Abstract
Simple Summary
Breast cancer (BC) is the major cause of cancer-related deaths in women worldwide. In addition to genetic diagnostics for variants in high-risk genes, there is a need for better risk stratification to target high-risk individuals. The polygenic risk score (PRS) has emerged as a valuable addition to help sorting women into different risk categories for BC development. This study aimed to evaluate the impact of adding a PRS, based on 313 genetic variants, to standard genetic testing for 382 German women with BC or a family history of the disease. By incorporating the PRS into risk prediction models, meaningful changes in 10-year risks were observed in 13.6% of individuals. Additionally, the inclusion of the PRS led to clinically significant changes in prevention recommendations for 12.0% of cases, supporting the use of the PRS for BC risk assessment in genetic counselling.
Abstract
Single nucleotide polymorphisms are currently not considered in breast cancer (BC) risk predictions used in daily practice of genetic counselling and clinical management of familial BC in Germany. This study aimed to assess the clinical value of incorporating a 313-variant-based polygenic risk score (PRS) into BC risk calculations in a cohort of German women with suspected hereditary breast and ovarian cancer syndrome (HBOC). Data from 382 individuals seeking counselling for HBOC were analysed. Risk calculations were performed using the Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm with and without the inclusion of the PRS. Changes in risk predictions and their impact on clinical management were evaluated. The PRS led to changes in risk stratification based on 10-year risk calculations in 13.6% of individuals. Furthermore, the inclusion of the PRS in BC risk predictions resulted in clinically significant changes in 12.0% of cases, impacting the prevention recommendations established by the German Consortium for Hereditary Breast and Ovarian Cancer. These findings support the implementation of the PRS in genetic counselling for personalized BC risk assessment.
Keywords: breast cancer, risk prediction, polygenic risk score, genetic counselling, hereditary breast and ovarian cancer syndrome
1. Introduction
Breast Cancer (BC) is the most prevalent form of cancer in women and the leading cause of cancer-related death among women worldwide [1]. To facilitate early detection of BC, improve chances of recovery, and decrease mortality rates, screening programs have been established [2,3]. However, current screening strategies, such as clinical examinations and mammograms, are vulnerable to overdiagnosis and overtreatment [4,5].
Personalized risk estimations of BC could help to improve prevention and screening programs by identifying women in risk categories who are most likely to benefit [6].
A positive family history is one of the primary risk factors for developing the disease [7]. Approximately 30% of all women with BC in Germany show a positive family history and fulfil the inclusion criteria of the German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC), making them eligible for extensive screening measures [3,8].
The genetic susceptibility to BC is determined by rare high-penetrance variants in BRCA1, BRCA2, PALB2, and TP53 [9,10,11], likewise rare moderate-risk variants (e.g., in ATM, CHEK2, and RAD51C) [12], as well as common low-risk variants, predominantly single nucleotide polymorphisms (SNPs) [13,14]. According to genome-wide association studies, each individual SNP can either slightly increase or decrease the risk of developing BC. When summarizing all SNPs in a polygenic risk score (PRS), the cumulative risk can be substantial [15,16]. Thus, the PRS can aid in stratifying women into different risk categories of developing BC. Moreover, the BC risk of women carrying a pathogenic variant (PV) in ATM, BRCA1/2, CHEK2, or PALB2 can also be refined using the PRS [17,18].
Belgium, France, Israel, Italy, the United Kingdom, and Spain are comparing personalized, risk-stratified BC screening to standard screening as part of the ongoing international MyPeBS study [19]. In Germany, however, the use of the PRS for risk prediction has not yet been incorporated into the daily practice of genetic counselling for familial BC.
The risk prediction is currently estimated by risk prediction algorithms [20] based on family history such as the Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm (BOADICEA) [21], which has been proven to identify women at high risk more accurately than other models [22] and has been prospectively validated [23,24]. Multiple studies presented that by combining the PRS with other known risk factors for BC risk stratification, discriminatory power between BC cases and controls can be improved [25,26,27,28].
Risk prediction tools such as the CanRisk [29] webtool using BOADICEA and a PRS based on 313 variants have recently been made accessible for healthcare professionals [30]. BOADICEA has also been updated to incorporate alternative PRSs and can be readily adapted to different PRSs in a manner that maintains consistency of the model [31].
In the present work, we examined the clinical value of the PRS for BC risk prediction based on personal medical history, family history, and PV carrier status in a cohort of 382 German women with suspected hereditary breast and ovarian cancer syndrome (HBOC).
We examined the extent to which the inclusion of the PRS leads to a relevant change in BC risk predictions. The impact of the PRS on BC risk prediction was then assessed by determining the possible change in clinical management, as stated in the guideline by the GC-HBOC. The guideline defines a specific group of women who are entitled to enrol in an intensified BC surveillance program if their calculated 10-year risk of developing BC is above 5% [8].
2. Materials and Methods
2.1. Study Cohort
We collected data from women from families with suspected HBOC and from families in which a PV in the causative genes for HBOC is already known from December 2020 to January 2023. All participants sought counselling for HBOC at the Institute of Human Genetics of the University of Leipzig, Germany, which is part of the Centre for Hereditary Breast and Ovarian Cancer Leipzig. A total of 382 women met the following eligibility criteria for this study:
-
(1)
Availability of genotyping data;
-
(2)
Availability of personal medical information (e.g., cancer diagnosis);
-
(3)
Available information on family history (e.g., family members with a cancer diagnosis);
-
(4)
Age range of 18 to 69 years;
-
(5)
The presence of a PV for which a risk calculation using CanRisk was possible in case of a positive carrier status.
Women with BC or OC were included if they met additional criteria:
-
(1)
Available information on tumour pathology;
-
(2)
Available clinical data such as age of first onset of disease;
-
(3)
Unilaterality in women with BC.
For individuals with unilateral BC, the risk of developing a contralateral tumour was calculated. Women with bilateral BC, ductal carcinoma in situ (DCIS), or pancreatic cancer could not be considered as they did not meet the requirements for BOADICEA computation [32]. All participants provided informed consent for their data to be used for research purposes. All individuals were anonymized.
2.2. Pedigree Collection and Initial Risk Assessment
The pedigrees included in this work were generated using the PhenoTips webtool [33] during the initial consultation. All participants’ pedigrees include at least three generations. The following data were available for each individual in the pedigrees:
-
(1)
Life status;
-
(2)
Year of birth and current age;
-
(3)
Cancer status.
The age of first onset of disease was known for all index patients.
All participants underwent an additional risk assessment using the checklist of The German Cancer Society [34,35]. The checklist is based on associated cases of HBOC in the maternal and paternal line, which are included with either single, double, or triple weighting while also considering the age of first onset of disease and the hormone receptor status in BC cases. Individuals with a score of ≥3 are considered to have a risk of carrying a PV of at least 10% and are thus eligible for genetic testing [36,37,38].
2.3. Molecular Genetics
All women from families with suspected HBOC underwent panel diagnostics including PRS testing. Some of the women, in whom a PV was known in the family, received the targeted testing within the framework of panel diagnostics including PRS testing, the rest received targeted testing by means of Sanger sequencing and additional PRS testing.
Genomic DNA was extracted from whole blood using MagCore Kit 101 and MagCore® instrument (RBC Bioscience, New Taipei City, Taiwan). DNA concentration was measured using NanoDrop™ 2000 (Thermo Scientific™, Waltham, MA, USA) and Qubit (Thermo Scientific™,Waltham, MA, USA). Next generation sequencing (NGS) after sample preparation using Twist Library Preparation EF Kit Twist Library Preparation EF Kit1, 2.0, and Twist Universal Adapter System—TruSeq Compatible, 96 Samples Plate A-D, enrichment using Twist Custom Panel, design name: Cancer_PRS_HUGV6; Twist Design ID: TE-96674869 (Twist Bioscience, South San Francisco, CA, USA), and sample identification using the Nimagen RC-PCR assay. Sequencing was conducted on a NextSeq500/550 Mid Output v2.5 kit (Illumina, San Diego, CA, USA; Sequencer: Illumina NextSeq550 (Illumina, San Diego, CA, USA). Mean coverage was at least 300×, and all target regions were covered 20×. Single nucleotide variants (SNVs) as well as copy number variants (CNVs) were detected within this setting. This analysis can be established in all labs familiar with NGS settings.
2.4. Variant Classification and Panel Sequencing
Analysis of the raw data was performed using the software Varfeed (Limbus, Rostock, Germany), and the variants (SNVs and CNVs) were annotated using the software Varvis (Limbus, Rostock, Germany). All variants were described in regard to GRCh37 (NM_000492.4) and classified according to the latest ACMG criteria [39]. The databases ClinVar [40], HGMD [41], and HerediCare [42] were used for classification based on the following considerations: gene and variant attributes, frequency in the general population (gnomAD [43]), (assumed) effect on protein function, in silico prediction tools (mainly CADD [44], SpliceAI-lookup [45]), conservation, and phenotype.
2.5. Targeted Sequencing for PRS Calculation
Raw reads were quality checked using fastqc [46], and remaining adapter sequences and bad quality data were removed using trimmomatic [47]. Processed data was aligned to hg19 using minimap2 [48], visual duplicates were marked with samtools [49]. Haplotypes for the BCAC-313 PRS model were called using freebayes, and positions with a coverage of <20× or conflicting haplotype signal were imputed using twice the allele frequency described in the BCAC-313 model [50]. The resulting haplotypes were used as an input to calculate the normalized z-score via the CanRisk API [29]. Resulting z-scores were combined with the related BOADICEA file to calculate the 5-year, 10-year, and lifetime risk for each participant.
2.6. Statistical Testing
The PRS values of the study cohort were tested for normal distribution using the Shapiro–Wilk test from the scipy package [51].
The study cohort was compared to the BCAC-313 model [52] cohort using the mean (mu) and standard deviation (sigma) provided by CanRisk. PRS distribution of the BCAC-313 cohort was modelled using the numpy package, and the comparison between the cohorts was conducted using the individual t-test from the scipy package (Supplementary Figure S1).
To test the effect of the PRS value on the 10-year risk calculation, the study cohort was filtered for women under the age of 50 without any known PVs in the 11 core genes, the cohort was divided in to high and low PRS individuals by comparison to the mean PRS value described for the BCAC-313 model. The effect of including or excluding the PRS value in the 10-year BC risk was calculated using the Mann–Whitney U test from the scipy package (Supplementary Figure S2).
3. Results
We included 382 women in our cohort. The mean age at time of evaluation was 45 years with an age range between 18 and 69 years. A total of 233 women (60.9%) were aged 18-49 years and 149 (39.1%) were aged 50 and older. At the time of analysis, 48.7% of women (186) had an invasive breast tumour, 7.9% (30) had ovarian cancer (OC), and 0.5% (2) were affected by both BC and OC. The average age at diagnosis for women affected by BC was 46 years with a range between 27 and 67 years. For OC cases, the mean age of diagnosis was 54 with a range between 22 and 67 years.
A total of 273 participants (71.5%) were evaluated for PVs in the 11 core genes ATM, BARD1, BRCA1, BRCA2, CDH1, CHEK2, BRIP1, PALB2, RAD51C, RAD51D, and TP53 via a multigene panel diagnostic [53]. A total of 109 women (28.5%) received targeted genetic testing via Sanger sequencing.
A total of 206 of 218 cases affected by cancer received a multigene panel diagnostic, revealing 39 carriers of a PV (35.8% of all PV carriers). Twelve cancer patients received targeted genetic testing, leading to detection of a further nine PV carriers (8.3% of all PV carriers). Among the 164 healthy participants, multigene panel analysis for 67 of them (40.8%) revealed 17 carriers of a PV (15.6% of all PV carriers), while targeted genetic testing in the further 97 individuals identified 44 additional PV carriers (40.3% of all PV carriers) (Table 1).
Table 1.
Characteristics of cohort; BC=breast cancer, OC=ovarian cancer, and PV=pathogenic variant.
| Individuals | |
|---|---|
| n | 382 |
| Age | |
| Mean | 45 |
| Range | 18–69 |
| Healthy | 164 |
| Affected | 218 |
| Affected by BC | 186 |
| Affected by OC | 30 |
| Affected by BC + OC | 2 |
| Age of onset of disease BC | |
| Mean | 46 |
| Range | 27–67 |
| Age of onset of disease OC | |
| Mean | 54 |
| Range | 22–67 |
| Array | |
| Multigene panel | 273 |
| Performed for healthy probands | 67 |
| Detected PVs | 17 |
| Performed for affected probands | 206 |
| Detected PVs | 39 |
| Sanger + PRS only | 109 |
| Performed for healthy probands | 97 |
| Detected PVs | 44 |
| Performed for affected probands | 12 |
| Detected PVs | 9 |
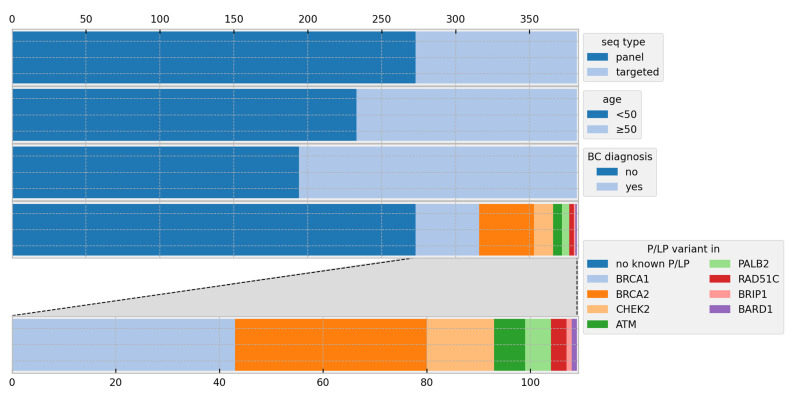
Most participants were carriers of PVs in either BRCA1 (11.3% of participants) or BRCA2 (9.7%). Other PVs were found in ATM (1.6%), BARD1 (0.3%), BRIP1 (0.3%), CHEK2 (3.4%), PALB2 (1.3%), and RAD51C (0.8%). (Figure 1)
Figure 1.
Cohort overview showing the proportions of participants regarding the sequencing methods, age distribution, breast cancer diagnosis, and pathogenic variants; BC = breast cancer, P/LP = pathogenic and likely pathogenic, and seq = sequencing.
3.1. Risk Calculations
The PRS (z-score) in our cohort is normally distributed with a mean of 0.45 (SD = 1.02, Shapiro–Wilk test p-value = 0.79) (Supplementary Figure S1).
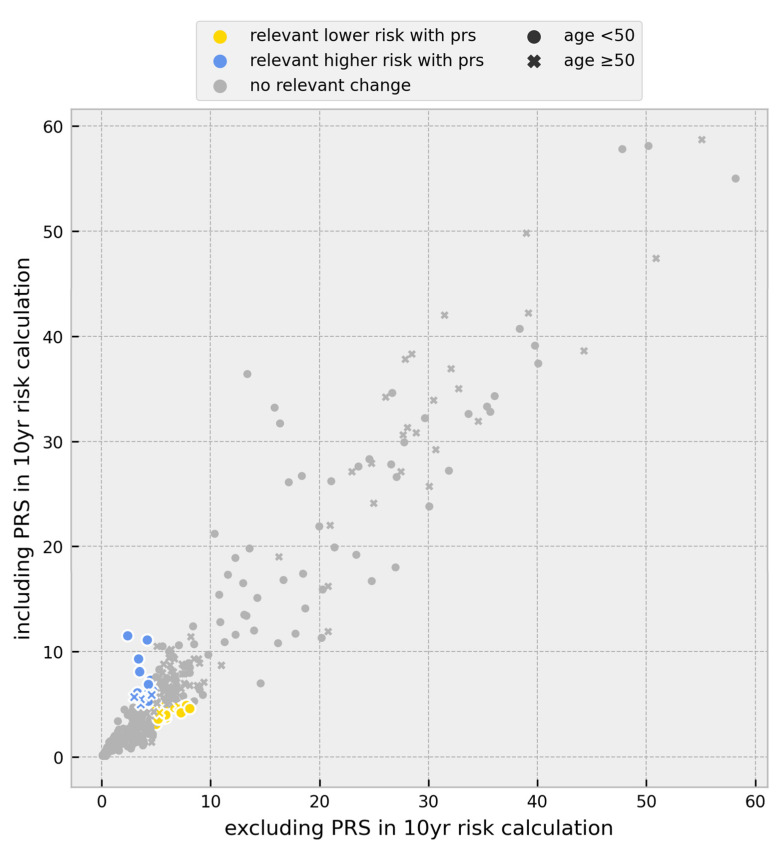
Through the inclusion of the PRS, the threshold of 5% in 10-year risk calculations was observed to be either surpassed or subordinated in 13.6% of all individuals. Among them, 8.1% surpassed the 5% threshold, defining them as women at high risk of developing BC according to the GC-HBOC. The basis for establishing the threshold is that the 10-year risk of ≥5% is approximately double the value of a 50-year-old woman from the general population [8]. A total of 5.5% fell below the 5% threshold, indicating a relevantly lower risk (Figure 2).
Figure 2.
Scatter plot of the change in individual breast cancer 10-year risk after inclusion of the PRS for all probands. The initial breast cancer 10-year risk based on age, pedigree information, and PV status was plotted against breast cancer 10-year risk including the 313-variant PRS in addition to initial breast cancer 10-year risk. Individuals surpassing the 5% threshold after PRS inclusion are shown as blue symbols. Individuals falling below the 5% threshold after PRS inclusion are shown as yellow symbols; PRS = polygenic risk score, and PV = pathogenic variant.
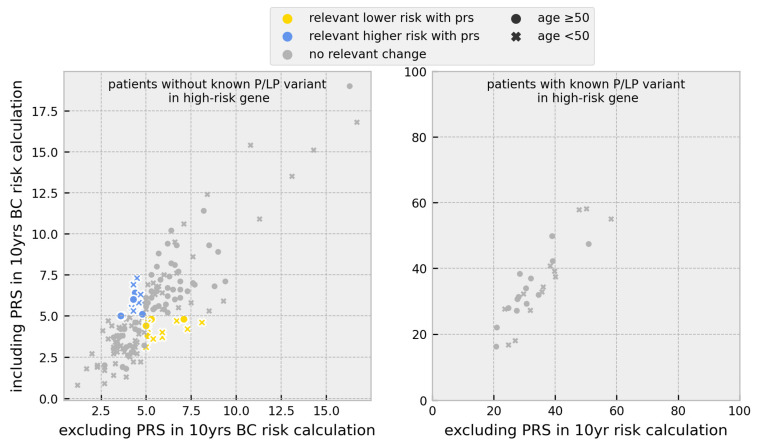
For 153 BC cases without any PVs, the inclusion of the PRS in 10-year risk calculations led to different risk stratification for 20.3%, with 11.1% surpassing the 5% threshold and 9.2% falling below. A total of 52.9% of cases exceeding the 5% threshold were under the age of 50.
No changes in risk stratification after including the PRS were observed in the 27 BC cases with PVs in high-risk genes (BRCA1, BRCA2, and PALB2) (Figure 3).
Figure 3.
Scatter plot of the change in individual breast cancer 10-year risk after inclusion of the PRS for all BC cases separated by PV carrier status; BC = breast cancer, P/LP = pathogenic and likely pathogenic, PRS = polygenic risk score, and PV = pathogenic variant.
The effect of including or excluding the PRS value in the 10-year BC risk calculation showed that individuals with lower PRS values than the mean described for the BCAC-313 model exhibit a significantly lower 10-year risk after incorporating the PRS into the analysis (p-value = 0.0011). In contrast, women with PRS values above the mean PRS value do not show a significantly higher 10-year risk (p-value = 0.31). However, it is important to note that for a specific individual, a higher PRS can still translate to a higher 10-year risk (Supplementary Figure S2).
3.2. Change in Prevention Management
The GC-HBOC considers risk calculations of the following groups for inclusion in the intensified breast cancer surveillance program [8,54,55]:
-
(1)
Healthy women under the age of 50 with unremarkable predictive multigene panel diagnostic results;
-
(2)
Healthy women under the age of 50 with unremarkable targeted genetic testing in moderate-risk genes;
-
(3)
Ovarian cancer patients under the age of 50 with unremarkable multigene panel diagnostic results;
-
(4)
Relatives of index patients with a PV detected by the multigene panel diagnostic. We did not include this group in our analyses.
If individuals included in one of these groups have a 10-year risk of ≥5% for developing BC, they are eligible for intensified screening to timely detect a potential breast tumour.
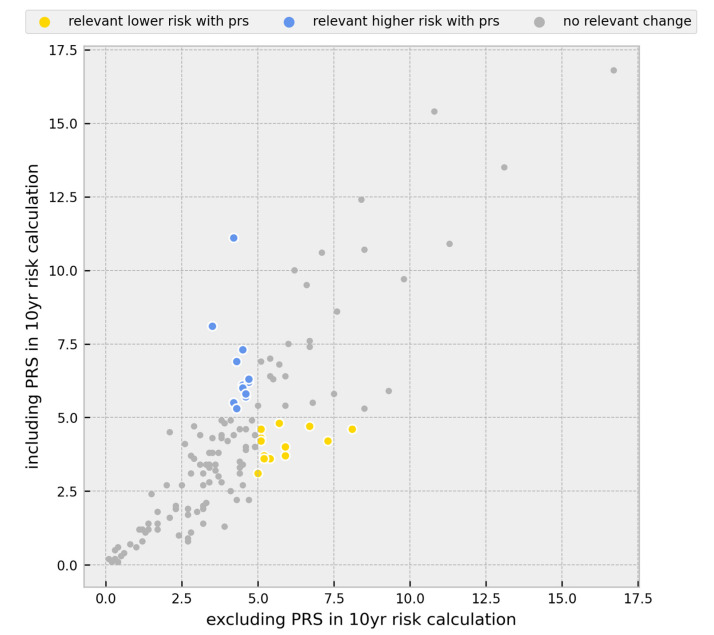
A total of 30 participants in our cohort met the characteristics of the first group. By including the PRS in 10-year risk estimations, 16.7% of these participants either exceeded or fell below the 5% threshold, indicating a change in clinical management. There are 49 individuals that can be attributed to the second group with relevant changes in 10-year risk predictions observed in 10.2% of cases. For the four women attributed to the third group, there were no changes. Overall, 12.0% of participants that can be assigned to one of the groups would require a change in prevention management (Figure 4).
Figure 4.
Scatter plot of the change in individual breast cancer 10-year risk after inclusion of the PRS for healthy women age < 50 with unremarkable predictive multigene panel diagnostic results, healthy women age < 50 with unremarkable targeted genetic testing in moderate-risk genes, and ovarian cancer patients age < 50 with unremarkable multigene panel diagnostic results. Individuals surpassing the 5%-risk threshold and consequent changes in clinical management are shown as blue dots. Individuals falling below the 5%-risk threshold and consequent changes in clinical management are shown as yellow dots; BC = breast cancer, and PRS = polygenic risk score.
4. Discussion
In this study, we demonstrated the impact of the 313-variant-based PRS on BOADICEA-based BC risk calculations in a cohort of 382 women, resulting in changes of BC risk stratification in 13.6% of all participants. These variations in risk assessment might have significant implications, e.g., for making informed decisions regarding preventive surgeries.
The majority of BC patients without a PV who have exceeded the 5% threshold after including the PRS in 10-year risk calculations are younger than 50 years (52.9%), indicating that this particular group would benefit from a more precise risk assessment due to the incorporation of the PRS. The association between PRS and BC risk decreasing with age was shown by Mavaddat et al. [13].
This is the first study conducted on the clinical application of the PRS in Germany. In a Dutch study, the impact of incorporating the PRS into risk calculations for 1331 non-BRCA1/2 carriers was investigated regarding screening procedures aligned with Dutch IKNL [56], UK NICE [57], and US NCCN [58] BC screening guidelines. The results revealed clinically significant shifts in 32.4%, 36.0%, and 25.7% of individuals (with 30% BC lifetime risk cut-off levels based on the IKNL and NICE BC screening guidelines) [59].
In our cohort, the inclusion of the PRS in BC risk predictions resulted in clinically relevant shifts leading to changes in prevention recommendations established by the GC-HBOC in 12.0% of participants that can be assigned to one of the groups defined by the consortium. This presents a different patient stratification from current clinical practice, in which solely family history is included in risk prediction.
The current susceptibility SNPs account for around 44% of the familial relative risk associated with common low-risk variants [60]. Recent genome-wide association studies have discovered new BC susceptibility loci [61,62], leaving a more extensive PRS to be expected in the future. By incorporating an expanded PRS in BC risk prediction, more accurate risk stratification can be achieved, resulting in a higher percentage of women transitioning to different risk categories, and leading to improved screening measures.
Based on the considerable utility of the PRS, a potential application beyond HBOC families as part of general cancer screening should be discussed. A meta study was conducted to assess the cost effectiveness of implementing the PRS for three prevalent cancer types (prostate, colorectal, and breast cancer). Out of the ten studies analysed, eight demonstrated cost effectiveness in the utilization of the PRS [63]. However, further prospective studies with a larger cohort and case control studies are needed to quantify the effect of incorporation of the PRS in general screening among the general population programs for the health care system.
The strengths of this study include good representation of families that are seen in genetic counselling. All participants provided thorough family and personal medical history, ensuring comprehensive data collection. The utilization of BOADICEA as a well-validated and comprehensive risk model, allows for accurate risk predictions in a familial setting [22].
Limitations
We had to assume the European ancestry based on the participants’ names. So far, the PRS is only validated for people of European ancestry [21], though some studies have indicated associations between a subset of the PRS and BC in Asian [64] and Latina/x/o [65] populations.
5. Conclusions
In summary, our findings support implementation of the PRS in genetic counselling, although it might present logistical challenges. By utilizing a reliable and comprehensive risk prediction model such as BOADICEA, pedigree-based family history, individual PRS, and molecular genetic analysis results can be combined easily, enabling the calculation of personalized BC risks, and therefore allowing for an improved clinical management in BC prevention.
Acknowledgments
We thank the teams of the Molecular Genetics Diagnostics Laboratory and of the Genetic Counselling Center of the Institute of Human Genetics as well as all participants and their family members. We also thank the German Consortium for Hereditary Breast and Ovarian Cancer.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15153938/s1, Figure S1: The distribution of the PRS in our cohort and that of the UK cohort. The PRS is normally distributed; PRS = polygenic risk score, Figure S2: Scatter plot of the change in individual breast cancer 10-year risk after inclusion of the PRS for all BC cases under the age of 50. Individuals are separated into high and low PRS values compared to the mean PRS value described for the BCAC-313 model; BC = breast cancer, BCAC = Breast Cancer Association Consortium, PRS = polygenic risk score; PV = pathogenic variant
Author Contributions
Conceptualization, S.S., J.H. and V.S.; Methodology, S.S., J.H., K.L. and S.D.; Formal Analysis, S.S. and S.D.; Data Curation, S.S.; Writing—Original Draft Preparation, S.S. and V.S.; Writing—Review and Editing, J.H. and S.D.; Visualization, S.D.; Supervision, J.H. and V.S.; Project Administration, J.H. and V.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of University of Leipzig (reference number 425-20-lk, 13 October 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Quante A.S., Strahwald B., Fischer C., Kiechle M. Kiechle Individualized risk of breast cancer—How should it be calculated, evaluated and discussed? Gynakologe. 2018;51:397–402. doi: 10.1007/s00129-018-4240-6. [DOI] [Google Scholar]
- 3.Deutsche Krebsgesellschaft. Deutsche Krebshilfe, and AWMF S3-Leitlinie Früherkennung, Diagnose, Therapie und Nachsorge des Mammakarzinoms, Version 4.4, Juni 2021. AWMF Registernummer: 032-045OL. 2021. [(accessed on 17 January 2023)]. Available online: https://www.leitlinienprogramm-onkologie.de/leitlinien/mammakarzinom/
- 4.Marmot M., Altman D.G., Cameron D.A., Dewar, Thompson S.G., Wilcox M. The benefits and harms of breast cancer screening: An independent review. Lancet. 2012;380:1778–1786. doi: 10.1016/S0140-6736(12)61611-0. [DOI] [PubMed] [Google Scholar]
- 5.Bleyer A., Welch H.G. Effect of Three Decades of Screening Mammography on Breast-Cancer Incidence. N. Engl. J. Med. 2012;367:1998–2005. doi: 10.1056/NEJMoa1206809. [DOI] [PubMed] [Google Scholar]
- 6.Mavaddat N., Michailidou K., Dennis J., Lush M., Fachal L., Lee A., Tyrer J.P., Chen T.-H., Wang Q., Bolla M.K., et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019;104:21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sokolova A., Johnstone K.J., McCart Reed A.E., Simpson P.T., Lakhani S.R. Histopathology. John Wiley and Sons Inc.; Hoboken, NJ, USA: 2022. Hereditary breast cancer: Syndromes, tumour pathology and molecular testing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quante A.S., Engel C., Kiechle M., Schmutzler R.K., Fischer C. Changes in risk calculation for the intensified surveillance programme of the German Consortium for Breast and Ovarian Cancer. Gynakologe. 2020;53:259–264. doi: 10.1007/s00129-020-04572-9. [DOI] [Google Scholar]
- 9.Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.-A., Mooij T.M., Roos-Blom M.-J., Jervis S., Van Leeuwen F.E., Milne R.L., Andrieu N., et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA—J. Am. Med. Assoc. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 10.Schon K., Tischkowitz M. Clinical implications of germline mutations in breast cancer: TP53. Breast Cancer Res. Treat. 2018;167:417–423. doi: 10.1007/s10549-017-4531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoniou A.C., Casadei S., Heikkinen T., Barrowdale D., Pylkäs K., Roberts J., Lee A., Subramanian D., De Leeneer K., Fostira F., et al. Breast-Cancer Risk in Families with Mutations in PALB2. N. Engl. J. Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Easton D.F., Pharoah P.D., Antoniou A.C., Tischkowitz M., Tavtigian S.V., Nathanson K.L., Devilee P., Meindl A., Couch F.J., Southey M., et al. Gene-Panel Sequencing and the Prediction of Breast-Cancer Risk. N. Engl. J. Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mavaddat N., Pharoah P.D.P., Michailidou K., Tyrer J., Brook M.N., Bolla M.K., Wang Q., Dennis J., Dunning A.M., Shah M., et al. Prediction of breast cancer risk based on profiling with common genetic variants. J. Natl. Cancer Inst. 2015;107:djv036. doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vachon C.M., Pankratz V.S., Scott C.G., Haeberle L., Ziv E., Jensen M.R., Brandt K.R., Whaley D.H., Olson J.E., Heusinger K., et al. The contributions of breast density and common genetic variation to breast cancer risk. J. Natl. Cancer Inst. 2015;107:dju397. doi: 10.1093/jnci/dju397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bahcall O.G. ICOGS collection provides a collaborative model. Nat. Genet. 2013;45:343. doi: 10.1038/ng.2592. [DOI] [PubMed] [Google Scholar]
- 16.Hall P., Easton D. Breast cancer screening: Time to target women at risk. Br. J. Cancer. 2013;108:2202–2204. doi: 10.1038/bjc.2013.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuchenbaecker K.B., McGuffog L., Barrowdale D., Lee A., Soucy P., Dennis J., Domchek S.M., Robson M., Spurdle A.B., Ramus S.J., et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J. Natl. Cancer Inst. 2017;109:djw302. doi: 10.1093/jnci/djw302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher S., Hughes E., Wagner S., Tshiaba P., Rosenthal E., Roa B.B., Kurian A.W., Domchek S.M., Garber J., Lancaster J., et al. Association of a Polygenic Risk Score with Breast Cancer among Women Carriers of High- And Moderate-Risk Breast Cancer Genes. JAMA Netw. Open. 2020;3:e208501. doi: 10.1001/jamanetworkopen.2020.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rouge-Bugat M.-E., Balleyguier C., Laurent N., Dautreppe A., Maillet L., Simon P., Fournet P., Scellier C., Menini T., Darmon E., et al. MyPeBS International randomized study comparing personalised, risk-stratified to standard breast cancer screening in women aged 40–70: Focus on recruitment strategy in France. Presse Médicale Open. 2022;3:100022. doi: 10.1016/j.lpmope.2022.100022. [DOI] [Google Scholar]
- 20.Louro J., Posso M., Boon M.H., Román M., Domingo L., Castells X., Sala M. A systematic review and quality assessment of individualised breast cancer risk prediction models. Br. J. Cancer. 2019;121:76–85. doi: 10.1038/s41416-019-0476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee A., Mavaddat N., Wilcox A.N., Msc A.P.C., Carver T., Hartley S., de Villiers C.B., Izquierdo A., Simard J., Schmidt M.K., et al. BOADICEA: A comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet. Med. 2019;21:1708–1718. doi: 10.1038/s41436-018-0406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhury P.P., Brook M.N., Hurson A.N., Lee A., Mulder C.V., Coulson P., Schoemaker M.J., Jones M.E., Swerdlow A.J., Chatterjee N., et al. Comparative validation of the BOADICEA and Tyrer-Cuzick breast cancer risk models incorporating classical risk factors and polygenic risk in a population-based prospective cohort of women of European ancestry. Breast Cancer Res. 2021;23:22. doi: 10.1186/s13058-021-01399-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X., Eriksson M., Czene K., Lee A., Leslie G., Lush M., Wang J., Dennis J., Dorling L., Carvalho S., et al. Prospective validation of the BOADICEA multifactorial breast cancer risk prediction model in a large prospective cohort study. J. Med. Genet. 2022;59:1196–1205. doi: 10.1136/jmg-2022-108806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakeman I.M.M., Rodríguez-Girondo M., Lee A., Ruiter R., Stricker B.H., Wijnant S.R.A., Kavousi M., Antoniou A.C., Schmidt M.K., Uitterlinden A.G., et al. Validation of the BOADICEA model and a 313-variant polygenic risk score for breast cancer risk prediction in a Dutch prospective cohort. Genet. Med. 2020;22:1803–1811. doi: 10.1038/s41436-020-0884-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shieh Y., Hu D., Ma L., Huntsman S., Gard C.C., Leung J.W.T., Tice J.A., Vachon C.M., Cummings S.R., Kerlikowske K., et al. Breast cancer risk prediction using a clinical risk model and polygenic risk score. Breast Cancer Res. Treat. 2016;159:513–525. doi: 10.1007/s10549-016-3953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dite G.S., MacInnis R.J., Bickerstaffe A., Dowty J.G., Allman R., Apicella C., Milne R.L., Tsimiklis H., Phillips K.-A., Giles G.G., et al. Breast cancer risk prediction using clinical models and 77 independent risk-associated SNPs for women aged under 50 years: Australian breast cancer family registry. Cancer Epidemiol. Biomark. Prev. 2016;25:359–365. doi: 10.1158/1055-9965.EPI-15-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maas P., Barrdahl M., Joshi A.D., Auer P.L., Gaudet M.M., Milne R.L., Schumacher F.R., Anderson W.F., Check D., Chattopadhyay S., et al. Breast Cancer Risk From Modifiable and Nonmodifiable Risk Factors Among White Women in the United States. JAMA Oncol. 2016;2:1295–1302. doi: 10.1001/jamaoncol.2016.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Rice M., Tworoger S.S., Rosner B.A., Eliassen A.H., Tamimi R.M., Joshi A.D., Lindstrom S., Qian J., Colditz G.A., et al. Addition of a polygenic risk score, mammographic density, and endogenous hormones to existing breast cancer risk prediction models: A nested case–control study. PLoS Med. 2018;15:e1002644. doi: 10.1371/journal.pmed.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.TCarver T., Hartley S., Lee A., Cunningham A.P., Archer S., de Villiers C.B., Roberts J., Ruston R., Walter F.M., Tischkowitz M., et al. Canrisk tool—A web interface for the prediction of breast and ovarian cancer risk and the likelihood of carrying genetic pathogenic variants. Cancer Epidemiol. Biomark. Prev. 2021;30:469–473. doi: 10.1158/1055-9965.EPI-20-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Archer S., De Villiers C.B., Scheibl F., Carver T., Hartley S., Lee A., Cunningham A.P., Easton D.F., McIntosh J.G., Emery J., et al. Evaluating clinician acceptability of the prototype CanRisk tool for predicting risk of breast and ovarian cancer: A multi-methods study. PLoS ONE. 2020;15:e0229999. doi: 10.1371/journal.pone.0229999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mavaddat N., Ficorella L., Carver T., Lee A., Cunningham A.P., Lush M., Dennis J., Tischkowitz M., Downes K., Hu D., et al. Incorporating Alternative Polygenic Risk Scores into the BOADICEA Breast Cancer Risk Prediction Model. Cancer Epidemiol. Biomark. Prev. 2023;32:422–427. doi: 10.1158/1055-9965.EPI-22-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.CanRisk Knowledge Base: Risk Calculations. [(accessed on 2 March 2023)]. Available online: https://canrisk.atlassian.net/wiki/spaces/FAQS/pages/131203073/Why+has+the+program+calculated+mutation+carrier+probabilities+but+not+cancer+risks.
- 33.Girdea M., Dumitriu S., Fiume M., Bowdin S., Boycott K.M., Chénier S., Chitayat D., Faghfoury H., Meyn M.S., Ray P.N., et al. PhenoTips: Patient phenotyping software for clinical and research use. Hum. Mutat. 2013;34:1057–1065. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 34.Deutsche Krebsgesellschaft Checkliste zur Erfassung einer familiären Belastung für Brust- und Eierstockkrebs. [(accessed on 22 February 2023)]. Available online: https://www.krebsgesellschaft.de/zertdokumente.html.
- 35.Rhiem K., Bücker-Nott H., Hellmich M., Fischer H., Ataseven B., Dittmer-Grabowski C., Latos K., Pelzer V., Seifert M., Schmidt A., et al. Benchmarking of a checklist for the identification of familial risk for breast and ovarian cancers in a prospective cohort. Breast J. 2019;25:455–460. doi: 10.1111/tbj.13257. [DOI] [PubMed] [Google Scholar]
- 36.Kast K., Rhiem K., Wappenschmidt B., Hahnen E., Hauke J., Bluemcke B., Zarghooni V., Herold N., Ditsch N., Kiechle M., et al. Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J. Med. Genet. 2016;53:465–471. doi: 10.1136/jmedgenet-2015-103672. [DOI] [PubMed] [Google Scholar]
- 37.Harter P., Hauke J., Heitz F., Reuss A., Kommoss S., Marmé F., Heimbach A., Prieske K., Richters L., Burges A., et al. Prevalence of deleterious germline variants in risk genes including BRCA1/2 in consecutive ovarian cancer patients (AGO-TR-1) PLoS ONE. 2017;12:e0186043. doi: 10.1371/journal.pone.0186043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engel C., Rhiem K., Hahnen E., Loibl S., Weber K.E., Seiler S., Zachariae S., Hauke J., Wappenschmidt B., Waha A., et al. Prevalence of pathogenic BRCA1/2 germline mutations among 802 women with unilateral triple-negative breast cancer without family cancer history. BMC Cancer. 2018;18:265. doi: 10.1186/s12885-018-4029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD®): 2003 Update. Human. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 42.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N. HerediCaRe: Documentation and IT Solution of a Specialized Registry for Hereditary Breast and Ovarian Cancer. Gesundheitswesen Suppl. 2021;83:S12–S17. doi: 10.1055/a-1658-0313. [DOI] [PubMed] [Google Scholar]
- 43.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kircher M., Witten D.M., Jain P., O’roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2019;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaganathan K., Panagiotopoulou S.K., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 46.Babraham Bioinformatics FastQC. [(accessed on 15 June 2023)]. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 47.Bolger A.M., Lohse M., Usadel B. Usadel Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrison E., Marth G. Haplotype-Based Variant Detection from Short-read Sequencing. 2012. [(accessed on 15 June 2023)]. Available online: http://arxiv.org/abs/1207.3907.
- 51.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.CanRisk Knowledge Base: What Variants are Used in the PRS? [(accessed on 22 June 2023)]. Available online: https://canrisk.atlassian.net/wiki/spaces/FAQS/pages/35979266/What+variants+are+used+in+the+PRS.
- 53.Rhiem K., Auber B., Briest S., Dikow N., Ditsch N., Dragicevic N., Grill S., Hahnen E., Horvath J., Jaeger B., et al. Consensus Recommendations of the German Consortium for Hereditary Breast and Ovarian Cancer. Breast Care. 2022;17:199–207. doi: 10.1159/000516376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bick U., Engel C., Krug B., Heindel W., Fallenberg E.M., Rhiem K., Maintz D., Golatta M., Speiser D., Rjosk-Dendorfer D., et al. High-risk breast cancer surveillance with MRI: 10-year experience from the German consortium for hereditary breast and ovarian cancer. Breast Cancer Res. Treat. 2019;175:217–228. doi: 10.1007/s10549-019-05152-9. [DOI] [PubMed] [Google Scholar]
- 55.Waha A., Versmold B., Kast K., Kiechle M., Ditsch N., Meindl A., Niederacher D., Hahnen E., Arnold N., Mundhenke C., et al. Konsensusempfehlung des Deutschen Konsortiums Familiärer Brust- und Eierstockkrebs zum Umgang mit Ergebnissen der Multigenanalyse. Tumor Diagn. Und Ther. 2018;39:187–193. doi: 10.1055/a-0574-4879. [DOI] [Google Scholar]
- 56.IKNL Richtlijn Borstkanker—Screening Buiten het Bevolkingsonderzoek. 2017. [(accessed on 31 May 2023)]. Available online: https://richtlijnendatabase.nl/richtlijn/borstkanker/screening/screening_buiten_het_bob/screening_buiten_het_bevolkingsonderzoek.html.
- 57.National Institute for Health and Care Excellence Familial Breast Cancer: Classification, Care and Managing Breast Cancer and Related Risks in People with a Family History of Breast Cancer Clinical Guideline Your Responsibility. 2013. [(accessed on 2 March 2023)]. Available online: www.nice.org.uk/guidance/cg164. [PubMed]
- 58.NCCN Clinical Practice Guidelines in Oncology; Breast Cancer Screening and Diagnosis. 2017. [(accessed on 31 May 2023)]. Available online: https://www.nccn.org/professionals/physician_gls/pdf/breast-screening.pdf. [DOI] [PubMed]
- 59.Lakeman I.M.M., Rodríguez-Girondo M.D.M., Lee A., Celosse N., Braspenning M.E., van Engelen K., van de Beek I., van der Hout A.H., García E.B.G., Mensenkamp A.R., et al. Clinical applicability of the Polygenic Risk Score for breast cancer risk prediction in familial cases Cancer genetics. J. Med. Genet. 2022;60:327–336. doi: 10.1136/jmg-2022-108502. [DOI] [PubMed] [Google Scholar]
- 60.Michailidou K., Lindstrom S., Dennis J., Beesley J., Hui S., Kar S., Lemacon A., Soucy P., Glubb D., Rostamianfar A., et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adedokun B., Du Z., Gao G., Ahearn T.U., Lunetta K.L., Zirpoli G., Figueroa J., John E.M., Bernstein L., Zheng W., et al. Cross-ancestry GWAS meta-analysis identifies six breast cancer loci in African and European ancestry women. Nat. Commun. 2021;12:4198. doi: 10.1038/s41467-021-24327-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H., kConFab Investigators. Ahearn T.U., Lecarpentier J., Barnes D., Beesley J., Qi G., Jiang X., O’mara T.A., Zhao N., et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat. Genet. 2020;52:572–581. doi: 10.1038/s41588-020-0609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dixon P., Keeney E., Taylor J.C., Wordsworth S., Martin R.M. Can polygenic risk scores contribute to cost-effective cancer screening? A systematic review. Genet. Med. 2022;24:1604–1617. doi: 10.1016/j.gim.2022.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho W.-K., Tan M.-M., Mavaddat N., Tai M.-C., Mariapun S., Li J., Ho P.-J., Dennis J., Tyrer J.P., Bolla M.K., et al. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nat. Commun. 2020;11:3833. doi: 10.1038/s41467-020-17680-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shieh Y., Fejerman L., Lott P.C., Marker K., Sawyer S.D., Hu D., Huntsman S., Torres J., Echeverry M., E Bohórquez M., et al. A Polygenic Risk Score for Breast Cancer in US Latinas and Latin American Women. J. Natl. Cancer Inst. 2020;112:590–598. doi: 10.1093/jnci/djz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.