Abstract
Background:
Colorectal cancer screening programs with collection of fecal samples may provide a platform for population-based gut microbiome-disease research. We investigated the impact of sample collection and storage methods on the accuracy and stability of the V3-V4 region of the 16S rRNA genes and bacterial quantity across seven different collection methods (i.e. no solution, two specimen collection cards and four types of fecal immunochemical test (FIT) used in four countries) among 19 healthy volunteers.
Methods:
Intraclass correlation coefficients (ICCs) were calculated for the relative abundance of the top three phyla, the most abundant genera, alpha-diversity metrics and the first principal coordinates of the beta-diversity matrices to estimate stability of microbial profiles after storage for 7 days at room temperature, 4°C, 30°C and after screening for the presence of occult blood in the stool, and accuracy compared to samples frozen immediately with no solution (i.e. the putative gold standard).
Results:
When compared to the putative gold standard, significant variation was observed for all collection methods, however, inter-individual variability was much higher than the variability introduced by the collection method. Stability ICCs were high (≥0.75) for FIT tubes that underwent colorectal cancer screening procedures, except for the relative abundance of Actinobacteria (0.65), and were lower for different FIT tubes stored at 30°C (range, 0.41–0.90) and at room temperature (range, 0.06–0.94).
Conclusions:
Paper-based collection cards and different types of FIT are acceptable tools for microbiome measurements.
Impact:
Our findings inform on the utility of commonly used fecal sample collection methods for developing microbiome-focused cohorts nested within screening programs.
Keywords: microbiome, colorectal cancer, screening programs, fecal immunochemical test, specimen collection cards
Introduction
Evidence on the role of the human microbiome in the development of chronic diseases such as obesity, diabetes and, potentially cancer, is growing (1–3). However, most of the current epidemiological literature is based on cross-sectional studies that used diverse methods (4). Most established prospective cohorts did not collect fecal samples and repeated, prospectively collected samples are likely necessary for advancing understanding of the relationship between the microbiome and chronic disease development. Colorectal cancer screening programs, which provide screening through fecal tests such as the fecal immunochemical test (FIT), may offer great potential for establishing population-based cohort studies with multiple fecal specimens and epidemiologic data.
The impact of fecal sample collection methods on gut microbiome parameters has been recently investigated (5–10). Microbial populations in fecal samples collected using fecal occult blood tests (FOBT) and FIT have been found to be stable at room temperatures for up to 4 to 7 days, with similar microbial communities compared to samples collected without an additive and frozen immediately. For example, in one study conducted among 52 healthy volunteers in the United States, all fecal sample collection methods yielded microbial data that appeared relatively reproducible, stable, and accurate, when compared to the putative gold standard, and provided evidence that these collection methods can be employed for microbiome analyses in population-based studies (8).
However, to potentially inform on the establishment of international studies based in colorectal cancer screening programs, additional methodologic work is needed to test FIT and specimen collection cards used in other countries where different FIT methods and screening procedures are employed. Therefore, we evaluated microbial stability in fecal specimens stored at room temperatures and accuracy of microbiome metrics from two different specimen collection cards (used in Afghanistan) and four different FIT tubes used in ongoing international colorectal cancer screening programs (in France and most European countries, Morocco, Turkey and Iran). Additionally, we investigated the impact of colorectal cancer screening procedures and alternative shipping temperatures (e.g., summer and winter temperatures) on microbiome accuracy and stability ascertained from FIT samples.
Methods
Study participants –
Nineteen healthy participants from the International Agency for Research on Cancer (IARC) personnel were recruited in Lyon, France. Eligible participants were at least 18 years of age or older and had not taken antibiotics in the past 3 months. This study was conducted in accordance with the Declaration of Helsinki. All participants provided written informed consent, and the study was approved by the IARC Ethics Committee. On average, participants were 40.2 years old and had a body mass index of 22.7 kg/m2. Most participants were female (78.9%), had no weight variation in the past six months (73.7%) and reported having a regular bowel movement at least once per day (94.8%) (Table S2 in supplementary material).
Fecal sample collection –
At recruitment, participants were provided with a fecal sample collection kit including all materials needed for sample collection at the workplace or at home. Participants were asked to provide a fecal sample in the fecal collection containers (Sarstedt, Nümbrecht, Germany) by filling the scoop contained in each tube. The participant collected the fecal sample at the workplace (n=9) or at home (n=10) and returned it to the study coordinator within a few hours after collection. The study coordinator then delivered it to the laboratory for immediate processing. The participants completed a questionnaire that was used to obtain information on the time and date of sample collection, typical bowel movements and general information.
Following the collection step, the fecal samples were manually homogenized and aliquoted for the different collection methods (Figure S1 in supplementary material). For each participant, fecal samples were aliquoted into two cryotubes without solution (considered to be the putative gold standard, average weight per cryotube 208.1 mg), two GenSaver specimen collection cards (Ahlstrom-Munksjö, Helsinki, Finland) used in Afghanistan, two GenCollect specimen collection cards (Ahlstrom-Munksjö, Helsinki, Finland) used in Afghanistan, four OC-Auto Sampling tubes (Eiken Chemical, Tokyo, Japan) used in France, two Hemotrust tubes (Biosynex, Illkirch-Graffenstaden, France) used in Morocco, two One-Step FOB tubes (Padyabteb, Tehran, Iran) used in Iran, and two Specimen Collection Container A tubes (Alfresa Pharma, Osaka, Japan) used in Turkey. For specimen collection cards, a disposable spatula was used to smear a small portion of the homogenised feces on each window of the cards; the flaps on the cards were closed and then each card was placed in a separate biohazard bag with desiccant. The FIT tubes were filled following the instructions provided by the different colorectal cancer screening programs. Specifically, the FIT probes were dipped into the homogenised fecal specimen and returned to the FIT tubes and were then shaken to mix as instructed. All steps were performed by the same laboratory technician.
Fecal sample storage –
The study samples are outlined in Table S1. After sample collection and processing, the two cryotubes and one of each sample type were immediately stored at −80°C. One of the OC-Auto Sampling tubes was placed in a blue mailer for colorectal cancer screening and mailed to the Reference Centre for Epidemiology and Cancer Prevention in Piemonte (CPO Piemonte) laboratory within 24 hours after processing. To mimic mailing during seasonal temperatures in France, one of the OC-Auto Sampling tubes was stored at a winter temperature (4°C) for 7 days in a refrigerator, returned to room temperature for at least 4 hours, and then frozen at −80°C. The remaining OC-Auto Sampling tubes was stored at a summer temperature (30°C) for 7 days in a water bath, returned to room temperature for at least 4 hours, and then frozen at −80°C. The remaining FIT tubes (i.e. Hemotrust, two One-Step FOB and Specimen Collection Container A tubes) were stored for 7 days at room temperature and then frozen at −80°C. The remaining half of the specimen collection cards (i.e. GenCollect and GenSaver cards) remained at room temperature in a closed cupboard from the date of collection until the date of DNA extraction (average 70.2 days).
On arrival at the CPO Piemonte laboratory, the FIT tubes were processed using standard colorectal cancer screening procedures (11). After testing for occult blood, the FIT tubes were immediately closed with parafilm and shipped at room temperature to the Micalis Institute (INRAE/AgroParisTech) in Jouy-en-Josas, France for 16S rRNA gene profiling. Upon receipt of all the specimens at the same time, the samples were removed from the tube using pliers and a sterile pipette, transferred to a 1.5 mL Eppendorf tube, shaken, and then two 250 μL aliquots were transferred to a sterile tube and frozen at −80°C.
DNA extraction, real-time quantitative PCR and 16SrRNA sequencing –
DNA extraction, real-time PCR and 16S rRNA sequencing methods are described in the supplementary material. Briefly, the samples were shipped to the Micalis Institute (INRAE/AgroParisTech) in Jouy-en-Josas, France and DNA was extracted with the PowerFecal DNA Isolation kit after the reception of the samples. The V3-V4 region of the 16S rRNA gene was PCR amplified and sequencing was performed with Illumina technology with MiSeq kit V2 2 × 250 bp. Quality control procedures and DNA concentrations available from the different sampling methods are described in the supplementary material (Tables S3 and S4).
Bioinformatics –
Data were stored on secured servers at INRAE MAIAGE (Jouy-en-Josas, France) and IARC (Lyon, France). Sequencing data were analyzed with Find, Rapidly, OTUs with Galaxy Solution (FROGS) v3.1.0 (12). Briefly, this pipeline included a pre-processing step where reads were merged with Paired-End Read Merger (PEAR) (13), dereplicated, and filtered according to their length, mismatches in primers with cutadapt (14), and N content. This step was followed by Swarm clustering (15) with an agglomeration distance of d = 3. Chimera detection was then performed using VSEARCH (16) before applying an Operational Taxonomic Unit (OTU) abundance filter (OTUs <0.005% of the total abundance are discarded). The most abundant sequence of each OTU was then affiliated with 100% similarity with blastn against the Silva v138 database (17). After processing, a total number of 8,239,370 sequences was found with an average number of 26,751 sequences per sample without any major deviation by sampling type. The average number of OTUs and genera per samples were 521 and 115 respectively. Two samples were discarded because of low sequencing depth and all remaining samples were rarefied to 7,144 reads per sample using R package phyloseq. Diversity metrics were then computed to represent the diversity of OTUs in each sample (alpha diversities: Shannon index, number of observed OTUs and Inverse Simpson index) and the differences between samples (beta diversities: Jaccard, Bray-Curtis, weighted and unweighted UniFrac distances).
Statistical analysis –
Statistical analyses were conducted using R, version 3.6.2 and the packages DESeq2, icc, phyloseq and vegan (18–21). Descriptive characteristics of study participants were based on the questionnaire provided by the participants. We performed visualization using multidimensional scaling (MDS) plots and all samples from a given participant tended to group together. To assess the impact of the protocol on the alpha diversity, comparison of measures of alpha diversity between each fecal collection method was performed and a linear mixed-effects models with the collection method as fixed effect and the participant as random effect was fitted to the data and used to calculate least-squares means of the alpha diversity metrics. Mean diversities for the collection methods were then compared using Tukey’s HSD tests. To estimate a distance-based coefficient of determination (R2) explained by participant and collection method from unweighted UniFrac, weighted UniFrac, Jaccard and Bray-Curtis distance matrices, permutational multivariate analysis of variance was performed (adonis() function, vegan package, R) (21). We calculated intraclass correlation coefficients (ICCs) using the variance components from a one-way ANOVA to evaluate the stability and accuracy of the different fecal collection methods. The ICCs were calculated based on (i) the square root of the relative abundances of the three most dominant phyla (Actinobacteria, Bacteroidetes, and Firmicutes) and the most abundant genera (Faecalibacterium, Bacteroides, UCG-002, Subdoligranulum, Roseburia, Eubacterium eligens group, Blautia, Christensenellaceae R-7 group, Ruminococcus), which were present in at least 50% of fecal samples with relative abundance of ≥0.1%, (ii) our three alpha diversity metrics, and (iii) the first multidimensional scaling axis, also called first principal coordinate (PC1), of our four beta diversity metrics. The first axis explained 8.2%, 15.8%, 17.6%, and 49.7% of the variability for Jaccard, Bray-Curtis, unweighted UniFrac and weighted UniFrac distances, respectively. To calculate accuracy ICCs, we compared one replicate of samples without solution frozen immediately (considered as the gold standard), selected randomly, to one sample from each of the other collection methods for each participant. To calculate stability ICCs at different temperatures and procedures for each fecal collection method, we compared one sample frozen immediately to one stored at different conditions for each participant. The 95% confidence interval was estimated using the ICCest() function from the R ICC package with default option confidence interval = “Smith”.
Results
Microbial alpha diversity by collection method –
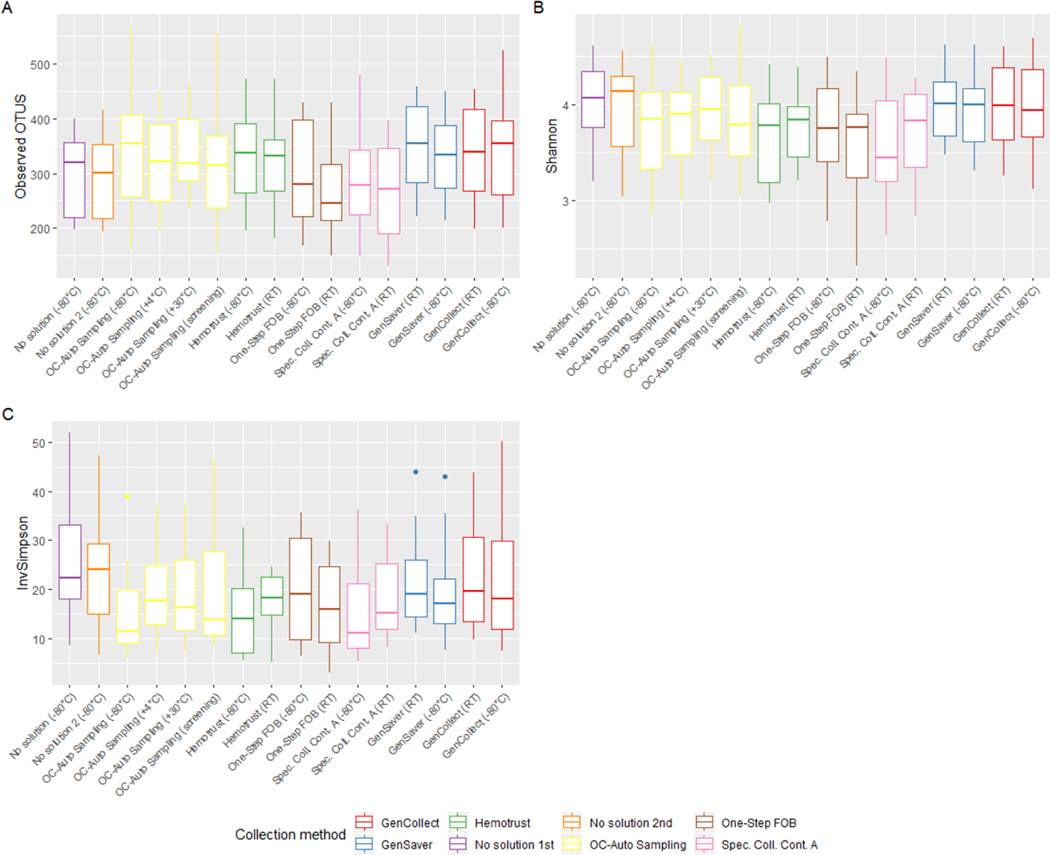
The Shannon index and inverse Simpson index values were, on average, highest for the immediately frozen samples without solution, (i.e. the putative gold standard) and the number of observed OTUs appeared highest in the specimen collection cards (Figure 1). However, the different storage conditions did not seem to have a statistically significant impact on these alpha diversity metrics. From the linear mixed-effects model, we found that compared to the immediately frozen samples without solution (296, 95% CI [257; 335]), the observed number of OTUs was significantly higher in GenSaver cards at room temperatures (347, 95% CI [308; 387], P = 0.01) and in OC-Auto Sampling FIT tubes at −80°C (344, 95% CI [305; 383], P = 0.03). Compared to the defined gold standard (4.03, 95% CI [3.81; 4.24]), the Shannon index was significantly lower in OC-Auto Sampling FIT tubes at −80°C (3.73, 95% CI [3.51; 3.94], P = 0.005), One-Step FOB tubes at −80°C and at room temperatures (3.76, 95% CI [3.54; 3.98], P=0.03 and 3.49, 95% CI [3.27; 3.72], P < 0.001, respectively), and in Specimen Collection Container A tubes at −80°C and at room temperatures (3.60, 95% CI [3.38, 3.82], P < 0.001 and 3.70, 95% CI [3.48; 3.91], P < 0.001, respectively).
Figure 1 –
Impact of specimen collection methods on microbial alpha diversity indexes.
Legend: Boxes represent the median and interquartile range for observed OTUs (A), Shannon index (B) and inverse Simpson index (C) by specimen collection method (n=302).
Percent variability explained by participant and collection method –
All ordinations suggested that the between participant variability was higher than the technical variability (Figure S2 in supplementary material). In multidimensional scaling (MDS) plots, each point represents one microbiome sample for 19 participants and seven collection methods; all samples from a given participant tended to group together (as shown by ellipses, one per participant). Additionally, based on four distance matrices (i.e. Jaccard, Bray-Curtis, unweighted UniFrac and weighted UniFrac), the overall variability in diversity was largely explained by between participants (55% to 79%) and only marginally by collection methods (4.8% to 14.8%, Figure S3). For example, based on the Bray-Curtis distance matrix, the protocol variability accounted for roughly 6.9% of the overall variability whereas the biological variability accounted for 79.0%.
Relative abundance comparisons –
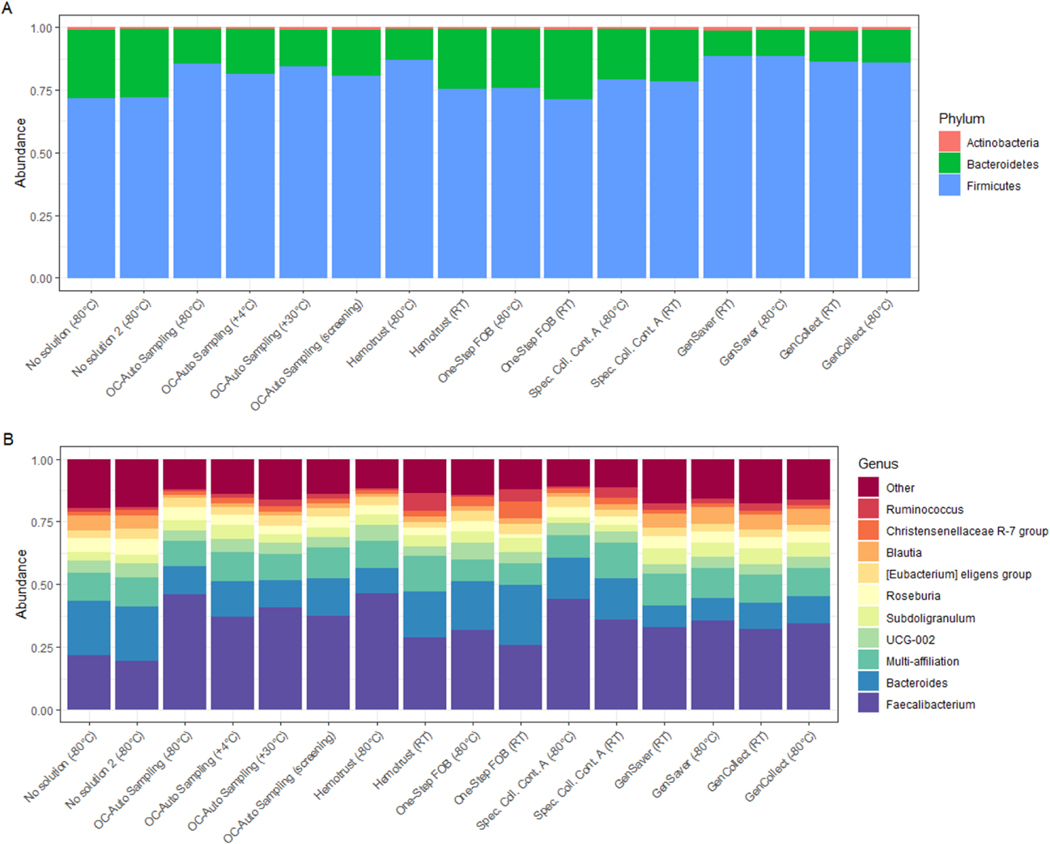
At the phylum level, the distributions of relative abundances of each phylum were consistent for all the collection methods; samples were mainly represented by Firmicutes followed by Bacteroidetes (Figure 2). At the genus level, when compared to the putative gold standard, the relative abundance of Faecalibacterium was greater in the other collection methods, especially in the OC-Auto Sampling FIT tubes. Consequently, when compared to gold standard, the relative abundance of other genera, such as Bacteroidetes or Blautia, was lower in other collections methods. However, there was also substantial inter-individual variability at the genus level (Figure S4 in supplementary material).
Figure 2 –
Variability of microbial phyla and prevalent genera obtained from the collection methods.
Legend: Bars represent the relative abundance of bacterial phylum (A) and genus (B) present in at least 50% of fecal samples with relative abundance of ≥0.1% in each collection methods (n=302).
Accuracy compared with putative gold standard –
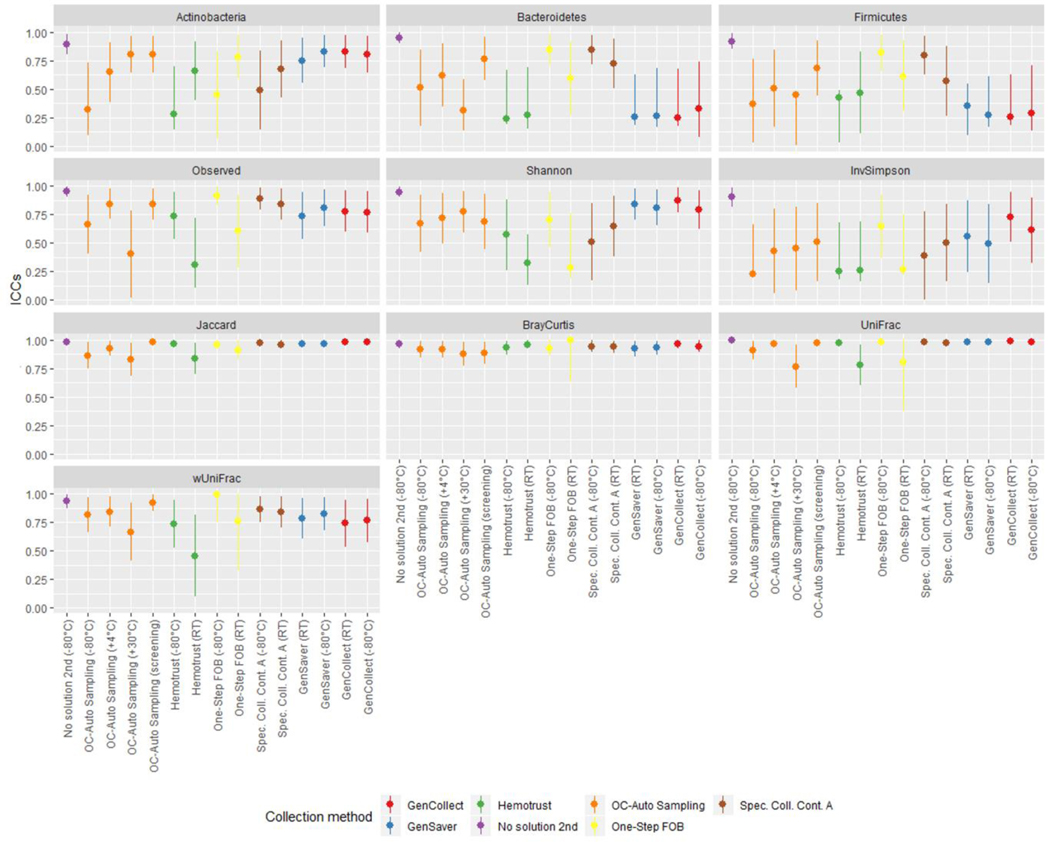
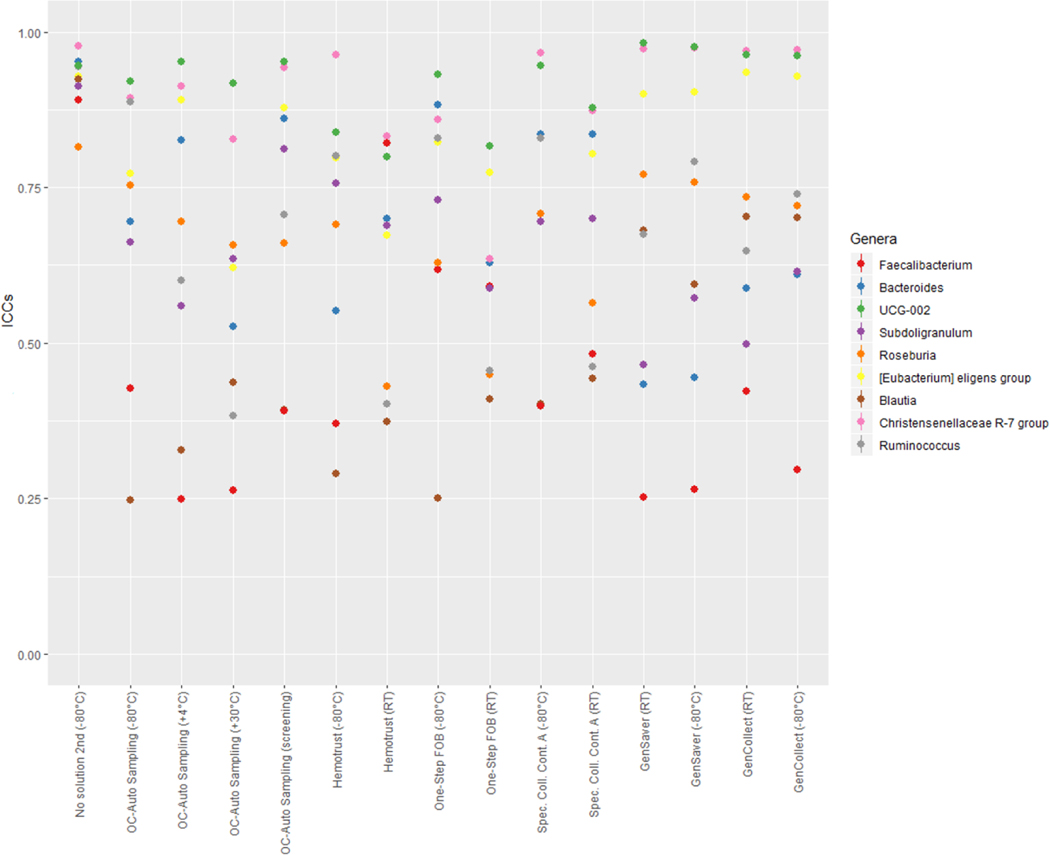
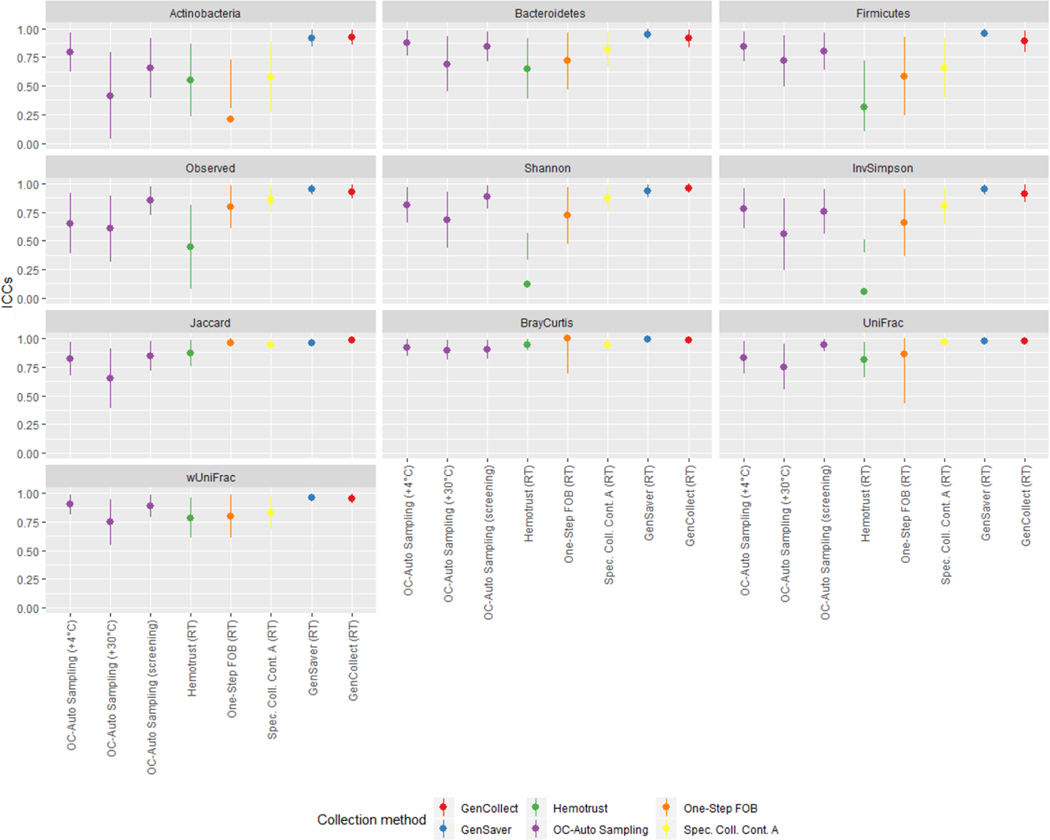
Samples collected without solution and immediately frozen at −80°C were considered as the putative gold standard and compared to samples collected using other methods in different storage conditions using ICCs (Figure 3, and Table S5 in supplementary material). Accuracy ICCs were variable for the relative abundance of Actinobacteria (range, 0.28–0.83), Bacteroidetes (range, 0.24–0.84), Firmicutes (range, 0.26–0.82) as well as for the inverse Simpson index (range, 0.23–0.73) and the weighted UniFrac (range, 0.46–0.93). Specifically, for the relative abundance of Actinobacteria, ICCs were ≥75% for OC-Auto Samples tubes stored at 30°C and those that went through screening, for One Step FOB tubes stored at room temperatures and for all the specimen collection cards. For the relative abundance of Bacteroidetes, ICCs were ≥75% for OC-Auto Samples tubes that went through screening, for One Step FOB tubes immediately frozen and for Specimen Collection Container A tubes immediately frozen. For the relative abundance of Firmicutes, ICCs were ≥75% for One Step FOB tubes immediately frozen and for Specimen Collection Container A tubes immediately frozen. ICCs were ≥75% for PC1 scores of weighted UniFrac for all the methods, except for OC-Auto Samples tubes stored at 30°C and for Hemotrust tubes stored at room temperatures. At the genus level, we did not detect any links between the accuracy differences and the Gram positive or Gram negative (Figure 4). As expected, genera that represent a phylogenetically narrow group of species, UGC-002 and Christenellaceae RT7 group, showed accuracies above 0.75. Conversely, ICCs were higher for observed OTUs (≥0.74) except for Hemotrust at room temperatures (0.31), OC-Auto Sampling tubes immediately frozen (0.66) and at 30°C (0.40), and One-Step FOB at room temperatures (0.60). Specimen collection cards showed the highest ICCs for the Shannon index (range, 0.79–0.88). ICCs were ≥75% for PC1 scores of Jaccard, Bray-Curtis and unweighted UniFrac distances for all collection methods. Additionally, technical variability was also quantified based on beta diversity distances between the two replicates of the putative gold standard and used as baseline to assess the accuracy of each method. For each participant, the distance between the putative gold standard and each collection method was computed (Figure S5 in supplementary material). All of those distances were slightly higher than the baseline, suggesting that the effect of the collection methods exceeds the technical variability.
Figure 3 –
The accuracy of phylum distribution and diversity metrics accuracy differ upon the collection method.
Legend: ICCs for accuracy of microbiome diversity metrics are represented of each fecal sample collection method compared to the gold-standard (samples with no solution, frozen immediately).
Figure 4 –
The accuracy of the relative abundance of the top ten genera differs upon the collection method.
Legend: ICCs for accuracy of the relative abundance of bacteria at the genus level are represented of each fecal sample collection method compared to the gold-standard (samples with no solution, frozen immediately). Gram-negative: Bacteroides, Subdoligranulum, Christensenellaceae R-7 group. Gram-positive: Faecalibacterium, Roseburia, Eubacterium eligens group, Blautia, Ruminococcus.
Stability –
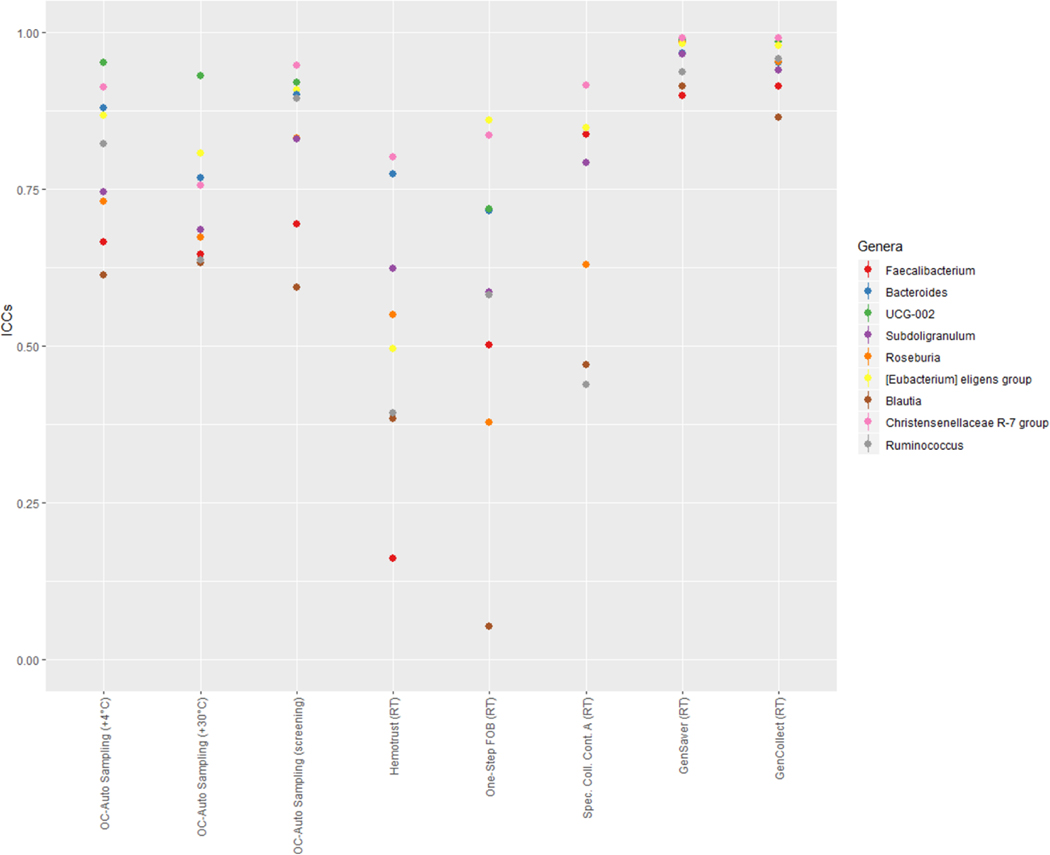
In each collection method, samples frozen immediately were compared to the samples stored in different conditions using ICCs (Figure 5, and Table S6 in supplementary material). When compared to specimen collection cards frozen directly, collection cards (i.e. GenCollect and GenSaver) kept at room temperatures showed high stability, with ICCs ≥89% for all seven metrics. Stability ICCs were generally lower and more variable in samples collected using Hemotrust tubes (range, 0.06–0.94). For One-Step FOB tubes the confidence intervals were wide, indicating high variability in stability at room temperatures. Specimen Collection Container A tubes at room temperatures showed stability ICCs ≥75% except for the relative abundance of Actinobacteria (0.58) and Firmicutes (0.66). Stability ICCs were lower and inconsistent for OC-Auto Sampling FIT tubes stored at 30°C (range, 0.41–0.90). However, stability ICCs were higher for OC-Auto Sampling FIT tubes stored at 4°C (≥0.75) except for the number of observed OTUs (0.65). Stability ICCs were also high for OC-Auto Sampling FIT tubes that went through colorectal cancer screening procedures, with ICCs ≥75%, except for the relative abundance of Actinobacteria (0.65). Interestingly, at the genus level, the stability remained very high, especially for Gencollect and Genesaver. Of note, the Blautia genus, including the homoacetogen Blautia hydrogenotrophica, was significantly lower at room temperature (Figure 2B and Figure 6). In addition, UGC-002, but also Faecalibacterium, Roseburia and Ruminococcus were also less stable when stored at 30°C.
Figure 5 –
Impact of storage conditions on the microbiota composition and diversity indexes.
Legend: ICCs for stability of microbiome diversity metrics of each fecal sample collection method stored in different conditions compared to those directly frozen. Gram-negative: Bacteroides, Subdoligranulum, Christensenellaceae R-7 group. Gram-positive: Faecalibacterium, Roseburia, Eubacterium eligens group, Blautia, Ruminococcus.
Figure 6 –
Impact of storage conditions on the most abundant genera.
Legend: ICCs for stability of the most abundant bacteria at the genus level of each fecal sample collection method stored in different conditions compared to those directly frozen.
Discussion
We compared microbiome stability and accuracy across different fecal sample collection methods used in ongoing colorectal cancer screening programs. We found that the overall variability in diversity was largely explained by differences between participants and less by the collection method. In addition, accuracy and stability ICCs were generally very high for PC1 of beta diversity matrices, except for OC-Auto Sampling tubes stored at 30°C and for Hemotrust tubes stored at room temperatures. Accuracy measures were very inconsistent for the relative abundance of the three phyla and alpha diversity, and in particular, very low for the inverse Simpson. This highlights the importance of using one consistent method for study comparisons. Overall, microbial profile stability was very high for specimen collection cards and seemed generally acceptable for FIT tubes, except for Hemotrust tubes. Colorectal cancer screening tests did not impact microbiome stability in FIT tubes, however, exposure to summer temperatures (i.e. >30°C) did influence stability. These results are informative for the development of future population-based cohorts with fecal sample collection within colorectal cancer screening programs.
As shown in prior studies, microbial composition and diversity were largely explained by between-participants differences and only marginally by the collection methods (8,9). Fecal specimen collection cards have been previously tested for microbial analysis and have shown moderate to excellent accuracy compared to the putative gold standard and stability at room temperatures (5,9,22–27). In this study, the specimen collection cards (i.e. GenCollect and GenSaver) stored at room temperatures for 10 weeks showed excellent stability when compared to the immediately frozen cards, although other studies detected lower amounts of DNA among fecal samples from humans and animals collected on FTA cards (Whatman) after several weeks (25,28). Also consistent with previous findings, we found that specimen collection cards tended to differ in bacterial taxa composition and OTUs than when using the putative gold standard (8,22,28), but we did not observed higher Shannon and inverse Simpson. Firmicutes, often spore formers, were the most represented phylum in samples collected with specimen collection cards, supporting the hypothesis that chemical cell lysis induced by the card matrix might be one explanation for these differences (22). Furthermore, Blautia genus, including the homoacetogen Blautia hydrogenotrophica, is also adequately represented in the collection cards, despite being highly sensitive to oxygen.
Fecal samples collected with FIT tubes have previously shown moderate to excellent accuracy compared to the gold standard and stability at room temperatures (10,29,30). However, the different types of FIT tubes in our study did not seem to perform equally. For the specimen cards and some of the FIT tubes, especially OC-Auto Sampling and Hemotrust tubes stored at −80°C, we detected significant differences in the relative abundance of phyla and genera with higher levels of Firmicutes and Faecalibacterium, including the anti-inflammatory butyrate producing Faecalibacterium prausnitzii, when compared to the gold standard, which support observations from a previous study (30). Similar to previous results (24,29), when compared to the gold-standard, FIT tubes showed good to excellent accuracy for beta diversity metrics, however, some FIT tubes, including OC-Auto Sampling, Hemotrust and Specimen Collection Container A tubes, revealed lower accuracy for alpha diversity metrics, especially those stored for 7 days at room temperatures or at 30°C.
When compared to those directly frozen, Hemotrust tubes stored at room temperature for 7 days showed poor stability for alpha diversity metrics and for One-Step FOB tubes the confidence intervals were wide, indicating high variability in stability at room temperature. In this study, we detected different stabilities at room temperature for Eubacterium eligens and Roseburia, other butyrate producers from the human gut microbiota. As OC-Auto Sampling tubes seemed to be less stable at 30°C, the collection and shipping of samples during high temperatures might have an impact on gut microbiome composition. These variations in accuracy and mostly in stability between the types of FIT tubes might be due to differences in DNA-stabilizing and anti-microbial properties of the solution inside the tubes, impacting the stabilization of DNA, prevention of bacterial growth, and preservation of microbial profiles.
This study has several limitations. First, we included principally female, healthy participants, which might limit the inference of our results to the general population. However, previous studies have found that stability and accuracy for comparison of samples that were frozen immediately without solution to other fecal sample collection methods were similar between different populations (7,8,10). Second, we used 16S rRNA gene sequencing to characterize the microbial composition, while other profiling methods such as whole-genome shotgun metagenomics are becoming more commonly employed in high-income settings (31). However, 16S rRNA gene sequencing remains the most affordable method to study the gut microbiome diversity, especially in the context of large epidemiologic cohorts. Additionally, in low-to-middle income countries, where sequencing technologies are not always available, these collection methods, and more specifically specimen collection cards, could be used to detect specific biomarker species that have been associated with diseases, such as Fusobacterium nucleatum or Parviromonas micra which have been associated with colorectal cancer development (4,32). Finally, freezing procedures might have an impact on relative abundance of Gram-positive and Gram-negative bacteria (33,34), and therefore, considering samples without solution frozen immediately as the gold standard method might be suboptimal. However, in the context of large population-based cohorts, immediate DNA extraction after defecation is likely unfeasible, and therefore, standardization of storage protocols is necessary.
Our study included many different collection methods, including two different specimen collection cards and four different FIT tubes, which are currently being used in ongoing colorectal cancer screening programs around the world. The different collection methods tested in this study have allowed us to highlight a number of important considerations for sample collections in large population settings such as acceptability from participants, processing safety, the volume of kit necessary, the storage logistics and cost. For instance, due to the rigidity of the tubes, the solution in OC-Auto Sampling tubes was not easy to sample safely, however, the amount of bacterial DNA obtained by qPCR indicated that sufficient material was available in these tubes. Based on qPCR data, the collected volume could be lowered from 500 microliters to 200 microliters to obtain sufficient DNA for several analyses and rendering it unnecessary to extract all of the liquid, which might be logistically challenging. Additionally, storage of specimen collection cards is easier and cheaper than tubes, which might help low-to-middle income countries to develop infrastructure for microbiome research. Finally, because of the small size of the kits, the acceptance is expected to be higher in the general population, compared to the gold standard, where provision of a whole fecal sample could make subjects uncomfortable and reluctant to participate. Furthermore, in this study, stability was assessed over the course of several days to several weeks at room temperature, but also for different conditions directly reflecting settings in colorectal cancer screening (i.e. shipping at different temperatures reflecting seasonal variation, colorectal cancer screening procedures including mailing and occult blood detection test). This is the first study in which the impact of colorectal cancer screening procedures on fecal samples using FIT tubes has been demonstrated. Importantly, OC-Auto Sampling FIT tubes that went through colorectal cancer screening procedures and tests had good stability, opening opportunities for establishing prospective cohorts within screening populations.
In conclusion, our study supports previous findings indicating that microbial data obtained from FIT tubes and specimen collection cards are relatively stable and accurate and may be appropriate methods to collect fecal samples for gut microbiome analysis in population-based cohort studies. Furthermore, our findings suggest that opportunistic collection of fecal samples in FIT tubes after colorectal cancer screening is feasible, thereby permitting the potential establishment of cohorts within such screening programs. Since different collection methods and high temperatures impact the stability and accuracy, it is important for future investigators, in the context of the implementation of large-scale epidemiologic studies, to coordinate their efforts and follow standardized protocols in order to accurately compare the microbiome between sites, groups or countries and be able to pool microbial data.
Supplementary Material
Acknowledgments
Disclaimer -
Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer / World Health Organization.
Funding -
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors; this research was jointly funded by IARC and INRAE.
Recognition of personal assistance -
The authors of this paper would like to thank Christophe Lallemand from IARC and Laurence Brazeau from INRAE for their help with laboratory work.
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
References
- 1.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26:5–11. [DOI] [PubMed] [Google Scholar]
- 2.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. [DOI] [PubMed] [Google Scholar]
- 3.Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. Br J Cancer. 2016;114:237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huybrechts I, Zouiouich S, Loobuyck A, Vandenbulcke Z, Vogtmann E, Pisanu S, et al. The Human Microbiome in Relation to Cancer Risk: A Systematic Review of Epidemiologic Studies. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha R, Chen J, Amir A, Vogtmann E, Shi J, Inman KS, et al. Collecting Fecal Samples for Microbiome Analyses in Epidemiology Studies. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2016;25:407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biol [Internet]. 2015. [cited 2018 Sep 28];16. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4674991/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogtmann E, Chen J, Kibriya MG, Chen Y, Islam T, Eunes M, et al. Comparison of Fecal Collection Methods for Microbiota Studies in Bangladesh. Appl Environ Microbiol. 2017;83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogtmann E, Chen J, Amir A, Shi J, Abnet CC, Nelson H, et al. Comparison of Collection Methods for Fecal Samples in Microbiome Studies. Am J Epidemiol. 2017;185:115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Zolnik CP, Qiu Y, Usyk M, Wang T, Strickler HD, et al. Comparison of Fecal Collection Methods for Microbiome and Metabolomics Studies. Front Cell Infect Microbiol. 2018;8:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd DA, Chen J, Vogtmann E, Hullings A, Song SJ, Amir A, et al. Reproducibility, stability, and accuracy of microbial profiles by fecal sample collection method in three distinct populations. PLOS ONE. Public Library of Science; 2019;14:e0224757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Segnan N, Senore C, Andreoni B, Azzoni A, Bisanti L, Cardelli A, et al. Comparing Attendance and Detection Rate of Colonoscopy With Sigmoidoscopy and FIT for Colorectal Cancer Screening. Gastroenterology. Elsevier; 2007;132:2304–12. [DOI] [PubMed] [Google Scholar]
- 12.Escudié F, Auer L, Bernard M, Mariadassou M, Cauquil L, Vidal K, et al. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics. Oxford Academic; 2018;34:1287–94. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. Oxford Academic; 2014;30:614–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–2. [Google Scholar]
- 15.Mahé F, Rognes T, Quince C, Vargas C de, Dunthorn M. Swarm v2: highly-scalable and high-resolution amplicon clustering. PeerJ. PeerJ Inc; 2015;3:e1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. PeerJ Inc; 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. Oxford Academic; 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package [Internet]. 2019. [cited 2020 Jan 31]. Available from: https://CRAN.R-project.org/package=vegan
- 22.Song SJ, Amir A, Metcalf JL, Amato KR, Xu ZZ, Humphrey G, et al. Preservation Methods Differ in Fecal Microbiome Stability, Affecting Suitability for Field Studies. mSystems. 2016;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominianni C, Wu J, Hayes RB, Ahn J. Comparison of methods for fecal microbiome biospecimen collection. BMC Microbiol. 2014;14:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rounge TB, Meisal R, Nordby JI, Ambur OH, de Lange T, Hoff G. Evaluating gut microbiota profiles from archived fecal samples. BMC Gastroenterol. 2018;18:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nechvatal JM, Ram JL, Basson MD, Namprachan P, Niec SR, Badsha KZ, et al. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Methods. 2008;72:124–32. [DOI] [PubMed] [Google Scholar]
- 26.Taylor M, Wood HM, Halloran SP, Quirke P. Examining the potential use and long-term stability of guaiac faecal occult blood test cards for microbial DNA 16S rRNA sequencing. J Clin Pathol. 2017;70:600–6. [DOI] [PubMed] [Google Scholar]
- 27.Tap J, Cools-Portier S, Pavan S, Druesne A, Öhman L, Törnblom H, et al. Effects of the long-term storage of human fecal microbiota samples collected in RNAlater. Sci Rep. 2019;9:601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hale VL, Tan CL, Knight R, Amato KR. Effect of preservation method on spider monkey (Ateles geoffroyi) fecal microbiota over 8weeks. J Microbiol Methods. 2015;113:16–26. [DOI] [PubMed] [Google Scholar]
- 29.Gudra D, Shoaie S, Fridmanis D, Klovins J, Wefer H, Silamikelis I, et al. A widely used sampling device in colorectal cancer screening programmes allows for large-scale microbiome studies. Gut. BMJ Publishing Group; 2019;68:1723–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baxter NT, Koumpouras CC, Rogers MAM, Ruffin MT, Schloss PD. DNA from fecal immunochemical test can replace stool for detection of colonic lesions using a microbiota-based model. Microbiome. 2016;4:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byrd DA, Sinha R, Hoffman KL, Chen J, Hua X, Shi J, et al. Comparison of Methods To Collect Fecal Samples for Microbiome Studies Using Whole-Genome Shotgun Metagenomic Sequencing. mSphere. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Löwenmark T, Löfgren-Burström A, Zingmark C, Eklöf V, Dahlberg M, Wai SN, et al. Parvimonas micra as a putative non-invasive faecal biomarker for colorectal cancer. Sci Rep. Nature Publishing Group; 2020;10:15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bahl MI, Bergström A, Licht TR. Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol Lett. 2012;329:193–7. [DOI] [PubMed] [Google Scholar]
- 34.Fouhy F, Deane J, Rea MC, O’Sullivan Ó, Ross RP, O’Callaghan G, et al. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PloS One. 2015;10:e0119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.