Abstract
Autonomous parvoviruses are tightly dependent on host cell factors for various steps of their life cycle. In particular, DNA replication and gene expression of the prototype strain of the minute virus of mice (MVMp) are closely linked to the onset of host cell DNA replication, pointing to the involvement of an S-phase-specific cellular factor(s) in parvovirus multiplication. The viral nonstructural protein NS-1 is absolutely required for parvovirus DNA replication and is able to transcriptionally regulate parvoviral and heterologous promoters. We previously showed that the promoter P4, which directs the transcription unit encoding the NS proteins, is activated at the onset of S phase. This activation is dependent on an E2F motif in the proximal region of promoter P4. An infectious MVM DNA clone was mutated in the E2F motif of P4. The wild type and the E2F mutant derivative were tested for their ability to produce progeny viruses after transfection of permissive cells. In the context of the whole MVMp genome, the E2F mutation abolished P4 induction in S phase and inactivated the infectious molecular clone, which failed to become amplified and generate progeny particles. The virus could be rescued when NS proteins were supplied in trans, showing that P4 hyperactivity in S is needed to reach a level of NS-1 expression that is sufficient to drive the viral replication cycle. These data show that E2F-mediated P4 activation at the early S phase is a limiting factor for parvovirus production. The primary barrier to parvovirus gene expression in G1 is thought to be promoter formation rather than activation, due to the poor conversion of the parental single-strand genome to a duplex form. The S dependence of P4 activation may therefore be a sign of the virus adaptation to life in the S-phase host cell. If the conversion block in G1 were to be leaky, the S induction of promoter P4 could be envisioned as a safeguard against the production of toxic NS proteins until cells reach the S phase and provide the full machinery for parvovirus replication.
Parvoviruses are a large family of viruses which infect animal species from insects to humans (27). The prototype strain of the parvovirus minute virus of mice (MVMp) belongs to the genus of autonomously replicating parvoviruses of vertebrates. MVM contains a linear, single-stranded (ss) genome of about 5 kb, which comprises two overlapping transcription units. One promoter, P4, directs the synthesis of a transcript whose spliced derivatives, R1 and R2, encode the nonstructural proteins NS-1 and NS-2, respectively. NS-1 and NS-2 are phosphoproteins which share their 85 N-terminal amino acids but differ in their C-terminal portion as a result of the splicing process (9). NS-1 is a multifunctional DNA-binding protein, endowed with ATPase, helicase, and nickase activities (7, 38). These functions are required for parvovirus DNA replication. Besides its replicative activities, NS-1 has been reported to have cytotoxic properties, notably in transformed cells, and to act as a transcriptional transregulator (36). Among others, NS-1 transactivates the second parvovirus promoter, P38, which regulates the expression of the transcription unit encoding the viral capsid proteins VP1 and VP2 (5, 20).
Because of their low genetic complexity, parvoviruses depend extensively on host cell factors for various steps of their life cycle (34). The availability of at least some of these factors appears to depend on cell proliferation. As already described for several other DNA viruses, parvovirus replication relies on cellular functions transiently expressed during the S phase of the cell cycle (25, 28, 32). However, parvoviruses can be distinguished from DNA tumor viruses by their inability to induce resting cells to initiate their DNA replication program (32). Hence, the onset of parvovirus multiplication is delayed until the host cells enter S phase on their own.
It has been shown that the uptake of parvoviruses occurs irrespective of the host cell position in the mitotic cycle, allowing parvoviruses to infect resting and proliferating cells with similar efficiencies (25, 28, 31). After virus uncoating, the input ss viral genome is converted into a double-stranded replicative form, providing a duplex template for mRNA transcription. The burst of viral protein production has been reported to be coupled with cell entry into S phase (9), suggesting that conversion of the MVM ss genome into a double-stranded DNA template (or a prior step following virus uptake and required to make the genome available for conversion) and/or expression of MVM genes might be S-phase-dependent events. There is indeed circumstantial evidence that the conversion of parvoviral genomic DNA is inefficient in G0/G1 cells (39). Furthermore, it has recently been shown that the MVM P4 promoter contains a DNA element that interacts in a cell-cycle-dependent manner with transcription factors of the E2F family, leading to a strong P4 activation at the very beginning of S phase (10). Therefore, several restrictions on parvovirus gene expression appear to occur prior to the G1/S transition. This regulation may benefit the virus by preventing the accumulation of NS proteins, which have cytotoxic effects and trigger capsid formation, until cells initiate their own DNA replication program and provide the right milieu for parvovirus DNA amplification.
The present work was focused on the putative promoter component of the dependence of parvoviruses on S phase. The S activation of promoter P4 has been demonstrated by means of artificial reporter constructs, hence the interest of first determining whether it also occurs in the context of the whole parvovirus DNA. Should this be the case, the question then arises whether this activation is necessary to the progress of the virus life cycle. These issues were addressed by mutating the E2F site of promoter P4 in an infectious MVM DNA clone and comparing the E2F mutant and original wild-type clones for their expression pattern and infectivity in transfected permissive cells. In contrast with the wild type, the E2F mutant was deficient in the induction of promoter P4-directed transcription in S phase and failed to replicate and generate progeny viruses unless NS proteins were supplied in trans. Likewise, the viral particles obtained from the E2F mutant DNA clone in the presence of ectopically expressed NS proteins were found to be noninfectious. These data indicate that P4 activation in S phase is a prerequisite for the occurrence of a productive virus cycle. The basal P4 activity appears to be insufficient to achieve the threshold level of NS protein expression required to drive parvovirus DNA replication.
MATERIALS AND METHODS
Cell culture and synchronization.
The established A9 and NBK cell lines were grown in Eagle’s minimum essential medium (MEM) supplemented with 5% aseptic fetal calf serum (FCS) at 37°C in a 5% CO2 incubator. Cells were arrested in G0 phase by cultivation for 72 h in MEM supplemented with 0.5% FCS (serum starvation) and synchronously released into a new mitotic cycle by addition of FCS to a final concentration of 20%.
Plasmids and cell transfection.
The infectious MVMp DNA clone, pMVM, was kindly provided by P. Tattersall (Yale University, New Haven, Conn.). The derivatives pMVM-E2F and pMVM-Ets, harboring mutations in the corresponding motifs of the proximal region of promoter P4, were produced by a previously described procedure (10). Briefly, the 185-nucleotide-long AflIII-NcoI fragment comprising the right arm of the 3′-terminal palindrome and downstream sequences up to the NS initiation codon was cloned into the SalI site of the pAlter vector (Promega), allowing subsequent site-directed mutagenesis with the altered sites system (Promega) according to the manufacturer’s recommendations. For this purpose, we used the oligonucleotides 5′-AAACCAAGGCATGAAAAGGAAG-3′ (mE2F) and 5′-AACCAAGGCGCGAAAATGAAGTGG-3′ (mEts), containing mutations (indicated in boldface) known to abolish the binding of transcription factors of the E2F (mE2F) and Ets (mEts) families, respectively. The mutated P4 fragments were substituted for the equivalent wild-type AflIII-NcoI fragment of plasmid pMVM, generating pMVM-E2F and pMVM-Ets constructs. ori− derivatives were obtained by deleting the 329-nucleotide PshAI-XbaI fragment of pMVM, pMVM-Ets, and pMVM-E2F, which comprises the right-end origin of replication, yielding pMVMori−, pMVM-Etsori−, and pMVM-E2Fori−. Cells were transfected by the Polybrene-dimethyl sulfoxide (30%) shock method (4, 14), with a total amount of 10 μg of plasmid DNA per transfection of 106 cells in a 100-mm petri dish.
Hirt extraction and hybridization of viral DNA.
Cells were lysed by addition of 2 ml of extraction buffer (10 mM Tris-HCl [pH 7.4], 10 mM EDTA, 0.6% sodium dodecyl sulfate [SDS]) per 100-mm petri dish, and viral DNA was isolated according to the Hirt procedure. Briefly, after treatment of the cell lysate with proteinase K (100 μg/ml) for 3 h at 37°C, protein and high-molecular-weight DNA were precipitated by addition of a 1/4 volume of 5 M NaCl, followed by incubation overnight on ice and centrifugation for 1 h at 12,000 rpm in a Sorvall SS34 rotor. The supernatant was precipitated by addition of 2.5 volumes of ethanol. The pellet was resuspended in 100 μl of TE buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA) and treated with RNase A (50 μg/ml) for 30 min at 37°C, followed by a further proteinase K treatment (100 μg/ml) for 30 min at 37°C. After phenol-extraction and ethanol precipitation, the DNA was recovered and resuspended in 100 μl of TE buffer. Samples (10 μl) were loaded on a 1% agarose gel and fractionated by electrophoresis in Tris-acetate-EDTA buffer at 80 mA for 3 h. After electrophoresis, DNA within the gel was denatured in 0.5 M NaOH–1.5 M NaCl for 30 min and neutralized in 1.5 M NaCl–0.5 M Tris-HCl (pH 7.4) for two times at 30 min each. Following transfer on a nitrocellulose membrane (Hybond C; Amersham), the DNA was immobilized by being baked for 1 h at 80°C in a dried atmosphere and hybridized with a radioactive probe that consisted of the VP protein-encoding fragment of MVMp DNA.
RNA extraction and RNase protection.
Total RNA was extracted by using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. The RNA was treated with RQ1 DNase (Promega) prior to RNase protection assays, in order to eliminate any contaminating DNA. To produce a radiolabeled antisense RNA probe, the P38 region of MVM DNA was amplified by PCR with the primers 5′-GGATCCTAATACGACTCACTATAGGGAGGCCCAGGTACTTGTAGCCAGGAG-3′ (containing the T7 promoter) and 5′-TGGCCCATGATTTGTGC-3′. The RNA probe was synthesized by means of the T7 RNA Maxi kit (Ambion), purified by 6% polyacrylamide–8 M urea gel electrophoresis, and eluted in hybridization buffer (80% formamide, 1 mM EDTA, 40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] [pH 6.4], 200 mM sodium acetate). For RNase protection assays, 10 μg of total RNA was hybridized overnight with the probe (106 cpm) in 20 μl of hybridization buffer at 50°C. After addition of 180 μl of buffer (10 mM Tris-HCl [pH 7.5], 5 mM EDTA, 200 mM sodium acetate) containing 2 U of RNase ONE (Promega), ss RNA was digested for 1 h at room temperature. The reaction was stopped by addition of 20 μl of 1 mg of tRNA per ml in 10% SDS. The protected fragments were precipitated by adding 550 μl of cold 100% ethanol, resuspended in 10 μl of loading buffer (80% deionized formamide, 1 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol, 0.1% SDS), and resolved by electrophoresis through a 6% polyacrylamide–8 M urea gel in Tris-borate-EDTA buffer. The labeled products were detected by autoradiography and quantified with a PhosphorImager.
DNA microinjection and filter assays.
Microinjection and DNA hybridization were performed as previously described (24). Briefly, about 5 × 102 A9 cells, adhering to a marked area of a 12-mm-diameter coverslip (CELLocate; Eppendorf), were microinjected each with 0.1 pl of plasmid DNA solution (10 μg/ml). Microinjection was performed with a Zeiss-AIS computer-controlled system. Coverslips were further incubated for 24 h in normal growth medium at 37°C. The cells were transferred and lysed on a nitrocellulose filter presaturated with 0.5 M NaOH–1.5 M NaCl. Filters were neutralized with 1.5 M NaCl–0.5 M Tris-HCl (pH 7.4), and DNA was immobilized by being baked for 1 h at 80°C in a dried atmosphere. The filters were hybridized with a radioactive DNA probe that consisted of the VP-encoding HindIII fragment of MVMp DNA.
Virus titration.
A9 or NBK indicator cells were infected in duplicate with different virus dilutions. Virus titers were determined either by plaque assay or by DNA hybridization assay, as described by Maxwell and Maxwell (21). For DNA hybridization, cells were transferred and lysed on a nitrocellulose filter at 20 h postinfection. The filters were hybridized with a radioactive MVM-specific DNA probe, as described above. For plaque assays, infected cell cultures in 60-mm dishes were overlaid with 2 ml of 0.75% agarose in a 1:1 mixture of phosphate-buffered saline (PBS)–Opti-MEM containing 5% FCS. After 6 days, plaques were visualized by means of a 2-ml overlay of 0.75% agarose in PBS containing 0.04% neutral red.
Protein analysis by Western blotting.
Extraction of total protein was performed according to the method of Kumar and Chambon (16) with previously described modifications (2). Briefly, A9 cells were harvested with a rubber policeman, collected in ice-cold PBS, and washed twice. The pellet was resuspended in 1.5 volumes of lysis buffer (20 mM HEPES [pH 7.9], 0.4 M NaCl, 25% glycerol, 1 mM EDTA, 2.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). After incubation on ice for 20 min, the lysate was frozen at −70°C, thawed on ice, and vigorously vortexed. After centrifugation (18,000 × g, 10 min, 4°C), the supernatant was recovered, frozen in liquid nitrogen, and subsequently used as whole-cell protein extract. Protein samples (20 μg) were fractionated by electrophoresis on a 12% polyacrylamide gel containing 1% SDS and transferred onto a Hybond ECL membrane (Amersham) with a Trans-Blot semidry electrophoretic transfer cell (Bio-Rad) according to the manufacturer’s instructions (200 mA for 1 h). The membranes were first incubated for 1 h in blocking buffer (5% powdered milk, 1% sodium caseinate in PBS) and then for 1 h with antibodies diluted in blocking buffer. Polyclonal antiserum raised against the C-terminal part of NS-1 (3) was used at a dilution of 1:2,500. Membranes were then washed three times for 5 min each in PBS containing 0.1% Tween 20 and further incubated for 1 h with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Dianova; 1:1,000 dilution in blocking buffer). All incubations were performed at room temperature. Immunocomplexes were detected with ECL reagent (Amersham) according to the manufacturer’s instructions.
RESULTS
Modification of an infectious MVMp DNA clone.
It has been shown in transient expression assays with reporter genes that the activity of promoter P4 is modulated in cis through motifs known to bind distinct cellular transcription factors. Among others, two recognition sites for Ets and E2F factors were identified within the proximal P4 region and found to contribute to promoter activity (10, 13). The E2F-binding site mediates the induction of promoter P4 at the very beginning of S phase; hence, its destruction reduces P4 activity by 80% in S phase but has no obvious transcriptional effect during other phases of the cell cycle (10). The Ets-binding site activates promoter P4 in a cell-cycle-independent manner, and its destruction reduces P4 activity by 40 to 50% during both G1 and S phases (10).
In order to investigate whether the S-phase-specific activation of promoter P4 has any physiological relevance to the MVM replication cycle, advantage was taken of an infectious MVM molecular clone obtained from P. Tattersall (Yale University) and designated pMVM. After transfection of permissive cells, the MVMp DNA component of the infectious molecular clone becomes excised from the plasmid backbone, replicates, and leads to production of progeny virus particles (15). The wild-type infectious molecular clone pMVM was modified by introducing point mutations within either the E2F- or, as a control, the Ets-binding site. The Ets-binding site was chosen as a reference since, in asynchronously growing cells, the Ets and E2F motifs contribute to a similar extent to the overall P4 promoter activity (12). The sequence modifications brought to the E2F and Ets elements, as given in Materials and Methods, have previously been shown to render the respective DNA motifs transcriptionally inactive (10, 13). The mutant clones, referred to as pMVM-E2F and pMVM-Ets, as well as the wild-type clone pMVM, were fully sequenced to verify that no further mutations had been introduced during the cloning process. Indeed, the sequences of the three molecular clones were identical, except for the changes introduced by site-directed mutagenesis.
Promoter P4 deprivation of a functional E2F-binding site suppresses the infectivity of the MVMp genomic clone.
Mouse (A9) and human (NBK) cells, which are both permissive for MVM, were transfected with the wild-type MVM DNA clone or its above-mentioned mutant derivatives. One week posttransfection, pMVM- and pMVM-Ets-inoculated cells showed a clear cytopathic effect, whereas no cytopathic effect was observed at this or later (up to 5 weeks) times in cultures transfected with pMVM-E2F. In order to quantify the amount of progeny viruses produced, cultures were harvested 7 days posttransfection and briefly sonicated to release progeny virions. Indicator cell cultures (A9 or NBK cells) were subsequently infected with serial dilutions of the virus suspensions obtained in this way, incubated for 48 h, transferred onto nitrocellulose membranes, and hybridized with radioactively labeled MVM DNA. Single cells in which the viral genome was transferred were then visualized by autoradiography, allowing the determination of the titers of replication-competent virus produced. As shown in Table 1, pMVM and pMVM-Ets gave rise to progeny virus bursts of similar size, irrespective of the cell line used for virus preparation and titration. In contrast, no progeny viruses were recovered after transfection with clone pMVM-E2F in both A9 and NBK cells. This indicated that the E2F motif mutation suppressed the infectivity of the MVMp genomic clone in both mouse and human cells, thereby accounting for the lack of cytopathogenicity observed after transfection with the E2F mutant.
TABLE 1.
Virus production after transfection of permissive cells with wild-type or mutant MVM DNA clonesa
| Indicator cell line | Virus titers (hybridization U/transfected culture) for producer cell line and virus DNA clone
|
|||||
|---|---|---|---|---|---|---|
| A9
|
NBK
|
|||||
| pMVM | pMVM-E2F | pMVM-Ets | pMVM | pMVM-E2F | pMVM-Ets | |
| A9 | 8 × 105 | ND | 9 × 105 | 4 × 105 | ND | 4 × 105 |
| NBK | 9 × 105 | ND | 9 × 105 | 8 × 105 | ND | 9 × 105 |
Cultures of 106 A9 or NBK cells were transfected with 10 μg of DNA of the wild-type (pMVM) or mutant (pMVM-E2F and pMVM-Ets) MVM genomic clones. Cells and supernatants were collected 7 days after transfection and sonicated in order to release progeny viruses. After elimination of cell debris, virus titers were determined by single-cell DNA hybridization, with A9 or NBK indicator cultures. Data shown are average values from five independent experiments (standard deviation < 10%). The background value detected with the replication-deficient MVM genomic clone (pMVMori−) was subtracted. ND, nondetectable (<3 U/transfected culture).
The E2F-binding site of promoter P4 is required for the strong induction of virus gene expression at the G1/S-phase transition.
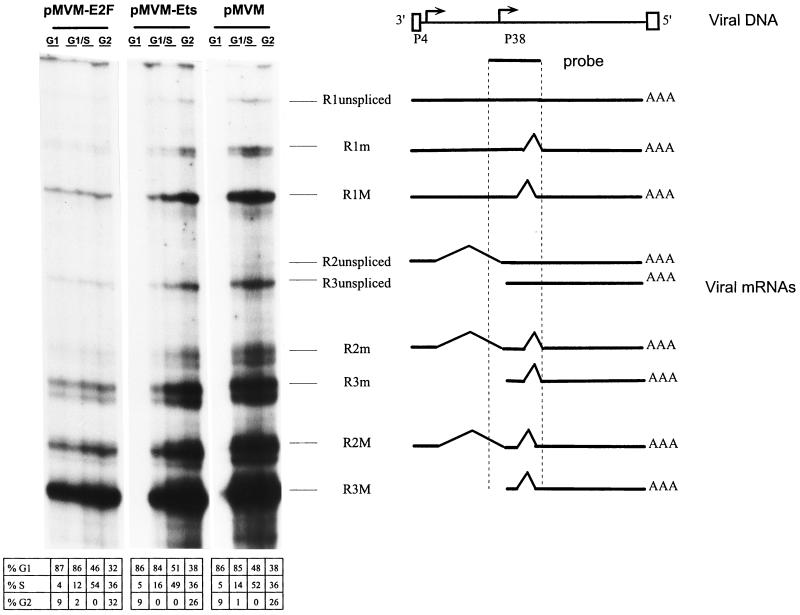
The parvovirus replication cycle is S phase dependent. In particular, viral transcripts only start to accumulate after host cells enter S phase (26). In reporter gene expression assays, promoter P4 was found to be activated in S phase through a functional E2F element (10). This prompted us to determine whether a similar regulation took place in the context of the whole parvoviral genome, by analyzing viral transcription as a function of the cell cycle after transfection of synchronized A9 cells with pMVM, pMVM-Ets, or pMVM-E2F DNA. To this end, A9 cells were serum starved by incubation for 72 h in 0.5% FCS, leading more than 95% of the cell population to arrest in G0 phase. G0-blocked cells were transfected with the different MVM DNA constructs and subsequently released from growth arrest by increasing the serum concentration to 20% at 24, 32, 36, and 44 h posttransfection. Total RNA was isolated at 48 h posttransfection when the majority of cells were in G1, G1/S, or G2 phase, corresponding to induction times with serum of 4, 12 to 16, and 24 h, respectively (Fig. 1, bottom part). The various viral mRNA species were then quantitated by an RNase protection assay. As illustrated in Fig. 1, G0 and G1 cells sustained a low level of transcription of all three genomic clones tested. It is noteworthy that the E2F mutation resulted in an increase in the level of P4-directed transcripts in G1, compared with the wild-type and Ets mutant promoters. This can probably be assigned to the known E2F-mediated repression of responsive promoters during the G1 phase (1). The wild-type and Ets mutant transfectants were further characterized by a striking increase in the accumulation of viral transcripts during the S phase. From the above-mentioned reporter gene expression assays, this induction of P4-derived R1 and R2 transcripts is likely to result, at least in part, from the E2F-dependent activation of promoter P4 at the G1/S transition (see below). The steady-state levels of P38-directed R3 transcripts were similarly enhanced in S phase. Since P38 is transactivated by the nonstructural protein NS-1 (5), which is expressed from the R1 transcript, these data indicated that functional NS-1 was indeed produced from both wild-type and Ets mutant MVM DNA in S-phase cells. As shown in Fig. 1, the MVM DNA clone carrying a point mutation within the E2F-binding site could be distinguished by its lack of responsiveness to the host cell cycle, in that S-phase transit was accompanied with little change in the steady-state level of viral transcripts. It therefore appeared that, in the context of the whole parvoviral genome, inactivation of the E2F-binding site of promoter P4 impaired the S-phase-specific burst of viral transcription directed by promoters P4 and P38.
FIG. 1.
Cell cycle dependence of viral mRNA accumulation in MVM DNA-transfected A9 cells. A9 cells arrested in G0 by serum starvation were transfected with the viral genomic clones pMVM, pMVM-Ets, and pMVM-E2F. Serum was added to a final concentration of 20% at 24 (G2), 32 to 36 (G1/S), and 44 (G1) h posttransfection. Total RNA was extracted at 48 h posttransfection and analyzed by RNase protection assay. The localization of the radiolabeled antisense RNA probe along the viral genome, as well as the different major (M) and minor (m) mRNA species, is depicted in the right panel. The distribution of the cell population according to mitotic cycle phase is given in the bottom part.
The lack of S-phase-specific gene expression from the E2F mutant correlates with its incompetence for DNA replication.
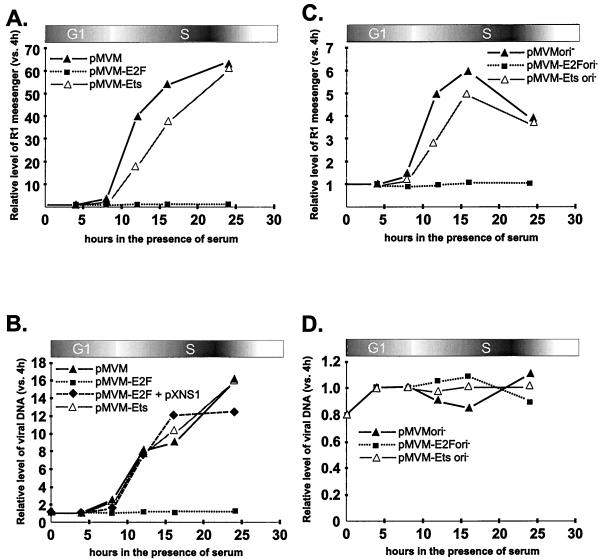
The strong enrichment in S-phase cells of transcripts generated from pMVM and pMVM-Ets may be due to P4 promoter activation and/or DNA template amplification, which are both known to be coupled with cell entry into S phase. The E2F motif may influence viral DNA transcription as well as replication. On the one hand, the E2F-binding site mediates the S induction of promoter P4 (10). On the other hand, though lying outside the minimal origin of DNA replication, the E2F motif is located in a region which is both in the vicinity of the left-end origin (8) and within the P4 promoter which directs the transcription unit encoding the NS-1 replicative protein (9). In order to assess the contribution of P4 activation to the E2F motif-dependent accumulation of parvoviral mRNAs during S phase, transcription and amplification of pMVM and pMVM-E2F clones were measured by RNase protection and Southern blotting assays, respectively, under conditions allowing—or not—viral DNA replication. To prevent MVM DNA replication, a deletion was introduced in the right-end origin of replication (37), giving rise to the ori− derivatives of pMVM and pMVM-E2F.
The replication-deficient pMVMori− clone was found to retain the capacity for enhanced viral mRNA yields in S phase, while the pMVM-E2Fori− construct failed to show this modulation (Fig. 2C). The lack of S induction of the E2F mutant form of P4 was not due to a general inactivation of the promoter, since the Ets mutation (which has previously been shown to reduce P4 activity [13]) had little effect on the magnitude of P4 stimulation in S phase (Fig. 2C). Therefore, these data indicated that the E2F-mediated activation of promoter P4 was directly responsible for the S-phase specificity of the induction of wild-type virus gene expression. Interestingly, the pMVMori− and pMVM-Etsori− clones achieved a much lower level of viral RNA induction in S phase, compared with the respective ori+ constructs (Fig. 2, compare panels A and C). Although an influence of the right end of MVM DNA on P4-directed gene expression cannot be ruled out, these data strongly suggested that S-phase-associated amplification of transcriptionally active viral DNA templates was also responsible to a large extent for the enhanced level of viral mRNAs during this phase of the cell cycle. In agreement with this possibility, pMVMori+ and pMVM-Etsori+ DNA became amplified at the G1/S transition, whereas the ori− derivatives did not (Fig. 2B, compare panels B and D). Interestingly, the enrichment of S cells in pMVM (ori+) transcription products was totally abolished by mutating the E2F element (Fig. 2A). This suggested that, besides its role in P4 induction in S phase, the E2F motif was required in a direct or indirect way for the S-associated amplification of MVM DNA templates. Indeed, the E2F mutant proved to be replication deficient irrespective of whether it contained a functional ori (Fig. 2B, compare panels B and D). It was concluded from these observations that the strong stimulation of viral mRNA accumulation in S-phase cells pretransfected with pMVM (ori+) resulted from the concerted action of P4 promoter induction and DNA template amplification through an E2F motif-dependent process(es). Thus, disruption of the E2F element appeared to have a dual negative effect on S-phase induction of parvovirus gene expression, by preventing not only the stimulation of P4-driven transcription of input genomic clones but also the further replication of transcriptionally active parvoviral DNA templates.
FIG. 2.
Quantification of viral mRNA and DNA production in MVM DNA-transfected A9 cells growing synchronously. G0-arrested cells were transfected with pMVM, pMVM-E2F, or pMVM-Ets DNA clones containing a functional (A and B) or defective (ori−) (C and D) right-hand origin of replication. Serum was added to a final concentration of 20% at 24, 32, 36, 40, and 44 h posttransfection, corresponding to 24, 16, 12, 8, and 4 h of serum induction, respectively. Total RNA and viral DNA were isolated at 48 h posttransfection and analyzed by RNase protection and Southern blotting assays, respectively. The amounts of viral replicative-form DNA (B and D) and R1 mRNA (A and C) were quantified by means of a PhosphorImager and are expressed as ratios of the values measured at a given time post-serum induction to the corresponding values at the 4-h point. For panel B, plasmid pXNS1 drove the expression of NS-1 proteins under the control of the constitutive cytomegalovirus early promoter (24) and was introduced together with pMVM-E2F by cell cotransfection. Shown are the average values from five independent experiments (standard deviation < 10%).
Competence of the MVM-E2F mutant for DNA replication can be restored by providing the NS-1 protein in trans.
The inability of the MVM-E2F DNA clone to become amplified might be due to the direct requirement of parvovirus DNA replication for an intact E2F-binding site, or to an insufficient level of production of NS-1 proteins in S phase. Indeed, the NS-1 protein is expressed from the transcription unit directed by the E2F-dependent promoter P4 and is indispensable for parvovirus DNA replication (9). If the amount of NS-1 were the limiting factor, it should be possible to rescue MVM-E2F DNA replication by supplying NS-1 in trans from a helper plasmid. In order to test this possibility, G0-arrested A9 cells were cotransfected with pMVM-E2F DNA and an NS-1-expressing plasmid (pXNS1 [24]) and analyzed by Southern blotting at various times after release into the cell cycle. Indeed, expression of NS proteins from the helper plasmid restored the ability of the E2F mutant to replicate its DNA to an extent which was similar to that achieved by wild-type MVM DNA (Fig. 2B). This result indicated that destruction of the E2F-binding site of promoter P4 prevented viral DNA amplification because of the failure of the E2F mutant to sustain a sufficient level of expression of the replicative NS-1 protein and not because of a cis requirement of DNA replication for the integrity of the E2F element. It is worth noting that the Ets mutation did not affect the extent of viral DNA amplification (Fig. 2B) while reducing the overall activity of promoter P4 (13), indicating that the contribution of the Ets motif to P4 activation in S phase was not limiting for NS-1 production in amounts sufficient for replication.
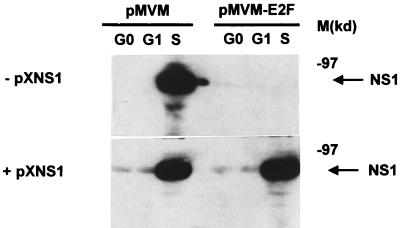
To substantiate this conclusion, pMVM and pMVM-E2F constructs were compared for their ability to produce NS-1 proteins in synchronized A9 cells, in the absence (Fig. 3, upper panel) or presence (Fig. 3, lower panel) of plasmid pXNS1. In the absence of pXNS1, no NS-1 expression could be detected during the G1 phase after cell transfection with either MVM DNA clone, while during the S phase, a clear-cut production of NS-1 proteins was achieved by pMVM but not by the pMVM-E2F mutant derivative. In the presence of pXNS1, low and high levels of NS-1 expression were observed in G1 and S phases, respectively, after transfection with either MVM DNA clone. Thus, the pXNS1 helper plasmid achieved a significant level of NS-1 production (corresponding to the G0/G1 signals seen in the bottom panel of Fig. 3), in contrast with the failure of pMVM-E2F alone to give rise to a detectable NS-1 yield even in S phase (Fig. 3, upper panel). These observations are consistent with the above statement that the helper plasmid compensated for a limitation on NS-1 expression due to the inactivation of the E2F motif of pMVM promoter P4. In cells transfected with wild-type pMVM, S phase was associated with a burst of NS-1 production (Fig. 3) which can in all likelihood be assigned to the activation of promoter P4 and the onset of parvovirus DNA amplification at the G1/S transition (see above). Since NS-1 expression from pXNS1 was not induced in S phase (data not shown), the NS-1 burst occurring when cells cotransfected with pXNS1 and pMVM-E2F reached S phase (Fig. 3, lower panel) can be assumed to arise from the latter clone. It therefore appeared that the basal NS-1 level brought about by plasmid pXNS1 was sufficient to allow the S-phase-dependent amplification of MVM-E2F DNA (Fig. 2B), leading the genomic clone to increase the overall NS-1 production.
FIG. 3.
Levels of NS-1 proteins in MVM DNA-transfected A9 cells released from serum starvation. pMVM (left)- or pMVM-E2F (right)-transfected cells were harvested at 4, 8, and 24 h after serum induction. Whole-cell protein extracts were prepared, and the NS-1 polypeptide was detected by Western blotting, with a polyclonal serum directed against the C-terminal region of the protein. M, molecular mass protein markers (in kilodaltons).
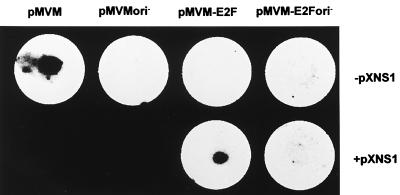
In order to further document the rescue of MVM-E2F replication by exogenously provided NS-1, microinjection experiments were carried out, allowing the administration of defined amounts of plasmid DNA into host cells. When pMVM-E2F was injected alone into A9 cells, no subsequent replication of the mutant MVM DNA was detected by cell hybridization, whereas wild-type pMVM gave a positive, ori-dependent signal under those conditions (Fig. 4, upper row). Yet, coinjection of pXNS1 with pMVM-E2F in a 1:2 ratio restored the ability of the mutant MVM genome to replicate in an ori-dependent fashion (Fig. 4, lower row). These results strongly argued for the failure of clone pMVM-E2F to replicate due to its incapacity to produce enough NS-1 proteins.
FIG. 4.
MVM DNA replication in cells microinjected with genomic DNA clones. Approximately 5 × 102 A9 cells from the central part of a coverslip were microinjected with the wild-type (pMVM) or mutant (pMVM-E2F) genomic DNA clone, in the absence (upper panel) or presence (lower panel) of the NS-1-expressing plasmid pXNS1. The ori− clones were deprived of a functional right-hand origin of replication. At 24 h postmicroinjection, cells were transferred onto a nitrocellulose filter and viral DNA amplification was revealed by hybridization with a radioactive probe corresponding to the VP region of MVMp DNA. The diameter of the filter is 15 mm.
An NS-1-producing plasmid complements the pMVM-E2F DNA clone for the production of progeny viruses.
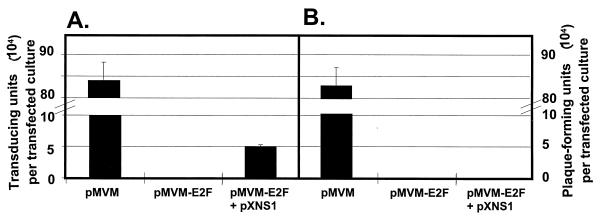
The mutant pMVM-E2F DNA clone was shown above to be impaired in its infectivity, i.e., in its ability to give rise to production of progeny particles (Table 1). On the other hand, the data presented in the above section indicated that an exogenous supply of NS-1 proteins was sufficient to rescue S-phase-specific replication and expression of MVM E2F mutant DNA. Altogether, these data prompted us to determine whether the formation of mutant virions from transfected pMVM-E2F DNA could be restored by providing NS-1 proteins in trans. To this end, A9 cells were transfected with pMVM-E2F DNA with or without the NS-expressing plasmid pXNS1 and analyzed for the formation of progeny virions. Virus production was quantified by both single-cell DNA hybridization (Fig. 5A) and plaque assays (Fig. 5B) with A9 indicator cells. The wild-type pMVM clone was used as a control and gave rise to a burst of virus particles that were fully infectious, as shown by their capacity for DNA replication (Fig. 5A) and propagation (Fig. 5B) in indicator cells. In agreement with the above data, the pMVM-E2F molecular clone was deficient in virus production, as measured by either method. However, this deficiency was partly overcome in the presence of the NS-1-expressing plasmid, which helped pMVM-E2F to generate particles capable of transferring the viral genome to indicator cells. As expected from their mutant genome, pMVM-E2F-derived virions proved to be defective since they were not able to propagate on their own (i.e., in the absence of exogenous NS-1 expression), as demonstrated by their failure to produce plaques in indicator cell cultures (Fig. 5B).
FIG. 5.
Rescue of virus formation from transfected pMVM-E2F DNA in the presence of exogenous NS-1 proteins. A9 cells were transfected with pMVM or the mutant derivative pMVM-E2F alone or in combination with the NS-1-expressing plasmid pXNS1, incubated for 7 days, and collected. Progeny viruses were released, and their titers were determined by single-cell hybridization (A) and plaque assay (B), with A9 indicator cells. While viral DNA transfer is detected by the hybridization assay, the full infectivity of progeny virions is revealed by the plaque assay.
DISCUSSION
Using reporter gene expression assays, we showed previously that promoter P4 is activated at the very beginning of S phase and that this S-specific induction requires a functional E2F-binding site (10). The generic name E2F designates heterodimeric transcription factors composed of two polypeptides belonging to the E2F and DP families, respectively (17, 18, 22). The transcriptional activity of E2F-DP dimers, often referred to as free E2F, is regulated in the course of the cell cycle by formation of higher-order complexes. In particular, interaction with members of the pocket protein family (pRb, p130, and p107) prevents E2F from activating responsive promoters. Binding of pocket proteins to E2F heterodimers (and hence the activity of the E2F transcription factor) is controlled by pocket protein phosphorylation, which appears to be primarily catalyzed by cell-cycle-regulated cyclin-dependent kinases (cdk’s) (11, 30). E2F proteins interact with cyclins and cdk’s either directly or through pocket proteins. After phosphorylation by cdk’s at the G1/S transition, pocket proteins can no longer bind to E2F proteins, leading to the release of transcriptionally active free E2F, which can transactivate target promoters at the onset of S phase (23). Accordingly, variations in P4 promoter activity throughout the cell cycle correlate with the binding of different complexes to the E2F recognition site. During G1 phase, this site is occupied by an E2F-p130 protein complex (10). In agreement with the ability of p130 to prevent E2F from activating transcription (35), only basal P4 activity is detected in G1. P4 activation at the G1/S transition is accompanied by replacement of the E2F-p130 complex by free E2F at the E2F-binding site, in keeping with the view that the latter are the transcriptionally active forms of E2F (1). Transient P4 hyperactivity during S phase coincides with maintenance of free E2F at this site and with the appearance of an additional higher-order complex containing p107, cyclin A, and cdk2 (10). This complex is thought to be involved in the later inhibition of E2F-mediated transactivation (1). All these complexes fade in late S and G2, when the P4 promoter reverts to its basal activity.
The above data were obtained under artificial conditions, i.e., after transfection of reporter gene constructs in which promoter P4 directs the expression of the firefly luciferase gene. In the present study using full-length MVM DNA clones, we extended these observations to the context of the whole parvoviral genome, by showing that promoter P4 is activated during S phase and that this induction requires a functional E2F-binding site. In previous reporter gene assays, promoter P4-directed expression showed a 10-fold activation when cells released from serum starvation entered S phase (10). Results obtained in the present work, by means of RNase protection assays, indicated that under conditions preventing viral DNA replication, S phase was associated with only a fivefold increase in the steady-state level of the P4-programmed viral mRNA R1. This quantitative variation between the S-induction levels is likely due to differences in the genes analyzed and the end points measured, knowing that additional modulations may take place at (post)transcriptional levels besides P4 activity. While moderate in the absence of concomitant viral DNA replication, the S-phase induction of MVM gene expression was much more pronounced when the viral genome was competent for replication, resulting in a 60-fold enrichment of R1 mRNAs in S-phase cells. This further enhancement can in all likelihood be assigned to the amplification of viral DNA (i.e., transcription templates), which indeed took place in S phase. Even when P4 promoter activity decreased in the middle of S phase, which was observed in reporter gene assays (10) and confirmed herein with replication-deficient mutants, the total amount of R1 mRNA still increased, presumably due to ongoing viral DNA replication. Besides its direct role in P4 induction at the G1/S transition, the E2F element of promoter P4 proved to be also required for the enhancement of MVM gene expression that is coupled with viral DNA replication in S phase. Consistently, disruption of the E2F motif prevented parvovirus DNA from becoming amplified in S cells. This defect could be complemented by an exogenous supply of NS-1 proteins, indicating that the replication incompetence of the E2F mutant resulted from its failure to produce sufficient amounts of the replicative NS-1 polypeptides. Thus, E2F appears to initiate a positive feedback loop in that it stimulates the production of the viral NS-1 protein (encoded by the P4-directed R1 transcript), which acts together with cellular factors to drive parvovirus DNA amplification, thereby increasing the pool of DNA templates that can be used for transcription. In this respect, the P4-E2F motif constitutes a key cis determinant of parvovirus multiplication, since its S-phase-coupled activating effect on promoter P4 is a prerequisite for the onset of virus DNA replication and the completion of a productive viral life cycle. Indeed, mutations abolishing the interaction of the P4-E2F element with cognate proteins were found to inactivate an infectious MVM DNA clone, yet mutant viruses could be produced provided that an exogenous source of NS-1 proteins was supplied in trans. Interestingly, an Ets-binding site mutant clone proved to be fully proficient in both DNA amplification and progeny virus production, while this site is known to take part in the overall activity of promoter P4 (present work and reference 13). Given that the E2F motif contributes to about 80% of P4 activity during the S phase (10), it seems that the Ets-dependent component of P4 activation is not limiting for the production of NS-1 proteins in amounts sufficient to trigger parvovirus replication.
Altogether, our data indicate that a threshold amount of NS-1 proteins has to accumulate to trigger virus replication in conjunction with cellular factors. This critical NS-1 production is achieved only in S phase since it is regulated at the transcriptional level by the S-controlled E2F motif of promoter P4. Given that parvoviruses rely on the host cell replication machinery for their own multiplication (9) and that the viral replicative protein NS-1 is endowed with cytotoxic activity (36), it may be advantageous to the virus to delay massive NS-1 production until cells become permissive for viral DNA replication by entering S phase. This might explain why NS-1 expression is placed under the control of an S-phase-dependent regulatory element. Yet, the present data provide no information on whether the restriction on parvovirus gene expression in G1 cells is primarily due to the E2F pathway or whether it would also occur if the E2F motif were replaced by a cell-cycle-independent, strong enhancer element. The latter possibility is supported by a recent claim that modified MVM viruses can be produced, the genome of which carries a substitution of a strong activating element for the proximal (E2F motif-containing) region of promoter P4 (33). The present experiments involved cell transfection with double-stranded full-length MVM DNA, thereby bypassing the early steps of the viral life cycle (virus uptake and uncoating and conversion of the ss genome into duplex replicative forms) which might also be early targets for limitations to virus gene expression under natural infection conditions. Whereas virus uptake appears to take place irrespective of the cell cycle (19), there is circumstantial evidence that conversion of the parental ss genome, and consequently the P4 sequences, to a duplex, and thus transcriptionally active, form is inefficient in G1 cells (39). This places a restriction on promoter formation prior to activation by transcription factors and argues for the view that the P4 promoter was selected to be highly active immediately after its formation, i.e., early in S phase, by acquiring an E2F site. Accordingly, the E2F-mediated P4 induction in S phase would be an adaptation to the S dependence of parvovirus replication rather than a determinant of this dependence. It should be stated, however, that the restriction of the DNA conversion reaction to S cells is difficult to demonstrate, given that only a small fraction of input ss DNA becomes converted to a duplex form in infected cells, and artifactual annealing of the major negative-strand virion genome with a minor fraction of encapsidated positive strands can take place after DNA extraction. In natural animal host populations, parvovirus infections are often persistent and asymptomatic, without obvious signs of virus gene expression. Yet, affected animals harbor infectious parvoviral DNA since induction of cell proliferation foci, e.g., through tumor formation or tissue regeneration, leads to the reactivation of viral DNA expression and replication and to the production of progeny viruses. At least part of persisting DNA may be in the ss conformation, accounting for its lack of expression. Furthermore, postconversion limitations to parvovirus gene expression may be involved. Indeed, it has recently been shown that infection of muscle cells with recombinant parvoviruses belonging to the group of adeno-associated viruses results in a long-term significant transgene expression (29), which implies that parvoviral ssDNA conversion to double-stranded forms occurs at least to some extent in quiescent tissues. It may thus be speculated that the E2F-element-mediated S-phase dependency of promoter P4 represents an additional safeguard against parvovirus gene expression (in particular, cytotoxic protein production) under these conditions. It also appears from the present work that a major component of the S-phase-associated increase in MVM gene expression is related to the amplification of viral DNA templates. The E2F-dependent activation of promoter P4 was found to be a prerequisite for MVM DNA amplification, through the involvement of the replicative NS-1 protein that is encoded by the P4-directed transcription unit. Yet, in order to amplify their DNA, parvoviruses also rely on the cellular replication machinery (including DNA polymerase and cofactors [6]), the full availability of which is likely to represent an additional factor confining virus multiplication to the S phase.
In conclusion, several limitations at both DNA replication and transcriptional levels appear to couple parvovirus multiplication with the S phase of host cells. While the cellular replication factors involved remain to be characterized, the present study led to the identification of E2F as the cellular transcription factor allowing the essential level of virus gene expression to be achieved specifically in S cells.
ACKNOWLEDGMENTS
We are indebted to Roger Fisher (DKFZ, Heidelberg, Germany) for help with the microinjection procedure and Tarig Bashir, François Fuks, Marianna Alunni-Fabbroni (DKFZ), and Bernhard Hirt (ISREC, Epalinges) for fruitful discussion and critical reading of the manuscript.
This work was supported by the Commission of the European Communities. L.D. and A.P. are fellows of the Commission of the European Communities.
REFERENCES
- 1.Beijersbergen R L, Bernards R. Cell cycle regulation by the retinoblastoma family of growth inhibitory proteins. Biochim Biophys Acta. 1996;1287:103–120. doi: 10.1016/0304-419x(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 2.Boeuf H, Reimund B, Jansen Durr P, Kedinger C. Differential activation of the E2F transcription factor by the adenovirus EIa and EIV products in F9 cells. Proc Natl Acad Sci USA. 1990;87:1782–1786. doi: 10.1073/pnas.87.5.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brockhaus K, Plaza S, Pintel D J, Rommelaere J, Salome N. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J Virol. 1996;70:7527–7534. doi: 10.1128/jvi.70.11.7527-7534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaney W G, Howard D R, Pollard J W, Sallustio S, Stanley P. High-frequency transfection of CHO cells using polybrene. Somat Cell Mol Genet. 1986;12:237–244. doi: 10.1007/BF01570782. [DOI] [PubMed] [Google Scholar]
- 5.Christensen J, Cotmore S F, Tattersall P. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J Virol. 1995;69:5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotmore S F, Christensen J, Nuesch J P, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2-3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotmore S F, Tattersall P. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 1994;13:4145–4152. doi: 10.1002/j.1460-2075.1994.tb06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrate. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 10.Deleu L, Fuks F, Spitkovsky D, Horlein R, Faisst S, Rommelaere J. Opposite transcriptional effects of cyclic AMP-responsive elements in confluent or p27KIP-overexpressing cells versus serum-starved or growing cells. Mol Cell Biol. 1998;18:409–419. doi: 10.1128/mcb.18.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dynlacht B D, Flores O, Lees J A, Harlow E. Differential regulation of E2F trans-activation by cyclin/cdk2 complexes. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 12.Faisst S, Perros M, Deleu L, Spruyt N, Rommelaere J. Mapping of upstream regulatory elements in the P4 promoter of parvovirus minute virus of mice. Virology. 1994;202:466–470. doi: 10.1006/viro.1994.1363. [DOI] [PubMed] [Google Scholar]
- 13.Fuks F, Deleu L, Dinsart C, Rommelaere J, Faisst S. ras oncogene-dependent activation of the P4 promoter of minute virus of mice through a proximal P4 element interacting with the Ets family of transcription factors. J Virol. 1996;70:1331–1339. doi: 10.1128/jvi.70.3.1331-1339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai S, Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kestler, J. Unpublished data.
- 16.Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 17.La Thangue N B. DRTF1/E2F: an expanding family of heterodimeric transcription factors implicated in cell-cycle control. Trends Biochem Sci. 1994;19:108–114. doi: 10.1016/0968-0004(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 18.La Thangue N B. E2F and the molecular mechanisms of early cell-cycle control. Biochem Soc Trans. 1996;24:54–59. doi: 10.1042/bst0240054. [DOI] [PubMed] [Google Scholar]
- 19.Linser P, Bruning H, Armentrout R W. Uptake of minute virus of mice into cultured rodent cells. J Virol. 1979;31:537–545. doi: 10.1128/jvi.31.2.537-545.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorson C, Burger L R, Mouw M, Pintel D J. Efficient transactivation of the minute virus of mice P38 promoter requires upstream binding of NS1. J Virol. 1996;70:834–842. doi: 10.1128/jvi.70.2.834-842.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maxwell I H, Maxwell F. A modified plaque assay and infected cell hybridization assay for wild-type and recombinant LuIII autonomous parvovirus. BioTechniques. 1994;16:876–881. [PubMed] [Google Scholar]
- 22.Mudryj M, Hiebert S W, Nevins J R. A role for the adenovirus inducible E2F transcription factor in a proliferation dependent signal transduction pathway. EMBO J. 1990;9:2179–2184. doi: 10.1002/j.1460-2075.1990.tb07387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuman E, Flemington E K, Sellers W R, Kaelin W G. Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pujol A, Deleu L, Nuesch J P, Cziepluch C, Jauniaux J C, Rommelaere J. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of nonstructural protein NS1. J Virol. 1997;71:7393–7403. doi: 10.1128/jvi.71.10.7393-7403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhode S L., III Replication process of the parvovirus H-1. I. Kinetics in a parasynchronous cell system. J Virol. 1973;11:856–861. doi: 10.1128/jvi.11.6.856-861.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoborg R V, Pintel D J. Accumulation of MVM gene products is differentially regulated by transcription initiation-RNA processing and protein stability. Virology. 1991;181:22–34. doi: 10.1016/0042-6822(91)90466-o. [DOI] [PubMed] [Google Scholar]
- 27.Siegl G. Biology and pathogenicity of autonomous parvoviruses. In: Berns K I, editor. The parvoviruses. New York, N.Y: Plenum Press; 1984. pp. 297–348. [Google Scholar]
- 28.Siegl G, Gautschi M. The multiplication of parvovirus LuIII in a synchronized culture system. II. Biochemical characteristics of virus replication. Arch Gesamte Virusforsch. 1973;40:119–127. doi: 10.1007/BF01242643. [DOI] [PubMed] [Google Scholar]
- 29.Snyder R O, Spratt S K, Lagarde C, Bohl D, Kaspar B, Sloan B, Cohen L K, Danos O. Efficient and stable adeno-associated virus-mediated transduction in the skeletal muscle of adult immunocompetent mice. Hum Gene Ther. 1997;8:1891–1900. doi: 10.1089/hum.1997.8.16-1891. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki Takahashi I, Kitagawa M, Saijo M, Higashi H, Ogino H, Matsumoto H, Taya Y, Nishimura S, Okuyama A. The interactions of E2F with pRB and with p107 are regulated via the phosphorylation of pRB and p107 by a cyclin-dependent kinase. Oncogene. 1995;10:1691–1698. [PubMed] [Google Scholar]
- 31.Tattersall P. Replication of the parvovirus MVM. I. Dependence of virus multiplication and plaque formation on cell growth. J Virol. 1972;10:586–590. doi: 10.1128/jvi.10.4.586-590.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tattersall P, Bratton J. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J Virol. 1983;46:944–955. doi: 10.1128/jvi.46.3.944-955.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tattersall P, D’Abramo A, Cotmore S F. VIIth International Parvovirus Workshop. 1997. Replication competent chimeric parvovirus dependent upon the HIV tat transactivator; p. 5.1. Heidelberg, Germany. [Google Scholar]
- 34.Tattersall P, Gardiner E M. Autonomous parvovirus-host-cell interactions. In: Tijssen P, editor. Handbook of parvoviruses. Vol. 1. Boca Raton, Fla: CRC Press; 1990. pp. 111–121. [Google Scholar]
- 35.Vairo G, Livingston D M, Ginsberg D. Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 36.Vanacker J M, Rommelaere J. Non-structural proteins of autonomous parvoviruses: from cellular effects to molecular mechanisms. Semin Virol. 1995;6:291–297. [Google Scholar]
- 37.Willwand K, Baldauf A Q, Deleu L, Mumtsidu E, Costello E, Beard P, Rommelaere J. The minute virus of mice (MVM) nonstructural protein NS1 induces nicking of MVM DNA at a unique site of the right-end telomere in both hairpin and duplex conformations in vitro. J Gen Virol. 1997;78:2647–2655. doi: 10.1099/0022-1317-78-10-2647. [DOI] [PubMed] [Google Scholar]
- 38.Wilson G M, Jindal H K, Yeung D E, Chen W, Astell C R. Expression of minute virus of mice major nonstructural protein in insect cells—purification and identification of ATPase and helicase activities. Virology. 1991;185:90–98. doi: 10.1016/0042-6822(91)90757-3. [DOI] [PubMed] [Google Scholar]
- 39.Wolter S, Richards R, Armentrout R W. Cell cycle-dependent replication of the DNA of minute virus of mice, a parvovirus. Biochim Biophys Acta. 1980;607:420–431. doi: 10.1016/0005-2787(80)90152-5. [DOI] [PubMed] [Google Scholar]