Abstract
A dicistronic minigenome containing the M-F gene junction was used to determine the role of the simian virus 5 (SV5) intergenic regions in transcription. The M-F junction differs from the other SV5 junctions by having a short M gene end U tract of only four residues (U4 tract) and a 22-base M-F intergenic sequence between the M gene end and F gene start site. Replacing the 22-base M-F intergenic region with nonviral sequences resulted in a minigenome template (Rep 22) that was defective in termination at the end of the M gene. Efficient M gene termination could be restored to the mutant Rep 22 template in either of two ways: by increasing the U tract length from four to six residues or by restoring a G residue immediately downstream of the wild-type (WT) U4 tract. In a dicistronic SH-HN minigenome, a U4-G combination was functionally equivalent to the naturally occurring SH U6-A gene end in directing SH transcription termination. In addition to affecting termination, the M-F intergenic region also influenced polymerase reinitiation. In the context of the WT U4-G M gene end, substituting nonviral sequences into the M-F intergenic region had a differential effect on F gene reinitiation, where some but not all nonviral sequences inhibited reinitiation. The inhibition of F gene reinitiation correlated with foreign sequences having a high C content. Deleting 6 bases or inserting 18 additional nucleotides into the middle of the 22-base M-F intergenic segment did not influence M gene termination or F gene reinitiation, indicating that M-F intergenic length per se is not a important factor modulating the SV5 polymerase activity. Our results suggest that the sequence diversity at an SV5 gene junction reflects specific combinations which may differentially affect SV5 gene expression and provide an additional level of transcriptional control beyond that which results from the distance of a gene from the 3′ end promoter.
For the nonsegmented negative-sense RNA viruses, transcription from the viral genome is thought to involve a sequential stop-start mechanism whereby monocistronic mRNAs are produced by termination of transcription at a 3′ upstream gene end followed by reinitiation at a downstream gene start site (reviewed in references 1 and 18). Sequences located at the junction between the tandemly linked viral genes contain important cis-acting signals that direct polymerase functions in transcription, including polyadenylation, transcription termination, and reinitiation at a downstream gene. The features of the viral gene junctions that control these functions of the viral polymerase are important for understanding the regulation of viral transcription.
The rhabdovirus and paramyxovirus gene junctions consist of three regions: the gene end of the 3′ upstream gene, a nontranscribed intergenic region, and the gene start site of the 5′ downstream gene (18). For some nonsegmented negative-sense RNA viruses such as vesicular stomatitis virus (VSV), human parainfluenza virus type 3 (HPIV-3), HPIV-1, and Sendai virus, the sequences at the gene junctions are highly conserved across the viral genome (6, 15, 26). In the case of VSV, the gene end regions contain an invariant 3′-AUACU7-5′ (genome sense) motif (26). The VSV gene end region contains a stretch of seven U residues that are thought to direct the viral polymerase to polyadenylate nascent mRNAs through a stuttering mechanism (28) and additional signals that promote termination of transcription (3, 13). Likewise, the sequences of the VSV intergenic regions are highly conserved, being usually composed of the dinucleotide 3′-GA-5′ (26). Reverse genetics experiments have identified a role for the conserved VSV intergenic region in controlling viral transcription. Alterations which change the sequence or the length of the intergenic GA dinucleotide can lead to defects in transcription termination and in some cases can affect reinitiation of transcription at a downstream gene start (2, 30). For HPIV-1, the conserved intergenic regions may be important for transcription termination. In HPIV-1-infected cells, there is elevated synthesis of an M-F readthrough mRNA (4a), and this correlates with a GAA-to-GCA change in the M-F intergenic trinucleotide.
The individual gene end and intergenic regions of the pneumovirus respiratory syncytial virus (RSV) genome are highly variable, differing in both sequence and overall length (7). By contrast to VSV, it has been reported that the RSV intergenic regions play no role in modulating the viral polymerase activities during transcription (16). This variability at viral gene junctions is also a property of those paramyxoviruses in the Rubulavirus genus (sequences compiled in reference 15), including HPIV-2, mumps virus (MuV), simian virus 41 (SV41), and the prototype member SV5.
The sequences at the SV5 gene junctions are highly diverse, including variations in the number of residues in the gene end U tract and in the sequence and overall length of the intergenic region (Fig. 1A). With the exception of the M-F junction, each of these diverse SV5 junctions directs efficient gene end termination and downstream gene reinitiation (21, 25). We have established a reverse genetics system whereby SV5 transcription is reconstituted in vivo from cDNA-derived components (25). A reverse genetics analysis of SV5 gene end sequences has identified a single G-to-A base substitution in the M gene end region which is responsible for the naturally occurring elevated M-F readthrough transcription (25). While these previous results have shown that the region located 3′ to the gene end U tract is an important cis-acting segment directing functions of the SV5 polymerase, the role of the highly diverse intergenic regions in modulating rubulavirus transcription has not been established.
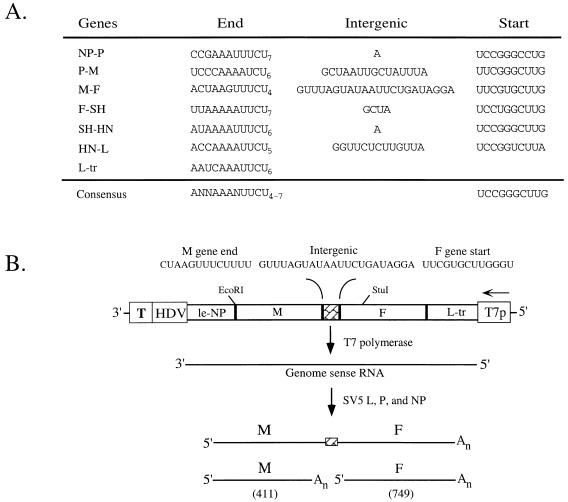
FIG. 1.
Sequences of SV5 gene junctions and structure of the M-F minigenome. (A) Sequences of SV5 gene junctions. The SV5 gene end, intergenic, and gene start sequences are listed as genomic RNA (3′ to 5′). Consensus sequences are estimates of the most frequent occurrence of a given nucleotide in each position. Sequences are from references 11 (HN), 12 (SH), (25) (NP and L), 22 (F), 29 (M), and 32 (P/V). L-tr, trailer/L gene. (B) Structure of the SV5 dicistronic M-F minigenome. The SV5 M-F minigenome cDNA is shown schematically as a rectangle, with a stippled box representing the 22-base M-F intergenic region. The sequence of the M gene end, M-F intergenic region, and F gene start is shown 3′ to 5′ (genome sense). The promoter for T7 RNA polymerase (T7p), the self-cleaving hepatitis delta virus ribozyme (HDV) and T7 terminator sequence (T), and restriction sites (EcoRI and StuI) in the cDNA clone that were used to construct mutants are indicated. The horizontal arrow indicates the direction of transcription by the T7 RNA polymerase to produce a genome-sense RNA. le-NP, NP gene/leader; L-tr, trailer/L gene; An, 3′ poly(A) region.
In the work presented here, we have used minigenomes containing an SV5 gene junction to analyze the role of the diverse intergenic regions in transcription. Our results show that alterations to the sequence of an SV5 intergenic region can lead to defects in termination and reinitiation of transcription, but the effect of these intergenic substitutions on viral transcription is highly dependent on the length of the upstream gene end U tract. Our results support the proposal that the diversity of sequences across the SV5 gene junctions reflects specific combinations of the variable U tract and intergenic regions which cooperate to create signals directing the functions of the viral polymerase.
MATERIALS AND METHODS
Cells and viruses.
Monolayer cultures of A549 cells were grown in Dulbecco’s modified Eagle’s medium containing 5% fetal calf serum. Vaccinia virus vTF7.3 (9) was grown and titered in CV-1 cells.
Construction of plasmids encoding SV5 minigenomes.
The construction of the M-F minigenome and the SH-HN minigenome have been described elsewhere (25). The orientation of DNA fragments encoding the minigenomes was such that the 5′-end trailer and 3′-end leader regions were flanked by the T7 RNA polymerase (T7pol) promoter and the hepatitis delta virus ribozyme sequences (24), respectively. The T7pol-derived genome-sense RNAs contain three additional 5′ G residues and an exact 3′ end due to ribozyme self-cleavage (23). Plasmid pMF2 was constructed such that T7pol transcription produces a genome-sense RNA which encodes the following sequence: 5′-terminal 302 bases from the trailer/L gene–484 bases from the 5′ end of the F gene–22-base F-M intergenic region–289 bases from the 3′ end of the M gene–3′-terminal 179 bases from the NP gene/leader region. For pSH-HN, T7pol transcription produces a genome-sense RNA which encodes the following sequence: 5′ trailer and last 270 bases of the L gene–first 562 bases from the 5′ end of the HN gene–intergenic A residue–last 266 bases from the end of the SH gene–the 3′-terminal 179 bases from the NP gene/leader.
Mutations were introduced into the M-F and SH-HN minigenomes by conventional PCR approaches as previously described (20), using Pwo polymerase (Boehringer Mannheim, Indianapolis, Ind.). AT nucleotides were added at the NP-M or the NP-SH junction as needed to maintain a total 6N-length genome as described previously (25). The M-F PCR products were digested with EcoRI and StuI and cloned into the corresponding sites in pMF2 (Fig. 1B). The SH-HN PCR products were digested with BstBI and EcoRI and cloned into the corresponding sites of pSH-HN. The nucleotide sequence of all PCR-derived DNA segments was determined and agreed with previous published sequences.
Analysis of in vivo transcription products.
RNA synthesized from the SV5 minigenomes was analyzed using the vaccinia virus T7 (vacT7) RNA polymerase system (9) as described previously (25). Briefly, 3.5-cm-diameter dishes of vTF7.3-infected A549 cells were transfected with 2 μg of minigenome plasmid, 1.5 μg of pGEM3-L, 0.4 μg of pGEM2-P, and 3.0 μg of pUC19-NP3A, using a cationic liposome reagent (27). In control samples where L plasmid was omitted, pBluescript (Stratagene) was used to normalize the overall concentration of DNA. Total intracellular RNA was isolated from cells by using Trizol reagent (Life Technologies) at 36 to 42 h postinfection. To isolate poly(A)+ RNA, total RNA samples were incubated with oligo(dT)-cellulose (New England Biolabs) in binding buffer (500 mM NaCl, 10 mM Tris-HCl [pH 7.5]) for 30 min with rocking. After washing with low-salt buffer (250 mM NaCl, 10 mM Tris-HCl [pH 7.5]), poly(A)+ RNA was eluted in elution buffer (10 mM Tris-HCl [pH 7.5]) and ethanol precipitated before analysis by Northern blotting (25). The amount of RNA analyzed for each sample was derived from an equivalent number of cells. Northern blots were hybridized with genome-sense 32P-labeled riboprobes corresponding to the following SV5 gene sequences: M (positions 1079 to 1265 [29]), F (160 to 407 [22]), SH (1 to 283 [12]), and HN (302 to 559 [11]). Quantitation of RNA transcription products was performed with PhosphoImager instrumentation and software (Molecular Dynamics). Appropriate background was subtracted by using lanes corresponding to minus L control samples. M gene termination and F gene reinitiation were calculated as the ratio of monocistronic mRNA to the sum of the monocistronic mRNA plus the readthrough M-F dicistronic mRNA product.
RESULTS
The sequence at the 3′ end of the SV5 M-F intergenic region is important for efficient termination at the end of the M gene.
We have previously established a reverse genetics system whereby SV5 transcription can be reconstituted in vivo from cDNA-derived components (25). A dicistronic M-F minigenome was constructed to contain the 3′ and 5′ regions of the SV5 genome linked by the gene junction which normally separates the genes encoding the viral membrane (M) protein and fusion (F) proteins (Fig. 1B). The dicistronic M-F minigenome serves as a template for transcription by the SV5 polymerase to produce a 411-base monocistronic M mRNA and a 749-base monocistronic F mRNA as well as a dicistronic M-F readthrough product (Fig. 1B) (25). To reconstitute viral transcription from the M-F minigenome, A549 cells were infected with a recombinant vaccinia virus expressing the T7 RNA polymerase (T7pol [9]) and then transfected with plasmids encoding the M-F minigenome and the viral proteins L, P, and NP. Poly(A)+ RNA was isolated from the infected-transfected cells and analyzed by Northern blotting with 32P-labeled riboprobes specific for mRNA from the M and F genes.
As shown in Fig. 2B, the M-F minigenome directed the synthesis of an mRNA that was detected with both M and F riboprobes, consistent with this species being an M-F readthrough transcript (wild type [WT]; lanes 1 and 7). We detected two smaller RNAs that correspond to monocistronic M and monocistronic F mRNAs, since they were detected only with the corresponding M- and F-specific riboprobes. Radioactivity contained in the SV5-specific mRNAs on the Northern blots was quantitated by PhosphorImager analysis. The extent of M gene termination or F gene reinitiation was calculated as the percentage of the total M-specific or F-specific poly(A)+ RNA detected as monocistronic M or F mRNA, respectively. In the work described here, the mean percentages of M gene termination and F gene reinitiation from 11 independent assays were 46 ± 5 and 50 ± 6, respectively, values which agree closely with those found for the M-F junction in SV5-infected A549 cells (25).
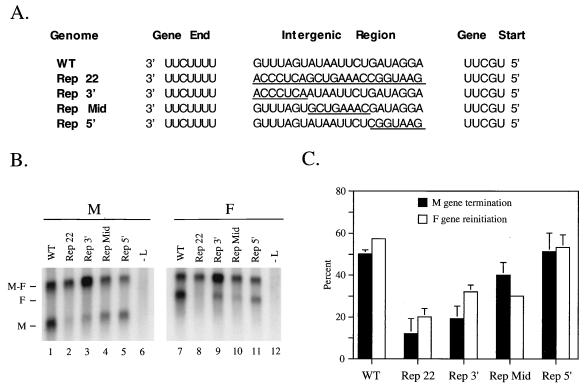
FIG. 2.
Substitutions in the 3′ end of the M-F intergenic region disrupt efficient M gene termination. (A) Gene junction sequences of minigenomes containing a WT or mutant M-F intergenic region. The WT intergenic region was replaced in its entirety (Rep 22), in the 3′ end (Rep 3′), in the middle (Rep Mid), or the 5′ end (Rep 5′) with nonviral sequences indicated by underlines. (B) Northern blot analysis of minigenome expression. VTF7.3-infected A549 cells were transfected with plasmids encoding genes for the viral proteins L, P, and NP, along with one of the M-F minigenome plasmids. Poly(A)+ RNA was analyzed by Northern blotting with 32P-labeled riboprobes specific for M or for F mRNA transcripts. The -L lanes are control samples in which the L plasmid was omitted from the transfection mix. Positions of the dicistronic M-F readthrough mRNA and the monocistronic M and F mRNAs are indicated. (C) Quantitation of M gene termination and F gene reinitiation from altered minigenomes. For each minigenome, the percentage of the total M mRNA that was detected as a gene end termination product (black bars) and the percentage of the total F mRNA detected in a monocistronic F mRNA (white bars) were determined from three independent experiments. Values represent the relative abundance of the mono- and dicistronic mRNAs. Lines above the bars indicate standard deviations from the mean.
To determine the role of the SV5 M-F intergenic region in viral transcription, the 22-base region between the M gene U tract and the F gene start site was replaced with a 22-base segment of nonviral sequence (Fig. 2A, Rep 22 minigenome). After expression in the vacT7 system, poly(A)+ RNA was isolated and analyzed by Northern blotting with M- and F-specific riboprobes. A representative blot is shown in Fig. 2B, and quantitation of results from three independent experiments is displayed in Fig. 2C. In the case of the Rep 22 minigenome, only 12% of the total M-specific mRNA was detected as a monocistronic species (Fig. 2B, lane 2), compared to the 50% monocistronic M mRNA directed by the WT M-F minigenome (lane 1). The vast majority of the M-specific mRNA synthesized from the Rep 22 template was found in the M-F dicistronic readthrough product, indicating that this sequence alteration had decreased termination at the M gene. In multiple experiments, the level of M-specific mRNA synthesized from the Rep 22 mutant was not significantly lower than that of the WT minigenome, suggesting that the defect in M gene termination was not due to an overall decrease in transcription levels. The Rep 22 substitution may have affected M mRNA polyadenylation or transcription termination or both of these events. Because our assay measures only the final product of these two tightly coupled polymerase functions, termination will be used here to describe both events. The Rep 22 minigenome also directed less monocistronic F message (lane 8) compared to the WT minigenome, which may reflect the requirement for termination at the upstream M gene end before reinitiation at the F gene start site (2, 3, 17). Taken together, these data indicate that changes in the sequence of the M-F intergenic region can result in defects in M gene termination and these changes may have either a direct or indirect effect on the reinitiation of transcription of the downstream F gene.
To locate sequences in the M-F intergenic region which are required for efficient termination and reinitiation, we constructed three minigenomes in which the 3′, middle, and 5′ regions of the M-F intergenic region were replaced with the corresponding nonviral sequences from the Rep 22 minigenome (Fig. 2A, Rep 3′, Rep Mid, and Rep 5′). The Rep 3′ minigenome was defective in directing M gene termination and F gene reinitiation to levels comparable to that for the Rep 22 minigenome (Fig. 2B, lanes 3 and 9). While the Rep Mid minigenome directed efficient M gene termination that was only slightly lower than that from the WT genome, the F gene reinitiation from this minigenome was still reduced to a level between that of Rep 22 and WT (lanes 4 and 10). By contrast, the 5′ Rep minigenome directed M gene termination and F gene reinitiation to levels that closely matched the WT level (lanes 5 and 11). These data indicate that changes in the sequences in the 3′ region of the M-F intergenic segment disrupt efficient M gene termination and that F gene reinitiation is reduced by a direct or indirect mechanism. By contrast, sequence changes in the 5′ region of the intergenic region do not affect M gene transcription termination.
The first nucleotide of the M-F intergenic region is important for efficient M gene termination, but only when linked to a short M gene U tract.
Four of the six SV5 intergenic regions begin with a G residue, while the remaining two contain a single A residue (Fig. 1A). We hypothesized that the Rep 22 minigenome was defective in directing M gene termination due to the lack of the endogenous G as the first nucleotide of the intergenic region. To test this hypothesis, we constructed a minigenome in which a G was substituted as the first nucleotide in the context of the mutant Rep 22 intergenic region to yield M U4-G Rep (M gene-four U residues-G residue) (Fig. 3A). When assayed with the vacT7 system, the level of M gene termination directed by M U4-G Rep closely matched that of the WT minigenome (Fig. 3B, lane 3). These data indicate that in the context of the WT M gene end region, the first nucleotide of the M-F intergenic region is an important factor in promoting efficient M gene termination. Interestingly, while efficient M gene termination was restored for the M U4-G Rep minigenome, the level of monocistronic F mRNA was not higher than that seen for the Rep 22 mutant (Fig. 3B, lane 9). An explanation for this result is presented below.
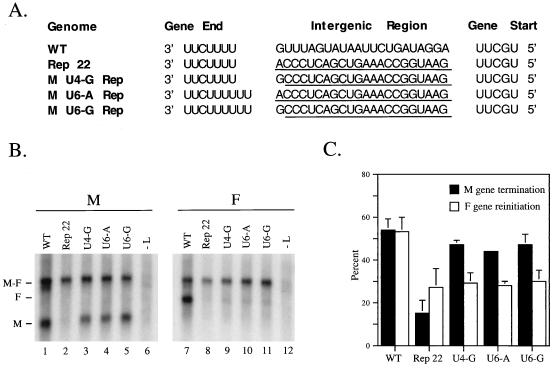
FIG. 3.
The first nucleotide of the M-F intergenic region is important for efficient M gene termination, but only in combination with a short U tract. (A) Genomic sequences of minigenomes containing a WT or mutant M-F intergenic region. The WT intergenic region was replaced with nonviral sequences (underlined) to create Rep 22. The first nucleotide of the Rep 22 intergenic region was restored to the WT G residue to create M U4-G Rep. In addition, mutants were constructed such that the M gene end U tract was extended to six residues in the context of a G (MU 6-G Rep) or an A (MU 6-A Rep) as the first nucleotide in the intergenic region, with the remaining intergenic sequences derived from Rep 22. The altered minigenomes were expressed and analyzed as described in the legend to Fig. 2. (B) Blot representative of typical results. (C) Percent M gene termination (black bars) and percent monocistronic F mRNA (white bars) determined from three independent experiments. Lines above the bars indicate standard deviations from the mean.
Our previous results showed that in the context of the WT M-F intergenic region, increasing the M gene end U tract from four to six bases had no effect on transcription termination (25). As shown in Fig. 1A, the SV5 intergenic regions which begin with an A residue are all preceded by U tracts composed of more than four residues. Thus, we hypothesized that a gene end containing a longer U tract could function when linked to either a G or an A as the first intergenic nucleotide, while a short U tract (e.g., the M gene end) requires a G to follow in order to promote efficient termination. To test this hypothesis, two additional Rep 22 mutant M-F minigenomes were constructed to contain an M gene end with a U tract of six residues followed by either an A or a G (M U6-A Rep and M U6-G Rep [Fig. 3A]). Northern blot analysis of the poly(A)+ RNAs synthesized from these genomes indicated that M gene termination was efficiently directed in both mutants (Fig. 3B, lanes 4 and 5). However, F gene reinitiation remained defective (lanes 10 and 11). Taken together, these data indicate that either of two combinations of the SV5 U tract-intergenic region can promote efficient M gene end termination: a U tract of four residues linked to a G in the first position of the intergenic region, or a U tract of six linked to an intergenic region starting with either a G or an A.
Nonviral intergenic sequences can inhibit reinitiation of transcription at a downstream gene start site.
The above results indicated that proper combinations of U tract length and the first base of an intergenic region could restore efficient M gene termination to the Rep mutant minigenomes, but each of the minigenomes still failed to direct reinitiation of transcription at the downstream F gene. Two possible explanations for this result are that the WT M-F intergenic region contains a positive-acting signal that is required to promote F gene reinitiation or that the Rep 22 sequences contain a negative-acting signal which inhibits reinitiation. To distinguish between these possibilities, a second M-F replacement mutant was constructed to contain a U4-G M gene end, followed by 14 nonviral bases that were different from Rep 22 and the seven nonviral Rep 5′ bases which were found to have no effect on polymerase functions (Rep G-14 [Fig. 4A]).
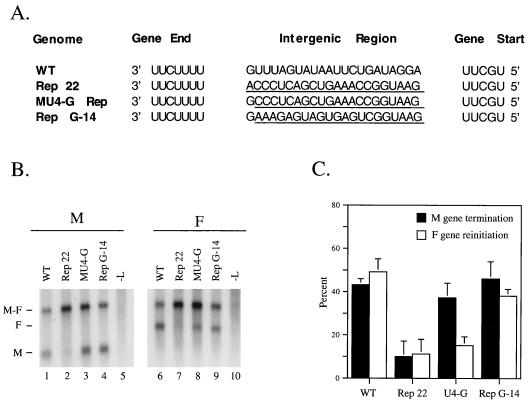
FIG. 4.
Nonviral intergenic sequences can inhibit reinitiation of transcription at a downstream gene start site. (A) Sequences of the WT, Rep 22, and M U4-G Rep intergenic regions. The minigenome Rep G-14 contains a G in the first position of the intergenic region followed by a 14-base nonviral sequence which differs from those contained in Rep 22. The minigenomes were expressed and analyzed as described in the legend to Fig. 2. (B) Representative blot. (C) Quantitation of the results from three independent experiments.
When analyzed with the vacT7 system, the Rep G-14 minigenome directed efficient M gene termination, as expected because of the U4-G M gene end combination (Fig. 4B, lane 4). Most importantly, the Rep G-14 mutant template also directed efficient reinitiation at the downstream F gene (lane 9), a result that differed significantly from the very low levels of monocistronic F synthesized from the M U4-G Rep minigenome (lane 8). These data suggest that the WT M-F intergenic region does not contain a sequence specific signal that is important for reinitiation of transcription at the downstream F gene. Rather, the Rep 22 replacement represents a nonviral nucleotide sequence which acts by an unknown mechanism to inhibit reinitiation by the SV5 polymerase. The Rep 22 sequence contains more C residues than the other SV5 intergenic segments, which could have a negative effect on reinitiation.
Termination and reinitiation at the M-F gene junction is not affected by changes in the length of the M-F intergenic region.
As with other members of the Rubulavirus genus (15), the SV5 intergenic regions vary in length, ranging from a single A residue (e.g., the NP-P junction [Fig. 1A]) to the 22-residue M-F gene junction. While the above results indicated that there was no specific sequence requirement in the M-F intergenic region other than the first G residue, the data did not address the role of intergenic length on viral transcription. To determine if changes in the length of the M-F intergenic region influenced polymerase function, we constructed minigenomes in which the middle of the WT M-F intergenic region was altered by the deletion of 6 bases or by the addition of either 6 or 18 bases (Fig. 5A). Alterations were designed to maintain an overall 6N-length genome, which we have shown to be important for efficient RNA replication (19). The minigenomes were expressed in the vacT7 system, and poly(A)+ RNAs were analyzed by Northern blotting with M- and F-specific riboprobes. As shown in Fig. 5B, each of the length-altered minigenomes directed M gene termination and F gene reinitiation at levels which were indistinguishable from that of the WT minigenome. These results indicate that the length of the M-F intergenic region per se is not an important factor governing the efficiency of M gene termination or F gene reinitiation. In addition, these data are consistent with the above proposal that other that the first G residue flanking the U tract, the SV5 M-F intergenic region does not contain sequence-specific signals important for directing polymerase functions.
FIG. 5.
Changes in the length of the M-F intergenic region do not affect M gene transcription termination or F gene reinitiation. The WT M-F minigenome was altered to contain a deletion of 6 bases (MF-6; solid line), insertion of 6 bases (MF+6; underline), or insertion of 18 bases (MF+18; underline and lowercase letters). (A) The minigenomes were expressed and analyzed as described in the legend to Fig. 2. (B) Quantitation of the results from three experiments.
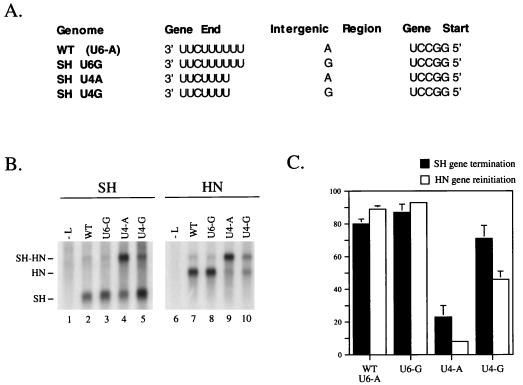
The first nucleotide of the SH-HN intergenic region is important for efficient SH gene termination, but only in combination with a short U tract.
A mutational analysis was carried out to determine if specific combinations of the SH U tract length and SH-HN intergenic nucleotide were required for efficient termination at the SH gene end and reinitiation at the downstream HN start site. The rationale in choosing the SV5 SH-HN junction was because this region of the genome is genetically and phenotypically different from the M-F gene junction described above: the SH-HN junction contains a six-residue U tract linked to a single intergenic A residue and directs efficient termination and reinitiation in SV5-infected cells (25). We have previously characterized a dicistronic minigenome which contains the SV5 SH-HN gene junction. As shown previously with the vacT7 system (25) and in Fig. 6B, ∼80% of the SH mRNA expressed from this SH-HN minigenome (U6-A) was the monocistronic form (lane 2) and ∼90% of the HN-specific mRNA resulted from reinitiation at the HN start site (lane 7). These two values closely match those found in the case of SV5-infected cells (25).
FIG. 6.
The first nucleotide of the SH-HN intergenic region is important for efficient SH gene termination, but only in combination with a short U tract. (A) SH-HN minigenomes were constructed to contain combinations of an SH gene end U4 or U6 tract followed by a single A or a single G as the intergenic region. Transcription from the minigenomes was analyzed as described in the legend to Fig. 2, except that poly(A)+ RNAs were analyzed by Northern blotting with 32P-labeled riboprobes specific for the SH or HN transcripts. (B) Representative blot. Positions of the dicistronic SH-HN readthrough mRNA and the monocistronic SH and HN mRNAs are indicated. (C) Percent SH gene termination (black bars) and percent monocistronic HN mRNA (white bars), determined from five independent experiments.
To test the hypothesis that a 5′ flanking G residue was required for efficient termination at an SV5 gene end with a short U tract, three mutant SH-HN minigenomes were constructed to contain combinations of a four or six residue U tract linked to either a G or an A (Fig. 6A). As shown in Fig. 6B (lanes 4 and 9), an SH-HN minigenome with a U4-A combination produced much less monocistronic SH and HN mRNAs (∼10 to 20%) and showed a corresponding increase in synthesis of the dicistronic SH-HN readthrough mRNA. The replacement of a G as the intergenic nucleotide in the case of the SH U4-G minigenome greatly increased termination efficiency at the SH gene end compared to the SH U4-A (lane 5), and these levels were similar to that of the WT minigenome (∼70% [Fig. 6C]). The level of monocistronic HN mRNA synthesized from the U4-G minigenome was reproducibly lower than that detected with the other minigenomes (lane 10; ∼45%, compared to ∼80% for WT [Fig. 6C]), suggesting that HN reinitiation was effected by the combined alterations to the U tract and intergenic nucleotide. These data on SH gene termination are consistent with the results from the above analysis of the M-F intergenic region and support the hypothesis that a long U tract at a gene end region can function efficiently when linked to either a G or an A as the first nucleotide of the intergenic. However, in the case of gene end regions with short U tracts (e.g., four U residues), efficient gene termination requires a flanking G as the first intergenic nucleotide.
DISCUSSION
The junctions between the SV5 genes are highly diverse, both in the number of residues in the gene end U tract and in the sequence and length of the intergenic regions (Fig. 1A). Yet with the exception of the naturally occurring high levels of M-F readthrough transcription, each of these diverse junctions directs efficient gene end termination and downstream gene reinitiation (21, 25). The goal of our work is to understand the relationship between the sequences of these diverse gene junctions and the common polymerase activities that they modulate. In this study, we have determined the effect on transcription of alterations to SV5 intergenic regions.
Our results indicate that a G residue as the first nucleotide of an SV5 intergenic region is important for efficient transcription termination, but only when linked to a short gene end U tract composed of four residues. The mechanism by which a U4-G gene end combination can be functionally equivalent to a U tract of six residues is not known. Four U residues may be at the lower limit of a functional U tract, and the intergenic G residue may act to increase polymerase stuttering to promote polyadenylation and termination. Our data support the proposal that combinations of the variable U tract and intergenic regions can create signals that differentially control the efficiency of SV5 polymerase activities.
For VSV, the conserved gene end U tract of seven residues is of the minimum length that will efficiently function in polyadenylation-termination (3). Remarkably, removing even a single U residue from this tract results in templates that direct high levels of readthrough products (3, 13). Our results indicate that SV5 differs from VSV in the stringency of the length requirement for the gene end U tract to function in termination. The WT SV5 M-F junction contains an M gene end U tract of only four residues and directs ∼50% readthrough transcription (21, 25). Importantly, our previous results showed that in the context of the WT M-F intergenic region, increasing the M gene end U tract from four to six or eight U residues did not increase M gene termination (25). The work described here provides an explanation for this previous result, since the U4-G combination is functionally equivalent to a longer U tract. The length of a U tract becomes a factor in SV5 transcription termination when a U4 tract is linked to an intergenic A residue, and this combination is not found at any SV5 gene junction. The flexibility in functional combinations of U tract length and intergenic nucleotides shown here for SV5 may be a feature of other paramyxoviruses. For example, the measles virus M, F, and L genes and the RSV NS1, NS2, F, and 22K genes contain short U tracts of only four residues (sequences listed in references 6 and 15), and for five of these seven examples, a G residue is found as the first intergenic nucleotide.
A 6-base deletion or an 18-base insertion in the middle of the M-F intergenic segment had no apparent effect on M gene termination or F gene initiation. While we have not established a lower size limit for a functional SV5 intergenic region, these data indicate that intergenic length per se is not a major factor in transcription across the diverse SV5 gene junctions. It has been speculated that the variations in length of the paramyxovirus intergenic regions serve to maintain the gene start sites in the proper context of the hexameric phase established by binding of NP to the genomic RNA (15). This hypothesis was not tested with the SV5 minigenomes described here, because changes in the length of the U tract or intergenic regions were compensated by second site alterations such that an overall 6N-length genome was maintained (19), and this maintained the hexamer phase of the F and HN gene start sites. Thus, it remains possible that the position of nucleotides within an NP-bound hexamer is important for both genome replication (5) and transcription (15).
With the exception of the U4-G gene end combination, our results indicate that the SV5 M-F intergenic region does not contain sequence specific signals that are necessary for directing termination and reinitiation. This is evident by the WT levels of M and F mRNAs synthesized from the Rep G-14 minigenome, which contains a 21-base nonviral M-F intergenic segment. Those intergenic regions which contain a single A residue also lack a sequence requirement for these polymerase functions, since in the context of the WT SH gene end, changing the intergenic A residue to either a C (not shown) or a G did not change the relative synthesis of monocistronic SH and HN mRNAs. In this regard, our results are similar to those found for RSV. Using dicistronic minigenomes that contain consensus gene end/poly(U) tract and gene start sites, it has been shown that the variable intergenic regions do not influence polymerase activity (16). Thus, the role of the diverse SV5 intergenic sequences in the viral life cycle is unknown. SV5 genomic RNA isolated from a persistently infected Vero cell line (4) shows no alterations in the M-F intergenic region (25a), suggesting that this sequence is conserved for some role in the virus life cycle which cannot be assayed by our vacT7 minigenome system. Work is in progress to determine the effect of changes in the M-F intergenic region on the growth of SV5 by using the full-length infectious clone (10).
Analysis of the M-F Rep 22 minigenome (Fig. 2 and 3) has shown that in some cases the sequence of an intergenic region can inhibit reinitiation at a downstream gene start site. A similar observation has been made for VSV minigenomes containing nonviral extensions of the intergenic region (31). No pattern was apparent in the foreign sequences, which would indicate why some intergenic regions allowed normal function of the VSV polymerase whereas others were defective. An analysis of the nucleotide composition in the intergenic regions of the rubulaviruses SV5, HPIV-2, SV41, and MuV suggests a basis for the inhibitory effect that some foreign sequences have on SV5 reinitiation. As shown in Table 1, the percent composition for the intergenic regions of the rubulavirus genomes is highest for A and U residues (27 and 43% overall for SV5) and is relatively low for C residues (11%). In support of a possible role for high C content inhibiting SV5 polymerase reinitiation, the Rep 22 intergenic sequence is 32% C. By contrast, the M-F minigenomes which also contained nonviral sequences but directed F gene reinitiation (e.g., the M-F length-altered mutants and Rep G-14) contained fewer C residues (5 to 18%). The efficiency of reinitiation at each of the SV5 gene junctions has not been determined and may differ between the SV5 gene junctions. Thus, while our data indicate that the SV5 intergenic regions do not contain a positive-acting signal that promotes reinitiation, it is possible that the inhibition of polymerase function seen with the Rep 22 intergenic sequence represents an extreme example of a negative-acting control mechanism.
TABLE 1.
Nucleotide compositions for intergenic regions of rubulavirus genomes
| Virus | Junctiona | % Composition
|
|||
|---|---|---|---|---|---|
| A | C | G | U | ||
| SV5 | NP-P | 100 | 0 | 0 | 0 |
| P-M | 27 | 13 | 13 | 47 | |
| M-F | 32 | 5 | 23 | 41 | |
| F-SH | 25 | 25 | 25 | 25 | |
| SH-HN | 100 | 0 | 0 | 0 | |
| HN-L | 8 | 15 | 23 | 54 | |
| All SV5 IG | 27 | 11 | 20 | 43 | |
| MU4-G Rep | 27 | 32 | 27 | 14 | |
| MF − 6 nt | 31 | 0 | 31 | 38 | |
| MF + 6 nt | 32 | 7 | 21 | 39 | |
| MF + 18 nt | 25 | 18 | 27 | 30 | |
| Rep G-14 | 36 | 5 | 41 | 18 | |
| HPIV-2 | All IG | 29 | 10 | 14 | 46 |
| SV41 | All IG | 29 | 17 | 20 | 34 |
| MuV | All IG | 43 | 7 | 21 | 29 |
For the nonsegmented negative-strand RNA viruses, the position of a viral gene relative to the 3′ end leader promoter is the major factor determining the level of transcription of viral mRNAs (e.g., references 14 and 33). It would be expected that for those viruses with conserved gene junctions, the activities of the viral polymerase would be uniform at each gene junction, and the polarity of transcription across the viral genome would result from a constant frequency of termination and reinitiation. Our results suggest that the diversity in sequence at the SV5 gene junctions reflects specific combinations which may differentially affect SV5 transcription. Thus, for viruses with nonconserved gene junctions, sequence diversity may provide an additional level of transcriptional control beyond that which results from the distance of a gene from the 3′-end promoter. In support of this speculation, two recombinant SV5 viruses which encode a green fluorescent protein gene between the viral HN and L genes show large differences in the relative level of green fluorescent protein expression, depending on the particular transcription control sequences engineered to flank the foreign gene (10). This finding raises the possibility that the diverse SV5 gene junctions have evolved as additional mechanisms to fine-tune the control of gene expression, of which, the cooperation between the short U tract and the intergenic region is one example.
ACKNOWLEDGMENTS
We thank Mike Keller and Doug Lyles and Sue Murphy for helpful comments on the manuscript.
This work was supported by NIH grant AI42023. Oligonucleotide synthesis was performed in the DNA Synthesis Core Laboratory of the Cancer Center of Wake Forest University, supported in part by NIH grant CA-12197.
REFERENCES
- 1.Banerjee A K, Barik S. Gene expression of vesicular stomatitis virus genome RNA. Virology. 1992;188:417–428. doi: 10.1016/0042-6822(92)90495-b. [DOI] [PubMed] [Google Scholar]
- 2.Barr J N, Whelan S P J, Wertz G W. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997;71:1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr J N, Whelan S P J, Wertz G W. Cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumgartner W, Krakowka S, Blakeslee J R. Persistent infection of Vero cells by paramyxoviruses. Intervirology. 1987;27:218–223. doi: 10.1159/000149987. [DOI] [PubMed] [Google Scholar]
- 4a.Bousse T, Takimoto T, Murti K G, Portner A. Elevated expression of the human parainfluenza virus type 1 F gene downregulates HN expression. Virology. 1997;232:44–52. doi: 10.1006/viro.1997.8524. [DOI] [PubMed] [Google Scholar]
- 5.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins P, Chancock R M, McIntosh K. Parainfluenza viruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Lippincott-Raven; 1996. pp. 1205–1241. [Google Scholar]
- 7.Collins P L, Dickens L E, Buckler-White A, Olmsted R A, Spriggs M K, Camargo E, Coelingh K V. Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc Natl Acad Sci USA. 1986;83:4594–4598. doi: 10.1073/pnas.83.13.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elango N, Varsanyi T M, Kovamees J, Norby E. Molecular cloning and characterization of six genes, determination of gene order and intergenic sequences and leader sequence of mumps virus. J Gen Virol. 1988;69:2893–2900. doi: 10.1099/0022-1317-69-11-2893. [DOI] [PubMed] [Google Scholar]
- 9.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;85:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He B, Paterson R G, Ward C D, Lamb R A. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 11.Hiebert S W, Paterson R G, Lamb R A. Hemagglutinin neuraminidase protein of the paramyxovirus simian virus 5: nucleotide sequence of the mRNA predicts an N-terminal membrane anchor. J Virol. 1985;54:1–6. doi: 10.1128/jvi.54.1.1-6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiebert S W, Paterson R G, Lamb R A. Identification and predicted sequence of a previously unrecognized small hydrophobic protein, SH, of the paramyxovirus simian virus 5. J Virol. 1985;54:744–751. doi: 10.1128/jvi.55.3.744-751.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang L N, Englund N, Pattnaik A K. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J Virol. 1998;72:1805–1813. doi: 10.1128/jvi.72.3.1805-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iverson L E, Rose J K. Localized attenuation and discontinued synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 15.Kolakofsky D, Pelet T, Garin D, Housmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo L, Fearns R, Collins P L. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J Virol. 1996;70:6143–6150. doi: 10.1128/jvi.70.9.6143-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo L, Grosfeld H, Cristina J, Hill M G, Collins P L. Effect of mutations in the gene-start and gene-end sequences motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J Virol. 1996;70:6892–6901. doi: 10.1128/jvi.70.10.6892-6901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B, Knipe D, Howley P, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1177–1204. [Google Scholar]
- 19.Murphy S K, Parks G D. Genome nucleotide lengths that are divisible by six are not essential but enhance replication of defective interfering RNAs of the paramyxovirus simian virus 5. Virology. 1997;232:145–157. doi: 10.1006/viro.1997.8530. [DOI] [PubMed] [Google Scholar]
- 20.Parks G D. Mapping of a region of the paramyxovirus L protein required for the formation of a stable complex with the viral phosphoprotein P. J Virol. 1994;68:4862–4872. doi: 10.1128/jvi.68.8.4862-4872.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paterson R G, Harris T J R, Lamb R A. Analysis and gene assignment of mRNAs of a paramyxovirus, simian virus 5. Virology. 1984;138:310–323. doi: 10.1016/0042-6822(84)90354-4. [DOI] [PubMed] [Google Scholar]
- 22.Paterson R G, Harris T J R, Lamb R A. Fusion protein of the paramyxovirus simian virus 5: nucleotide sequence of the mRNA predicts a highly hydrophobic glycoprotein. Proc Natl Acad Sci USA. 1984;81:6706–6710. doi: 10.1073/pnas.81.21.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattnaik A K, Ball L A, Legrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 24.Perrotta A T, Been M D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature (London) 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 25.Rassa J C, Parks G D. Molecular basis for naturally occurring elevated readthrough transcription across the M-F junction of the paramyxovirus SV5. Virology. 1998;247:274–286. doi: 10.1006/viro.1998.9266. [DOI] [PubMed] [Google Scholar]
- 25a.Rassa, J. C., and G. D. Parks. Unpublished observation.
- 26.Rose J K. Complete intergenic and flanking gene sequences from the genome of vesicular stomatitis virus. Cell. 1980;19:415–421. doi: 10.1016/0092-8674(80)90515-2. [DOI] [PubMed] [Google Scholar]
- 27.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 28.Schubert M, Keene J D, Herman R C, Lazzarini R A. Site on the vesicular stomatitis virus genome specifying polyadenylation and the end of the L gene mRNA. J Virol. 1980;34:550–559. doi: 10.1128/jvi.34.2.550-559.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheshberadaran H, Lamb R A. Sequence characterization of the membrane protein gene of paramyxovirus simian virus 5. Virology. 1990;176:234–243. doi: 10.1016/0042-6822(90)90248-p. [DOI] [PubMed] [Google Scholar]
- 30.Stillman E A, Whitt M A. Mutational analysis of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stillman E A, Whitt M A. The length and sequence composition of vesicular stomatitis virus intergenic regions affect mRNA levels and the site of transcription initiation. J Virol. 1998;72:5565–5572. doi: 10.1128/jvi.72.7.5565-5572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas S M, Lamb R A, Paterson R G. Two mRNAs that differ by two non templated nucleotides encode the amino co-terminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wertz G W, Perepelitsa V P, Ball L A. Gene arrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc Natl Acad Sci USA. 1998;95:3501–3506. doi: 10.1073/pnas.95.7.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]