Abstract
The sequence of the complete genome of ovine enzootic nasal tumor virus, an exogenous retrovirus associated exclusively with contagious intranasal tumors of sheep, was determined. The genome is 7,434 nucleotides long and exhibits a genetic organization characteristic of type B and D oncoviruses. Enzootic nasal tumor virus is closely related to the Jaagsiekte sheep retrovirus and to sheep endogenous retroviruses.
Animal models of retrovirus-induced tumors of the lymphoreticular system are numerous and have contributed greatly to our understanding of oncogenesis (reviewed in reference 10). However, animal models of retrovirus-associated epithelial tumors are restricted to mouse mammary tumors, sheep pulmonary adenomatosis (SPA), and enzootic nasal tumors (ENT) of sheep and goats. Of these, ENT and SPA are also important for economic and animal welfare considerations in affected flocks. Both have long incubation periods, but once clinical signs become apparent the diseases are invariably fatal.
There is compelling evidence that SPA (also known as Jaagsiekte or ovine pulmonary carcinoma) is caused by Jaagsiekte sheep retrovirus (JSRV) (reviewed in reference (15), although classical approaches to investigate the presence of an etiological role for this virus continue to be hampered by the lack of a cell culture system to propagate JSRV. The relationship between ENT and its associated retrovirus, enzootic nasal tumor virus (ENTV), is less well characterized. The presence of the retrovirus was first shown by electron microscopy in intranasal tumors of sheep (7, 23, 37) and goats (4, 5, 35) and in nasal fluid produced by these tumors. Antigenic cross-reactivity of ENTV with Mason-Pfizer monkey virus (MPMV) p27 (6, 31) and recombinant JSRV capsid protein (7) indicated that ENTV has a type D retroviral capsid. However, although the complete genome of JSRV has been sequenced (38), the data for ENTV is limited to two small regions of the gag gene (3, 29). The close homology between JSRV and sheep endogenous retroviral sequences (SERVs) (14, 38) was a problem for the development of JSRV-specific PCRs. This was finally resolved by the identification of small areas of sequence difference between JSRV and SERVs (1, 26). A similar approach is required to develop sensitive ENTV-specific detection techniques in order to elucidate the role of ENTV in disease. This is particularly important because ENTV, like JSRV, does not replicate in tissue culture.
Here we report the cloning and sequencing of the complete ENTV genome from an ovine case of ENT and investigate its relationship to JSRV, SERVs, and other retroviruses. This data is a vital step in the study of this animal model of epithelial cell-derived tumors and opens the way to further studies to elucidate the pathogenesis and oncogenic mechanisms of this virus.
MATERIALS AND METHODS
Source of material.
Nasal fluid from a sheep with naturally occurring ENT was collected, clarified, and concentrated as described previously (6). The resultant pellet was resuspended as a 100-fold concentrate in TNE buffer (0.01 M Tris-Cl [pH 7.5], 0.1 M NaCl, 0.001 M EDTA). Samples of ENT and kidney were collected from this sheep at necropsy and snap frozen in liquid nitrogen. Tissue samples were stored at −70°C. DNA was extracted from these tissues by the method of Wu et al. (36). The sheep was verified to be negative for SPA by macroscopic and microscopic pathologic testing at necropsy.
Density gradient fractionation.
ENTV was purified from 200 μl of 100-fold-concentrated nasal fluid by isopycnic centrifugation on 20 to 55% (wt/wt) sucrose gradients (16). Fractions (0.5 ml) were collected, and fractions with densities between 1.17 and 1.19 g/ml were pooled, diluted in TNE buffer and centrifuged at 100,000 × g for 1 h at 4°C. The pellet was resuspended in 0.5 ml of solution D (4 M guanidinium thiocyanate, 25 mM sodium citrate [pH 7.0], 0.5% [wt/vol] sarcosyl, 0.1 M 2-mercaptoethanol) for RNA preparation.
RNA preparation and reverse transcription.
Total RNA was extracted from pooled sucrose gradient fractions by the acid guanidinium thiocyanate-phenol-chloroform method (2), without adding carrier RNA or treating with DNase. The RNA was resuspended in 15 μl of 1 mM EDTA. cDNA was synthesized by random-primed or oligo(dT)-primed reverse transcription with the Moloney murine leukemia virus (H-) riboclone cDNA synthesis system (Promega). The presence of ENTV cDNA was confirmed by gag-PCR (3).
Construction of cDNA libraries.
EcoRI linkers were ligated to the cDNA by using the Riboclone EcoRI adapter ligation system (Promega). Excess adapters were removed from the ligation product with Sephacryl-400 as directed by the manufacturer. All of the cDNA-linker product was ligated to lambda arms by using the λZapII-EcoRI-calf intestinal alkaline phosphatase cloning kit as described by the manufacturer (Stratagene). Gigapack II Gold packaging extract (Stratagene) was used for packaging.
Recombinant clones were selected initially with JSRV clones as probes (pJS382 [38], pJS107 [38], and pLG1 [26]) and later by using ENTV clones from the library. The selected clones were excised by using plating strain SOLR, which converts lambda ZapII to Bluescript SK(−) plasmid and allows for blue-white screening for inserts.
PCR.
Gaps in the sequence were filled by PCR with primers chosen from the available ENTV sequence data. The positions of the PCR fragments relative to the ENTV genome are shown in Fig. 1. In each case, PCR products were pooled from at least four independent reactions, gel purified, and cloned into pGEMT (Promega) or pCR2.1 (Invitrogen) as specified by the manufacturers. Product a was amplified from cDNA made from ENTV RNA prepared as described above. Products b, c, and d were obtained by amplification of ENTV provirus from nasal tumor DNA.
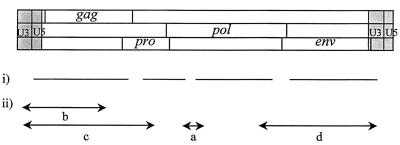
FIG. 1.
Schematic diagram of the ENTV provirus showing (i) sequence derived from cDNA library clones and (ii) PCR products generated to complete the sequencing of ENTV.
(i) PCR a.
Product a was amplified from ENTV cDNA with primers ATGGGAGCCCTACAACCTGG (nucleotides [nt] 3113 to 3131 [Fig. 2]) and CACGAGGATACAAGGAGAAACC (reverse of nt 3542 to 3572 [Fig. 2]). The PCR was carried out in 2.5 mM MgCl2–10 mM Tris (pH 9.2)–50 mM KCl–20 mM (NH4)2SO4–200 μM each deoxynucleoside triphosphate (dNTP)–0.125 μM each primer–1.25 U of Taq. A total of 45 cycles were used, each consisting of 1 min at 94°C, 30 s at 59°C, and 1 min at 72°C.
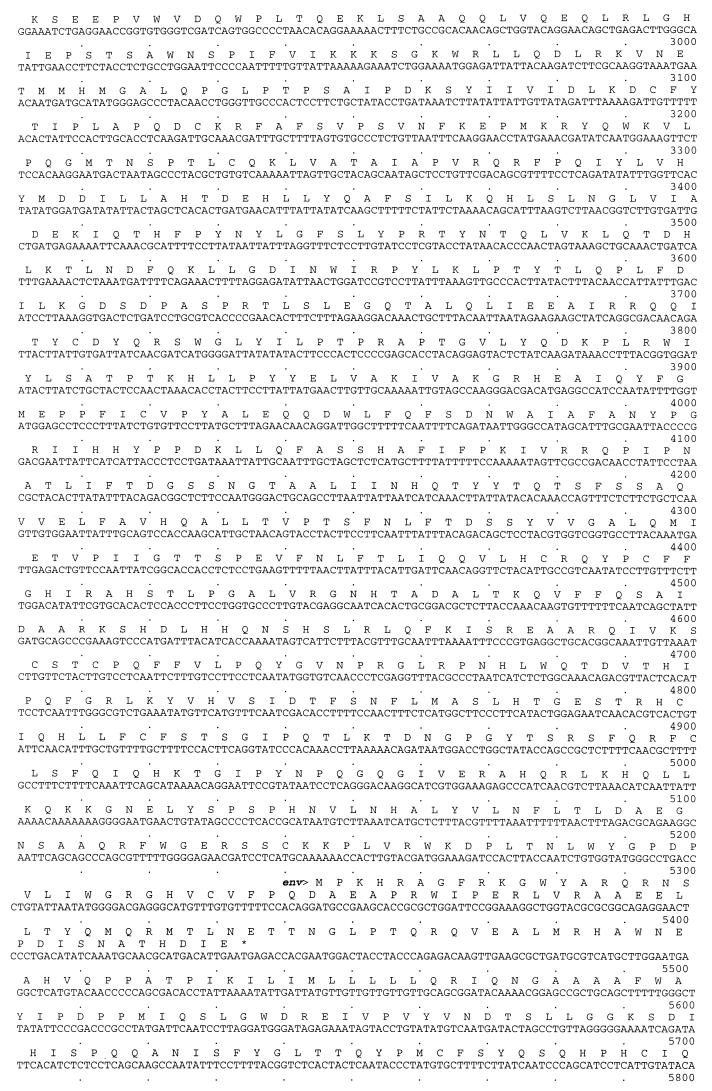
FIG. 2.
Complete nucleotide sequence of the ENTV genome together with the deduced amino acid sequences of the open reading frames. The limits of U3, R, and U5 are indicated by arrows. The inverted repeats (IR), primer binding site (PBS), and polypurine tract (PPT) are indicated by underlines. The consensus protease cleavage site for env SU and TM is indicated by ∇.
(ii) PCR b.
A 250-ng portion of nasal tumor DNA was used as template for the first round of PCR with primers PL1 (TCAGGAAGTCTTAAGAGCTTTTAG) (nt 7267 to 7290 [Fig. 2]), and PG1 (ATACTGCAGCYCGATGGCCAG) (reverse of nt 1802 to 1822 [Fig. 2]). A 2-μl volume of the product of this PCR was used in the second round of amplification with primers PL2 (GATGTCTTTTGGTTTTGCAACATG) (nt 7297 to 7320 [Fig. 2]) and PG1. The same conditions were used for both the first and second rounds of PCR as follows: 1.5 mM MgCl2–50 mM KCl–10 mM Tris-HCl (pH 8.3)–100 μM each dNTP–10 pmol of each primer–1.25 U of Taq polymerase (Boehringer). The PCR cycles were 94°C for 4 min, then 30 cycles at 94°C for 40 s, 63°C for 40 s, and 72°C for 90 s plus a progressive increment at 72°C of 10 s per cycle from cycle 11 onward, and a final extension of 72°C for 7 min. A tumor-specific product of approximately 2 kb was confirmed to originate from ENTV by demonstration of the gag PstI site previously shown to be a molecular marker for ENTV (reference 3 and data not shown).
(iii) PCR c.
Fragment c was amplified from nasal tumor DNA with primers PL3 (ATGATCTCAAGTTACTTAACTTGC) (nt 4859 to 4880 [Fig. 2]) and NT4R (CTGTTCTGGATTGGTAGTTTGG) (reverse of nt 7376 to 7399 [Fig. 2]). The PCR conditions were 100 ng of template DNA–50 mM KCl–10 mM Tris-HCl (pH 8.3)–2 mM MgCl2–12.5 pmol of each primer–200 μM each dNTP–1 U of Taq polymerase. A total of 40 cycles of 94°C for 1 min, 59°C for 1 min, and 72°C for 2 min were performed, with a final extension of 5 min at 72°C.
(iv) PCR d.
Fragment d was amplified with PCR primers NTG4F (CTCATGGCTTCCCTTCATACTG) and PL4 (AAGCAAGTTAAGTAACTTGAGATC) with nasal tumor DNA as the template. The PCR conditions were 100 ng of template DNA–50 mM KCl–10 mM Tris-HCl (pH 8.3)–1.5 mM MgCl2–12.5 pmol of each primer–200 μM each dNTP–1 U of Taq polymerase. A total of 40 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 2.5 min were performed, with a final extension of 5 min at 72°C.
Sequence analysis and phylogeny.
Each clone was sequenced in the forward and reverse orientations with a Li-Cor automated sequencer. The clones were ordered initially by alignment with JSRV by using the Genetics Computer Group (GCG) Bestfit program (13). The sequence data were compiled with the GCG fragment assembly package (13). The sequence of ENTV was confirmed by sequencing at least two overlapping clones. Sequences obtained by PCR amplification were confirmed by sequencing at least three independent clones. As would be expected for sequences of an uncloned virus population, albeit isolated from a single animal, some differences were found. Where differences occurred, the consensus sequence is shown.
The GCG programs Eclustalw and Clustree were used for phylogenetic analysis. Percent divergence figures between all pairs of sequences were calculated and used to generate unrooted phylogenetic trees by the neighbor-joining method of Saitou and Nei (30).
Nucleotide sequence accession numbers.
The nucleotide sequence of the complete genome of ovine ENTV will appear in the EMBL Nucleotide Sequence Database under accession no. Y16627.
RESULTS
A total of 39 clones were selected by screening of the ENTV cDNA libraries. Sequencing of these clones generated more than 75% of the ENTV genome. The remaining gaps, as identified by alignment with the JSRV genome, were generated by PCR (Fig. 1). Gap a in the pol gene was filled by PCR on cDNA from virus purified from nasal fluid from an animal with ENT. The sequencing gaps remaining at either end of the virus were amplified by PCR from nasal tumor genomic DNA, taking advantage of the fact that the long terminal repeat (LTR) is duplicated in the provirus (product b in Fig. 1). ENTV-specific U3 primers were designed by selecting regions of difference between the clones from the library and the published sequences of endogenous sheep retroviruses (1, 26) and used to amplify the region from U3 across U5 into gag. The LTR sequence obtained from these clones allowed the identification of a unique region in the ENTV LTR, from which more efficient exogenous specific PCR primers, PL3 and PL4, were chosen. These primers, together with primers from within pro or pol, respectively, allowed amplification from tumor DNA of products c and d, which included the two remaining sequence gaps. Control reactions, with kidney DNA from the same animal as the template, verified that the PCRs amplified only exogenous ENTV and not endogenous sequences. We have previously reported the presence of two restriction enzyme sites, PstI and AatII, within gag which are specific for ENTV (3). These occur at bases 1691 to 1696 and 1757 to 1762, respectively, in the full-length sequence of ENTV (Fig. 2). No JSRV or endogenous sequences that have these specific sites have been reported. Both these sites were present in all our clones spanning this region, which is further evidence that these clones were not derived from endogenous sequences. Furthermore, none of the clones contained the JSRV-specific ScaI site in gag (26).
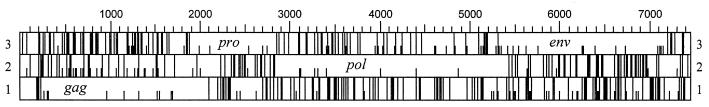
The complete sequence of ovine ENTV is shown in Fig. 2. The genome is 7,434 nt long. It has a genetic organization typical of type B and D retroviruses in that it has gag, pro, pol, and env genes (Fig. 3). No further open reading frames are present in ENTV. Comparison with other retroviruses suggest that pro and pol are expressed as gag-pro and gag-pro-pol precursors generated by ribosomal frameshifting and that env transcripts are generated by splicing.
FIG. 3.
Genetic organization of ENTV. Stop codons in the three reading frames are represented by long vertical bars; methionine residues are represented by short vertical bars.
LTR.
The LTR of ENTV is 374 bases long and is bound by the inverted-repeat sequences GCAG (nt 122 to 125) and CTGC (nt 7172 to 7175) and flanked by the primer binding site (nt 127 to 142) and the polypurine tract (nt 7154 to 7170). U3 is 249 bp, R is 13 bp, and U5 is 112 bp. U3 is defined as the region between R and the polypurine tract that contains the initiation site for synthesis of the positive strand of viral DNA. Like JSRV, ENTV uses tRNA1,2Lys as the primer for reverse transcriptase (RT) to initiate minus-strand synthesis. In the DNA form of retroviral LTRs, the U3 region contains cis-acting sequences necessary for viral replication and regulatory signals for retroviral transcription. Although the ENTV LTR must interact with cellular factors, no recognized consensus sequences for transcription factor binding sites were identifiable. Experiments with expression constructs involving the LTR linked to a reporter gene must be performed before the functional regions of the ENTV LTR can be dissected.
Leader sequence.
The untranslated region between the primer binding site and the start of gag is likely to contain the splice donor site for generation of subgenomic (env) transcripts, as well as the packaging signal which specifies incorporation of genome RNA into virions. Alignment with JSRV suggests that the splice donor site is after base 189.
Viral proteins. (i) gag.
ENTV gag (nt 256 to 2094) encodes a 613-amino-acid (aa) polypeptide. The boundaries between the Gag proteins were assigned, as for JSRV (38), by alignment with MPMV, squirrel monkey retrovirus (SMRV), and mouse mammary tumor virus (MMTV) sequences. The ENTV capsid protein (CA) is encoded by nt 1027 to 1677, and the nucleic acid binding protein is encoded by nt 1678 to 1995. There are two sequences of Cys-X2-Cys-X4-His-X4-Cys, separated by 13 residues, in the nucleic acid binding protein. This is typical of a zinc finger domain and is thought to mediate nucleocapsid binding to the genomic RNA.
(ii) pro.
The pro open reading frame (nt 1878 to 2858) encodes a 326-residue polypeptide. It is likely to be expressed as a gag-pro fusion polypeptide generated by ribosomal frameshifting, as has been shown for MMTV (17). The exact site and relative efficiency of this frameshifting event remain to be elucidated. The protein encoded by pro is thought to comprise two domains, a pseudoprotease (or protease-like) domain which exhibits a dUTPase activity, as has been suggested for other retroviruses by both computer analysis (22) and functional studies (9), and an active protease (Pr). The sequence Leu-Asp-Thr-Gly, indicative of the core amino acid sequence of cellular aspartyl protease, is encoded by nt 2469 to 2480.
(iii) pol.
The pol open reading frame (nt 2825 to 5437) encodes a peptide of 870 aa giving a predicted molecular mass of 99 kDa. pol is likely to be translated as a fused gag-pro-pol polypeptide, again generated by ribosomal frameshifting. pol encodes the RT and integrase domains. The putative RT active site is the Tyr-Met-Asp-Asp sequence encoded by nt 3401 to 3412 in ENTV.
(iv) env.
The env open reading frame (nt 5346 to 7196), at its start, overlaps pol, and the env transcript is likely to be generated by splicing. Like JSRV, ENTV contains two possible ATG start codons 7 aa apart, but the second is thought to be the initiation codon used since it is preceded by a predicted splice acceptor sequence. Retroviral env open reading frames encode a polypeptide precursor which is cleaved twice, first following the peptide signal sequence and second following an internal proteolytic cleavage site, which generates the surface (SU) and transmembrane (TM) envelope proteins. In ENTV, env encodes a polypeptide precursor of 617 aa (predicted molecular mass, 68 kDa). A hydrophobic region (encoded by nt 5526 to 5573) is thought to be the peptide signal sequence, and cleavage probably occurs cotranslationally at the carboxyl-terminal end of this sequence. A consensus proteolytic cleavage site RPKR (encoded by nt 6468 to 6479) generates a SU protein of 302 aa and a TM protein of 239 aa. A 21-aa hydrophobic region (encoded by nt 6994 to 7046) is likely to be the transmembrane domain that anchors the TM protein in the membrane. This gives a predicted cytoplasmic domain of 50 aa and an extracellular domain of 182 aa.
The predicted molecular mass of the env proteins from the sequence data alone are 34 kDa for SU and 27 kDa for TM. However, the actual molecular masses will be larger due to glycosylation. The motif Asn-X-Ser/Thr, which can serve as a site for N-linked glycosylation, is found nine times in SU and twice in TM.
Comparison with JSRV.
As shown in Table 1, the genes of ENTV and JSRV are very similar, with an overall amino acid similarity of greater than 95%. The biggest differences occur at the 3′ end of env and in the U3 LTR region. In addition, the open reading frame ORFX postulated for JSRV contains two stop codons in ENTV and therefore does not appear to be an open reading frame in ENTV.
TABLE 1.
Percentage nucleotide identity and amino acid identity and similarity between ENTV and JSRV.
| Region | Nucleic acid similarity (%) | Amino acid similarity (%) | Amino acid identity (%) |
|---|---|---|---|
| gag | 89.7 | 96.7 | 95.0 |
| pro | 95.2 | 97.2 | 96.3 |
| pol | 92.3 | 98.0 | 96.9 |
| env | 85.5 | 95.2 | 89.0 |
| SU | 87.9 | 96.3 | 93.4 |
| TM | 81.9 | 93.2 | 81.9 |
| LTR | 75.7 | ||
| U5 | 82.9 | ||
| R | 100 | ||
| U3 | 72.6 |
The 3′ end of env is the most divergent of the coding regions of JSRV and ENTV. The viruses are 96% identical in SU and 93% identical in TM at the amino acid level. The TM protein can be further subdivided into external, transmembrane, and intracytoplasmic domains, which exhibit 98, 81, and 75% amino acid similarity, respectively (86, 80, and 63% nucleotide identity). Sequencing of six independent PCR-derived clones and two cDNA clones across this site confirmed that there is nucleotide sequence divergence and not a frameshift mutation at the 3′ end of env. Many of the amino acid substitutions are conservative, suggesting that there are some functional restraints on mutation of this protein.
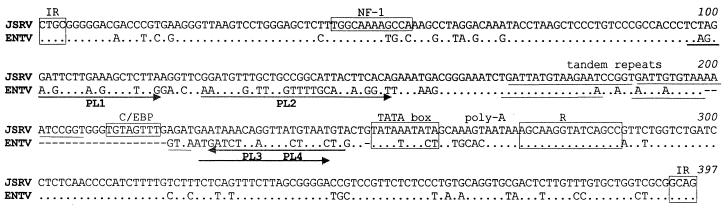
Alignment of the LTR of ENTV with that of JSRV shows that they are quite similar (75.7% similarity), although JSRV has an insertion of 21 bp in U3 compared to ENTV. Several regions contain a cluster of base differences. These regions are also the most divergent from SERV sequences and for this reason were chosen for PCR primers. Interestingly, ENTV does not appear to contain the same regulatory sequences as JSRV. For example, the NF-1 site (TGGCA4GCCA) seen in JSRV (38) (Fig. 4, bases 44 to 55) is interrupted by the insertion of a T in ENTV (TGGCA4TGCCA). Also, the 19-bp tandem repeat identified in JSRV (38) is shorter in ENTV (15 bp). Finally, the C/EBP sequence (Fig. 4, bases 211 to 218) is not found in ENTV.
FIG. 4.
Alignment of the LTR sequences of ENTV and JSRV. The JSRV sequence shown is that of York et al. (38). Sequences of the LTR primers are underlined. Gaps in alignment are shown by dashes; dots represents identity.
Comparison with other retroviruses.
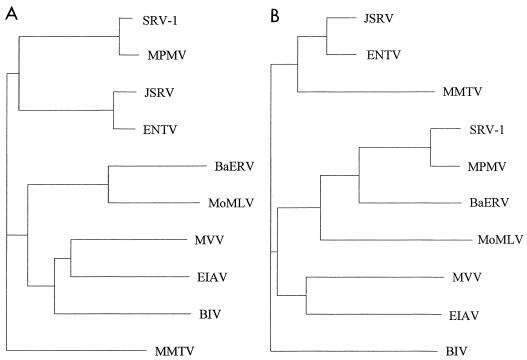
Construction of a phylogenetic tree based on comparison of gag genes of a range of different retroviruses (Fig. 5A) shows that ENTV and JSRV group most closely with MPMV. However, if env sequences are compared (Fig. 5B), ENTV and JSRV are most similar to MMTV. (The tree shown is for SU, but comparison of env TM regions generates a very similar tree.) These results confirm that ENTV has a type B envelope and a type D capsid.
FIG. 5.
Phylogenetic trees of (A) gag and (B) env SU nucleotide sequences of ENTV, baboon endogenous retrovirus (BaERV) (18), bovine immunodeficiency virus (BIV) (11), equine infectious anemia virus (EIAV) (19), JSRV (38), MMTV (24), simian SRV-1 type D retrovirus (SRV-1) (27), MPMV (33), maedi-visna virus (MVV) (28), and Moloney murine leukemia virus (MoMLV) (32). The sequences were aligned by using GCG Eclustalw, and unrooted trees were constructed by the neighbor-joining method of Saitou and Nei (30) (Clustree). Horizontal branch lengths are proportional to evolutionary distances as estimated by percent divergence.
Construction of a phylogenetic tree based on LTR sequences (data not shown) shows that ENTV is most closely related to JSRV and SERVs. These have only approximately 43% identity to the type D retroviruses examined (simian SRV-1 type D retrovirus [SRV-1] and MPMV) and less than 33% identity to any of the other retroviruses listed in Fig. 5. Although this degree of identity is in the same range as that for comparison between the gag and env genes compared above, the LTR sequences from diverse retroviruses do not generate a reliable alignment and do not aid the identification of functional regions of the LTR.
DISCUSSION
This is the first report of the full sequence of ENTV. Determination of the sequence has enabled comparisons to be made between ENTV and other viruses. This confirms that ENTV is a chimeric typeB/D retrovirus and that it is distinct from the related virus JSRV and from the endogenous retroviral sequences found in the sheep genome. Work currently under way suggests that ovine ENTV is also distinct from caprine ENTV (25a). It is hoped that similarities and differences between members of this family of viruses may give an insight into the mechanism of cellular transformation and factors determining cellular tropism.
Although ENTV and JSRV are distinct viruses associated with discrete pathologic findings, translation and alignment of the open reading frames suggests a very high degree of homology between them (Table 1). The similarity between the coding regions of these viruses is in agreement with the evidence that antisera against CA proteins of ENTV and JSRV are cross-reactive. Antisera against other proteins have not been tested, but the degree of sequence similarity between JSRV and ENTV genes suggests that the proteins will all be antigenically cross-reactive.
Results for the few regions of SERVs that have been sequenced suggest that there is also a high degree of similarity between any putative expressed endogenous sequences and ENTV or JSRV. Although these JSRV and ENTV related endogenous sequences have not been linked to any pathologic findings, they may play an important role in allowing these viruses to persist in the host and eventually induce tumours. By reverse transcription-PCR, SERV transcripts have been detected in all sheep tissues tested, but protein has not been detected, probably due to the limited sensitivity of detection methods for proteins compared to nucleic acids. If these SERV sequences are expressed during ontogeny, it is likely that sheep would become immunotolerant to them and therefore to the related exogenous viral proteins, as has been hypothesized previously (25, 38). The surprisingly small number of sequence variations observed in our clones from the population of ENTVs in a single case of ENT supports the hypothesis that this virus is not subject to immune pressure. The earlier sequence data from two regions of ENTV gag (3, 29) also contains few changes from the ENTV sequence presented in this paper.
A putative open reading frame named ORFX was described in JSRV (29), and it has been suggested that this may encode an oncogenic product. In ENTV, the region homologous to ORFX contains two stop codons and therefore is not an open reading frame. Since it is likely that these two viruses have the same oncogenic mechanism, we therefore argue against ORFX playing any role in cellular transformation.
Apart from ORFX, the main differences between ENTV and JSRV occur in the LTR and the env TM subunit, suggesting that one or both of these regions must be important in determining viral tropism. The role of the cytoplasmic domain of env is thought to be important in envelope processing (21) and in ensuring the incorporation of the envelope glycoproteins into the budding virus (8, 12). It is the SU region of the retroviral envelope that interacts with the cell surface receptor and therefore is largely responsible for directing cell tropism at the surface binding level. Therefore, it is surprising to find that it is the TM and intracytoplasmic domains of env which differ the most between these two viruses. The change in the 3′ end of env could be of little significance compared to that of U3 and may be a by-product of a single recombinational event in the 3′ end of the ancestral type B/D retrovirus, which led to the divergence of JSRV and ENTV. However, it has been shown that alterations in the TM cytoplasmic domain can alter the pathogenicity of retroviruses (20, 39), and so a role for this domain in affecting the pathogenicity of ENTV and JSRV cannot be ruled out.
While a number of transcription factor binding sites are identifiable in JSRV, they could not be shown in the ENTV LTR. The differences in the LTRs of these two viruses are likely to affect the replication potential and therefore the pathogenicity of these viruses and may reflect the presence of different transcription factors in the cells that they infect. Sequences in the LTR that regulate transcription in specific cell types may also contribute to the ability of the retrovirus to transform these same cell types, as has been shown for murine leukemia virus (34). Thus, an understanding of transcriptional control of ENTV may also contribute to elucidating the mechanism of viral oncogenesis. We are currently characterizing the LTR function of these viruses.
In conclusion, we have presented, for the first time, the complete sequence of ENTV. Analysis of the sequence places this virus as another member of a group of B/D-type retroviruses, which also includes JSRV and SERVs. The disease pathology associated with ENTV has some similarities to that associated with JSRV in that both are tumors derived from secretory epithelial cells. However, the diseases are clearly independent and affect different target organs. A virus species is defined as “a polythetic class of viruses that constitutes a replicating lineage and occupies a particular ecological niche” (International Committee on the Taxonomy of Viruses). Thus, it can be argued that JSRV and ENTV are distinct viral species.
The main sequence differences between JSRV and ENTV reside in ORFX, the U3 LTR, and the 3′ end of env. Whether it is these differences and/or more subtle sequence differences that direct the tropism of these viruses requires further investigation.
The availability of the sequence of ENTV is a significant step that opens the way for the development of ENTV-specific detection techniques, which are vital to the further study of the distribution and pathogenesis of this virus and to the development of strategies to eradicate ENT.
ACKNOWLEDGMENTS
This work was supported by CEC contract AIR3-CT94-084, BBSRC grant RO6361, SOAFED grant ROAME MRI/027/95, and Spanish grant CICYT AGF96-0535-C02-01.
We thank I. D. Bennett for the sequencing and P. Harrison for critical discussion of the manuscript.
REFERENCES
- 1.Bai J, Zhu R Y, Stedman K, Cousens C, Carlson J, Sharp J M, DeMartini J C. Unique long terminal repeat U3 sequences distinguish exogenous Jaagsiekte sheep retroviruses associated with ovine pulmonary carcinoma from endogenous loci in the sheep genome. J Virol. 1996;70:3159–3168. doi: 10.1128/jvi.70.5.3159-3168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 3.Cousens C, Minguijon E, Garcia M, Ferrer L M, Dalziel R G, Palmarini M, De las Heras M, Sharp J M. PCR-based detection and partial characterization of a retrovirus associated with contagious intranasal tumors of sheep and goats. J Virol. 1996;70:7580–7583. doi: 10.1128/jvi.70.11.7580-7583.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De las Heras M, Garcia de Jalon J A, Sharp J M. Pathology of enzootic intranasal tumour in thirty-eight goats. Vet Pathol. 1991;28:474. doi: 10.1177/030098589102800603. [DOI] [PubMed] [Google Scholar]
- 5.De las Heras M, Sharp J M, Garcia de Jalon J A, Dewar P. Enzootic nasal tumour of goats: demonstration of a type D-related retrovirus in nasal fluids and tumours. J Gen Virol. 1991;72:2533–2535. doi: 10.1099/0022-1317-72-10-2533. [DOI] [PubMed] [Google Scholar]
- 6.De las Heras M, Sharp J M, Ferrer L M, Garcia de Jalon J A, Cebrian L M. Evidence for a type D-like retrovirus in enzootic nasal tumour of sheep. Vet Rec. 1993;132:441. doi: 10.1136/vr.132.17.441. [DOI] [PubMed] [Google Scholar]
- 7.De las Heras M, Minguijón E, Ferrer L M, Ortín A, Dewar P, Cebrian L M, Pascual Z, García L, García de Jalón J A, Sharp J M. Naturally occurring enzootic nasal tumours of sheep in Spain: pathology and associated retrovirus. Eur J Vet Pathol. 1998;4:1–5. [Google Scholar]
- 8.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4765. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elder J H, Lerner D L, Hasselkus-Light C S, Fontenot D R, Hunter E, Luciw P A, Montelaro R C, Phillips T R. Distinct subsets of retroviruses encode dUTPase. J Virol. 1992;66:1791–1794. doi: 10.1128/jvi.66.3.1791-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan H. Retroviruses and their role in cancer. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Academic Press, Inc.; 1994. pp. 313–362. [Google Scholar]
- 11.Garvey K J, Oberste M S, Elser J E, Braun M J, Gonda M A. Nucleotide sequence and genome organization of biologically active proviruses of the bovine immunodeficiency-like virus. Virology. 1990;175:391–409. doi: 10.1016/0042-6822(90)90424-p. [DOI] [PubMed] [Google Scholar]
- 12.Gebhardt A, Bosch J V, Ziemiecki A, Friis R R. Rous sarcoma virus p19 and gp35 can be chemically cross-linked to high molecular weight complexes. An insight into virus assembly. J Mol Biol. 1984;174:297–317. doi: 10.1016/0022-2836(84)90340-1. [DOI] [PubMed] [Google Scholar]
- 13.Genetics Computer Group. Program manual for the Wisconsin package, version 8. Madison, Wis: GCG, Inc.; 1994. [Google Scholar]
- 14.Hecht S J, Carlson J O, DeMartini J C. Analysis of a type D retroviral capsid gene expressed in ovine pulmonary carcinoma and present in both affected and unaffected sheep genomes. Virology. 1994;202:480–484. doi: 10.1006/viro.1994.1366. [DOI] [PubMed] [Google Scholar]
- 15.Hecht J S, Sharp J M, DeMartini J C. Retroviral aetiopathogenesis of ovine pulmonary carcinoma: a critical appraisal. Br Vet J. 1996;152:395–409. doi: 10.1016/s0007-1935(96)80034-0. [DOI] [PubMed] [Google Scholar]
- 16.Herring A J, Sharp J M, Scott F M, Angus K W. Further evidence for a retrovirus as the aetiological agent of sheep pulmonary adenomatosis (Jaagziekte) Vet Microbiol. 1983;8:237–249. doi: 10.1016/0378-1135(83)90076-7. [DOI] [PubMed] [Google Scholar]
- 17.Jacks T, Townsley K, Varmus H E, Majors J. Two efficient ribosomal frameshifting events are required for synthesis of MMTV gag-related polyproteins. Proc Natl Acad Sci USA. 1987;84:4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato S, Matsuo K, Nishimura N, Takahashi N, Takano T. The entire nucleotide sequence of baboon endogenous virus DNA: A chimeric genome structure of murine type C and type D retrovirus. Jpn J Genet. 1987;62:127–137. [Google Scholar]
- 19.Kawakami T, Sherman L, Dahlberg J, Gazit A, Yaniv A, Tronick S R, Aaronson S A. Nucleotide sequence analysis of equine infectious anemia virus proviral. DNA Virology. 1987;158:300–312. doi: 10.1016/0042-6822(87)90202-9. [DOI] [PubMed] [Google Scholar]
- 20.Kodama T, Wooley D P, Naidu Y M, Kestler H W, Daniel M D, Li Y, Desrosiers R C. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh W C, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 22.McGeoch D J. Protein sequence comparisons show that the “pseudoproteases encoded by the poxviruses and certain retoviruses belong to the deoxyuridine triphosphatase family. Nucleic Acids Res. 1990;18:4105–4100. doi: 10.1093/nar/18.14.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKinnon A O, Thorsen J, Hayes M A, Misener C R. Enzootic nasal adenocarcinoma of sheep in Canada. Can Vet J. 1982;23:88–94. [PMC free article] [PubMed] [Google Scholar]
- 24.Moore R, Dixon M, Smith R E, Peters G, Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987;61:480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortín A, Minguijón E, Dewar P, García M, Ferrer L M, Palmarini M, Gonzalez L, Sharp J M, De las Heras M. Lack of a specific immune response against a recombinant capsid protein of Jaagsiekte sheep retrovirus in sheep and goats naturally affected by enzootic nasal tumouror sheep pulmonary adenomatosis. Vet Immunol Immunopathol. 1998;61:229–237. doi: 10.1016/s0165-2427(97)00149-9. [DOI] [PubMed] [Google Scholar]
- 25a.Ortín, A. Unpublished data.
- 26.Palmarini M, Cousens C, Dalziel R G, Bai J, DeMartini J C, Sharp J M. The exogenous form of jaagsiekte retrovirus (JSRV) is specifically associated with a contagious lung cancer of sheep. J Virol. 1996;70:1618–1623. doi: 10.1128/jvi.70.3.1618-1623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Power M D, Marx P A, Bryant M L, Gardner M B, Barr P J, Luciw P A. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome retrovirus. Science. 1986;231:1567–1572. doi: 10.1126/science.3006247. [DOI] [PubMed] [Google Scholar]
- 28.Querat G, Audoly G, Sonigo P, Vigne R. Nucleotide sequence analysis of SA-OMVV, a visna-related ovine lentivirus: phylogenetic history of lentiviruses. Virology. 1990;175:434–447. doi: 10.1016/0042-6822(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 29.Rosati S, Kwang J, Tolari F, Keen J. Characterisation of enzootic nasal tumor virus capsid antigen. Vet Microbiol. 1996;53:261–269. doi: 10.1016/s0378-1135(96)01218-7. [DOI] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Sharp J M, Herring A J. Sheep pulmonary adenomatosis: demonstration of a protein which cross-reacts with the major core proteins of Mason-Pfizer monkey virus and mouse mammary tumour virus. J Gen Virol. 1983;64:2323–2327. doi: 10.1099/0022-1317-64-10-2323. [DOI] [PubMed] [Google Scholar]
- 32.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 33.Sonigo P, Barker C S, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 34.Tupper J C, Chen H, Hays E F, Bristol G C, Yoshimura F K. Contributions to transcriptional activity and to viral leukemogenicity made by sequences within and downstream of the MCF13 murine leukemia virus enhancer. J Virol. 1992;66:7080–7088. doi: 10.1128/jvi.66.12.7080-7088.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitellozzi G, Mughetti L, Palmarini M, Mandara M T, Mechelli L, Sharp J M, Manocchio I. Enzootic intranasal tumour of goats in Italy. J Vet Med Ser B. 1993;40:459–468. doi: 10.1111/j.1439-0450.1993.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 36.Wu Q, Chen M, Buchwald M, Phillips R A. A simple, rapid method for isolation of high quality genomic DNA from animal tissues. Nucleic Acids Res. 1995;23:5087–5088. doi: 10.1093/nar/23.24.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yonemichi H, Ohgi Y, Fujimoto K, Okado M, Onuma T, Mikami T. Intranasal tumour of the ethmoid olfactory mucosa in sheep. Am J Vet Res. 1978;39:1599–1606. [PubMed] [Google Scholar]
- 38.York D F, Vigne R, Verwoerd D W, Querat G. Nucleotide sequence of the Jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J Virol. 1992;66:4930–4939. doi: 10.1128/jvi.66.8.4930-4939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Zhu L, Benedict C A, Chen D, Anderson W F, Cannon P M. Functional domains in the retroviral transmembrane protein. J Virol. 1998;72:5392–5398. doi: 10.1128/jvi.72.7.5392-5398.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]