Abstract
Simple Summary
Cancer is a global health problem with high personal and economic burden worldwide. The reprogramming of metabolism experienced by carcinomas offers a large set of enzymes that could be exploited to prevent cancer growth and metastasis. This review emphasizes the interplay between mitochondrial ATP synthase and its physiological inhibitor, the ATPase Inhibitory Factor 1 (IF1), in the metabolic reprogramming of OXPHOS in cancer cells to an enhanced glycolytic phenotype. We highlight the cell-type specificity by which the ATP synthase/IF1 axis exerts its activity as a tumor promotor or signals an anti-metastatic phenotype. Moreover, the implication of the ATP synthase/IF1 axis in cell death, and as a promising target for cancer therapy, is also stressed. We think that investigations aimed at characterizing the posttranscriptional mechanisms that regulate the activity of ATP synthase/IF1 axis will provide additional promising biomarkers for effective treatment of the disease.
Abstract
Cancer poses a significant global health problem with profound personal and economic implications on National Health Care Systems. The reprograming of metabolism is a major trait of the cancer phenotype with a clear potential for developing effective therapeutic strategies to combat the disease. Herein, we summarize the relevant role that the mitochondrial ATP synthase and its physiological inhibitor, ATPase Inhibitory Factor 1 (IF1), play in metabolic reprogramming to an enhanced glycolytic phenotype. We stress that the interplay in the ATP synthase/IF1 axis has additional functional roles in signaling mitohormetic programs, pro-oncogenic or anti-metastatic phenotypes depending on the cell type. Moreover, the same axis also participates in cell death resistance of cancer cells by restrained mitochondrial permeability transition pore opening. We emphasize the relevance of the different post-transcriptional mechanisms that regulate the specific expression and activity of ATP synthase/IF1, to stimulate further investigations in the field because of their potential as future targets to treat cancer. In addition, we review recent findings stressing that mitochondria metabolism is the primary altered target in lung adenocarcinomas and that the ATP synthase/IF1 axis of OXPHOS is included in the most significant signature of metastatic disease. Finally, we stress that targeting mitochondrial OXPHOS in pre-clinical mouse models affords a most effective therapeutic strategy in cancer treatment.
Keywords: cancer, OXPHOS, Warburg effect, mitochondrial ATP synthase, ATPase inhibitory factor 1, metabolic reprogramming, metastasis, RNA binding proteins, mitohormesis, cell death
1. Introduction
Mitochondria are central hubs in cellular physiology integrating cellular metabolism, bioenergetics, the execution of cell death and signaling through different effectors like Ca2+, reactive oxygen species (mtROS), mtDNA and different metabolites [1,2,3,4]. Given the fundamental roles played by the organelle, mitochondrial dysfunction is found in many different pathologies that include metabolic syndrome, neurodegeneration, inflammation, cardiovascular and rare diseases, cancer and normal physiological aging [5,6,7,8,9].
In the cancer field, Otto Warburg was first to describe, almost 100 years ago [10], that tumor cells and embryonic tissues have an exacerbated glucose consumption in the presence of oxygen. This observation led him to suggest, in agreement with the Pasteur effect [11], that tumors and embryonic tissues should exhibit an altered mitochondrial function to sustain their abnormal aerobic glycolysis [12]. His hypothesis was heatedly debated and almost abandoned by the outbreak of Molecular Biology and the discovery of oncogenes. In fact, at the beginning of this century, six hallmarks of the cancer phenotype were defined which included self-sufficiency in growth signals, insensitivity to anti-growth signals, limitless replicative potential, tissue invasion and metastasis, sustain angiogenesis and evading apoptosis [13]. However, the reprogramming of energy metabolism was not listed as a cancer hallmark at that stage. It was a decade later that a review by the same authors included two additional hallmarks of cancer: the avoidance of immune surveillance and the reprogramming of metabolism [14,15]. The renascence of Warburg’s postulates in the cancer field was spurred by the implementation of Positron Emission Tomography (PET) in clinical oncology, using the radionuclide 18F-dexoxyglucose (18FDG) [16,17,18], which represents the bed-side translation of Warburg’s metabolic observation regarding the glucose avidity of tumors [19]. The cancer imaging technique stimulated metabolically oriented “omic” investigations of human carcinomas that opened up new windows of hope in cancer patients for its potential in diagnostic, staging, treatment and follow up of the disease [20,21,22,23,24,25]. In this review, we summarize the role of the mitochondrial ATP synthase and the role of its inhibitor protein, the ATPase Inhibitory Factor 1 (IF1), in metabolic rewiring of cancer and its progression to emphasize the relevance that the engine that controls cellular life and death has in this field of investigation.
2. Metabolic Rewiring and Cancer Progression
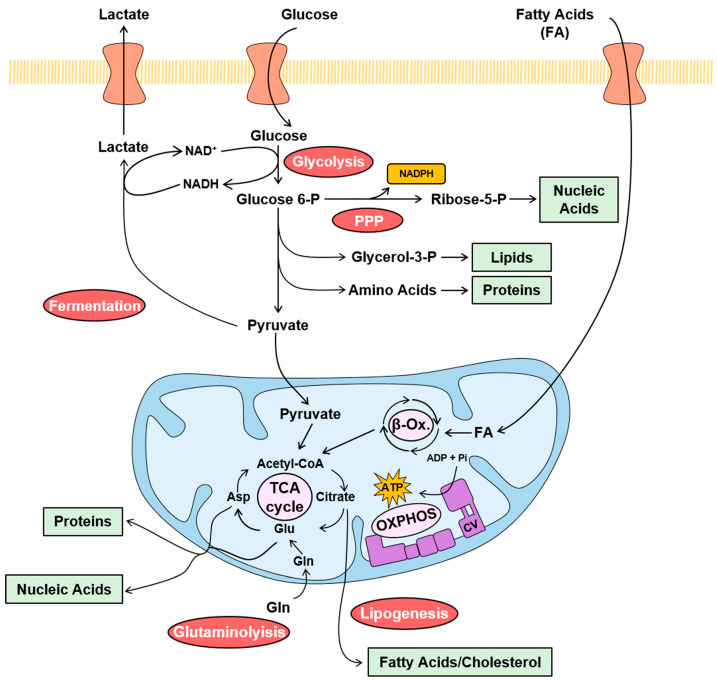
Carcinomas undergo a coordinated reprogramming of their metabolic pathways to satisfy the demand for high energy and for the building blocks that are required to sustain cellular proliferation [21,22,26,27,28,29]. The metabolic reprogramming experienced by cancer is generally known as the Warburg effect, and it is characterized by the sharp increase in the rates of glycolysis and of the fermentation of pyruvate into lactate, which allows the regeneration of NAD+ for glycolysis to proceed at high rates (Figure 1). Glycolysis and fermentation do not require oxygen to function. Concurrent with the hyperactivation of glycolysis and fermentation, the oxidation of pyruvate coupled to oxidative phosphorylation (OXPHOS) in mitochondria, which is strictly dependent on the availability of oxygen, is marginally increased and/or partially inhibited in cancer cells, to spare the backbone of carbon skeletons for the synthesis of precursors (Figure 1) [26,29,30,31]. In fact, the citrate formed in the TCA cycle is exported to the cytoplasm for providing the acetyl-CoA that is used for the synthesis of fatty acids and cholesterol (Figure 1) [32]. However, cancer cells replenish carbon skeletons in the TCA cycle in the form of glutamate by increasing the metabolism of glutamine (Figure 1) [33]. Simultaneously, the metabolism of glucose through the pentose phosphate pathway (PPP) is also significantly augmented, to produce the ribose that is used for building nucleotides and producing the NADPH that is used in biosynthetic processes and in the maintenance of the cellular redox state (Figure 1) [34]. Consistent with the reprogramming of metabolism in carcinomas, the sharp increase in different enzymes of glycolysis provide biomarkers that predict a bad patient prognosis [35], whereas, for example, the silencing of glycolytic enzymes in glioblastoma (GBM) xenografts dramatically increase the survival of mice [36]. In contrast, the loss of mitochondrial proteins involved in pyruvate oxidation, β-oxidation and OXPHOS predicts bad patient prognosis [35,37,38,39], supporting the theory that a diminished metabolic activity of the organelle compromises survival. Likewise, the overexpression of enzymes of metabolic pathways that are induced in carcinomas, such as glutamine metabolism [35,40,41] and the biosynthesis of fatty acids [35,42,43,44,45], predicts bad patient prognosis.
Figure 1.
Metabolic pathways in cancer. The color-coded scheme illustrates the major cancer-promoted changes in relevant metabolic pathways observed in human carcinomas. Glycolysis, fermentation, pentose phosphate pathway (PPP), lipogenesis and glutaminolysis are enhanced (red circles) in carcinomas. However, the mitochondrial oxidation of pyruvate, β-oxidation (β-Ox.), tricarboxylic acid (TCA) cycle and oxidative phosphorylation (OXPHOS) are marginally increased (pink circles) or diminished when compared to the activation of metabolic pathway in the cytoplasm. Glucose enters the cell by specific transporters and is metabolized at high rate through glycolysis to provide precursors such as glycerol-3-P and amino acids for lipid and protein synthesis (green box), respectively. Pyruvate is reduced to lactate in fermentation to regenerate NAD+ to allow glycolysis to proceed and is exported from the cell. In addition, glucose is catabolized through the PPP to obtain NADPH (orange box), for biosynthetic processes and the maintenance of the cellular redox state, and ribose-5-P, for nucleic acid biosynthesis (green box). In the mitochondria, pyruvate is oxidized to generate acetyl-CoA that feeds the TCA cycle. Fatty acids (FA) oxidation also feeds acetyl-CoA to the TCA cycle. Citrate in the TCA cycle is drained to the cytoplasm for the biosynthesis of FA and cholesterol. Glutaminolysis feeds the TCA cycle with glutamate. The electrons obtained in the oxidation of pyruvate and FA are transferred to the electron transport chain system for the synthesis of the ATP by OXPHOS (purple).
The question is, what are the underlying mechanisms that promote metabolic reprogramming in carcinomas? The general idea is that oncogene-driven metabolic changes could explain the metabolic rewiring in cancer, since the activation of oncogenes and/or the inhibition of tumor suppressors (Myc, Akt, p53, HIF-1α, APC/C-Cdh1) certainly affect energy metabolism and could account for changes in metabolic flux [26,46,47,48]. However, the Warburg phenotype is reversible, and could be acquired with or without any oncogenic alteration as demonstrated by adaptation of cancer cells to changing environments [49,50,51,52,53,54,55,56,57,58]. Indeed, the induction of proliferation in normal non-cancer cells or the adaption of cancer cells to different milieu where the tumors develop [50,51,52,59], provide examples in this regard. Overall, we believe that the rewiring of energy metabolism represents a reversible adaptive phenotypic response of the cancer cell to the induction of proliferation, hypoxia and/or other agents of the tumor microenvironment, that results in the hyperstimulation of glycolysis and fermentation affecting mitochondrial OXPHOS in a cell-type specific manner. Energy metabolism provides a therapeutic target with great potential for the treatment of cancer patients, since it is a trait of the disease that is reversible and amenable to modifications by inhibitors of the enzymes that steer metabolism [20,60,61,62,63,64,65,66].
3. The Mitochondrial ATP Synthase/IF1 Axis
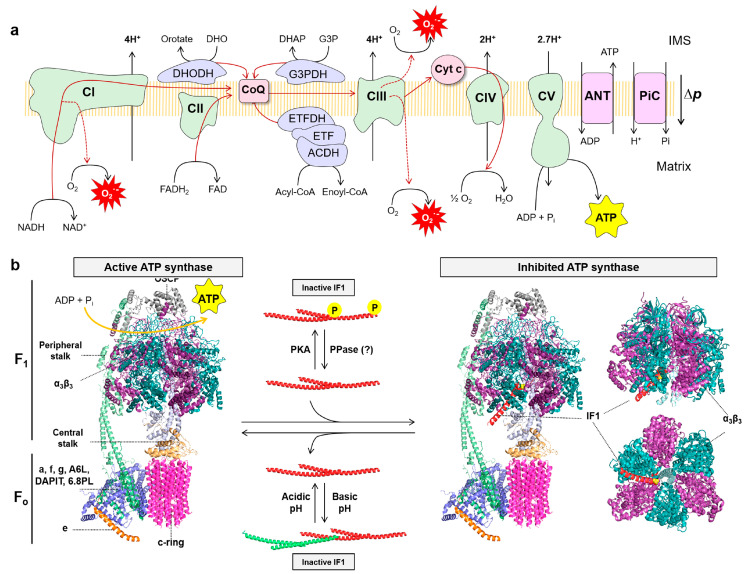
The OXPHOS system integrates the complexes of the electron transport chain (ETC) (complex I–IV), a variety of different dehydrogenases, the ATP synthase, two diffusible electron carriers such as CoQ and cytochrome c, and substrate transporters in the inner mitochondrial membrane (IMM) (Figure 2a) [30,67,68]. Respiratory complexes of the ETC, and a variety of FAD-dependent dehydrogenases that feed electrons directly to CoQ, transfer electrons obtained in biological oxidations in the form of NADH and FADH2 down to molecular oxygen, to generate the proton electrochemical gradient responsible for the proton-motive force (∆p) across the IMM (Figure 2a). The ATP synthase utilizes ∆p by coupling the backflow of protons into the matrix of the organelle for the generation of ATP from ADP and inorganic phosphate (Pi) (Figure 2a).
Figure 2.
The mitochondrial OXPHOS system: (a) The OXPHOS system consists of five protein complexes: complexes I–IV (CI–IV) of the electron transport chain (ETC) and complex V (CV) or ATP synthase. In addition, there are mobile electron carriers such as coenzyme Q (CoQ) and cytochrome c (cyt c), as well as membrane transporters that include among others the pyruvate carrier (not shown), adenine nucleotide translocase (ANT) and the phosphate carrier (PiC). Other FAD-dependent dehydrogenases transfer electrons to CoQ, such as glycerol-3-phosphate dehydrogenase (G3PDH), dihydroorotate dehydrogenase (DHOH), acyl-CoA dehydrogenase complex (ACDH), electron transfer flavoprotein (ETF) and electron transfer flavoprotein dehydrogenase (ETFDH). The electron transfer from NADH and FADH2 to molecular oxygen (O2), the final electron acceptor, builds a proton-motive force (∆p) by the pumping of protons through complexes I, III and IV, which is used by ATP synthase to produce ATP; (b) Structure of monomeric bovine active ATP synthase (left) and bound to the N-terminal inhibitory fragment of IF1 in the inhibited ATP synthase state (right). The soluble F1 domain is composed of α3β3 subunits (purple/blue) and the central stalk (γ subunit, light blue, and δ, ε subunits, light yellow), while the Fo domain is formed by a ring of 8 c subunits (pink) and a subunit (light blue). These two domains are linked by a peripheral stalk, composed of b, d, F6 subunits (light green), and the oligomycin sensitivity-conferring protein (OSCP; grey). The supernumerary subunits include e (orange), f, g, A6L, the protein associated with diabetes in insulin-sensitive tissues (DAPIT), and the 6.8 kDa proteolipid (6.8PL) (light blue). Right, the interaction between the N-terminal inhibitory fragment of IF1 (red) and the α3β3 and γ subunits is shown. The Ala14 residue of IF1 (Ser14 in human and mouse IF1) is highlighted in yellow. The active ATP synthase is able to synthetize ATP using ADP and Pi. When IF1 is bound it is inhibited and no longer synthetizes nor hydrolyzes ATP. Regulation of the ATP synthase is exerted by IF1. IF1 can be inactivated by forming oligomers that mask the inhibitory regions that bind to the F1 domain when the matrix pH is above neutrality. However, in mitochondrial de-energization conditions the pH drops below neutrality and IF1 becomes activated. On the other hand, IF1 can be phosphorylated in S39 by PKA preventing its interaction with the ATP synthase. IF1 has a short-half life and there is no evidence yet for the existence of an IF1 protein phosphatase (PPase). Molecular reconstruction from PDB: 6ZPO, 1OHH and 1GMJ. Images created using the PyMOL molecular graphics system.
The ATP synthase, a protein complex composed of 18 different proteins in humans, drives the production of ATP using the free energy stored in the proton-motive force (Figure 2a,b) [69]. It consists of two main domains, the Fo embedded in the IMM and the F1 domain projected into the matrix, containing the α and β subunits of the catalytic core (Figure 2b) [70]. The Fo and F1 domains are linked together by a central (subunit γ, ε and δ) and a peripheral (subunit b, d, OSCP and F6) stalk (Figure 2b). The protons are imported into the matrix through subunit a causing the rotation of the c-ring in the Fo domain. The central stalk transfers this rotational energy to the α3β3 subunits in the F1 domain, leading to the synthesis of ATP (Figure 2b). The majority of ATP synthase subunits are encoded in nuclear genes with the exception of two hydrophobic subunits (subunit a and 8 or A6L) that are encoded in the mitochondrial genome [70]. However, the ATP synthase is a reversible engine. In situations where the ∆p collapses, such as hypoxia/anoxia, the ATP synthase acts in reverse to maintain the ∆p at the expense of hydrolyzing ATP.
The ATP synthase has a small inhibitory protein, the ATPase Inhibitory Factor 1 (IF1), that for long time has been considered an inhibitor only (unidirectional) of the hydrolytic activity of the enzyme by binding the αβ interface in the F1 domain (Figure 2b) [71,72]. IF1 is a highly conserved protein among mammalian species and is encoded in the nuclear ATP5IF1 gene [73]. In the last decade, we and others have demonstrated that IF1 not only inhibits the ATPase activity of the enzyme, but also inhibits its ATP synthetic activity [74,75,76,77,78], promoting the re-emergence of IF1 as the natural physiological inhibitor of both ATP synthase activities, mostly by results in genetic mouse models of loss and gain of function of Atp5if1 [79,80,81,82,83,84].
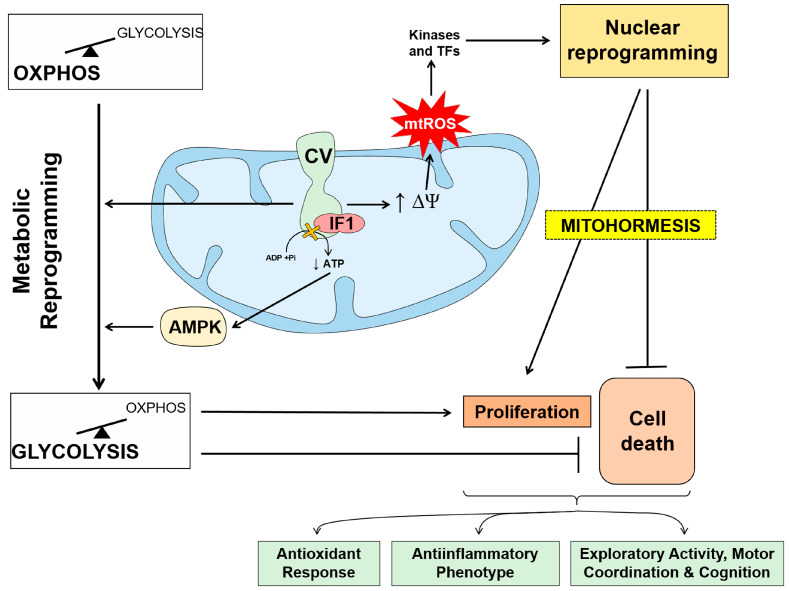
The IF1-mediated inhibition of ATP synthase results in partial blockade of the import of protons into mitochondria, the limitation of OXPHOS activity and the metabolic reprogramming of the cells to an enhanced glycolysis (Figure 3) [74,75,84,85]. Moreover, the blockage in proton import also promotes mitochondrial hyperpolarization and the subsequent production of ROS (mtROS) at respiratory complexes of the ETC (Figure 3) [75,82]. The generated mtROS modify the activity of protein kinases and of transcription factors that signal to the nucleus different programs that allow adaptation of the cell to changing cues (Figure 3) [75,79,80,81,82]. In some mouse tissues, the IF1-mediated activation of mtROS signaling promotes long-lasting metabolic and molecular cytoprotective mechanisms that allow cells to withstand subsequent insults (Figure 3) [79,80,81], a process known as mitohormesis [1,2,3,86]. However, in other mouse tissues which are naturally devoid of IF1 [87], the overexpression of IF1 is detrimental [80,88,89]. In sharp contrast, the overexpression of IF1 in neurons protects from excitotoxic insults [79] and increases the exploratory activity, motor coordination and cognition of mice (Figure 3) [82]. Similarly, the overexpression of IF1 in intestinal epithelial cells provides protection against inflammation (Figure 3) [81], whereas IF1 ablation results in a pro-inflammatory phenotype of fatal consequences [83]. Overall, these results emphasized the relevance of the ATP synthase/IF1 axis in signaling and controlling tissue-specific and non-cell autonomous cell fate decisions.
Figure 3.
The role of ATP synthase/IF1 axis in metabolic reprogramming and signaling. Binding of IF1 to the ATP synthase limits the production of ATP and promotes metabolic reprogramming towards an enhanced glycolytic phenotype by allosteric regulation of the enzymes of glycolysis and the activation of AMPK. Inhibition of ATP synthase prevents the backflow of protons into the matrix increasing the mitochondrial membrane potential (∆Ψm). The increase in ∆Ψm leads to an increase in the generation of mitochondrial reactive oxygen species (mtROS) at the electron transport chain that signal to the nucleus through kinases and transcription factors. Nuclear reprogramming promotes the activation of cellular responses that include proliferation, prevention of cell death allowing adaptation of the cell to changing cues in mitohormetic responses. The cellular signaling response mediated by IF1 overexpression is cell type specific in mouse tissues and promotes antioxidant responses, anti-inflammatory phenotypes and motor coordination and cognition. In contrast, IF1 overexpression in skeletal muscle and heart is deleterious.
Besides its function in energy provision and in signaling, the ATP synthase also plays a structural role in the IMM by forming dimers and oligomers to shape cristae at its rims (Figure 4a,b) [70]. Recent cryo-EM structures of mammalian tetrameric ATP synthases (Figure 4b) [90,91] and isobaric quantitative Protein Interaction Reporter (iqPIR) cross-linking technologies [84], have uncovered the structural role that IF1 plays in oligomerization and inhibition of ATP synthase, and in reprogramming of the tissue to an enhanced glycolysis [84]. Indeed, the overexpression of IF1 in forebrain neurons [82] and cardiomyocytes [84] or of the active mutant version of IF1 (H49K) in liver [80] and skeletal muscle [89] of transgenic mice promotes the oligomerization and inhibition of ATP synthase.
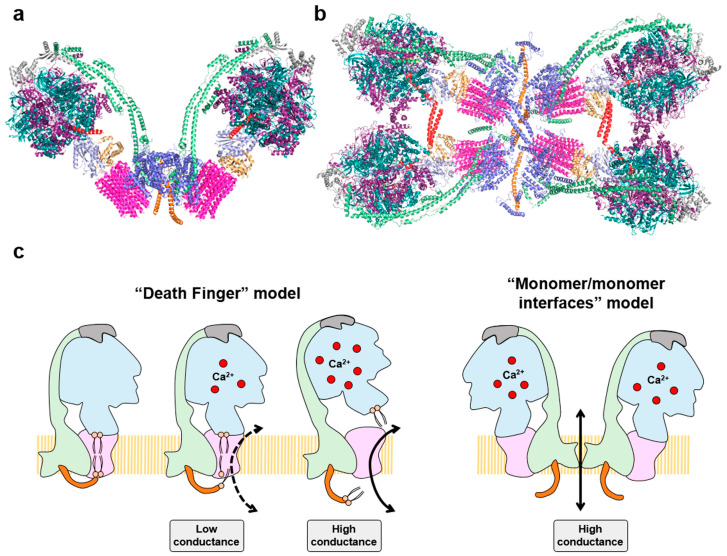
Figure 4.
Oligomeric assemblies of the ATP synthase and the permeability transition pore (PTP): (a) Structure of bovine dimeric ATP synthase promotes the curvature of the inner mitochondrial membrane at its tips. Molecular reconstruction from PDB: 7AJD; (b) Cryo-electron microscopy structure of porcine IF1-inhibited ATP synthase tetramers viewed from the matrix side. The IF1 dimers (red) bind to two adjacent antiparallel ATP synthase dimers. Molecular reconstruction from PDB: 6J5K. Images created using the PyMOL molecular graphics system. Both in a, and b, the color coding of the subunits is the same as in Figure 2; (c) Two major models explain the participation of the ATP synthase in the PTP. The “death finger” model proposes that Ca2+ binding to β subunit causes a rearrangement in the F1 domain increasing its rigidity that is transmitted through OSCP (grey) to the peripheral stalk (green) and to the e subunit (orange). Subunit e exerts a pulling force on the outer lipids that plug the center of the c-ring, leading to the formation of a channel that allows the entrance of ions. Mild Ca2+ levels lead to closed and open states in a low-conductance mode of opening (“flickering”) in the PTP. However, when Ca2+ increases the channel is more prone to opening, promoting complete displacement of inner lipids in the c-rotor and of the central stalk of the ATP synthase, committing cells to death by the high-conductance PTP. In the “monomer/monomer interfaces” model, the Ca2+ induced conformational changes in dimers of the ATP synthase destabilize the dimeric assembly opening a high conductance pore in the interface of the dimer (subunits e and g).
Moreover, the ATP synthase is also a critical component in the regulation and execution of cell death since its components [92] and activity [93,94,95] are required for the efficient execution of cell death. In fact, the ATP synthase is structurally and functionally implicated in permeability transition (mPT) [96,97,98]. mPT is exerted through a high-conductance channel in the IMM, known as the Permeability Transition Pore (PTP) [99,100,101]. PTP opening leads to mitochondrial swelling and cell death and is regulated by Ca2+ and/or cyclophilin D-dependent structural changes on ATP synthase [97,102], whereas it is inhibited by cyclosporine A (CsA) [103,104]. The ATP synthase is postulated as a key component of the PTP [96,97,105,106,107,108], although its participation in mPT has been questioned [109,110]. Nowadays, the participation of the ATP synthase in PTP is undisputed after the reunification of the different alternatives that implicated the enzyme in the “death finger” and “monomer/monomer interfaces” models of the PTP (Figure 4c) [96,97,98,102]. In both models, when Ca2+ binds the β subunit it causes a rearrangement in the F1 domain increasing its rigidity [111], that is transmitted through OSCP, a subunit placed on top of the F1 domain, to the peripheral stalk and to the e subunit (Figure 4c). Because of this structural change, in the “death finger” model subunit e exerts a pulling force on the outer lipids that plug the center of the c-ring, leading to the formation of a channel that allows the entrance of ions (Figure 4c) [91,96,97,98]. Under physiological levels of matrix Ca2+, the PTP oscillates between closed and open states in a low-conductance mode of channel opening, representing the “flickering” modality of the PTP (Figure 4c) [96,97]. However, as matrix Ca2+ increases, the channel is more prone to openings, promoting complete displacement of inner lipids in the c-rotor and of the central stalk of the ATP synthase, committing cells to death by the high-conductance PTP (Figure 4c) [97,98]. In the alternative “monomer/monomer interfaces” model, the Ca2+-induced conformational changes trigger partial changes in the monomer-monomer interfaces (e/g subunits) of ATP synthase dimers, leading to the formation of the high-conductance PTP (Figure 4c) [97,98].
Interestingly, we and others have demonstrated that the expression of IF1 prevents cell death [75,112,113] and promotes in vivo the oligomerization of ATP synthase in mouse livers, hearts, skeletal muscles and neurons [80,82,84,89], in agreement with cryo-EM studies that show that dimers of IF1 bind two antiparallel dimers of the ATP synthase in a tetrameric inhibited structure of the enzyme (Figure 4b) [90,91]. Hence, it is tempting to suggest that the IF1-mediated oligomerization of the ATP synthase prevents the Ca2+-induced conformational changes in the enzyme that result in PTP opening and cell death. In fact, recent findings in isolated mitochondria of intestinal epithelial cells of IF1-ablated mice confirmed that IF1 is necessary to oligomerize and inhibit a fraction of ATP synthase [83]. This mechanism confers resistance to the Ca2+-induced PTP opening [83], supporting the theory that IF1-mediated oligomeric states of the ATP synthase act as a structural brake to prevent cell death.
4. ATP Synthase and the Bioenergetic Signature of Cancer
As previously mentioned, the high rates of pyruvate fermentation observed in cancer cells under aerobic conditions lead Warburg to suggest that carcinomas had an impaired bioenergetic activity of mitochondria [114]. However, while the upregulation of glycolysis in carcinomas is nowadays out of the question, the alteration of mitochondrial bioenergetics is still a matter of debate, despite the obvious evidence linking the down-regulation of mitochondrial metabolism and bioenergetics in cancer progression [20,35,51,115,116,117,118,119,120]. Perhaps the debate emerges due to the metabolic behavior of some cancer cells when growing in culture [121], differences in the mitochondrial proteome and activity in different tissues [122,123], the heterogeneity of the microenvironment of carcinomas [124,125,126,127,128] and/or the finding that metastatic disease requires an enhanced expression of OXPHOS proteins [129,130,131,132,133,134,135]. In any case, mitochondrial structure and its proteomic composition is certainly altered in a large number of carcinomas [115,136,137,138,139] and, as recently reviewed [35], many of the mitochondrial proteins involved in OXPHOS and in other mitochondrial activities remain unaltered or inhibited when compared to their non-tumor adjacent tissues (NAT). In general, the loss of expression of mitochondrial proteins correlates with a bad prognosis for the patients [20,35]. Overall, these results suggest that OXPHOS is diminished, especially when compared to the sharp induction of glycolysis and other metabolic pathways (Figure 1). In this scenario, the ATP synthase, that integrates the bioenergetic and death-signaling functions of mitochondria, is called to play a relevant function in cancer onset and progression.
Indeed, the down-regulation of the catalytic subunit of mitochondrial ATP synthase (β-F1-ATPase) is observed in a large number of human carcinomas [20,35,115,140], either when expressed in absolute levels or normalized relative to other mitochondrial proteins, such as Hsp60 and/or to the glycolytic GAPDH. In fact, the quotient of the expression level of these three biomarkers β-F1-ATPase/Hsp60/GAPDH offers a bioenergetic cellular (BEC) index that informs about the overall capacity of mitochondria in the tissue or carcinoma [26,115]. Moreover, the BEC index is essentially the same in lung, breast and esophageal carcinomas despite the large differences in the expression of these proteins in the corresponding non-tumor adjacent tissues (NAT) [141]. These findings suggest that transformations blur the tissue-specific differences in energy metabolism to converge on the same metabolic phenotype that sustains tumor growth. Interestingly, a low expression of β-F1-ATPase and a low BEC index in the carcinoma predicts poor overall (OS) and disease free survival (DFS) for colon cancer patients [115], suggesting that the downregulation of OXPHOS contributes to tumor progression [26,115,142]. The relevance of a low BEC index in predicting a poor prognosis for patients bearing different types of carcinomas (breast, colon, lung, melanoma, ovarian, gallbladder) and leukemias has been extensively documented [16,35,39,115,142,143,144,145,146,147,148,149,150]. Furthermore, the BEC index significantly correlates with migration speed and in predicting drug responses across the NCI-60 cell line panel [151], as well as in predicting the response to different targeted therapies in preclinical mouse models bearing different human carcinomas [20,39,51,60,61,62,120,152,153].
Remarkably, a high-throughput quantitative analysis of 27 biomarkers of metabolism—using Reverse Phase Protein Array (RPPA) technology, in a cohort of 128 tumors and the corresponding paired NAT of patients bearing lung adenocarcinomas (LUAD)— confirmed that the BEC index is significantly diminished in the carcinomas, suggesting that OXPHOS activity is limited in LUAD [20], in agreement with previous reports [16,23,145]. In fact, discrimination of a tumor from NAT was accomplished by the signature of six metabolic proteins analyzed [20]. Remarkably, five out of the six proteins in the signature are mitochondrial proteins, strongly suggesting that the mitochondrial proteome is a most relevant target in metabolic reprogramming of LUAD [20].
Interestingly, and despite the content of β-F1-ATPase is not increased in the whole cohort of LUAD studied [20], the analysis revealed that the subgroup of patients bearing carcinomas with the highest content of β-F1-ATPase had a worse prognosis when compared to the low content subgroup of patients [20], in agreement with a previous report [66]. Similar findings have been reported in large cohorts of breast [144] and melanoma [146] patients suggesting that, despite there being an overall limitation of OXPHOS in the carcinomas, mitochondrial bioenergetics is required for metastatic disease and cancer progression (see below) [131,154,155].
The downregulation of the mitochondrial ATP synthase in cancer can be mediated at the expression and activity levels [30]. The expression of the catalytic subunit of ATP synthase (β-F1-ATPase) in mammalian liver is complex, including the subcellular localization of its mRNA in a structure that is frequently found attached to the outer mitochondrial membrane [156,157]. β-F1-ATPase mRNA localization and translation requires at least two cis-acting elements and a large set of RNABPs [156,158] and the 3′-UTR of the mRNA that acts as a translational enhancer sequence [159,160,161,162]. Consistently, the expression of β-F1-ATPase in prevalent human carcinomas, in differentiation and during the cell cycle [51,159,163,164,165,166,167], is also controlled at the level of translation by a set of mRNA-binding proteins that upon binding the 3′UTR of β-F1-ATPase mRNA inhibit its translation [30,160,166]. Remarkably, some of the mechanisms of translational control of β-F1-ATPase are conserved between liver hepatocarcinogenesis [168] and rat liver development [159,163,169].
An affinity purification approach of RNABPs using as bait the full-length β-F1-ATPase mRNA allowed the identification of nine bona fide mRNA binding proteins [170]. Among them, G3BP1 (Ras-GTPase-Activating Protein SH3-Domain-Binding Protein) was found to interact both in vivo and in vitro with the 3′UTR of β-F1-ATPase mRNA, to promote inhibition of the synthesis of the protein by preventing mRNA translation [30,166,170]. Interestingly, G3BP1 is overexpressed in breast carcinomas [171] and its overexpression predicts shorter time for recurrence of the disease [30]. Likewise, G3BP1 is also overexpressed in gastric cancer representing an independent prognostic factor of poor prognosis [172]. These results stress the relevance that translational control exerted by RNABPs has in the biogenesis of ATP synthase, and hence, in the bioenergetic signature and function of mitochondria.
In addition, translational control of β-F1-ATPase mRNA (β-mRNA) is also exerted by miR-127-5p that targets the 3′UTR of the mRNA and significantly reduces its translational efficiency without affecting β-mRNA abundance [173]. Interestingly, this mechanism of translational control for the expression of mitochondrial ATP synthase is operative in fetal human and rat livers but not in oncogenesis [173].
Furthermore, both in Acute Myeloid Leukemia and Chronic Myeloid Leukemia the downregulated expression of β-F1-ATPase is exerted by hypermethylation of the promoter of the ATP5F1B gene, limiting the availability of β-F1-ATPase mRNA in cancer cells [150,174]. Overall, these findings just uncover the “tip of an iceberg” that represents the scarcely studied field of translational control in regulating the biogenesis of mitochondria and of the ATP synthase, which has paramount implications in the tissue-specific biogenesis of the organelle and in cancer progression.
5. ATPase Inhibitory Factor 1, the Physiological Inhibitor of the ATP Synthase
As mentioned above, the control of the overall ATP synthase activity is not only exerted at the protein level, but also by regulation of the content and activity of IF1, its inhibitor protein. For many years, it has been considered that under normal physiological situations, i.e., when the mitochondrial matrix pH is above neutrality, IF1 forms inactive oligomers by masking the inhibitory regions that bind to the F1 domain of the enzyme (Figure 2b) [175,176]. Only when mitochondria become de-energized, and matrix pH drops below neutrality, IF1 is considered to become activated by depolymerization and binds to the ATP synthase to inhibit its hydrolase activity (Figure 2b) [175,176]. pH regulation of IF1 activity involves protonation/deprotonation of five conserved histidine residues that play a relevant role in structural changes and oligomerization [175,177]. Interestingly, the IF1-H49K mutant retains its inhibitory activity on the enzyme even at pHs above 6.7, both in vitro [74,85,175,177] and in vivo under basal physiological conditions of mitochondria [79,80,89].
Our studies in metabolic reprograming in human carcinomas revealed that normal lung, colon and breast tissues express negligible quantities of IF1 [85] but showed a sharp increase in the expression of the protein in the carcinomas and in cultured cancer cells [74,75,85]. These findings raised the reasonable hypothesis that IF1 might represent a sort of mitochondrial “oncogene” [178], not only to control the hydrolytic activity of the ATP synthase, as it was postulated many years ago, but also its ATP synthetic activity. In other words, the overexpression of IF1 could be mediating the partial inhibition of OXPHOS contributing to the metabolic reprogramming experienced in oncogenesis. Indeed, the overexpression of IF1 in several cancer cells [74,75,77,78,85], mouse models of gain of function of IF1 [79,80,81,82,84,89] and pharmacologic in vivo approaches [76], amply demonstrated that IF1 overexpression inhibits the ATP synthetic activity of ATP synthase in isolated mitochondria, diminished tissue ATP levels, activated AMPK and promoted the metabolic rewiring of cells and tissues to an enhanced glycolytic phenotype, i.e., induced the Warburg phenotype. Conversely, silencing or knocking out IF1 in cancer cells [20,179] and mouse models of loss of function of IF1 [82,83] resulted in and enhanced activity of the ATP synthase.
Stem cells, iPSC and cancer cells have a predominant glycolytic phenotype whereas OXPHOS prevails in differentiated cells [180,181]. Interestingly, IF1 also plays a relevant role in metabolic reprogramming during differentiation of human mesenchymal stem cells (hMSC) and in the maintenance/acquisition of stemness [182]. In fact, hMSC also present high levels of IF1, altered mitochondrial structure and molecular composition, low rates of OXPHOS and an enhanced glycolytic metabolism [182]. However, when hMSC cells are induced to differentiate into osteocytes, the expression of IF1 is lost by an enhanced rate of degradation of the protein that is concurrent with the biogenesis of OXPHOS proteins, the increase in oxygen consumption rates and the partial inhibition of glycolysis [181,182]. On the contrary, nuclear reprogramming of somatic cells to induce pluripotency (iPSC) promotes the downregulation of OXPHOS concurrently with the activation of glycolysis [183] and the re-expression of high cellular levels of IF1 [184]. Altogether, these findings emphasize the relevance of IF1 in metabolic reprogramming in cancer and pluripotent stem cells.
Remarkably, the results in tissues of mouse models with regulated expression of IF1 further suggest that mitochondria in non-stressed physiological conditions of some cellular types such as cardiomyocytes [76], neurons [82] and intestinal epithelial cells [83] contain a fraction of IF1-bound and -inhibited ATP synthase, in agreement with recent cryo-EM studies of mammalian heart ATP synthases [90,91]. We suggest that the IF1-inhibited ATP synthase in these tissues, besides regulating the output of ATP as a function of energy demand, is involved in controlling mitochondrial signaling, structure and cell death as discussed in a previous section.
The regulation of IF1 activity as an inhibitor of ATP synthase is also exerted by covalent modification of the protein by phosphorylation (Figure 2b). Phosphoproteomic studies indicated that serine residues of IF1 are phosphorylated in cancer cells [185,186,187]. Indeed, we showed that IF1 is phosphorylated in several serine residues in cells in cultures and in mouse hearts in vivo in response to cAMP-dependent activation of PKA [76]. Site-specific phosphodeficient and phosphomimetic IF1 serine mutants revealed that the PKA-mediated phosphorylation of IF1 in S39, but not in other serine residues, prevented the interaction of IF1 with ATP synthase and enhanced both the ATP synthetic and hydrolytic activities of the enzyme (Figure 2b) [76]. Interestingly, it should be noted that tissues that contain high expression levels of IF1 present both phospho- and dephosphorylated IF1 [76,87], consistent with the existence of two pools of ATP synthase in their mitochondria, active and IF1-bound inhibited enzyme [76].
Remarkably, recent findings have demonstrated that the binding of kynurenic acid to the GPR35 orphan receptor prevents the phosphorylation of IF1 in human and mouse cardiomyocytes to prevent ATP hydrolysis as an anti-ischemic ATP-conservation mechanism [188,189]. Interestingly, these authors further confirmed that dephosphorylated IF1 binds to ATP synthase and promotes enzyme oligomerization [188], in agreement with recent cryo-EM structures of the oligomeric inhibited enzyme [84,90,91] and our results in mouse models expressing IF1 [79,80,81,82,83,89]. These results further suggest that the content of dephospho-IF1 controls the fraction of oligomeric and inhibited ATP synthase and hence, that the phosphorylation/dephosphorylation of IF1 represents a most relevant mechanism for a fine and rapid adjustment of the function of the OXPHOS system in different physiological situations [73,76].
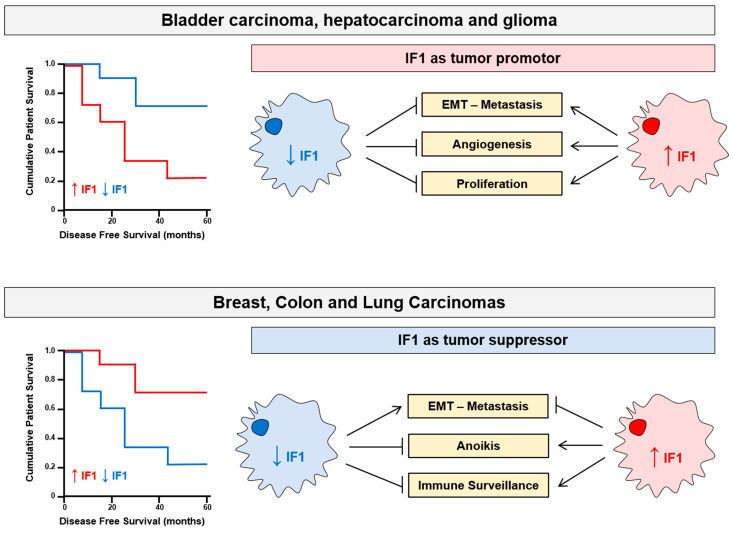
Indeed, the phosphorylation of IF1 has been observed when there is an increase in energy demand in the heart in vivo or in cells in cultures when they are forced to increase OXPHOS [76]. Likewise, IF1 is phosphorylated when cells progress through the OXPHOS-dependent G1-phase of the cell cycle [76]. In contrast, hypoxia and progression through the reductive phases of the cell cycle (S/G2/M) promote dephosphorylation of IF1, the reduction of OXPHOS and an enhanced glycolytic flux [76]. Consistently, dephosphorylated IF1 is the prevalent form of the inhibitor protein in lung, colon, and breast carcinomas, contributing to the inhibition of ATP synthase and the reprogramming of metabolism to an enhanced glycolysis and fermentation [76]. As previously mentioned, and besides controlling energy metabolism, the fraction of IF1-bound inhibited ATP synthase in the carcinomas is also fulfilling additional functions through signaling a pro-oncogenic phenotype, by stimulating proliferation/invasion/metastasis (Figure 5) [75,80,88,190,191], or an anti-oncogenic phenotype, by preventing metastatic disease and immune surveillance (Figure 5) (see next section) [20,85,147,179,192].
Figure 5.
The role of IF1 as tumor promotor or tumor suppressor. In bladder carcinomas, hepatocarcinomas and gliomas, high expression levels of IF1 (red) predict a poor prognosis for the patients when compared to carcinomas with low levels of IF1 (blue). In these carcinomas, IF1 acts as a tissue-specific tumor promotor facilitating proliferation, angiogenesis, epithelial mesenchymal transition (EMT) and metastasis. In contrast, in breast, colon and lung carcinomas, high expression levels of IF1 (red) predict a good prognosis for the patients when compared to carcinomas with low levels of IF1 (blue). In these carcinomas, IF1 acts as a tissue-specific tumor suppressor preventing epithelial mesenchymal transition (EMT) and metastasis by favoring cell death upon cellular detachment and immune surveillance.
Besides the mechanisms that regulate the fraction of IF1 that is active as an inhibitor of the ATP synthase discussed above (Figure 2b), the mechanisms that control the overall mitochondrial content of IF1 are also of relevance. As reviewed elsewhere [73], the ATP5IF1 gene interacts with c-Myc, c-Fos, NFκB and HIF-1α transcription factors. However, it is remarkable that in the early phase of cell reprogramming of somatic cells into pluripotency, c-Myc is the transcription factor that triggers a sharp increase in IF1 expression, precipitating the metabolic program necessary for cellular reprogramming [184].
Most available evidence suggests that regulation of IF1 expression is exerted at post-transcriptional levels by controlling the rates of its synthesis and degradation [182]. In fact, the increase in protein content of IF1 in carcinomas [85] or its sharp reduction during cellular differentiation [182], are exerted in the absence of relevant changes in the abundance of IF1-mRNA. In fact, the IF1 protein has a very high turnover rate (2–3 h), both in cancer cells [85] and in differentiated osteocytes [182], which is much faster than the turnover of many other subunits of ATP synthase in cancer cells (18 h) [193]. IF1 degradation is mediated by several mitochondrial serine proteases [85,182] and metalloproteases [194], but the specific genes involved have not been identified yet [182].
In addition, IF1-mRNA is also subjected to tissue-specific translation control by RNABPs [30,195]. In fact, both in the heart [196] and in the liver [197] knockdown of LRPPRC (leucine rich pentatricopeptide repeat containing protein), a RNABP that is involved in post-transcriptional regulation of mtDNA expression [198], severely reduces the activity and assembly of ATP synthase. In the heart, the inhibition an assembly deficit of ATP synthase is associated with the overexpression of IF1 [196]. Interestingly, LRPPRC specifically interacts with IF1-mRNA [30] and its over-expression in human and mouse cells significantly diminished the expression of IF1, increasing the rate of cellular ATP production [87]. Contrariwise, the silencing of LRPPRC in human and mouse cells augmented IF1 expression and reduced the ATP synthesis rate of the cells [87]. Interestingly, LRPPRC is tissue-specifically expressed [87] and the studies of its relevance in IF1-mRNA translation certainly deserves further investigations that will contribute to uncovering the tissue-specific mechanism that controls the activity of ATP synthase.
6. Tissue-Specific Activity of IF1: Tumor Promotor and Tumor Suppressor
A fundamental characteristic of IF1 is that it is a tissue-specific and species-specific expressed protein in mouse and human tissues [87]. Human normal tissues such as the heart, brain, kidney, stomach, endometrium and liver have a high IF1 content, whereas breast, colon, and lung epithelia contain negligible levels of IF1 [85,87]. Likewise, in human carcinomas the expression of IF1 is also variable [85]. Whereas oncogenesis in epithelial cells of the endometrium, stomach and kidney does not promote an increase in IF1 expression, carcinomas in the colon, lung and breast show a very sharp increase in the content of IF1 [85].
In agreement with the pro-oncogenic role of IF1, studies in human hepatocellular carcinomas (HCC) showed that high levels of IF1 predict worse overall and progression free survival for the patients (Figure 5) [88]. Mechanistically, IF1 activates the non-canonical NFκB pathway, driving angiogenesis and metastasis through the activation of SNAI1 and VEGF [88]. Consistent with these findings, transgenic mice overexpressing in the liver the constitutively active mutant IF1-H49K promoted the inhibition of OXPHOS and a state of metabolic preconditioning guided by the activation of the stress kinases AMPK and p38 MAPK [80]. Diethylnitrosamine-induced liver carcinogenesis in IF1-H49K transgenic mice contributed to an enhanced liver carcinogenesis by augmenting proliferation and apoptotic resistance of carcinoma cells [80]. Likewise, the overexpression of IF1 in bladder carcinomas [190] and in gliomas [191] also predict a worse prognosis for the patients and a shorter time to disease recurrence (Figure 5). Mechanistically, the silencing of IF1 diminished the rates of proliferation, migration and invasive capacities of the cells, preventing epithelial mesenchymal transition (EMT) (Figure 5) [88,190,191]. In contrast, the overexpression of IF1 in these cancer cells promoted angiogenesis and activation of EMT, increasing the expression of E-cadherin and diminishing that of vimentin, to promote their metastatic capacity (Figure 5) [88,190,191].
However, in breast [85,147], colon [85,179] and lung [20] carcinomas, high levels of IF1 expression correlate with a good prognosis for the patients (Figure 5), suggesting that in these tissues the expression of IF1 acts as a tumor suppressor. We have investigated the molecular basis of the tumor-suppressor function of high IF1 expression in breast, colon and lung cancer cells, by means of the generation of stable cancer cell lines that overexpress or are devoid of IF1 [20,147,179]. In these studies, cancer cells overexpressing IF1 restrain OXPHOS, enhance glycolysis and trigger the production of a mtROS signal that activates transcription of the NFκB promoter which results in protecting cells from death-inducing agents [20,75,85,147,179]; in other words, it would initially appear that IF1 is favoring a phenotype prone to cancer progression. However, the overexpression of IF1 in these cancer cells also promotes a transcriptome that indicates a more epithelial phenotype when compared to IF1-ablated or knockdown cells (Figure 5) [20,147,179]. In fact, we showed that IF1 overexpressing cancer cells proliferate, migrate and invade less than cancer cells with low expression levels or that are devoid of IF1 (Figure 5) [20,147,179]. Furthermore, in the case of colon and lung cancer cell lines, the overexpression of IF1 renders the cells prone to anoikis [20,179], the form of cell death upon cellular detachment, presumably because of the limitation of OXPHOS in the metastatic cell. In addition, colon spheroids of IF1-silenced cells have an increased ability to escape from immune surveillance by Natural Killer cells (Figure 5) [179], thus favoring metastasis. As a result, tumor development in mouse xenografts using colon and lung cancer cells confirmed that IF1-silenced or IF1-ablated cells are more tumorigenic and metastatic in vivo than IF1-overexpressing cells [20,179]. Overall, despite that a high IF1 level confers to breast, colon and lung cancer cells a more glycolytic phenotype, it restrains tumor growth and metastatic disease by repressing OXPHOS and thus underpinning the better prognosis of the cancer patients that bear tumors with high expression levels of IF1 (Figure 5). However, it is reasonable to suggest that the tumor suppression function afforded by the mitochondrial dose of IF1 in these types of carcinomas also depends on the nuclear reprogramming exerted by the activation of specific kinases and transcription factors mediated by mtROS signaling generated as a result of the IF1-mediated inhibition of the ATP synthase (Figure 3). In this regard, it should be noted that the activation of these cellular programs is different depending on the cell type as shown in tissue-specific mouse models of loss and gain of function of IF1 [79,80,81,84,89].
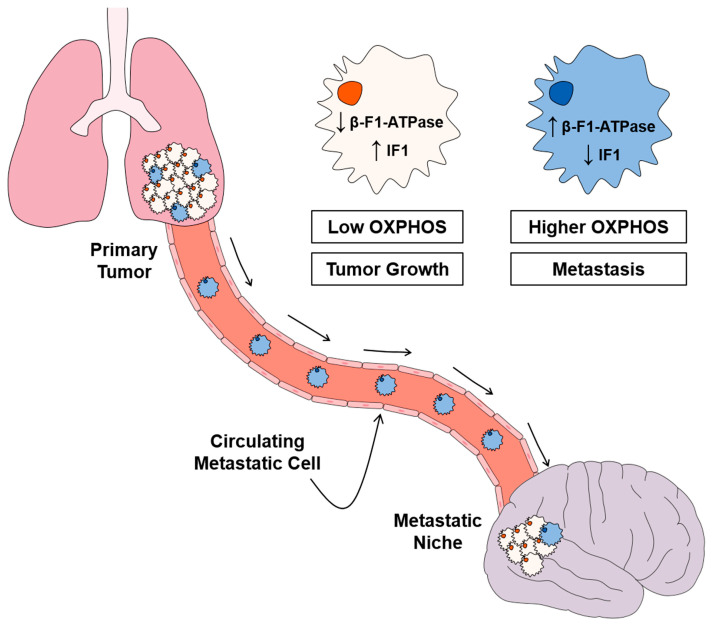
As mentioned above, OXPHOS is required for metastatic disease (Figure 6) [20,129,130,131,133,147,179] and, consistently, melanoma, breast and lung carcinomas [20,66,144,146] with the highest expression level of β-F1-ATPase predict higher chances of disease recurrence for the patients (Figure 6). Interestingly, in the RPPA study of LUAD carcinomas, machine learning provided a highly significant signature of metastasis (p = 3.17 10−19) that included the expression of β-F1-ATPase, IF1, superoxide dismutase (SOD2), thioredoxin (TRX), peroxiredoxin 6 (PRX6) and 4-hydroxynonenal (4-HNE) modification of proteins [20]. Remarkably, the signature included the two biomarkers of OXPHOS. A high β-F1-ATPase represented a bad independent predictor of OS (HR (95% CI) = 4.524(0.955–21.43), p = 0.05) whereas a high IF1 expression level represented a good independent predictor of recurrence of the disease (HR (95% CI) = 0.11(0.025–0.51), p = 0.005) (Figure 6) [20]. In agreement with these results, it has been reported that cancer cells with higher invasive capacity experience an increase in mitochondrial respiration as well as ATP production (Figure 6) [131]. Moreover, metastatic breast cancer cells have lower IF1 levels when compared to the primary tumors [199], suggesting that metastatic cells rely on OXPHOS when compared to cancer cells in the primary tumor (Figure 6) [131].
Figure 6.
The ATP synthase/IF1 axis in metastatic disease. A working scheme summarizes the interplay of the two biomarkers of OXPHOS (β-F1-ATPase and IF1) involved in the metastatic signature of lung adenocarcinomas [20]. Low and high OXPHOS cells coexist in the primary tumor. Low OXPHOS cells (light orange cells; low of levels of β-F1-ATPase and high levels of IF1) promote the growth of the primary tumor through an enforced glycolysis. Cells with higher OXPHOS (blue cells; high of levels of β-F1-ATPase and low levels of IF1) prime metastatic disease [131]. Metastatic disease requires higher OXPHOS cells because they are less vulnerable to cell death upon cellular detachment from the primary tumor [20,179], and are able to colonize the metastatic niche since they have higher chances to scape immune surveillance [179].
Overall, we believe that therapeutic approaches targeting the ATP synthase/IF1 axis and hence, the activity of OXPHOS in carcinomas, should take into consideration the tissue in which the carcinoma develops.
7. Mitochondria: A Promising Target for Cancer Treatment
Novel therapeutic approaches based on personalized medicine are required to minimize the social and economic burden caused by cancer. The enzymes that control metabolism are promising targets to combat progression of the disease [9,14,26,200,201]. In this regard, several glycolytic inhibitors have been proposed and some of them are actually in clinical trials [26,51,152,201,202,203,204]. Mitochondrial metabolism also offers other effective targets to restrain tumor growth [201,203,205,206,207,208]. In this regard, antibiotics such as doxycycline that target the mitochondrial ribosome and inhibit translation prevent mitochondrial biogenesis which is required for the clonal expansion and survival of cancer stem cells (CSCs) [209]. Doxycycline has also been used in combination with other anticancer therapies [210,211,212,213] and their effectiveness lies in their ability to prevent metastasis by inhibiting the growth of CSCs. Interestingly, a Phase II clinical pilot study has demonstrated that doxycycline reduces the expression of stemness markers limiting the CSC burden in early breast cancer patients [214]. Tri-phenyl-phosphonium (TPP)-related compounds also offer novel strategies for cancer treatment, as they inhibit CSC metastasis either as stand-alone [215] or in combination with an inhibitor of mitochondrial translation [216]. Moreover, cyanine dyes also inhibit mitochondrial metabolism suppressing the growth CSCs impeding metastasis in vivo [217]. Other examples are those afforded by metformin [218] and phenformin [219,220] that promote the inhibition of mitochondrial OXPHOS and reduce the risk of cancer in diabetic patients, increasing their rates of survival. More recently, the FDA-approved bedaquiline, which promotes the downregulation of the γ subunit of the ATP synthase, inhibits mitochondrial ATP production and metastasis in vivo [221,222]. These results emphasize the importance of drug repurposing strategies to accelerate clinical translation of drugs for treatment of cancer—in other words, the use of drugs already available and with acceptable known side effects in humans to improve the quality of life of cancer patients [223,224].
In this line, we recently found that nebivolol, a third-generation β1-blocker, halts colon and breast tumor growth in vivo by a severe inhibition of OXPHOS [60]. Mechanistically, nebivolol binds to β1-adrenergic receptors in cancer cells, causing the increase in IF1 content bound to the ATP synthase and promoting its inhibition. Concurrently, nebivolol prevents the phosphorylation of NDUFS7, a Complex I subunit, which promotes partial inhibition of the activity of the enzyme [60]. In addition, nebivolol also arrests proliferation of endothelial cells by blocking β1-adrenergic receptors impeding tumor angiogenesis [60]. Hence, nebivolol treatment promotes the arrest of colon, breast, lung and squamous cell carcinomas [20,60,225] by promoting a metabolic and redox crisis in cancer cells [60]. We propose that nebivolol is a very strong candidate to be repurposed in “basket trials”, those in which cancer patients with carcinomas from different origins are included. Specially, to prevent cancer recurrence and metastasis after conventional cancer therapies that render cancer cells OXPHOS-dependent [226,227,228,229].
Moreover, OXPHOS is not the only target in mitochondria to combat cancer. In fact, enzymes of fatty acid oxidation, such as carnitine palmitoyltransferase 1 (CPT1), electron transfer flavoprotein subunit α (ETFA) and hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit α (HADHA), are increased in LUAD [20], suggesting that the oxidation of fatty acids is required for cancer progression and might provide a target for therapy. Indeed, inhibitors of fatty acid oxidation have been demonstrated to be promising drugs to be repurposed in the case of leukemia cells [230], breast [231] and colon carcinomas [232]. Hence, it is possible to design therapies that simultaneously target two metabolic pathways of mitochondria. Indeed, we have targeted lung adenocarcinomas in mice by restraining the assimilation of fatty acids with orlistat, or its oxidation in mitochondria with etomoxir, in both cases in combination with nebivolol [20]. With this approach, we demonstrated that the combined treatment provided better welfare and extended the lifespan of mice greater than conventional treatment with cisplatin [20]. Overall, these results highlight mitochondria as a gold standard target for the effective treatment of a large diversity of carcinomas.
8. Conclusions and Future Directions
Despite major genetic advances in the understanding of cancer, cancer patients still require the development of innovative therapeutic strategies matched to the phenotype of their tumors to halt progression of the disease. The accumulated evidence indicates that the metabolic reprogramming experienced in cancer provides new biomarkers that, alone or in combination, could be exploited to halt the disease in a successful personalized medicine. In this review, we have summarized the different functions that the mitochondrial ATP synthase and its inhibitor protein, IF1, play in cellular biology and in cancer progression. We have overviewed the mechanism by which the ATP synthase/IF1 axis contributes to metabolic reprogramming to an enhanced glycolytic phenotype, both in cancer cells and in the maintenance of stemness, and its potential both as biomarkers of prognosis and as targets for therapy. Moreover, we have highlighted how the ATP synthase/IF1 axis contributes to the signaling of cell-type specific programs that allow the adaptation of the cell/organisms to different changing cues, and finally, how the ATP synthase/IF1 axis also participates in preventing the execution of cell death and hence, in therapeutic resistance of the carcinomas. We have emphasized that the relative low activity of mitochondrial metabolic pathways, such as OXPHOS and β-oxidation in lung adenocarcinomas, contribute to cancer progression. In fact, the analysis of the expression of enzymes of metabolism revealed that the alteration of mitochondria is a most evident target in lung adenocarcinomas, and both the catalytic subunit of ATP synthase (β-F1-ATPase) and IF1, provide two most relevant biomarkers of metastatic disease. Actually, and depending on the type of carcinoma, the expression levels of IF1 define its activity as a tumor suppressor or as an oncogene. Although the glycolytic phenotype is needed for tumor growth, metastatic disease requires the activity of OXPHOS, i.e., relatively high β-F1-ATPase expression in the carcinoma. However, the activity of ATP synthase is counterbalanced by enhanced expression levels of IF1 that exert tumor suppressor activities in some carcinomas. Hence, targeting mitochondrial ATP synthase and IF1 holds great promise for overcoming therapeutic resistance and impeding metastatic disease. We have emphasized the mechanisms that control the expression and activity of β-F1-ATPase and IF1 in cancer, which show similarities with those operating in development and differentiation, to highlight the relevance that post-transcriptional mechanisms play in the regulation of the mitochondrial phenotype of mammalian tissues and carcinomas. Unfortunately, this is a field that is scarcely investigated, but that could illuminate new biomarkers and therapeutic targets for cancer treatment. Finally, we summarize recent preclinical findings of the potential that OXPHOS and β-oxidation have as targets to prevent cancer progression. We think that research focused on the ATP synthase/IF1 axis will accelerate translation of energy metabolism in more effective and personalized cancer therapies.
Acknowledgments
We thank current and past members of the lab for helpful discussions.
Author Contributions
S.D.-Z. and J.M.C. contributed to the conception, design and writing of the paper. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The work was supported by grants from MINECO (PID2019-108674RB-100), CIBERER-ISCIII (CB06/07/0017) and Fundación Ramón Areces, Spain. SDZ was supported by a predoctoral fellowship from FPI-MINECO, Fondo Social Europeo.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Esparza-Molto P.B., Nuevo-Tapioles C., Cuezva J.M. Regulation of the H(+)-ATP synthase by IF1: A role in mitohormesis. Cell. Mol. Life Sci. 2017;74:2151–2166. doi: 10.1007/s00018-017-2462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandel N.S. Evolution of Mitochondria as Signaling Organelles. Cell Metab. 2015;22:204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Shadel G.S., Horvath T.L. Mitochondrial ROS Signaling in Organismal Homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spinelli J.B., Haigis M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018;20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunnari J., Suomalainen A. Mitochondria: In sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Dai L., Li D. Mitophagy in neurological disorders. J. Neuroinflamm. 2021;18:297. doi: 10.1186/s12974-021-02334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roth K.G., Mambetsariev I., Kulkarni P., Salgia R. The Mitochondrion as an Emerging Therapeutic Target in Cancer. Trends Mol. Med. 2020;26:119–134. doi: 10.1016/j.molmed.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miwa S., Kashyap S., Chini E., von Zglinicki T. Mitochondrial dysfunction in cell senescence and aging. J. Clin. Investig. 2022;132:e158447. doi: 10.1172/JCI158447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vyas S., Zaganjor E., Haigis M.C. Mitochondria and Cancer. Cell. 2016;166:555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warburg O. The Metabolism of Tumors. Arnold Constable; London, UK: 1930. [Google Scholar]
- 11.Lehninger A. Biochemistry. Worth Publishers; New York, NY, USA: 1970. [Google Scholar]
- 12.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Rios F., Sanchez-Arago M., Garcia-Garcia E., Ortega A.D., Berrendero J.R., Pozo-Rodriguez F., Lopez-Encuentra A., Ballestin C., Cuezva J.M. Loss of the mitochondrial bioenergetic capacity underlies the glucose avidity of carcinomas. Cancer Res. 2007;67:9013–9017. doi: 10.1158/0008-5472.CAN-07-1678. [DOI] [PubMed] [Google Scholar]
- 17.Rigo P., Paulus P., Kaschten B.J., Hustinx R., Bury T., Jerusalem G., Benoit T., Foidart-Willems J. Oncological applications of positron emission tomography with fluorine-18 fluorodeoxyglucose. Eur. J. Nucl. Med. 1996;23:1641–1674. doi: 10.1007/BF01249629. [DOI] [PubMed] [Google Scholar]
- 18.Mankoff D.A., Eary J.F., Link J.M., Muzi M., Rajendran J.G., Spence A.M., Krohn K.A. Tumor-specific positron emission tomography imaging in patients: [18F] fluorodeoxyglucose and beyond. Clin. Cancer Res. 2007;13:3460–3469. doi: 10.1158/1078-0432.CCR-07-0074. [DOI] [PubMed] [Google Scholar]
- 19.Ortega A.D., Sanchez-Arago M., Giner-Sanchez D., Sanchez-Cenizo L., Willers I., Cuezva J.M. Glucose avidity of carcinomas. Cancer Lett. 2009;276:125–135. doi: 10.1016/j.canlet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Torresano L., Santacatterina F., Dominguez-Zorita S., Nuevo-Tapioles C., Nunez-Salgado A., Esparza-Molto P.B., Gonzalez-Llorente L., Romero-Carraminana I., Nunez de Arenas C., Sanchez-Garrido B., et al. Analysis of the metabolic proteome of lung adenocarcinomas by reverse-phase protein arrays (RPPA) emphasizes mitochondria as targets for therapy. Oncogenesis. 2022;11:24. doi: 10.1038/s41389-022-00400-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R., Green D.R. Metabolic reprogramming and metabolic dependency in T cells. Immunol. Rev. 2012;249:14–26. doi: 10.1111/j.1600-065X.2012.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasan K., Werner M., Chandel N.S. Mitochondrial Metabolism as a Target for Cancer Therapy. Cell Metab. 2020;32:341–352. doi: 10.1016/j.cmet.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J.Y., Zhang C., Wang X., Zhai L., Ma Y., Mao Y., Qian K., Sun C., Liu Z., Jiang S., et al. Integrative Proteomic Characterization of Human Lung Adenocarcinoma. Cell. 2020;182:245–261.e17. doi: 10.1016/j.cell.2020.05.043. [DOI] [PubMed] [Google Scholar]
- 24.Ding Z., Wang N., Ji N., Chen Z.S. Proteomics technologies for cancer liquid biopsies. Mol. Cancer. 2022;21:53. doi: 10.1186/s12943-022-01526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Chen F., Chandrashekar D.S., Varambally S., Creighton C.J. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat Commun. 2022;13:2669. doi: 10.1038/s41467-022-30342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuezva J.M., Ortega A.D., Willers I., Sanchez-Cenizo L., Aldea M., Sanchez-Arago M. The tumor suppressor function of mitochondria: Translation into the clinics. Biochim. Biophys. Acta. 2009;1792:1145–1158. doi: 10.1016/j.bbadis.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Vander Heiden M.G., Lunt S.Y., Dayton T.L., Fiske B.P., Israelsen W.J., Mattaini K.R., Vokes N.I., Stephanopoulos G., Cantley L.C., Metallo C.M., et al. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb. Symp. Quant. Biol. 2013;76:325–334. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- 28.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 29.DeBerardinis R.J., Chandel N.S. We need to talk about the Warburg effect. Nat Metab. 2020;2:127–129. doi: 10.1038/s42255-020-0172-2. [DOI] [PubMed] [Google Scholar]
- 30.Esparza-Molto P.B., Cuezva J.M. Reprogramming oxidative phosphorylation in cancer: A role for RNA binding proteins. Antioxid. Redox Signal. 2020;33:927–945. doi: 10.1089/ars.2019.7988. [DOI] [PubMed] [Google Scholar]
- 31.Paul S., Ghosh S., Kumar S. Tumor glycolysis, an essential sweet tooth of tumor cells. Semin. Cancer Biol. 2022;86:1216–1230. doi: 10.1016/j.semcancer.2022.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Zaidi N., Swinnen J.V., Smans K. ATP-citrate lyase: A key player in cancer metabolism. Cancer Res. 2012;72:3709–3714. doi: 10.1158/0008-5472.CAN-11-4112. [DOI] [PubMed] [Google Scholar]
- 33.Jin J., Byun J.K., Choi Y.K., Park K.G. Targeting glutamine metabolism as a therapeutic strategy for cancer. Exp. Mol. Med. 2023;55:706–715. doi: 10.1038/s12276-023-00971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patra K.C., Hay N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torresano L., Nuevo-Tapioles C., Santacatterina F., Cuezva J.M. Metabolic reprogramming and disease progression in cancer patients. Biochim. Et Biophys. Acta. Mol. Basis Dis. 2020;1866:165721. doi: 10.1016/j.bbadis.2020.165721. [DOI] [PubMed] [Google Scholar]
- 36.Sanzey M., Abdul Rahim S.A., Oudin A., Dirkse A., Kaoma T., Vallar L., Herold-Mende C., Bjerkvig R., Golebiewska A., Niclou S.P. Comprehensive analysis of glycolytic enzymes as therapeutic targets in the treatment of glioblastoma. PLoS ONE. 2015;10:e0123544. doi: 10.1371/journal.pone.0123544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Z., Lu J., Han L., Wang X., Man Q., Liu S. Prognostic significance of two lipid metabolism enzymes, HADHA and ACAT2, in clear cell renal cell carcinoma. Tumour Biol. 2016;37:8121–8130. doi: 10.1007/s13277-015-4720-4. [DOI] [PubMed] [Google Scholar]
- 38.Wang J., Liu F., Ao P., Li X., Zheng H., Wu D., Zhang N., She J., Yuan J., Wu X. Correlation of PDK1 expression with clinicopathologic features and prognosis of hepatocellular carcinoma. Onco Targets Ther. 2016;9:5597–5602. doi: 10.2147/OTT.S110646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin P.C., Lin J.K., Yang S.H., Wang H.S., Li A.F., Chang S.C. Expression of beta-F1-ATPase and mitochondrial transcription factor A and the change in mitochondrial DNA content in colorectal cancer: Clinical data analysis and evidence from an in vitro study. Int. J. Color. Dis. 2008;23:1223–1232. doi: 10.1007/s00384-008-0539-4. [DOI] [PubMed] [Google Scholar]
- 40.Liu G., Zhu J., Yu M., Cai C., Zhou Y., Yu M., Fu Z., Gong Y., Yang B., Li Y., et al. Glutamate dehydrogenase is a novel prognostic marker and predicts metastases in colorectal cancer patients. J. Transl. Med. 2015;13:144. doi: 10.1186/s12967-015-0500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin L., Chun J., Pan C., Kumar A., Zhang G., Ha Y., Li D., Alesi G.N., Kang Y., Zhou L., et al. The PLAG1-GDH1 Axis Promotes Anoikis Resistance and Tumor Metastasis through CamKK2-AMPK Signaling in LKB1-Deficient Lung Cancer. Mol. Cell. 2018;69:87–99.e7. doi: 10.1016/j.molcel.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li K., Zhang C., Chen L., Wang P., Fang Y., Zhu J., Chen S., Du J., Shen B., Wu K., et al. The role of acetyl-coA carboxylase2 in head and neck squamous cell carcinoma. PeerJ. 2019;7:e7037. doi: 10.7717/peerj.7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osugi J., Yamaura T., Muto S., Okabe N., Matsumura Y., Hoshino M., Higuchi M., Suzuki H., Gotoh M. Prognostic impact of the combination of glucose transporter 1 and ATP citrate lyase in node-negative patients with non-small lung cancer. Lung Cancer. 2015;88:310–318. doi: 10.1016/j.lungcan.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Wang D., Yin L., Wei J., Yang Z., Jiang G. ATP citrate lyase is increased in human breast cancer, depletion of which promotes apoptosis. Tumour Biol. 2017;39:1010428317698338. doi: 10.1177/1010428317698338. [DOI] [PubMed] [Google Scholar]
- 45.Cai Y., Wang J., Zhang L., Wu D., Yu D., Tian X., Liu J., Jiang X., Shen Y., Zhang L., et al. Expressions of fatty acid synthase and HER2 are correlated with poor prognosis of ovarian cancer. Med. Oncol. 2015;32:391. doi: 10.1007/s12032-014-0391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu S., Balakrishnan A., Bok R.A., Anderton B., Larson P.E., Nelson S.J., Kurhanewicz J., Vigneron D.B., Goga A. (13)C-Pyruvate Imaging Reveals Alterations in Glycolysis that Precede c-Myc-Induced Tumor Formation and Regression. Cell Metab. 2011;14:131–142. doi: 10.1016/j.cmet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeBerardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 48.Hu Y., Lu W., Chen G., Wang P., Chen Z., Zhou Y., Ogasawara M., Trachootham D., Feng L., Pelicano H., et al. K-ras(G12V) transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res. 2012;22:399–412. doi: 10.1038/cr.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonnet S., Archer S.L., Allalunis-Turner J., Haromy A., Beaulieu C., Thompson R., Lee C.T., Lopaschuk G.D., Puttagunta L., Bonnet S., et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 50.Michelakis E.D., Webster L., Mackey J.R. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br. J. Cancer. 2008;99:989–994. doi: 10.1038/sj.bjc.6604554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Arago M., Chamorro M., Cuezva J.M. Selection of cancer cells with repressed mitochondria triggers colon cancer progression. Carcinogenesis. 2010;31:567–576. doi: 10.1093/carcin/bgq012. [DOI] [PubMed] [Google Scholar]
- 52.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 53.Schulz T.J., Thierbach R., Voigt A., Drewes G., Mietzner B., Steinberg P., Pfeiffer A.F., Ristow M. Induction of oxidative metabolism by mitochondrial frataxin inhibits cancer growth: Otto Warburg revisited. J. Biol. Chem. 2006;281:977–981. doi: 10.1074/jbc.M511064200. [DOI] [PubMed] [Google Scholar]
- 54.Fantin V.R., St-Pierre J., Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 55.McFate T., Mohyeldin A., Lu H., Thakar J., Henriques J., Halim N.D., Wu H., Schell M.J., Tsang T.M., Teahan O., et al. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J. Biol. Chem. 2008;283:22700–22708. doi: 10.1074/jbc.M801765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hong X., Zhong L., Xie Y., Zheng K., Pang J., Li Y., Yang Y., Xu X., Mi P., Cao H., et al. Matrine Reverses the Warburg Effect and Suppresses Colon Cancer Cell Growth via Negatively Regulating HIF-1alpha. Front. Pharmacol. 2019;10:1437. doi: 10.3389/fphar.2019.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cruz-Bermudez A., Laza-Briviesca R., Vicente-Blanco R.J., Garcia-Grande A., Coronado M.J., Laine-Menendez S., Alfaro C., Sanchez J.C., Franco F., Calvo V., et al. Cancer-associated fibroblasts modify lung cancer metabolism involving ROS and TGF-beta signaling. Free Radic. Biol. Med. 2019;130:163–173. doi: 10.1016/j.freeradbiomed.2018.10.450. [DOI] [PubMed] [Google Scholar]
- 58.Fu Y., Liu S., Yin S., Niu W., Xiong W., Tan M., Li G., Zhou M. The reverse Warburg effect is likely to be an Achilles’ heel of cancer that can be exploited for cancer therapy. Oncotarget. 2017;8:57813–57825. doi: 10.18632/oncotarget.18175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramanathan A., Wang C., Schreiber S.L. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc. Natl. Acad. Sci. USA. 2005;102:5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nuevo-Tapioles C., Santacatterina F., Stamatakis K., Nunez de Arenas C., Gomez de Cedron M., Formentini L., Cuezva J.M. Coordinate beta-adrenergic inhibition of mitochondrial activity and angiogenesis arrest tumor growth. Nat Commun. 2020;11:3606. doi: 10.1038/s41467-020-17384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mascaraque M., Delgado-Wicke P., Nuevo-Tapioles C., Gracia-Cazana T., Abarca-Lachen E., Gonzalez S., Cuezva J.M., Gilaberte Y., Juarranz A. Metformin as an Adjuvant to Photodynamic Therapy in Resistant Basal Cell Carcinoma Cells. Cancers. 2020;12:668. doi: 10.3390/cancers12030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mascaraque-Checa M., Gallego-Rentero M., Nicolas-Morala J., Portillo-Esnaola M., Cuezva J.M., Gonzalez S., Gilaberte Y., Juarranz A. Metformin overcomes metabolic reprogramming-induced resistance of skin squamous cell carcinoma to photodynamic therapy. Mol. Metab. 2022;60:101496. doi: 10.1016/j.molmet.2022.101496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobbelstein M., Moll U. Targeting tumour-supportive cellular machineries in anticancer drug development. Nat Rev. Drug Discov. 2014;13:179–196. doi: 10.1038/nrd4201. [DOI] [PubMed] [Google Scholar]
- 64.Pollak M. Overcoming Drug Development Bottlenecks with Repurposing: Repurposing biguanides to target energy metabolism for cancer treatment. Nat. Med. 2014;20:591–593. doi: 10.1038/nm.3596. [DOI] [PubMed] [Google Scholar]
- 65.Galluzzi L., Kepp O., Vander Heiden M.G., Kroemer G. Metabolic targets for cancer therapy. Nat Rev. Drug Discov. 2013;12:829–846. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 66.Amoedo N.D., Sarlak S., Obre E., Esteves P., Begueret H., Kieffer Y., Rousseau B., Dupis A., Izotte J., Bellance N., et al. Targeting the mitochondrial trifunctional protein restrains tumor growth in oxidative lung carcinomas. J. Clin. Investig. 2021;131:e133081. doi: 10.1172/JCI133081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cogliati S., Enriquez J.A., Scorrano L. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem. Sci. 2016;41:261–273. doi: 10.1016/j.tibs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 68.Wittig I., Schagger H. Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochim. Biophys. Acta. 2009;1787:672–680. doi: 10.1016/j.bbabio.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 69.Boyer P.D. The ATP synthase. A splendid molecular machine. Annu. Rev. Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 70.Kuhlbrandt W. Structure and Mechanisms of F-Type ATP Synthases. Annu. Rev. Biochem. 2019;88:515–549. doi: 10.1146/annurev-biochem-013118-110903. [DOI] [PubMed] [Google Scholar]
- 71.Pullman M.E., Monroy G.C. A Naturally Occurring Inhibitor of Mitochondrial Adenosine Triphosphatase. J. Biol. Chem. 1963;238:3762–3769. doi: 10.1016/S0021-9258(19)75338-1. [DOI] [PubMed] [Google Scholar]
- 72.Walker J.E. The ATP synthase: The understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 73.Garcia-Bermudez J., Cuezva J.M. The ATPase Inhibitory Factor 1 (IF1): A master regulator of energy metabolism and of cell survival. Biochim. Biophys. Acta. 2016;1857:1167–1182. doi: 10.1016/j.bbabio.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 74.Sanchez-Cenizo L., Formentini L., Aldea M., Ortega A.D., Garcia-Huerta P., Sanchez-Arago M., Cuezva J.M. Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J. Biol. Chem. 2010;285:25308–25313. doi: 10.1074/jbc.M110.146480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Formentini L., Sánchez-Aragó M., Sánchez-Cenizo L., Cuezva J.M. The mitochondrial ATPase Inhibitory Factor 1 (IF1) triggers a ROS-mediated retrograde pro-survival and proliferative response. Mol. Cell. 2012;45:731–742. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 76.Garcia-Bermudez J., Sanchez-Arago M., Soldevilla B., Del Arco A., Nuevo-Tapioles C., Cuezva J.M. PKA Phosphorylates the ATPase Inhibitory Factor 1 and Inactivates Its Capacity to Bind and Inhibit the Mitochondrial H(+)-ATP Synthase. Cell Rep. 2015;12:2143–2155. doi: 10.1016/j.celrep.2015.08.052. [DOI] [PubMed] [Google Scholar]
- 77.Kahancova A., Sklenar F., Jezek P., Dlaskova A. Regulation of glucose-stimulated insulin secretion by ATPase Inhibitory Factor 1 (IF1) FEBS Lett. 2018;592:999–1009. doi: 10.1002/1873-3468.12991. [DOI] [PubMed] [Google Scholar]
- 78.Kahancova A., Sklenar F., Jezek P., Dlaskova A. Overexpression of native IF1 downregulates glucose-stimulated insulin secretion by pancreatic INS-1E cells. Sci. Rep. 2020;10:1551. doi: 10.1038/s41598-020-58411-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Formentini L., Pereira M.P., Sanchez-Cenizo L., Santacatterina F., Lucas J.J., Navarro C., Martinez-Serrano A., Cuezva J.M. In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning. EMBO J. 2014;33:762–778. doi: 10.1002/embj.201386392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Santacatterina F., Sanchez-Cenizo L., Formentini L., Mobasher M.A., Casas E., Rueda C.B., Martinez-Reyes I., Nunez de Arenas C., Garcia-Bermudez J., Zapata J.M., et al. Down-regulation of oxidative phosphorylation in the liver by expression of the ATPase inhibitory factor 1 induces a tumor-promoter metabolic state. Oncotarget. 2016;7:490–508. doi: 10.18632/oncotarget.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Formentini L., Santacatterina F., Nunez de Arenas C., Stamatakis K., Lopez-Martinez D., Logan A., Fresno M., Smits R., Murphy M.P., Cuezva J.M. Mitochondrial ROS Production Protects the Intestine from Inflammation through Functional M2 Macrophage Polarization. Cell Rep. 2017;19:1202–1213. doi: 10.1016/j.celrep.2017.04.036. [DOI] [PubMed] [Google Scholar]
- 82.Esparza-Molto P.B., Romero-Carraminana I., Nunez de Arenas C., Pereira M.P., Blanco N., Pardo B., Bates G.R., Sanchez-Castillo C., Artuch R., Murphy M.P., et al. Generation of mitochondrial reactive oxygen species is controlled by ATPase inhibitory factor 1 and regulates cognition. PLoS Biol. 2021;19:e3001252. doi: 10.1371/journal.pbio.3001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dominguez-Zorita S., Romero-Carraminana I., Santacatterina F., Esparza-Molto P.B., Simo C., Del-Arco A., Nunez de Arenas C., Saiz J., Barbas C., Cuezva J.M. IF1 ablation prevents ATP synthase oligomerization, enhances mitochondrial ATP turnover and promotes an adenosine-mediated pro-inflammatory phenotype. Cell Death Dis. 2023;14:413. doi: 10.1038/s41419-023-05957-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou B., Caudal A., Tang X., Chavez J.D., McMillen T.S., Keller A., Villet O., Zhao M., Liu Y., Ritterhoff J., et al. Upregulation of mitochondrial ATPase inhibitory factor 1 (ATPIF1) mediates increased glycolysis in mouse hearts. J. Clin. Investig. 2022;132:e155333. doi: 10.1172/JCI155333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanchez-Arago M., Formentini L., Martinez-Reyes I., Garcia-Bermudez J., Santacatterina F., Sanchez-Cenizo L., Willers I.M., Aldea M., Najera L., Juarranz A., et al. Expression, regulation and clinical relevance of the ATPase inhibitory factor 1 in human cancers. Oncogenesis. 2013;2:e46. doi: 10.1038/oncsis.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yun J., Finkel T. Mitohormesis. Cell Metab. 2014;19:757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Esparza-Molto P.B., Nuevo-Tapioles C., Chamorro M., Najera L., Torresano L., Santacatterina F., Cuezva J.M. Tissue-specific expression and post-transcriptional regulation of the ATPase inhibitory factor 1 (IF1) in human and mouse tissues. FASEB J. 2019;33:1836–1851. doi: 10.1096/fj.201800756R. [DOI] [PubMed] [Google Scholar]
- 88.Song R., Song H., Liang Y., Yin D., Zhang H., Zheng T., Wang J., Lu Z., Song X., Pei T., et al. Reciprocal activation between ATPase inhibitory factor 1 and NF-kappaB drives hepatocellular carcinoma angiogenesis and metastasis. Hepatology. 2014;60:1659–1673. doi: 10.1002/hep.27312. [DOI] [PubMed] [Google Scholar]
- 89.Sanchez-Gonzalez C., Nuevo-Tapioles C., Herrero Martin J.C., Pereira M.P., Serrano Sanz S., Ramirez de Molina A., Cuezva J.M., Formentini L. Dysfunctional oxidative phosphorylation shunts branched-chain amino acid catabolism onto lipogenesis in skeletal muscle. EMBO J. 2020;39:e103812. doi: 10.15252/embj.2019103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gu J., Zhang L., Zong S., Guo R., Liu T., Yi J., Wang P., Zhuo W., Yang M. Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science. 2019;364:1068–1075. doi: 10.1126/science.aaw4852. [DOI] [PubMed] [Google Scholar]
- 91.Pinke G., Zhou L., Sazanov L.A. Cryo-EM structure of the entire mammalian F-type ATP synthase. Nat. Struct. Mol. Biol. 2020;27:1077–1085. doi: 10.1038/s41594-020-0503-8. [DOI] [PubMed] [Google Scholar]
- 92.Gross A., Pilcher K., Blachly-Dyson E., Basso E., Jockel J., Bassik M.C., Korsmeyer S.J., Forte M. Biochemical and genetic analysis of the mitochondrial response of yeast to BAX and BCL-X(L) Mol. Cell. Biol. 2000;20:3125–3136. doi: 10.1128/MCB.20.9.3125-3136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Santamaria G., Martinez-Diez M., Fabregat I., Cuezva J.M. Efficient execution of cell death in non-glycolytic cells requires the generation of ROS controlled by the activity of mitochondrial H+-ATP synthase. Carcinogenesis. 2006;27:925–935. doi: 10.1093/carcin/bgi315. [DOI] [PubMed] [Google Scholar]
- 94.Dey R., Moraes C.T. Lack of oxidative phosphorylation and low mitochondrial membrane potential decrease susceptibility to apoptosis and do not modulate the protective effect of Bcl-x(L) in osteosarcoma cells. J. Biol. Chem. 2000;275:7087–7094. doi: 10.1074/jbc.275.10.7087. [DOI] [PubMed] [Google Scholar]
- 95.Matsuyama S., Xu Q., Velours J., Reed J.C. The Mitochondrial F0F1-ATPase proton pump is required for function of the proapoptotic protein Bax in yeast and mammalian cells. Mol. Cell. 1998;1:327–336. doi: 10.1016/S1097-2765(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 96.Mnatsakanyan N., Park H.A., Wu J., He X., Llaguno M.C., Latta M., Miranda P., Murtishi B., Graham M., Weber J., et al. Mitochondrial ATP synthase c-subunit leak channel triggers cell death upon loss of its F1 subcomplex. Cell Death Differ. 2022;29:1874–1887. doi: 10.1038/s41418-022-00972-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bernardi P., Carraro M., Lippe G. The mitochondrial permeability transition: Recent progress and open questions. FEBS J. 2021;289:7051–7074. doi: 10.1111/febs.16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bonora M., Giorgi C., Pinton P. Molecular mechanisms and consequences of mitochondrial permeability transition. Nat. Rev. Mol. Cell Biol. 2022;23:266–285. doi: 10.1038/s41580-021-00433-y. [DOI] [PubMed] [Google Scholar]
- 99.Hunter D.R., Haworth R.A. The Ca2+-induced membrane transition in mitochondria. I. The protective mechanisms. Arch. Biochem. Biophys. 1979;195:453–459. doi: 10.1016/0003-9861(79)90371-0. [DOI] [PubMed] [Google Scholar]
- 100.Kinnally K.W., Campo M.L., Tedeschi H. Mitochondrial channel activity studied by patch-clamping mitoplasts. J. Bioenerg. Biomembr. 1989;21:497–506. doi: 10.1007/BF00762521. [DOI] [PubMed] [Google Scholar]
- 101.Petronilli V., Szabo I., Zoratti M. The inner mitochondrial membrane contains ion-conducting channels similar to those found in bacteria. FEBS Lett. 1989;259:137–143. doi: 10.1016/0014-5793(89)81513-3. [DOI] [PubMed] [Google Scholar]
- 102.Gerle C. Mitochondrial F-ATP synthase as the permeability transition pore. Pharmacol. Res. 2020;160:105081. doi: 10.1016/j.phrs.2020.105081. [DOI] [PubMed] [Google Scholar]
- 103.Baines C.P., Kaiser R.A., Purcell N.H., Blair N.S., Osinska H., Hambleton M.A., Brunskill E.W., Sayen M.R., Gottlieb R.A., Dorn G.W., et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 104.Crompton M., Ellinger H., Costi A. Inhibition by cyclosporin A of a Ca2+-dependent pore in heart mitochondria activated by inorganic phosphate and oxidative stress. Biochem. J. 1988;255:357–360. [PMC free article] [PubMed] [Google Scholar]
- 105.Giorgio V., von Stockum S., Antoniel M., Fabbro A., Fogolari F., Forte M., Glick G.D., Petronilli V., Zoratti M., Szabo I., et al. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc. Natl. Acad. Sci. USA. 2013;110:5887–5892. doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alavian K.N., Beutner G., Lazrove E., Sacchetti S., Park H.A., Licznerski P., Li H., Nabili P., Hockensmith K., Graham M., et al. An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proc. Natl. Acad. Sci. USA. 2014;111:10580–10585. doi: 10.1073/pnas.1401591111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bonora M., Bononi A., De Marchi E., Giorgi C., Lebiedzinska M., Marchi S., Patergnani S., Rimessi A., Suski J.M., Wojtala A., et al. Role of the c subunit of the FO ATP synthase in mitochondrial permeability transition. Cell Cycle. 2013;12:674–683. doi: 10.4161/cc.23599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mnatsakanyan N., Llaguno M.C., Yang Y., Yan Y., Weber J., Sigworth F.J., Jonas E.A. A mitochondrial megachannel resides in monomeric F1FO ATP synthase. Nat Commun. 2019;10:5823. doi: 10.1038/s41467-019-13766-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He J., Ford H.C., Carroll J., Ding S., Fearnley I.M., Walker J.E. Persistence of the mitochondrial permeability transition in the absence of subunit c of human ATP synthase. Proc. Natl. Acad. Sci. USA. 2017;114:3409–3414. doi: 10.1073/pnas.1702357114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carroll J., He J., Ding S., Fearnley I.M., Walker J.E. Persistence of the permeability transition pore in human mitochondria devoid of an assembled ATP synthase. Proc. Natl. Acad. Sci. USA. 2019;116:12816–12821. doi: 10.1073/pnas.1904005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Giorgio V., Burchell V., Schiavone M., Bassot C., Minervini G., Petronilli V., Argenton F., Forte M., Tosatto S., Lippe G., et al. Ca(2+) binding to F-ATP synthase beta subunit triggers the mitochondrial permeability transition. EMBO Rep. 2017;18:1065–1076. doi: 10.15252/embr.201643354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Faccenda D., Tan C.H., Seraphim A., Duchen M.R., Campanella M. IF1 limits the apoptotic-signalling cascade by preventing mitochondrial remodelling. Cell Death Differ. 2013;20:686–697. doi: 10.1038/cdd.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Galber C., Fabbian S., Gatto C., Grandi M., Carissimi S., Acosta M.J., Sgarbi G., Tiso N., Argenton F., Solaini G., et al. The mitochondrial inhibitor IF1 binds to the ATP synthase OSCP subunit and protects cancer cells from apoptosis. Cell Death Dis. 2023;14:54. doi: 10.1038/s41419-023-05572-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. doi: 10.1126/science.124.3215.269. [DOI] [PubMed] [Google Scholar]
- 115.Cuezva J.M., Krajewska M., de Heredia M.L., Krajewski S., Santamaria G., Kim H., Zapata J.M., Marusawa H., Chamorro M., Reed J.C. The bioenergetic signature of cancer: A marker of tumor progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- 116.Wang X., Moraes C.T. Increases in mitochondrial biogenesis impair carcinogenesis at multiple levels. Mol. Oncol. 2011;5:399–409. doi: 10.1016/j.molonc.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.D’Errico I., Salvatore L., Murzilli S., Lo Sasso G., Latorre D., Martelli N., Egorova A.V., Polishuck R., Madeyski-Bengtson K., Lelliott C., et al. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1alpha) is a metabolic regulator of intestinal epithelial cell fate. Proc. Natl. Acad. Sci. USA. 2011;108:6603–6608. doi: 10.1073/pnas.1016354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xing F., Luan Y., Cai J., Wu S., Mai J., Gu J., Zhang H., Li K., Lin Y., Xiao X., et al. The Anti-Warburg Effect Elicited by the cAMP-PGC1alpha Pathway Drives Differentiation of Glioblastoma Cells into Astrocytes. Cell Rep. 2017;18:468–481. doi: 10.1016/j.celrep.2016.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.LaGory E.L., Wu C., Taniguchi C.M., Ding C.C., Chi J.T., von Eyben R., Scott D.A., Richardson A.D., Giaccia A.J. Suppression of PGC-1alpha Is Critical for Reprogramming Oxidative Metabolism in Renal Cell Carcinoma. Cell Rep. 2015;12:116–127. doi: 10.1016/j.celrep.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu W., Beck B.H., Vaidya K.S., Nash K.T., Feeley K.P., Ballinger S.W., Pounds K.M., Denning W.L., Diers A.R., Landar A., et al. Metastasis suppressor KISS1 seems to reverse the Warburg effect by enhancing mitochondrial biogenesis. Cancer Res. 2014;74:954–963. doi: 10.1158/0008-5472.CAN-13-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ikari R., Mukaisho K.I., Kageyama S., Nagasawa M., Kubota S., Nakayama T., Murakami S., Taniura N., Tanaka H., Kushima R.P., et al. Differences in the Central Energy Metabolism of Cancer Cells between Conventional 2D and Novel 3D Culture Systems. Int. J. Mol. Sci. 2021;22:1805. doi: 10.3390/ijms22041805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mootha V.K., Bunkenborg J., Olsen J.V., Hjerrild M., Wisniewski J.R., Stahl E., Bolouri M.S., Ray H.N., Sihag S., Kamal M., et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/S0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 123.Fernandez-Vizarra E., Enriquez J.A., Perez-Martos A., Montoya J., Fernandez-Silva P. Tissue-specific differences in mitochondrial activity and biogenesis. Mitochondrion. 2011;11:207–213. doi: 10.1016/j.mito.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 124.Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E., Walford G.A., Sugiana C., Boneh A., Chen W.K., et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim J., DeBerardinis R.J. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab. 2019;30:434–446. doi: 10.1016/j.cmet.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Labuschagne C.F., Cheung E.C., Blagih J., Domart M.C., Vousden K.H. Cell Clustering Promotes a Metabolic Switch that Supports Metastatic Colonization. Cell Metab. 2019;30:720–734. doi: 10.1016/j.cmet.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hensley C.T., Faubert B., Yuan Q., Lev-Cohain N., Jin E., Kim J., Jiang L., Ko B., Skelton R., Loudat L., et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Momcilovic M., Jones A., Bailey S.T., Waldmann C.M., Li R., Lee J.T., Abdelhady G., Gomez A., Holloway T., Schmid E., et al. In vivo imaging of mitochondrial membrane potential in non-small-cell lung cancer. Nature. 2019;575:380–384. doi: 10.1038/s41586-019-1715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.R., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vazquez F., Lim J.H., Chim H., Bhalla K., Girnun G., Pierce K., Clish C.B., Granter S.R., Widlund H.R., Spiegelman B.M., et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.LeBleu V.S., O’Connell J.T., Gonzalez Herrera K.N., Wikman H., Pantel K., Haigis M.C., de Carvalho F.M., Damascena A., Domingos Chinen L.T., Rocha R.M., et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014;16:992–1003, 1001-1015. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Viale A., Pettazzoni P., Lyssiotis C.A., Ying H., Sanchez N., Marchesini M., Carugo A., Green T., Seth S., Giuliani V., et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sancho P., Burgos-Ramos E., Tavera A., Bou Kheir T., Jagust P., Schoenhals M., Barneda D., Sellers K., Campos-Olivas R., Grana O., et al. MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 134.Martinez-Reyes I., Diebold L.P., Kong H., Schieber M., Huang H., Hensley C.T., Mehta M.M., Wang T., Santos J.H., Woychik R., et al. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Mol. Cell. 2016;61:199–209. doi: 10.1016/j.molcel.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Basu S., Gnanapradeepan K., Barnoud T., Kung C.P., Tavecchio M., Scott J., Watters A., Chen Q., Kossenkov A.V., Murphy M.E. Mutant p53 controls tumor metabolism and metastasis by regulating PGC-1alpha. Genes Dev. 2018;32:230–243. doi: 10.1101/gad.309062.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pedersen P.L. Tumor mitochondria and the bioenergetics of cancer cells. Prog. Exp. Tumor Res. 1978;22:190–274. doi: 10.1159/000401202. [DOI] [PubMed] [Google Scholar]
- 137.Arismendi-Morillo G. Electron microscopy morphology of the mitochondrial network in gliomas and their vascular microenvironment. Biochim. Biophys. Acta. 2011;1807:602–608. doi: 10.1016/j.bbabio.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 138.Seyfried T.N., Flores R.E., Poff A.M., D’Agostino D.P. Cancer as a metabolic disease: Implications for novel therapeutics. Carcinogenesis. 2014;35:515–527. doi: 10.1093/carcin/bgt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Elliott R.L., Jiang X.P., Head J.F. Mitochondria organelle transplantation: Introduction of normal epithelial mitochondria into human cancer cells inhibits proliferation and increases drug sensitivity. Breast Cancer Res. Treat. 2012;136:347–354. doi: 10.1007/s10549-012-2283-2. [DOI] [PubMed] [Google Scholar]
- 140.Isidoro A., Martinez M., Fernandez P.L., Ortega A.D., Santamaria G., Chamorro M., Reed J.C., Cuezva J.M. Alteration of the bioenergetic phenotype of mitochondria is a hallmark of breast, gastric, lung and oesophageal cancer. Biochem. J. 2004;378:17–20. doi: 10.1042/bj20031541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Acebo P., Giner D., Calvo P., Blanco-Rivero A., Ortega A.D., Fernandez P.L., Roncador G., Fernandez-Malave E., Chamorro M., Cuezva J.M. Cancer abolishes the tissue type-specific differences in the phenotype of energetic metabolism. Transl. Oncol. 2009;2:138–145. doi: 10.1593/tlo.09106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Aldea M., Clofent J., Nunez de Arenas C., Chamorro M., Velasco M., Berrendero J.R., Navarro C., Cuezva J.M. Reverse phase protein microarrays quantify and validate the bioenergetic signature as biomarker in colorectal cancer. Cancer Lett. 2011;311:210–218. doi: 10.1016/j.canlet.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 143.Sheffer M., Bacolod M.D., Zuk O., Giardina S.F., Pincas H., Barany F., Paty P.B., Gerald W.L., Notterman D.A., Domany E. Association of survival and disease progression with chromosomal instability: A genomic exploration of colorectal cancer. Proc. Natl. Acad. Sci. USA. 2009;106:7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Isidoro A., Casado E., Redondo A., Acebo P., Espinosa E., Alonso A.M., Cejas P., Hardisson D., Fresno Vara J.A., Belda-Iniesta C., et al. Breast carcinomas fulfill the Warburg hypothesis and provide metabolic markers of cancer prognosis. Carcinogenesis. 2005;26:2095–2104. doi: 10.1093/carcin/bgi188. [DOI] [PubMed] [Google Scholar]
- 145.Cuezva J.M., Chen G., Alonso A.M., Isidoro A., Misek D.E., Hanash S.M., Beer D.G. The bioenergetic signature of lung adenocarcinomas is a molecular marker of cancer diagnosis and prognosis. Carcinogenesis. 2004;25:1157–1163. doi: 10.1093/carcin/bgh113. [DOI] [PubMed] [Google Scholar]
- 146.Najera L., Alonso-Juarranz M., Garrido M., Ballestin C., Moya L., Martinez-Diaz M., Carrillo R., Juarranz A., Rojo F., Cuezva J.M., et al. Prognostic implications of markers of the metabolic phenotype in human cutaneous melanoma. Br. J. Dermatol. 2019;181:114–127. doi: 10.1111/bjd.17513. [DOI] [PubMed] [Google Scholar]
- 147.Garcia-Ledo L., Nuevo-Tapioles C., Cuevas-Martin C., Martinez-Reyes I., Soldevilla B., Gonzalez-Llorente L., Cuezva J.M. Overexpression of the ATPase Inhibitory Factor 1 Favors a Non-metastatic Phenotype in Breast Cancer. Front. Oncol. 2017;7:69. doi: 10.3389/fonc.2017.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hjerpe E., Egyhazi Brage S., Carlson J., Frostvik Stolt M., Schedvins K., Johansson H., Shoshan M., Avall-Lundqvist E. Metabolic markers GAPDH, PKM2, ATP5B and BEC-index in advanced serous ovarian cancer. BMC Clin. Pathol. 2013;13:30. doi: 10.1186/1472-6890-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun J., Yang Z.L., Miao X., Zou Q., Li J., Liang L., Zeng G., Chen S. ATP5b and beta2-microglobulin are predictive markers for the prognosis of patients with gallbladder cancer. J. Mol. Histol. 2015;46:57–65. doi: 10.1007/s10735-014-9597-9. [DOI] [PubMed] [Google Scholar]
- 150.Li R.J., Zhang G.S., Chen Y.H., Zhu J.F., Lu Q.J., Gong F.J., Kuang W.Y. Down-regulation of mitochondrial ATPase by hypermethylation mechanism in chronic myeloid leukemia is associated with multidrug resistance. Ann. Oncol. 2010;7:1506–1514. doi: 10.1093/annonc/mdp569. [DOI] [PubMed] [Google Scholar]
- 151.Yizhak K., Le Devedec S.E., Rogkoti V.M., Baenke F., de Boer V.C., Frezza C., Schulze A., van de Water B., Ruppin E. A computational study of the Warburg effect identifies metabolic targets inhibiting cancer migration. Mol. Syst. Biol. 2014;10:744. doi: 10.15252/msb.20145746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sanchez-Arago M., Cuezva J.M. The bioenergetic signature of isogenic colon cancer cells predicts the cell death response to treatment with 3-bromopyruvate, iodoacetate or 5-fluorouracil. J. Transl. Med. 2011;9:19. doi: 10.1186/1479-5876-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yang H., Cui Y., Zhu Y. Comprehensive analysis reveals signal and molecular mechanism of mitochondrial energy metabolism pathway in pancreatic cancer. Front. Genet. 2023;14:1117145. doi: 10.3389/fgene.2023.1117145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fiorillo C., Schena C.A., Quero G., Laterza V., Pugliese D., Privitera G., Rosa F., Schepis T., Salvatore L., Di Stefano B., et al. Challenges in Crohn’s Disease Management after Gastrointestinal Cancer Diagnosis. Cancers. 2021;13:574. doi: 10.3390/cancers13030574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Martinez-Reyes I., Cardona L.R., Kong H., Vasan K., McElroy G.S., Werner M., Kihshen H., Reczek C.R., Weinberg S.E., Gao P., et al. Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature. 2020;585:288–292. doi: 10.1038/s41586-020-2475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ricart J., Egea G., Izquierdo J.M., San Martin C., Cuezva J.M. Subcellular structure containing mRNA for beta subunit of mitochondrial H+-ATP synthase in rat hepatocytes is translationally active. Pt 2Biochem. J. 1997;324:635–643. doi: 10.1042/bj3240635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lithgow T., Cuezva J.M., Silver P.A. Highways for protein delivery to the mitochondria. Trends Biochem. Sci. 1997;22:110–113. doi: 10.1016/S0968-0004(97)01007-4. [DOI] [PubMed] [Google Scholar]
- 158.Egea G., Izquierdo J.M., Ricart J., San Martín C., Cuezva J.M. mRNA encoding the beta-subunit of the mitochondrial F1-ATPase complex is a localized mRNA in rat hepatocytes. Biochem. J. 1997;322:557–565. doi: 10.1042/bj3220557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Izquierdo J.M., Cuezva J.M. Control of the translational efficiency of beta-F1-ATPase mRNA depends on the regulation of a protein that binds the 3’ untranslated region of the mRNA. Mol. Cell. Biol. 1997;17:5255–5568. doi: 10.1128/MCB.17.9.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Willers I.M., Isidoro A., Ortega A.D., Fernandez P.L., Cuezva J.M. Selective inhibition of beta-F1-ATPase mRNA translation in human tumours. Biochem. J. 2010;426:319–326. doi: 10.1042/BJ20091570. [DOI] [PubMed] [Google Scholar]
- 161.Izquierdo J.M., Cuezva J.M. Internal-ribosome-entry-site functional activity of the 3’-untranslated region of the mRNA for the beta subunit of mitochondrial H+-ATP synthase. Biochem. J. 2000;346:849–855. doi: 10.1042/bj3460849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Di Liegro C.M., Bellafiore M., Izquierdo J.M., Rantanen A., Cuezva J.M. 3’-untranslated regions of oxidative phosphorylation mRNAs function in vivo as enhancers of translation. Pt 1Biochem. J. 2000;352:109–115. doi: 10.1042/bj3520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Izquierdo J.M., Ricart J., Ostronoff L.K., Egea G., Cuezva J.M. Changing patterns of transcriptional and post-transcriptional control of beta-F1-ATPase gene expression during mitochondrial biogenesis in liver. J. Biol. Chem. 1995;270:10342–10350. doi: 10.1074/jbc.270.17.10342. [DOI] [PubMed] [Google Scholar]
- 164.Valcarce C., Navarrete R.M., Encabo P., Loeches E., Satrustegui J., Cuezva J.M. Postnatal development of rat liver mitochondrial functions. The roles of protein synthesis and of adenine nucleotides. J. Biol. Chem. 1988;263:7767–7775. doi: 10.1016/S0021-9258(18)68565-5. [DOI] [PubMed] [Google Scholar]
- 165.Zhang Y., Chen Y., Gucek M., Xu H. The mitochondrial outer membrane protein MDI promotes local protein synthesis and mtDNA replication. EMBO J. 2016;35:1045–1057. doi: 10.15252/embj.201592994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Willers I.M., Cuezva J.M. Post-transcriptional regulation of the mitochondrial H(+)-ATP synthase: A key regulator of the metabolic phenotype in cancer. Biochim. Biophys. Acta. 2011;1807:543–551. doi: 10.1016/j.bbabio.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 167.Martinez-Diez M., Santamaria G., Ortega A.D., Cuezva J.M. Biogenesis and Dynamics of Mitochondria during the Cell Cycle: Significance of 3’UTRs. PLoS ONE. 2006;1:e107. doi: 10.1371/journal.pone.0000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.De Heredia M.L., Izquierdo J.M., Cuezva J.M. A conserved mechanism for controlling the translation of beta-F1-ATPase mRNA between the fetal liver and cancer cells. J. Biol. Chem. 2000;275:7430–7437. doi: 10.1074/jbc.275.10.7430. [DOI] [PubMed] [Google Scholar]
- 169.Luis A.M., Izquierdo J.M., Ostronoff L.K., Salinas M., Santaren J.F., Cuezva J.M. Translational regulation of mitochondrial differentiation in neonatal rat liver. Specific increase in the translational efficiency of the nuclear-encoded mitochondrial beta-F1-ATPase mRNA. J. Biol. Chem. 1993;268:1868–1875. doi: 10.1016/S0021-9258(18)53935-1. [DOI] [PubMed] [Google Scholar]
- 170.Ortega A.D., Willers I.M., Sala S., Cuezva J.M. Human G3BP1 interacts with beta-F1-ATPase mRNA and inhibits its translation. J. Cell Sci. 2010;123:2685–2696. doi: 10.1242/jcs.065920. [DOI] [PubMed] [Google Scholar]
- 171.Barnes C.J., Li F., Mandal M., Yang Z., Sahin A.A., Kumar R. Heregulin induces expression, ATPase activity, and nuclear localization of G3BP, a Ras signaling component, in human breast tumors. Cancer Res. 2002;62:1251–1255. [PubMed] [Google Scholar]
- 172.Min L., Ruan Y., Shen Z., Jia D., Wang X., Zhao J., Sun Y., Gu J. Overexpression of Ras-GTPase-activating protein SH3 domain-binding protein 1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2015;67:677–688. doi: 10.1111/his.12695. [DOI] [PubMed] [Google Scholar]
- 173.Willers I.M., Martínez-Reyes I., Martínez-Diez M., Cuezva J. miR-127-5p targets the 3’UTR of human β-F1-ATPase mRNA and inhibits its translation. Biochim. Biophys. Acta-Bioenerg. 2012;1817:838–848. doi: 10.1016/j.bbabio.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 174.Xiao X., Yang J., Li R., Liu S., Xu Y., Zheng W., Yi Y., Luo Y., Gong F., Peng H., et al. Deregulation of mitochondrial ATPsyn-beta in acute myeloid leukemia cells and with increased drug resistance. PLoS ONE. 2013;8:e83610. doi: 10.1371/journal.pone.0083610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cabezon E., Butler P.J., Runswick M.J., Walker J.E. Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J. Biol. Chem. 2000;275:25460–25464. doi: 10.1074/jbc.M003859200. [DOI] [PubMed] [Google Scholar]
- 176.Cabezon E., Butler P.J., Runswick M.J., Carbajo R.J., Walker J.E. Homologous and heterologous inhibitory effects of ATPase inhibitor proteins on F-ATPases. J. Biol. Chem. 2002;277:41334–41341. doi: 10.1074/jbc.M207169200. [DOI] [PubMed] [Google Scholar]
- 177.Schnizer R., Van Heeke G., Amaturo D., Schuster S.M. Histidine-49 is necessary for the pH-dependent transition between active and inactive states of the bovine F1-ATPase inhibitor protein. Biochim. Biophys. Acta. 1996;1292:241–248. doi: 10.1016/0167-4838(95)00208-1. [DOI] [PubMed] [Google Scholar]
- 178.Sanchez-Arago M., Formentini L., Garcia-Bermudez J., Cuezva J.M. IF1 reprograms energy metabolism and signals the oncogenic phenotype in cancer. Cell Cycle. 2012;11:2963–2964. doi: 10.4161/cc.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gonzalez-Llorente L., Santacatterina F., Garcia-Aguilar A., Nuevo-Tapioles C., Gonzalez-Garcia S., Tirpakova Z., Toribio M.L., Cuezva J.M. Overexpression of Mitochondrial IF1 Prevents Metastatic Disease of Colorectal Cancer by Enhancing Anoikis and Tumor Infiltration of NK Cells. Cancers. 2019;12:22. doi: 10.3390/cancers12010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tormos K.V., Anso E., Hamanaka R.B., Eisenbart J., Joseph J., Kalyanaraman B., Chandel N.S. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab. 2011;14:537–544. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen C.T., Shih Y.R., Kuo T.K., Lee O.K., Wei Y.H. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26:960–968. doi: 10.1634/stemcells.2007-0509. [DOI] [PubMed] [Google Scholar]
- 182.Sanchez-Arago M., Garcia-Bermudez J., Martinez-Reyes I., Santacatterina F., Cuezva J.M. Degradation of IF1 controls energy metabolism during osteogenic differentiation of stem cells. EMBO Rep. 2013;14:638–644. doi: 10.1038/embor.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Folmes C.D., Nelson T.J., Martinez-Fernandez A., Arrell D.K., Lindor J.Z., Dzeja P.P., Ikeda Y., Perez-Terzic C., Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Prieto J., Seo A.Y., Leon M., Santacatterina F., Torresano L., Palomino-Schatzlein M., Gimenez K., Vallet-Sanchez A., Ponsoda X., Pineda-Lucena A., et al. MYC Induces a Hybrid Energetics Program Early in Cell Reprogramming. Stem Cell Rep. 2018;11:1479–1492. doi: 10.1016/j.stemcr.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sharma K., D’Souza R.C., Tyanova S., Schaab C., Wisniewski J.R., Cox J., Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 186.Zhou H., Di Palma S., Preisinger C., Peng M., Polat A.N., Heck A.J., Mohammed S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 2013;12:260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 187.Christensen G.L., Kelstrup C.D., Lyngso C., Sarwar U., Bogebo R., Sheikh S.P., Gammeltoft S., Olsen J.V., Hansen J.L. Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Mol. Cell. Proteom. 2010;9:1540–1553. doi: 10.1074/mcp.M900550-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wyant G.A., Yu W., Doulamis I.P., Nomoto R.S., Saeed M.Y., Duignan T., McCully J.D., Kaelin W.G., Jr. Mitochondrial remodeling and ischemic protection by G protein-coupled receptor 35 agonists. Science. 2022;377:621–629. doi: 10.1126/science.abm1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Nesci S. GPR35, ally of the anti-ischemic ATPIF1-ATP synthase interaction. Trends Pharmacol. Sci. 2022;43:891–893. doi: 10.1016/j.tips.2022.09.003. [DOI] [PubMed] [Google Scholar]
- 190.Wei S., Fukuhara H., Kawada C., Kurabayashi A., Furihata M., Ogura S., Inoue K., Shuin T. Silencing of ATPase Inhibitory Factor 1 Inhibits Cell Growth via Cell Cycle Arrest in Bladder Cancer. Pathobiology. 2015;82:224–232. doi: 10.1159/000439027. [DOI] [PubMed] [Google Scholar]
- 191.Wu J., Shan Q., Li P., Wu Y., Xie J., Wang X. ATPase inhibitory factor 1 is a potential prognostic marker for the migration and invasion of glioma. Oncol. Lett. 2015;10:2075–2080. doi: 10.3892/ol.2015.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhang C., Min L., Liu J., Tian W., Han Y., Qu L., Shou C. Integrated analysis identified an intestinal-like and a diffuse-like gene sets that predict gastric cancer outcome. Tumour Biol. 2016;37:16317–16335. doi: 10.1007/s13277-016-5454-7. [DOI] [PubMed] [Google Scholar]
- 193.Garcia-Aguilar A., Martinez-Reyes I., Cuezva J.M. Changes in the turnover of the cellular proteome during metabolic reprogramming: A role for mtROS in proteostasis. J. Proteome Res. 2019;18:3142–3155. doi: 10.1021/acs.jproteome.9b00239. [DOI] [PubMed] [Google Scholar]
- 194.Shen L., Zhi L., Hu W., Wu M.X. IEX-1 targets mitochondrial F1Fo-ATPase inhibitor for degradation. Cell Death Differ. 2009;16:603–612. doi: 10.1038/cdd.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Garcia-Aguilar A., Cuezva J.M. A Review of the Inhibition of the Mitochondrial ATP Synthase by IF1 in vivo: Reprogramming Energy Metabolism and Inducing Mitohormesis. Front. Physiol. 2018;9:1322. doi: 10.3389/fphys.2018.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mourier A., Ruzzenente B., Brandt T., Kuhlbrandt W., Larsson N.G. Loss of LRPPRC causes ATP synthase deficiency. Hum. Mol. Genet. 2014;23:2580–2592. doi: 10.1093/hmg/ddt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cuillerier A., Honarmand S., Cadete V.J.J., Ruiz M., Forest A., Deschenes S., Beauchamp C., Consortium L., Charron G., Rioux J.D., et al. Loss of hepatic LRPPRC alters mitochondrial bioenergetics, regulation of permeability transition and trans-membrane ROS diffusion. Hum. Mol. Genet. 2017;26:3186–3201. doi: 10.1093/hmg/ddx202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ruzzenente B., Metodiev M.D., Wredenberg A., Bratic A., Park C.B., Camara Y., Milenkovic D., Zickermann V., Wibom R., Hultenby K., et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kurbasic E., Sjostrom M., Krogh M., Folkesson E., Grabau D., Hansson K., Ryden L., Waldemarson S., James P., Nimeus E. Changes in glycoprotein expression between primary breast tumour and synchronous lymph node metastases or asynchronous distant metastases. Clin. Proteom. 2015;12:13. doi: 10.1186/s12014-015-9084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Weinberg S.E., Chandel N.S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Martinez-Outschoorn U.E., Peiris-Pages M., Pestell R.G., Sotgia F., Lisanti M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 202.Hay N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer. 2016;16:635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Luengo A., Gui D.Y., Vander Heiden M.G. Targeting Metabolism for Cancer Therapy. Cell Chem. Biol. 2017;24:1161–1180. doi: 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li L., Liang Y., Kang L., Liu Y., Gao S., Chen S., Li Y., You W., Dong Q., Hong T., et al. Transcriptional Regulation of the Warburg Effect in Cancer by SIX1. Cancer Cell. 2018;33:368–385. doi: 10.1016/j.ccell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 205.Birsoy K., Wang T., Chen W.W., Freinkman E., Abu-Remaileh M., Sabatini D.M. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wheaton W.W., Weinberg S.E., Hamanaka R.B., Soberanes S., Sullivan L.B., Anso E., Glasauer A., Dufour E., Mutlu G.M., Budigner G.S., et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Shi Y., Lim S.K., Liang Q., Iyer S.V., Wang H.Y., Wang Z., Xie X., Sun D., Chen Y.J., Tabar V., et al. Gboxin is an oxidative phosphorylation inhibitor that targets glioblastoma. Nature. 2019;567:341–346. doi: 10.1038/s41586-019-0993-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Leanza L., Romio M., Becker K.A., Azzolini M., Trentin L., Manago A., Venturini E., Zaccagnino A., Mattarei A., Carraretto L., et al. Direct Pharmacological Targeting of a Mitochondrial Ion Channel Selectively Kills Tumor Cells In Vivo. Cancer Cell. 2017;31:516–531. doi: 10.1016/j.ccell.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 209.Lamb R., Ozsvari B., Lisanti C.L., Tanowitz H.B., Howell A., Martinez-Outschoorn U.E., Sotgia F., Lisanti M.P. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget. 2015;6:4569–4584. doi: 10.18632/oncotarget.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lamb R., Fiorillo M., Chadwick A., Ozsvari B., Reeves K.J., Smith D.L., Clarke R.B., Howell S.J., Cappello A.R., Martinez-Outschoorn U.E., et al. Doxycycline down-regulates DNA-PK and radiosensitizes tumor initiating cells: Implications for more effective radiation therapy. Oncotarget. 2015;6:14005–14025. doi: 10.18632/oncotarget.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.De Francesco E.M., Maggiolini M., Tanowitz H.B., Sotgia F., Lisanti M.P. Targeting hypoxic cancer stem cells (CSCs) with Doxycycline: Implications for optimizing anti-angiogenic therapy. Oncotarget. 2017;8:56126–56142. doi: 10.18632/oncotarget.18445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.De Francesco E.M., Bonuccelli G., Maggiolini M., Sotgia F., Lisanti M.P. Vitamin C and Doxycycline: A synthetic lethal combination therapy targeting metabolic flexibility in cancer stem cells (CSCs) Oncotarget. 2017;8:67269–67286. doi: 10.18632/oncotarget.18428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ozsvari B., Magalhaes L.G., Latimer J., Kangasmetsa J., Sotgia F., Lisanti M.P. A Myristoyl Amide Derivative of Doxycycline Potently Targets Cancer Stem Cells (CSCs) and Prevents Spontaneous Metastasis, without Retaining Antibiotic Activity. Front. Oncol. 2020;10:1528. doi: 10.3389/fonc.2020.01528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Scatena C., Roncella M., Di Paolo A., Aretini P., Menicagli M., Fanelli G., Marini C., Mazzanti C.M., Ghilli M., Sotgia F., et al. Doxycycline, an Inhibitor of Mitochondrial Biogenesis, Effectively Reduces Cancer Stem Cells (CSCs) in Early Breast Cancer Patients: A Clinical Pilot Study. Front. Oncol. 2018;8:452. doi: 10.3389/fonc.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ozsvari B., Sotgia F., Lisanti M.P. Exploiting mitochondrial targeting signal(s), TPP and bis-TPP, for eradicating cancer stem cells (CSCs) Aging. 2018;10:229–240. doi: 10.18632/aging.101384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ozsvari B., Sotgia F., Lisanti M.P. First-in-class candidate therapeutics that target mitochondria and effectively prevent cancer cell metastasis: Mitoriboscins and TPP compounds. Aging. 2020;12:10162–10179. doi: 10.18632/aging.103336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sargiacomo C., Stonehouse S., Moftakhar Z., Sotgia F., Lisanti M.P. MitoTracker Deep Red (MTDR) Is a Metabolic Inhibitor for Targeting Mitochondria and Eradicating Cancer Stem Cells (CSCs), with Anti-Tumor and Anti-Metastatic Activity In Vivo. Front. Oncol. 2021;11:678343. doi: 10.3389/fonc.2021.678343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sterne J. Treatment of diabetes mellitus with N,N-dimethylguanylguanidine (LA. 6023, glucophage) Therapie. 1959;14:625–630. [PubMed] [Google Scholar]
- 219.Janzer A., German N.J., Gonzalez-Herrera K.N., Asara J.M., Haigis M.C., Struhl K. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:10574–10579. doi: 10.1073/pnas.1409844111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Veiga S.R., Ge X., Mercer C.A., Hernandez-Alvarez M.I., Thomas H.E., Hernandez-Losa J., Ramon Y.C.S., Zorzano A., Thomas G., Kozma S.C. Phenformin-Induced Mitochondrial Dysfunction Sensitizes Hepatocellular Carcinoma for Dual Inhibition of mTOR. Clin. Cancer Res. 2018;24:3767–3780. doi: 10.1158/1078-0432.CCR-18-0177. [DOI] [PubMed] [Google Scholar]
- 221.Fiorillo M., Scatena C., Naccarato A.G., Sotgia F., Lisanti M.P. Bedaquiline, an FDA-approved drug, inhibits mitochondrial ATP production and metastasis in vivo, by targeting the gamma subunit (ATP5F1C) of the ATP synthase. Cell Death Differ. 2021;28:2797–2817. doi: 10.1038/s41418-021-00788-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Fiorillo M., Ozsvari B., Sotgia F., Lisanti M.P. High ATP Production Fuels Cancer Drug Resistance and Metastasis: Implications for Mitochondrial ATP Depletion Therapy. Front. Oncol. 2021;11:740720. doi: 10.3389/fonc.2021.740720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sleire L., Forde H.E., Netland I.A., Leiss L., Skeie B.S., Enger P.O. Drug repurposing in cancer. Pharmacol. Res. 2017;124:74–91. doi: 10.1016/j.phrs.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 224.Gyawali B., Booth C.M. Cancer treatments should benefit patients: A common-sense revolution in oncology. Nat. Med. 2022;28:617–620. doi: 10.1038/s41591-021-01662-6. [DOI] [PubMed] [Google Scholar]
- 225.Chen Q., Jiang H., Wang Z., Cai L.Y., Jiang Y.C., Xie L., Zhou Y., Zeng X., Ji N., Shen Y.Q., et al. Adrenergic Blockade by Nebivolol to Suppress Oral Squamous Cell Carcinoma Growth via Endoplasmic Reticulum Stress and Mitochondria Dysfunction. Front. Pharmacol. 2021;12:691998. doi: 10.3389/fphar.2021.691998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Lee K.M., Giltnane J.M., Balko J.M., Schwarz L.J., Guerrero-Zotano A.L., Hutchinson K.E., Nixon M.J., Estrada M.V., Sanchez V., Sanders M.E., et al. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Metab. 2017;26:633–647.e637. doi: 10.1016/j.cmet.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lee M., Hirpara J.L., Eu J.Q., Sethi G., Wang L., Goh B.C., Wong A.L. Targeting STAT3 and oxidative phosphorylation in oncogene-addicted tumors. Redox Biol. 2018;25:101073. doi: 10.1016/j.redox.2018.101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sica V., Bravo-San Pedro J.M., Stoll G., Kroemer G. Oxidative phosphorylation as a potential therapeutic target for cancer therapy. Int. J. Cancer. 2020;146:10–17. doi: 10.1002/ijc.32616. [DOI] [PubMed] [Google Scholar]
- 229.Zhao Z., Mei Y., Wang Z., He W. The Effect of Oxidative Phosphorylation on Cancer Drug Resistance. Cancers. 2022;15:62. doi: 10.3390/cancers15010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Samudio I., Harmancey R., Fiegl M., Kantarjian H., Konopleva M., Korchin B., Kaluarachchi K., Bornmann W., Duvvuri S., Taegtmeyer H., et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J. Clin. Investig. 2010;120:142–156. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Camarda R., Zhou A.Y., Kohnz R.A., Balakrishnan S., Mahieu C., Anderton B., Eyob H., Kajimura S., Tward A., Krings G., et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat. Med. 2016;22:427–432. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Iwamoto H., Abe M., Yang Y., Cui D., Seki T., Nakamura M., Hosaka K., Lim S., Wu J., He X., et al. Cancer Lipid Metabolism Confers Antiangiogenic Drug Resistance. Cell Metab. 2018;28:104–117.e5. doi: 10.1016/j.cmet.2018.05.005. [DOI] [PubMed] [Google Scholar]