Abstract
The human cytomegalovirus (HCMV) gCIII complex contains glycoprotein H (gH; gpUL75), glycoprotein L (gL; gpUL115), and glycoprotein O (gO; gpUL74). To examine how gH, gL, and gO interact within HCMV-infected cells to assemble the tripartite complex, pulse-chase experiments were performed. These analyses demonstrated that gH and gL associate by the end of the pulse period to form a disulfide dependent gH-gL complex. Subsequently, the gH-gL complex interacts with a 100-kDa precursor form of gO to form a 220-kDa precursor of the mature gH-gL-gO complex that contains a 125-kDa form of gO. The 220-kDa precursor complex (pgCIII) was sensitive to treatment with endoglycosidase H (endo H), while the mature gCIII complex was essentially resistant to digestion with this enzyme, suggesting that formation of pgCIII complex occurs in the endoplasmic reticulum (ER) and is processed to mature gH-gL-gO (gCIII) in a post-ER compartment. While the N-linked glycans on the 100-kDa form of gO were modified to endo H-resistant states as the 125-kDa gO formed, additional posttranslational modifications were detected on gO. These processing alterations were non-N-linked oligosaccharide modifications that could not be accounted for by phosphorylation or by O-glycosylation of the type sensitive to O-glycanase. Of gH, gL, gO, and the various complexes that they form, only the mature form of the complex was detectable at the infected cell membrane, as judged by surface biotinylation studies.
Human cytomegalovirus (HCMV), the largest of the human herpesviruses, has a glycoprotein coding capacity unparalleled by other viruses. Laboratory strains, such as AD169, contain at least 57 open reading frames (ORFs) with the predictive features of glycoproteins, while clinical isolates, such as Toledo, contain an additional 13 ORFs that may also encode glycoproteins (5, 6). This tremendous glycoprotein coding capacity implies a large array of glycoproteins, some of which are likely functionally redundant, while others are proposed to play specialized functional roles tailored to replication and pathogenic features in the biology of HCMV infection. It is noteworthy, therefore, that few glycoprotein gene products have been characterized with respect to biosynthesis within infected cells and incorporation into the virion. To date, only one envelope glycoprotein, glycoprotein B (gB; gpUL55), has been intensively characterized at this level (reviewed in reference 3).
The gCIII complex is one of three high-molecular-mass, disulfide-dependent glycoprotein complexes found in the HCMV envelope (11). For many years, the only known component of the 240-kDa gCIII complex was the glycoprotein H (gH) homolog (8, 11, 26, 27). Based on the proposed function of HCMV gH (19, 24, 26), this glycoprotein complex likely has an indispensable role in viral fusion events. Subsequently, the HCMV glycoprotein L (gL) homolog (UL115) (18, 29) was also found to be a constituent of the gCIII complex (15, 21). Recently, a third viral gene product was confirmed to be a member of the gCIII complex (15, 21). We have reported the identification of this third component as the product of the HCMV UL74 ORF, which we designated glycoprotein O (gO) (16). Thus, it is now known that the gCIII is a heterotrimeric glycoprotein complex composed of the products of three distinct HCMV genes, UL75 (gH), UL115 (gL), and UL74 (gO).
The purpose of this study was to characterize the biosynthesis of gO and to define the sequence of events that leads to the formation of the mature complex containing all three proteins. Pulse chase analysis revealed a precursor-product relationship between a 100-kDa endoglycosidase H (endo H)-sensitive form of gO and the diffusely migrating, endo H-resistant 125-kDa form of gO. The 125-kDa gO, which is the form found in mature gCIII, has additional posttranslational modifications that could not be attributed to N-glycosylation or phosphorylation, nor could they be identified as O-linked glycosylation. Our study also revealed that the tripartite gH-gL-gO complex assembles in two distinct steps. First, gH and gL associate via disulfide bonding with very rapid kinetics to form a gH-gL complex. Subsequently, the 100-kDa form of gO associates with gH-gL to form a 220-kDa high-mannose-decorated precursor gCIII (pgCIII) complex, which is processed to the mature gCIII complex in a post-endoplasmic reticulum (ER) compartment. Of the various forms of gH, gL, gO, and their associated complexes, only the mature gCIII complex was detectable at the plasma membrane of infected cells. Together, these data form the basis for an understanding of the pathway required for formation of the tripartite gH-gL-gO complex.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Human fibroblast (IF) cells were cultured as previously described (7). The AD169 strain of HCMV was grown and titered as previously described (7). Monoclonal antibodies 14-4b, 27-78, and 7-17, generously supplied by W. Britt, and polyclonal antibody 26388, kindly provided by A. Minson, were described previously (2, 4, 15). The generation of antibody 6824 and of the anti-UL74 (gO) serum was also described previously (7, 16).
Radiolabeling of infected cell proteins.
Steady-state labeling of HCMV-infected IF cells was performed as previously described (15). For 32P labeling, infected cells were incubated for 16 h in medium containing 10% fetal bovine serum and [32P]orthophosphate (500 mCi/ml; Amersham). For pulse-chase labeling, IF cells were infected at a multiplicity of infection of approximately 3. At 3 days postinfection, cells were starved for 1 h in methionine-cysteine-deficient medium and then pulse-labeled with 300 μCi of [35S]methionine-cysteine (NEN) per ml for 20 min. Cell monolayers were rinsed with phosphate-buffered saline (PBS) and chased in cell culture medium supplemented with 10% fetal bovine serum and a 100-fold concentration of cold methionine. At various time intervals, the cells were harvested, lysed, and subjected to immunoprecipitation.
Immunoprecipitations.
Immunoprecipitations were performed as previously described (15). For sequential immunoprecipitations, the antibody-antigen complexes from the primary immunoprecipitations were eluted from the protein A beads by incubation in 2% sodium dodecyl sulfate (SDS)–50 mM dithiothreitol (DTT) at 95°C for 3 min. The eluted protein solutions were diluted up to 1 ml with lysis buffer (for a final concentration of 0.2% SDS–2.5 mM DTT), the second immunoprecipitating antibody was added, and the immunoprecipitation proceeded as usual. Immunoprecipitates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). The resultant gels were dried and imaged with a GS-525 Molecular Imager (Bio-Rad).
Enzymatic digestions.
For digestion with endo H (Boehringer Mannheim), immunoprecipitated proteins were eluted from the protein A beads in 0.5% SDS at 95°C for 3 min. An equal volume of 100 mM sodium citrate (pH 5.5) was added to the eluted protein solution. Each sample was divided into two aliquots, one receiving 10 mU of enzyme and the other receiving an equal volume of PBS. Digestion was allowed to proceed for 16 h at 37°C. For digestion with peptide:N-glycosidase F (PNGase; Boehringer Mannheim), immunoprecipitated proteins were eluted from the protein A beads in a solution of 0.45% SDS in PBS at 95°C for 3 min. The eluted protein solution was diluted twofold with PBS and made 50 mM EDTA and 1% Nonidet P-40. Each sample was split into two aliquots, one receiving 2 U of enzyme and the other receiving an equal volume of PBS. Digestion was allowed to proceed for 16 h at 37°C. For digestion with neuraminidase and O-glycosidase (Boehringer Mannheim), immunoprecipitated proteins were eluted from the protein A beads in a solution of 0.45% SDS in PBS (pH 6.3) at 95°C for 3 min. The eluted protein solution was diluted twofold with PBS (pH 6.3) and made 1% in Nonidet P-40. Each sample was split into two aliquots, one receiving 15 mU of neuraminidase and the other receiving an equal volume of PBS (pH 6.3). After a 4-h incubation at 37°C, 3 mU of O-glycosidase or an equal volume of PBS (pH 6.3) was added to the appropriate samples, and incubation proceeded for an additional 12 h.
Immunoblotting.
Immunoblotting was performed as previously described (15). In brief, proteins were resolved by SDS-PAGE (with and without reducing agents) and electrotransferred to nitrocellulose (Millipore) for immunoblotting. Primary antibodies were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit antibodies (Pierce). Streptavidin conjugated to HRP (SA-HRP; (Vector Laboratories) was used for detection of biotinylated proteins. Renaissance Western blot chemiluminesence reagent (NEN) was used to detect the HRP conjugates.
Cell surface biotinylation.
Biotinylation was performed essentially as described previously (16). In brief, infected cell monolayers were washed with PBS-MC (PBS supplemented with 1 mM MgCl2 and 0.1 mM CaCl2) and chilled to 4°C. EZ-link sulfo-NHS-LC biotin (Pierce) in PBS-MC (2 mg/ml) was added, and the cells were incubated at 4°C for 1 h. The biotin solution was removed, and cells were washed extensively with PBS-MC. To quench the biotinylation reaction, 10 mM glycine in PBS-MC was added to the cells for 10 min at 4°C. The glycine solution was removed, and the cells were washed extensively with PBS-MC.
RESULTS
Kinetics of intracellular association of gH, gL, and gO.
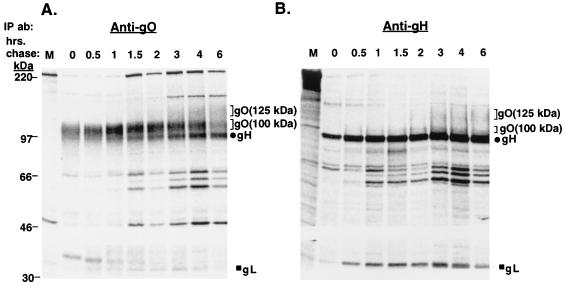
To characterize the biosynthesis of gO in comparison to one of its complex partners, gH, pulse-chase labeling experiments were performed. HCMV-infected cell proteins were immunoprecipitated by anti-gO or anti-gH antibodies and analyzed by reducing SDS-PAGE (Fig. 1). In immunoprecipitations with the anti-gO antibody (Fig. 1A), a 100-kDa protein was recovered in the pulse sample (0 h of chase). Concurrent with the decrease in abundance of the 100-kDa protein at later chase times was the appearance of a diffusely migrating 125-kDa species. The migration pattern of this 125-kDa protein was similar to that of the 125-kDa gO found in the gCIII complex (15, 16, 21). Immunoblotting of the immunoprecipitated 125- and 100-kDa species revealed that both of these proteins were reactive with the anti-gO antibody, confirming their identities as forms of gO (data not shown). Given the order of appearance of these two forms, it is likely that the 100-kDa gO is a precursor of the 125-kDa gO. The processing of the 100-kDa gO to the 125-kDa gO was relatively slow, requiring nearly 6 h of chase. In addition to these two forms of gO, two other coprecipitating proteins of approximately 86 and 34 kDa were detected beginning at 1 to 2 h of chase. The electrophoretic mobilities of these coprecipitating species suggested they represent gH and gL, respectively.
FIG. 1.
Pulse-chase analysis of HCMV-infected cells (reducing SDS-PAGE). HCMV-infected cells were pulse-labeled and chased for various intervals up to 6 h. Cell lysates were immunoprecipitated and subjected to reducing SDS-PAGE. (A) Immunoprecipitations with the anti-gO antibody; (B) immunoprecipitations with the anti-gH antibody 14-4b. IP ab, immunoprecipitating antibody.
Pulse-chase analysis using an anti-gH antibody revealed a similar pattern of immunoprecipitated proteins (Fig. 1B). At 0 h of chase, gH (86 kDa) was readily apparent. An approximately 34-kDa protein, likely representing gL, was also coprecipitated with gH during the pulse period. At approximately 1 h of chase, a faint 100-kDa protein migrating directly above gH was seen; by 2 to 3 h of chase, a 125-kDa coprecipitating protein reminiscent of the 100- and 125-kDa forms of gO seen in Fig. 1A was evident.
Confirmation of the identities of the coprecipitating proteins.
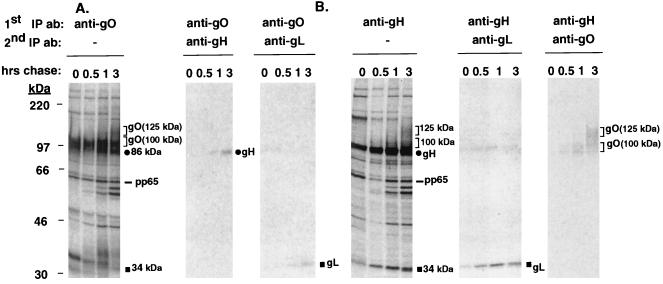
Both the anti-gO and anti-gH antibodies coprecipitated a number of proteins. The mobilities of most of the coimmunoprecipitating proteins were suggestive, but not definitive proof, of their identities. To verify the identities of the coimmunoprecipitating species, pulse-chase samples were first immunoprecipitated with anti-gO or anti-gH antibodies; proteins recovered in the precipitate were released and reimmunoprecipitated with anti-gH, -gL, or -gO antibodies. As shown in Fig. 2A, the 86- and 34-kDa proteins that coprecipitated with the 100-kDa form of gO by 1 h of chase were gH and gL, respectively. When the anti-gH antibody was used as the initial precipitating antibody (Fig. 2B), the 34-kDa coprecipitating protein was verified as gL, confirming the conclusion that these two proteins associate with very rapid kinetics. In addition, the 100- and 125-kDa proteins detected in the anti-gH immunoprecipitates were confirmed as the 100- and 125-kDa forms of gO (Fig. 2B).
FIG. 2.
Sequential immunoprecipitations of the pulse-chase samples. HCMV-infected cells were pulse-labeled and chased for various intervals up to 3 h. Cell lysates were immunoprecipitated first with either anti-gO or anti-gH antibodies (1st IP ab). The immunoprecipitated proteins were eluted from the protein A beads by incubation in 2% SDS–50 mM DTT at 95°C, diluted in radioimmunoprecipitation assay buffer, and subjected to secondary immunoprecipitation with anti-gH antibody 6824, anti-gL antibody 26388, or the anti-gO antibody (2nd IP ab). (A) Proteins immunoprecipitated with the anti-gO antibody, which were reimmunoprecipitated by anti-gH or anti-gL antibodies. (B) Proteins immunoprecipitated with the anti-gH antibody 14-4b, which were reimmunoprecipitated by anti-gO or anti-gL antibodies.
Besides the gH, gL, and gO proteins identified in the pulse-chase analyses, other coprecipitating proteins of unknown identity were observed (Fig. 1 and 2). A closely migrating series of four bands between 56 and 66 kDa was detected at later chase times in both anti-gO and anti-gH immunoprecipitations (Fig. 1 and 2). An antibody specific for the HCMV pp65 tegument protein was reactive with the slowest-migrating (ca. 66-kDa) protein but with none of the other faster-migrating proteins (data not shown). Also, in anti-gO immunoprecipitations, a coprecipitating protein of approximately 34 kDa was prominent at early chase times and chased into a more diffusely migrating form of ca. 36 kDa, suggestive of a glycosylated protein (Fig. 1A and 2A). Based on the mobility and the possible glycosylated nature of this protein, it was originally hypothesized that it represented gL; however, the gL antibody was not reactive with these 34- to 36-kDa proteins (Fig. 2A). Efforts are now under way to characterize these unidentified coprecipitating proteins.
Kinetics of formation of disulfide-dependent complexes.
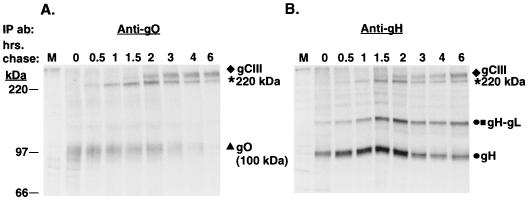
The reduced pulse-chase immunoprecipitations demonstrated the very rapid association of gH and gL (at 0 h of chase), whereas the three proteins (100-kDa gO, gH, and gL) were not coprecipitated until ca. 1 h postsynthesis. To examine the corresponding disulfide-dependent complexes that formed as a result of these various interactions between gH, gL, and gO, immunoprecipitates of the pulse-chase samples were analyzed under nonreducing conditions (Fig. 3). In the anti-gO immunoprecipitates, the 100-kDa gO was evident at 0 h of chase (Fig. 3A). Between 0.5 and 1 h of chase, a species migrating slightly above the 220-kDa protein standard was seen. This species (labeled as 220 kDa in the figures) reached maximum levels by 2 h of chase and subsequently waned over the course of the chase. By 1.5 to 2 h of chase, a species migrating slower than the 220-kDa species was evident. This species was previously demonstrated to be the mature gCIII complex, known to contain gH, gL, and 125-kDa gO (15, 16). Consistent with this finding was the appearance of the 125-kDa form of gO at chase times when the mature gCIII complex was observed (compare Fig. 1 and 3). Of interest is the fact that at no time is free, non-disulfide-bonded 125-kDa gO observed, suggesting it is exclusively contained in the gCIII complex. This is in contrast to gH. Figure 3B shows that gH and a 115-kDa gH-gL dimer (15) were evident in the pulse sample. While the signal strength of these species waned at later chase times, they did not decrease appreciably or to the same degree as for gO. The 220-kDa species was recovered in anti-gH precipitates as well, followed by the appearance of mature gCIII complex at ca. 2 h of chase.
FIG. 3.
Pulse-chase analysis of HCMV-infected cells (nonreducing SDS-PAGE). HCMV-infected cells were pulse-labeled and chased for various intervals up to 6 h. Cell lysates were immunoprecipitated and subjected to nonreducing SDS-PAGE. (A) Immunoprecipitations with the anti-gO antibody; (B) Immunoprecipitations with the anti-gH antibody 14-4b. IP ab, immunoprecipitating antibody.
The 220-kDa species is composed of gH, gL, and the 100-kDa form of gO.
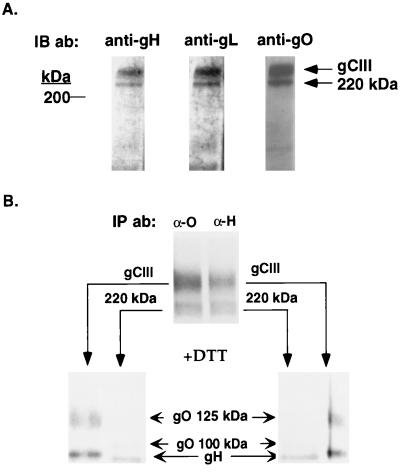
The unidentified 220-kDa species appeared at chase times when gH, gL, and the 100-kDa gO were known to interact (Fig. 1 and 2), suggesting that the 220-kDa species may represent a disulfide-linked complex of these three glycoproteins. Two approaches were used to determine the composition of the 220-kDa species. First, unreduced lysates of HCMV-infected cells were immunoblotted with anti-gH, anti-gL, or anti-gO antibodies (Fig. 4A). Similar to the mature gCIII complex, the 220-kDa species was reactive with antibodies to all three glycoproteins, demonstrating the presence of gH, gL, and some form of gO. We also performed excision/reduction experiments. For this analysis, the immunoprecipitated 220-kDa species was excised from a nonreducing SDS-polyacrylamide gel, reduced, and subjected to a second SDS-PAGE to resolve the separated components of this complex (Fig. 4B). This analysis revealed that the 220-kDa gCIII contained the 100-kDa form of gO, in contrast to the mature gCIII, which contained the 125-kDa form of gO.
FIG. 4.
Analysis of the composition of the 220-kDa species. (A) HCMV-infected cell lysates were subjected to nonreducing SDS-PAGE, electroblotted to nitrocellulose, and probed with anti-gH antibody 6824, anti-gL antibody 26388, or the anti-gO antibody. IB ab, immunoblotting antibody. (B) Excision/reduction analysis of mature gCIII and the 220-kDa species. 35S-labeled HCMV-infected cell lysates were immunoprecipitated with either the anti-gO antibody (α-O) or the anti-gH antibody 14-4b (α-H) and subjected to nonreducing SDS-PAGE. The upper panel shows the resultant autoradiogram of these immunoprecipitations (positions of gCIII and the 220-kDa species are indicated at the sides). gCIII and the 220-kDa species were excised from products of both immunoprecipitations and subjected to reducing SDS-PAGE. The identities of the resolved proteins are marked between the two lower panels.
Endo H sensitivity of gH, gL, gO, and their associated complexes.
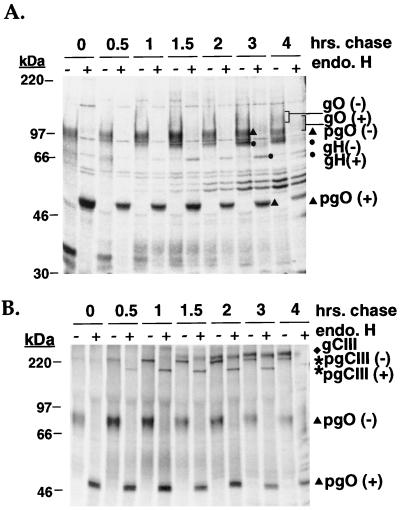
The pulse-chase analyses suggest a stepwise association of gCIII. First, gH and gL associate, followed by their interaction with a 100-kDa form of gO to form the 220-kDa pgCIII complex. Subsequently, the mature form of the gCIII complex, containing gH, gL, and the 125-kDa gO, is evident. The rapid initial association of gH and gL argues that the formation of gH-gL complex occurs in the ER, but the subcellular site where the pgCIII or mature gCIII complex forms is unclear. To investigate whether the pgCIII and/or mature gCIII complex forms in the ER, immunoprecipitate of pulse-chase samples with the anti-gO antibody were digested with endo H, which cleaves high-mannose N-linked glycans from the polypeptide backbone. Sensitivity to endo H is indicative of ER localization, while endo H resistance is evidence of glycoproteins whose N-linked glycans were processed to a complex form in the Golgi complex. Figure 5A shows the digested immunoprecipitated proteins analyzed by reducing SDS-PAGE. The 100-kDa form of gO shifts in electrophoretic mobility to an approximately 54-kDa species in the presence of endo H that correlates well with the predicted polypeptide backbone mass of gO (16). The mobility of the 125-kDa form of gO, seen at later chase times, was only slightly affected by endo H, suggesting that it contained primarily complex-type glycans. These data suggest that the 100-kDa form of gO is a high-mannose precursor of the processed 125-kDa gO. Similarly, the coprecipitating gH, evident by 1 h of chase, was also shifted upon endo H treatment to an approximately 78-kDa species, correlating well with the predicted peptide backbone mass of gH. An endo H-resistant form of gH was apparent at 3 to 4 h of chase. The digestion pattern of gL could not be accurately assessed due to the low level of incorporation of radioactive label and the presence of the unidentified 30- to 35-kDa coprecipitating species. We conducted the same experiment but analyzed the samples under nonreducing conditions (Fig. 5B). The pgCIII complex shifted in mobility upon endo H digestion, demonstrating the presence of high-mannose oligosaccharides and consistent with our finding that the 100-kDa gO is contained in this species. In contrast, the mature form of the complex was largely unaffected by treatment with endo H. This observation is in agreement with the results of Li et al., who demonstrated that endo H treatment of virion-derived gCIII caused only a slight increase in SDS-PAGE mobility (21). These results suggest that gH, gL, and the 100-kDa gO (pgO in Fig. 5) associate to form a 220-kDa pgCIII complex in the ER. The pgCIII complex is processed in the Golgi apparatus to yield the mature form of the gCIII complex, consisting of gH, gL, and the 125-kDa gO, which contain complex N-linked glycans.
FIG. 5.
Endo H digestion of pulse-chase samples. HCMV-infected cells were pulse-labeled and chased for various intervals up to 4 h. Cell lysates were immunoprecipitated with the anti-gO antibody, and the immunoprecipitated proteins were incubated in the presence or absence of endo H as outlined in Materials and Methods. − and + denote positions of the proteins after no treatment and after treatment with endo H, respectively. The digested proteins were resolved by SDS-PAGE under reducing (A) and nonreducing (B) conditions. pgO, precursor gO.
Posttranslational modification of the 125-kDa gO.
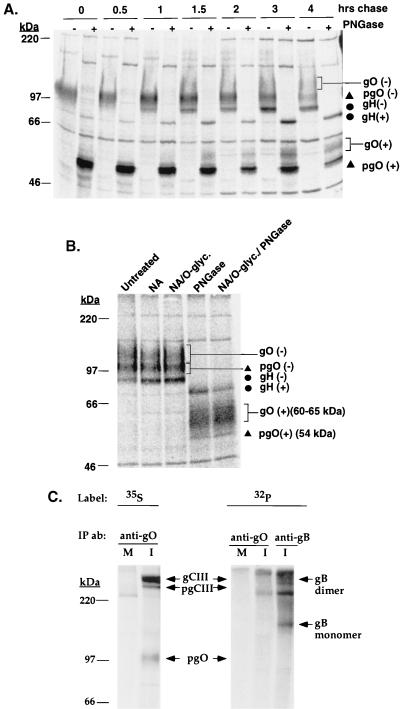
To further characterize the posttranslational modifications associated with gCIII, proteins from anti-gO immunoprecipitations of pulse-chase samples were digested with PNGase to remove all N-linked oligosaccharides. If gO contained exclusively N-linked oligosaccharides, PNGase digestion would be expected to shift the 125-kDa gO to a 54-kDa form (Fig. 6A). As expected, this treatment shifted the mobility of the 100-kDa pgO to 54 kDa. In contrast, the 125-kDa gO did not shift to the 54-kDa polypeptide backbone species but rather shifted to a diffusely migrating species of approximately 60 to 65 kDa. This finding suggests that 125-kDa gO has a non-N-linked oligosaccharide modification accounting for ca. 5 to 10 kDa of mass. The diffuse migration pattern of the 60- to 65-kDa species suggested glycosylation such as O-linked glycans. Analysis of the gO amino acid sequence by the NetOGlyc 2.0 prediction program (12–14) indicated a strong predilection for O-glycosylation of this glycoprotein. To test for the presence of O-linked glycans and sialic acid on the 125-kDa gO, anti-gO immunoprecipitates were digested with neuraminidase, neuraminidase followed by O-glycosidase, or a combination of neuraminidase, O-glycosidase, and PNGase. The mobility of the 125-kDa gO was slightly altered by neuraminidase alone but not significantly affected by digestion with O-glycosidase (Fig. 6B). However, it should be noted that the specificity of O-glycosidase is limited to cleaving only the disaccharide unit Gal-β(1-3)-GalNAc from O-linked oligosaccharides (9) and thus may not cleave the types of O-linked glycans that might be present on gO. To test whether a phosphorylation event could account for this non-N-linked modification on the 125-kDa gO, HCMV-infected cells were metabolically labeled with [32P]orthophosphate. Immunoprecipitations of these cell lysates did not reveal the phosphorylation of either the 220-kDa pgCIII or mature gCIII complex, although gB, which is known to be phosphorylated (10, 25), did incorporate the 32P label (Fig. 6C).
FIG. 6.
(A) PNGase digestion of pulse-chase samples. HCMV-infected cells were pulse-labeled and chased for various intervals up to 4 h. Cell lysates were immunoprecipitated with the anti-gO antibody, and the immunoprecipitated proteins were incubated in the presence or absence of PNGase as outlined in Materials and Methods. − and + denote positions of the proteins after no treatment and after treatment with PNGase, respectively. The digested proteins were resolved by reducing SDS-PAGE. (B) O-glycosidase and neuraminidase digestion of the 125-kDa gO. 35S-labeled HCMV-infected lysates were immunoprecipitated by the anti-gO antibody, and the immunoprecipitated proteins were digested with neuraminidase (NA), neuraminidase followed by O-glycosidase (NA/O-glyc.), neuraminidase followed by O-glycosidase and PNGase (NA/O-glyc./PNGase), or PNGase alone (PNGase) or were untreated. The digested proteins were resolved by reducing SDS-PAGE. (C) Immunoprecipitations of [32P]orthophosphate-labeled HCMV lysates. Mock infected (M) and HCMV-infected (I) cells were metabolically labeled with either [32P]orthophosphate or [35S]methionine-cysteine. Lysates of these labeled cells were immunoprecipitated with either the anti-gO antibody or a cocktail of anti-gB antibodies 7-17 and 27-78. Immunoprecipitated proteins were resolved by nonreducing SDS-PAGE.
The gCIII complex is the predominant form of gH, gL, or gO at the cell surface.
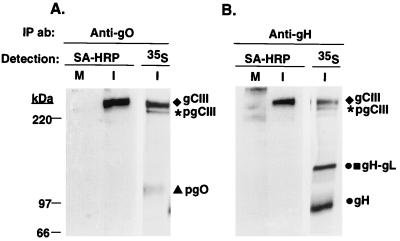
To determine which forms of gH, gL, gO, and their associated complexes were found at the plasma membrane of HCMV-infected cells, cell surface proteins were biotinylated and immunoprecipitated with anti-gO or anti-gH antibodies. Immunoprecipitated proteins were resolved by nonreducing SDS-PAGE, and the biotinylated species were detected with SA-HRP. Figure 7 shows that the gCIII complex was the major biotinylated species in immunoprecipitations with either anti-gO or anti-gH antibodies. In contrast, neither the gH-gL complex nor the pgCIII complex appeared to be biotinylated, suggesting that they were not present at the plasma membrane of infected cells.
FIG. 7.
Cell surface biotinylation of HCMV-infected cells. Mock-infected (M) and HCMV-infected (I) cells were surface biotinylated (described in Materials and Methods), lysed, and immunoprecipitated with either the anti-gO antibody or anti-gH antibody 14-4b. The immunoprecipitated proteins were resolved by nonreducing SDS-PAGE, electroblotted, and probed with SA-HRP. As controls, anti-gO and anti-gH immunoprecipitations of 35S-labeled HCMV-infected lysates were also resolved by nonreducing SDS-PAGE and imaged.
DISCUSSION
The HCMV gCIII is a multicomponent envelope glycoprotein complex that contains the HCMV gH and gL homologs as well as the product of the UL74 gene, gO. The function of the gCIII complex in the viral life cycle is not well defined. Functional studies of HCMV gH and other herpesvirus gH homologs have implicated this glycoprotein in the direct fusion events that occur during entry and cell-to-cell spread of virus, although the specific molecular role of gH is not clear. In the case of HCMV gH, its role in fusion may be facilitated by engagement of a cellular receptor (19, 20). However, this cellular protein has not been genetically characterized. Studies of the gL homologs have clearly demonstrated a chaperone-type function that is generally required for the proper processing and targeting of its partner, gH (8, 17, 18, 22, 28, 29, 31). Whether gL has any additional function(s) is not known. As for HCMV gO and its homologs in other betaherpesviruses, it is possible only to speculate on potential functions in the absence of any direct evidence. As a prerequisite to functional studies of the tripartite complex and its components, it is crucial to have a thorough knowledge of the intracellular processing pathway that these three proteins undergo to produce mature functional complex.
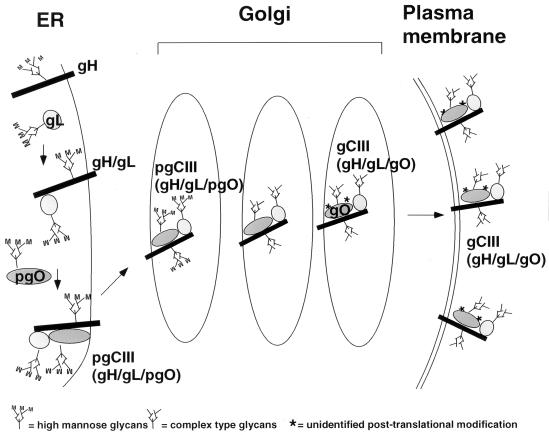
Heterotrimeric viral glycoprotein complexes composed of three distinct gene products are exceedingly rare. To date, only one other tripartite viral glycoprotein complex, the Epstein-Barr virus gH-gL-gp42 complex, has been described (23). In this study, we have examined the intracellular formation and processing of a tripartite viral glycoprotein complex. The results obtained have yielded a model for the biosynthesis of the tripartite gH-gL-gO complex (Fig. 8). Shortly after or coincident with their translation in the ER, gH and gL quickly associate to form a 115-kDa disulfide-dependent gH-gL dimer. Within 60 min of gH-gL formation, a precursor form of gO associates with gH-gL to form the 220-kDa pgCIII complex. Based on the presence of high-mannose N-linked oligosaccharides on pgIII, the formation of the tripartite precursor likely occurs in the ER. By 1.5 to 2 h of chase, mature gH-gL-gO complex becomes apparent, which corroborates earlier time course studies of this complex (1, 21). The subsequent processing of precursor to the mature form of the complex likely occurs in post-ER compartments since the mature form contains mainly complex-type glycans and a 125-kDa form of gO. Our analysis revealed that 125-kDa gO contains additional non-N-linked modifications that we were unable to identify. The presence of O-linked glycosylation on gO is still a possibility, considering the limited type of sugar moieties recognized and cleaved by O-glycosidase (9). Also, other, less common posttranslational modifications, such as sulfation, may be present on the 125-kDa gO. Further analysis of the 125-kDa gO will be required to address this matter. Once these post-ER processing steps have been completed, the mature gH-gL-gO complex is competent to traffic to the plasma membrane. It should be noted that this model assumes a stoichiometric ratio of 1:1:1 for the glycoproteins constituting the gCIII complex, although such a ratio remains to be formally demonstrated, as does the precise molecular mass of the complex.
FIG. 8.
Model of the intracellular formation and processing of the tripartite gH-gL-gO HCMV envelope complex. For detailed description of the model, see the text.
Although this model provides a basic framework for understanding the intracellular processing of the gH-containing complex, more detailed analyses will be required to answer additional questions concerning the associations of gH, gL, and gO. Specifically, it is not known how interdependent gH, gL, and gO are for their processing and targeting. It has been well documented that gH requires association with gL for proper processing of gH’s N-linked glycans and for targeting of the resulting gH-gL complex to the plasma membrane (17, 18, 22, 24, 28, 29, 31). Coexpression of gH and gL in the absence of other HCMV gene products has been demonstrated to be sufficient for transport of gH-gL complex to the cell surface, implying that gH and gL do not require any additional viral gene products for targeting to the plasma membrane (18, 24, 29). Our analyses also revealed the formation of a gH-gL complex in HCMV-infected cells, but interestingly, this gH-gL complex was not detectable at the cell surface. This finding suggests that the gH-gL complex is unable to reach the cell surface without coexpression of gO in HCMV-infected cells. Currently, our laboratory is generating a gO-deficient HCMV strain to address the intricate nature of the gH, gL, and gO interactions. Likewise, it is not known whether proper processing of gO requires gH and/or gL. Also, gO may be dependent on association with gH for its membrane localization, as preliminary evidence suggests that gO may be a soluble, rather than membrane-spanning, protein (data not shown). An in-depth characterization of gO is now under way.
Another intriguing issue involves the form(s) of the gH-containing complex(es) in the virion. Li et al. have reported that in addition to the three-protein complex, free uncomplexed forms of gH exist in the viral envelope (21). Although our study did not investigate virion-associated forms of gH, gL, and gO, we did demonstrate that mature form of the complex was the predominant form of gH at the cell surface, while no uncomplexed forms of gH were detectable. Our findings do not necessarily rule out the possibility that uncomplexed forms of gH are present in other subcellular membranes, in particular those that may serve as sites of envelopment for HCMV. In fact, our pulse-chase analyses did demonstrate the persistence of free gH and gH-gL dimer throughout a 6-h chase. It is interesting that recent studies of the Epstein-Barr virus gH-gL-gp42 complex have suggested that the viral envelope contains both bipartite gH-gL complexes and tripartite gH-gL-gp42 complexes (30). Additional in-depth investigation of gH, gL, gO, and their intracellular and virion-associated complexes will be required for a complete understanding of their ramifications with respect to the biology of HCMV.
ACKNOWLEDGMENT
This study was supported in part by Public Health Service grant AI-34998.
REFERENCES
- 1.Bogner E, Reschke M, Reis B, Britt W, Radsak K. Recognition of compartmentalized intracellular analogs of glycoprotein H of human cytomegalovirus. Arch Virol. 1992;126:67–80. doi: 10.1007/BF01309685. [DOI] [PubMed] [Google Scholar]
- 2.Britt W J. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology. 1984;135:369–378. doi: 10.1016/0042-6822(84)90193-4. [DOI] [PubMed] [Google Scholar]
- 3.Britt W J, Mach M. Human cytomegalovirus glycoproteins. Intervirology. 1996;39:401–412. doi: 10.1159/000150510. [DOI] [PubMed] [Google Scholar]
- 4.Britt W J, Vulger L, Butfiloski E J, Stephens E B. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J Virol. 1990;64:1079–1085. doi: 10.1128/jvi.64.3.1079-1085.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cha T-A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 7.Compton T. An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J Virol. 1993;67:3644–3648. doi: 10.1128/jvi.67.6.3644-3648.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cranage M P, Smith G L, Bell S E, Hart H, Brown C, Bankier A T, Tomlinson P, Barrell B G, Minson T C. Identification and expression of a human cytomegalovirus glycoprotein with homology to the Epstein-Barr virus BXLF2 product, varicella-zoster virus gpIII, and herpes simplex virus type 1 glycoprotein H. J Virol. 1988;62:1416–1422. doi: 10.1128/jvi.62.4.1416-1422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endo Y, Kobata A. Partial purification and characterization of an endo-alpha-N-acetylgalactosaminidase from the culture of medium of Diplococcus pneumoniae. J Biochem (Tokyo) 1976;80:1–8. doi: 10.1093/oxfordjournals.jbchem.a131240. [DOI] [PubMed] [Google Scholar]
- 10.Fish K N, Soderberg-Naucler C, Nelson J A. Steady-state plasma membrane expression of human cytomegalovirus gB is determined by the phosphorylation state of Ser900. J Virol. 1998;72:6657–6664. doi: 10.1128/jvi.72.8.6657-6664.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gretch D R, Kari B, Rasmussen L, Gehrz R C, Stinski M F. Identification and characterization of three distinct families of glycoprotein complexes in the envelopes of human cytomegalovirus. J Virol. 1988;62:875–881. doi: 10.1128/jvi.62.3.875-881.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen J E, Lund O, Engelbrecht J, Bohr H, Nielsen J O. Prediction of O-glycosylation of mammalian proteins: specificity patterns of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase. Biochem J. 1995;308(Pt. 3):801–813. doi: 10.1042/bj3080801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen J E, Lund O, Nielsen J O, Brunak S. O-GLYCBASE: a revised database of O-glycosylated proteins. Nucleic Acids Res. 1996;24:248–252. doi: 10.1093/nar/24.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen J E, Lund O, Tolstrup N, Gooley A A, Williams K L, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconi J. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- 15.Huber M T, Compton T. Characterization of a novel third member of the human cytomegalovirus glycoprotein H-glycoprotein L complex. J Virol. 1997;71:5391–5398. doi: 10.1128/jvi.71.7.5391-5398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber M T, Compton T. The human cytomegalovirus UL74 gene encodes the third component of the glycoproteinH-glycoprotein L-containing envelope complex. J Virol. 1998;72:8191–8197. doi: 10.1128/jvi.72.10.8191-8197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson A C, Johnson D C. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992;66:2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaye J F, Gompels U A, Minson A C. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J Gen Virol. 1992;73:2693–2698. doi: 10.1099/0022-1317-73-10-2693. [DOI] [PubMed] [Google Scholar]
- 19.Keay S, Baldwin B. Anti-idiotype antibodies that mimic gp86 of human cytomegalovirus inhibit viral fusion but not attachment. J Virol. 1991;65:5124–5728. doi: 10.1128/jvi.65.9.5124-5128.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keay S, Merigan T C, Rasmussen L. Identification of cell surface receptors for the 86-kilodalton glycoprotein of human cytomegalovirus. Proc Natl Acad Sci USA. 1989;86:10100–10103. doi: 10.1073/pnas.86.24.10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Nelson J A, Britt W J. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J Virol. 1997;71:3090–3097. doi: 10.1128/jvi.71.4.3090-3097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Buranathai C, Gross C, Hutt-Fletcher L M. Chaperone functions common to nonhomologous EBV gl and VSV gL proteins. J Virol. 1997;71:1667–1670. doi: 10.1128/jvi.71.2.1667-1670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, Turk S M, Hutt-Fletcher L M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milne R S, Paterson D A, Booth J C. Human cytomegalovirus glycoprotein H/glycoprotein L complex modulates fusion-from-without. J Gen Virol. 1998;79(Pt.4):855–865. doi: 10.1099/0022-1317-79-4-855. [DOI] [PubMed] [Google Scholar]
- 25.Norais N, Hall J A, Gross L, Tang D, Kaur S, Chamberlain S H, Burke R L, Marcus F. Evidence for a phosphorylation site in cytomegalovirus glycoprotein gB. J Virol. 1996;70:5716–5719. doi: 10.1128/jvi.70.8.5716-5719.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pachl C, Probert W S, Hermsen K M, Masiarz F R, Rasmussen L, Merigan T C, Spaete R R. The human cytomegalovirus strain Towne glycoprotein H gene encodes glycoprotein p86. Virology. 1989;169:418–426. doi: 10.1016/0042-6822(89)90167-0. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen L, Nelson R, Kelsall D, Merigan T. Murine monoclonal antibody to a single protein neutralizes the infectivity of human cytomegalovirus. Proc Natl Acad Sci USA. 1984;81:876–880. doi: 10.1073/pnas.81.3.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spaete R R, Perot K, Scott P I, Nelson J A, Stinski M F, Pachl C. Coexpression of truncated human cytomegalovirus gH with the UL115 gene product or the truncated human fibroblast growth factor receptor results in the transport of gH to the cell surface. Virology. 1993;193:853–861. doi: 10.1006/viro.1993.1194. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Kenyon W J, Li Q, Mullberg J, Hutt-Fletcher L M. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J Virol. 1998;72:5552–5558. doi: 10.1128/jvi.72.7.5552-5558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaswen L R, Stephens E B, Davenport L C, Hutt-Fletcher L M. Epstein-Barr glycoprotein gp85 associates with the BKRF2 gene product and is incompletely processed as a recombinant protein. Virology. 1993;195:387–396. doi: 10.1006/viro.1993.1388. [DOI] [PubMed] [Google Scholar]