Abstract
Reovirus outer-capsid proteins μ1, ς3, and ς1 are thought to be assembled onto nascent core-like particles within infected cells, leading to the production of progeny virions. Consistent with this model, we report the in vitro assembly of baculovirus-expressed μ1 and ς3 onto purified cores that lack μ1, ς3, and ς1. The resulting particles (recoated cores, or r-cores) closely resembled native virions in protein composition (except for lacking cell attachment protein ς1), buoyant density, and particle morphology by scanning cryoelectron microscopy. Transmission cryoelectron microscopy and image reconstruction of r-cores confirmed that they closely resembled virions in the structure of the outer capsid and revealed that assembly of μ1 and ς3 onto cores had induced rearrangement of the pentameric λ2 turrets into a conformation approximating that in virions. r-cores, like virions, underwent proteolytic conversion to particles resembling native ISVPs (infectious subvirion particles) in protein composition, particle morphology, and capacity to permeabilize membranes in vitro. r-cores were 250- to 500-fold more infectious than cores in murine L cells and, like virions but not ISVPs or cores, were inhibited from productively infecting these cells by the presence of either NH4Cl or E-64. The latter results suggest that r-cores and virions used similar routes of entry into L cells, including processing by lysosomal cysteine proteinases, even though the former particles lacked the ς1 protein. To examine the utility of r-cores for genetic dissections of μ1 functions in reovirus entry, we generated r-cores containing a mutant form of μ1 that had been engineered to resist cleavage at the δ:φ junction during conversion to ISVP-like particles by chymotrypsin in vitro. Despite their deficit in δ:φ cleavage, these ISVP-like particles were fully competent to permeabilize membranes in vitro and to infect L cells in the presence of NH4Cl, providing new evidence that this cleavage is dispensable for productive infection.
Mammalian orthoreoviruses (reoviruses) serve as useful models to study the entry of nonenveloped animal viruses into their host cells. Reovirus virions comprise eight proteins ranging from 12 to 600 in copy number and arranged in two concentric icosahedral capsids (see references 26, 28, and 42 for reviews). The segmented double-stranded RNA genome and several components of the virus-bound transcriptase are enclosed within the inner capsid. The inner capsid and structures within it are not thought to play a role in viral entry except in constituting the major part of the payload delivered into the cytoplasm by the penetration machinery housed in the outer capsid (40). The outer capsid is formed primarily by N-myristoylated protein μ1 (76 kDa; 600 copies), which is the putative membrane penetration protein of reoviruses (20, 21, 32, 39, 41, 55). The μ1 protein is found in virions mostly as fragments μ1N (4 kDa) and μ1C (72 kDa), which are thought to arise by autolysis (41, 54). In this report, the term “μ1” is used to indicate full-length μ1 protein and all of its fragments unless individual fragments are specified. Pentamers of protein λ2 (144 kDa; 60 copies) substitute the μ1 lattice around the icosahedral fivefold axes. Protein ς3 (41 kDa; 600 copies), the major surface protein of virions, adopts icosahedral positions through its close interactions with underlying μ1 subunits (15). By virtue of these interactions, ς3 plays critical roles in μ1 assembly into progeny particles (33, 37, 49), and in regulating the conformational status and exposure of μ1 (see below). Protein ς1 (50 kDa; 36 copies) forms a trimeric cell attachment fiber at each fivefold axis (31, 52).
The requirement for proteolytic cleavage of outer-capsid proteins to allow reovirus particles to enter host cells is well documented. In brief, when virions are used to infect cultured cells, ς3 is degraded and μ1/μ1C is cleaved at the δ:φ junction by one or more lysosomal cysteine proteinase at early times postinfection (1, 9, 11, 29, 53). Compounds like NH4Cl and E-64 that block proteolysis of ς3 and μ1/μ1C within cells prevent viral infection (1, 9, 29, 53). However, treatment of virions with proteinases (e.g., chymotrypsin [CHT] or trypsin [TRY]) in vitro can generate infectious subvirion particles (ISVPs) that lack ς3 and contain μ1/μ1C as cleaved μ1δ/δ and φ fragments (5, 27, 39) and that can bypass the requirement for active lysosomal proteinase(s). The latter property of ISVPs strongly suggests that proteolysis is needed to activate particles for subsequent steps in infection such as membrane penetration. Additional evidence for this model is the capacity of ISVPs but not virions to permeabilize membranes in vitro (9, 21, 32, 38, 55). Recent evidence indicates that cleavage of μ1/μ1C at the δ:φ junction during viral entry into cultured cells is dispensable for membrane penetration and infection (9; also see this study), suggesting that ς3 is the outer-capsid protein that must undergo cleavage (24).
Membrane penetration by ISVP-like particles results in cytoplasmic delivery of a particle which has yet to be fully characterized in terms of protein composition (7, 26, 28, 42, 46) but which is known to have been activated to transcribe the 10 particle-bound genome segments into full-length mRNAs. This primary transcriptase particle (7, 26, 40, 46, 50) shares at least some properties with cores, which are transcriptionally active particles generated by in vitro proteolytic digestion of virions (7, 27, 47). Cores are distinguished from ISVPs in that they have lost proteins μ1 and ς1 in addition to ς3 and contain the λ2 turrets in a distinct conformation from that in virions and ISVPs (6, 14, 15, 46). This conformational change in λ2 may be essential for viral transcription because it opens a large central channel in the turret through which viral mRNAs are thought to exit the transcribing particle (15, 58). Particles resembling cores are also generated from newly synthesized proteins within infected cells and are thought to be the precursors upon which outer-capsid proteins μ1, ς3, and ς1 are assembled to produce mature virions (33, 37, 49, 57).
In this report, we describe particles generated by stoichiometric assembly of baculovirus-expressed outer-capsid proteins μ1 and ς3 onto reovirus cores. These particles, which we term recoated cores (r-cores), were distinguishable from cores and closely resembled native virions according to various physicochemical, biochemical, and structural criteria, including the conformation of their λ2 turrets. In addition, r-cores were 250- to 500-fold more infectious than core particles and, like virions but not ISVPs or cores, appeared to require the activity of lysosomal cysteine proteinases for infection of L cells, even though they lacked the ς1 cell attachment protein. We also generated and characterized particles containing a mutant form of μ1 with an alteration that greatly inhibited δ:φ cleavage. These experiments provided new evidence that cleavage at the δ:φ junction during viral entry is dispensable for membrane penetration and reovirus infection in cultured cells. The current findings, along with other recent work describing an approach for recoating ISVPs with ς3 (24), demonstrate the utility of recoated subvirion particles for molecular-genetic studies of reovirus entry. In addition, because μ1-ς3 assembly onto cores in vitro likely resembles this process as it occurs in infected cells, the generation of r-cores can be used to analyze the molecular determinants of reovirus outer-capsid assembly.
MATERIALS AND METHODS
Cells and viruses.
Spinner-adapted murine L cells were grown in suspension in Joklik’s modified minimal essential medium (Irvine Scientific, Irvine, Calif.) supplemented to contain fetal bovine serum (2%), neonatal bovine serum (2%) (HyClone Laboratories, Logan, Utah), and penicillin (100 U/ml)-streptomycin (100 μg/ml) (Irvine Scientific). Type 1 Lang (T1L) was the reovirus used in this study. Plaque assays to determine the infectivities of reovirus preparations were performed as described previously (18). Spodoptera frugiperda Sf21 and Trichoplusia ni Tn High Five insect cells (Invitrogen, Carlsbad, Calif.) were grown in TC-100 medium (Gibco BRL, Gaithersburg, Md.) supplemented to contain heat-inactivated fetal bovine serum (10%).
Virions, ISVPs, and cores.
Purified T1L virions were obtained as described elsewhere (39). Virion buffer contains 150 mM NaCl, 10 mM MgCl2, and 10 mM Tris (pH 7.5). ISVPs were prepared by digestion of virions with Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK)-treated CHT (Sigma) as described elsewhere (39). T1L cores were prepared as follows. Virions present in extracts of reovirus-infected L cells clarified by extraction with Freon (39) were pelleted by centrifugation at 25,000 rpm and 5°C for 2 h in a Beckman SW28 rotor. The virion pellet was resuspended in virion buffer to a concentration in excess of 8 × 1013 particles/ml and digested with CHT (200 μg/ml) for 2 h at 37°C. Reactions were terminated by addition of phenylmethylsulfonyl fluoride (PMSF; Sigma) to 1 to 5 mM. The resulting cores were purified by banding on two successive preformed CsCl gradients (ρ = 1.25 to 1.50 g/cm3) and were dialyzed extensively against virion buffer. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining (see below) were routinely used to confirm that ς3, μ1, and ς1 proteins were not present in detectable amounts in the core preparations used to produce r-cores. Particle concentrations were estimated by A260 (13, 51).
Expression of μ1 and ς3.
The original T1L M2 clone used in this study was generated by Simon Noble (43). Briefly, the cDNA was produced from viral transcripts (19) by using avian myeloblastosis virus reverse transcriptase (Gibco BRL) and a primer (5′-TGTGCCTGCATCCCTTAACC-3′) annealed to the 3′ end of the M2 mRNA. The cDNA was then used as a template for second-strand synthesis using the DNA polymerase I Klenow fragment (New England Biolabs, Beverly, Mass.) and a primer (5′-GCTAATCTGCTGACCGTCACTC-3′) that annealed to the 3′ end of the cDNA. After polynucleotide kinase treatment (New England Biolabs), the resulting double-stranded DNA was ligated into the SmaI site of the pBluescriptII-KS vector (Stratagene, La Jolla, Calif.) to generate pBKS-M2L. The T1L S4 gene was cloned into the EcoRI site of the pcDNAI-Amp vector (Invitrogen) as described previously (48).
To generate a recombinant baculovirus expressing both μ1 and ς3 proteins, the T1L M2 and S4 genes were subcloned into the pFastbacDUAL vector (Gibco BRL), using the NotI and HindIII sites for M2 and the SmaI and XhoI sites for S4 to generate pFbD-M2L-S4L. This cloning strategy positioned the M2 and S4 genes for transcription from the baculovirus polyhedrin and p10 promoters, respectively. The dual clone was then used to generate a recombinant baculovirus per the Bac-to-Bac system (Gibco BRL). Baculoviruses were propagated in Sf21 cells following infection with a multiplicity of infection (MOI) of 0.5 to 1 PFU/cell. To produce μ1 and ς3 in large amounts, Tn High Five cells were infected with fourth-passage virus stocks at an MOI of 5 to 10 PFU/cell, and cells were harvested at 48 to 66 h postinfection. 35S-labeled μ1 and ς3 were produced as described elsewhere (19). Cytoplasmic extracts of baculovirus-infected cells were prepared by lysis with Triton X-100 as described previously (19) except that lysis reactions did not contain RNase inhibitors and were supplemented with Complete proteinase-inhibitor cocktail (4%; Boehringer Mannheim, Indianapolis, Ind.).
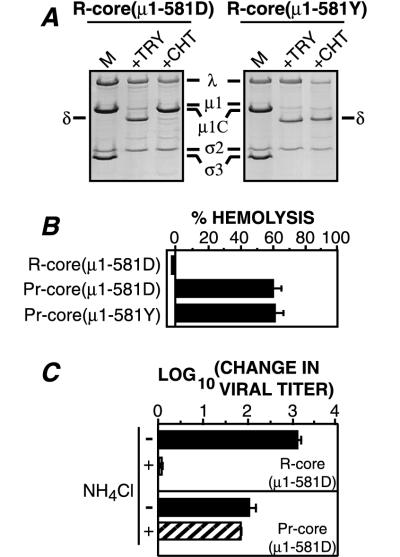
After having completed the experiments for this study, we learned by automated DNA sequencing of both strands of our original T1L M2 clone (at the University of Wisconsin Biotechnology Center DNA Facility) that it encoded two amino acid changes (P344L and L359F) relative to a published M2 sequence for the Fields lab clone of reovirus T1L (25). Subsequent experiments using a T1L M2 clone in which we modified those nucleotides to match the published sequence demonstrated that the amino acid changes had little or no effect on the generation of r-cores, their specific infectivities in L cells, or their capacities to undergo conversion to ISVP-like particles in vitro, to infect L cells in the absence or presence of NH4Cl, and to hemolyze erythrocytes (RBCs) in vitro, as shown for the original T1L M2 clone in Fig. 1, 5, 6, and 8 and Tables 1 and 2. Because the cryoelectron microscopy and three-dimensional (3-D) image reconstruction results presented in this study had been done for r-cores generated with the original T1L M2 clone, only data obtained with that clone have been presented in the figures.
FIG. 1.
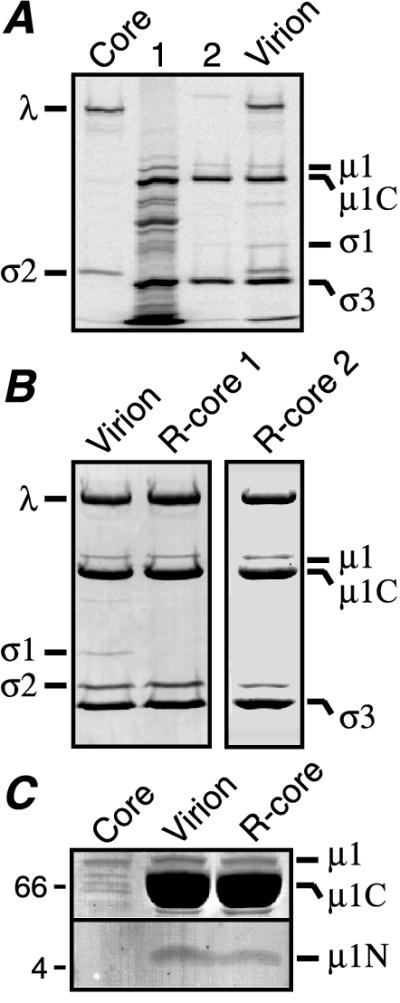
In vitro assembly of baculovirus-expressed μ1 and ς3 proteins onto reovirus cores. (A) [35S]methionine/cysteine-labeled cytoplasmic extract prepared from 7.5 × 107 Tn High Five cells infected with a μ1- and ς3-expressing recombinant baculovirus was incubated with unlabeled reovirus cores (5 × 1012 particles). The resulting r-core particles were purified on two successive CsCl gradients. Both the extract (lane 1) and the purified r-cores (lane 2) were subjected to SDS-PAGE and phosphor imaging. 35S-labeled cores (Core) and virions (Virion) were included as markers to indicate positions of the viral proteins. (B) Two independent preparations of purified r-cores (R-core 1 and R-core 2), generated as described above except by using unlabeled insect cell extract, were examined by SDS-PAGE and Coomassie blue staining. A lane of purified virions was included for comparison. (C) Purified cores, virions, and r-cores were examined by tricine-SDS-PAGE (44) on a 10 to 16% acrylamide gradient gel and Coomassie blue staining. Only those portions of the gel containing μ1 and its fragments are shown. Positions of molecular weight markers are indicated by Mr (103) at the left.
FIG. 5.
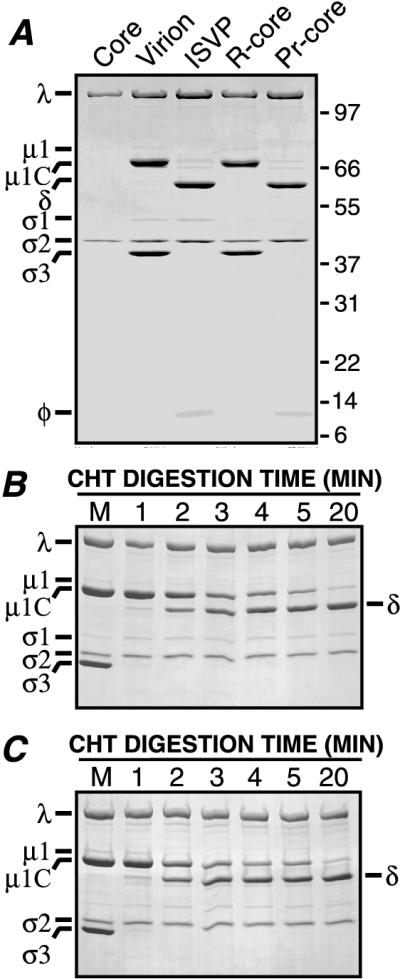
Proteolytic processing of r-cores to pr-cores by CHT. Viral proteins were visualized by SDS-PAGE and Coomassie blue staining. (A) Purified virions or r-cores were treated with CHT for 1 h, and the resulting ISVPs or pr-cores were purified by centrifugation through CsCl density gradients. Viral proteins were resolved on a 5 to 20% acrylamide gradient gel (39). Positions of molecular weight markers are indicated by Mr (103) at the right. A lane of purified cores was included for comparison. (B) Purified virions were treated with CHT for specified times. A marker lane of untreated virions was included for comparison (M). (C) Same as panel B except that purified r-cores were used instead of virions.
FIG. 6.
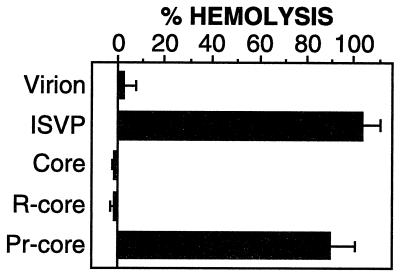
Capacity of pr-cores to induce lysis of RBCs in vitro. Bovine calf RBCs (final concentration, 3% [vol/vol]; Colorado Serum Co., Denver, Colo.) were incubated with purified virions, r-cores, cores, or CHT treatment mixtures containing ISVPs or pr-cores (3 × 1012 particles/ml) at 37°C for 40 min in the presence of CsCl (200 mM). Reactions were terminated by removal onto ice, and RBCs were pelleted by centrifugation at 300 × g for 5 min. The extent of hemolysis in each reaction was determined by measuring A415 of the supernatant and was expressed as a percentage (hemolysis by distilled water = 100%). Each bar represents the mean ± SD from three trials.
FIG. 8.
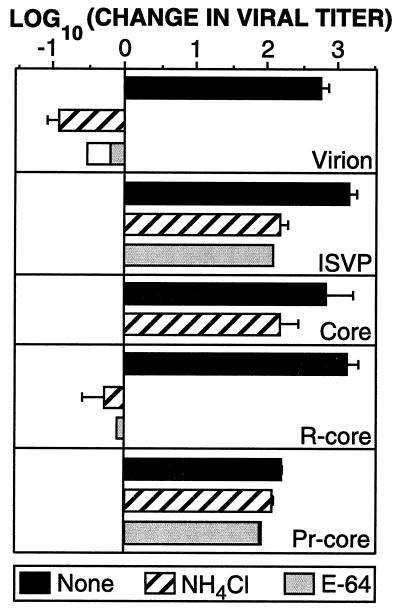
Capacity of r-cores to replicate in the absence and presence of inhibitors of viral infectivity. Purified virions, cores, r-cores, or CHT treatment mixtures containing ISVPs or pr-cores were used to infect L-cell monolayers at an MOI of 0.01 PFU/cell in the absence of inhibitor (solid bars) or presence of 20 mM NH4Cl (hatched bars) or 300 μM E-64 (gray bars) in the growth medium. Infectious titers at times 0 and 24 h were measured by plaque assay. Each bar represents the mean [log10 (PFU/ml)t = 24 h − log10 (PFU/ml)t = 0] ± SD derived from three independent experiments for no inhibitor and NH4Cl treatments. Overlaid bars represent values from two experiments for E-64 treatment.
TABLE 1.
Quantitation of μ1C and ς3 proteins in r-coresa
| Particle type | Prepn | λ/μ1Cb | ς3/μ1Cb |
|---|---|---|---|
| Virion | 1 | 0.67 ± 0.02 | 0.78 ± 0.03 |
| 2 | 0.59 ± 0.03 | 0.77 ± 0.06 | |
| r-core | 1 | 0.67 ± 0.03 | 0.76 ± 0.02 |
| 2c | 0.68 ± 0.03 | 0.81 ± 0.03 | |
| 3 | 0.65 ± 0.03 | 0.72 ± 0.06 | |
| r-core(μ1-581D) | 1 | 0.60 ± 0.06 | 0.73 ± 0.06 |
See text for descriptions of r-cores.
Volume-based intensities of the λ, μ1C, and ς3 protein bands were determined by densitometry of Coomassie blue-stained SDS-polyacrylamide gels on which the proteins from purified particles were separated. Band ratios were calculated from three different lanes for each particle preparation. Mean ± SD of the three values is reported.
TABLE 2.
Infectivity of purified r-coresa
| Particle type | Prepnb | P/PFUc | Relative P/PFUd |
|---|---|---|---|
| Core | A | 1.1 × 109 | 1.0 |
| B | 5.0 × 108 | 1.0 | |
| r-core | A-1 | 2.1 × 106 | 530 |
| A-2 | 2.3 × 106 | 480 | |
| B-1 | 1.4 × 106 | 360 | |
| B-2e | 1.1 × 106 | 450 | |
| r-core(μ1-581D) | B-1 | 1.9 × 106 | 260 |
| Virion | 1 | 150 | 7.3 × 106 |
| 2 | 110 | 9.7 × 106 |
See text for descriptions of r-cores.
Each core preparation is designated with a capital letter. Each r-core preparation is designated with a letter, representing the core preparation from which it was derived, followed by the preparation number.
Particle (P) per PFU values for each preparation were obtained by dividing P per milliliter by PFU per milliliter and are presented as an average of three determinations (SD < 0.10 log10 unit for all values). Concentrations of virion and core preparations were estimated by A260 (13, 51). Concentrations of r-core preparations were determined by densitometry of Coomassie blue-stained SDS-polyacrylamide gels compared with virion standards. Viral infectivity was measured by plaque assay (18).
Defined as the ratio of P/PFU value of a preparation of cores to P/PFU value of a matching core or r-core preparation. Relative P/PFU values of virion preparations were calculated by using the PFU-per-milliliter value of core preparation A.
r-cores and pr-cores.
To prepare r-cores, insect cell cytoplasmic extracts containing both μ1 and ς3 were incubated with purified T1L cores at a ratio of 1.5 × 107 cell equivalents per 1012 cores for 2 h at 37°C. Reaction mixtures were then loaded atop 14-ml step gradients, each containing a preformed CsCl gradient (ρ = 1.25 to 1.55 g/cm3, 12 ml) and a 2-ml sucrose cushion (20% [wt/vol]), and gradients were centrifuged for 2 to 16 h in a Beckman SW28 rotor at 25,000 rpm and 5°C. r-cores were recovered as an optically homogeneous band and were further purified by loading onto a preformed CsCl gradient (ρ = 1.25 to 1.45 g/cm3, 4 ml) and centrifugation overnight in a Beckman SW50.1 rotor at 40,000 rpm and 5°C. The harvested particles were dialyzed extensively against virion buffer. Particle concentrations were estimated by densitometry of Coomassie blue-stained SDS-polyacrylamide gels (see below) loaded with a dilution series of purified T1L virions. To prepare proteinase-treated r-cores (pr-cores), r-cores were digested with CHT and purified in the manner described for ISVPs (39).
SDS-PAGE and densitometry.
Samples were subjected to SDS-PAGE as described elsewhere (9). Proteins were visualized by using Coomassie brilliant blue R-250 (Sigma). Gels loaded with radiolabeled proteins were dried onto filter paper and visualized by phosphor imaging (Molecular Dynamics, Sunnyvale, Calif.). Ten percent acrylamide gels were used unless otherwise specified. To estimate relative amounts of proteins from Coomassie blue-stained gels, wet gels were scanned with a laser densitometer (Molecular Dynamics), and volume-based intensities of the protein bands were determined by using the ImageQuant program (Molecular Dynamics). A calibration curve of intensity of the λ protein band versus particle concentration estimated by A260 was generated with a dilution series of purified T1L virions, and the concentration of each purified r-core and pr-core preparation was determined from its λ band intensity. The λ band intensities of these samples were always within the range of values provided by dilutions of purified virions, and at least two lanes of each sample were used in estimating its concentration.
Cryoelectron microscopy and 3-D image reconstructions.
Sample preparation, high-resolution scanning cryoelectron microscopy (cryo-SEM), image capture, and processing for publication were done as described elsewhere (8). For high-resolution transmission cryoelectron microscopy (cryo-TEM), purified particles were embedded in vitreous ice, and micrographs were recorded at a nominal magnification of ×38,000, using standard low-dose procedures on a Philips CM200 microscope (3); 228 particles were selected from four micrographs (defocus values of 1.6, 2.0, 2.2, and 2.3 μm) and analyzed with image-processing techniques for icosahedral particles (2, 17). The particle orientations were evenly distributed throughout the asymmetric unit, as evidenced by all inverse eigenvalues being <1.0 and at least 99% being <0.1 (17). The final 3-D reconstruction was calculated at 24-Å resolution.
Buoyant density measurements.
The buoyant density of r-cores was determined by CsCl equilibrium density centrifugation. Purified 35S-labeled r-cores (4,000 cpm) were loaded onto a preformed CsCl gradient (ρ = 1.25 to 1.46 g/cm3, 12 ml), and centrifuged in a Beckman SW41 rotor for 16 to 20 h at 25,000 rpm and 5°C. Gradients were fractionated, and the amount of radioactivity in each fraction was measured with a Beckman LS-233 scintillation counter. The density of each fraction was determined from its refractive index. A single symmetrical peak of radioactivity was obtained, and the density at the apex of this peak was taken as the buoyant density of r-core particles.
Proteinase treatment mixtures containing ISVPs or pr-cores.
The ISVPs and pr-cores used in the hemolysis, endpoint infectivity, and neutralization experiments (see below) were not purified on gradients. Instead, proteinase treatment mixtures containing ISVPs or pr-cores were used as follows. Purified virions or r-cores (1 × 1012 to 5 × 1012 particles/ml) were incubated with the specified proteinase (CHT or Nα-p-tosyl-l-sulfonyl phenylalanyl chloromethyl ketone [TPCK]-treated TRY [Sigma]) (200 μg/ml) at 37°C. Unless indicated otherwise, digestion reactions were terminated at 20 min by removal onto ice and addition of PMSF (1 to 5 mM) (for CHT) or TLCK (0.70 mM) (for TRY). After further incubation on ice (≥20 min), proteinase treatment mixtures were used immediately. Proteinase treatment mixtures containing pr-cores(μ1-581D) or pr-cores(μ1-581Y) in Fig. 9 were generated as follows. Sequencing-grade CHT (Boehringer Mannheim) was treated with TLCK (150 μM) for 20 min at room temperature to inactivate any residual TRY. r-cores (4 × 1012 particles/ml) were incubated with this TLCK-treated CHT or with TPCK-treated TRY for 10 min at 37°C, and digestion reactions were terminated by removal onto ice and addition of PMSF or TLCK, respectively.
FIG. 9.
Generation of pr-cores(μ1-581D) and measurement of their capacity to induce lysis of RBCs and to initiate infection. (A) Purified r-cores(μ1-581D) or r-cores(μ1-581Y) were incubated with TLCK-treated CHT or TPCK-treated TRY to generate pr-cores(μ1-581D) or pr-cores(μ1-581Y), respectively. Viral proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining. A marker lane of untreated r-cores(μ1-581D) or r-cores(μ1-581Y) was loaded onto each gel for comparison (M). (B) Purified r-cores(μ1-581D) or CHT treatment mixtures containing pr-cores(μ1-581D) or pr-cores(μ1-581Y) (2 × 1012 particles/ml) were incubated with bovine calf RBCs at 37°C for 30 min in the presence of CsCl, and the extent of hemolysis induced by each virus preparation was measured as described for Fig. 6. Each bar represents the mean ± SD from three trials except in the case of r-cores(μ1-581D) where two trials were performed, and the average of both is shown. The extent of hemolysis was lower than that in Fig. 6 because a lower concentration of particles was used. (C) The capacity of purified r-cores(μ1-581D) or CHT treatment mixtures containing pr-cores(μ1-581D) to replicate in L cells over a 24 h-period in the absence (solid bars) or presence (hatched bars) of 20 mM NH4Cl was determined from three independent experiments as described for Fig. 8. An MOI of 0.01 PFU/cell was used.
Hemolysis experiments.
Hemolysis experiments were performed as described elsewhere (9).
Endpoint experiments for infectivity.
Growth of reovirus particles over a 24-h period in L cells was monitored in the absence or presence of NH4Cl (20 mM) or E-64 (300 μM) essentially as described previously (9). Briefly, L-cell monolayers in 2-dram vials (5 × 105 per vial) were infected with virus (100 μl; MOI = 0.01 PFU/cell) at 4°C, and attachment was allowed to proceed for 1 h at 4°C. At this time, the inoculum was removed, and growth medium (500 μl) containing no inhibitor, NH4Cl, or E-64 was added. Vials were frozen at −80°C (time zero samples) or placed at 37°C (24-h samples). At 24 h, all samples were freeze-thawed twice and titrated by plaque assay. Viral growth after 24 h was measured as log10 (PFU/ml)t = 24 h − log10 (PFU/ml)t = 0 ± standard deviation (SD).
Neutralization experiments.
A core-specific rabbit antiserum was generated by Simon Noble, using heat-inactivated reovirus T1L cores (43). The capacity of T1L ς1-specific monoclonal antibody (MAb) 5C6 (56) or the core-specific antiserum to neutralize infections of L cells by reovirus particles was determined as follows. Virus (1,000 PFU/ml) was incubated with antibody (1 μg/ml for 5C6, 1/1,000 dilution for the core-specific antiserum) in phosphate-buffered saline supplemented with 2 mM MgCl2 at 37°C for 1 h. The virus-antibody mixture (100 μl) was then added to an L-cell monolayer and allowed to attach for 1 h at room temperature, after which a standard plaque assay protocol was followed (18). The extent of neutralization was expressed as the percent plaque survival from the ratio of the number of PFU obtained in the presence of antibody to the number of PFU obtained in its absence.
Site-directed mutagenesis.
Plasmid pBKS-M2L was the template for site-directed mutagenesis using the QuikChange protocol (Stratagene). The complementary mutagenic primers 5′-TCAACTCGAGACTGGGGATGGTGTACGGATATT-3′ and 5′-AATATCCGTACACCATCCCCAGTCTCGAGTTG A-3′ (nucleotide changes underlined) were used. The missense mutation changed tyrosine 581 to aspartate, while the silent mutation created an XhoI restriction site. Mutant clones were generated and identified by screening for the addition of an XhoI site at the appropriate location in the plasmid. The presence of the desired mutations and the absence of second-site mutations was confirmed by sequencing putative mutant clones from nucleotides 1090 to 2116. The MluI-AgeI restriction fragment (nucleotides 1124 to 1875) containing the mutations was subcloned into pFbD-M2L-S4L for production of recombinant baculovirus.
Computer software.
SDS-polyacrylamide gels were scaled uniformly and adjusted for optimal brightness and contrast in Photoshop 4.0 (Adobe Systems, San Jose, Calif.). All figures were produced in Illustrator 7.0 (Adobe).
RESULTS
Stoichiometric assembly of baculovirus-expressed μ1 and ς3 proteins onto reovirus cores.
A cytoplasmic extract of [35S]methionine/cysteine-labeled μ1 and ς3 was prepared from insect cells infected with a recombinant baculovirus designed to express both proteins (see Materials and Methods). This extract was incubated with unlabeled reovirus cores at 37°C for 2 h, and reovirus particles in the reaction mixture were separated from unbound proteins by banding in two successive CsCl density gradients. SDS-PAGE and phosphor imaging of the banded material revealed the presence of polypeptides that comigrated with proteins μ1, μ1C, and ς3 (Fig. 1A). Coomassie blue staining of the same gels revealed that the banded material also contained reovirus core proteins (data not shown). Other 35S-labeled proteins from the insect cell extract were not associated with the particle band (Fig. 1A), suggesting that μ1/μ1C and ς3 had bound specifically to cores. In contrast to the results obtained with extracts containing both μ1/μ1C and ς3, neither μ1 nor ς3 bound to cores upon incubation of these particles with extracts containing either protein alone (10, 23).
Core particles with bound μ1/μ1C and ς3, generated as described above except by using nonradiolabeled proteins, contained approximately stoichiometric amounts of μ1C and ς3 according to quantitations from SDS-polyacrylamide gels (Fig. 1B; Table 1). We termed these particles r-cores. In addition, r-cores and virions contained similar amounts of both full-length μ1 (Fig. 1B and C) and its N-terminal fragment μ1N (Fig. 1C). Cleavage of μ1 to μ1N and μ1C is a putatively autoproteolytic step that requires μ1-ς3 complex formation (30, 41, 54) and may play a role in assembly of μ1 and ς3 onto reovirus particles (41, 49). Thus, r-cores closely resembled native virions in protein composition except that r-cores lacked the cell attachment protein ς1 (Fig. 1B) because it is absent from reovirus cores (Fig. 1A) and was not added back exogenously.
To provide additional evidence that r-cores and virions contained similar amounts of μ1 and ς3, we measured the buoyant density of purified r-cores by centrifugation through CsCl density gradients. As expected, the density of r-cores (1.363 ± 0.004 g/cm3; n = 3) was very similar to that of virions (1.36 g/cm3) and lower than that of cores (1.43 to 1.44 g/cm3) (40, 51). Addition of a vast excess of insect cell extract to cores did not produce particles exceeding virions in their levels of μ1C and ς3 or possessing a density lower than that of virions (data not shown). However, r-core-like particles containing substoichiometric amounts of μ1C and ς3 and possessing a buoyant density between those of r-cores and cores could be generated by reducing the amount of extract added to cores (10).
While the r-cores described above were generated with T1L cores, T1L μ1, and T1L ς3, large numbers of r-cores have also been produced from type 3 Dearing (T3D) cores, T1L μ1, and T1L ς3 as well as from T1L cores, T3D μ1, and T1L ς3 (16). Thus, the core-recoating phenomenon is generalizable to at least one other strain of mammalian reovirus and is likely to permit in vitro reassortment of cores and μ1 and ς3 proteins from various reovirus strains for use in genetic studies.
r-cores resemble virions in outer-capsid structure as determined by both cryo-SEM and cryo-TEM and image reconstruction.
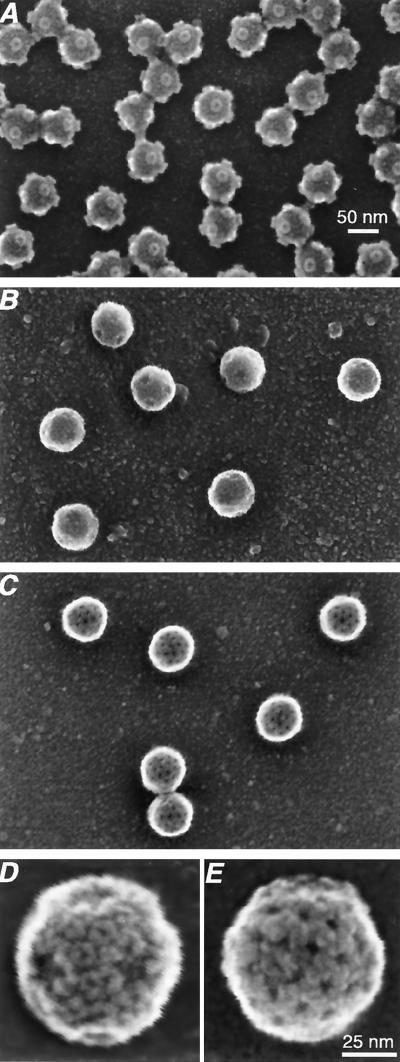
To determine whether the stoichiometric assembly of baculovirus-expressed μ1 and ς3 onto cores regenerated an authentic outer capsid, purified preparations of virions and r-cores were subjected to cryo-SEM (8, 24). When observed at low magnification, r-cores (Fig. 2B) appeared as monolayers of discrete, generally spherical particles. These particles had a distinct appearance from cores (Fig. 2A) which are smaller in diameter and possess protruding turrets composed of protein λ2 at the fivefold axes (8). Images of individual r-cores at higher magnification revealed ring-like projections distributed over most of the particle surface, including partial rings adjacent to the fivefold axes (Fig. 2B and D). Similar features were seen in cryo-SEM images of virions and attributed to complete and partial hexamers of protein ς3 (8). These findings suggest that r-cores are a homogeneous population of particles that possess a ς3 layer very similar to that in virions.
FIG. 2.
Cryo-SEM of purified r-cores and pr-cores. Low-magnification (×150,000) images of cores (A), r-cores (B), and pr-cores (C) and high-magnification (×500,000) images of r-cores (D) and pr-cores (E) are shown. The particles in panels D and E are oriented near an axis of twofold symmetry. The scale bar in panel A applies to panels A to C, while that in panel E applies to panels D to E.
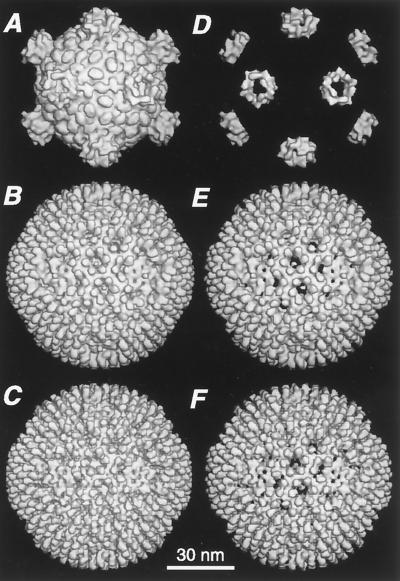
r-cores were visualized at higher resolution in three dimensions by using cryo-TEM techniques and image reconstruction procedures for icosahedral particles. The 3-D reconstruction calculated to 24 Å revealed that r-cores (Fig. 3C and F) closely resembled virions (Fig. 3B and E) in outer-capsid structure. The 600 finger-like projections at the surface of r-cores and virions represent 600 molecules of ς3 (15). In both r-cores and virions, these molecules are arranged into complete hexamers surrounding each P3 channel and partial hexamers (four subunits) surrounding each P2 channel according to the rules of T=13(laevo) icosahedral symmetry (35). One ς3 molecule per incomplete hexamer contacts the λ2 subunit(s) projecting into the P2 channel in both particle types. In addition, radial sections demonstrated the similar organization of the ς3 and μ1 layers in r-cores and virions (data not shown). The essentially identical appearance and organization of outer-capsid features in both cryo-SEM images of individual r-cores and virions and 3-D reconstructions calculated from cryo-TEM images of multiple particles supports the conclusion that r-cores are structurally similar to virions.
FIG. 3.
Image reconstructions of r-cores from cryo-TEM images. The reconstruction of r-cores (C and F; 24 Å) is compared with previously reported reconstructions of cores (A and D; 32 Å) and virions (B and E; 28 Å) (15). (A to C) Surface-shaded views down a twofold axis of symmetry for each particle. (D to F) Same views as in panels A to C except that the reconstructions were radially cropped to remove all densities below 321 Å, thereby isolating features attributable to outer-capsid proteins μ1, ς3, ς1, and λ2 in the display.
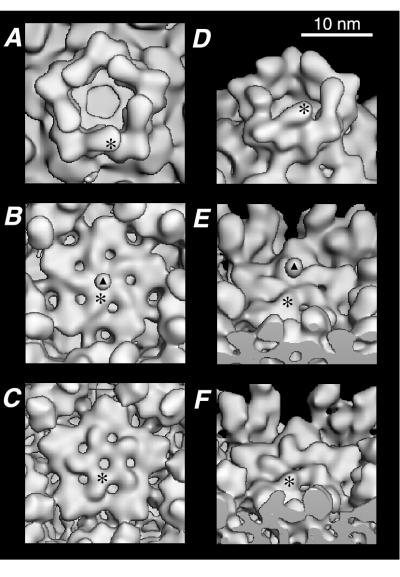
r-cores and virions contain similar conformational states of the pentameric λ2 turret.
Each pentameric turret formed by five subunits of protein λ2 undergoes a dramatic conformational rearrangement upon conversion of virions or ISVPs to cores in vitro (15). In the virion and ISVP, the turret is capped by a five-pointed “flower” that consists of five petal-like structures, each contributed by a λ2 monomer (Fig. 4B and E). The ς1 fiber, apparent as a bead-like density suspended above each fivefold axis, is thought to make contact with λ2 at the center of the flower (Fig. 4B and E). During conversion of ISVPs to cores, each petal moves upward and clockwise about a putative hinge in λ2, opening a central channel through which reovirus mRNAs are thought to be extruded in the transcribing core (15, 58) (Fig. 4A, B, D, and E). According to the cryo-TEM-derived image reconstruction of r-cores (Fig. 4C and F), assembly of μ1 and ς3 onto cores in vitro appears to have induced reversal of this conformational change, with each λ2 petal rotating downward and counterclockwise to close the central channel and to generate a structure that closely resembles the central flower present in virions. One difference between virions and r-cores in the structure of the λ2 turret is the absence in r-cores of the density attributed to ς1, consistent with the fact that these particles lack ς1 (Fig. 1). Another difference is the presence of a small pore (∼25 Å in diameter) at the center of the turret in r-cores but not virions (Fig. 4B, C, E, and F). We propose that this pore is occupied by the base of the ς1 fiber in virions. The significance of other minor differences between the virion and r-core reconstructions, including the slightly higher radial projection of the petals forming the central flower in r-cores, remains unclear.
FIG. 4.
Close-up surface-shaded views of the λ2 turret in r-cores. The reconstructions of r-cores (C and F), cores (A and D), and virions (B and E) are shown in close-up for a representative λ2 turret. Asterisks indicate one of the five petal-like elements that form the central λ2 flower in virions and r-cores and then move outward and upward during conversion of virions to cores. Triangles in panels B and E indicate the drop-like density in virions attributed to ς1 that is absent in the r-core reconstruction. (A to C) Top views. (D to F) Views generated by tilting images by 45° about the horizontal axis. Images were sectioned so as to permit a clear view of the λ2 turret.
r-cores undergo conversion to ISVP-like particles upon proteinase treatment in vitro.
Virions can undergo stepwise disassembly of the outer capsid to generate ISVPs when treated with CHT or TRY under defined conditions in vitro (5, 27, 39). During such treatments, virion-bound ς3 protein is degraded, while μ1/μ1C is cleaved into stable fragments μ1δ/δ (63/59 kDa; N-terminal portion of μ1/μ1C) and φ (13 kDa; C-terminal portion of μ1/μ1C) (Fig. 5A) (39). Treatment of r-cores with CHT (Fig. 5A) or TRY (see Fig. 9A) generated particles termed pr-cores (proteinase-treated r-cores) that resembled ISVPs in protein composition. In pr-cores as in ISVPs (39), the μ1δ/δ and φ fragments remained particle bound after purification in CsCl gradients (Fig. 5A). Virions (4, 32, 39) and r-cores underwent proteolytic processing of ς3 and μ1/μ1C with similar kinetics, in that the former protein was rapidly degraded while the latter was cleaved more slowly at the δ:φ junction (Fig. 5B and C). These results strongly suggest that the conformations of μ1 and ς3 in r-cores and virions are similar.
To provide evidence that pr-cores resemble ISVPs not only in protein composition but also in particle morphology, pr-cores and ISVPs were subjected to cryo-SEM. Low-resolution images of pr-cores (Fig. 2C) revealed fields of discrete particles with a uniform spherical appearance similar to that of ISVPs (8) but distinct from that of r-cores (Fig. 2B) or cores (Fig. 2A). Examination of individual pr-core particles at higher magnification (Fig. 2E) revealed that pr-cores lacked the rings of protein ς3 present in r-cores (Fig. 2B and D). Instead, the surface of pr-cores was similar to that of ISVPs (8), including a linear meshwork percolated by large channels. This meshwork present at the ISVP surface has been attributed to the icosahedral lattice formed by μ1 (8, 15). The close correspondence in the surface features of pr-cores and ISVPs strongly suggests that r-cores and pr-cores contain a native μ1 lattice.
Capacity of pr-cores to lyse RBCs.
The unique capacity of ISVPs to permeabilize lipid bilayers in vitro (9, 21, 32, 38, 55) is likely related to their role in penetration of host cell membrane(s) during reovirus infection. Since pr-cores resembled ISVPs in protein composition (Fig. 5) and particle morphology (Fig. 2), we surmised that they too might possess the capacity to permeabilize lipid bilayers. To test this hypothesis, we examined the capacity of purified virions, cores, r-cores, and CHT treatment mixtures containing ISVPs or pr-cores to lyse RBCs in vitro. In the presence of NaCl, none of these particle types lysed RBCs (data not shown) (9, 38). However, when NaCl was replaced by CsCl, which is thought to accelerate conformational change(s) in viral proteins required for membrane permeabilization (9, 10, 55), ISVPs but not virions or cores were induced to mediate hemolysis, as expected (Fig. 6) (9, 38). The ISVP-like pr-cores were also induced to mediate hemolysis, but the virion-like r-cores were not (Fig. 6). These results indicate that the in vitro assembly of μ1 and ς3 onto cores restores the protein machinery required for reovirus particles to interact with and permeabilize membranes in vitro. This machinery is inactive in r-cores and virions but can be activated to permeabilize membranes following limited in vitro proteolysis of these particles to pr-cores and ISVPs, respectively.
Infectivity of r-cores.
Since r-cores behaved like virions in their capacity to permeabilize membranes upon proteolytic conversion to ISVP-like particles (pr-cores), we speculated that r-cores may have a greater capacity than cores to penetrate host cell membrane(s) and initiate productive infection. To test this possibility, we measured the specific infectivities of purified virions, r-cores, and cores in murine L cells (Table 2). r-cores were 200- to 500-fold more infectious than cores on a per-particle basis, indicating that the outer-capsid proteins μ1 and ς3 added to cores play functional roles in reovirus infections of L cells. However, r-cores were still only 10−4 times as infectious as virions. We speculate that the low specific infectivity of r-cores at least partly reflects that these particles lack protein ς1 (Fig. 1, 3, 4, and 5) and so attach to cells very inefficiently (10).
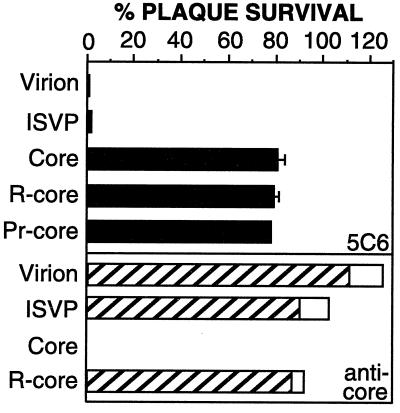
Because the core preparations used in this study were derived from virions by proteolytic digestion in vitro, we were concerned that the infectivity of cores, and r-cores generated from them, might be attributable to virions or ISVPs present as contaminants. To investigate this possibility, we measured the capacity of T1L ς1-specific MAb 5C6 (56) or a core-specific antiserum (43) to neutralize purified preparations of virions, cores, r-cores, and CHT treatment mixtures containing ISVPs or pr-cores (Fig. 7). In agreement with previous findings (56), 5C6 neutralized the infectivity of virions and ISVPs in L cells. In contrast, core, r-core, and pr-core preparations were not neutralized by 5C6, suggesting that the infectivity of these preparations arises from the ς1-independent infection of L cells by each of these types of particles. In support of the findings with 5C6, preparations of cores, but not virions or ISVPs, were neutralized by the core-specific antiserum (Fig. 7). Thus, the infectivity of cores (and r-cores derived from them) is not explained by residual virions or ISVPs that may be present in the core preparations. In addition, the capacity of r-cores to resist neutralization by the core-specific antiserum (Fig. 7) again distinguishes them from the cores from which they were derived.
FIG. 7.
Capacity of ς1-specific and core-specific antibodies to neutralize infectivity of cores and r-cores. Purified virions, cores, r-cores, or CHT treatment mixtures containing ISVPs or pr-cores were incubated with a ς1-specific MAb (5C6), a core-specific antiserum (anticore), or phosphate-buffered saline. Virus-antibody mixtures were used to infect L-cell monolayers, and infectious titers were measured by plaque assay. Neutralization of infectivity was expressed as the percentage of plaques remaining after antibody treatment. Each bar represents the mean plaque survival ± SD derived from three independent experiments for 5C6 treatment. Overlaid bars represent values from two experiments with the core-specific antiserum.
Requirement for lysosomal acidic pH and cysteine-proteinase activity during infections with r-cores.
Virions cannot replicate in cultured cells treated with the weak base NH4Cl or the cysteine-proteinase inhibitor E-64 (1, 9, 53). One mechanism through which both compounds appear to act is the blockade of entry-related cleavages of ς3 and μ1/μ1C mediated by acid-dependent lysosomal cysteine proteinase(s). In contrast to their effects on infections by virions, NH4Cl and E-64 do not block infections by ISVPs because these particles have already undergone the cleavages of ς3 and μ1/μ1C in vitro (1, 9, 40, 53) (Fig. 8). To determine whether infections by r-cores also require proteolysis of ς3 and/or μ1/μ1C, we tested the capacity of purified virions, cores, r-cores, and CHT-treatment mixtures containing ISVPs or pr-cores to replicate in L cells in the absence or presence of NH4Cl and E-64. r-cores, like virions and unlike their precursor cores, could not replicate in the presence of either compound (Fig. 8). In contrast, CHT-generated pr-cores were, like ISVPs, resistant to the effects of either compound (Fig. 8). Thus, r-cores resemble virions in their requirement for proteolysis during entry. These findings, together with the shared capacity of ISVPs and pr-cores to permeabilize lipid bilayers in vitro, argue that virions and r-cores use a similar mechanism and pathway for penetration of host cell membranes, even though r-cores lack the ς1 cell attachment protein.
Mutant μ1/μ1C(581D) protein bound to r-cores is poorly cleaved at the δ:φ junction by CHT.
A recent study from one of our laboratories (9) used dpSVPs, novel subvirion particles lacking ς3 but containing μ1/μ1C that remains mostly uncleaved at the δ:φ junction, to provide evidence that cleavage at the δ:φ junction during viral entry is dispensable for reovirus infections of cultured cells. However, since dpSVPs were generated by treating virions with a combination of proteinase and detergent, we were concerned that the outer capsid in dpSVPs may have been altered by the detergent in a way that allowed these particles to circumvent a requirement for the δ:φ cleavage. To investigate this possibility, we used r-cores to decouple the ς3 and μ1/μ1C cleavages without the use of detergent. Because CHT cleaves μ1/μ1C after tyrosine 581 (39), we reasoned that mutating this residue in an appropriate fashion should significantly reduce cleavage at the δ:φ junction by CHT. Accordingly, we generated r-cores containing mutant μ1/μ1C(581D) (tyrosine 581→aspartate) and wild-type ς3. r-cores(μ1-581D) resembled r-cores(μ1-581Y) in protein composition (Table 1; Fig. 9A), specific infectivity (Table 2), and capacity to replicate in L cells (Fig. 9C), indicating that the Y581D mutation in μ1 neither interferes with stoichiometric assembly of μ1 and ς3 onto cores nor prevents those proteins from assuming conformations required for function.
When r-cores(μ1-581D) were incubated with TLCK-treated CHT in vitro, protein ς3 was rapidly degraded, but cleavage of protein μ1/μ1C(581D) was greatly reduced from that seen with μ1/μ1C(581Y) (Fig. 9A). In contrast, particle-bound μ1/μ1C(581D) protein was as susceptible as μ1/μ1C(581Y) to cleavage by TRY (Fig. 9A), a proteinase which was previously shown to cleave virion-bound μ1/μ1C after arginine 584 (39). The μ1δ/δ fragments in TRY-generated pr-cores(μ1-581Y) and pr-cores(μ1-581D) showed similar mobilities in SDS-polyacrylamide gels as ISVP-bound μ1δ/δ (data not shown), suggesting that TRY cleaves at a similar position in r-core-bound μ1/μ1C as well. Thus, the Y581D mutation in particle-bound μ1/μ1C specifically inhibits its cleavage by CHT, with no evident effect on the conformation of other sequences surrounding the δ:φ cleavage site.
Capacity of CHT-generated pr-cores(μ1-581D) to lyse RBCs and initiate infection.
To assess the role of the μ1/μ1C cleavage in membrane permeabilization by ISVP-like particles, we compared the capacities of CHT treatment mixtures containing pr-cores(μ1-581D) or pr-cores(μ1-581Y) to mediate hemolysis. The two particle types lysed RBCs to similar extents (Fig. 9B) even though they differed greatly in the extent to which the μ1/μ1C protein had been cleaved at the δ:φ junction by CHT (Fig. 9A). These results indicate that r-cores can permeabilize membranes in vitro even when nearly all of the 600 copies of μ1/μ1C per particle remain uncleaved at the δ:φ junction.
The capacity of CHT-generated pr-cores(μ1-581D) to lyse RBCs in vitro suggested that these particles might also be competent to penetrate host cell membranes when allowed to infect cells under conditions that block cleavage at the δ:φ junction in vivo. To test this hypothesis, we measured the growth of CHT-generated pr-cores(μ1-581D) in L cells, in the absence or presence of NH4Cl. Cells were treated with NH4Cl because this compound is known to abrogate the δ:φ cleavage during infections with dpSVPs (9). CHT-generated pr-cores(μ1-581D) (Fig. 9C), like pr-cores(μ1-581Y) (Fig. 8 and 9C), replicated to normal levels in the presence of NH4Cl. These findings are consistent with the capacity of dpSVPs to replicate in cultured cells in the presence of NH4Cl (9) and provide additional evidence that cleavage at the δ:φ junction during entry into cells is dispensable for productive infection.
DISCUSSION
Assembly of the reovirus outer capsid.
Events in reovirus-infected cells leading to assembly of outer-capsid proteins μ1, ς3, and ς1 into progeny virions remain to be fully elucidated. Biochemical and genetic data suggest (i) that proteins μ1 and ς3 form hetero-oligomeric complexes in solution (22, 30, 49, 54, 59) and (ii) that these complexes are later assembled onto core-like precursor particles (33, 37, 49, 57). Findings in this report support element ii of this model by providing the first direct evidence that μ1 and ς3 proteins can spontaneously assemble onto purified core particles in stoichiometric amounts. In addition, the findings that assembly of μ1 and ς3 onto cores was obtained with insect cell extracts containing both proteins (Fig. 1), but not either protein alone (10, 23), are consistent with element i, namely, that μ1 and ς3 must form complexes in solution before assembly onto core-like particles. The insect cell extracts used to recoat cores appear to have contained μ1-ς3 complexes, as judged by the cleavage of μ1 to fragments μ1N and μ1C within the extracts (Fig. 1) (54), and the cosedimentation of baculovirus-coexpressed μ1 and ς3 in sucrose gradients (10). In contrast, most μ1 remained uncleaved in insect cell extracts containing μ1 and not ς3 (10). Important assembly-related questions that may yet be addressed with r-cores include which amino acids in μ1-ς3 complexes and components of the core need to interact during assembly of the outer capsid.
Implications of the λ2 conformational change induced by assembly of μ1 and ς3 onto cores.
Assembly of μ1 and ς3 onto cores essentially reverses the dramatic conformational change in λ2 that occurs during generation of these cores from virions or ISVPs (Fig. 3 and 4). Protein μ1 makes extensive contacts with the walls of the λ2 turret in virions and ISVPs, and just as the loss of μ1-λ2 contacts during conversion of ISVPs to cores may induce the λ2 conformational change (14, 15), the regeneration of μ1-λ2 contacts during recoating of cores may trigger its reversal. Conversion of ISVPs to cores also results in activation of the virus-bound transcriptase (see references 42 and 45 for reviews). Since the central channel generated by the λ2 conformational change is thought to be necessary for the egress of nascent mRNA molecules from the transcribing core (15, 58), closure of this channel in r-cores might be expected to shut off core transcription. Preliminary results indicate that r-cores are inactive at transcription (16). Since μ1 and ς3 are thought to be assembled onto nascent core-like particles in the infected cell (see above), this inactivation of viral transcription upon μ1-ς3 assembly may play a role in viral maturation within cells.
In virions and ISVPs, the base of the ς1 cell attachment fiber appears to contact λ2 at the center of the flower that caps the turret (15) (Fig. 4). Thus, this part of λ2 is thought to contain the ς1-binding site, which is destroyed upon conversion of ISVPs to cores (15). Creation of a similar flower-like structure atop the λ2 turret upon assembly of μ1 and ς3 onto cores in vitro suggests that the ς1-binding site may have been restored in r-cores (Fig. 3 and 4). Indeed, preliminary results suggest that ς1 can be added to cores along with μ1 and ς3 (10). We speculate that subtle rearrangements in λ2 upon assembly of ς1 may eliminate the remaining small differences between virions and r-cores in the conformation of the turret.
Roles of outer-capsid proteins in reovirus-membrane interactions.
The outer-capsid protein μ1, which is the major surface protein of ISVPs, is an important determinant of the capacity of ISVPs to permeabilize membranes in vitro (9, 21, 32, 38, 55). In this study we showed that cores, which cannot lyse RBCs, gain the capacity to do so after addition of proteins μ1 and ς3 and subsequent removal of ς3 (Fig. 6). This new result most simply suggests that particle-bound μ1 protein itself mediates hemolysis. However, the outer-capsid proteins ς1 and λ2 are also exposed at the ISVP surface, and each of these proteins is either lost from the particle (ς1) (15, 18) or altered in conformation (λ2) (15) during conversion of ISVPs to cores with associated loss in hemolytic activity. Hence, ς1 and λ2 might also be involved in membrane permeabilization by ISVPs. The capacity of pr-cores to hemolyze RBCs in a ς1-independent fashion (Fig. 6) provides strong evidence that ς1 is in fact not required for membrane disruption in vitro. More experiments are needed to assess the precise roles of μ1 and λ2 in membrane permeabilization by ISVPs.
Infectivity of cores and r-cores in L cells.
While reovirus cores are generally described to be noninfectious, we found that they possess a very low but reproducible infectivity in L cells (∼10−7 times that of virions on a per-particle basis) (Table 2). Moreover, the results of neutralization experiments (Fig. 7) indicate that a large proportion of this infectivity cannot be attributed to virions or ISVPs that may be contaminating the core preparation. Thus, cores can apparently initiate infection of L cells, albeit inefficiently, even though they lack the outer-capsid proteins ς1 and μ1/ς3 involved in cell attachment and membrane penetration, respectively. Entry into cells by cores, unlike that by virions, does not require processing of particle-bound proteins by exogenous proteinases (Fig. 8). Addition of the reovirus membrane penetration proteins (μ1/ς3) to cores, to generate r-cores as in this study, not only increased their infectivity to a substantial degree (Table 2) but also restored the requirement for proteolytic processing during viral entry (Fig. 8). Preliminary results indicate that addition of the reovirus cell attachment protein (ς1) to cores as well provides an additional large enhancement to their infectivity (10). Thus, the outer-capsid proteins μ1/ς3 and ς1 appear not to be absolutely required for reovirus infectivity but rather enhance that of an intrinsically infectious core particle (36) by allowing it to reach the cytoplasm of a host cell more efficiently. The infectious nature of transcriptionally active subvirion particles lacking cell attachment and membrane penetration proteins has also been noted for members of two other genera of the Reoviridae family: rotaviruses (12) and orbiviruses (34).
r-cores as tools for studies of viral entry.
In a recent report from our laboratories (24), we described recoating genetics as a useful new approach for molecular-genetic studies of the roles of outer-capsid protein ς3 in reovirus entry. In that approach, the properties of ISVPs recoated in vitro with baculovirus-expressed ς3 proteins are analyzed to determine the effects of engineered alterations in ς3. For example, in the previous report, sequence determinants of a difference in ς3 cleavage between reoviruses T1L and T3D were localized to a C-terminal portion of ς3 by using ISVPs recoated with a panel of ς3 chimeras (24). In the current study, we extended this general approach to include recoating of cores with baculovirus-expressed μ1 and ς3 proteins and its use for molecular-genetic studies of the roles of μ1 in reovirus entry. New evidence in this study for the dispensability of the δ:φ cleavage in entry using cores recoated with a mutant μ1 protein (Fig. 9) provided a concrete demonstration of the expanded utility of recoating genetics. The generation and analysis of r-cores containing other mutant and chimeric forms of μ1 is a major focus of current studies (10, 16).
ACKNOWLEDGMENTS
We thank Simon Noble for generating the original T1L M2 clone used in this study as well as the core-specific antiserum. We thank S. J. Harrison for excellent technical support and the other members of our laboratories for helpful discussions. We also thank D. L. Farsetta, C. L. Luongo, and J. Jané-Valbuena for critical reviews of preliminary versions of the manuscript.
This work was supported by NIH grants AI39533 (to M.L.N.), GM33050 and AI35212 (to T.S.B.), and AI32139 (to L.A.S.); research grants from the Lucille P. Markey Charitable Trust (to the Wisconsin Institute for Molecular Virology and the Purdue Structural Biology Center); NIH research technology grant RR-00570 (to the Wisconsin Integrated Microscopy Resource); DARPA research contract MDA 972-97-1-0005 (to M.L.N.); and American Cancer Society research grant RPG-98-12701-MBC (to L.A.S.). K.C. was additionally supported by a predoctoral fellowship from the Howard Hughes Medical Institute. S.B.W. was additionally supported by the Purdue Biophysics Training Grant and a Purdue Research Foundation Fellowship. C.M.C. was additionally supported by a Wisconsin/Hilldale Research Fellowship and a Wisconsin Biochemistry Mary Shine Peterson Research Scholarship. M.L.N. received additional support as a Shaw Scientist from the Milwaukee Foundation.
REFERENCES
- 1.Baer G S, Dermody T S. Mutations in reovirus outer-capsid protein ς3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J Virol. 1997;71:4921–4928. doi: 10.1128/jvi.71.7.4921-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker T S, Cheng R H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 3.Baker T S, Drak J, Bina M. Reconstruction of the three-dimensional structure of simian virus 40 and visualization of the chromatin core. Proc Natl Acad Sci USA. 1988;85:422–426. doi: 10.1073/pnas.85.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodkin D K, Nibert M L, Fields B N. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J Virol. 1989;63:4676–4681. doi: 10.1128/jvi.63.11.4676-4681.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borsa J, Copps T P, Sargent M D, Long D G, Chapman J D. New intermediate subviral particles in the in vitro uncoating of reovirus virions by chymotrypsin. J Virol. 1973;11:552–564. doi: 10.1128/jvi.11.4.552-564.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borsa J, Long D G, Sargent M D, Copps T P, Chapman J D. Reovirus transcriptase activation in vitro: involvement of an endogenous uncoating activity in the second stage of the process. Intervirology. 1974;4:171–188. doi: 10.1159/000149856. [DOI] [PubMed] [Google Scholar]
- 7.Borsa J, Sargent M D, Lievaart P A, Copps T P. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology. 1981;111:191–200. doi: 10.1016/0042-6822(81)90664-4. [DOI] [PubMed] [Google Scholar]
- 8.Centonze V E, Chen Y, Severson T F, Borisy G G, Nibert M L. Visualization of individual reovirus particles by low-temperature, high-resolution scanning electron microscopy. J Struct Biol. 1995;115:215–225. doi: 10.1006/jsbi.1995.1046. [DOI] [PubMed] [Google Scholar]
- 9.Chandran K, Nibert M L. Protease cleavage of reovirus capsid protein μ1/μ1C is blocked by alkyl sulfate detergents, yielding a new type of infectious subvirion particle. J Virol. 1998;72:467–475. doi: 10.1128/jvi.72.1.467-475.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandran, K., and M. L. Nibert. Unpublished data.
- 11.Chang C T, Zweerink H J. Fate of parental reovirus in infected cell. Virology. 1971;46:544–555. doi: 10.1016/0042-6822(71)90058-4. [DOI] [PubMed] [Google Scholar]
- 12.Chen D, Ramig R F. Rescue of infectivity by in vitro transcapsidation of rotavirus single-shelled particles. Virology. 1993;192:422–429. doi: 10.1006/viro.1993.1057. [DOI] [PubMed] [Google Scholar]
- 13.Coombs K M. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology. 1998;243:218–228. doi: 10.1006/viro.1998.9061. [DOI] [PubMed] [Google Scholar]
- 14.Drayna D, Fields B N. Activation and characterization of the reovirus transcriptase: genetic analysis. J Virol. 1982;41:110–118. doi: 10.1128/jvi.41.1.110-118.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dryden K A, Wang G, Yeager M, Nibert M L, Coombs K M, Furlong D B, Fields B N, Baker T S. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farsetta, D. L., K. Chandran, and M. L. Nibert. Unpublished data.
- 17.Fuller S D, Butcher S J, Cheng R H, Baker T S. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J Struct Biol. 1996;116:45–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 18.Furlong D B, Nibert M L, Fields B N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillian A L, Nibert M L. Amino terminus of reovirus nonstructural protein ςNS is important for ssRNA binding and nucleoprotein complex formation. Virology. 1998;240:1–11. doi: 10.1006/viro.1997.8905. [DOI] [PubMed] [Google Scholar]
- 20.Hazelton P R, Coombs K C. The reovirus mutant tsA279 has temperature-sensitive lesions in the L2 and M2 genes: the M2 gene is associated with decreased viral protein production and blockade in transmembrane transport. Virology. 1995;207:46–58. doi: 10.1006/viro.1995.1050. [DOI] [PubMed] [Google Scholar]
- 21.Hooper J W, Fields B N. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J Virol. 1996;70:459–467. doi: 10.1128/jvi.70.1.459-467.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huismans H, Joklik W K. Reovirus-coded polypeptides in infected cells: isolation of two native monomeric polypeptides with affinity for single-stranded and double-stranded RNA, respectively. Virology. 1976;70:411–424. doi: 10.1016/0042-6822(76)90282-8. [DOI] [PubMed] [Google Scholar]
- 23.Jané-Valbuena, J., and M. L. Nibert. Unpublished data.
- 24.Jané-Valbuena J, Nibert M L, Spencer S M, Walker S B, Baker T S, Chen Y, Centonze V E, Schiff L A. Reovirus virion-like particles obtained by recoating infectious subvirion particles with baculovirus-expressed ς3 protein: an approach for analyzing ς3 functions during viral entry. J Virol. 1999;73:2963–2973. doi: 10.1128/jvi.73.4.2963-2973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayasuriya A K A. Ph.D. thesis. Cambridge, Mass: Harvard University; 1991. [Google Scholar]
- 26.Joklik W K. Recent progress in reovirus research. Annu Rev Genet. 1985;19:537–575. doi: 10.1146/annurev.ge.19.120185.002541. [DOI] [PubMed] [Google Scholar]
- 27.Joklik W K. Studies on the effect of chymotrypsin on reovirions. Virology. 1972;49:700–715. doi: 10.1016/0042-6822(72)90527-2. [DOI] [PubMed] [Google Scholar]
- 28.Joklik W K, Roner M R. What reassorts when reovirus genome segments reassort? J Biol Chem. 1995;270:4181–4184. doi: 10.1074/jbc.270.9.4181. [DOI] [PubMed] [Google Scholar]
- 29.Kothandaraman S, Hebert M C, Raines R T, Nibert M L. No role for pepstatin A-sensitive acidic proteinases in reovirus infections of L or MDCK cells. Virology. 1998;251:264–272. doi: 10.1006/viro.1998.9434. [DOI] [PubMed] [Google Scholar]
- 30.Lee P W, Hayes E C, Joklik W K. Characterization of anti-reovirus immunoglobulins secreted by cloned hybridoma cell lines. Virology. 1981;108:134–146. doi: 10.1016/0042-6822(81)90533-x. [DOI] [PubMed] [Google Scholar]
- 31.Lee P W, Hayes E C, Joklik W K. Protein sigma 1 is the reovirus cell attachment protein. Virology. 1981;108:156–163. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- 32.Lucia-Jandris P, Hooper J W, Fields B N. Reovirus M2 gene is associated with chromium release from mouse L cells. J Virol. 1993;67:5339–5345. doi: 10.1128/jvi.67.9.5339-5345.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPhillips T H, Ramig R F. Extragenic suppression of temperature-sensitive phenotype in reovirus: mapping suppressor mutations. Virology. 1984;135:428–439. doi: 10.1016/0042-6822(84)90198-3. [DOI] [PubMed] [Google Scholar]
- 34.Mertens P P, Burroughs J N, Walton A, Wellby M P, Fu H, O’Hara R S, Brookes S M, Mellor P S. Enhanced infectivity of modified bluetongue virus particles for two insect cell lines and for two Culicoides species. Virology. 1996;217:582–593. doi: 10.1006/viro.1996.0153. [DOI] [PubMed] [Google Scholar]
- 35.Metcalf P, Cyrklaff M, Adrian M. The three-dimensional structure of reovirus obtained by cryo-electron microscopy. EMBO J. 1991;10:3129–3136. doi: 10.1002/j.1460-2075.1991.tb04874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moody M D, Joklik W K. The function of reovirus proteins during the reovirus multiplication cycle: analysis using monoreassortants. Virology. 1989;173:437–446. doi: 10.1016/0042-6822(89)90556-4. [DOI] [PubMed] [Google Scholar]
- 37.Morgan E M, Zweerink H J. Reovirus morphogenesis. Corelike particles in cells infected at 39 degrees with wild-type reovirus and temperature-sensitive mutants of groups B and G. Virology. 1974;59:556–565. doi: 10.1016/0042-6822(74)90465-6. [DOI] [PubMed] [Google Scholar]
- 38.Nibert M L. Ph.D. thesis. Cambridge, Mass: Harvard University; 1993. [Google Scholar]
- 39.Nibert M L, Fields B N. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J Virol. 1992;66:6408–6418. doi: 10.1128/jvi.66.11.6408-6418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nibert M L, Furlong D B, Fields B N. Mechanisms of viral pathogenesis. Distinct forms of reoviruses and their roles during replication in cells and host. J Clin Investig. 1991;88:727–734. doi: 10.1172/JCI115369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nibert M L, Schiff L A, Fields B N. Mammalian reoviruses contain a myristoylated structural protein. J Virol. 1991;65:1960–1967. doi: 10.1128/jvi.65.4.1960-1967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nibert M L, Schiff L A, Fields B N. Reoviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1557–1596. [Google Scholar]
- 43.Noble, S., and M. L. Nibert. Unpublished data.
- 44.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 45.Shatkin A J, Kozak M. Biochemical aspects of reovirus transcription and translation. In: Joklik W K, editor. The Reoviridae. New York, N.Y: Plenum Press; 1983. pp. 79–106. [Google Scholar]
- 46.Shatkin A J, LaFiandra A J. Transcription by infectious subviral particles of reovirus. J Virol. 1972;10:698–706. doi: 10.1128/jvi.10.4.698-706.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shatkin A J, Sipe J D. RNA polymerase activity in purified reoviruses. Proc Natl Acad Sci USA. 1968;61:1462–1469. doi: 10.1073/pnas.61.4.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shepard D A, Ehnstrom J G, Schiff L A. Association of reovirus outer capsid proteins ς3 and μ1 causes a conformational change that renders ς3 protease sensitive. J Virol. 1995;69:8180–8184. doi: 10.1128/jvi.69.12.8180-8184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shing M, Coombs K M. Assembly of the reovirus outer capsid requires μ1/ς3 interactions which are prevented by misfolded ς3 protein in temperature-sensitive mutant tsG453. Virus Res. 1996;46:19–29. doi: 10.1016/s0168-1702(96)01372-x. [DOI] [PubMed] [Google Scholar]
- 50.Silverstein S C, Astell C, Levin D H, Schonberg M, Acs G. The mechanisms of reovirus uncoating and gene activation in vivo. Virology. 1972;47:797–806. doi: 10.1016/0042-6822(72)90571-5. [DOI] [PubMed] [Google Scholar]
- 51.Smith R E, Zweerink H J, Joklik W K. Polypeptide components of virions, top component and cores of reovirus type 3. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 52.Strong J E, Leone G, Duncan R, Sharma R K, Lee P W K. Biochemical and biophysical characterization of the reovirus cell attachment protein ς1: evidence that it is a homotrimer. Virology. 1991;184:23–32. doi: 10.1016/0042-6822(91)90818-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sturzenbecker L J, Nibert M, Furlong D, Fields B N. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J Virol. 1987;61:2351–2361. doi: 10.1128/jvi.61.8.2351-2361.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tillotson L, Shatkin A J. Reovirus polypeptide ς3 and N-terminal myristoylation of polypeptide μ1 are required for site-specific cleavage to μ1C in transfected cells. J Virol. 1992;66:2180–2186. doi: 10.1128/jvi.66.4.2180-2186.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tosteson M T, Nibert M L, Fields B N. Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc Natl Acad Sci USA. 1993;90:10549–10552. doi: 10.1073/pnas.90.22.10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Virgin H W, IV, Mann M A, Fields B N, Tyler K L. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J Virol. 1991;65:6772–6781. doi: 10.1128/jvi.65.12.6772-6781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu P, Miller S E, Joklik W K. Generation of reovirus core-like particles in cells infected with hybrid vaccinia viruses that express genome segments L1, L2, L3, and S2. Virology. 1993;197:726–731. doi: 10.1006/viro.1993.1648. [DOI] [PubMed] [Google Scholar]
- 58.Yeager M, Weiner S, Coombs K M. Transcriptionally active reovirus core particles visualized by electron cryo-microscopy and image reconstruction. Biophys J. 1996;70:A116. [Google Scholar]
- 59.Yue Z, Shatkin A J. Regulated, stable expression and nuclear presence of reovirus double-stranded RNA-binding protein ς3 in HeLa cells. J Virol. 1996;70:3497–3501. doi: 10.1128/jvi.70.6.3497-3501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]