Abstract
Polymer models serve as useful tools for studying the formation and physical properties of biomolecular condensates. In recent years, the interface dividing the dense and dilute phases of condensates has been discovered to be closely related to their functionality, but the conformational preferences of the constituent proteins remain unclear. To elucidate this, we perform molecular simulations of a droplet formed by liquid‒liquid phase separation of homopolymers, as a surrogate model for the prion-like low-complexity domains. By systematically analyzing the polymer conformations at different locations in the droplet, we find that the chains become compact at the droplet interface compared to the droplet interior. Further, segmental analysis revealed that the end sections of the chains are enriched at the interface to maximize conformational entropy, and are more expanded than the middle sections of the chains. We find that the majority of chain segments lie tangential to the droplet surface and only the chain ends tend to align perpendicular to the interface. These trends also hold for the natural proteins FUC LC and LAF-1 RGG, which exhibit more compact chain conformations at the interface compared with the droplet interior. Our findings provide important insights into the interfacial properties of biomolecular condensates and highlight the value of using simple polymer physics models to understand the underlying mechanisms.
Keywords: Liquid‒liquid phase separation, membraneless organelles, interfacial behavior, molecular simulations
Membraneless organelles or biomolecular condensates formed through liquid‒liquid phase separation (LLPS) have been widely reported in various cellular functions, including gene expression, signal transduction, stress response, and the assembly of macromolecular complexes1–5. Examples of such condensates include the nucleolus, Cajal bodies, P bodies, and stress granules. Intrinsically disordered proteins (IDPs) play an important role in the formation of biomolecular condensates through LLPS6–11. Due to the numerous similarities between IDPs and synthetic polymers, classical polymer-based models offer a powerful approach for investigating the conformations12, dynamics13, and phase behavior of IDPs14–17. In particular, such models have been extensively used to reveal the sequence-dependent conformations of IDPs in the dense and dilute phases of condensates18–27.
In recent years, there has been increasing recognition of the important role played by the interfaces dividing the dense and dilute phases of biomolecular condensates. Folkmann et al. have reported that the IDP assemblies at the interface of PGL-3 droplets reduced the surface tension, thus preventing droplet coarsening. Kelley et al. reported that amphiphilic proteins on the surface of condensates acted like surfactants, thus regulating the size and structure of condensates28. Further, the interface can affect the interactions between biomolecular condensates and other biomolecules within the cell29. Böddeker et al. demonstrated that cytoskeletal filaments had a nonspecific affinity for stress granule interfaces, which explained the distinct enhancement of tubulin density around granules30. Lipiński et al. reported that the condensate interface can serve as a nucleation site promoting the aggregation of amyloidogenic proteins31. Inspired by the critical role of interfaces in various functionalities, much research has focused on determining mesoscopic interfacial properties such as surface tension13,32,33, surface adsorption34, and electrochemical properties35, which are deemed important for regulating biological functions29,36. For example, it was shown that the condensates could form an autophagosome only if the surface tension is lower than a critical value37. The film formed by protein adsorption at the air/water interface has a low interfacial elasticity at the protein’s isoelectric pH because of the compact structure38.
However, the microscopic conformations of individual IDPs at interfaces remain largely unclear, even though these exposed polymers largely dictate the surface properties. . For example, surfaces grafted by a synthetic polymer such as PNIPAM have been shown to result in a four-fold difference in the Young’s modulus between the swollen and collapsed states,39 and the surface wettability is stronger for polymer interfaces in stretched states than in collapsed states40. For biomolecular condensates, the chain conformations can be important for their functions like the regulation of condensates conformation on mRNA decapping41. To shed light on this question, Farag et al. recently studied the interface conformations of biomolecular condensates using lattice-based Monte Carlo simulations42,43.They reported that the overall dimensions of prion-like low-complexity domains and homopolymers varied within the condensates, being most expanded at the interface and preferring to be oriented perpendicular to it. These pioneering studies open up avenues for detailed characterization of the segmental-level conformational properties and their contribution to the chain-level conformational preferences at the condensate interfaces, which remain elusive.
In this letter, we report chain-level and segmental-level conformations of IDPs at the condensate interface from molecular simulations of hydrophobic homopolymers and two naturally occurring IDPs. We use an off-lattice polymer model, where each residue (“monomer”) is represented by a spherical bead in an implicit solvent (model details are provided as Supporting Information (SI)). This model is a good approximation for numerous IDPs, such as prion-like low-complexity domains42. Recently, we successfully used hydrophobic homopolymers as a reference to establish the biophysics of phase separation of IDPs, revealing that distributed interactions better stabilize the condensed phase than localized interactions24. To understand the basic principles governing the chain conformations in the droplet, we focus here on the hydrophobic homopolymers and include the results for the natural IDPs in the SI.
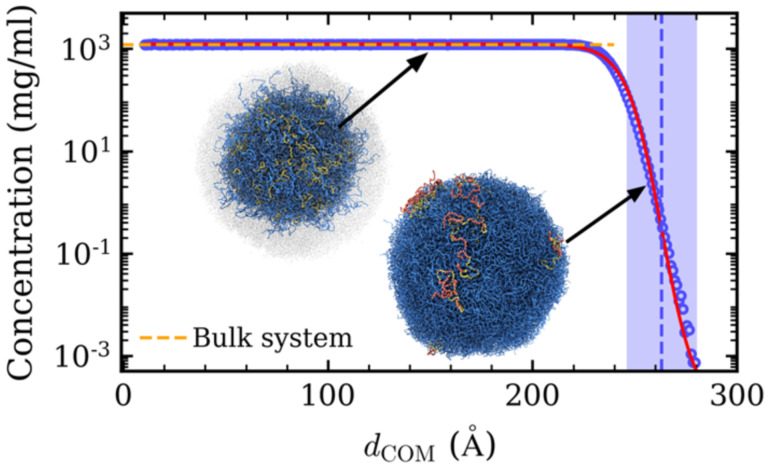
Due to the hydrophobic nature of the employed model IDP, it quickly formed a spherical condensate of radius R = 249Å, which is defined as the position where the concentration drops to half the value near the droplet center (Fig. 1). Drawing upon previous research on the liquid-vapor interface in slabs44, the radial concentration profile is fit to a hyperbolic tangent functional form
| (1) |
where dCOM is the distance from the droplet’s center of mass, c(dCOM) is the radial monomer concentration, cdense and cdilute are the monomer concentrations of the dense phase and dilute phase, respectively, dMID is the midpoint of the interface, and δ is the width of the interface. By fitting the computed radial concentration curve to (Eq. 1), the interfacial region was defined (Fig. 1). To establish a bulk reference without interface effects, we simulated a concentrated homopolymer solution in a cubic box at a constant pressure of P = 0 atm. With increasing dCOM, the concentration in the droplet interior remained constant (equal to the expected concentration in the bulk system), while it monotonically decreased at the interface. In this case, the dilute phase concentration is equal to 0 mg⁄ml, as all chains are inside the droplet.
Fig. 1.
Concentration as a function of distance from the droplet’s center of mass (dCOM). The horizontal dashed line represents the concentration in the bulk system. The red line is the fitted curve for the simulation data (symbols). The purple shaded area is the interface region. The vertical dashed line represents the middle of the interface. The inset shows simulation snapshots of the droplet interior and interface. The yellow chains in the interior snapshot are at a distance of dCOM = 150 Å. The chains colored yellow and red in the interface snapshot have their center-of-mass in the interface, with the red beads representing segments in the interface.
After determining the droplet interface, we studied the conformations of polymer chains within the interface and dense phase of the droplet. For this purpose, we computed the radius of gyration of the chains45
| (2) |
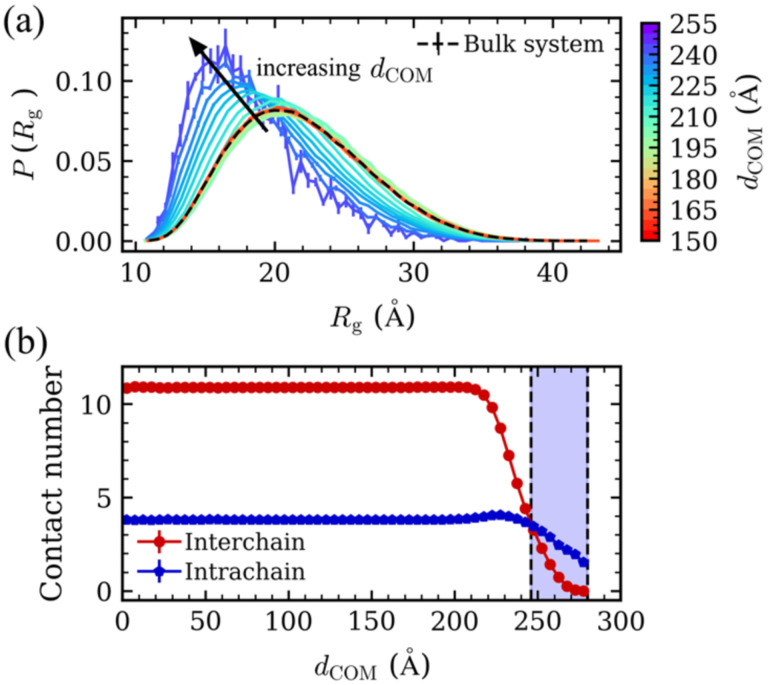
where the sum goes over the N monomers in the chain, and ri is the distance from the ith monomer to the chain’s center of mass (COM). We find that the corresponding Rg distribution in the bulk system (Fig. 2) was significantly broader, as compared to that of the single chain (Fig. S1), highlighting the preference of chains to expand inside the condensate, as expected from Flory’s ideality hypothesis46. Next, we analyzed the distribution of Rg at different positions within the droplet. Based on the distance between the chain’s COM and the droplet’s COM (dCOM), we assigned the polymer chains into different positional bins (Fig. 2a). We found that when dCOM ≤ 210 Å, where the local monomer concentration is equal to the bulk system concentration (Fig. 1), the Rg distributions overlapped with the bulk distribution, indicating the consistency between the interior of the droplet and the bulk phase. With increasing dCOM, the peak of the Rg distribution shifted gradually to smaller Rg values (Fig. 2a), demonstrating a continuous transition from expanded conformations to more compact conformations when moving from the interior of the droplet towards the interface. This result is also supported by the average number of interchain and intrachain contacts for each monomer (Fig. 2 b), which decreased monotonically with increasing dCOM, because of the decreasing monomer concentration (Fig. 1). At the interface, the number of average intrachain contacts was larger than the number of interchain contacts, which means that the chains interacted primarily with themselves and thus were more compact than in the droplet interior, where interchain interactions dominated.
Fig. 2.
(a) Distribution of radius of gyration (Rg). The dashed line represents the Rg distribution in a bulk system. The color gradient from red to purple corresponds to the distribution of Rg for chains with dCOM ranging from 150 to 255 Å. (b) Average contact number per monomer as a function of dCOM for the interchain and intrachain contacts. The vertical dashed lines represent the interface boundaries.
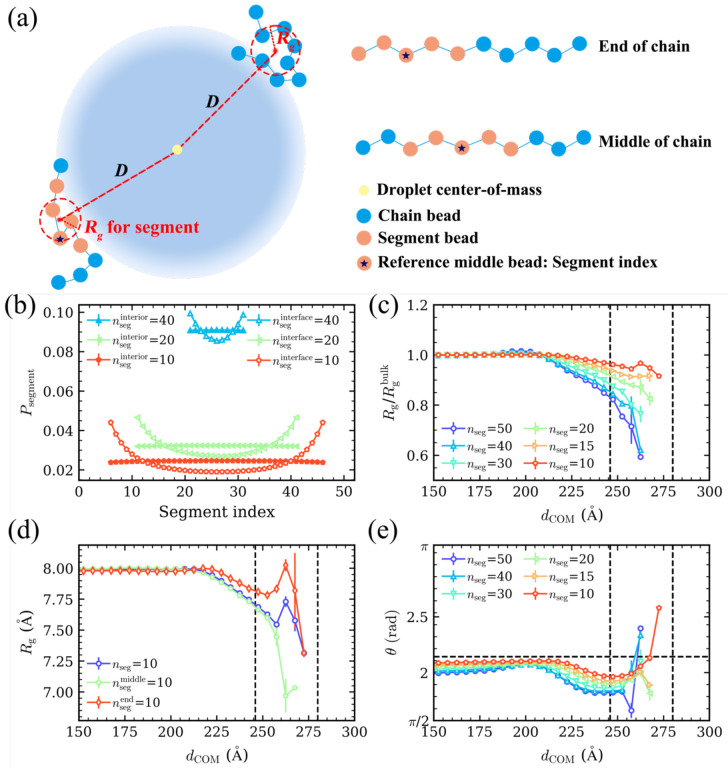
Our observation of compact chain-level conformations at the interface led us to investigate how segments within a chain contributed to its compaction, following previous calculations for the end-to-end vector of grafted polymers47 and Rg of polymer thin films48. We first characterized the distribution of chain segments of different length nseg at the droplet interface (Fig. 3a,b). The middle bead of the segments for a given nseg serves as the reference bead, the index of which we refer to as segment index. Note that a segment index closer to 1 or 50 corresponds to a segment closer to the chain termini, and the curves are symmetrical because the homopolymer chains do not have any “heads” or “tails”. We found that the highest probability always occurred at the position closest to the chain’s ends at the interface, regardless of the length of the segment, suggesting that the chain terminals were more inclined to distribute at the interface compared to the middle of the chains. This is in stark contrast to the droplet interior, where we found the same probabilities for segments at different relative positions within the chain (Fig. 3b). The enrichment of chain ends at the interface can be understood by considering the loss in conformational entropy incurred by placing a monomer at the interface, which is smaller for the end monomers compared to any other monomer.
Fig. 3.
Segmental analysis. (a) Schematic diagram of the Rg calculation method for chains and their segments. Blue beads represent the chain monomers, while orange beads represent a specific segment. (b) Probability distribution of the segments for different segment lengths (nseg) at the interface (hollow symbols) and in the droplet interior (solid symbols). The segment index is represented by the index of middle bead of each segment (reference bead). When nseg is even, the larger index between the two middle beads is considered as the segment index. (c) Normalized average Rg of the different length segments with respect to dCOM. The black dashed lines represent the boundaries of the interface. (d) Rg of segments consisting of 10 monomers as a function of dCOM. The purple line represents for the average Rg for all segments in the chain of that length. The green and red lines represent the segments located at the middle and end of the chains, respectively. (e) Angle (θ) of the different length segments with respect to dCOM. The horizontal dashed line represents the average angle for isotropic distribution of segments.
We next asked if the spatial preference of the chain segments at the droplet interface vs its interior had an effect on the segmental-level conformations. For this purpose, we calculated the average Rg for the segments of different lengths nseg as a function of distance dCOM between their COM and the droplet’s COM (Fig. 3c). We found that the Rg at the interface is smaller than that in the droplet interior for all nseg considered. For nseg > 15, Rg monotonically decreased as the droplet interface is approached. For nseg = 15, Rg no longer decreased in the interface region. Interestingly, for nseg = 10, we found that Rg marginally increased in the middle of the interface region, following which it decreased again. To explain this nonmonotonic trend in Rg for the short segments at the interface, we analyzed Rg of different nseg located only at the chain ends and compared it with that in the middle of the chain. For longer segments nseg ≥ 20, the segmental Rg monotonically decreased from the droplet interior to the interface, regardless of the position of the segments (Fig. S2). However, for shorter segments nseg ≤ 15 (Figs. 3d and S2), we found that the Rg of the segments located at the chain ends exhibited a nonmonotonic trend in the interface region: we observed an increase in segmental Rg for dCOM > 250 Å, attaining the maximum value in the middle of the interface region, following which it continued to decrease. Surprisingly, the Rg of shorter segments located in the middle of the chains monotonically decreased throughout the interface region. These observations indicate that the chain termini are the only contributors to the observed expansion of short segments (Fig. 3c). In summary, although the whole chain is more compact at the interface, short terminal segments are more expanded than middle segments at the interface and even slightly more expanded than the short segments in the droplet interior.
Given the stark differences in the global and local conformations of the chains in the interface region, we next investigated their chain-level and segmental-level orientations. We defined the angle θ between the segment-to-droplet COM vector and the segment end-to-end vector to characterize the orientation of the chains in the interface region (Fig. S3a). The positions of segments are again defined through dCOM. When the angle is close to π⁄2, the segment lies tangential to the droplet surface, whereas an angle close to π corresponds to an orientation perpendicular to the droplet surface. For different segment lengths (nseg = 10, 15, 20, 30, 40) and the whole chain (nseg = 50), the angles show an increasing trend at the interface (Fig. 3e). This increase primarily originates from the segments near the chain ends (Fig. S3b) rather than segments in the middle of the chains (Fig. S3c). This observation can be explained by considering the end-to-end distance (Re) (Fig. S4). The smallest angle for segments (Fig. S3a) can be estimated as:
| (3) |
where R is the distance between the terminal bead of the segment and the droplet COM, and Re is the end-to-end distance of the segment. At the interface, R is much larger than Re, so that θmin becomes close to π⁄2 (i.e., tangential orientation). πIf segments are oriented randomly, the average angle should be close to π − 1 (calculation shown in SI). We found that the angles of segments located in the middle of chains were between π⁄2 and π − 1, indicating that these segments lied predominantly tangential to the surface (Fig. S3c). While the angles of end segments located in the droplet interior were between π⁄2 and π − 1, end segments near the interface exhibited a tendency to form angles larger than π − 1 (Fig. S3b). However, these segments remained closer to the angle representing isotropic segments, π − 1, than perpendicular segments, π. These results demonstrate that despite the tendency of chain ends to align perpendicular to the surface, the majority of chain segments remain primarily tangential to the droplet surface.
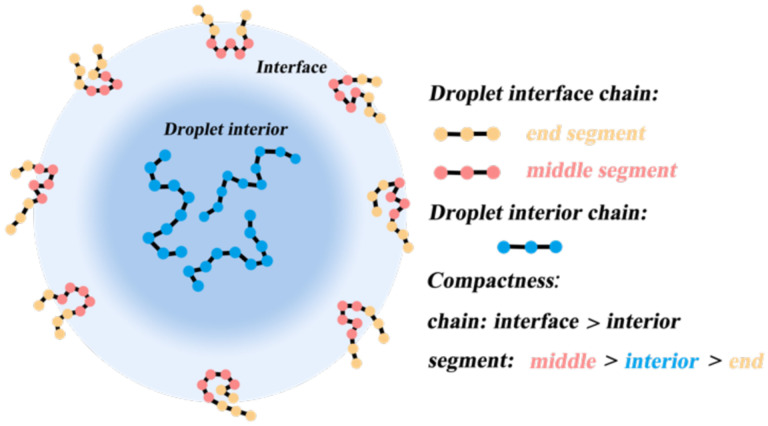
Based on our investigation of size and orientation of chain segments at different positions in the droplet, we propose a model for homopolymer conformations in condensates as depicted in Fig. 4. Compared with the droplet interior, which we found to be consistent with the bulk phase, the distribution of Rg in the interface shifted to smaller values, indicating more compact chain conformations at the droplet interface. Through our characterization of the conformational preferences of segments of different lengths at different positions, we have discovered that segments located at the chains’ ends slightly expand more in the droplet interface compared to the droplet interior, while the middle segments of the chains are more collapsed in the interface region than in the droplet interior. These end segments are enriched at the interface to minimize the entropy loss during LLPS. At the interface, the chain ends exhibit a tendency to align perpendicularly to the interface, but the majority of the chains are still parallel to the surface. We speculate that these conformational changes originate from the change in the local environment surrounding the chains: Compared to the interior of the droplet, the lower monomer concentration at the interface leads to a decrease in the (effective) solvent quality, thus resulting in chain collapse. Similar compact chain conformations at the interface have been reported previously for hydrophobic homopolymers49–51.
Fig. 4.
Schematic of homopolymer conformations at the droplet interface. Polymer chains are more compact at the interface than in the droplet interior. Compared to the interior of the droplet, the chain ends are more expanded, while the middle segments of the chains are more collapsed at the interface. The chain ends exhibit a tendency to be perpendicular to the droplet surface, whereas the majority of chains lie predominantly tangential to the droplet surface.
Our findings illustrate the conformational preferences at the condensate interface using a homopolymer model, which can also be observed for naturally occurring IDPs. For FUS LC and LAF-1 RGG (Fig. S5), chains are more collapsed at the interface compared with the expanded conformations in the droplet interior. In future work, quantitatively describing the relationship between conformations and interfacial properties will be highly beneficial for our understanding of the biological functions of biomolecular condensates.
Supplementary Material
Acknowledgments
This material is based on the research supported by the National Institute of General Medical Science of the National Institutes of Health under the grant R01GM136917 and the Welch Foundation under the grant A-2113-20220331. A.N. acknowledges funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through Project 470113688. We gratefully acknowledge the computational resources provided by the Texas A&M High Performance Research Computing (HPRC) to complete this work.
Footnotes
Supporting Information
The details of the model and simulation, method of average angle calculation, sequences for natural protein (FUS LC and LAF-1 RGG), probability distribution of Rg, average Rg, Re, angle of different segments at different positions and Rg of natural proteins.
REFERENCES
- (1).Hyman A. A.; Weber C. A.; Jülicher F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30 (1), 39–58. 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- (2).Banani S. F.; Lee H. O.; Hyman A. A.; Rosen M. K. Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18 (5), 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Alberti S.; Gladfelter A.; Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176 (3), 419–434. 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Dignon G. L.; Best R. B.; Mittal J. Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem. 2020, 71 (1), 53–75. 10.1146/annurev-physchem-071819-113553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lyon A. S.; Peeples W. B.; Rosen M. K. A Framework for Understanding the Functions of Biomolecular Condensates across Scales. Nat. Rev. Mol. Cell Biol. 2021, 22 (3), 215–235. 10.1038/s41580-020-00303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Uversky V. N.; Kuznetsova I. M.; Turoverov K. K.; Zaslavsky B. Intrinsically Disordered Proteins as Crucial Constituents of Cellular Aqueous Two Phase Systems and Coacervates. FEBS Lett. 2015, 589 (1), 15–22. 10.1016/j.febslet.2014.11.028. [DOI] [PubMed] [Google Scholar]
- (7).Brady J. P.; Farber P. J.; Sekhar A.; Lin Y.-H.; Huang R.; Bah A.; Nott T. J.; Chan H. S.; Baldwin A. J.; Forman-Kay J. D.; Kay L. E. Structural and Hydrodynamic Properties of an Intrinsically Disordered Region of a Germ Cell-Specific Protein on Phase Separation. Proc. Natl. Acad. Sci. 2017, 114 (39), E8194–E8203. 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Martin E. W.; Mittag T. Relationship of Sequence and Phase Separation in Protein Low-complexity Regions. Biochemistry 2018, 57 (17), 2478–2487. 10.1021/acs.biochem.8b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Dignon G. L.; Zheng W.; Kim Y. C.; Best R. B.; Mittal J. Sequence Determinants of Protein Phase Behavior from a Coarse-Grained Model. PLOS Comput. Biol. 2018, 14 (1), e1005941. 10.1371/journal.pcbi.1005941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Schuster B. S.; Dignon G. L.; Tang W. S.; Kelley F. M.; Ranganath A. K.; Jahnke C. N.; Simpkins A. G.; Regy R. M.; Hammer D. A.; Good M. C.; Mittal J. Identifying Sequence Perturbations to an Intrinsically Disordered Protein That Determine Its Phase-Separation Behavior. Proc. Natl. Acad. Sci. 2020, 117 (21), 11421–11431. 10.1073/pnas.2000223117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Mohanty P.; Kapoor U.; Sundaravadivelu Devarajan D.; Phan T. M.; Rizuan A.; Mittal J. Principles Governing the Phase Separation of Multidomain Proteins. Biochemistry 2022, 61 (22), 2443–2455. 10.1021/acs.biochem.2c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Dignon G. L.; Zheng W.; Best R. B.; Kim Y. C.; Mittal J. Relation between Single-Molecule Properties and Phase Behavior of Intrinsically Disordered Proteins. Proc. Natl. Acad. Sci. 2018, 115 (40), 9929–9934. 10.1073/pnas.1804177115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Devarajan D. S.; Wang J.; Nikoubashman A.; Kim Y. C.; Mittal J. Sequence-Dependent Material Properties of Biomolecular Condensates. bioRxiv May 12, 2023, p 2023.05.09.540038. 10.1101/2023.05.09.540038. [DOI] [Google Scholar]
- (14).Brangwynne C. P.; Tompa P.; Pappu R. V. Polymer Physics of Intracellular Phase Transitions. Nat. Phys. 2015, 11 (11), 899–904. [Google Scholar]
- (15).Soranno A. Physical Basis of the Disorder-Order Transition. Arch. Biochem. Biophys. 2020, 685, 108305. 10.1016/j.abb.2020.108305. [DOI] [PubMed] [Google Scholar]
- (16).Welles R. M.; Sojitra K. A.; Garabedian M. V.; Xia B.; Regy R. M.; Gallagher E. R.; Mittal J.; Good M. C. Determinants of Disordered Protein Co-Assembly Into Discrete Condensed Phases. bioRxiv March 12, 2023, p 2023.03.10.532134. 10.1101/2023.03.10.532134. [DOI] [PubMed] [Google Scholar]
- (17).Dignon G. L.; Zheng W.; Kim Y. C.; Mittal J. Temperature-Controlled Liquid–Liquid Phase Separation of Disordered Proteins. ACS Cent. Sci. 2019, 5 (5), 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Das R. K.; Pappu R. V. Conformations of Intrinsically Disordered Proteins Are Influenced by Linear Sequence Distributions of Oppositely Charged Residues. Proc. Natl. Acad. Sci. 2013, 110 (33), 13392–13397. 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sawle L.; Ghosh K. A Theoretical Method to Compute Sequence Dependent Configurational Properties in Charged Polymers and Proteins. J. Chem. Phys. 2015, 143 (8), 085101. 10.1063/1.4929391. [DOI] [PubMed] [Google Scholar]
- (20).Lin Y.-H.; Forman-Kay J. D.; Chan H. S. Theories for Sequence-Dependent Phase Behaviors of Biomolecular Condensates. Biochemistry 2018, 57 (17), 2499–2508. 10.1021/acs.biochem.8b00058. [DOI] [PubMed] [Google Scholar]
- (21).Devarajan D. S.; Rekhi S.; Nikoubashman A.; Kim Y. C.; Howard M. P.; Mittal J. Effect of Charge Distribution on the Dynamics of Polyampholytic Disordered Proteins. Macromolecules 2022, 55 (20), 8987–8997. 10.1021/acs.macromol.2c01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ghosh K.; Huihui J.; Phillips M.; Haider A. Rules of Physical Mathematics Govern Intrinsically Disordered Proteins. Annu. Rev. Biophys. 2022, 51, 355–376. 10.1146/annurev-biophys-120221-095357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bauer D. J.; Stelzl L. S.; Nikoubashman A. Single-Chain and Condensed-State Behavior of HnRNPA1 from Molecular Simulations. J. Chem. Phys. 2022, 157 (15), 154903. 10.1063/5.0105540. [DOI] [PubMed] [Google Scholar]
- (24).Rekhi S.; Sundaravadivelu Devarajan D.; Howard M. P.; Kim Y. C.; Nikoubashman A.; Mittal J. Role of Strong Localized vs Weak Distributed Interactions in Disordered Protein Phase Separation. J. Phys. Chem. B 2023, 127 (17), 3829–3838. 10.1021/acs.jpcb.3c00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Yu M.; Heidari M.; Mikhaleva S.; Tan P. S.; Mingu S.; Ruan H.; Reinkemeier C. D.; Obarska-Kosinska A.; Siggel M.; Beck M.; Hummer G.; Lemke E. A. Visualizing the Disordered Nuclear Transport Machinery in Situ. Nature 2023, 617 (7959), 162–169. 10.1038/s41586-023-05990-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yuan X.; W. Hatch H.; C. Conrad J.; B. Marciel A.; C. Palmer J. PH Response of Sequence-Controlled Polyampholyte Brushes. Soft Matter 2023, 19 (23), 4333–4344. 10.1039/D3SM00447C. [DOI] [PubMed] [Google Scholar]
- (27).Perry S. L. Phase Separation: Bridging Polymer Physics and Biology. Curr. Opin. Colloid Interface Sci. 2019, 39, 86–97. 10.1016/j.cocis.2019.01.007. [DOI] [Google Scholar]
- (28).Kelley F. M.; Favetta B.; Regy R. M.; Mittal J.; Schuster B. S. Amphiphilic Proteins Coassemble into Multiphasic Condensates and Act as Biomolecular Surfactants. Proc. Natl. Acad. Sci. 2021, 118 (51), e2109967118. 10.1073/pnas.2109967118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gouveia B.; Kim Y.; Shaevitz J. W.; Petry S.; Stone H. A.; Brangwynne C. P. Capillary Forces Generated by Biomolecular Condensates. Nature 2022, 609 (7926), 255–264. 10.1038/s41586-022-05138-6. [DOI] [PubMed] [Google Scholar]
- (30).Böddeker T. J.; Rosowski K. A.; Berchtold D.; Emmanouilidis L.; Han Y.; Allain F. H. T.; Style R. W.; Pelkmans L.; Dufresne E. R. Non-Specific Adhesive Forces between Filaments and Membraneless Organelles. Nat. Phys. 2022, 18 (5), 571–578. 10.1038/s41567-022-01537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Lipiński W. P.; Visser B. S.; Robu I.; Fakhree M. A. A.; Lindhoud S.; Claessens M. M. A. E.; Spruijt E. Biomolecular Condensates Can Both Accelerate and Suppress Aggregation of α-Synuclein. Sci. Adv. 2022, 8 (48), eabq6495. 10.1126/sciadv.abq6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Qin J.; Priftis D.; Farina R.; Perry S. L.; Leon L.; Whitmer J.; Hoffmann K.; Tirrell M.; de Pablo J. J. Interfacial Tension of Polyelectrolyte Complex Coacervate Phases. ACS Macro Lett. 2014, 3 (6), 565–568. 10.1021/mz500190w. [DOI] [PubMed] [Google Scholar]
- (33).Mitrea D. M.; Chandra B.; Ferrolino M. C.; Gibbs E. B.; Tolbert M.; White M. R.; Kriwacki R. W. Methods for Physical Characterization of Phase-Separated Bodies and Membrane-Less Organelles. J. Mol. Biol. 2018, 430 (23), 4773–4805. 10.1016/j.jmb.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Erkamp N. A.; Farag M.; Qian D.; Sneideris T.; Welsh T. J.; Ausserwöger H.; Weitz D. A.; Pappu R. V.; Knowles T. P. J. Adsorption of RNA to Interfaces of Biomolecular Condensates Enables Wetting Transitions. bioRxiv January 13, 2023, p 2023.01.12.523837. 10.1101/2023.01.12.523837. [DOI] [Google Scholar]
- (35).Dai Y.; Chamberlayne C. F.; Messina M. S.; Chang C. J.; Zare R. N.; You L.; Chilkoti A. Interface of Biomolecular Condensates Modulates Redox Reactions. Chem 2023, 9 (6), 1594–1609. 10.1016/j.chempr.2023.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Bratek-Skicki A.; Van Nerom M.; Maes D.; Tompa P. Biological Colloids: Unique Properties of Membraneless Organelles in the Cell. Adv. Colloid Interface Sci. 2022, 310, 102777. 10.1016/j.cis.2022.102777. [DOI] [PubMed] [Google Scholar]
- (37).Agudo-Canalejo J.; Schultz S. W.; Chino H.; Migliano S. M.; Saito C.; Koyama-Honda I.; Stenmark H.; Brech A.; May A. I.; Mizushima N.; Knorr R. L. Wetting Regulates Autophagy of Phase-Separated Compartments and the Cytosol. Nature 2021, 591 (7848), 142–146. 10.1038/s41586-020-2992-3. [DOI] [PubMed] [Google Scholar]
- (38).Burgess D.; Sahin N. O. Interfacial Rheological and Tension Properties of Protein Films. J. Colloid Interface Sci. 1997, 189 (1), 74–82. 10.1006/jcis.1997.4803. [DOI] [Google Scholar]
- (39).Besford Q. A.; Uhlmann P.; Fery A. Spatially Resolving Polymer Brush Conformation: Opportunities Ahead. Macromol. Chem. Phys. 2023, 224 (1), 2200180. 10.1002/macp.202200180. [DOI] [Google Scholar]
- (40).Chang B.; Zhang B.; Sun T. Smart Polymers with Special Wettability. Small 2017, 13 (4), 1503472. 10.1002/smll.201503472. [DOI] [PubMed] [Google Scholar]
- (41).Tibble R. W.; Depaix A.; Kowalska J.; Jemielity J.; Gross J. D. Biomolecular Condensates Amplify MRNA Decapping by Biasing Enzyme Conformation. Nat. Chem. Biol. 2021, 17 (5), 615–623. 10.1038/s41589-021-00774-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Farag M.; Cohen S. R.; Borcherds W. M.; Bremer A.; Mittag T.; Pappu R. V. Condensates Formed by Prion-like Low-Complexity Domains Have Small-World Network Structures and Interfaces Defined by Expanded Conformations. Nat. Commun. 2022, 13 (1), 7722. 10.1038/s41467-022-35370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Farag M.; Borcherds W. M.; Bremer A.; Mittag T.; Pappu R. V. Phase Separation in Mixtures of Prion-Like Low Complexity Domains Is Driven by the Interplay of Homotypic and Heterotypic Interactions. bioRxiv March 16, 2023, p 2023.03.15.532828. 10.1101/2023.03.15.532828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Kuo I. W.; Mundy C. J.; Eggimann B. L.; McGrath M. J.; Siepmann J. I.; Chen B.; Vieceli J.; Tobias D. J. Structure and Dynamics of the Aqueous Liquid− Vapor Interface: A Comprehensive Particle-Based Simulation Study. J. Phys. Chem. B 2006, 110 (8), 3738–3746. [DOI] [PubMed] [Google Scholar]
- (45).Fixman M. Radius of Gyration of Polymer Chains. J. Chem. Phys. 1962, 36 (2), 306–310. [Google Scholar]
- (46).Flory P. J. Statistical Mechanics of Chain Molecules; Hanser: Munich, 1989. [Google Scholar]
- (47).Midya J.; Rubinstein M.; Kumar S. K.; Nikoubashman A. Structure of Polymer-Grafted Nanoparticle Melts. ACS Nano 2020, 14 (11), 15505–15516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Doruker P.; Mattice W. L. Simulation of Polyethylene Thin Films on a High Coordination Lattice. Macromolecules 1998, 31 (4), 1418–1426. 10.1021/ma971322z. [DOI] [Google Scholar]
- (49).Jiang S.; Luo C.; Lu Y. Multilayered Nature in Crystallization of Polymer Droplets Studied by MD Simulations: Orientation and Entanglement. Polymer 2023, 268, 125696. 10.1016/j.polymer.2023.125696. [DOI] [Google Scholar]
- (50).Kadoya N.; Arai N. Size Dependence of Static Polymer Droplet Behavior from Many-Body Dissipative Particle Dynamics Simulation. Phys. Rev. E 2017, 95 (4), 043109. 10.1103/PhysRevE.95.043109. [DOI] [PubMed] [Google Scholar]
- (51).Berressem F.; Scherer C.; Andrienko D.; Nikoubashman A. Ultra-Coarse-Graining of Homopolymers in Inhomogeneous Systems. J. Phys. Condens. Matter 2021, 33 (25), 254002. 10.1088/1361-648X/abf6e2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.