria | Pakistan/ France | Yes/Yes/Yes | Yes/Yes | L | Campbell et al. 1997; Verpy et al. 2001 |
|
| USH1C | DFNB18A | 605242 | 11p14–15.1 | India | China | Yes | No | L | Jain et al. 1998; Ouyang et al. 2002 |
|
| OTOG | DFNB18B | 604487 | 11p15.1 | Netherlands/ Spain | Netherlands/ Spain | No/No | No/No | L | Schraders et al. 2012 | |
| TECTA | DFNB21 | 602574 | 11q | Lebanon | Lebanon | Yes | Yes | L | Mustapha et al. 1998a; Mustapha et al. 1998b |
|
| OTOA | DFNB22 | 607038 | 16p12.2 | Palestine | Palestine | Yes | Yes | L | Zwaenepoel et al. 2002 | |
| PCDH15 | DFNB23 | 605514 | 10p11.2-q21 | Pakistan | Pakistan | Yes | Yes | L | Ahmed et al. 2003b | |
| RDX | DFNB24 | 179410 | 11q23 | Pakistan | Pakistan | Yes | Yes | L | Khan et al. 2007 | |
| GRXCR1 | DFNB25 | 613283 | 4p13 | Pakistan/ Netherlands | Pakistan/ Netherlands | Yes/Yes | Yes/Yes | L | Schraders et al. 2010a | |
| GAB1 | DFNB26d | 604439 | 4q31 | Pakistan | Pakistan | Yes | Yes | L | E |
Riazuddin et al. 2000; Yousaf et al. 2018 |
| TRIOBP | DFNB28 | 609761 | 22q13 | Palestine | Palestine | Yes | Yes | L | Shahin et al. 2006 | |
| CLDN14 | DFNB29 | 605608 | 21q22 | Pakistan | Pakistan | Yes | Yes | L | Wilcox et al. 2001 | |
| MYO3A | DFNB30 | 606808 | 10p11.1 | Iraq | Iraq | Yes | Yes | L | Walsh et al. 2002 | |
| WHRN | DFNB31 | 607928 | 9q32-q34 | Palestine | Palestine | Yes | Yes | L | Mustapha et al. 2002; Mburu et al. 2003 |
|
| CDC14A | DFNB32 | 603504 | 1p13.3–22.1 | Tunisia | Iran | Yes | Yes | L | E | Masmoudi et al. 2003; Delmaghani et al. 2016 |
| ESRRB | DFNB35 | 602167 | 14q24.1–24.3 | Pakistan | Turkey | Yes | Yes | L | Ansar et al. 2003; Collin et al. 2008 |
|
| ESPN | DFNB36 | 606351 | 1p36.3 | Pakistan | Pakistan | Yes | Yes | L | Naz et al. 2004 | |
| MYO6 | DFNB37 | 600970 | 6q13 | Pakistan | Pakistan | Yes | Yes | L | Ahmed et al. 2003a | |
| HGF | DFNB39 | 142409 | 7q21.1 | Pakistan | Pakistan/ India | Yes | Yes/Yes | L | Wajid et al. 2003; Schultz et al. 2009 |
|
| ILDR1 | DFNB42 | 609739 | 3q13.31-q22.3 | Pakistan | Pakistan | Yes | Yes | L | Aslam et al. 2005; Borck et al. 2011 |
|
| ADCY1 | DFNB44 | 103072 | 7p14.1-q11.22 | Pakistan | Pakistan | Yes | Yes | L | E | Ansar et al. 2004; Santos-Cortez et al. 2014 |
| CIB2 | DFNB48 | 605564 | 15q23-q25.1 | Pakistan | Pakistan | Yes | Yes | L | Ahmad et al. 2005; Riazuddin et al. 2012 |
|
| MARVELD2 | DFNB49 | 610572 | 5q12.3-q14.1. | Pakistan | Pakistan | Yes | Yes | L | Ramzan et al. 2005; Riazuddin et al. 2006 |
|
| COL11A2 | DFNB53 | 120290 | 6p21.3 | Iran | Iran | Yes | Yes | L | Chen et al. 2005 | |
| PJVK | DFNB59 | 610219 | 2q31.1-q31.3 | Iran | Iran | Yes | Yes | L | Delmaghani et al. 2006 | |
| SLC26A5 | DFNB61d | 604943 | 7q22.1 | USAe | USAe | Yes | Yes | NAf | NAf | Liu et al. 2003 |
| LRTOMT | DFNB63 | 612414 | 11q13.2-q13.4 | Iran | Iran | Yes | Yes | L | Du et al. 2008 | |
| DCDC2 | DFNB66 | 605755 | 6p21.2–22.3 | Tunisia | Tunisia | Yes | Yes | E | Grati et al. 2015 | |
| LHFPL5 | DFNB67 | 609427 | 6p21.31 | Tunisia | Pakistan/ India | Yes | Yes/Yes | L | Tlili et al. 2005; Shabbir et al. 2006 |
|
| S1PR2 | DFNB68 | 605111 | 19p13.2 | Pakistan | Pakistan | Yes | Yes | L | E | Santos et al. 2006; Santos-Cortez et al. 2016 |
| BSND | DFNB73 | 606412 | 1p32.3 | Pakistan | Pakistan | Yes | Yes | L | Riazuddin et al. 2009 | |
| MSRB3 | DFNB74 | 613719 | 12q14.2-q15 | Pakistan | Pakistan | Yes | Yes | L | Waryah et al. 2009; Ahmed et al. 2011 |
|
| SYNE4 | DFNB76 | 615535 | 19q13.12 | Iraq | Iraq | Yes | Yes | L | Horn et al. 2013 | |
| LOXHD1 | DFNB77 | 613072 | 18q12-q21 | Iran | Iran | Yes | Yes | L | Grillet et al. 2009 | |
| TPRN | DFNB79 | 613354 | 9q34.3 | Pakistan | Pakistan | Yes | Yes | L | N |
Khan et al. 2010; Rehman et al. 2010 |
| PTPRQ | DFNB84A | 603317 | 12q21.31 | Palestine | Netherlands/ Moroccan | Yes | No/Yes | L | Schraders et al. 2010b; Shahin et al. 2010 |
|
| OTOGL | DFNB84B | 614925 | 12q21.31 | Palestine | Netherlands/ Turkey | Yes | No/Yes | L | E | Shahin et al. 2010; Yariz et al. 2012 |
| TBC1D24 | DFNB86 | 613577 | 16p13.3 | Pakistan | Pakistan | Yes | Yes | L | E | Ali et al. 2012; Rehman et al. 2014 |
| ELMOD3 | DFNB88 | 615427 | 2p12-p11.2 | Pakistan | Pakistan | Yes | Yes | L | E | Jaworek et al. 2013 |
| KARS1 | DFNB89 | 601421 | 16q21-q23.2 | Pakistan | Pakistan | Yes | Yes | L | E |
Basit et al. 2011; Santos-Cortez et al. 2013 |
| SERPINB6 | DFNB91 | 173321 | 6p25 | Turkey | Turkey | Yes | Yes | L | Sırmacı et al. 2010) | |
| CABP2 | DFNB93 | 607314 | 11q12.3–11q13.2 | Iran | Iran | Yes | Yes | L | N | Tabatabaiefar et al. 2011; Schrauwen et al. 2012 |
| NARS2 | DFNB94 | 612803 | 11q13.4-q14.1 | Pakistan | Pakistan | Yes | Yes | L | E | Simon et al. 2015 |
| MET | DFNB97 | 164860 | 7q31.2-q31.31 | Pakistan | Pakistan | Yes | Yes | L | E | Mujtaba et al. 2015 |
| TSPEAR | DFNB98 | 612920 | 21q22.3-qter | Iran | Iran | Yes | Yes | L | E | Delmaghani et al. 2012 |
| TMEM132E | DFNB99 | 616178 | 17q12 | China | China | Yes | Yes | E | Li et al. 2015 | |
| PPIP5K2 | DFNB100 | 611648 | 5q13.2-q23.2 | Pakistan | Pakistan | Yes | Yes | L | E | Yousaf et al. 2018b |
| GRXCR2 | DFNB101 | 615762 | 5q32 | Pakistan | Pakistan | Yes | Yes | L | E | Imtiaz et al. 2014 |
| EPS8 | DFNB102 | 600206 | 12p12.3 | Algeria | Algeria | Yes | Yes | E | Behlouli et al. 2014 | |
| CLIC5 | DFNB103 | 607293 | 6p21.1 | Turkey | Turkey | Yes | Yes | L | Seco et al. 2015 | |
| FAM65B | DFNB104 | 611410 | 6p22.3 | Turkey | Turkey | Yes | Yes | E | Diaz-Horta et al. 2014 | |
| EPS8L2 | DFNB106 | 614988 | 11p15.5 | Algeria | Algeria | Yes | Yes | E | Dahmani et al. 2015 | |
| WBP2 | DFNB107 | 606962 | 17q25.1 | China | China | No | No | N | Buniello et al. 2016 | |
| ROR1 | DFNB108 | 602336 | 1p31.3 | Turkey | Turkey | Yes | Yes | E | Diaz-Horta et al. 2016 | |
| ESRP1 | DFNB109d | 612959 | 8q22.1 | USAe | USAe | No | No | E | Rohacek et al. 2017 | |
| COCH | DFNB110d | 603196 | 14q12 | Belgians from Morocco | Belgians from Morocco | Yes | Yes | N | Janssens de Varebeke et al. 2018 | |
| MPZL2 | DFNB111d | 604873 | 11q23.3 | Netherlands | Netherlands | Yes | Yes | L | E | Wesdorp et al. 2018 |
| BDP1 | DFNB112d | 607012 | 5q13.2 | Qatar | Qatar | Yes | Yes | L | E | Girotto et al. 2013 |
| CEACAM16 | DFNB113 | 614591 | 19q13.31-q13.32 | Iran | Iran | Yes | Yes | E | Booth et al. 2018 | |
| GRAP | DFNB114 | 604330 | 17p11.2 | Turkey | Turkey | Yes | Yes | E | Li et al. 2019 | |
| SPNS2 | DFNB115d | 612584 | 17p13.2 | USAe/UK | USAe/UK | No/No | No/No | E | Ingham et al. 2019 | |
| CLDN9 | DFNB116d | 615799 | 16p13.3 | Turkey | Turkey | Yes | Yes | G | Sineni et al. 2019 | |
| CLRN2 | DFNB117d | 618988 | 4p15.32 | Iran | Iran | Yes | Yes | L | E | Vona et al. 2021 |
Abbreviations: L=Linkage analyses and/or homozygosity mapping, NGS=next generation sequencing, N=custom/targeted NGS gene panel, G=whole genome sequencing, E=exome sequencing, NA=Not Applicable/none of these methods were used (see footnotes ‘f’ for this Table)
The complete references are provided in Supplementary Information.
There is evidence that may suggest some of the GJB6 variants are causal through their influence on GJB2 (Rodrigues-Paris et al. 2011).
Articles are printed in the same volume in Nature Genetics.
MYO15A was identified through the study of non-consanguineous families from the genetic isolate of Bengkala, Bali, that has a prevalence of 2% for the c.6337A>T:p.Ile2113Phe, and two consanguineous Indian families.
Provisional loci on OMIM.
Most likely of European ancestry.
Liu et al. 2003 performed a direct screen of a candidate gene in human patients.
Analysis of Next Generation Sequence data to identify ARNSHI genes
In the past decade, exome sequencing (ES) and whole genome sequencing (WGS) have rapidly become more accessible, cost-effective, and currently are the preferred method of studying families that segregate Mendelian traits (Figure 2). Identified variants can be annotated using, for example, ANNOVAR which allows annotation of a large number of bioinformatic tools such as Combined Annotation Dependent Depletion (CADD) (Liu et al. 2016), population specific allele frequencies from The Genome Aggregation Database (gnomAD) (Genome Aggregation Database Consortium et al. 2020), variant classification information from ClinVar (Landrum et al. 2018), and custom datasets such as large-scale screens for HI in mice (Ingham et al. 2019). These annotations have proven very effective in aiding in the identification of causal variants.
Copy number (CNVs), a type of Structural Variant (SV), can also be identified using NGS. Other SVs, such as translocations and gene fusions, often seen in large sizes (50 bases to >1kilobases) have been harder to identify although, currently evolving strategies for their identification have seen some success in cancer and neurological diseases using long-read sequencing, as opposed to the standard short read sequencing currently commonly used for exomes. Certain ARNSHI genes are known to commonly harbor pathogenic CNVs, such as OTOA and STRC (Shearer et al. 2014).
Considering all the above advantages, research strategies for ARNSHI cohorts have increasingly taken the approach of analyzing exome sequence data without performing linkage analysis. The trend is increasingly observed for genes identified from 2014 onwards (Table 1). With the development of more effective variant calling tools, and further accessibility to cost-effective ES, more samples from individuals with HI are being exome sequenced in search of novel, rare coding variants with potentially high impact on function.
Prior to the analysis of sequence data, the most plausible inheritance models for the pedigrees should be determined. For ARNSHI, variants are generally filtered retaining those with an minor allele frequency (MAF) of, for example, ≤0.005 in every population that are either homozygous or potentially compound heterozygous, with the exception of a few known population enriched variants with a higher MAF, such as p.Gly12fs in GJB2 (Chakchouk et al. 2019). When variants detected via ES are potentially compound heterozygotes, the genotypes of the parents are necessary to determine if the variants are in trans or cis. In rare occasions, parental genotyping may show that one of the ARNSI variants has arisen de novo, in which phasing of variants may require additional follow-up (e.g., long-read sequencing, ddPCR, etc). For each family, if DNA samples from all family members have not undergone NGS, the segregation of the identified variant with the affection status of the family members needs to be verified using, for example, Sanger sequencing. Lastly, due to the high interaction among proteins of the ear sensory epithelia and hair cells, digenic and polygenic inheritance have also been described for HI (Schrauwen et al. 2018; Khalil et al. 2020). The definition of digenic inheritance in the literature can be variable, but for HI, genes have been often reported in a classic digenic model, for example, where two trans heterozygous variants in two genes are required for the expression of a phenotype (Schrauwen et al. 2018).
Linkage analysis and homozygosity mapping in the NGS era
Although linkage analysis can also be performed using either exome or whole genome sequence data, this type of analysis is rarely performed to localize causal variants (Wang et al. 2015). Linkage analysis is sometimes performed to analyze the candidate causal variants that were identified through WGS and ES and using genotype data for all available informative pedigree members to provide statistical evidence of potential involvement of the variant in ARNSHI etiology. Since the introduction of NGS, homozygosity mapping has been performed using WGS and ES data and may give clues to the regions where the causal variant lies (Wakeling et al. 2019).
Overview of ARNSHI loci and genes identified and the role of consanguinity
There are over 100 ARNSHI loci (designated by DFNB followed by a number), but for the purpose of this article we will concentrate on those for which a gene has been identified and have DFNB and OMIM (McKusick 2007) numbers (Table 1). There are several reasons for concentrating on loci for which the gene has been discovered: 1) for loci for which the gene is unknown, the mapped region may be incorrect; 2) more than one DFNB number is sometimes assigned to the same locus/gene; and 3) the family used to identify the DFNB locus was later determined to have a syndromic HI. We only report here on genes with an OMIM number because the validity of these genes has been assessed. Additional information on gene-disease relations can be obtained by accessing ClinGen (Rehm et al. 2015), a consortium focused on curating the strength of gene-disease relations including ARNSHI genes. Although not discussed here, there have also been several candidate ARNSHI genes reported for which additional evidence is needed to irrefutably link them to ARNSHI.
Three years after the first ARNSHI locus, DFNB1A, was mapped to chr13q12.11 in two consanguineous Tunisian pedigrees (Guilford et al. 1994), the first ARNSHI gene, GJB2 was identified in a large consanguineous Pakistani family (Kelsell et al. 1997). The global prevalence of GJB2 variants leading to ARNSHI is 21.3% (Chan and Chang 2014). Inarguably, GJB2 is the major cause of congenital ARNSHI worldwide and there are regional specific enrichment of certain pathogenic variants, for example, p.Gly12fs in Europe (Zelante et al. 1997), p.Trp24X in the Indian subcontinent (Santos et al. 2005), and p.Arg143Trp in Ghana (Adadey et al. 2020).
The successful identification of ARNSHI genes primarily in Pakistani families (41%; N=30) is due to the high rates of consanguinity where ~60% of all marriages are between first cousins (can be as high as 70% in certain remote provinces), relatively large family sizes (3.5 live births per female), and dedicated local scientists (Ullah et al. 2017). Other countries where novel ARNSHI genes have been frequently first reported include: Iran (12.3%; N=9); Turkey (10.9%; N=8); India (10.9%; N=8); and the Netherlands (8.2%; N=6). (Table 2). Furthermore, 16 (21.9%; 16/73) ARNSHI genes were first identified through the study of families from more than one population. Overall, 93% (68/73) of the ARNSHI loci and 92% (67/73) of the ARNSHI genes were first identified in consanguineous families. The most recent report of a new ARNSHI gene, CLRN2, was identified through the study of a consanguineous Iranian family (Vona et al. 2021). Besides GJB2, a number of genes that were discovered in consanguineous families have been shown to also play a role in ARNSHI in outbred populations, for example, SLC26A4 (Chen et al. 2016), CDH23 (Astuto et al. 2002), and STRC (Vona et al. 2015).
Table 2: Countries used to identify ARNSHI genes and loci.
Number and percentage of ARNSHI loci and genes identified by country. When families from more than one country were used to identify a locus or gene, all countries are included and therefore the number of loci/genes does not sum to 73.
| Country | Number of genes identified (%) | Number of loci identified* (%) |
|---|---|---|
| Algeria | 2 (2.7) | 2 (2.7) |
| China | 3 (4.1) | 2 (2.7) |
| France | 1 (1.4) | 0 (0) |
| India | 8 (10.9) | 5 (6.8) |
| Indonesia | 1 (1.4) | 1 (1.4) |
| Iraq | 2 (2.7) | 2 (2.7) |
| Iran | 9 (12.3) | 8 (10.9) |
| Lebanon | 2 (2.7) | 2 (2.7) |
| Moroccoa | 2 (2.7) | 2 (2.7) |
| Netherlands | 6 (8.2) | 3 (4.1) |
| Palestine | 4 (5.4) | 6 (8.2) |
| Pakistan | 30 (41.1) | 26 (35.6) |
| Qatar | 1 (1.4) | 1 (1.4) |
| Spain | 2 (2.7) | 2 (2.7) |
| Syria | 0 (0) | 2 (2.7) |
| Tunisia | 2 (2.7) | 5 (6.8) |
| Turkey | 8 (10.9) | 7 (9.6) |
| United Kingdom | 1 (1.4) | 1 (1.4) |
| United Statesb | 3 (4.1) | 3 (4.1) |
For which the gene has been identified
Includes Belgians from Morocco.
Most likely of European ancestry
The role of non-consanguineous families in identification of ARNSHI genes cannot be discounted. For example, DFNB18A was identified in a consanguineous Indian family but the gene for this locus, MYO7A was identified through the study of a non-consanguineous Chinese family. Three ARNSHI loci were first mapped in non-consanguineous families, and the corresponding genes identified in the same families: OTOG (DFNB18B) in a Dutch and a Spanish family; WBP2 (DFNB107) in a Chinese family; and ESRP1 (DFNB109) and SPNS2 (DFNB115) both in separate European-American families (Table 1). Although the ARNSHI locus (DFNB3) was mapped through the study of non-consanguineous families, the gene, MYO15A was identified by studying both non-consanguineous Balinese families that were used to map DFNB3 and consanguineous Indian families (Table 1).
The introduction of NGS expedited novel ARNSHI gene identification
In 2009 a seminal article was published, showing proof of principle of detecting causal variants for Mendelian traits using ES (Ng et al. 2010). Since 2010, 32 novel ARNSHI genes (43.8%; N=32/73) have been discovered using NGS (Table 1).
NGS was first used in 2010 to identify, TPRN (DFNB79), in a consanguineous Pakistani family using a custom targeted capture to interrogate the DFNB79 interval. DFNB79 was mapped using linkage analysis via microsatellite markers in a consanguineous Pakistani family and this family was also used to discover TPRN (Khan et al. 2010). In 2012, the next three ARNSHI genes were identified using NGS (Table 1). CABP2 was discovered by studying three consanguineous Iranian families using a custom capture array that targeted the genes in the DFNB93 interval that was previously identified using SNPs and linkage. TSPEAR (DFNB98) was identified via linkage analysis and ES using DNA samples obtained from a consanguineous Iranian family. Lastly, OTOGL (DFNB84B) was discovered by performing linkage analysis using SNP markers in a consanguineous Turkish family followed by ES.
A number of ARNSHI genes were identified through performing linkage mapping followed by ES in consanguineous families (Table 1). More recently, this evolved to the use of ES only, without prior linkage analysis. The first example being the identification of EPS8 in a consanguineous Algerian family (Behlouli et al. 2014). Although most ARNSHI genes identified are still predominantly through the study of consanguineous pedigrees, NGS has facilitated the ability to perform gene identification in outbred families, for example, WBP2 and ESRP1 (Buniello et al. 2016; Rohacek et al. 2017) that mainly have ARNSHI due to compound heterozygous variants. As described above, outbred families provide little linkage information and therefore in most circumstances they cannot be used to map the ARNSHI locus to a genetic region, which was necessary for the positional candidate approach. With the advent of NGS, it has become possible to perform variant filtering and identify causal genes without prior knowledge of the genetic region containing the causal variant. Currently, the detection of multiple families with variants in the same gene, which may include smaller families, is considered important in establishing gene-disease validity in Mendelian disease (DiStefano et al. 2019). The detection of multiple families and individuals are aided by decreasing NGS costs and resources such as GeneMatcher that connect scientists with interests in the same gene (Sobreira et al. 2015).
ES advanced gene identification for ARNSHI loci that were mapped and remained undiscovered after a decade, for example, GAB1 (DFNB26), CDC14A (DFNB32), ADCY1 (DFNB44), and S1PR2 (DFNB68) (Table 1). Notably, the same consanguineous Pakistani pedigree was used to map DFNB26 (Riazuddin et al. 2000) and identify the gene for this locus, GAB1, 18 years later (Yousaf et al. 2018).
Since the identification of TSPEAR in 2012, >60% of all ARNSHI genes were discovered utilizing ES. Around 37% of all genes (27/73) were identified before exome sequence data were first used in analysis of ARNSHI. Although linkage studies and homozygosity mapping continue to be valuable tools in gene identification, since 2014, 9 genes (12.3%) were identified without the need for the former (Table 1).
With the decreasing costs of WGS, it can be readily implemented to identify novel ARNSHI genes. In 2019, CLDN9 was the first ARNSHI gene identified using WGS (Sineni et al. 2019). In comparison to exome data, whole genome sequence data provides more uniform read depth coverage, accurate copy number variant evaluation, and an assessment of the entire genome. However, it remains difficult to interpret single nucleotide variants and smaller insertions and deletions outside of coding regions.
The utility of RNA NGS to prioritize and identify ARNSHI genes
NGS also advanced the possibility to study the inner ear transcriptome, and more recently single cell RNA sequencing (scRNA-seq) analyses have become more easily accessible. scRNA-seq, as the name indicates, involves the sequence analysis of RNAs per single cell in a tissue. This can be done via fluorescence-activated cell sorting (FACS), microfluidics (chip or droplet methods), and other techniques. Microfluidic partitioning technologies have recently advanced this field significantly as they allow barcoding of RNAs per single cell and the possibility to perform massive parallel scRNA sequencing in various tissues, including inner ear tissues (Kolla et al. 2020).
scRNA-seq has been especially helpful in studying cells of the cochlear and vestibular epithelium during inner ear development. Recently, Kolla et al. (2020) characterized the developing mouse inner ear sensory epithelium using massive parallel scRNA-seq, a resource which the authors made publicly available for other researchers as well. This dataset can be accessed by other scientists to quickly check spatiotemporal expression of novel human candidate genes in the different cell types of the inner ear sensory epithelium, without performing immunohistochemistry and in situ hybridization experiments. This study also found genes that were previously not known to be expressed in mouse hair or prosensory cells, i.e., Rprm, Cd164l2, Ccer2, and Gng8.
scRNA-seq and RNA-seq have also been useful in detecting genes with low expression levels in certain tissues. The first transcriptome of rat cochleae aided in discovering genes that were previously not known to be expressed in certain developmental stages (Cho et al. 2002). More recently, scRNA-seq data from inner hair, outer hair and Dieter’s cells facilitated the annotation of new exons in Mendelian HI genes (Ranum et al. 2019).
Unfortunately, expression data on human inner ear tissues is limited due to the challenges in obtaining tissue. One study adopts NGS to study tissues from the human cochleae and the vestibular system obtained during trans-labyrinthine and trans-cochlear approaches of tumors to the skull base (Schrauwen et al. 2016). Another study profiled microRNA expression in the developing human cochleovestibular nerve and otic vesicles via NGS (Chadly et al. 2018). Further studies of human tissues utilizing scRNA-seq are important to gain insight into inner ear specific transcripts and their spatial expression pattern. One such ongoing study created a single cell map of the developing human cochlea (Yu et al. 2019).
Last, open access tools such as gEAR (Orvis et al. 2020) aid in the exploration and visualization of inner ear expression data generated from various independent research groups and help to prioritize potential novel human HI genes. Other databases with useful inner ear or early craniofacial expression data include the Shared Harvard Inner-Ear Laboratory Database (SHIELD) (Shen et al. 2015), and The Gene expression Omnibus (GEO) (Barrett et al. 2012).
Animal models in the aid of ARNSHI identification
Animal models of hearing loss have been instrumental in the validation of human ARNSHI genes, as a crucial aid in the understanding of the function of these genes in the hearing system, and also to identify candidate genes which can be used to screen human families for causal variation. One such example is Gipc3, a gene in which variants underlie progressive sensorineural HI and audiogenic seizures in mice (Charizopoulou et al. 2011). Once identified as a HI gene in mice, screening of human families revealed its human ortholog GIPC3 was the underlying cause of ARNSHI DFNB15 (Charizopoulou et al. 2011). Other examples in which known animal HI genes aided in human ARNSHI gene identification include MYO15A (Wang et al. 1998) and S1PR2 (Santos-Cortez et al. 2016).
Future directions
Current genetic diagnostic testing for HI using exome or custom capture has a ~37–39% diagnostic rate in the United States (Sloan-Heggen et al. 2016; Sheppard et al. 2018). The diagnostic rate differs based on family history and ancestry. For patients from the US, the diagnostic rates depended on family history of HI and the mode of inheritance: 50% ADHI; 41% ARHI; and 37% no family history. The diagnostic success rate by ancestry in one US study was for Europeans 38%; Asians 63%; Middle Easterners 72%; and Africans 26% (Sloan-Heggen et al. 2016). These differences in successful rates by ancestry are impacted by the genetic diversity of the population and the studies on HI that have been performed. For ARNSHI, gnomAD was interrogated by ancestry to evaluate the frequencies of known pathogenic and likely pathogenic variants (Chakchouk et al. 2019). For Latinos and African/African-Americans, the prevalence of HI due to known ARNSHI variants is 26.1 and 5.2 affected per 100,000 individuals, respectively, which is much lower than 96.9 affected per 100,000 individuals for Ashkenazi Jews. This low prevalence might be attributable to the fact that most genetic studies on HI include few Latinos or individuals of African ancestry (Mittal et al. 2018).
The understanding of the genetic spectrum of causal variation in ARNSHI is crucial in diagnostic testing. Studying ethnically/racially diverse populations is important to discover novel genes and variants, since some may be ancestry specific. In the context of ARNSHI, there are populations with high consanguinity that have not been studied but may be informative to identify novel ARNSHI genes, for example, Sudan (63% consanguinity and 4.3 live births per woman), Mauritania (47% consanguinity and 4.79 live births per woman), and isolated Egyptian Nubians (80% consanguinity and 3.3 live births per woman) (Saha et al. 1990; Tadmouri et al. 2009; Anwar et al. 2014; Romdhane et al. 2019). Studies of some populations may also be limited due to geopolitics and/or inaccessibility to the scientific community, for example, Sudan and North Korea. Overall, there is an imminent need to study diverse populations, especially those from sub-Saharan Africa, to improve our understanding of the genetic spectrum of ARNSHI.
Genomic technologies are constantly improving in accuracy and affordability. Newer technologies such as long-read sequencing may be able to capture complex structural variants or regions of the genome not well assessed via short-read sequencing and improve pathogenic variant discovery.
Last, the interrogation of multi-omic datasets including transcriptomics, epigenomics, proteomics, and metabolomics will improve our assessment of possible pathogenic variation. In addition, large-scale animal model phenotyping projects and databases, such as The Zebrafish Information Network (ZFIN; for zebrafish), FlyBase (for drosophila melanogaster), and the International Mouse Phenotyping Consortium (IMPC) and Mouse Genome Informatics (MGI), also provide helpful links to animal HI genes to potential aid in identifying human ARNSHI genes.
In conclusion, NGS has progressed the identification of novel ARNSHI genes in the last decade. Consanguineous pedigrees remain invaluable resources to identify new ARNSHI genes. To further advance our knowledge on the genetic architecture of ARNSHI, research should focus on novel sequencing techniques, and the study of diverse and understudied populations such as Africa.
Supplementary Material
Funding
Support was obtained from the National Institute of Deafness and other Communication Disorders grants R01 DC01165, DC003594, and DC016593.
Footnotes
Declarations:
Code Availability
Not Applicable
Conflicts of Interest
The authors declare no conflict of interest.
Availability of data and material
Not Applicable
References
- Adadey SM, Wonkam-Tingang E, Twumasi Aboagye E, et al. (2020) Connexin Genes Variants Associated with Non-Syndromic Hearing Impairment: A Systematic Review of the Global Burden. Life 10:258. 10.3390/life10110258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar WA, Khyatti M, Hemminki K (2014) Consanguinity and genetic diseases in North Africa and immigrants to Europe. Eur J Public Health 24:57–63. 10.1093/eurpub/cku104 [DOI] [PubMed] [Google Scholar]
- Astuto LM, Bork JM, Weston MD, et al. (2002) CDH23 Mutation and Phenotype Heterogeneity: A Profile of 107 Diverse Families with Usher Syndrome and Nonsyndromic Deafness. Am J Hum Genet 71:262–275. 10.1086/341558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, et al. (2012) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41:D991–D995. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basit S, Lee K, Habib R, et al. (2011) DFNB89, a novel autosomal recessive nonsyndromic hearing impairment locus on chromosome 16q21-q23.2. Hum Genet 129:379–385. 10.1007/s00439-010-0934-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlouli A, Bonnet C, Abdi S, et al. (2014) EPS8, encoding an actin-binding protein of cochlear hair cell stereocilia, is a new causal gene for autosomal recessive profound deafness. Orphanet J Rare Dis 9:55. 10.1186/1750-1172-9-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, Ingham NJ, Lewis MA, et al. (2016) Wbp2 is required for normal glutamatergic synapses in the cochlea and is crucial for hearing. EMBO Mol Med 8:191–207. 10.15252/emmm.201505523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadly DM, Best J, Ran C, et al. (2018) Developmental profiling of microRNAs in the human embryonic inner ear. PLOS ONE 13:e0191452. 10.1371/journal.pone.0191452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaib H, Place C, Salem N, et al. (1996) A Gene Responsible for a Sensorineural Nonsyndromic Recessive Deafness Maps to Chromosome 2p22–23. Hum Mol Genet 5:155–158. 10.1093/hmg/5.1.155 [DOI] [PubMed] [Google Scholar]
- Chakchouk I, Zhang D, Zhang Z, et al. (2019) Disparities in discovery of pathogenic variants for autosomal recessive non-syndromic hearing impairment by ancestry. Eur J Hum Genet 27:1456–1465. 10.1038/s41431-019-0417-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DK, Chang KW (2014) GJB2-associated hearing loss: Systematic review of worldwide prevalence, genotype, and auditory phenotype: Systematic Review of Cx-26-Associated Hearing Loss. The Laryngoscope 124:E34–E53. 10.1002/lary.24332 [DOI] [PubMed] [Google Scholar]
- Charizopoulou N, Lelli A, Schraders M, et al. (2011) Gipc3 mutations associated with audiogenic seizures and sensorineural hearing loss in mouse and human. Nat Commun 2:201. 10.1038/ncomms1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Dong C, Wang Q, et al. (2016) Targeted Next-Generation Sequencing Successfully Detects Causative Genes in Chinese Patients with Hereditary Hearing Loss. Genet Test Mol Biomark 20:660–665. 10.1089/gtmb.2016.0051 [DOI] [PubMed] [Google Scholar]
- Cho Y, Gong T-WL, Stöver T, et al. (2002) Gene Expression Profiles of the Rat Cochlea, Cochlear Nucleus, and Inferior Colliculus. JARO - J Assoc Res Otolaryngol 3:54–67. 10.1007/s101620010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin RWJ, Kalay E, Tariq M, et al. (2008) Mutations of ESRRB Encoding Estrogen-Related Receptor Beta Cause Autosomal-Recessive Nonsyndromic Hearing Impairment DFNB35. Am J Hum Genet 82:125–138. 10.1016/j.ajhg.2007.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiStefano MT, Hemphill SE, Oza MA, et al. (2019) ClinGen expert clinical validity curation of 164 hearing loss gene–disease pairs. Genet Med 21:2239–2247. 10.1038/s41436-019-0487-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman TB, Liang Y, Weber JL, et al. (1995) A gene for congenital, recessive deafness DFNB3 maps to the pericentromeric region of chromosome 17. Nat Genet 9:86–91. 10.1038/ng0195-86 [DOI] [PubMed] [Google Scholar]
- Genome Aggregation Database Consortium, Karczewski KJ, Francioli LC, et al. (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford P, Arab SB, Blanchard S, et al. (1994) A non–syndromic form of neurosensory, recessive deafness maps to the pericentromeric region of chromosome 13q. Nat Genet 6:24–28. 10.1038/ng0194-24 [DOI] [PubMed] [Google Scholar]
- Ingham NJ, Pearson SA, Vancollie VE, et al. (2019) Mouse screen reveals multiple new genes underlying mouse and human hearing loss. PLOS Biol 17:e3000194. 10.1371/journal.pbio.3000194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irshad S, Santos R, Muhammad D, et al. (2005) Localization of a novel autosomal recessive non-syndromic hearing impairment locus DFNB55 to chromosome 4q12-q13.2: Hearing impairment locus DFNB55. Clin Genet 68:262–267. 10.1111/j.1399-0004.2005.00492.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsell DP, Dunlop J, Stevens HP, et al. (1997) Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 387:80–83. 10.1038/387080a0 [DOI] [PubMed] [Google Scholar]
- Khalil A, Karroum SB, Barake R, et al. (2020) Post-lingual non-syndromic hearing loss phenotype: a polygenic case with 2 biallelic mutations in MYO15A and MITF. BMC Med Genet 21:1. 10.1186/s12881-019-0942-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SY, Riazuddin S, Shahzad M, et al. (2010) DFNB79: reincarnation of a nonsyndromic deafness locus on chromosome 9q34.3. Eur J Hum Genet 18:125–129. 10.1038/ejhg.2009.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolla L, Kelly MC, Mann ZF, et al. (2020) Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat Commun 11:2389. 10.1038/s41467-020-16113-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570. 10.1126/science.2884728 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247. 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, et al. (2018) ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46:D1062–D1067. 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wu C, Li C, Boerwinkle E (2016) dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat 37:235–241. 10.1002/humu.22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick VA (2007) Mendelian Inheritance in Man and Its Online Version, OMIM. Am J Hum Genet 80:588–604. 10.1086/514346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Patel AP, Nguyen D, et al. (2018) Genetic basis of hearing loss in Spanish, Hispanic and Latino populations. Gene 647:297–305. 10.1016/j.gene.2018.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton NE (1955) Sequential Tests for the Detection of Linkage’. 42 [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, et al. (2010) Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 42:30–35. 10.1038/ng.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvis J, Gottfried B, Kancherla J, et al. (2020) gEAR: gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. Bioinformatics [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1983) Linkage analysis and family classification under heterogeneity. Ann Hum Genet 47:311–320. 10.1111/j.1469-1809.1983.tb01001.x [DOI] [PubMed] [Google Scholar]
- Ott J, Wang J, Leal SM (2015) Genetic linkage analysis in the age of whole-genome sequencing. Nat Rev Genet 16:275–284. 10.1038/nrg3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum PT, Goodwin AT, Yoshimura H, et al. (2019) Insights into the Biology of Hearing and Deafness Revealed by Single-Cell RNA Sequencing. Cell Rep 26:3160–3171.e3. 10.1016/j.celrep.2019.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL, Berg JS, Brooks LD, et al. (2015) ClinGen — The Clinical Genome Resource. N Engl J Med 372:2235–2242. 10.1056/NEJMsr1406261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Castelein CM, Ahmed ZM, et al. (2000) Dominant modifier DFNM1 suppresses recessive deafness DFNB26. Nat Genet 26:431–434. 10.1038/82558 [DOI] [PubMed] [Google Scholar]
- Rohacek AM, Bebee TW, Tilton RK, et al. (2017) ESRP1 Mutations Cause Hearing Loss due to Defects in Alternative Splicing that Disrupt Cochlear Development. Dev Cell 43:318–331.e5. 10.1016/j.devcel.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romdhane L, Mezzi N, Hamdi Y, et al. (2019) Consanguinity and Inbreeding in Health and Disease in North African Populations. Annu Rev Genomics Hum Genet 20:155–179. 10.1146/annurev-genom-083118-014954 [DOI] [PubMed] [Google Scholar]
- Saha N, Hamad RE, Mohamed S (1990) Inbreeding Effects on Reproductive Outcome in a Sudanese Population. Hum Hered 40:208–212. 10.1159/000153932 [DOI] [PubMed] [Google Scholar]
- Santos R, Wajid M, Pham T, et al. (2005) Low prevalence of Connexin 26 (GJB2) variants in Pakistani families with autosomal recessive non-syndromic hearing impairment: GJB2 variants in Pakistan. Clin Genet 67:61–68. 10.1111/j.1399-0004.2005.00379.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Cortez RLP, Faridi R, Rehman AU, et al. (2016) Autosomal-Recessive Hearing Impairment Due to Rare Missense Variants within S1PR2. Am J Hum Genet 98:331–338. 10.1016/j.ajhg.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Chakchouk I, Acharya A, et al. (2018) Novel digenic inheritance of PCDH15 and USH1G underlies profound non-syndromic hearing impairment. BMC Med Genet 19:122. 10.1186/s12881-018-0618-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Hasin-Brumshtein Y, Corneveaux JJ, et al. (2016) A comprehensive catalogue of the coding and non-coding transcripts of the human inner ear. Hear Res 333:266–274. 10.1016/j.heares.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelow D, Schuelke M, Hildebrandt F, Nurnberg P (2009) HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res 37:W593–W599. 10.1093/nar/gkp369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer A, Kolbe DL, Azaiez H, et al. (2014) Copy number variants are a common cause of non-syndromic hearing loss. Genome Med 6:37. 10.1186/gm554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Scheffer DI, Kwan KY, Corey DP (2015) SHIELD: an integrative gene expression database for inner ear research. Database 2015:bav071. 10.1093/database/bav071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard S, Biswas S, Li MH, et al. (2018) Utility and limitations of exome sequencing as a genetic diagnostic tool for children with hearing loss. Genet Med 20:1663–1676. 10.1038/s41436-018-0004-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineni CJ, Yildirim-Baylan M, Guo S, et al. (2019) A truncating CLDN9 variant is associated with autosomal recessive nonsyndromic hearing loss. Hum Genet 138:1071–1075. 10.1007/s00439-019-02037-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Bierer AO, Shearer AE, et al. (2016) Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet 135:441–450. 10.1007/s00439-016-1648-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJH, Van Camp G (1999) Non-syndromic hearing impairment: gene linkage and cloning. Int J Pediatr Otorhinolaryngol 49:S159–S163. 10.1016/S0165-5876(99)00153-6 [DOI] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Valle D, Hamosh A (2015) GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum Mutat 36:928–930. 10.1002/humu.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadmouri GO, Nair P, Obeid T, et al. (2009) Consanguinity and reproductive health among Arabs. Reprod Health 6:17. 10.1186/1742-4755-6-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah MA, Husseni AM, Mahmood SU (2017) Consanguineous marriages and their detrimental outcomes in Pakistan: an urgent need for appropriate measures. Int J Community Med Public Health 5:1. 10.18203/2394-6040.ijcmph20175757 [DOI] [Google Scholar]
- Vona B, Doll J, Hofrichter MAH, Haaf T (2020) Non-syndromic hearing loss: clinical and diagnostic challenges. 13 [Google Scholar]
- Vona B, Hofrichter MAH, Neuner C, et al. (2015) DFNB16 is a frequent cause of congenital hearing impairment: implementation of STRC mutation analysis in routine diagnostics. Clin Genet 87:49–55. 10.1111/cge.12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vona B, Mazaheri N, Lin S-J, et al. (2021) A biallelic variant in CLRN2 causes non-syndromic hearing loss in humans. Hum Genet 140:915–931. 10.1007/s00439-020-02254-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos B, Noll D, Pigeon M, et al. (2019) Risk factors for hearing loss in children: a systematic literature review and meta-analysis protocol. Syst Rev 8:172. 10.1186/s13643-019-1073-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeling MN, Laver TW, Wright CF, et al. (2019) Homozygosity mapping provides supporting evidence of pathogenicity in recessive Mendelian disease. Genet Med 21:982–986. 10.1038/s41436-018-0281-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A (1998) Association of Unconventional Myosin MYO15 Mutations with Human Nonsyndromic Deafness DFNB3. Science 280:1447–1451. 10.1126/science.280.5368.1447 [DOI] [PubMed] [Google Scholar]
- Wang GT, Zhang D, Li B, et al. (2015) Collapsed haplotype pattern method for linkage analysis of next-generation sequence data. Eur J Hum Genet 23:1739–1743. 10.1038/ejhg.2015.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga S, Grati M, Cohen-Salmon M, et al. (1999) A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat Genet 21:363–369. 10.1038/7693 [DOI] [PubMed] [Google Scholar]
- Yousaf R, Ahmed ZM, Giese APJ, et al. (2018) Modifier variant of METTL13 suppresses human GAB1–associated profound deafness. J Clin Invest 128:1509–1522. 10.1172/JCI97350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KS, Frumm SM, Park JS, et al. (2019) Development of the Mouse and Human Cochlea at Single Cell Resolution. Bioinformatics [Google Scholar]
- Zelante L, Gasparini P, Estivill X (1997) Connexin26 mutations associated with the most common form of non- syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum Mol Genet 6:1605–1609. 10.1093/hmg/6.9.1605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not Applicable