Abstract
Herpes simplex virus infection is initiated by VP16, a viral transcription factor that activates the viral immediate-early (IE) genes. VP16 does not recognize the IE gene promoters directly but instead forms a multiprotein complex with Oct-1 and HCF-1, a ubiquitous nuclear protein required for progression through the G1 phase of the cell cycle. The functional significance of recruiting HCF-1 to the VP16-induced complex is not understood. Here we describe the identification of a second HCF-like protein, designated HCF-2. HCF-2 is smaller than HCF-1 but shares three regions of strong amino acid sequence homology, including the β-propeller domain required for association with VP16. HCF-2 is expressed in many tissues, especially the testis, and shows a more dynamic pattern of subcellular localization than HCF-1. Although HCF-2 associates with VP16 and can support complex assembly with Oct-1 and DNA, it is significantly less efficient than HCF-1. A similar preference is shown by LZIP, a cellular counterpart of VP16. Analysis of chimeric proteins showed that differences between the fifth and sixth kelch repeats of the β-propeller domains from HCF-1 and HCF-2 dictate this selectivity. These results reveal an unexpected level of specificity in the recruitment of HCF-1 to the VP16-induced complex, paralleling the preferential selection of Oct-1 rather than the closely related POU domain protein Oct-2. Implications for regulation of the viral life cycle are discussed.
The lytic cycle of herpes simplex viruses (HSVs) involves an elaborate cascade of gene expression, initiated by the viral transcriptional activator VP16 (reviewed in references 9, 26, and 35). VP16 (also known as Vmw65 or αTIF) is a structural component of the virion that is released into the infected cell. After translocation to the nucleus, VP16 associates in a sequential manner with two cellular proteins, first the host cell factor HCF (also referred to as C1, VCAF, or CFF) and then the POU domain protein Oct-1 (8, 18, 33). Together these three proteins form the VP16-induced complex on a specific DNA sequence—the TAATGARAT motif—found upstream of each of the viral immediate-early (IE) promoters (3). Once VP16 is recruited to the IE gene promoters, it initiates high levels of transcription by virtue of its potent activation domain (28, 36).
HCF comprises a family of polypeptides derived from a >2,000-amino-acid precursor through proteolytic processing (19, 20, 41). Cleavage occurs at a series of six centrally located 26-amino-acid repeats (called HCFPRO repeats) producing multiple amino- and carboxy-terminal fragments that remain tightly but noncovalently associated following cleavage (19, 43). VP16 interacts with a discrete amino-terminal domain (HCFVIC) composed of six kelch-like repeats that folds as a six-bladed β-propeller. The HCF β-propeller is sufficient for promoting VP16-induced complex assembly in vitro and in vivo (40), although the carboxy terminus may also contribute to complex formation (21).
The precise cellular function of HCF is not known; however, analysis of tsBN67 cells, a temperature-sensitive hamster cell line, has shown that HCF is required for cell cycle progression. At the nonpermissive temperature, tsBN67 cells undergo a G0/G1 arrest and can reenter the proliferative cycle, even after several days in the arrested state induced by lowering the temperature. This reversible block to proliferation is due to a single proline-to-serine change in the β-propeller domain of tsBN67 HCF (10). The missense mutation does not significantly alter the stability or processing of HCF but prevents association between HCF and VP16. Because a single-point mutation in HCF disrupts both transactivation by VP16 and cell proliferation, it is likely that VP16 mimics the interaction between HCF and a cellular protein required for progression through the G1 phase of the cell cycle (10, 40). This hypothesis is strengthened by the fact that HCF has been conserved in evolution: HCF from insects and nematodes can readily support VP16-induced complex formation (18, 39). One such cellular target of HCF is the ubiquitous basic leucine zipper transcription factor known as LZIP or Luman (7, 23). LZIP together with a related Drosophila bZIP protein called dCREB-A/BBF-2 contains a short tetrapeptide motif (the HCF-binding motif [HBM]) that is found in VP16. Point mutations in the HBMs of both VP16 and LZIP prevent binding to HCF (7, 23, 24), and short peptides derived from VP16 that span this motif act as potent inhibitors of VP16-induced complex formation (12, 30, 44). Both LZIP and dCREB-A function as potent transcriptional activators, suggesting that HCF plays a role in cellular as well as viral transcription (1, 23, 24, 32).
Here we report the identification of a second human HCF protein, designated HCF-2. HCF-2 and the original HCF (now HCF-1) share two extensive regions of amino acid homology, corresponding to the β-propeller and self-association domains at the amino terminus and the self-association domain at the carboxy terminus. Although HCF-2 can associate with VP16 and promote VP16-induced complex formation on a viral TAATGARAT element, it is much less efficient that HCF-1. Thus, despite the sequence similarity, HCF-2 is unlikely to function as a physiological target of HSV. We have mapped the critical differences between the β-propeller domains of HCF-1 and HCF-2 to the fifth and sixth kelch repeats. Finally, we show that HCF-2 cannot substitute for HCF-1 in complementation of the tsBN67 cell proliferation defect and instead acts as a growth inhibitor when overexpressed. These findings suggest that HSV has specifically targeted the unique growth-promoting functions of HCF-1.
MATERIALS AND METHODS
Identification of HCF-2 expressed sequence tags.
The National Center for Biotechnology Information expressed sequence tag (EST) database (dbest) was searched by using the BLAST version 2.0 algorithm (blastn). Additional searches were performed by using the sequence of the first HCF-2 EST clone (GenBank accession no. AA421368) and a composite sequence assembled by using MacVector software (Oxford Molecular Group). In addition to the human cDNA clones, seven ESTs derived from the murine HCF-2 cDNA were identified (GenBank accession no. AA013895, AA2266375, AA492986, AA615766, AU019838, AI020559, and AI159365). The composite murine sequence (921 nucleotides) predicts a protein fragment that is 92% identical and 99% similar to the carboxy-terminal 232 residues of human HCF-2.
Isolation of HCF-2 cDNAs and Northern blotting.
Additional cDNAs were isolated by screening a mixed oligo(dT) and random-primed human adult testis library (Clontech) with a 32P-labeled 533-bp random-primed fragment of HCF-2 cDNA (corresponding to nucleotides 1246 to 1778). This fragment was generated by PCR amplification from an EST clone (GenBank accession no. AA421368) by using the HCF-2-specific primers 5′-GTCAGGATGGACCCTCACAGAC-3′ and 5′-GCCACTGGATTTGAAGGAGTC-3′. DNA was prepared from positive phages by using LambdaSorb phage absorbent (Promega). Northern blot analysis was performed by using the labeled cDNA fragment described above to probe a blot of human tissue poly(A)+ RNAs (2 μg/lane) as instructed by the manufacturer (Clontech). Following autoradiography, the blot was stripped and reprobed with a 1,078-bp HCF-1 cDNA fragment (corresponding to nucleotides 1194 to 2271 of our published cDNA sequence, GenBank accession no. L20010).
Construction of HCF-2 expression vectors.
Fragments encoding either the β-propeller domain (residues 2 to 373) or complete open reading frame (ORF; residues 2 to 792) were generated by amplification using high-fidelity PCR (Expand high-fidelity PCR system; Boehringer Mannheim). Unique XbaI and BamHI sites were included at the 5′ and 3′ ends, respectively, and the fragments were introduced between the corresponding sites in the mammalian expression vector pCGN (34). The full-length ORF was constructed from two amplified fragments that were fused by using a unique internal TaqI restriction site. The cDNA clone used as the template for amplification of the carboxy-terminal end of the HCF-2 ORF contained a C-to-T change (position 2185 in composite cDNA) relative to other HCF-2 cDNAs and changes a proline residue to a serine. Whether this reflects a reverse transcription error or a polymorphism is unknown; however, because this residue is a proline in all other HCF proteins and is therefore likely to be important for function, we modified the sequence to code for a proline by using site-directed mutagenesis according to the QuikChange protocol (Stratagene). The sequences of all PCR-generated fragments were verified by DNA sequence analysis.
To generate swaps between portions of the β-propeller domains of HCF-1 and HCF-2, we engineered a BglII restriction site at a conserved position near the beginning of HCFKEL5 (residues Arg-255 and Ser-256 in HCF-1 and residues Arg-245 and Ser-246 in HCF-2). The mutagenesis primer pairs (HCF-1, 5′-GGCGCCTCTTCCTAGATCTCTCCACTCGGCAACCACCATCG-3′ and its complement; HCF-2, 5′-GGGACAGTGCCACTTCCAAGATCTCTTCATACAGCCAGTGTTATAGG-3′ and its complement) were used to introduce the BglII site (underlined). The amino acid sequences are unchanged by these mutations.
Yeast two-hybrid interaction assay.
The polylinkers of the Gal4 DNA-binding domain (DBD) expression vector pGBT9 and Gal4 activation domain (AD) expression plasmid pGAD424 were modified to include a T7 epitope tag and unique SpeI restriction site, creating pYDBT and pYADT respectively. XbaI-BamHI fragments encoding the HCF-1 and HCF-2 β-propeller domains, as well as wild-type VP16ΔC (residues 5 to 411), mutant VP16ΔC (VP16ΔC E361A/385Ala3), and the amino terminus of human LZIP, were then subcloned between the unique SpeI and BamHI sites of the appropriate yeast vector. The resulting DBD and AD plasmids were cotransformed into the reporter strain Y190 containing Gal4-responsive his3 and lacZ reporter genes. The cDNA fragment encoding the full-length ORF of human LZIP was generated by PCR using an EST cDNA clone (GenBank accession no. R14706; Genome Systems Inc.) as a template. The amplified fragment was verified by DNA sequencing.
Transfections, coimmunoprecipitations, immunoblotting, and electrophoretic mobility shift assays.
Human 293T cells were transfected with Lipofectamine (Life Technologies), using 20 μl of lipid reagent per 6-cm-diameter dish. Whole-cell nuclear extracts were prepared after 40 to 48 h by lysing cells in high-salt lysis buffer (420 mM KCl, 10 mM Tris-HCl [pH 7.9], 5% glycerol, 0.25% Nonidet P-40, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.2 mM sodium vanadate, 50 μM sodium fluoride, 1 mM dithiothreitol). Nuclei were extracted at 4°C for 30 min and removed by centrifugation. For immunoprecipitations, 100 μl of extract was incubated with 2.5 μl of hemagglutinin (HA)-specific antibody (12CA5)-coupled protein G-agarose beads at 4°C for 1 h. The beads were washed four times in 1 ml of wash buffer (200 mM KCl, 10 mM Tris-HCl [pH 7.9], 5% glycerol, 0.5 mM EDTA) before separation by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Immunoblotting was performed by semidry transfer and detected by enhanced chemiluminescence (SuperSignal; Pierce). The anti-HA antibody and anti-T7 antibody (Novagen) were diluted 1:5,000 and 1:10,000, respectively. Electrophoretic mobility shift assays were performed as described previously (40, 41); complex formation was performed at 30°C, and electrophoresis was carried out at room temperature. Luciferase reporter assays were performed under standard conditions. Extracts were prepared in a commercial lysis buffer (Promega, Inc.) and measured with a LB9507 luminometer (EG&G Berthold, Inc.).
Immunofluorescence.
HeLa cells were seeded onto sterile coverslips and transfected with 1.5 μg of each HCF-1 or HCF-2 expression plasmid by using Lipofectamine (Life Technologies). After 36 h, the cells were fixed in 3.7% paraformaldehyde for 30 min, washed twice in phosphate-buffered saline (PBS), and permeabilized for 10 min with PBS containing 0.3% Triton X-100 at room temperature. The samples were then washed three times with PBS and once in PBS containing 1% fetal bovine serum (FBS). The coverslips were incubated for 1 h at 4°C with the anti-T7 antibody (diluted 1:600 in PBS with 1% FBS and 300 μg of bovine serum albumin per ml), washed four times in PBS with 0.1% Triton X-100, and washed once with PBS with 1% FBS. Coverslips were incubated with the secondary antibody (fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G diluted 1:1,000) for an additional 1 h. Fluorescence was observed with a Nikon fluorescence microscope.
Complementation of the tsBN67 proliferation defect.
Subconfluent tsBN67 cells were incubated at 33.5°C for 20 h and transfected with 2 μg of each HCF expression vector together with 2 μg of pSV2neo by using Lipofectamine (Life Technologies). The DNA mixes were sterilized by ethanol precipitation prior to transfection. After 2 days at 33.5°C, transfected cells were split into two 15-cm-diameter dishes, and Geneticin (0.8 mg/ml) was included in the media to select for stable transfectants. One dish was maintained at 33.5°C, and the other was shifted to 39.5°C. Colonies on the plates incubated at 39.5°C were counted 1.5 to 2 weeks after transfection. Protein extracts were prepared from cells maintained at 33.5°C and assayed by immunoblotting. Colonies were visualized by staining with 0.5% crystal violet in 80% methanol.
Nucleotide sequence accession number.
The sequence of the human HCF-2 cDNA has been deposited in the GenBank database under accession no. AF117210.
RESULTS
Isolation of cDNAs encoding HCF-2.
Sequences corresponding to the amino-terminal self-association domain of HCF-1 (see Fig. 2A) were used to search the National Center for Biotechnology Information dbest, using the BLAST 2.0 program tblastn. We identified an EST sequence (GenBank accession no. AA421368) that encoded a hypothetical peptide with extensive amino acids homology to the self-association domain and the carboxy-terminal portion of the β-propeller domain of HCF-1 but which differed significantly at the nucleotide level. Additional searches of dbest identified additional overlapping EST sequences as well seven clones encoding the murine homologue (see Materials and Methods). Based on the presence of two HCF signature motifs, we designated this new protein HCF-2, following our original nomenclature for HCF-1 (41).
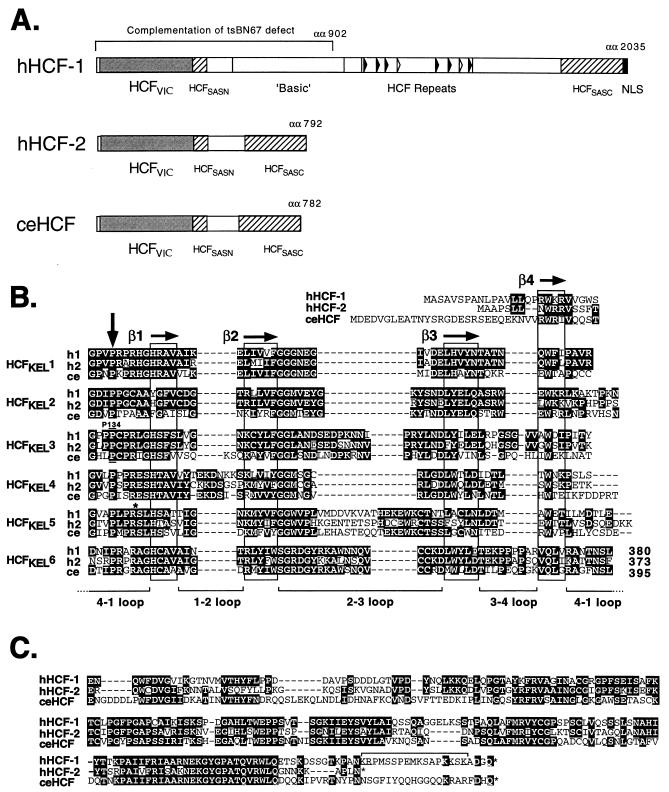
FIG. 2.
HCF proteins share a common architecture. (A) The primary structures of human HCF-1 and HCF-2 as well as nematode HCF (GenBank accession no. U61948; references 17 and 21) are shown schematically. The amino-terminal β-propeller (HCFVIC) domains are shown as shaded boxes. The eight HCFPRO repeats of HCF-1 are indicated by open (nonfunctional) and filled (functional) arrowheads. The amino- and carboxy-terminal self-association domains (HCFSASN and HCFSASC) are shown as hatched boxes. The nuclear localization signal (NLS) of HCF-1 is indicated by a filled box at the extreme carboxy terminus. The first 902 amino acid (αα) residues of HCF-1 comprising the β-propeller, HCFSASN domain, and a poorly defined basic region are required to complement the tsBN67 defect. (B) Comparison of the six kelch repeats which make up the β-propeller (HCFVIC) domains of human HCF-1 and HCF-2 (hHCF-1 and hHCF-2) and C. elegans HCF (ceHCF). Identical residues are highlighted in black. Following the crystal structure of galactose oxidase, each kelch repeat is predicted to fold into four β-strands (β1 and β4 [boxed]) forming one β-sheet or blade of the β-propeller. Residue Pro-134 of HCF-1 (h1), which is mutated in tsBN67 cells, lies in the 4-1 loop connecting HCFKEL2 and HCFKEL3. This position is conserved in HCF-2 (h2) and in nematode HCF(ce), emphasizing its importance in domain function. An asterisk near the beginning of HCFKEL5 marks the location (Arg-Ser) of an engineered BglII site used to generate chimeric versions of the β-propeller. (C) Comparison of the carboxy-terminal conserved regions of HCF-1, HCF-2, and C. elegans HCF. The bipartite nuclear localization signal of HCF-1 (bracketed) lies at the extreme carboxy terminus of the polypeptide. HCF-2 lacks an equivalent cluster of basic residues.
To determine a complete ORF, we screened an adult human testis library, using a portion of the original cDNA clone as a probe. This library was chosen because the tissue origins for the HCF-2 EST collection revealed a significant bias towards testis-derived libraries (see below). From 6.5 × 105 recombinants, we isolated a total of 22 positive clones. Several independent clones were sequenced, yielding a 2,583-bp composite cDNA sequence, predicting a single ORF encoding 792 amino acids. No alternative mRNA splicing products were identified by this analysis. The predicted amino acid sequence is shown in Fig. 1. The assignment of the first methionine remains tentative until additional 5′ sequences containing an in-frame stop codon have been identified. The HCF-1 and HCF-2 sequences are strikingly different at the nucleotide level, most notably in overall nucleotide composition. The HCF-1 ORF is very G+C rich (63%) compared to that of HCF-2 (43% G+C), and this may explain the lack of cross-hybridization during our initial cloning and analysis of HCF cDNAs (41, 42).
FIG. 1.
Amino acid sequence of human HCF-2. The amino- and carboxy-terminal blocks of homology to HCF-1 and nematode HCF are boxed.
At the amino and carboxy ends of the predicted HCF-2 protein, there are extensive regions of homology to mammalian HCF-1 (10, 17, 39) and to a Caenorhabditis elegans HCF homologue (17). These conserved regions (boxed in Fig. 1) correspond to the β-propeller domain required for interaction with VP16 and LZIP (40) and the amino- and carboxy-terminal self-association domains. This conserved arrangement of functional domains is shown schematically in Fig. 2A. The β-propeller domains of HCF-1 and HCF-2 (364 residues) are 69% identical and 83% similar, the amino-terminal self-association domains (43 residues) are 54% identical and 77% similar, and the carboxy-terminal self-association domains (192 residues) are 59% identical and 72% similar. Alignments of the β-propeller domain and conserved carboxy-terminal domain sequences from HCF-1, HCF-2, and nematode HCF are shown in Fig. 2B and C, respectively. Within the β-propeller domain, sequence identity is high across the entire domain, except for the 4-1 loop of HCFKEL4 and the 2-3 loop of HCFKEL5, which have diverged in all three proteins. Most striking is the conservation of the proline residue at the fourth position of each repeat (indicated by a vertical arrow in Fig. 2B). In HCF-1 this includes proline 134, which is mutated to serine in tsBN67 HCF, resulting in a conditional cell cycle arrest. The remarkable conservation of this proline emphasizes its importance in domain function. In terms of general structure, nematode HCF more closely resembles HCF-2 (Fig. 2A). The predicted nematode HCF polypeptide (782 residues) is considerably closer in size to HCF-2 (792 residues) than HCF-1 (>2,000 residues). At the amino acid sequence level, however, the two mammalian proteins are equally similar to the invertebrate counterpart, suggesting that HCF-1 and HCF-2 arose by gene duplication subsequent to the separation of vertebrate and nematode ancestors.
The two conserved regions in HCF-2 are separated by 201 amino acids without obvious similarity to HCF-1 or other proteins. Most notably, HCF-2 lacks the basic region of HCF-1, which is required for complementation of the tsBN67 defect (40). This spacer region is relatively serine/threonine rich (28%), as is also the case in C. elegans HCF. In addition, the predicted HCF-2 polypeptide sequence does not contain the HCFPRO repeats that characterize HCF-1 (19, 43), suggesting that HCF-2 is not subject to proteolytic processing. This is confirmed by expression of an epitope-tagged version of the complete HCF-2 ORF in human 293T cells, yielding a single ∼95- to 100-kDa polypeptide species that presumably corresponds to the predicted 87-kDa primary translation product (data not shown and Fig. 5B).
FIG. 5.
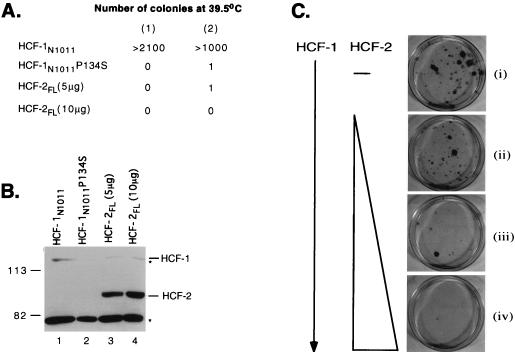
HCF-2 cannot complement the tsBN67 cell proliferation defect. Hamster tsBN67 cells were stably transfected with the HCF-1 or HCF-2 expression plasmids indicated. Following transfection, the cells were incubated at 39.5°C with G418 for 1.5 to 2 weeks, and complementation was scored as the number of colonies of proliferating cells. (A) Numbers of rescued colonies resulting from transfection with 5 μg of pCGNHCF-1N1011, 5 μg of pCGNHCF-1N1011P134S, and either 5 or 10 μg of pCGNHCF-2FL. The results from two independent experiments, (1) and (2), are given. (B) Expression of the HA-tagged proteins was monitored by immunoblotting. Extracts were prepared from duplicate cultures maintained at 33.5°C with G418, resolved on an SDS–7% polyacrylamide gel, and probed with an antibody against the HA epitope tag. The relative mobility of prestained molecular weight markers (Bio-Rad), given in kilodaltons, is indicated on the left. Two nonspecific cross-hybridizing bands are indicated with asterisks. (C) Coexpression of HCF-2FL severely inhibits complementation by wild-type HCF-1. tsBN67 cells were transfected with 1 μg of pCGNHCF-1N902 alone (dish i) or together with 2.5 μg (dish ii), 5 μg (dish iii), or 10 μg (dish iv) of pCGTHCF-2FL. The total amount of expression vector was normalized to 20 μg, using pCMV-lacZ. Representative plates are shown, and proliferating colonies were stained with 0.5% crystal violet.
HCF-2 is expressed in many tissues and at high levels in the testis.
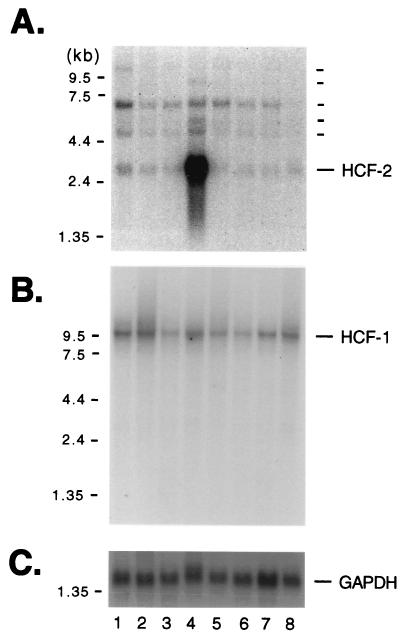
To examine the expression patterns of HCF-2, we analyzed poly(A)+ RNA from various human tissues for the presence of transcripts that hybridize with an HCF-2 cDNA probe. The result of this experiment is shown in Fig. 3. We chose a 0.5-kb fragment from the unique central region of HCF-2 as the probe to avoid the possibility of cross-hybridization with HCF-1. As shown in Fig. 3A, we observed a complex pattern of hybridization under stringent conditions. Three transcripts (3.2, 4.9, and 7 kb) were detected in all of the tissues except peripheral blood, in which only the 3.2-kb transcript was evident (Fig. 3A, lane 8). The 3.2-kb transcript is extremely abundant in testis (lane 4) and is likely to account for the cDNA described in this study. An additional >10-kb species was detected in spleen tissue (lane 1), and in testis additional 6- and 9-kb species were evident (lane 4), raising the possibility that alternative versions of the HCF-2 mRNA or even additional HCF-like genes exist. The same blot was stripped and reprobed with an HCF-1 cDNA fragment (Fig. 3B). As we have previously reported (41, 42), the HCF-1 cDNA detects a single transcript species of ∼10 kb, present at similar levels in all tissues examined.
FIG. 3.
HCF-2 is expressed in multiple tissues and is abundant in the testis. Northern blot analysis of HCF-2 and HCF-1 mRNA. Poly(A)+ RNA (2 μg) from normal human tissue was probed with an HCF-2-specific probe (A); the blot was then stripped and reprobed with an HCF-1-specific probe (B) or a GAPDH-specific probe (C). The tissues analyzed were spleen (lane 1), thymus (lane 2), prostate (lane 3), testis (lane 4), ovary (lane 5), small intestine (lane 6), mucosal lining of the colon (lane 7), and peripheral blood leukocytes (lane 8). Positions of size markers are shown to the left of each panel.
HCF-2 shows a more dynamic pattern of subcellular localization than HCF-1.
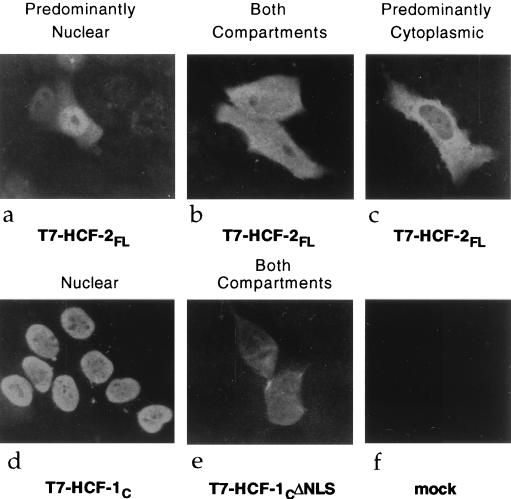
HCF-1 is an exclusively nuclear protein (19), and correct localization is dependent on bipartite signal that lies close to the carboxy terminus of the precursor polypeptide (3a). Despite extensive homology within the carboxy terminus of HCF-2, there are no equivalent basic clusters (Fig. 2C). This observation suggested the interesting possibility that HCF-2 is a cytoplasmic protein or is targeted to the nucleus by a different signal. To examine the subcellular localization of HCF-2, we transiently transfected human HeLa cells with an expression vector encoding full-length HCF-2. A bacteriophage T7 gene 10 epitope tag was included at the amino terminus to allow detection of the recombinant protein. Subcellular localization was determined by indirect immunofluorescence using an anti-T7 monoclonal antibody. Figure 4 shows the immunofluorescence pattern of representative cells from an unsynchronized population of transfected cells. Considerable cell-by-cell heterogeneity was observed with HCF-2 (Fig. 4a to c), ranging from a predominantly nuclear staining pattern (Fig. 4a) to a predominantly cytoplasmic staining pattern (Fig. 4c). Many cells were intermediate, showing staining in both compartments (Fig. 4b). This heterogeneity was in striking contrast to uniform staining patterns of two HCF-1 controls: the carboxy terminus of HCF-1 (T7-HCF-1C [Fig. 4d]), which localized to the nucleus of all transfected cells, and a version of the HCF-1 carboxy terminus lacking the nuclear localization signal (T7-HCF-1CΔNLS [Fig. 4e]) which was exclusively cytoplasmic. These results indicate that HCF-2, in contrast to HCF-1, can occupy multiple cellular compartments.
FIG. 4.
Subcellular localization of HCF-2, determined by immunofluorescence analysis of HeLa cells transfected with expression plasmids encoding HCF-2FL (a to c), HCF-1C (d), and HCF-1CΔNLS (e). Thirty-six hours after transfection, cells were fixed and probed with an anti-T7 monoclonal antibody followed by a fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G secondary antibody.
HCF-2 does not complement the tsBN67 cell proliferation defect.
Conditional loss of HCF-1 function in tsBN67 cells leads to a reversible G1/G0 cell cycle arrest, and this defect can be complemented by stable expression of an amino-terminal fragment (residues 1 to 902) from wild-type HCF-1 (10, 40). To determine whether HCF-2 could complement the tsBN67 cell proliferation defect, we transfected tsBN67 cells with expression vectors encoding full-length HCF-2 (HCF-2FL) and measured colony formation at 39.5°C, the nonpermissive temperature. The results of this experiment are summarized in Fig. 5. As a positive control, we expressed the amino terminus of HCF-1 (residues 1 to 1011; HCF-1N1011), which we have shown is sufficient to complement the tsBN67 defect (40); as a negative control, we expressed the tsBN67 version of this fragment (HCF-1N1011P134S). We used selection for G418 resistance to eliminate untransfected cells. As shown in Fig. 5A, stable expression of the wild-type HCF-1 amino terminus gave rise to a large number of colonies at the nonpermissive temperature, indicative of efficient rescue of the temperature-sensitive defect. In contrast, expression of either full-length HCF-2 (HCF-2FL) or the mutant version of HCF-1 (HCF-1N1011P134S) gave rise to few or no rescued colonies. Expression of HCF-2FL was confirmed by immunoblotting extracts from a parallel cultures maintained at 33.5°C (Fig. 5B, lanes 3 and 4). The levels of HCF-2 protein (lanes 3 and 4) were in fact substantially higher than the levels of wild-type HCF-1N1011 (lane 1). These results indicate that complementation of the tsBN67 proliferation defect requires one or more functions that are specific to HCF-1.
We next examined whether coexpression of HCF-2 could interfere with complementation by wild-type HCF-1 (Fig. 5C). Cultures of tsBN67 cells were stably transfected with an HCF-1 expression plasmid (HCF-1N902; dish i) or with the HCF-1 plasmid together with increasing amounts HCF-2 expression plasmid (dishes ii to iv). HCF-1N902 and HCF-1N1011 complement with similar efficiency (40), and we chose the smaller protein simply because it is easier to detect by immunoblotting. As before, complementation was scored by examining the number of G418-resistant colonies produced at the nonpermissive temperature. To avoid the effects of promoter competition, the total amount of cytomegalovirus-driven expression plasmid was held constant by using a vector expressing β-galactosidase. As with HCF-1N1011, cultures transfected with HCF-1N902 (dish i) gave rise to many rescued colonies. This number was noticeably reduced by cotransfection with a 2.5-fold excess of full-length HCF-2 expression plasmid (dish ii) and essentially abolished with a 5- or 10-fold excess of HCF-2 (dishes iii and iv). This result indicates that full-length HCF-2 can function as a potent inhibitor of HCF-1 dependent cell proliferation.
HCF-2 supports VP16-induced complex formation.
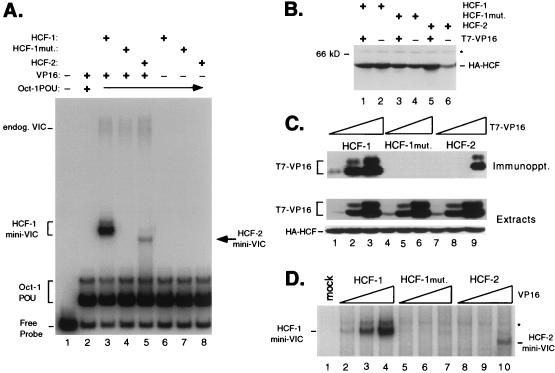
The extensive amino acid sequence conservation throughout the β-propeller domains of HCF-1 and HCF-2 suggested that HCF-2 may be able to interact with HSV VP16 and support VP16-induced complex formation. To address this, we coexpressed the HCF-2 β-propeller domain (residues 1 to 373) together with VP16 in transiently transfected 293T cells and assayed for VP16-induced complex formation by using an electrophoretic mobility shift assay. As controls, we transfected expression plasmids encoding wild-type and mutant (tsBN67) versions of the HCF-1 β-propeller (HCF-1N380 and HCF-1N380P134S, respectively). Figure 6A shows the results of this analysis. Whole-cell extracts were prepared from the transfected cells and mixed with a labeled DNA probe containing a VP16-responsive TAATGARAT element derived from the HSV ICP0 promoter together with Escherichia coli-expressed Oct-1 POU domain protein. Wild-type HCF-1 gave rise to a robust VP16-induced complex (Fig. 6A, lane 3, labeled HCF-1 mini-VIC), while P134S HCF failed to support complex formation (lane 4). HCF-2 was also able to support VP16-induced complex formation (lane 5), giving rise to a complex (labeled HCF-2 mini-VIC) with a gel mobility slightly faster than that of the complex formed by HCF-1. The distinctive mobility of the HCF-2-containing complex may reflect a slightly different shape, as the HCF-1 and HCF-2 β-propeller domains differ in length by only seven amino acids and migrate with the same mobility in a denaturing gel (Fig. 6B). This result demonstrates that HCF-2 is capable of interacting with VP16 and promoting the assembly of a VP16-induced complex that includes the Oct-1 POU domain and DNA. The most striking difference between HCF-1 and HCF-2 was in the relative abundance of the VP16-induced complex (compare lanes 3 and 5). Although equivalent amounts of HCF-1 and HCF-2 proteins were present in these extracts (as determined by immunoblotting [data not shown]), HCF-1 produced an approximately 10-fold-stronger shift. This result suggests that HCF-2 may associate with VP16 less efficiently or may be compromised in a subsequent step of VP16-induced complex assembly.
FIG. 6.
The β-propeller domain of HCF-2 is capable of supporting VP16-induced complex formation. (A) HCF polypeptides were coexpressed with VP16ΔC by transfection of 293T cells. Whole-cell extracts were prepared after 40 h and assayed for stabilization of the VP16-induced complex in an electrophoretic mobility shift assay. Lane 1 contains probe alone, and lanes 2 to 5 contain probe with Oct-1 POU domain proteins and the following cell extracts: mock transfection (lane 2), wild-type HCF-1N380 and VP16ΔC (lane 3), HCF-1N380 P134S and VP16ΔC (lane 4), HCF-2N373 and VP16ΔC (lane 5), wild-type HCF-1N380 alone (lane 6), HCF-1N380P134S alone (lane 7), and HCF-2N373 alone (lane 8). The positions of the free probe, Oct-1 POU domain complex, and VP16-induced complex containing native human HCF-1 (endog. VIC) or truncated HCF (HCF-1 or HCF-2 mini-VIC) are indicated. mut., mutant. (B) Immunoblot of extracts used for panel A. Extracts were resolved on an SDS–10% polyacrylamide gel, transferred to nitrocellulose, and immunoblotted with a monoclonal antibody against HA (12CA5). The size of a molecular weight marker is indicated on the left, and a nonspecific cross-reacting protein is indicated with an asterisk. (C) HCF-VP16 coimmunoprecipitation. Direct interaction between HCF-2 and VP16ΔC was examined by coimmunoprecipitation assay. Extracts were prepared from 293T cells transfected with 10 ng, 100 ng, and 1.0 μg of T7-tagged VP16ΔC expression plasmid together with a constant 3 μg of expression plasmids encoding HA-tagged wild-type HCF-1N380 (lanes 1 to 3), P134S HCF-1N380 (lanes 4 to 6), and HCF-2N373 (lanes 7 to 9). Extracts were precipitated with an anti-HA antibody, and VP16 was detected by immunoblotting with the anti-T7 epitope tag antibody. Direct immunoblotting of the extracts (lower two panels) showed that all HA-tagged HCF polypeptides were expressed at equivalent levels and that the expression of T7-tagged VP16 was proportional to amount of input plasmid. (D) Transfected cells extracts used for panel C were assayed for VP16-induced complex formation by electrophoretic mobility shift assay. Labeled TAATGARAT-containing probe and the Oct-1 POU domain were mixed with the following transfected cell extracts: mock transfection (lane 1), HCF-1N380 with 10 ng (lane 2), with 100 ng (lane 3), and with 1.0 μg of VP16ΔC expression plasmid (lane 4), HCF-1N380P134S with 10 ng (lane 5), with 100 ng (lane 6), and with 1.0 μg of VP16ΔC expression plasmid (lane 7), and HCF-2 with 10 ng (lane 8), with 100 ng (lane 9), and with 1.0 μg of VP16ΔC expression plasmid (lane 10). For clarity, only the VP16-induced complexes produced by recombinant HCF (labeled HCF-1 and HCF-2 mini-VIC) are shown. A nonspecific complex is indicated with an asterisk.
VP16 selectively recruits HCF-1 rather than HCF-2.
To determine whether HCF-1 and HCF-2 differ in the ability to associate with VP16, we used a coimmunoprecipitation assay to determine the relative affinity of each β-propeller domain for VP16. Human 293T cells were cotransfected with increasing amounts of VP16 expression vector (10 ng, 100 ng, and 1 μg) together with a constant amount of HCF-1, HCF-1, P134S, and HCF-2 expression plasmids. A representative experiment is shown in Fig. 6B. The HCF polypeptides were tagged with an influenza virus HA epitope, and VP16 was tagged with the T7 epitope. HA-tagged HCF polypeptides were recovered by immunoprecipitation using an anti-HA monoclonal antibody linked to agarose beads. As we have described previously (40), VP16 could be readily recovered with the wild-type HCF-1 β-propeller (Fig. 6B, lanes 1 to 3) but not by a version of HCF-1 carrying the tsBN67 point mutation (lanes 4 to 6). Recovery by HCF-1 was proportional to VP16 expression and was detectable at 10 ng of VP16 expression plasmid (lane 1).
Coimmunoprecipitation of VP16 by HCF-2 was detected only at the highest level of VP16 expression (1.0 μg of VP16 plasmid [lane 9]), and recovery was similar to that of HCF-1 with 100 ng of VP16 expression plasmid, indicating at least a 10-fold-lower relative affinity. The same extracts were assayed for VP16-induced complex assembly in an electrophoretic mobility shift assay (Fig. 6C). Consistent with the coimmunoprecipitation results, complex formation by HCF-2 was detected only in the presence of maximum levels of VP16 (lane 10) and at a level similar to that seen at the midpoint of the HCF-1 titration (lane 3). These results indicate that the reduced level of VP16-induced complex formation by HCF-2 is likely a consequence of a lower relative affinity for VP16.
The cellular bZIP protein LZIP also discriminates between HCF-1 and HCF-2.
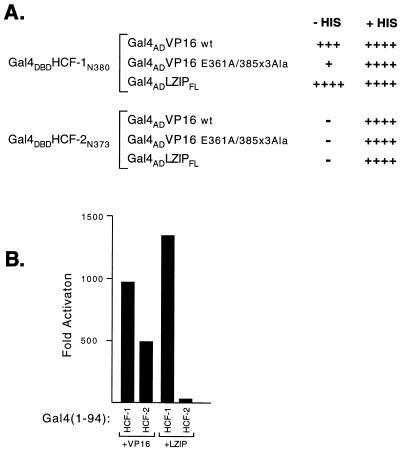
In addition to VP16, HCF-1 has been shown to associate with a ubiquitous cellular bZIP protein known as LZIP or Luman (7, 23). This interaction is mediated by the β-propeller domain of HCF-1 and a tetrapeptide sequence known as the HBM present in both VP16 and LZIP (7, 24). Point mutagenesis studies have revealed a strong parallel in the way that LZIP and VP16 recognize HCF-1 and suggest that LZIP may also interact less effectively with HCF-2. We addressed this question by using a yeast two-hybrid assay in which the HCF-1 and HCF-2 β-propeller domains were expressed as Gal4 DBD fusion proteins, while VP16 and LZIP were expressed as Gal4 AD fusions. Interaction was measured by activation of a GAL1-HIS3 reporter gene, allowing growth in the absence of histidine. These results are summarized in Fig. 7A. Wild-type VP16 and LZIP both interacted with wild-type HCF-1N380, as indicated by growth on both plates. In contrast, HCF-2 did not interact with VP16 or LZIP in this assay. As a control, a mutant version of VP16 (VP16-E361A/385Ala3) failed to interact with either bait, confirming that the interaction is specific and that neither Gal4 DBD fusion activated the GAL1-HIS3 reporter on its own. This result indicates that LZIP can also clearly discriminate between the similar β-propeller domains of HCF-1 and HCF-2, interacting strongly with HCF-1 and not at all with HCF-2.
FIG. 7.
LZIP interacts with HCF-1 but not HCF-2. (A) Yeast two-hybrid interaction assays with the β-propeller domains of HCF-1 and HCF-2. A yeast GAL1-HIS3 reporter strain was transformed with expression plasmids encoding the HCF-1 or HCF-2 β-propeller domain fused to the Gal4 DBD together with a plasmid encoding either wild-type (wt) VP16, mutant VP16 (VP16 E361A/385Ala3), or human LZIP (LZIPFL) each fused to the Gal4 AD. Transformants were incubated for 3 days at 30°C in both the presence and the absence of histidine. Growth without histidine (− His) demonstrates activation of the GAL1-HIS3 reporter gene, and growth with histidine (+ His) demonstrates that the expression plasmids were not toxic to the cells; 20 mM 3-amino-1,2,4-triazole (Sigma) was added to the media to suppress the low-level HIS3 expression observed in the presence of the Gal4 DBD fusions alone. (B) One-hybrid assay in transiently transfected 293T cells. A Gal4-responsive luciferase reporter gene (p5×Gal-E1B-luc) was transfected into 293T cells by electroporation together with 1 μg of pCGNGal(1-94)HCF-1N380 or pCGNGal(1-94)HCF-2N373 and 500 ng of pUC119, pCGTVP16+C, or pCGTLZIPN154, as indicated. Values are the average of duplicate experiments. Fold activation was calculated relative to the activity of each Gal4 fusion cotransfected with pUC119.
In addition to the yeast two-hybrid assay, we examined the interaction between HCF-2 and LZIP by using an one-hybrid assay in transfected mammalian cells. In this assay, either the amino terminus of LZIP or full-length VP16 is recruited to a reporter promoter through association with the HCF-1 or HCF-2 β-propeller domain fused to residues 1 to 94 of the Gal4 DBD. 293T cells were transfected by electroporation and after 30 h assayed for luciferase activity. These results are shown in Fig. 7B. Cotransfection of Gal4–HCF-1N380 with either VP16 or LZIP resulted in strong activation of the reporter gene. In contrast, cotransfection of Gal4–HCF-2N373 with VP16 gave a more modest level of activation, consistent with the reduced affinity measured by immunoprecipitation, while LZIP barely activated above background levels. Gal4–HCF-1N380 and Gal4–HCF-2N373 alone did not activate the reporter (data not shown). These results show that HCF-2 interacts weakly with VP16 and essentially fails to interact with LZIP.
Sequences within HCFKEL repeats 4 and 5 determine the differential recognition of VP16 and LZIP.
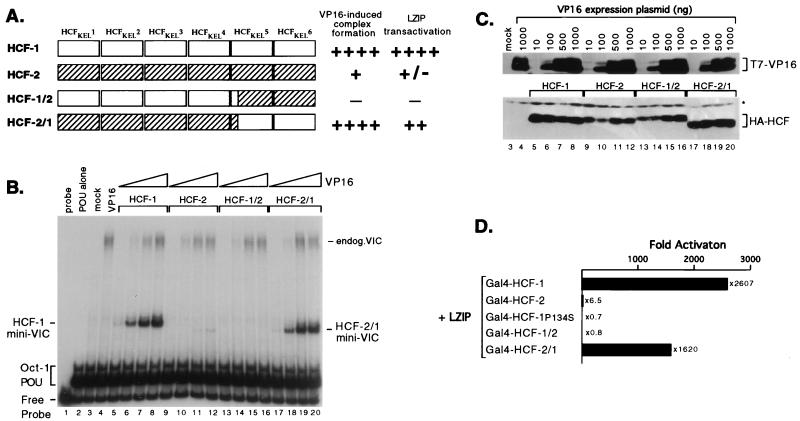
To probe the mechanism of selective recruitment in more detail, we generated a pair of exchanges within the HCF-1 and HCF-2 β-propellers: swapping the more divergent HCFKEL5 and HCFKEL6 repeats (Fig. 2B). These chimeric proteins are shown schematically in Fig. 8A. A precise exchange was achieved by engineering a unique BglII restriction site at a conserved Arg-Ser dipeptide near the beginning of HCFKEL5 (indicated with an asterisk in Fig. 2B). The modified parental HCF-1(Bgl) and HCF-2(Bgl) proteins, together with the reciprocal swaps HCF-1/2(Bgl) and HCF-2/1(Bgl), were coexpressed in transiently transfected 293T cells together with increasing amounts of VP16 expression plasmid. Extracts were prepared and tested for VP16-induced complex formation by gel mobility shift assay (Fig. 8B). Expression of each epitope-tagged polypeptide was confirmed by immunoblotting of the same extracts (Fig. 8C). As shown already, the HCF-1 β-propeller (Fig. 8B, lanes 5 to 8) gave rise to a robust VP16-induced complex (mini-VIC) that was detectable even at the lowest concentration of VP16 (Fig. 8B, lane 5). In contrast, HCF-2 promoted very weak VP16-induced complex formation with a characteristic small increase in gel mobility (lanes 9 to 12). Less HCF-2 than HCF-1 was expressed (Fig. 8C; compare lanes 5 to 8 with lanes 9 to 12), slightly overemphasizing the difference in complex-forming ability. The two chimeric proteins showed markedly different complex-forming activities. The HCF-1/2 chimera was essentially inactive (lanes 13 to 16), while the HCF-2/1 chimera formed a strong complex similar to that formed by HCF-1 (compare lanes 5 to 8 and 17 to 20) and was detectable at the lowest concentration of VP16 (lane 17). Interestingly, the VP16-induced complex formed by the HCF-2/1 chimera migrated with the characteristic faster mobility of HCF-2. These results show that important determinants for VP16-induced complex formation lie in the fifth and sixth kelch repeats.
FIG. 8.
Sequences near the carboxy terminus of the HCF-1 β-propeller are required for high-affinity interactions with VP16 and LZIP. (A) Schematic representation of the parental and chimeric versions of the HCF-1 and HCF-2 β-propeller domains generated by using an engineered BglII site at analogous positions in HCF-1 (residues Arg-255/Ser-256) and HCF-2 (residues Arg-245/Ser-246). (B) 293T cells were transiently transfected with 3 μg of each HA-tagged β-propeller domain expression plasmid together with 10 ng, 100 ng, 500 ng, or 1.0 μg of T7-tagged VP16ΔC expression plasmid. The HCF expression plasmids were pCGNHCF-1N380 (lanes 5 to 8), pCGNHCF-2N373 (lanes 9 to 12), pCGNHCF-2/1 (lanes 13 to 16), and pCGNHCF-2/1 (lanes 17 to 20). Lanes 3 and 4 are extracts from mock-transfected cells or cells transfected with 1 μg of pCGTVP16ΔC. Extracts were prepared after 30 h and assayed by gel mobility shift assay using a labeled ICP0 probe and bacterial Oct-1 POU domain. The positions of the free probe, Oct-1 POU domain complex, and VP16-induced complex containing native human HCF-1 (endog. VIC) or truncated HCFs (HCF-1 or HCF-2 mini-VIC) are indicated. (C) The extracts shown in panel B were resolved on an SDS–10% polyacrylamide gel and immunoblotted with anti-T7 and anti-HA antibodies (upper and lower panels, respectively). For simplicity, numbering corresponds to the lanes in panel B. A nonspecific cross-reacting band is indicated with an asterisk. (D) Mammalian cell one-hybrid assay. Human 293T cells were transfected by electroporation with 500 ng of Gal4-responsive luciferase reporter gene (p5xGal-E1B-luc), 500 ng of pCGTLZIPN154 and 750 ng of pCGNGal(1-94)HCF-1N380, pCGNGal(1-94)HCF-2N373, pCGNGal(1-94)HCF-1N380P134S, pCGNGal(1-94)HCF-1/2, or pCGNGal(1-94)HCF-2/1. Luciferase activity was determined 24 h after transfection and plotted in terms of relative activation compared to the reporter cotransfected with pCGTLZIPN154 alone. Values are the average of duplicate experiments.
As shown above, LZIP exhibits an even stronger preference for HCF-1 than does VP16. Using the mammalian one-hybrid assay, we examined whether the addition of HCFKEL5 and HCFKEL6 to the HCF-2 β-propeller would allow LZIP to interact with the HCF-2 β-propeller. This experiment is shown in Fig. 8D. As before, LZIP functioned as a very potent activator (2,600-fold stimulation) when tethered to the reporter promoter through association with Gal4–HCF-1 and did not activate when coexpressed with Gal4–HCF-2, indicating an inability to interact with the HCF-2 β-propeller. The specificity of the assay was confirmed by using the tsBN67 point mutant Gal4–HCF-1 P134S, which also failed to support transactivation. Consistent with the restored association with VP16, LZIP interacted strongly with the HCF-2/1 chimeric β-propeller (1,600-fold activation), activating to a level that was approximately half that of Gal4–HCF-1. Expression of each Gal4 fusion protein was confirmed by immunoblotting (data not shown). These results show that LZIP also discriminates between HCF-1 and HCF-2 by recognizing differences in the fifth and sixth kelch repeats and that LZIP can interact efficiently with an HCF-2-derived β-propeller that contains HCF-1 versions of HCFKEL5 and HCFKEL6.
DISCUSSION
HCF (now HCF-1) was first purified and cloned by virtue of its association with the HSV transactivator VP16 (19, 20, 41). Subsequent analysis of cDNA clones and characterization of the genomic locus of HCF-1 in both humans and mice did not uncover evidence for other closely related genes (5, 6, 17, 42). In this study, we describe a new HCF protein, designated HCF-2, which we identified by database searching using the amino acid sequence of a functionally defined domain. This represents the first evidence that HCF-1 belongs to a family of related proteins. Based on size and organization, the human HCF-2 protein appears more similar to C. elegans HCF than to HCF-1. Both HCF-2 and nematode HCF lack HCFPRO repeats and contain a relatively short nonconserved sequence separating the two self-association domains. Within the conserved domains, however, nematode HCF exhibits equivalent homology to HCF-1 and HCF-2, suggesting the mammalian proteins arose from a comparatively recent gene duplication event. The HCFPRO repeats, which so far are known only for HCF-1, may thus represent a recent addition to a more ancient basic HCF architecture.
Northern blotting showed that the HCF-2 gene is transcribed at low levels in many tissues, and a number of different transcripts were detected. Most strikingly, there is a marked accumulation of the 3.2-kb transcript in the testis. This finding was corroborated through dot blot analysis of poly(A)+ RNA derived from 50 human tissue sources, in which no other tissue showed an equivalently strong hybridization signal (data not shown). In addition, 42% of the unique human HCF-2 EST clones present in GenBank were from testis-derived cDNA libraries. The significance of high expression in the testis is unknown but may reflect a specific role in the specialized series of cell divisions that take place during spermatogenesis or in the regulation of meiosis (reviewed in references 4 and 11).
HCF-2 shows a more complex pattern of subcellular localization than HCF-1. Using an unsynchronized population of cells, we found that exogenously expressed HCF-2 distributed dynamically between the nuclear and cytoplasmic compartments. The striking heterogeneity did not correlate with the level of expression and instead may have resulted from the asynchrony of the population, with individual transfected cells caught at different points in the cell cycle. Analysis of synchronized cells will address this interesting possibility. Shuttling between compartments could provide a general mechanism for regulating the transcriptional activity of HCF-2. For example, studies of the E2F family have shown that while E2F-1, -2, and -3 are predominantly nuclear throughout the cell cycle, E2F-4 and -5 are nuclear from G0 until mid-G1 and then relocate to the cytoplasm in late G1, S, and G2 phases. This coincides with reduced transactivation by E2F-4 as cells pass the restriction point (25, 37). Two mechanisms that could account for the cytoplasmic localization of HCF-2 are the lack of a conventional basic nuclear localization signal and the presence of a weak nuclear export signal. We have no evidence for the latter and favor a model in which nuclear localization of HCF-2 is achieved through regulated association with other cellular proteins that are specifically targeted to the nucleus. Coexpression of HCF-2FL with either the amino or carboxy terminus of HCF-1 did not alter its localization, suggesting that HCF-1 does not provide this function (data not shown).
The association of VP16 and LZIP with HCF is mediated by a short tetrapeptide, the HBM. This motif recognizes a less-well defined but evolutionarily conserved interaction surface provided by the β-propeller domain of HCF-1 (7, 24). Although the HCF-2 β-propeller supports VP16-induced complex assembly in vitro, it is inefficient compared to the HCF-1 β-propeller. This strong preference for HCF-1 was also observed for full-length VP16 and HCF proteins (data not shown). With LZIP, the difference in association is even more marked, suggesting that HCF-2 cannot interact with LZIP under physiological conditions. We have exploited this sharp functional difference to begin to map critical residues within the β-propeller domain involved in recognition of the HBM. This analysis has shown that important determinants lie towards the carboxy-terminal end of the HCF β-propeller. Transfer of HCFKEL5 and HCFKEL6 from HCF-1 to the HCF-2 β-propeller confers an HCF-1 like specificity. HCFKEL5 is notable in being the least conserved of the six repeats, especially within the 2-3 and 4-1 loops. Mutagenesis studies have already shown that residues in HCFKEL2, HCFKEL3, and HCFKEL4 are important for association with both VP16 and LZIP (7), indicating that HCFKEL5 and HCFKEL6 do not alone constitute the interaction surface. Instead, we suspect that these repeats or more precisely the hypervariable loops within HCFKEL5, function as the determinant of specificity, possibly by recognizing the equally variable sequences flanking the core HBM tetrapeptide. The further reduced activity of the HCF-1/2 chimera compared to HCF-2 may indicate additional differences in HCFKEL1 through 4 that partially compensate for the deficiency in HCFKEL5 and -6. The availability of two functionally distinct HCF proteins will allow us to address this further. Additional swaps within HCFKEL5 and HCFKEL6 should identify the amino acid differences responsible for the selective association and, when combined with targeted mutagenesis, ultimately define a docking surface for the HBM.
We suspect that there are additional cellular HBM-containing transcription factors, analogous to LZIP, that preferentially associate with HCF-2 rather than HCF-1. Although the functional significance of recruiting an HCF molecule to a site-specific activator is not fully understood, the existence of a family of proteins with slightly different specificity offers an opportunity for regulation. For example, it may allow cells in the testis to use HCF-2 for a specific task without compromising the housekeeping function of HCF-1 in regulating cell proliferation. Screens to identify HBM-containing proteins that associate with HCF-2 are in progress. Our observation that HCF-2 functions as an inhibitor of tsBN67 complementation by HCF-1 suggests the two proteins share at least one common target. It is conceivable that HCF-2 uses its conserved carboxy terminus to sequester proteins used by HCF-1. Once recruited to HCF-2, these factors would become unavailable for use by site-specific activators such as LZIP. Accumulation of HCF-2 in the cytoplasm would enhance any inhibitory effect, by sequestering associated proteins in a separate compartment of the cell. Measuring the relative abundance of endogenous HCF-2 protein in a given cell type must await the generation of specific antibodies, but in principle, simple fluctuations in the relative levels of HCF-1 and HCF-2 could provide a mode of regulation.
In summary, our results indicate that HSVs have evolved a mechanism to preferentially target one member of an emerging family of HCF proteins. This association may be very significant for the viral life cycle and is strongly reminiscent of the preferential recruitment of Oct-1 to the VP16-induced complex rather than the closely related Oct-2 protein (8, 18, 33). Discrimination is achieved in the case of Oct-1 through recognition of a single glutamic acid residue on the solvent-exposed surface of the Oct-1 homeodomain (22, 27). VP16 plays a key role in launching the lytic cycle during natural infections (2, 31), and a lack of VP16 function may act as a signal for the virus to establish a latent or quiescent infection (16). HCF-1 may be the favored target because of its role in promoting G1 progression. Many DNA viruses require G1- or S-phase-specific components of the host cell for their replication (15), and although HSV can infect resting cells, there is growing evidence that cellular G1- and S-phase functions are induced early in infection (13, 14, 29, 38). HCF-1 activity may be required for the synthesis of these essential G1/S factors.
We favor the view that the VP16-induced complex functions as both a sensor and a switch, first gauging the physiological status of the infected cell and then, through activation of the viral IE genes, selecting the lytic pathway. This hypothesis is now strengthened by the fact that VP16 selectively recruits not one but two members of multiprotein families, HCF-1 and Oct-1, both of which have been implicated in the control of cell proliferation. The combined presence of functional HCF-1 and Oct-1 proteins may indicate to the virus that the cellular environment is favorable for lytic growth.
ACKNOWLEDGMENTS
We thank Stavros Giannakopoulos for help with Northern blotting and Michael Garabedian and Muktar Mahajan for reagents and assistance with the yeast assays. Thanks go to Rich Freiman, Michael Garabedian, David Ron, Bob Schneider, and Naoko Tanese for discussion and insightful comments on the manuscript.
This work was supported by funds from the Department of Microbiology at NYU School of Medicine, a development award from the Kaplan Comprehensive Cancer Center (R000), and an institutional award from the American Cancer Society (IRG-14-39).
REFERENCES
- 1.Abel T, Bhatt R, Maniatis T. A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev. 1992;6:466–480. doi: 10.1101/gad.6.3.466. [DOI] [PubMed] [Google Scholar]
- 2.Ace C I, McKee T A, Ryan J M, Cameron J M, Preston C M. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989;63:2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.apRhys C M, Ciufo D M, O’Neill E A, Kelly T J, Hayward G S. Overlapping octamer and TAATGARAT motifs in the VF65-response elements in herpes simplex virus immediate-early promoters represent independent binding sites for the cellular nuclear factor III. J Virol. 1989;63:2798–2812. doi: 10.1128/jvi.63.6.2798-2812.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Boutros, M., A. Wilson, and W. Herr. Unpublished data.
- 4.de Rooij D G. Stem cells in the testis. Int J Exp Pathol. 1998;79:67–80. doi: 10.1046/j.1365-2613.1998.t01-1-00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frattini A, Chatterjee A, Faranda S, Sacco M G, Villa A, Herman G E, Vezzoni P. The chromosomal localization and the HCF repeats of human host cell factor (HCFC1) are conserved in the mouse homologue. Genomics. 1996;32:277–280. doi: 10.1006/geno.1996.0116. [DOI] [PubMed] [Google Scholar]
- 6.Frattini A, Faranda S, Redolfi E, Zucchi I, Villa A, Patrosso M C, Strina D, Susani L, Vezzoni P. Genomic organization of the human VP16 accessory protein, a house keeping gene (HCFC1) mapping to Xq28. Genomics. 1994;23:30–35. doi: 10.1006/geno.1994.1455. [DOI] [PubMed] [Google Scholar]
- 7.Freiman R N, Herr W. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerster T, Roeder R G. A herpesvirus transactivator protein interacts with transcription factor OTF-1 and other cellular proteins. Proc Natl Acad Sci USA. 1988;85:6347–6351. doi: 10.1073/pnas.85.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goding C R, O’Hare P. Herpes simplex virus Vmw65-octamer binding protein interaction: a paradigm for combinatorial control of transcription. Virology. 1989;173:363–367. doi: 10.1016/0042-6822(89)90548-5. [DOI] [PubMed] [Google Scholar]
- 10.Goto H, Motomura S, Wilson A C, Freiman R N, Nakebeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–37. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 11.Gromoll J, Wessels J, Rosiepen G, Brinkworth M H, Weinbauer G F. Expression of mitotic cyclin B1 is not confined to proliferating cells in the rat testis. Biol Reprod. 1997;57:1312–1319. doi: 10.1095/biolreprod57.6.1312. [DOI] [PubMed] [Google Scholar]
- 12.Haigh A, Greaves R, O’Hare P. Interference with the assembly of a virus-host transcription complex by peptide competition. Nature. 1990;344:257–259. doi: 10.1038/344257a0. [DOI] [PubMed] [Google Scholar]
- 13.Hilton M J, Mounghane D, McLean T, Contractor N V, O’Neil J, Carpenter K, Bachenheimer S L. Induction by herpes simplex virus of free and heteromeric forms of E2F transcription factor. Virology. 1995;213:624–638. doi: 10.1006/viro.1995.0034. [DOI] [PubMed] [Google Scholar]
- 14.Hossain A, Holt T, Ciacci-Zanella J, Jones C. Analysis of cyclin-dependent kinase activity after herpes simplex virus type 2 infection. J Gen Virol. 1997;78:3341–3348. doi: 10.1099/0022-1317-78-12-3341. [DOI] [PubMed] [Google Scholar]
- 15.Knipe D M. Virus-host cell interactions. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 273–299. [Google Scholar]
- 16.Kosz-Vnenchak M, Jacobson J, Coen D M, Knipe D M. Evidence for a novel regulatory pathway for herpes simplex virus gene expression in trigeminal ganglion neurons. J Virol. 1993;67:5383–5393. doi: 10.1128/jvi.67.9.5383-5393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kristie T M. The mouse homologue of the human transcription factor C1 (host cell factor). Conservation of forms and function. J Biol Chem. 1997;272:26749–26755. doi: 10.1074/jbc.272.42.26749. [DOI] [PubMed] [Google Scholar]
- 18.Kristie T M, LeBowitz J H, Sharp P A. The octamer-binding proteins form multi-protein-DNA complexes with the HSV αTIF regulatory protein. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 20.Kristie T M, Sharp P A. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus α-trans-induction factor (VP16) J Biol Chem. 1993;268:6525–6534. [PubMed] [Google Scholar]
- 21.LaBoissiere S, Walker S, O’Hare P. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol Cell Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai J-S, Cleary M A, Herr W. A single amino acid exchange transfers VP16-induced positive control from Oct-1 to the Oct-2 homeo domain. Genes Dev. 1992;6:2058–2065. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- 23.Lu R, Yang P, O’Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cell factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu R, Yang P, Padmakumar S, Misra V. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, Luman, in its interaction with HCF. J Virol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller H, Moroni M C, Vigo E, Petersen B O, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 27.Pomerantz J L, Kristie T M, Sharp P A. Recognition of the surface of a homeo domain protein. Genes Dev. 1992;6:2047–2057. doi: 10.1101/gad.6.11.2047. [DOI] [PubMed] [Google Scholar]
- 28.Sadowski I, Ma J, Triezenberg S J, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 29.Schang L M, Phillips J, Schaffer P A. Requirement for cellular cyclin-dependent kinases in herpes simplex virus replication and transcription. J Virol. 1998;72:5626–5637. doi: 10.1128/jvi.72.7.5626-5637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simmen K A, Newell A, Robinson M, Mills J S, Canning G, Handa R, Parkes K, Borkakoti N, Jupp R. Protein interactions in the herpes simplex type 1 VP16-induced complex: VP16 peptide inhibition and mutational analysis of the host cell factor requirements. J Virol. 1997;71:3886–3894. doi: 10.1128/jvi.71.5.3886-3894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smiley J R, Duncan J. 1814 linker insertion mutant. J. Virol. 71:6191–6193. 1997. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 produces a phenotype similar to that of the. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smolik S M, Rose R E, Goodman R H. A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol Cell Biol. 1992;12:4123–4131. doi: 10.1128/mcb.12.9.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stern S, Tanaka M, Herr W. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989;341:624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 35.Thompson C C, McKnight S L. Anatomy of an enhancer. Trends Genet. 1992;8:232–236. [Google Scholar]
- 36.Triezenberg S J, Kingsbury R C, McKnight S L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 37.Verona R, Moberg K, Estes S, Starz M, Vernon J P, Lees J A. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilcock D, Lane D P. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature. 1991;349:429–431. doi: 10.1038/349429a0. [DOI] [PubMed] [Google Scholar]
- 39.Wilson A C, Cleary M A, Lai J-S, LaMarco K, Peterson M G, Herr W. Combinatorial control of transcription: the herpes simplex virus VP16-induced complex. Cold Spring Harbor Symp Quant Biol. 1993;58:167–178. doi: 10.1101/sqb.1993.058.01.021. [DOI] [PubMed] [Google Scholar]
- 40.Wilson A C, Freiman R N, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell-cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson A C, LaMarco K, Peterson M G, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 42.Wilson A C, Parrish J E, Massa H F, Nelson D L, Trask B J, Herr W. The gene encoding the VP16-accessory protein HCF (HCFC1) resides in human Xq28 and is highly expressed in fetal tissues and the adult kidney. Genomics. 1995;25:462–468. doi: 10.1016/0888-7543(95)80046-o. [DOI] [PubMed] [Google Scholar]
- 43.Wilson A C, Peterson M G, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 44.Wu T J, Monokian G, Mark D F, Wobbe C R. Transcriptional activation by herpes simplex virus type 1 VP16 in vitro and its inhibition by oligopeptides. Mol Cell Biol. 1994;14:3484–3493. doi: 10.1128/mcb.14.5.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]