ABSTRACT
Clinical diagnosis of several neurodegenerative disorders based on clinical phenotype is challenging due to its heterogeneous nature and overlapping disease manifestations. Therefore, the identification of underlying genetic mechanisms is of paramount importance for better diagnosis and therapeutic regimens. With the emergence of next-generation sequencing, it becomes easier to identify all gene variants in the genome simultaneously, with a system-wide and unbiased approach. Presently various bioinformatics databases are maintained on discovered gene variants and phenotypic indications are available online. Since individuals are unique in their genome, evaluation based on their genetic makeup helps evolve the diagnosis, counselling, and treatment process at the personal level. This article aims to briefly summarize the utilization of next-generation sequencing in deciphering the genetic causes of Alzheimer’s disease and address the limitations of whole genome and exome sequencing.
Keywords: Neurodegenerative Diseases, High-Throughput Nucleotide Sequencing, Alzheimer Disease, Exome Sequencing, Whole Genome Sequencing
RESUMO
O diagnóstico clínico de vários distúrbios neurodegenerativos com base no fenótipo clínico é difícil devido à sua natureza heterogênea e às manifestações da doença que se sobrepõem. Portanto, a identificação dos mecanismos genéticos subjacentes é de suma importância para um melhor diagnóstico e regimes terapêuticos. Com o surgimento do sequenciamento de próxima geração, o diagnóstico se tornou mais acessível com uma abordagem imparcial em todo o sistema para identificar simultaneamente todas as variantes de genes no genoma. Atualmente, vários bancos de dados de bioinformática sobre variantes genéticas descobertas e indicações fenotípicas estão disponíveis online. Uma vez que os indivíduos são únicos em seu genoma, a avaliação com base em sua composição genética ajudou na evolução do processo de diagnóstico, aconselhamento e tratamento em nível pessoal. Este artigo teve como objetivo resumir brevemente a utilização do sequenciamento de próxima geração para decifrar as causas genéticas da doença de Alzheimer (DA) e abordar as limitações do sequenciamento completo do genoma e do exoma.
Palavras-chave: Doenças Neurodegenerativas, Sequenciamento de Nucleotídeos em Larga Escala, Doença de Alzheimer, Sequenciamento do Exoma, Sequenciamento Completo do Genoma
NEXT-GENERATION SEQUENCING
Sequencing methods, such as Maxam-Gilbert’s chemical degradation method, were first employed to get fragments sequencing, but were soon replaced by the first-generation sequencing techniques such as Sanger’s chain-termination method, due to its hazardous chemical use and toxicity. Sanger sequencing was the popular rapid method used in the late 1900s for the Human Genome Project (HGP) and was also employed to deduce the gene variants responsible for causing disorders 1,2 . These techniques were efficient, but candidate gene selection and sequencing costs were challenging and time-consuming.
By the early 2000s, high-throughput next-generation sequencing (NGS) technologies were developed, making diagnosis easier and hassle-free. NGS is a combination of biology, statistics and information technology that allows massive parallel sequencing of genomes within a relatively short period of time. It achieves tremendous success in microbial genetics, monogenic diseases and complex diseases such as cancer genomics and other multifactorial syndromes. Recently, neurology also adopted the NGS techniques along with other imaging and biochemical methods to gain more expertise in identifying the variants causing disorders 3 . NGS can also be used to study deoxyribonucleic acid (DNA) methylation, protein DNA interaction, ribonucleic acid (RNA) study (RNA-Seq) etc. 4 .
There are three NGS approaches currently employed namely whole-genome sequencing (WGS), whole-exome sequencing (WES), and gene-targeted panels.
WHOLE GENOME SEQUENCING
WGS sequences the whole genome together. It helps to uncover variation in any part of the human genome, including coding, noncoding, and mitochondrial DNA (mtDNA) regions. WGS is considered the best option once DNA variations outside protein-coding regions can affect gene activity and protein production, potentially leading to genetic disorders 4 . It also helps to gather more information on an unknown or partially-known disorder and to discover the genomic instabilities leading to complex disorders 5 . It becomes easier to predict any specific variation running in the linage or genetic pool leading to specific phenotypes through various genome-wide association studies (GWAS). In the Encyclopedia of DNA Elements (ENCODE) project, one can see that not only coding regions but also non-coding regions are responsible for causing different complex traits 6 . WGS allows for the detection of copy number variations (CNVs), gross chromosomal abnormalities, and intergenic, regulatory and deep intronic variants, leading to a higher diagnostic yield. In the neurogenetics field, the WGS was first successfully used for the identification of a causative coding mutation in an autosomal recessive neurodegenerative Charcot-Marie Tooth disease 7 . Whole-genome methylation-specific studies can provide important information on how epigenetic and environmental factors alter gene expression.
WHOLE EXOME SEQUENCING
WES sequences only the exons or protein-coding parts of genes. It is seen that most known disease-causing mutations (∼85%) occur in exons of the gene, hence WES is widely used among clinicians and academics. It is targeted only to exons; therefore, considered a cost-effective method that demands less storage volume (∼4–5 Gb per exome) and reduced time consumption for analysis 8 . WES offers comprehensive coverage and increased sequencing depth which helps in identifying single nucleotide variants (SNVs) and small insertions/deletions (indels) for population genetics, genetic disease research, and cancer studies. It provides a better platform for detecting mutations running in a family using trio analysis which enables couples to plan their family in a better and healthier way. Through the exome enrichment strategy, we can get a more precise view of gene regulation which includes untranslated regions (UTRs) and microRNAs (miRNA). With WES, there are chances of incidental findings, which can give a valuable insight to the existing knowledge of the disease condition and its pathogenesis in various disorders 9 . It helps modify disease diagnosis steps and treatment strategies better.
Gene-targeted panels or custom panels sequence only a few genes that are particularly linked to a specific disorder. Gene-targeted panels are observed to be highly effective in the diagnosis of genetic diseases. It is often very small (250 Kb to 5 Mb) in size thus bringing down sequencing requirements and helping in answering distinct scientific questions quickly. It is an economic and suitable application for finding a particular disease or disorder. However, this approach is limited when it comes to complex neurodegenerative disorders.
NEXT-GENERATION SEQUENCING WORKFLOW
There are different techniques and pipelines used in sequencing genomes, depending upon the demands at a specific time. But all the methods notably follow three steps in NGS i.e., library preparation, sequencing, and data analysis. The DNA/RNA is extracted first from the tissue sample, then a quality control (QC) check is done to ensure its purity and quantity by ultraviolet (UV) spectrophotometer and fluorometric methods 10 .
Template preparation is the prime step in NGS workflow, where the DNA/complementary DNA (cDNA) library is prepared by fragmenting into numerous small coting by physical, enzymatic, and chemical methods, and attaching adaptors to both ends. These libraries are then amplified either by emulsion PCR (ePCR) in ion torrent sequencing or cluster formation by bridge PCR (bPCR) in Illumina sequencing in different customized sizes and prepared for sequencing. The sequenced library can be directly used for whole-genome analysis or undergo a targeted enrichment process for whole-exome analysis and targeted gene panel testing 11 .
Most clinical sequencing is performed on different types of instruments such as Illumina sequencers including the HiSeq, MiSeq, NexSeq, Pacific Biosciences, Ion Torrent series of machines including the IonPGM, IonProton, and IonS5, and others 12 . The data generated after sequencing is analyzed using different pipelines and software packages. The results obtained will be interpreted based on the requirement of analysis using various sets of bioinformatics tools.
ALZHEIMER’S DISEASE
Neurodegenerative disorder (NDD), as the name suggests, is a disorder in which cells of the central nervous system stop working or die. They are classified and diagnosed based on clinical features such as physical signs, symptom-onset, and disease course. Alzheimer’s disease (AD) is one of the most common NDDs characterized by dementia that typically begins with subtle mild cognitive impairment (MCI), gradually becomes severe and, finally, leads to total impairment of mental functions. It is commonly seen in the aging population and is becoming a significant cause of socio-economic burden worldwide. Neuropathologic findings mainly extracellular β-amyloid plaques and intraneuronal neurofibrillary tangles (containing tau protein that accumulate in vulnerable brain regions) are the hallmark of AD 13 . Initially, damage occurs in the hippocampus and the entorhinal cortex (memory-forming part of the brain). It then leads to seizure of neuronal function and loose connections of neurons, and gradually to shrinkage of brain parts.
As of 2021, more than 50 million people were affected by dementia worldwide, and this number is estimated to triple to 152 million by 2050 as the world’s population ages 14 . From 1990 to 2019, the incidence and prevalence of AD and other dementias increased 147.95% and 160.84%, respectively 15 .
Four subtypes are identified in AD so far. Familial or Early-Onset Alzheimer’s Disease (EOAD) constitutes less than 2% of total AD; neurological and depressive behaviors are early symptoms of EOAD 16 . Mutations in amyloid precursor protein (APP), presenin 1 (PSEN1), and presenin 2 (PSEN2), discovered through linkage studies, are the genes predominantly responsible for causing EOAD (Table 1). EOAD is referred to as “Mendelian AD” due to the almost complete penetrance and mostly autosomal-dominant mode of transmission of implicated DNA sequence changes 17 .
Table 1. Common genes in early onset Alzheimer’s disease.
| Genes | Function of protein* | Pathway of disease | Inheritance | Chromosomal location† |
|---|---|---|---|---|
| PSEN1 | Subunit of gamma-γ-secretase complex; integral membrane protein processing | AβPP Metabolism | AD | 14q24.2 |
| PSEN2 | Subunit of gamma-γ-secretase complex; integral membrane protein processing | AβPP Metabolism | AD | 1q42.13 |
| APP | Transmembrane protein; neural growth and repair | AβPP Metabolism | AD | 21q21.3 |
Abbreviations: PSEN1: presenin 1; PSEN2: presenin 2; APP: amyloid precursor protein; AD: autosomal dominant; AβPP: amyloid-β protein precursor.
With the advent of NGS, clinicians can better trace diseases at molecular level. AD and its associated genes have been researched extensively 18–22 . In the late 1900s and early 2000s, many genes were found to cause Alzheimer’s phenotype through GWAS. In 2003, the first GWAS were initiated in AD and, in 2007, it was published a meta-analysis of AD susceptibility genes. An AD database was then created, called AlzGene (http://www.alzgene.org) 23 .
An estimated 52 pathogenic mutations are identified in the APP gene; most of them are positioned in the vicinity of the β and γ-secretase cleavage sites (exons 16 and 17). Different mutations in V717I/G/F/L and E693K/Q/G/Del residues of APP make them mutation hotspots in the APP gene.
Mutations in exons 5, 6, 7, and 8 of the PSEN1 gene account for 70% of all identified mutations. Five different mutations of PSEN1 residue 143 (I143V/F/N/T/M, encoded by exon 5) are identified, making I143 residue a mutation hotspot 24 . It has been discerned that the PSEN1 variant (p.Thr291Pro), found in an individual presenting with spastic paraplegia, can later precede dementia onset in PSEN1-related familial AD 25 .
Several candidate gene approaches and GWAS have been performed to identify new genes related to AD (Table 2). Late-onset AD (LOAD) is reported to be caused by multiple gene; semantic dementia and conceptual formation deficit progress in LOAD. Around 90–95% of AD cases are attributed to sporadic mutations 26 . AD overlaps with other disease-related pathways such as Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), Huntington disease (HD) 27 .
Table 2. Common genes in late onset Alzheimer’s disease.
| Genes | Function of protein* | Pathway of disease | Cause | Chromosomal location† |
|---|---|---|---|---|
| APOE | Redistribution of lipids | Cholesterol metabolism | SNP | 19q13.2 |
| CR1 | Receptor of complement C3b protein that binds Aβ, mediates innate immunity | Immune response | SNP | 1q32.2 |
| CLU | Apoptosis and clearance of cellular debris, lipid transport and inflammation | Cholesterol, immune metabolism | SNP | 8p21.1 |
| ABCA7 | Transportation of phospholipids and phagocytosis | Cholesterol metabolism | SNP/haplodeficiency | 19p13.3 |
| PICALM | Synaptic neurotransmitter release and intracellular trafficking | Endocytosis | SNP | 11q14.2 |
| TREM2 | Inflammatory response | Loss of function (missense) mutation | 6p21.1 | |
| SORL1 | Vessel trafficking and cargo sorting | Endocytosis | SNP/nonsense and missense mutation; somatic mutations | 11q24.1 |
| ADAM10 | Mediates integral membrane protein cleavage | AβPP Metabolism | Mutations | 15q21.3 |
| BIN1 | Endocytosis, inflammation, calcium homeostasis and apoptosis | Tau Pathology | Mutations | 2q14.3 |
Abbreviations: APOE: apolipoprotein E; SNP: single nucleotide polymorphism.
Apolipoprotein E (APOE) e4 allele on chromosome 19, identified using Sanger and family-based approaches, significantly contributes to AD diagnosis in homozygous (APOE e4/e4) and heterozygous (APOE e3/e4) conditions. APOE e4 alleles are strongly associated with AD risk and contribute to various functional abnormalities, neurotoxicity, mitochondrial dysfunction, and cerebrovascular defects 28 .
Various studies have been conducted targeting ABCA7, BIN1, CLU, CR1, MS4A6A, EPHA1, CD2AP, and PICALM in different genetic pools. Few pathogenic mutations such as splice site, stop mutation, and frameshift deletions were identified suggesting a loss-of-function mechanism associated with LOAD 29 . Several missense mutations were found, of which most variants were classified as of uncertain significance due to the lack of functional studies.
A well-known mutation in TREM2 [R47H], identified as causing partial loss of function, contributes to Aβ accumulation by attenuating microglial-mediated Aβ clearance 30 . The clinical phenotype of mutations in FTD genes, including GRID2IP, WDR76, GRN, MAPT, and C9ORF72, can be clinically indistinguishable from typical AD 31 . Rare variants in the MAPT gene were found to be associated with AD in patients without ApoE e4 and tau pathology 32 . Loss-of-function or null variants in the SORL1 gene is a significant genetic risk factor for AD, as the truncated protein may result in disruption of its ability to bind APP 33 .
Homozygous and compound heterozygous VWA2 mutations mimic autosomal recessive inheritance in sporadic AD cases 34 . A missense variant p.Asp238Glu in UNC13B showed segregation within two families of Puerto Rican ancestry and was overrepresented in the AD cases 35 .
A family-based study showed a genome-wide significant linkage peak in 9p21 which overlapped with an AD linkage region. Novel genome-wide significant (GWS) AD-associated non-synonymous variants were identified, as well as a protective variant in PLCG2 (p.P522R), a risk variant in ABI3 (p.S209F), and a novel variant in TREM2 (p.R62H). These genes are highly expressed in microglia and highlight an immune-related protein-protein interaction network enriched for previously identified AD risk genes 36 .
Familial segregation in PLD3 (V232M) was seen, suggesting that PLD3 influences APP metabolism, such that overexpression leads to lower Aβ levels while knock-down of PLD3 leads to increased levels of Aβ 37 . The MUC6 VNTR repeat expansion influences AP2A2 gene expression involved in clathrin-coated vesicle function and is associated with AD pathogenesis, particularly tau proteinopathy 38 .
The GGC repeat expansion of NOTCH2NLC gene leads to neuronal intranuclear inclusion disease (NIID) and was also observed in family members affected by AD and Parkinson’s disease 39 . A rare nonsynonymous variant in the SHARPIN gene, p.Gly186Arg, is potentially associated with increased risk of LOAD. It leads to aberrant cellular localization of the variant protein and attenuates the activation of NF-κB, a central mediator of inflammatory and immune responses 40 .
Individuals with Down syndrome (trisomy 21) developed the AD neuropathologic hallmarks after the age of 40 years, due to overexpression of APP on chromosome 21 and the resultant overproduction of β-amyloid in the brains of people’s trisomy for this gene 41 .
Somatic (non-inherited) mtDNA mutations and mitochondrial dysfunction are thought to be important drivers of ageing and age-related neurodegenerative diseases such as AD 42 . The sequencing of OGDH, DLST, and DLD genes, encoding alpha-ketoglutarate dehydrogenase complex (αKGDHc) subunits, identified a likely pathogenic [R263H] mutation in the DLD gene associated with AD 43 .
Few gender-based studies have been conducted on AD disorder; nevertheless, it was found that females are at higher risk. APOE e4 females may show increased levels of AD pathology, more compromised brain network integrity, and/or accelerated longitudinal decline at a given level of AD pathology than males 44 . Greater hippocampal electroencephalograph disruption and memory impairment were seen in female ACE1 [R1279Q] KI mice, compared to males, suggesting a mechanism for higher AD risk in women 45 .
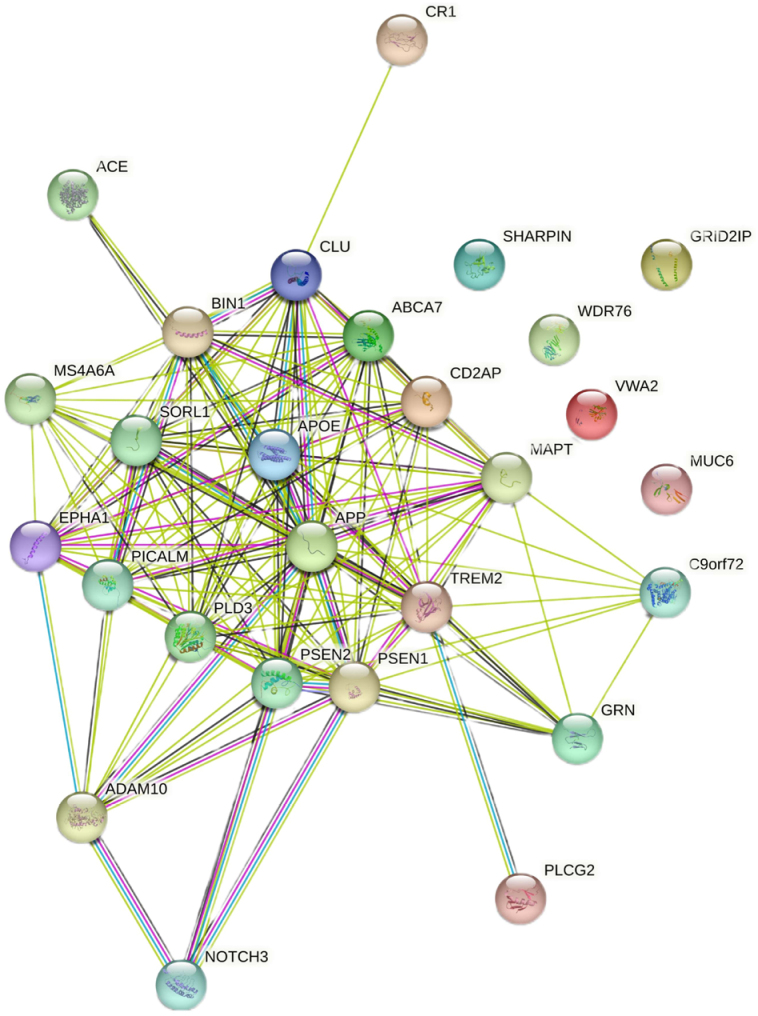
Despite these recent advances in AD genomics, a significant part of the genetic contribution to AD remains unexplained. Further functional studies are required to examine mutation-specific expressions and understand the mechanisms by which the mutations lead to disease 46 . Figure 1 presents the interaction between the genes associated with Alzheimer’s disease. The functional links between these genes are identified and documented by experimental, biochemical, and expressional studies in scientific literature.
Figure 1. Interaction between the genes associated with Alzheimer’s disease.
CHALLENGES OF WHOLE-EXOME SEQUENCING AND WHOLE-GENOME SEQUENCING
The high complexity of NGS workflow and result interpretation are the major challenges encountered in WES and WGS. Most variants are inevitably detected in every individual tested and it is essential to provide a comprehensive clinical interpretation for these variants with a long time invested 47 . This highlights the fact that the cost for providing clinical WGS/WES is likely to remain high even as sequencing costs fall. Every step of the NGS assay requires thorough validation, therefore the sample undergoes quality checks under standard guidelines. Sequencing errors such as low depth, low alternate allele frequency, low coverage region, etc. occurring due to technical limitations, may lead to a missed variant or a false-positive result.
In genome analysis, variant calling is affected by many factors. First, polymorphic region — a region with multiple variants is scattered throughout the region and is known as a “confetti effect”. Any variant calling in this region can be challenging. Second, homopolymer repeat regions — tracts of repeated small nucleotide sequences together, which are skipped or cannot be picked up by sequencing. Third, strand bias — it occurs when reads aligned to a reference are biased towards the forward or reverse strands. It is common around exon boundaries, particularly for WES with a high chance of a false positive variant or a missed variant 48 . Fourth, low depth of coverage — it means that the number of reads covering a region is few. Sometimes, false calls can be made by assuming polymorphic variant as a rare significant variant 49 . In this case, Sanger sequencing can be used to validate the variant as it tends to provide qualitative results, differently from NGS. Sanger sequencing is ideal for sequencing homogeneous samples that include one template, one gene, or one region.
Most genes referred above have 100% coverage in NGS sequencing. Few genes, such as PICALM, CD2AP and ADAM10, and CR1, are not fully covered due to segmental duplication site (pseudogenes) of polymorphic low covered regions, present in the genes (Table 3) 50 .
Table 3. Average gene coverage and sequencing depth expected in sequencing for Alzheimer’s disease genes.
| Genes | Average coverage (%) | Average depth |
|---|---|---|
| APP | 100 | 105.73 |
| PSEN1 | 98.89 | 111.32 |
| PSEN2 | 100 | 110.2 |
| APOE | 100 | 92.4 |
| BIN1 | 100 | 121.66 |
| CR1 | 62.73 | 70.58 |
| CLU | 100 | 119.45 |
| ABCA7 | 100 | 117.71 |
| PICALM | 93.12 | 83.1 |
| TREM2 | 100 | 123.81 |
| SORL1 | 99.88 | 101.53 |
| ADAM10 | 96.01 | 83.47 |
| CD2AP | 93.58 | 80.23 |
| MS4A6A | 99.74 | 101.22 |
| EPHA1 | 100 | 123.04 |
| GRID2IP | 99.79 | 113.99 |
| GRN | 100 | 139.3 |
| MAPT | 100 | 185.73 |
| C9ORF72 | 96.79 | 76.9 |
| NOTCH3 | 99.77 | 151.88 |
| PLCG2 | 99.86 | 127.72 |
| PLD3 | 100 | 113.51 |
| ACE | 99.91 | 112.06 |
| WDR76 | 99.87 | 106.15 |
| VWA2 | 100 | 127.97 |
| MUC6 | 100 | 374.11 |
| SHARPIN | 100 | 132.06 |
Abbreviations: APP: Amyloid Beta A4 Precursor Protein; PSEN1: Presenilin 1; PSEN2: Presenilin 2; APOE: Apolipoprotein E; BIN1: Bridging Integrator 1; CR1: Complement Component Receptor 1; CLU: Clusterin; ABCA7: ATP-Binding Cassette, Subfamily A, Member 7; PICALM: Phosphatidylinositol-Binding Clathrin Assembly Protein; TREM2: Triggering Receptor Expressed on Myeloid Cells 2; SORL1: Sortilin-Related Receptor; ADAM10: A Disintegrin And Metalloproteinase Domain 10; CD2AP: CD2-Associated Protein; MS4A6A: Membrane-Spanning 4-Domains, Subfamily A, Member 6A; EPHA1: Ephrin Receptor EphA1; GRID2IP: GRID2-Interacting Protein 1; GRN: Granulin Precursor; MAPT: Microtubule-Associated Protein TAU; C9ORF72: Chromosome 9 Open Reading Frame 72; NOTCH3: Notch Receptor 3; PLCG2: Phospholipase C, Gamma-2; PLD3: Phospholipase D Family, Member 3; ACE: Angiotensin I-Converting Enzyme; WDR76: WD Repeat-Containing Protein 76; VWA2: Von Willebrand Factor A Domain-Containing Protein 2; MUC6: Mucin 6, Gastric; SHARPIN: Shank-Associated RH Domain Interactor.
In a targeted-panel sequencing, the clinical importance of the genes and selective enrichment of targeted-genomic areas for NGS are the primary concerns. The selection of suitable target capture approaches and sequencing methods are crucial in yielding good quality results. This is determined by several factors such as the sample type (fresh, frozen, or formalin-fixed paraffin-embedded [FFPE]), quantity and quality of DNA or RNA routinely available 51 .
As it is evident that every variation cannot be classified as pathogenic, a thorough validation is required before sending a final report to the patient. Research is a continuous process and functional studies on different variations can lead to upgrading or downgrading a variant classification. A revision of the variant in reports must be done timely so that clinicians can design and provide adequate treatment to patients.
Failure to state the authenticity of large deletions or duplications in genes (copy number variations) can lead to serious disorders. Therefore, cross-confirmatory tests are recommended to ascertain the CNV and its effect on the patient’s phenotype, so that treatments can be planned accordingly 52 .
In conclusion, over the past couple of decades, high-throughput genome technologies have changed the genetic landscape of AD. NGS combined with other molecular advances, such as omics data, biochemical and functional studies, can now provide scientists with the ability to gain a comprehensive view of molecular disease pathways.
NGS assists in deducing the gain or loss of function in genes responsible for causing AD. Moreover, recent advances in NGS and its analysis have helped detect and confirm short tandem deletions and duplications. Several modifications to the current technology have been made on a day-to-day basis so that people can yield maximum benefit from NGS and help them lead a better and quality life. In the near future, both hypothesis-free (whole-genome, whole-exome) and hypothesis-driven (targeted-exome) NGS approaches will probably disentangle much of the disease genetics. Despite the few limitations stated above, it can pave the way for developing novel therapeutics and designing the right treatment on an individual level, called personalized medicine or precision medicine, which can effectively prevent or halt the progression of this devastating disease.
Footnotes
This study was conducted by the NGS laboratory, MedGenome labs Pvt. Ltd., Bangalore, India.
Funding: none.
REFERENCES
- 1.Heather JM, Chain B. The sequence of sequencers: the history of sequencing DNA. Genomics. 2016;107(1):1–8. doi: 10.1016/j.ygeno.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. The third revolution in sequencing technology. Trends Genet. 2018;34(9):666–681. doi: 10.1016/j.tig.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Shademan B, Avci CB, Nikanfar M, Nourazarian A. Application of next-generation sequencing in neurodegenerative diseases: opportunities and challenges. Neuromolecular Med. 2021;23(2):225–235. doi: 10.1007/s12017-020-08601-7. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y, Ruivenkamp CAL, Hoffer MJV, Vrijenhoek T, Kriek M, van Asperen CJ, et al. Next-generation diagnostics: gene panel, exome, or whole genome? Hum Mutat. 2015;36(6):648–655. doi: 10.1002/humu.22783. [DOI] [PubMed] [Google Scholar]
- 5.Choudhury A, Ramsay M, Hazelhurst S, Aron S, Bardien S, Botha G, et al. Whole-genome sequencing for an enhanced understanding of genetic variation among South Africans. Nat Commun. 2017;8(1):2062–2062. doi: 10.1038/s41467-017-00663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jayadev S, Smith CO, Bird TD. Neurogenetics: five new things. Neurol Clin Pract. 2011;1(1):41–48. doi: 10.1212/CPJ.0b013e31823c0f5f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao HM, Hong JS. Why neurodegenerative diseases are progressive: uncontrolled inflammation drives disease progression. Trends Immunol. 2008;29(8):357–365. doi: 10.1016/j.it.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foo JN, Liu JJ, Tan EK. Whole-genome and whole-exome sequencing in neurological diseases. Nat Rev Neurol. 2012;8(9):508–517. doi: 10.1038/nrneurol.2012.148. [DOI] [PubMed] [Google Scholar]
- 10.Singer J, Ruscheweyh HJ, Hofmann AL, Thurnherr T, Singer F, Toussaint NC, et al. NGS-pipe: a flexible, easily extendable and highly configurable framework for NGS analysis. Bioinformatics. 2018;34(1):107–108. doi: 10.1093/bioinformatics/btx540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hess JF, Kohl TA, Kotrová M, Rönsch K, Paprotka T, Mohr V, et al. Library preparation for next generation sequencing: a review of automation strategies. Biotechnol Adv. 2020;41:107537–107537. doi: 10.1016/j.biotechadv.2020.107537. [DOI] [PubMed] [Google Scholar]
- 12.Thankachan A, Thomas B. A study of next generation sequencing data, workflow, application and platform comparison; Series: Materials Science and Engineering. International Conference on Recent Advancements and Effectual Researches in Engineering Science and Technology (RAEREST); Kerala State, India. Apr 20-21, 2018. [DOI] [Google Scholar]
- 13.Lopez JAS, González HM, Léger GC. Alzheimer’s disease. Handb Clin Neurol. 2019;167:231–255. doi: 10.1016/B978-0-12-804766-8.00013-3. [DOI] [PubMed] [Google Scholar]
- 14.Porsteinsson AP, Isaacson RS, Knox S, Sabbagh MN, Rubino I. Diagnosis of early Alzheimer’s disease: clinical practice in 2021. J Prev Alzheimers Dis. 2021;8(3):371–386. doi: 10.14283/jpad.2021.23. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Feng X, Sun X, Hou N, Han F, Liu Y. Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990-2019. Front Aging Neurosci. 2022;14:937486–937486. doi: 10.3389/fnagi.2022.937486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bature F, Guinn BA, Pang D, Pappas Y. Signs and symptoms preceding the diagnosis of Alzheimer’s disease: a systematic scoping review of literature from 1937 to 2016. BMJ Open. 2017;7(8):e015746. doi: 10.1136/bmjopen-2016-015746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertram L. Next generation sequencing in Alzheimer’s disease. Methods Mol Biol. 2016;1303:281–297. doi: 10.1007/978-1-4939-2627-5_17. [DOI] [PubMed] [Google Scholar]
- 18.Hippius H, Neundörfer G. The discovery of Alzheimer’s disease. Dialogues Clin Neurosci. 2003;5(1):101–108. doi: 10.31887/DCNS.2003.5.1/hhippius. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savage JC, Picard K, González-Ibáñez F, Tremblay ME. A brief history of microglial ultrastructure: distinctive features, phenotypes, and functions discovered over the past 60 years by electron microscopy. Front Immunol. 2018;9:803–803. doi: 10.3389/fimmu.2018.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jack CR, Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moloney CM, Lowe VJ, Murray ME. Visualization of neurofibrillary tangle maturity in Alzheimer’s disease: a clinicopathologic perspective for biomarker research. Alzheimers Dement. 2021;17(9):1554–1574. doi: 10.1002/alz.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weller J, Budson A. Current understanding of Alzheimer’s disease diagnosis and treatment. F1000Res. 2018;7:F1000–F1000. doi: 10.12688/f1000research.14506.1. Faculty Rev-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 24.Zou Z, Liu C, Che C, Huang H. Clinical genetics of Alzheimer’s disease. Biomed Res Int. 2014;2014:291862–291862. doi: 10.1155/2014/291862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chelban V, Breza M, Szaruga M, Vandrovcova J, Murphy D, Lee CJ, et al. Spastic paraplegia preceding PSEN1-related familial Alzheimer’s disease. Alzheimers Dement (Amst) 2021;13(1):e12186. doi: 10.1002/dad2.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Sidorenko J, Couvy-Duchesne B, Marioni RE, Wright MJ, Goate AM, et al. Risk prediction of late-onset Alzheimer’s disease implies an oligogenic architecture. Nat Commun. 2020;11(1):4799–4799. doi: 10.1038/s41467-020-18534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gan L, Cookson MR, Petrucelli L, La Spada AR. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat Neurosci. 2018;21(10):1300–1309. doi: 10.1038/s41593-018-0237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9(2):106–118. doi: 10.1038/nmeurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagyinszky E, Youn YC, An SSA, Kim SY. The genetics of Alzheimer’s disease. Clin Interv Aging. 2014;9:535–551. doi: 10.2147/CIA.S51571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Condello C, Yuan P, Grutzendler J. Microglia-mediated neuroprotection, TREM2, and Alzheimer’s disease: evidence from optical imaging. Biol Psychiatry. 2018;83(4):377–387. doi: 10.1016/j.biopsych.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harms M, Benitez BA, Cairns N, Cooper B, Cooper P, Mayo K, et al. C9orf72 hexanucleotide repeat expansions in clinical Alzheimer disease. JAMA Neurol. 2013;70(6):736–741. doi: 10.1001/2013.jamaneurol.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin SC, Pastor P, Cooper B, Cervantes S, Benitez BA, Razquin C, et al. Pooled-DNA sequencing identifies novel causative variants in PSEN1, GRN and MAPT in a clinical early-onset and familial Alzheimer’s disease Ibero-American cohort. Alzheimers Res Ther. 2012;4(4):34–34. doi: 10.1186/alzrt137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raghavan NS, Brickman AM, Andrews H, Manly JJ, Schupf N, Lantigua R, et al. Whole-exome sequencing in 20,197 persons for rare variants in Alzheimer’s disease. Ann Clin Transl Neurol. 2018;5(7):832–842. doi: 10.1002/acn3.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoogmartens J, Hens E, Engelborghs S, Vandenberghe R, De Deyn PP, Cacace R, et al. Contribution of homozygous and compound heterozygous missense mutations in VWA2 to Alzheimer’s disease. Neurobiol Aging. 2021;99:100.e17–100.e23. doi: 10.1016/j.neurobiolaging.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Rajabli F, Feliciano-Astacio BE, Cukier HN, Wang L, Griswold AJ, Hamilton-Nelson KL, et al. Linkage of Alzheimer disease families with Puerto Rican ancestry identifies a chromosome 9 locus. Neurobiol Aging. 2021;104:115.e1–115.e7. doi: 10.1016/j.neurobiolaging.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet. 2017;49(9):1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505(7484):550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson PT, Fardo DW, Katsumata Y. The MUC6/AP2A2 locus and its relevance to Alzheimer’s disease: a review. J Neuropathol Exp Neurol. 2020;79(6):568–584. doi: 10.1093/jnen/nlaa024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian Y, Wang JL, Huang W, Zeng S, Jiao B, Liu Z, et al. Expansion of human-specific GGC repeat in neuronal intranuclear inclusion disease-related disorders. Am J Hum Genet. 2019;105(1):166–176. doi: 10.1016/j.ajhg.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asanomi Y, Shigemizu D, Miyashita A, Mitsumori R, Mori T, Hara N, et al. A rare functional variant of SHARPIN attenuates the inflammatory response and associates with increased risk of late-onset Alzheimer’s disease. Mol Med. 2019;25(1):20–20. doi: 10.1186/s10020-019-0090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Head E, Powell D, Gold BT, Schmitt FA. Alzheimer’s disease in Down syndrome. Eur J Neurodegener Dis. 2012;1(3):353–364. [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava S. The mitochondrial basis of aging and age-related disorders. Genes (Basel) 2017;8(12):398–398. doi: 10.3390/genes8120398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Csaban D, Pentelenyi K, Toth-Bencsik R, Illes A, Grosz Z, Gezsi A, et al. The role of the rare variants in the genes encoding the alpha-ketoglutarate dehydrogenase in Alzheimer’s disease. Life (Basel) 2021;11(4):321–321. doi: 10.3390/life11040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nebel RA, Aggarwal NT, Barnes LL, Gallagher A, Goldstein JM, Kantarci K, et al. Understanding the impact of sex and gender in Alzheimer’s disease: a call to action. Alzheimers Dement. 2018;14(9):1171–1183. doi: 10.1016/j.jalz.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuddy LK, Prokopenko D, Cunningham EP, Brimberry R, Song P, Kirchner R, et al. Aβ-accelerated neurodegeneration caused by Alzheimer’s-associated ACE variant R1279Q is rescued by angiotensin system inhibition in mice. Sci Transl Med. 2020;12(563):eaaz2541–eaaz2541. doi: 10.1126/scitranslmed.aaz2541. https://doi.org/scitranslmed.aaz2541 . [DOI] [PubMed] [Google Scholar]
- 46.Neuner SM, Tcw J, Goate AM. Genetic architecture of Alzheimer’s disease. Neurobiol Dis. 2020;143:104976–104976. doi: 10.1016/j.nbd.2020.104976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meienberg J, Bruggmann R, Oexle K, Matyas G. Clinical sequencing: is WGS the better WES? Hum Genet. 2016;135(3):359–362. doi: 10.1007/s00439-015-1631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20(10):1122–1130. doi: 10.1038/gim.2017.247. [DOI] [PubMed] [Google Scholar]
- 49.Seaby EG, Ennis S. Challenges in the diagnosis and discovery of rare genetic disorders using contemporary sequencing technologies. Brief Funct Genomics. 2020;19(4):243–258. doi: 10.1093/bfgp/elaa009. [DOI] [PubMed] [Google Scholar]
- 50.Silva R, Burch JB. Evidence that chicken CR1 elements represent a novel family of retroposons. Mol Cell Biol. 1989;9(8):3563–3566. doi: 10.1128/mcb.9.8.3563-3566.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto T, Shimojima K, Ondo Y, Imai K, Chong PF, Kira R, et al. Challenges in detecting genomic copy number aberrations using next-generation sequencing data and the eXome Hidden Markov Model: a clinical exome-first diagnostic approach. Hum Genome Var. 2016;3:16025–16025. doi: 10.1038/hgv.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luthra R, Chen H, Roy-Chowdhuri S, Singh RR. Next-generation sequencing in clinical molecular diagnostics of cancer: advantages and challenges. Cancers (Basel) 2015;7(4):2023–2036. doi: 10.3390/cancers7040874. [DOI] [PMC free article] [PubMed] [Google Scholar]