Abstract
Human monocytes and macrophages are persistent reservoirs of human immunodeficiency virus (HIV) type-1. Persistent HIV infection of these cells results in increased levels of NF-κB in the nucleus secondary to increased IκBα, IκBβ, and IκBɛ degradation, a mechanism postulated to regulate viral persistence. To characterize the molecular mechanisms regulating HIV-mediated degradation of IκB, we have sought to identify the regulatory domains of IκBα targeted by HIV infection. Using monocytic cells stably expressing different transdominant molecules of IκBα, we determined that persistent HIV infection of these cells targets the NH2 but not the COOH terminus of IκBα. Further analysis demonstrated that phosphorylation at S32 and S36 is necessary for HIV-dependent IκBα degradation and NF-κB activation. Of the putative N-terminal IκBα kinases, we demonstrated that the Iκκ complex, but not p90rsk, is activated by HIV infection and mediates HIV-dependent NF-κB activation. Analysis of viral replication in cells that constitutively express IκBα negative transdominant molecules demonstrated a lack of correlation between virus-induced NF-κB (p65/p50) nuclear translocation and degree of viral persistence in human monocytes.
The Rel family of transcription factors plays an important role in the transactivation of several viral genes, including those of human immunodeficiency virus (HIV) type 1 (HIV-1) (25, 38). HIV-1 replication is regulated, in part, at the transcriptional level through the interplay of viral regulatory proteins with cellular transcription factors interacting with the viral long terminal repeat (LTR) (39). Since the identification and functional characterization of NF-κB cis-acting sequences within the HIV LTR (38), multiple studies have addressed the essential or dispensable role that this transcription factor plays in the reactivation of HIV from a true latent state and in the control of viral persistence (1, 10, 27, 31, 54, 55). Unfortunately, these studies have yielded conflicting results as to the role of NF-κB in these two steps of the viral life cycle in infected host cells. Differences in the type of host cells studied, HIV strain or genetic constructs used, and methodological approaches may explain these conflicting results.
Understanding the potential impact of NF-κB on the regulation of HIV latency has again become a priority, as recent studies suggest that NF-κB controls the reactivation of latent HIV in T cells from HIV-infected patients undergoing highly active antiretroviral therapy (19). An additional reservoir of HIV, separate from that of T cells harboring latent HIV, are cells of the monocyte lineage in which persistent viral replication is observed (36). During all stages of HIV infection, tissue macrophages provide a unique viral reservoir. In these cells, HIV persistently replicates in the absence of cytopathicity, escapes immune surveillance, and spreads via cell-to-cell contact (reviewed in reference 36). The important role of macrophages in AIDS pathogenesis has prompted the investigation of the molecular mechanisms which regulate HIV-1 persistence in these immune cells; one of these mechanisms is thought to be NF-κB dependent. Human macrophages express a constitutive level of NF-κB in the nuclei in the absence of exogenous cellular activation (25). This constitutive pool of nuclear NF-κB may be sufficient to allow for the initiation of HIV transcription immediately following infection. In addition, NF-κB may be required to further sustain persistent HIV replication, as multiple studies have demonstrated that persistent HIV replication in human macrophages or monocytes further upregulates NF-κB activity (2, 34, 40, 43, 48). However, the mechanisms by which HIV infection induces the activation of NF-κB in cells of the monocyte lineage remains unknown. Their identification would greatly enhance the understanding of this process and allow future testing of whether inhibition of the virus-induced activation of NF-κB may decrease viral persistence in cells of the monocyte lineage, hence eliminating an important reservoir of HIV replication in infected patients.
NF-κB is a heterodimeric protein composed of different combinations of members of the Rel family of transcription factors. A well-characterized form of NF-κB is a heterodimer of p50 and Rel-A (p65) (reviewed in references 3 and 4). In the majority of cells studied, NF-κB is anchored in the cytosol by an inhibitory protein, IκB. An extensively studied IκB molecule, IκBα, has previously been shown to physically interact with NF-κB and to mask the nuclear localization signal of p50 and Rel-A (6). Following cell activation by one of an array of extracellular stimuli, IκBα undergoes a hyperphosphorylation event that renders the inhibitory molecule susceptible to degradation (7, 13, 47). This process results in the release of NF-κB, which undergoes nuclear translocation and drives gene transcription. Significant advances in the understanding of the molecular mechanisms and the structure-function of the phosphorylation and degradation of IκBα have recently been made. IκBα is constitutively phosphorylated at its COOH terminus by protein kinase-casein kinase II (PK-CK2) (5, 33, 35, 45). While the exact function of this phosphorylation is poorly understood, it appears that phosphorylation at the COOH terminus may play a role in the constitutively rapid protein turnover of IκBα in resting cells, thus potentially favoring a low degree of continuous NF-κB translocation. On the contrary, the N terminus contains two series (S32 and S36) which are required for stimulus-dependent phosphorylation (8, 9, 11, 15, 46, 49, 50, 52) by specific kinases, such as the ones present in the Iκκ complex (Iκκα and Iκκβ) or p90rsk (12, 16, 20, 24, 37, 42, 44, 53, 57). Phosphorylation at these sites primes IκBα to undergo ubiquitination and subsequent degradation by the proteosome.
Our group has previously determined that a mechanism whereby HIV infection results in an increase in the nuclear translocation of NF-κB involves modification and enhancement of IκBα turnover (34). The half-life of IκBα in HIV-infected cells is reduced by at least 50% compared to that in uninfected cells, and this fact directly correlates with increased levels of the nuclear pool of NF-κB in HIV-infected cells. That IκBα is the target of persistent HIV infection in monocytic cells has been further confirmed by other groups (14, 27); one of those groups further demonstrated that inhibition of IκBα degradation with proteosome inhibitors decreases HIV-induced NF-κB activation (27). What remain to be elucidated are the molecular mechanisms whereby HIV infection targets IκBα. Potential mechanisms regulated by HIV infection could target the COOH terminus of IκBα, favoring an enhanced “basal” turnover of this inhibitor molecule by activating PK-CK2 or the proteolytic machinery. Alternatively, HIV infection could result in the activation of other IκBα kinases that target S32 and S36, thus continuously priming IκBα to be degraded via the proteosome. Lastly, HIV could target other regulatory sites of IκBα or even other molecules, such as Rel-A, that could result in the dissociation of NF-κB from IκBα, thus rendering IκBα less stable.
To investigate these possibilities, we have used a cell model of monocytic cells in which persistent HIV replication results in NF-κB activation and a variety of genetically modified tagged IκBα molecules can be constitutively overexpressed. Our results indicate that HIV infection targets the NH2 terminus of IκBα, specifically S32 and S36, causing the enhanced degradation of IκBα and hence increased NF-κB nuclear translocation. The Iκκ complex kinase activity is selectively activated and is shown to mediate increased NF-κB activation in HIV-infected cells. In addition, we demonstrate that HIV-mediated NF-κB activation is not necessary to maintain viral persistence in monocytic cells.
MATERIALS AND METHODS
Reagents and antibodies.
Tumor necrosis factor (TNF) was purchased from Genzyme (Cambridge, Mass.) and stored in aliquots at −70°C. Cycloheximide was purchased from Sigma (St. Louis, Mo.) and stored at −20°C. Calpain inhibitor I (N-acetyl-Leu-Leu-norleucinal or ALLN) was purchased from Boehringer Mannheim Biochemicals (Indianapolis, Ind.), solubilized in ethyl alcohol, and stored in aliquots at −20°C. Bay 11-7082 (41) was purchased from Biomol (Plymouth Meeting, Pa.), solubilized in ethyl alcohol, and stored at −20°C. G418 was purchased from Calbiochem-Novabiochem Corporation (La Jolla, Calif.), solubilized in RPMI medium, and stored in aliquots at −20°C.
The expression of the Flag-tagged IκB constructs was monitored with an anti-Flag monoclonal antibody (Kodak, New Haven, Conn.). To control for equal loading of proteins in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis, an anti–β-actin polyclonal antibody (Sigma) and an anti-p90rsk antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) were used. Polyclonal anti-IκBα serum was generated with a glutathione S-transferase (GST)–MAD3 fusion protein (34). The viral envelope protein gp120 was detected with an anti-gp120 polyclonal antibody (Center for Biologics Evaluation and Research, Food and Drug Administration, Bethesda, Md.). The identity of the complexes binding DNA in the gel shift assays was determined with polyclonal antibodies against the different members of the Rel family (Santa Cruz Biotechnology). Antibodies against p90rsk, Iκκα, Iκκβ, Raf-1, and NF-κB were purchased from Santa Cruz Biotechnology.
DNA constructs.
pCMV2-FLAG-IκBα-wt consisted of the full-length “wild-type” sequence of human IκBα (26) cloned into the SmaI-HindIII sites of pCMV2FLAG (Kodak) to generate N-terminally Flag-tagged IκBα-wt (Flag-IκBα-wt). Flag-IκBα-wt was used as a template for subsequent mutations and deletions by PCR-based techniques. Flag-IκBα-ΔN consisted of an N-terminal deletion lacking the first 37 amino acids. This construct was generated with the sense primer wt-FLAG (5′CGGAATTCATGGACTACAAAGACGAT3′) and the antisense primer wt-B (5′GGAATTCCTCATAACGTCAGACGCTG3′). EcoRI sites were created upstream and downstream of the coding sequence. Flag-IκBα-ΔC consisted of a C-terminal deletion lacking the last 40 amino acids and was generated with the sense primer wt-FLAG and the antisense primer ΔC (5′GCGAATTCTCAAAGGTTTTCTAGTGTC3′). This construct contained an EcoRI site downstream of the coding sequence. Flag-IκBα-2N consisted of the full-length sequence of IκBα-wt in which S32 and S36 were mutated to alanine residues. To generate these mutations, a sense primer with the sequence 5′GACGCAGGCCTGGACGCAATG3′ and an antisense primer with the sequence 5′CATTGCGTCCAGGCCTGCGTC3′ were used. Flag-IκBα-4C was created by mutation of S283, S288, S293, and T291 to alanine residues in the PEST sequence (35), cloning into the HindIII-EcoRI site of pCMV2FLAG, and then PCR amplifying with the sense primer wt-FLAG and the antisense primer wt-B. Flag-IκBα-wt, Flag-IκBα-ΔN, Flag-IκBα-ΔC, Flag-IκBα-2N, and Flag-IκBα-4C were then digested and cloned into the EcoRI site of SFFV-Neo under the transcriptional regulation of the Friend spleen focus-forming virus (SFFV) 5′ LTR (22). All of the cloning was verified by DNA sequencing.
Plasmid κB-luc contains three tandem copies of the κB motif of the HIV LTR cloned upstream of the minimal conalbumin-luciferase (con-luc) promoter reporter gene. Plasmid pBLCAT 2 is a mammalian reporter vector designed for the expression of chloramphenicol acetyltransferase (CAT) in mammalian cells transcribed by the minimal thymidine kinase (TK) promoter (Promega, Madison, Wis.). Plasmids Iκκα wt and kinase dead were kind gifts from Alain Israel, Institute Pasteur, Paris, France. Plasmids Iκκβ wt and kinase dead were obtained from M. Roth (Tularik, San Jose, Calif.). Iκκα kinase dead was generated by mutation of aspartic acid 144 to asparagine. Iκκβ kinase dead was created by mutation of lysine 44 to alanine. pcDNA3-Iκκ expression vectors were generated by cloning the cDNA of wild-type Iκκα or Iκκβ or its respective mutant into the cytomegalovirus expression vector pcDNA3 (Invitrogen).
Gene transfection and generation of cell lines.
The U937 promonocytic cell line was purchased from the American Type Culture Collection and grown in RPMI 1640 supplemented with 5% heat-inactivated fetal bovine serum (Intergen), 1% glutamine, and 1% penicillin-streptomycin. To generate IκBα-expressing cell lines, 107 freshly thawed and exponentially growing U937 cells were resuspended in RPMI 1640 and electroporated with 20 μg of previously linearized DNA by use of a BTX cell electroporator at 250 V for 10 ms. U937 cells electroporated without DNA were used as controls. At 24 h after transfection, cells were resuspended in selection medium containing 5% fetal bovine serum and 700 μg of G418 per ml. After 3 to 4 weeks, upon the incipient growth of neomycin-resistant bulk cultures, cells were cloned by limiting dilution (30). Stable integration and expression of the transfected genes within each monoclonal population were verified by serial passages of the cultures in the absence of the selective antibiotic and by immunoblotting with anti-Flag antibodies.
Separate clones expressing equal levels of Flag-IκBα constructs were selected, and their CD4 surface expression was verified by flow cytometry analysis. Thereafter, three clones expressing each of the Flag-IκBα constructs were pooled, and exponentially growing cells were mock or HIV infected. The level of expression of each of the tagged IκBα constructs was confirmed before and during the period of HIV infection by immunoblotting of cytosolic extracts with anti-Flag antibodies.
Transient transfection of U937 cells was performed as follows. A total of 107 exponentially growing U937 cells were incubated with 4 μg of the con-luc or κB-con-luc reporter construct, 6 μg of the pDNA3-Iκκ construct, 4 μg of the pBLCAT2 reporter, and 300 μg of DEAE-dextran (Pharmacia, Piscataway, N.J.) per ml for 90 min at room temperature. Dimethyl sulfoxide (10%) was then added for 3 min, followed by extensive washing and plating at 0.5 × 107/cells/ml. Two days later, cells were harvested. Luciferase levels were measured with the Promega luciferase assay system, and CAT activity was measured with a CAT enzyme-linked immunosorbent assay kit (Boehringer).
HIV infection and measurement of HIV replication.
U937 cells expressing SFFV, Flag-IκBα-wt, Flag-IκBα-ΔN, Flag-IκBα-ΔC, Flag-IκBα-2N, and Flag-IκBα-4C were infected with the HIV LAV-Bru strain as previously described (2, 34, 40). Briefly, 107 exponentially growing U937 cells were sedimented by low-speed centrifugation and resuspended overnight in 10 ml of infective supernatant containing 360 ng of p24 per ml. Mock-infected cells were used as a control. After 24 h, cells were extensively washed and resuspended in culture medium. Cells were passaged twice a week at 0.25 × 106 cells/ml and used from day 30 through day 90 postinfection. During this period, cell supernatants were collected, precleared by centrifugation at 1,500 rpm for 5 min at 4°C, and stored for future analysis of HIV p24 antigen content by an enzyme-linked immunosorbent assay (Coulter-Immunotech Immunology, Westbrook, Maine). At least eight consecutive infections were used for each of these experiments. All the cell lines studied maintained HIV persistence and 100% viability during the study period, except for the U937 clones expressing Flag-IκBα-ΔC, which maintained viability and normal growth while uninfected which underwent immediate and massive cytopathicity upon HIV infection in four consecutive attempts. Therefore, a U937 cell line expressing Flag-IκBα-ΔC could not support a persistent HIV infection. In addition, in some experiments, HIV gp120 expression was determined by immunoblotting with anti-gp120 antibodies.
Nuclear and cytosolic extracts, electrophoretic mobility shift assays, and immunoblotting.
Nuclear and cytosolic extracts were prepared by a modification of the method of Dignam et al. (17). A total of 107 cells were washed with ice-cold phosphate-buffered saline and then with buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl). Cells were then lysed for 10 min on ice in the same buffer containing 0.1% Nonidet P-40, 0.5 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, and 2 μg of pepstatin per ml. After centrifugation, cells were washed twice with buffer A. Nuclei were pelleted by centrifugation, lysed by resuspension in 25 μl of buffer C (20 mM HEPES, 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, DTT, PMSF, aprotinin, leupeptin, pepstatin) and rotated at 4°C for 30 min. After centrifugation, the supernatants were diluted in 50 μl of buffer D (20 mM HEPES, 20% glycerol, 0.05 M KCl, 0.2 M EDTA, DTT, PMSF, aprotinin, leupeptin, pepstatin) and stored at −70°C.
For electrophoretic mobility shift assays, 6 μg of nuclear extract was incubated with a [γ-32P]ATP-labeled double-stranded NF-κB oligonucleotide probe in 15 μl of DNA binding buffer for 15 min at room temperature as previously described (2, 34, 40). Components of the HIV-induced DNA binding protein complexes were identified by incubation of the extract with specific polyclonal antibodies against p50 and Rel-A prior to addition of the labeled probe. The resulting protein-DNA complexes were resolved on a 5% polyacrylamide gel and visualized by autoradiography.
To characterize the level of expression of the Flag-IκBα constructs in uninfected and infected cells, 40 μg of cytosolic protein was analyzed by SDS–10% PAGE. Proteins were transferred to Immobilon-P membranes (Millipore) by standard procedures and blotted with an anti-Flag monoclonal antibody, followed by incubation with rabbit anti-mouse immunoglobulin G (Pierce) and then horseradish peroxidase (Amersham, Buckinghamshire, England). Immunoreactive proteins were detected with an ECL Western blotting detection kit (Amersham). β-Actin and p90rsk were used as internal controls for equal loading in all experiments.
Preparation of recombinant IκBα.
The IκBα-MAD3 cDNA (26) plasmid was obtained from Cetus Corporation and was used as a template for subsequent PCR amplification.
The amino-terminal IκBα-MAD3 (positions 1 to 54) sequence was amplified with wild-type primer A (5′CGGGATCCATGTTCCAGGCGGCCGAG3′) as the sense primer, creating a BamHI site upstream of the coding sequence, and wild-type primer B (5′GGAATTCCTCAGCGGATCTCCTGCAGCT3′) as the antisense primer, creating an EcoRI site downstream of the coding sequence. An S32/36A double mutant was amplified from the full-length cDNA by use of primers to create alanines at S32 and S36. Following digestion with BamHI-EcoRI, these sequences were ligated into pGEX-KG (derived from pGEX-2T, from Pharmacia, Piscataway, N.J.). These constructs were transformed into Escherichia coli DH5α cells, which were grown exponentially. After 60 min of stimulation with isopropylthiogalactopyranoside (Sigma), cells were lysed. Proteins were isolated by affinity chromatography on glutathione-bonded 4% cross-linked agarose (Sigma). The purity of GST-IκBα (positions 1 to 54) containing the first 54 amino acids of IκBα and GST-IκBα (positions 1 to 54) containing S32/36A was analyzed by SDS–10% PAGE and subsequent Coomassie blue staining. The purity of both proteins was greater than 90%.
Immunoprecipitation of IκBα kinases and in vitro kinase assays.
Whole-cell extracts were prepared for immunoprecipitation and in vitro kinase assays as follows. Aliquots of 107 exponentially growing U937 cells were washed twice with cold phosphate-buffered saline, resuspended in lysis buffer containing 40 mM Tris-HCl (pH 8), 0.3 M NaCl, 0.1% Nonidet P-40, 6 mM EDTA, 6 mM EGTA, 10 mM NaF, 10 mM p-nitrophenyl phosphate (PNPP), 10 mM β-glycerolphosphate, 300 μM sodium orthovanadate, 1 mM DDT, 2 μM PMSF, 10 μg of aprotinin per ml, 1 μg of leupeptin per ml, and 1 μg of pepstatin per ml, and incubated on ice. Cells were then centrifuged at 12,000 × g for 15 min at 4°C. The resultant supernatant contained total cellular proteins, which were quantitated with a Bio-Rad protein assay.
For immunoprecipitation of the Iκκ complex, p90rsk, or Raf-1, 100 μg of cell extract was incubated with anti-Iκκα, anti-Iκκβ, anti-p90rsk, or anti–Raf-1 antibodies for 1 h at 4°C, after which protein A-agarose beads (Life Technologies, Gaithersburg, Md.) were added for 1 h. The beads were then washed three times with 0.5 M NaCl-based lysis buffer, followed by one wash with a buffer containing 50 mM Tris-HCl (pH 7.4) and 40 mM NaCl. The washed beads were then incubated in 15 μl of kinase buffer (20 mM HEPES [pH 7.4], 2 mM MgCl, 2 mM MnCl, 10 μM ATP, 10 mM NaF, 10 mM PNPP, 10 mM β-glycerolphosphate, 300 μM sodium orthovanadate, 2 μM PMSF, 10 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml, 1 mM DTT) with 2 μg of GST-IκBα (positions 1 to 54) or GST-IκBα (positions 1 to 54) containing S32/36A and 0.1 μCi of [γ-32P]ATP. The kinase reaction was performed for 30 min at 30°C, and samples were resolved by SDS-PAGE, transferred to Immobilon-P membranes, and exposed to film.
RESULTS
Increased degradation of Flag-IκBα-wt in HIV-infected cells.
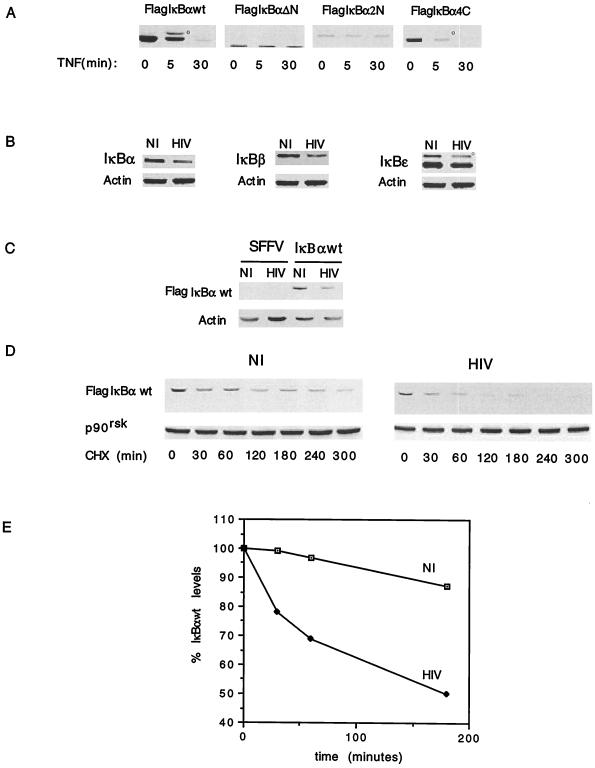
To confirm the expression and functionality of the Flag-IκBα constructs, pooled clones expressing equal levels of Flag-IκB constructs were treated or not treated with TNF, followed by the analysis of the cytosolic extracts by SDS-PAGE and immunoblotting with anti-Flag antibodies. As shown in Fig. 1A, TNF stimulation led to the rapid hyperphosphorylation and subsequent degradation of Flag-IκBα-wt. In contrast, Flag-IκBα-ΔN and Flag-IκBα-2N were refractory to TNF-induced hyperphosphorylation and subsequent degradation. Flag-IκBα-4C behaved similarly to Flag-IκBα-wt in that it was susceptible to TNF-mediated hyperphosphorylation and degradation. These results confirm that the constitutively overexpressed Flag-IκBα molecules are regulated as previously described for native IκBα and highlight the functional relevance of the N terminus containing S32 and S36 in TNF-induced IκBα hyperphosphorylation and degradation.
FIG. 1.
Functional characterization of Flag-IκBα molecules in U937 cells. (A) Pooled clones of U937 cells expressing the different Flag-IκBα constructs were stimulated with TNF for different time periods, and the cell lysates were analyzed by immunoblotting with anti-Flag antibodies. The hyperphosphorylated form of IκBα is indicated by a small circle. (B) Immunoblotting of cell lysates from mock-infected (NI) or HIV-infected (HIV), SFFV-expressing U937 cells with anti-IκBα, anti-IκBβ, anti-IκBɛ, and antiactin antibodies. The hyperphosphorylated form of IκBɛ is indicated by a small circle. (C) Immunoblotting of cell lysates from mock-infected (NI) or HIV-infected (HIV), SFFV- or Flag-IκBα-wt-expressing U937 cells with anti-Flag and antiactin antibodies. (D) The half-life of Flag-IκBα-wt was estimated by immunoblotting of cell lysates from mock-infected (NI) or HIV-infected (HIV), Flag-IκBα-wt-expressing U937 cells treated with cycloheximide (CHX) for different periods of time with anti-Flag antibodies. Equal protein loading was calculated by immunoblotting the same membrane with anti-p90rsk antibody. (E) The half-life of Flag-IκBα-wt was calculated by measuring with a densitometer the disintegrations per minute of Flag-IκBα-wt and normalizing them to those for p90rsk from each experimental time point shown in panel D.
As expected (34), persistent HIV infection of U937 cells resulted in decreased cytosolic levels of native IκBα. Moreover, IκBβ and IκBɛ protein levels were also significantly decreased in HIV-infected cells compared to uninfected cells (Fig. 1B).
Having determined that overexpressed Flag-IκBα constructs function similarly to native IκBα upon stimulation with known inducers of NF-κB and that HIV infection of U937 cells results in decreased steady-state levels of endogenous IκB, we next investigated whether Flag-IκBα-wt is also a target of HIV infection. Immunoblotting of cytosolic fractions from mock- and HIV-infected cells expressing Flag-IκBα-wt was performed with anti-Flag antibodies. U937 cells transfected with the parental empty retrovirus vector (SFFV) were also mock or HIV infected and used as controls. As shown in Fig. 1C, the steady-state protein levels of Flag-IκBα-wt were decreased in the cytosolic fractions of HIV-infected cells compared to mock-infected cells, confirming that HIV infection decreases the cytosolic levels of Flag-IκBα and indicating that tagged IκBα constructs can be used to study the regulatory domain(s) targeted by persistent HIV infection in monocytes.
Having previously demonstrated that the decreased level of native IκBα is a result of the enhanced rate of IκBα degradation in persistently HIV-infected monocytes, we investigated whether this process also accounted for the decreased level of Flag-IκBα-wt in infected cells. The half-life of Flag-IκBα-wt was estimated by immunoblotting Flag-IκBα-wt from cytosolic fractions from mock- and HIV-infected cells treated for different time periods with cycloheximide. As shown in Fig. 1D, the turnover of Flag-IκBα-wt was increased in HIV-infected cells compared to mock-infected cells. The half-lives of Flag-IκBα-wt calculated from Fig. 1D were found to be approximately 60 min in HIV-infected cells and 128 min in uninfected cells (Fig. 1E).
The NH2 terminus but not the PEST sequence present in the COOH terminus of IκBα is necessary for IκBα degradation by HIV infection.
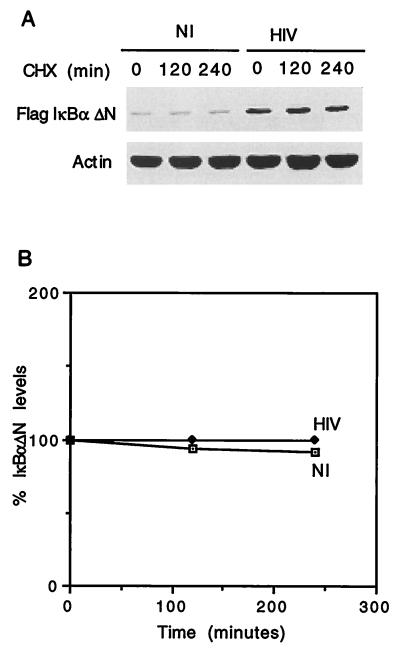
To characterize which of the regulatory domains of IκBα is targeted by HIV infection, we first focused on the NH2-terminal domain of IκBα. We analyzed the turnover and half-life of Flag-IκBα-ΔN. U937 cells stably transfected with the empty retrovirus vector (SFFV), Flag-IκBα-wt, or Flag-IκBα-ΔN were mock or HIV infected. The half-lives of these constructs were measured by analyzing the levels of the Flag-IκBα constructs in cytosolic extracts from cell cultures treated for different time periods with cycloheximide (as for Fig. 1). As shown in Fig. 2A, Flag-IκBα-ΔN was very stable not only in mock-infected but also in HIV-infected U937 cells, with the resulting half-lives being estimated at greater than 4 h (Fig. 2B). The enhanced stability of Flag-IκBα-ΔN in both mock- and HIV-infected cells contrasts with the more rapid turnover of Flag-IκBα-wt in mock-infected cells and even more rapid turnover in HIV-infected cells (Fig. 1D and Fig. 2A). These results indicate that the increased degradation of IκBα that ensues in HIV-infected cells appears to be dependent on the NH2-terminal domain of the molecule. In addition, these results highlight the potential relevance of this IκBα domain in the regulation of the basal turnover of IκBα in unstimulated transformed cells.
FIG. 2.
Deletion of the first 37 amino acids of IκBα conveys resistance to HIV-mediated degradation. (A) Mock-infected (NI) or HIV-infected (HIV), Flag-IκBα-ΔN-expressing U937 cells were treated with cycloheximide (CHX) for different time periods, after which cell lysates were analyzed by immunoblotting with anti-Flag or antiactin antibodies. (B) The half-life of Flag-IκBα-ΔN was calculated as described in the legend to Fig. 1E.
Previous studies have demonstrated that mutation of S283, S288, T291, and S293 to alanines eliminates the constitutive phosphorylation of IκBα mediated by PK-CK2 and may influence the turnover of IκBα (5, 33, 35, 45). Based on this information, we analyzed the turnover and half-life of Flag-IκBα-4C in mock- or HIV-infected U937 cells and compared them to those of Flag-IκBα-wt. In mock-infected U937 cells, the basal turnover of Flag-IκBα-4C was slightly longer than that of Flag-IκBα-wt (Fig. 3), suggesting a potential role of the C-terminal amino acids S283, S288, T291, and S293 in the basal turnover of IκBα in unstimulated monocytic cells. The half-life of Flag-IκBα-4C was shorter in HIV-infected cells than in mock-infected cells but was similar to the half-life of Flag-IκBα-wt in HIV-infected cells (Fig. 3). These results demonstrate that the amino acids present in the PEST sequence of IκBα are not involved in the HIV-mediated degradation and turnover of IκBα.
FIG. 3.
Mutation of the phosphoamino acids present in the PEST sequence does not alter the HIV-mediated degradation of IκBα. (A) Mock-infected (NI) or HIV-infected (HIV), Flag-IκBα-4C- or Flag-IκBα-wt-expressing U937 cells were treated for different time periods with cycloheximide (CHX), and cell lysates were analyzed by immunoblotting with anti-Flag and anti-p90rsk antibodies. The lysates used for detecting p90rsk levels were the same as those from Flag-IκBα-4C-expressing U937 cells. Similar results were obtained with Flag-IκBα-wt-expressing U937 cell lysates. (B) The half-lives of Flag-IκBα-4C (left panel) and Flag-IκBα-wt (right panel) were calculated as described in the legend to Fig. 1E.
HIV-induced degradation of Flag-IκBα requires phosphorylation at the NH2-terminal residues S32 and S36.
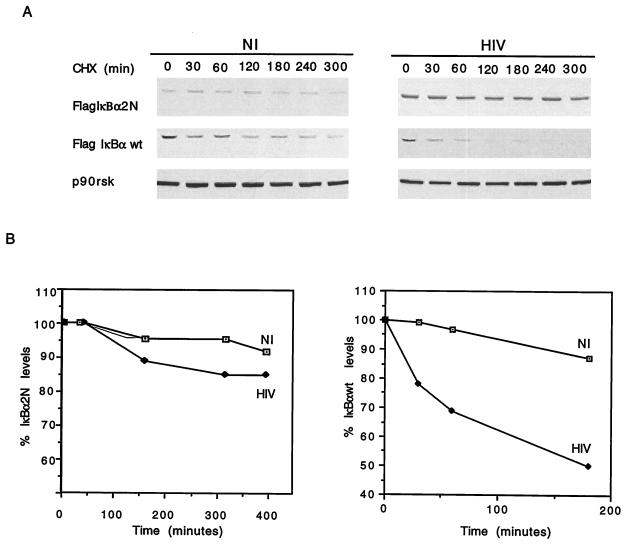
Several studies have identified S32 and S36 as targets of inducible IκBα kinases (8, 9, 11, 15, 46, 49, 50, 52). As shown in Fig. 1A, mutation of S32 and S36 to alanines yields an IκBα construct that is refractory to the hyperphosphorylation and subsequent degradation triggered by TNF in U937 cells. Having identified the NH2-terminal domain of IκBα as a target of HIV-induced degradation, we next questioned whether S32 and S36 could be the amino acids that are targeted by HIV infection. For this, we investigated the half-life and turnover of Flag-IκBα-2N in mock- and HIV-infected U937 cells and compared them to the half-life and turnover of Flag-IκBα-wt. Following the same experimental design as that used for Fig. 2 and 3, we observed that in mock-infected cells, mutation of S32 and S36 to alanines significantly prolonged the half-life of IκBα (greater than 5 h) compared to the more rapid turnover of Flag-IκBα-wt (Fig. 4). This very low rate of basal degradation of Flag-IκBα-2N is similar to that observed for Flag-IκBα-ΔN (Fig. 2). Relevant to the focus of this study, we demonstrate that the half-life of Flag-IκBα-wt is significantly reduced in HIV-infected cells compared to mock-infected cells and that Flag-IκBα-2N was refractory to HIV-mediated IκBα degradation (Fig. 4). These results confirm that S32 and S36 are the IκBα amino acids targeted by persistent HIV infection to result in enhanced degradation of IκBα.
FIG. 4.
Mutation of S32 and S36 of IκBα abrogates HIV-mediated IκBα degradation. (A) Mock-infected (NI) or HIV-infected (HIV), Flag-IκBα-2N- or Flag-IκBα-wt-expressing U937 cells were treated with cycloheximide (CHX) for different time periods, after which cell lysates were analyzed by SDS-PAGE and immunoblotted with anti-Flag and anti-p90rsk antibodies. The p90rsk lysates were the same as those from Flag-IκBα-2N-expressing U937 cells. Similar results were obtained with Flag-IκBα-wt-expressing U937 cell lysates. (B) The half-lives of Flag-IκBα-2N and Flag-IκBα-wt were calculated as described in the legend to Fig. 1E.
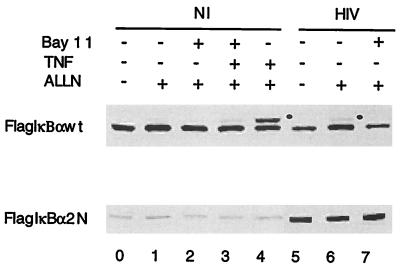
S32 and S36 are the targets of IκBα kinases that are activated by a variety of stimuli, such as inflammatory cytokines, and phosphorylation at these amino acids renders IκBα susceptible to degradation by the proteosome (8, 9, 11, 12, 15, 16, 24, 29, 37, 42, 43, 46, 49, 50, 52, 53, 57). To investigate whether HIV infection results in the hyperphosphorylation of IκBα, mock- or HIV-infected U937 cells expressing Flag-IκBα-wt were treated with the proteosome inhibitor ALLN for 3 h, after which cytosolic extracts were separated by SDS-PAGE and immunoblotted with anti-Flag antibodies. To control for the accurate detection of hyperphosphorylated IκBα, mock-infected Flag-IκBα-wt-expressing U937 cells were treated or not treated with TNF and/or a pharmacological inhibitor previously shown to inhibit the TNF-induced hyperphosphorylation of IκBα (Bay 11-7082) (41). As shown in Fig. 5 (upper panel), a more slowly migrating form of Flag-IκBα-wt was observed in mock-infected, TNF-treated cells, specifically in the presence of ALLN (lanes 3 and 4). In HIV-infected Flag-IκBα-wt-expressing U937 cells, a more slowly migrating form of Flag-IκBα-wt was observed only when ALLN was used (compare lanes 6 with lane 5). These effects are dependent on the presence of S32 and S36 in the Flag-IκBα construct, as their mutation to alanines abrogated both TNF-induced and HIV-dependent IκBα hyperphosphorylation (Fig. 5, lower panel). Altogether, these results indicate that the enhanced degradation of IκBα that is observed in HIV-infected monocytes is a result of specific hyperphosphorylation of IκBα at S32 and S36. Whether the differences in the kinetics of IκBα hyperphosphorylation at S32 and S36 between transient stimuli, such as TNF, and chronic stimuli, such as persistent HIV infection, are due to the use of different IκBα kinases or simply different upstream control mechanisms is currently unknown.
FIG. 5.
HIV infection of U937 cells induces hyperphosphorylation of IκBα which is dependent on S32 and S36. Mock-infected (NI) or HIV-infected (HIV), Flag-IκBα-wt- or Flag-IκBα-2N-expressing U937 cells were treated (+) or not treated (−) with Bay 11-7082 (Bay 11), TNF, or ALLN, after which the cell lysates were analyzed by SDS-PAGE and immunoblotted with anti-Flag antibodies. The supershifted hyperphosphorylated IκBα form of Flag-IκBα-wt is indicated by a bullet.
The Iκκ complex mediates HIV-dependent IκBα degradation and NF-κB activation.
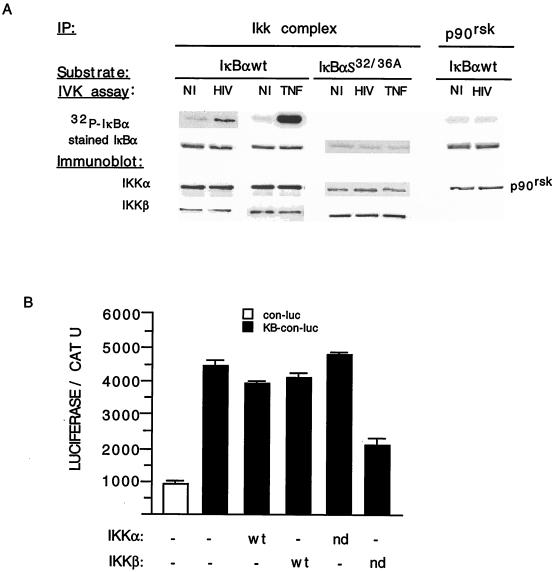
Two kinases in the Iκκ complex (Iκκα and Iκκβ) have recently been shown to phosphorylate S32 and S36 of IκBα and to be the targets of inflammatory cytokines, such as TNF and interleukin 1 (12, 16, 29, 37, 42, 53, 57). Having identified S32 and S36 as the regulatory amino acids of IκBα which are targeted by HIV, we questioned whether the Iκκ complex is activated by HIV infection and mediates the increased levels of nuclear NF-κB activation in infected cells. Mock-infected and persistently HIV-infected U937 cells were lysed, followed by immunoprecipitation of the Iκκ complex, p90rsk, or Raf-1. The kinase activities of these immunoprecipitates were analyzed in an in vitro kinase reaction with GST-IκBα (positions 1 to 54) or GST-IκBα (positions 1 to 54) containing S32/36A as a substrate. In HIV-infected samples, increased IκBα kinase activity was present in the Iκκ complex immunoprecipitate but not in the p90rsk (Fig. 6A) or the Raf-1 (data not shown) immunoprecipitate. This kinase activity was specific for S32 and S36, as their mutation eliminated the basal and HIV-induced Iκκ complex activity. Also, as shown in Fig. 6A, there was no difference in the amounts of Iκκ complex immunoprecipitated with anti-Iκκα antibodies in mock- and HIV-infected U937 cells, thus eliminating the possibility that HIV infection simply increases the pool of Iκκ kinases. These data indicate that HIV infection activates the Iκκ complex, resulting in phosphorylation at S32 and S36.
FIG. 6.
The Iκκ complex but not p90rsk mediates the HIV-induced activation of NF-κB. (A) In vitro kinase assay of Iκκ and p90rsk. Immunoprecipitates (IP) from mock-infected (NI), mock-infected and TNF treated (TNF), and HIV-infected (HIV), SFFV-expressing U937 cells were lysed, and the Iκκ complex and p90rsk were immunoprecipitated with anti-Iκκα and anti-p90rsk antibodies, respectively. Immunoprecipitates were analyzed in an in vitro kinase (IVK) assay with recombinant protein IκBα-wt (positions 1 to 54) or IκBα S32/36A (positions 1 to 54) as a substrate (32P-IκBα). The membrane was subsequently stained with Coomassie blue (stained IκBα) or immunoblotted (Immunoblot) with anti-Iκκα, anti-Iκκβ, or anti-p90rsk antibodies. (B) SFFV-expressing, HIV-infected U937 cells were transiently transfected with con-luc (□) or κB-con-luc (■) together with expression vectors for wild-type (WT) or negative dominant (nd) forms of Iκκα or Iκκβ and a TK CAT reporter gene. Luciferase units were normalized to CAT units. The NF-κB luciferase activity of uninfected, SFFV-expressing cells was similar to that of con-luc in HIV-infected cells, and none of the Iκκα or Iκκβ (wt or nd) expression vectors modified the basal level of plasmid κB-luc activity in uninfected cells (data not shown). This experiment is representative of three additional ones. Each transfection point was determined in duplicate, and error bars indicate ± standard deviations.
The potential relevance of the Iκκ complex in mediating the HIV-dependent activation of NF-κB was further analyzed in transient transfection experiments. Transcription from an NF-κB-dependent luciferase reporter gene was analyzed with both mock- and HIV-infected U937 cells in the presence or absence of wild-type or dominant negative forms of Iκκα and Iκκβ. A minimal TK promoter driving the expression of CAT was used to normalize for transfection efficiency differences that might be present between mock- and HIV-infected cells. The results of these experiments demonstrated that the increased NF-κB activity that is observed in HIV-infected cells is reduced by an Iκκβ dominant negative expression vector but not by a dominant negative form of Iκκα or the wild-type form of either kinase (Fig. 6B). Altogether, these studies demonstrate that the Iκκ complex is activated by HIV infection and mediates virus-induced IκBα hyperphosphorylation and NF-κB activation.
HIV-1 replication in U937 cells expressing different transdominant mutants of IκBα.
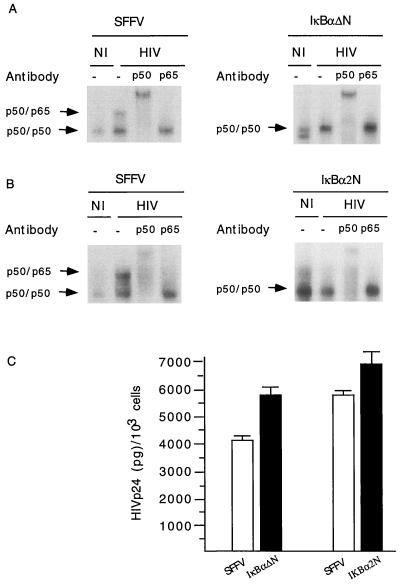
Having determined that S32 and S36 of IκBα are required for HIV-mediated IκBα degradation, we next questioned whether U937 cells expressing Flag-IκBα-ΔN or Flag-IκBα-2N (constructs that are refractory to HIV-dependent degradation) would inhibit HIV-mediated NF-κB activation, and if so, whether this inhibition would result in decreased viral replication. Nuclear extracts and cell-free supernatants were obtained from mock- or HIV-infected cells stably transfected with the SFFV vector, Flag-IκBα-ΔN or Flag-IκBα-2N at the same time as the cytosolic fractions were analyzed to determine the half-lives of the respective IκBα constructs (Fig. 2 and 4). Nuclear extracts were analyzed by a gel shift assay with an oligonucleotide containing NF-κB DNA binding motifs, and viral replication was monitored by measuring p24 levels in culture supernatants. As shown in Fig. 7A and B, left panels, HIV infection of SFFV-expressing U937 cells led to nuclear translocation of a DNA binding protein complex composed of p50 and Rel-A (p65); this finding was not observed in HIV-infected cells expressing either Flag-IκBα-ΔN or Flag-IκBα-2N (Fig. 7A and B, right panels). These observations directly correlate with the inability of these two Flag-IκBα constructs to undergo HIV-mediated degradation, as demonstrated in Fig. 2 and 4.
FIG. 7.
Genetic interference with HIV-mediated NF-κB activation does not result in reduced HIV replication. (A) Gel shift assays of nuclear extracts from mock-infected (NI) or HIV-infected (HIV), SFFV- or Flag-IκBα-ΔN-expressing U937 cells. Antibodies against p50 (p50) or p65 (p65) were added to the gel shift assay. The corresponding molecular complex is indicated. (B) Same panel A, except that nuclear extracts from SFFV-expressing U937 cells were compared in parallel to those from Flag-IκBα-Δ-2N-expressing U937 cells. (C) The HIV p24 content in supernatants of HIV-infected, SFFV-expressing U937 cells (□) or cells expressing Flag-IκBα-ΔN (■) and Flag-IκBα-2N (■) was calculated in duplicate. This experiment is representative of two additional ones. Error bars indicate standard deviations.
The levels of HIV replication in the cells expressing SFFV, Flag-IκBα-ΔN, or Flag-IκBα-2N were then analyzed by measuring HIV p24 levels in supernatants from the same cultures as those used to study the IκBα half-life (Fig. 2 and 4) and NF-κB nuclear translocation (Fig. 7A and B). The results of these experiments indicated that there was no significant reduction in the levels of p24 in supernatants of U937 cells expressing Flag-IκBα-ΔN or Flag-IκBα-2N compared to supernatants of control cultures (SFFV expressing) (Fig. 7C).
DISCUSSION
Using an HIV-susceptible promonocytic cell line which can support persistent viral replication, we have determined that the IκBα residues S32 and S36 and the Iκκ complex (12, 16, 29, 37, 42, 53, 57) are required to mediate HIV-dependent IκBα degradation and, hence, NF-κB activation. The identification of a transdominant negative IκBα molecule which is refractory to HIV-dependent degradation and thus is capable of blocking the HIV-mediated activation of NF-κB extends and confirms previous studies from our group indicating that IκBα is a target molecule and that persistent HIV infection leads to increased NF-κB activation (34). In addition, it provides supporting data that HIV-mediated NF-κB (p50/p65) activation is not necessary to support viral persistence in the U937 monocytic cell line.
The use of pooled clones of monocytic cells that constitutively express genetically modified IκBα constructs has proven to be a valuable tool with which to study the role of NF-κB replication in HIV persistence. Different from punctual stimuli (inflammatory cytokines or transient expression of human T-cell leukemia virus type 1 tax), the activation of NF-κB by HIV infection is dependent on the establishment of viral persistence, achieved only after 6 to 10 days of viral infection (2, 34, 40). Due to this unique virus-host cell interaction, the experimental approaches which can be utilized to address the mechanisms by which HIV activates NF-κB have been significantly limited. Previous attempts have used nonmonocytic cell lines which are highly susceptible to gene transfection, such as 293 or COS-7 cells (28, 54, 55). However, such studies, rather than focusing on HIV persistence in stimulating NF-κB activation, have addressed the role of IκBα in controlling the reactivation of HIV from latency or in inhibiting the initiation of viral replication.
The use of our genetically modified monocytic cells has allowed us to address the mechanism(s) by which HIV leads to NF-κB activation and then to study the role of IκBα in controlling viral persistence. To ensure the relevance of this model, significant efforts were made to verify the maintenance of stable IκBα expression and CD4 expression and the functionality of the tagged overexpressed IκBα clones throughout the infections (months). In addition, as was the case for the Flag-IκBα-2N construct, experiments were repeated with each individual clone separately to verify that the results obtained with a pool of three clones were not due to the overgrowth of a single clone. The level of expression of Flag-IκBα-ΔN and Flag-IκBα-2N was significantly higher in HIV-infected cells than in uninfected cells. This result may be due to increased transcription from the SFFV retrovirus promoter in HIV-infected U937 cells. As both the IκBα N-terminal deletion and the IκBα N-terminal mutation are refractory to HIV-induced degradation, over time the steady-state Flag-IκBα-ΔN and Flag-IκBα-2N protein levels may lead to an increase in the levels of the transgene.
The reduced half-life of IκBα observed in HIV-infected cells differs significantly from the very short half-life of IκBα observed following TNF stimulation. This observation initially led to the hypothesis that sites other than S32 and S36 are targeted by HIV infection. However, the finding that S32 and S36 are required for enhanced IκBα turnover in HIV-infected cells, together with the observation that the Iκκ complex (12, 16, 29, 37, 42, 53) is activated and mediates NF-κB activation in HIV-infected cells, demonstrates a shared utilization of this kinase complex and this IκBα domain in mediating NF-κB activation by unrelated stimuli. What accounts for the significant difference in IκBα half-lives with two separate stimuli, i.e., TNF (5 to 10 min) and HIV (50 to 60 min), which share the same IκBα regulatory domain and kinase complex, is unknown. We have previously demonstrated that within an HIV-infected U937 cell culture, ≥90% of cells express intracytoplasmic HIV p24, thus excluding the possibility that a small subpopulation that is actively HIV infected results in a dilution effect (34).
From the available data, we conclude that it is a lower degree of Iκκ complex activation by HIV infection that correlates with the smaller amount of hyperphosphorylated IκBα and the slower IκBα turnover in HIV-infected cells than in TNF-treated cells. Whether the lower degree of Iκκ activation is secondary to the utilization of different secondary messengers that lie upstream of Iκκ is unknown. It is also plausible that HIV infection targets regulatory processes that control the basal level of Iκκ activity rather than its “inducible” activity. Recent data indicate that protein phosphatase 2A (PP2A) dephosphorylates Iκκα, resulting in a decrease in its kinase activity (16) and explaining the NF-κB-activating function of the PP2A inhibitor okadaic acid (16). It is theoretically possible that HIV infection inhibits PP2A, resulting in a higher “basal” Iκκ activity which is separate from the TNF-inducible Iκκ activity. Previous studies from our group have identified p21ras (21) and the atypical protein kinase C isoforms ξ and ι (20) as essential components of NF-κB activation mediated by HIV infection. Whether these secondary messengers target the Iκκ complex is unknown, but recent advances in the characterization of Iκκ complex regulation will now enable the study of the role of these secondary messengers in HIV-induced NF-κB activation and their linkage to the activation of the Iκκ complex.
The apparent lack of dependence of viral persistence on HIV-mediated NF-κB (p50/p56) activation is a significant conclusion from this study. While several groups, including ours, have consistently demonstrated that persistent HIV infection of monocytic cells results in the selective activation of NF-κB (p50/p65) (2, 34, 40, 43), it has not been possible to clearly demonstrate that the HIV-dependent activation of the p50/p65 heterodimer is necessary to maintain viral persistence in such cells. Attempts to test this question have been made with proteosome inhibitors (14, 27). These compounds were shown to inhibit HIV-dependent IκBα degradation and, hence, p50/p65 heterodimer nuclear translocation, which correlated with a reduction in HIV replication. Because proteosome inhibitors may inhibit a variety of additional cell functions and, potentially, specific steps of the HIV cycle, the role of HIV-dependent activation of NF-κB in regulating viral persistence in monocytic cells remains to be fully clarified. The use of genetic approaches such as the one described in this study allows for more specific inhibition of NF-κB. Interestingly, overexpression of transdominant mutants of Flag-IκBα is sufficient to inhibit the nuclear translocation of additional NF-κB (p50/p65) complexes that may result from the enhanced HIV-dependent degradation of IκBβ and/or IκBɛ, indicating that an IκBα negative dominant molecule overrides the functional impact of the two other IκB molecules, at least in HIV-infected monocytic cells.
Our results indicate an apparent dispensable role of HIV-triggered NF-κB (p50/p65) activation in maintaining viral persistence in U937 cells. Whereas it is still possible that HIV-induced NF-κB (p50/p65) activation is indeed involved in controlling viral persistence, inhibition of this mechanism may have allowed for the utilization of complementary mechanisms to maintain viral persistence in the absence of constitutive or HIV-induced nuclear translocation of the p50/p65 heterodimer. Other members of the NF-κB family or alternative transcription factors, such as Sp1, may be constitutively present in the nuclei of host cells or selectively activated by HIV infection (51) and thus may compensate for the lack of nuclear translocation of p50/p65 dimers observed in the clones expressing IκBα negative transdominant molecules. Rel-B nuclear translocation is thought to be refractory to IκBα inhibition (18, 32). Therefore, either the constitutive presence of Rel-B in nuclei or its potential nuclear translocation following HIV infection might serve to compensate for a lack of HIV-induced p50/p65 in the nuclei of infected cells expressing IκBα negative transdominant molecules. While our gel shift assay experiments with NF-κB DNA concatemers did not demonstrate any NF-κB DNA binding activity in HIV-infected cells expressing IκBα S32 and S36 mutants, the detection of DNA binding activity of Rel-B may have been elusive, as previously suggested. Infection of U937 cells which express an IκBα S32/36A mutant with HIV provirus lacking the NF-κB cis-acting motifs could help clarify the potential role of nuclear proteins which could bind and regulate transcription through the NF-κB cis-acting sequences (23).
While the above hypothesis can be adequately tested with U937 cells, it is ultimately necessary to test the role of the HIV-mediated activation of NF-κB in regulating viral persistence within true physiological host cells, such as human macrophages. In these cells, persistent HIV infection results in the continuous activation of NF-κB (34); thus, it is mandatory to test whether its inhibition alters viral persistence in these host cells. Unfortunately, the current lack of specific inhibitors of IκBα phosphorylation at S32 and S36 and the limitation of applying genetic approaches, such as those used here with U937 cells, to primary human macrophages preclude the conclusion that the observations derived from promonocytic cells apply to human macrophages.
With the identification of the Iκκ complex and IκBα S32 and S36 as targets of persistent HIV infection in monocytes, HIV infection can now be added to the growing list of NF-κB activators that utilize this recently identified complex of N-terminal IκBα kinases. In addition, differences in the degree of Iκκ activation and hence in IκBα turnover between a “chronic” stimulus, such as persistent HIV infection, and other, more “punctual” ones suggest that there may be different means of activating the Iκκ complex within the same cell. Lastly, using genetically modified IκBα constructs, we have been able to demonstrate that the HIV-mediated activation of NF-κB is not necessary to maintain viral persistence. Thus, future efforts should be directed at exploring the complementary role of other NF-κB family members or additional transcription factors in regulating viral persistence in human macrophages as an important cell reservoir of HIV infection.
REFERENCES
- 1.Alcamí J, Laín de Lera T, Folgueira L, Pedraza M-A, Jacqué J-M, Bachelerie F, Noriega A R, Hay R T, Harrich D, Gaynor R B, Virelizier J-L, Arenzana-Seisdedos F. Absolute dependence on κB responsive elements for initiation and TAT-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J. 1995;14:1552–1560. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachelerie F, Alcami J, Arenzana-Seisdedos F, Virelizier J-L. HIV enhancer activity perpetuated by NF-κB induction on infection of monocytes. Nature (London) 1991;350:709–712. doi: 10.1038/350709a0. [DOI] [PubMed] [Google Scholar]
- 3.Baeuerle P A, Baltimore D. NF-κB ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 4.Baeuerle P A, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 5.Barroga C F, Stevenson J K, Schwarz E M, Verma I M. Constitutive phosphorylation of IκBα by casein kinase II. Proc Natl Acad Sci USA. 1995;92:7637–7641. doi: 10.1073/pnas.92.17.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S. IκB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 7.Beg A A, Finco T S, Nantermet P V, Baldwin A S. Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of IκBα: a mechanism of NF-κB activation. Mol Cell Biol. 1993;13:3301–3310. doi: 10.1128/mcb.13.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of IκBα proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 10.Chen B K, Feinberg M B, Baltimore D. The κB sites in the human immunodeficiency virus type 1 long terminal repeat enhance virus replication yet are not absolutely required for viral growth. J Virol. 1997;71:5495–5504. doi: 10.1128/jvi.71.7.5495-5504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteosome pathway. Genes Dev. 1995;9:1586–1587. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z J, Parent L, Maniatis T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 13.Cordle S R, Donald R, Reed M A, Hawiger J. Lipopolysaccharide induces phosphorylation of MAD3 and activation of c-rel and related NF-κB proteins in human monocytic THP-1 cells. J Biol Chem. 1993;268:11803–11810. [PubMed] [Google Scholar]
- 14.DeLuca C, Roulston A, Koromilas A, Wainberg M A, Hiscott J. Chronic human immunodeficiency virus type 1 infection of myeloid cells disrupts the autoregulatory control of the NF-κB/Rel pathway via enhanced IκBα degradation. J Virol. 1996;70:5183–5193. doi: 10.1128/jvi.70.8.5183-5193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiDonato J, Mercurio F, Rosette C, Wu-Li J, Suyang H, Ghosh S, Karin M. Mapping of the inducible IκB phosphorylation sites that signal its ubiquitination and degradation. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 17.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira V, Tarantino N, Körner M. Discrimination between RelA and RelB transcriptional regulation by a dominant negative mutant of IκBα. J Biol Chem. 1998;273:592–599. doi: 10.1074/jbc.273.1.592. [DOI] [PubMed] [Google Scholar]
- 19.Finzl D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 20.Folgueira L, McElhinny J A, Bren G D, MacMorran W S, Diaz-Meco M T, Moscat J, Paya C V. Protein kinase C-ξ mediates NF-κB activation in human immunodeficiency virus-infected monocytes. J Virol. 1996;70:223–231. doi: 10.1128/jvi.70.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folgueira L, Algeciras A, MacMorran W S, Bren G D, Paya C V. The ras-raf pathway is activated in human immunodeficiency virus-infected monocytes and participates in the activation of NF-κB. J Virol. 1996;70:2332–2338. doi: 10.1128/jvi.70.4.2332-2338.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuhlbrigge R C, Fine S M, Unanue E R, Chaplin D D. Expression of membrane interleukin 1 by fibroblasts transfected with murine pro-interleukin 1α cDNA. Proc Natl Acad Sci USA. 1988;85:5649–5653. doi: 10.1073/pnas.85.15.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuminori H, Tanaka H, Hirano Y, Hiramoto M, Handa H, Makino I, Scheidereit C. Functional interference of Sp1 and NF-κB through the same DNA binding site. Mol Cell Biol. 1998;18:1266–1274. doi: 10.1128/mcb.18.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghoda L, Lin X, Green W C. The 90-kDa ribosomal S6 kinase (pp90rsk) phosphorylates the N-terminal regulatory domain of IκBα and stimulates its degradation in vitro. J Biol Chem. 1997;272:21281–21288. doi: 10.1074/jbc.272.34.21281. [DOI] [PubMed] [Google Scholar]
- 25.Griffin G E, Leung K, Folks T M, Kunkel S, Nabel G J. Activation of HIV gene expression during monocyte differentiation by induction of NF-κB. Nature. 1989;339:70–73. doi: 10.1038/339070a0. [DOI] [PubMed] [Google Scholar]
- 26.Haskill D, Beg A A, Tompkins S M, Morris J S, Yurochko A D, Sampson-Johannes A, Mondal K, Ralph P, Baldwin A S., Jr Characterization of an immediate-early gene induced in adherent monocytes that encodes IκB-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- 27.Jacqué J-M, Fernández B, Arenzana-Seisdedos F, Thomas D, Baleux F, Virelizier J L, Bachelerie F. Permanent occupancy of the human immunodeficiency virus type 1 enhancer by NF-κB is needed for persistent viral replication in monocytes. J Virol. 1996;70:2930–2938. doi: 10.1128/jvi.70.5.2930-2938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon H, Pelletier N, DeLuca C, Genin P, Cisternas S, Lin R, Wainberg M A, Hiscott J. Inducible expression of IκBα repressor mutants interferes with NF-κB activity and HIV-1 replication in Jurkat T cells. J Biol Chem. 1998;273:7431–7440. doi: 10.1074/jbc.273.13.7431. [DOI] [PubMed] [Google Scholar]
- 29.Lee F S, Hagler J, Chen Z J, Maniatis T. Activation of the IκBα kinase complex by MEKK1, a kinase of the JNK pathway. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 30.Lefkovits I, Waldmann H. Limiting dilution analysis of cells in the immune system. Cambridge, England: Cambridge University Press; 1979. [Google Scholar]
- 31.Leonard J, Parrott C, Buckler-White A J, Turner W, Ross E K, Martin M A, Rabson A B. The NF-κB binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J Virol. 1989;63:4919–4924. doi: 10.1128/jvi.63.11.4919-4924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lernbecher T, Kistler B, Wirth T. Two distinct mechanisms contribute to the constitutive activation of relB in lymphoid cells. EMBO J. 1994;13:4060–4069. doi: 10.1002/j.1460-2075.1994.tb06723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol. 1996;16:1401–1409. doi: 10.1128/mcb.16.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McElhinny J A, MacMorran W S, Bren G D, Ten R M, Israel A, Paya C V. Regulation of IκBα and p105 in monocytes and macrophages persistently infected with human immunodeficiency virus. J Virol. 1995;69:1500–1509. doi: 10.1128/jvi.69.3.1500-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McElhinny J A, Trushin S A, Bren G D, Chester N, Paya C V. Casein kinase II phosphorylates IκBα at S-283, S-288, S-293, and T-291 and is required for its degradation. Mol Cell Biol. 1996;16:899–906. doi: 10.1128/mcb.16.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meltzer M S, Skillman D R, Hoover D L, Hanson B D, Turpin J A, Chester Kalter D, Gendelman H E. Macrophages and the human immunodeficiency virus. Immunol Today. 1990;11:217–223. doi: 10.1016/0167-5699(90)90086-o. [DOI] [PubMed] [Google Scholar]
- 37.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Wu Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. Iκκ-1 and Iκκ-2: cytokine-activated IκB kinases essential for NFκB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 38.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature (London) 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 39.Oh S H I, Gaynor R B. Intracellular factors involved in gene expression of human retroviruses. In: Levy J A, editor. The Retroviridae. 4th ed. New York, N.Y: Plenum Press; 1995. pp. 97–187. [Google Scholar]
- 40.Paya C V, Ten R M, Bessia C, Alcami J, Hay R T. NF-κB-dependent induction of the NF-κB p50 subunit gene promoter underlies self-perpetuation of human immunodeficiency virus transcription in monocytic cells. Proc Natl Acad Sci USA. 1992;89:7826–7830. doi: 10.1073/pnas.89.16.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce J W, Schoenleber R, Jesmok G, Best J, Moore S A, Collins T, Gerritsen M E. Novel inhibitors of cytokine-induced IκBα phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effect in vivo. J Biol Chem. 1997;272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 42.Régnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 43.Roulston A, D’Addario M, Boulerice F, Caplan S, Wainberg M A, Hiscott J. Induction of monocyte differentiation and NF-κB-like activities by human immunodeficiency virus 1 infection of myelomonoblastic cells. J Exp Med. 1992;175:751–752. doi: 10.1084/jem.175.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schouten G J, Vertegaal A C O, Whiteside S T, Israel A, Toebes M, Dorsman J C, van der Eb A J, Zantema A. IκBα is a target for the mitogen-activated 90 kDa ribosomal S6 kinase. EMBO J. 1997;16:3133–3144. doi: 10.1093/emboj/16.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarz E M, Antwerp D V, Verma I M. Constitutive phosphorylation of IκBα by casein kinase II occurs preferentially at serine 293: requirement for degradation of free IκBα. Mol Cell Biol. 1996;16:3554–3559. doi: 10.1128/mcb.16.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun S-C, Elwood J, Greene W C. Both amino- and carboxyl-terminal sequences within IκBα regulate its inducible degradation. Mol Cell Biol. 1996;16:1058–1065. doi: 10.1128/mcb.16.3.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun S-C, Elwood J, Beraud C, Greene W C. Human T-cell leukemia type 1 tax activation of NF-κB/Rel involves phosphorylation and degradation of IκBα- and RelA (p65)-mediated induction of the c-rel gene. Mol Cell Biol. 1994;14:7377–7384. doi: 10.1128/mcb.14.11.7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzan, M., D. Salaun, C. Neuveut, B. Spire, I. Hirsch, P. de Bouteiller, G. Querat, and J. Sire. Induction of NF-κB during monocyte differentiation by HIV type 1 infection. J. Immunol. 146:377–383. [PubMed]
- 49.Traenckner E B-M, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. Phosphorylation of human IκBα on serines 32 and 36 controls IκBα proteolysis and NFκB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Antwerp D J, Verma I M. Signal-induced degradation of IκBα: association with NF-κB and the PEST sequence in IκBα are not required. Mol Cell Biol. 1996;16:6037–6045. doi: 10.1128/mcb.16.11.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, Mukherjee S, Jia F, Narayan O, Zhao L-J. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and transactivation of viral long terminal repeat. J Biol Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 52.Whiteside S T, Ernst M K, LeBail O, Laurent-Winter C, Rice N, Israel A. N- and C-terminal sequences control degradation of MAD3/IκBα in response to inducers of NF-κB activity. Mol Cell Biol. 1995;15:5339–5345. doi: 10.1128/mcb.15.10.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IκB kinase-β: NFκB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 54.Wu B-Y, Woffendin C, Duckett C S, Ohno T, Nabel G J. Regulation of human retroviral latency by the NF-κB/IκBα family: inhibition of human immunodeficiency virus replication by IκB through a Rev-dependent mechanism. Proc Natl Acad Sci USA. 1995;92:1480–1484. doi: 10.1073/pnas.92.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu B-Y, Woffendin C, MacLachlan I, Nabel G J. Distinct domains of IκBα inhibit human immunodeficiency virus type 1 replication through NF-κB and Rev. J Virol. 1997;71:3161–3167. doi: 10.1128/jvi.71.4.3161-3167.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin M-J, Christerson L B, Yamamoto Y, Kwak Y-T, Xu S, Mercurio F, Barbosa M, Cobb M H, Gaynor R B. HTLV-1 Tax protein binds to Mekk1 to stimulate IκB kinase activity and NF-κB activation. Cell. 1998;93:875–884. doi: 10.1016/s0092-8674(00)81447-6. [DOI] [PubMed] [Google Scholar]
- 57.Zandi E, Torhwarf D M, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (Iκκ) contains two kinase subunits, Iκκα and Iκκβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]