Abstract
Because immunologic classification of human immunodeficiency virus type 1 (HIV) might be more relevant than genotypic classification for designing polyvalent vaccines, studies were undertaken to determine whether immunologically defined groups of HIV (“immunotypes”) could be identified. For these experiments, the V3 region of the 120-kDa envelope glycoprotein (gp120) was chosen for study. Although antibodies (Abs) to V3 may not play a major protective role in preventing HIV infection, identification of a limited number of immunologically defined structures in this extremely variable region would set a precedent supporting the hypothesis that, despite its diversity, the HIV family, like the V3 region, might be divisible into immunotypes. Consequently, the immunochemical reactivities of 1,176 combinations of human anti-V3 monoclonal Abs (MAbs) and V3 peptides, derived from viruses of several clades, were studied. Extensive cross-clade reactivity was observed. The patterns of reactivities of 21 MAbs with 50 peptides from clades A through H were then analyzed by a multivariate statistical technique. To test the validity of the mathematical approach, a cluster analysis of the 21 MAbs was performed. Five groups were identified, and these MAb clusters corresponded to classifications of these same MAbs based on the epitopes which they recognize. The concordance between the MAb clusters identified by mathematical analysis and by their specificities supports the validity of the mathematical approach. Therefore, the same mathematical technique was used to identify clusters within the 50 peptides. Seven groups of peptides, each containing peptides from more than one clade, were defined. Inspection of the amino acid sequences of the peptides in each of the mathematically defined peptide clusters revealed unique “signature sequences” that suggest structural motifs characteristic of each V3-based immunotype. The results suggest that cluster analysis of immunologic data can define immunotypes of HIV. These immunotypes are distinct from genotypic classifications. The methods described pave the way for identification of immunotypes defined by immunochemical and neutralization data generated with anti-HIV Env MAbs and intact, viable HIV virions.
Within three years of isolation of the human immunodeficiency virus type 1 (HIV) from patients in North America and Western Europe, the genetic diversity of HIV was recognized as a consistent feature, manifesting itself in the constant and variable regions of the 120-kDa envelope glycoprotein (gp120) of the virus (48). With further virus isolations from patients around the world and extensive sequencing, HIV strains were grouped into genotypes, or clades, based on sequence clustering patterns (41). To date, these sequence analyses have revealed at least 10 major clades, designated A through I, in the major group (group M) and a still unknown number of clades in the outlier group (group O) (24, 25, 30, 32, 40, 42). The extensive variability of HIV is now recognized as having a critical impact on diagnosis, therapy, and prevention (11).
The issue of HIV diversity is currently being revisited from the point of view of the human immune response to this virus family. It is clear from previous studies that HIV genotypes do not generally correspond to serotypes defined on the basis of immunochemical or neutralizing activity (4, 16, 17, 29, 36, 44, 45, 47, 53), although data reported by Mascola et al. suggest that clade E viruses constitute an immunologically distinct subtype within group M (33). Clearly, however, much more extensive work is needed to determine if immunologically related groups (immunotypes) of HIV can be defined and whether they will be more relevant than genotypes for the design of a vaccine.
In fact, both sequence data and immunochemical data, when analyzed by various mathematical approaches, suggest that serotypes do indeed exist and that they do not appear to correlate with clades. Two groups independently studied the amino acid sequences and serologic characteristics of the V3 portion of gp120 (4, 28, 47); the results of these studies suggested that there are rational alternatives to the genotypic classification of HIV and that these newly defined groups contain viruses from multiple clades. An initial study by Korber et al. using V3 sequence data employed protein similarity-based cluster analysis (28). These studies of the V3 sequences of 302 viruses from clades A through F suggested that 14 clusters could be observed. While some clusters contained viruses from only a single clade (e.g., clade D or E), other clusters contained representatives of multiple clades. Moreover, clades A and C were found to have identical or highly similar V3 amino acid sequences, and the D clade sequences were found to possess the most radically divergent set of V3 loop sequences. Additional studies using a subtype-specific enzyme-linked immunosorbent assay (ELISA) with 321 HIV-positive sera from patients in 10 countries and 19 V3 peptides from clades A through F, followed by cluster analysis of the serologic data, revealed five to nine serologic groups, some of which contained a single clade (e.g., A or D), while others contained representatives of multiple clades (4, 47). These studies, and others (29, 36, 44, 45, 53), reveal the existence of HIV epitopes shared by viruses belonging to different clades and suggest the existence of HIV immunotypes. However, with the exception of the serologic studies conducted with defined peptides (4, 47), experiments using polyclonal sera provide only limited information about the identity of the shared epitopes.
Monoclonal antibodies (MAbs) provide the specificity needed to map shared epitopes. Indeed, human MAbs to the V3 region have revealed shared epitopes by their broad intra- and interclade reactivity (9, 16, 17, 20, 37, 43, 55). Extensive cross-clade reactivity is also exhibited by MAbs to the CD4 binding domain, to the C terminus of gp120, and to regions on gp41 (6, 9, 10, 27, 37, 49, 50, 55). Cross-clade reactivity of these MAbs also suggests that immunotypes may exist and that the epitopes defining the immunotypes can be identified.
To test the hypothesis that HIV immunotypes can be identified, a panel of 21 human anti-V3 MAbs was studied immunochemically for cross-reactivity to V3 peptides derived from the sequences of viruses of group M (clades A through H) and group O. Extensive cross-clade reactivity was observed. The patterns of interaction were then analyzed mathematically, revealing groups of V3 loops that cluster together on the basis of their profiles of reactivity with the MAbs. Each of these clusters contains peptides from more than one clade, demonstrating that the immunotypic clusters do not correspond to cladal groups. Moreover, peptides belonging to the same cluster possess characteristic patterns of amino acids, i.e., signature sequences, which distinguish them from peptides of other clusters. These studies of the V3 loop provide the basis for further investigation of immunochemically and functionally defined HIV immunotypes based on examination of intact virions with MAbs derived from subjects infected with diverse viruses and MAbs specific for several envelope epitopes.
MATERIALS AND METHODS
MAbs.
Twenty-one immunoglobulin G anti-V3 MAbs were derived from cell lines made from the peripheral blood of patients infected with HIV-1. Production of these human MAbs has been described in detail (19). With the exception of MAb 1324E, all MAbs came from the cells of subjects in the United States infected with clade B. MAb 1324E came from the cells of a subject infected in Thailand with clade E (16). Selection of the heterohybridomas producing the anti-V3 MAbs was performed by screening with V3MN (14, 19, 20), gp120IIIB (14), V3RF (17), V3E (16), or oligomeric gp140 from HIV451 (18, 51, 52). An additional anti-V3 MAb, 1108, was derived by screening cells with a 23-mer peptide, peptide 987, which is the best mimeotope of MAb 447-52D (26). Peptide 987 was supplied by A. Conley (Merck Research Laboratories, West Point, Pa.) and contains the GPGR motif, which is the core epitope of MAb 447-52D (26), within a random amino acid sequence generated in a random phage display library. MAb 447-52D is referred to hereafter as MAb 447.
Peptides.
Fifty-six 19- to 30-mer peptides which span the tips of the V3 loops of viruses from group M (clades A through H) and group O were synthesized. Eleven V3 peptides, representing the sequences of MN, SF2, SC, NY/5, RF, WMJ2, CDC4, BRU, SF33, MAL and ELI, were purchased from Intracel, Inc. (Cambridge, Mass.); these peptides were purified by high-performance liquid chromatography (HPLC) and found to have a purity of >80%. All peptides from Intracel contained cysteine residues at the N terminus. One peptide, D687, was synthesized by Genemed Biotechnologies, Inc. (South San Francisco, Calif.); it was synthesized by using the standard 9-fluorenylmethoxycarbonyl (Fmoc) technology, purified by C-18 reverse phase HPLC, identified by mass spectroscopy, and found to have a purity of >80%. Peptide 987 was provided by A. Conley (see above) and was previously described (26). The peptide that represents is the consensus sequence of the V3 loop of clade E was synthesized by C. Fiol, Colorado State University (Fort Collins, Colo.), and was used as a preparation which was 68% pure as judged by HPLC analysis. The V3 peptides from clades G and H were purchased from Princeton BioMolecules Corp. (Columbus, Ohio); purity as assessed by HPLC, amino acid analysis, and mass spectroscopy was >85%. The remaining peptides from clades A, C, D, E, and F were synthesized by Lawrence Loomis-Price at the H. M. Jackson Foundation (Rockville, Md.) by standard solid-phase methods with an ABI 433 automated peptide synthesizer (Applied Biosystems, Foster City, Calif.) and were >70% pure as assessed by HPLC with the exceptions of peptides D3MA959 and 12233, both clade C, whose purities were 53 and 50%, respectively. None of the N- or C-terminal amino acids were derivatized, and none of the peptides were cyclic.
ELISA.
A standard peptide ELISA which has been described previously was used (15, 17). Briefly, V3 peptides were coated onto plastic Immulon 2HB plates (Dynex Technologies, Inc., Chantilly, Va.) at 1 μg/ml. Plates were blocked for 1 h at 37°C with 2.5% bovine serum albumin in phosphate-buffered saline and then washed three times with phosphate-buffered saline containing 0.05% Tween 20 (pH 7.4). Subsequently, each human MAb, at 10 μg/ml, was added and incubated for 1.5 h at 37°C. After subsequent washing, the plates were incubated with alkaline phosphatase-conjugated goat anti-human immunoglobulin G (γ-chain specific) (Zymed, Inc., South San Francisco, Calif.). Color was developed with the substrate p-nitrophenyl phosphate. Plates were read at 410 nm. Negative controls consisted of V3 peptide-coated wells reacted with an irrelevant human MAb, 670-D, specific for a linear epitope in the C5 region of gp120 (55).
A panel of 21 anti-V3 MAbs and 50 peptides (exclusive of the six clade O peptides) was run four times, and all sets of data were analyzed. Because of the close concordance of the data sets, the results of only a single set are shown in this presentation. The panel of 21 MAbs with the six clade O V3 peptides was run only once. Because of the lack of reactivity between these MAbs and the clade O V3 peptides (see below), these latter data were not included in the mathematical analyses.
Mathematical analyses.
The programs used to analyze the optical density (OD) data generated by studying the ELISA reactivities of the 21 MAbs with the 50 V3 peptides were written in the S language, which is available as part of the interactive statistical package SPLUS (Mathsoft, Inc.) originated from the AT&T Bell Labs. The procedure used has been described in detail in reference 44 and is based on the principle of grouping Abs and antigens with similar specificities and reactivities. Basically, the profile of reactivities of each MAb with all 50 peptides was compared for similarities to the profile of each of the other MAbs. Assignment to individual groupings was made by comparing reactivity profiles; those patterns closest to one another were grouped together. This was accomplished by calculating the distance between each pair of profiles. This distance was the total absolute difference in reactivity between two profiles across the panel. The procedures for clustering the MAbs and for clustering the peptides were performed by using the same panel of data because it provides the answers to both of the questions being addressed, namely, which MAbs have similar reactivity profiles and which peptides have similar reactivity profiles.
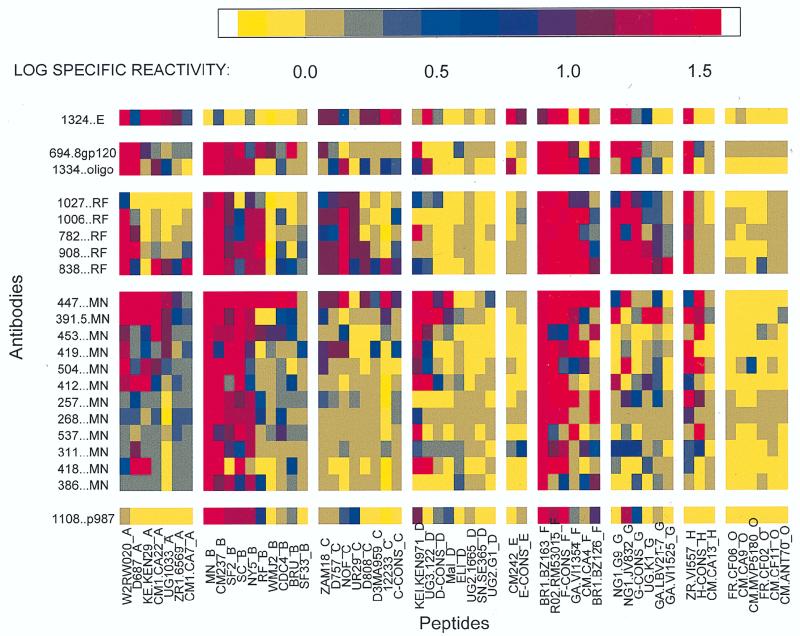
Immunoreactivity was assessed by using ELISA data that were treated as continuous variables and were normalized by calculating the log specific reactivity (LSR), which was calculated as log (ODexperimental MAb + peptide/ ODcontrol MAb + peptide) for each MAb and peptide combination. (For the LSRs, see Fig. 1, where the columns display the reactivities of the V3 peptides and the rows display the reactivities of the human anti-V3 MAbs studied.)
FIG. 1.
ELISA results of the reactivity of each V3 peptide with each of the human anti-V3 MAbs. The data are displayed as a matrix in which the spectrum of color shown at the top of the figure corresponds to the LSR, which is defined as log (ODexperimental MAb + peptide/ODcontrol MAb + peptide) for each of the 1,176 combinations of MAbs and peptides. The MAbs are listed vertically, and the suffix of each denotes the peptide or protein with which it was selected (see text). The peptides are listed horizontally, and each suffix denotes the clade of the virus from which the peptide was derived.
From the LSR shown for each MAb and peptide combination was subtracted the average LSR of all MAbs with that peptide and the average LSR of all peptides with that MAb. Then, the grand average LSR of all MAbs and peptides was added; this produced a double-centered 21 by 50 data matrix of specific interactions. Each of the 21 MAbs was then modeled as a single point in 50 dimensions. The data were subsequently projected to a four-dimensional subspace by using the singular value decomposition of the matrix. This produced a version of the matrix, one from which nonstructural variation (noise) had been removed. The relationships among the 21 MAbs were then depicted by clustering them hierarchically (bottom up) to produce a complete dendrogram, and the Hartigan algorithm (see below) was applied to identify the approximate number of clusters into which the 21 MAbs fell. The identical procedure was followed for analyzing the 50 peptides and identifying the members and number of peptide clusters.
The following modifications of the mathematical analysis, described in detail in reference 44, were made: unweighted, rather than weighted, averages were calculated; a bottom-up hierarchical clustering procedure was used; and the number of clusters was identified with a modification of Hartigan’s algorithm (22). This algorithm is based on an F statistic which compares within-cluster variability to between-cluster variability. The modification consists of the sequential use of this F statistic to increase the number of clusters until the sequence of F statistics fails to exceed a critical threshold. For a discussion of the use of such algorithms, see reference 35.
RESULTS
Reactivity of 21 anti-V3 MAbs with 56 V3 peptides from group M (clades A through H) and group O.
Twenty-one human anti-V3 MAbs were reacted with 56 V3 peptides representing sequences derived from viruses in groups M (clades A through H) and O. The results of the ELISA reactions between these 1,176 MAb and peptide pairs are shown in Fig. 1 and are expressed as the log (ODexperimental MAb + peptide/ODcontrol MAb + peptide); for clarity, the LSR values have been replaced by a spectrum of colors corresponding to the degree of reactivity observed. Each row shows the reactivity of a single MAb with the 56 peptides tested. The first row shows data acquired with the anti-V3E MAb 1324E (16). The rest of the rows show data acquired with anti-V3 MAbs derived from the cells of clade B-infected subjects and selected with clade B reagents. Each column of Fig. 1 shows the reactivity of a single peptide with the 21 MAbs tested. The peptides are grouped both according to clade, indicated by the last letter of the peptide designation, and in decreasing order of reactivity. Each “cell” in the figure represents the reactivity of the particular MAb and peptide combination tested. The results shown in Fig. 1 indicate several features, as follows.
(i) Anti-V3 MAbs display exceptionally broad cross-clade reactivity. Indeed, cross-clade reactivity is the rule rather than the exception. Every MAb reacted with peptides from more than one clade, and only peptides from group O failed to react significantly with any of the MAbs. The lack of reactivity of the group O V3 peptides is attributable to their extreme divergence in sequence from the V3 region of group M viruses and is consistent with previously published results describing the serologic features that distinguish groups M and O (34, 39, 45).
(ii) The anti-V3E MAb 1324E was able to distinguish certain peptides that the subtype B-derived MAbs could not. For example, MAb 1324E reacted with peptides from clade E (among others) but reacted weakly with peptides from clade B. In contrast, the anti-V3 MAbs derived from clade B-infected individuals reacted with peptides from clade B (among others) but failed to react with peptides from clade E. In addition, MAb 1324E distinguished between clade B and clade F peptides, whereas anti-V3B MAbs did not.
(iii) The most extensive reactivities of the anti-V3B MAbs are with peptides from clades A, B, and F.
(iv) Peptides from a single clade display divergent reactivities. For example, about half of the clade B peptides react well with the 20 anti-V3B MAbs while the others react poorly. This suggests that immunologic polymorphism within individual clades exists, as has previously been suggested by Morgado et al. (38).
These results correct the widely accepted misconception, derived from early studies of limited numbers of clade B lab strains, that V3 antibodies are type specific (21, 46). The data presented in Fig. 1, as well as previously published results which were derived from immunochemical and functional studies showing broad cross-reactivity of anti-V3 MAbs within clade B and between the various clades of HIV (9, 13, 14, 16, 17, 20, 23, 55), demonstrate that anti-V3 antibodies are broadly cross-reactive rather than being type specific.
To ascertain what other reactivity patterns that are difficult to observe in this particular two-dimensional representation of the data exist in the data set, the data matrix in Fig. 1 was subjected to the mathematical analyses described in Materials and Methods. The analyses were performed by comparing profiles of immunoreactivity (i) to detect clusters of related MAbs, i.e., MAbs within this set of anti-V3 reagents which have similar patterns of reactivity, and (ii) to detect immunologically defined clusters of related peptides, i.e., peptides with similar patterns of immunoreactivity. Each pattern of reactivity can be seen by scanning each row (for each MAb profile) or each column (for each peptide profile). MAbs with similar profiles formed individual MAb clusters; peptides with similar profiles formed individual peptide clusters.
Cluster analysis of the MAbs.
If the mathematical techniques used in this study were appropriate, there should be a concordance between the MAb groupings that were defined mathematically on the basis of binding activity and the groupings that were defined by MAb specificity. Therefore, MAb clusters were identified mathematically and subsequently examined with respect to the peptide or protein used to select each MAb and the epitopes for which the MAbs within each cluster were specific.
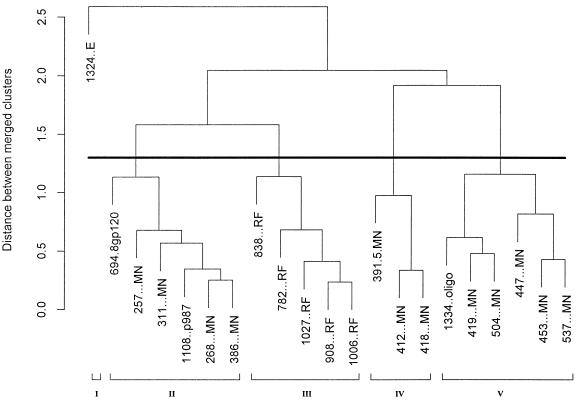
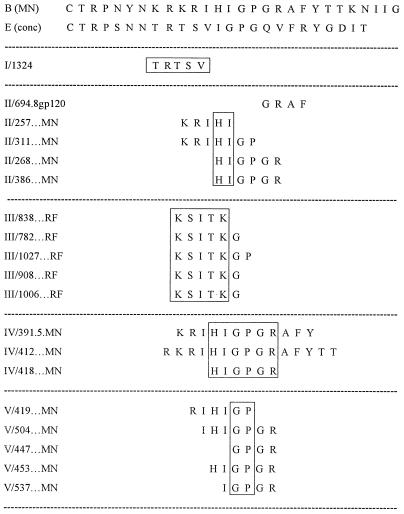
The results of the mathematical clustering of the MAbs are displayed as a dendrogram in Fig. 2. The suffix of each MAb listed in Fig. 2 identifies the peptide or protein with which the MAb was selected, as follows: MAbs 694.8 and 1334 were selected for reactivities with gp120IIIB and oligomeric gp140451, respectively (14, 18); MAbs designated with the RF suffix were selected for reactivity with V3RF (17); MAbs shown with the MN suffix were selected for reactivity with V3MN (14, 19, 20); and MAb 1108 was selected with peptide 987, a mimeotope for the anti-V3 MAb 447 (26). Nineteen of the 21 MAbs that were tested had been characterized with respect to their core epitopes (16, 17, 19, 20); these are shown in Fig. 3, where the boxes enclose the amino acids within the core epitopes of the MAbs in each cluster which are shared by all or most of the MAbs in that cluster. Without the information from the cluster analysis, most of the MAbs selected on V3MN could not have been separated into three distinct groups (clusters II, IV, and V), since many of the core epitopes of MAbs in these clusters display considerable overlap. The members of these MAb clusters do not, however, simply recognize the limited primary sequences shown in the boxed areas of Fig. 3, as the antibody combining sites recognize more than these few core amino acids. Thus, while the MAbs generally reacted with peptides containing their respective core epitopes, reactivity was often noted when the core epitope was not present and was occasionally lacking when the core epitope was present. For example, MAb 447 reacted with 14 of 16 peptides containing its core epitope, GPGR; it also reacted with many peptides lacking this sequence (for instance, those containing GPGQ), and it failed to react in 2 of 16 instances with GPGR-containing peptides. This phenomenon has been noted previously (7, 43) and underscores the critical role played in antibody recognition by conformation-conferred shapes, even within so-called linear epitopes. Similarly, it can be seen that some MAbs with identical core epitopes, e.g., MAbs 268 and 453, fall into different clusters. This, too, suggests that the core epitope only partially describes each MAb’s reactivity and that interactions between the MAb and regions outside of the core epitope contribute to the binding energy and the reactivity pattern of each MAb. (The role of sequence variation in the V3 loop, its impact on reactivity with the MAbs, and how the structures of the V3 epitopes vary independent of clade are discussed below.)
FIG. 2.
Dendrogram of the 21 anti-V3 MAbs showing the immunologic relationships defined by their reactivities with 50 V3 peptides. The dark horizontal line denotes the point in the tree at which Hartigan’s algorithm delineates the most appropriate number of clusters—five in this case. Each cluster is numbered at the bottom of the figure. The vertical axis shows the distance between merged clusters measured in mean-squared LSR units.
FIG. 3.
Core epitopes of anti-V3 MAbs grouped by mathematically defined clusters. The core epitopes of 19 of the 21 MAbs used in these studies have been identified (16, 17, 20). These MAbs are shown grouped according to the MAb cluster into which they fall; this is noted as the roman numeral preceding each MAb designation and is based on the analysis shown in Fig. 2. The core epitope for each MAb is shown. For orientation, the sequences of the V3 loops of the consensus sequences of V3MN [B (MN)] and of V3E [E (conc)] are presented. The boxes denote the amino acids within the core epitopes of the MAbs in each cluster which are shared by all or most of the MAbs in that cluster.
The interrelationships of the MAbs are displayed as a dendrogram in Fig. 2, and by using Hartigan’s algorithm, the number of MAb clusters was identified as five. It can readily be seen that the single MAb derived from the cells of a clade E-infected individual and selected with the V3E consensus peptide (MAb 1324E) forms a distinct cluster (cluster I in Fig. 2 and 3). The five MAbs selected with V3RF, which recognize an epitope to the left of the tip of the V3 loop, also form their own cluster (cluster III in Fig. 2 and 3). As can be seen from the dendrogram in Fig. 2, the MAbs in cluster III are joined by a short branch to the MAbs of cluster II. Thus, cluster II is more closely related to cluster III than it is to cluster I, IV, or V. The amino acids with which most MAbs in clusters II and III react are immediately to the left of the tip of the V3 loop, as shown in Fig. 3. Similarly, clusters IV and V are most closely related (Fig. 2), and the amino acids with which the MAbs in clusters IV and V react are at the tip of the loop: cluster IV MAbs react with a broad region encompassing the entire crown of the loop, whereas cluster V MAbs react with a narrower band of amino acids at the tip of the loop (Fig. 3).
Cluster analysis of the peptides.
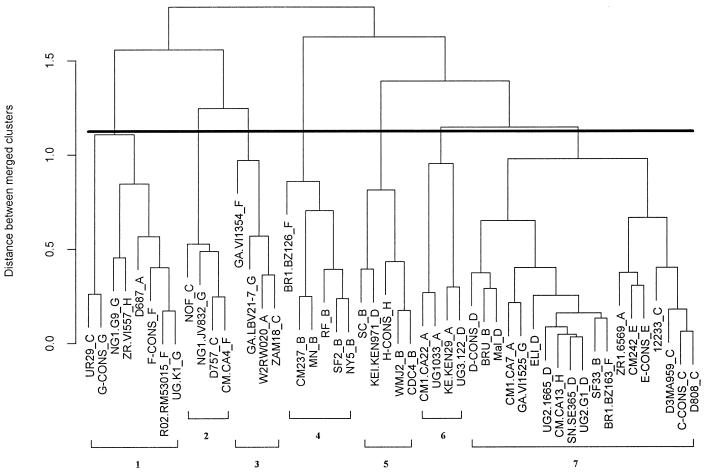
The correlation of mathematically defined MAb clusters with their immunochemical characteristics supports the use of the mathematical approach used to identify biologically relevant clusters within the data matrix shown in Fig. 1. Consequently, this matrix was also analyzed for the presence of immunologically defined clusters of peptides. Only the 50 peptides from group M viruses (clades A through H) were included in the analysis, since the group O peptides reacted poorly or not at all with the 21 MAbs studied here. Figure 4 shows the dendrogram derived from the mathematical analysis conducted to seek clusters among the 50 peptides. To identify the number of peptide clusters, a modified version of Hartigan’s algorithm was again used (22). The exact number of clusters is extremely sensitive to minor variations in the data; however, the breakpoint in the curve generated by the algorithm (data not shown) suggests that seven clusters exist. While this number may change as new MAbs are added to the panel, examination of the seven clusters, as defined here, revealed that each cluster contains peptides from more than one clade, a finding that is consistent with earlier work showing that HIV serotypes identified based on the reactivity of V3 peptides with polyclonal HIV-positive sera do not correlate with clades (4).
FIG. 4.
Dendrogram of 50 peptides showing their immunologic relationships defined by reactivities with the 21 anti-V3 MAbs. The dark horizontal line denotes the point in the tree at which Hartigan’s algorithm delineates the most appropriate number of clusters—seven in this case. Each cluster is numbered at the bottom of the figure. The vertical axis shows the distance between merged clusters measured in mean-squared LSR units. The clade of the virus from which each peptide was derived is shown as the last letter in each peptide designation.
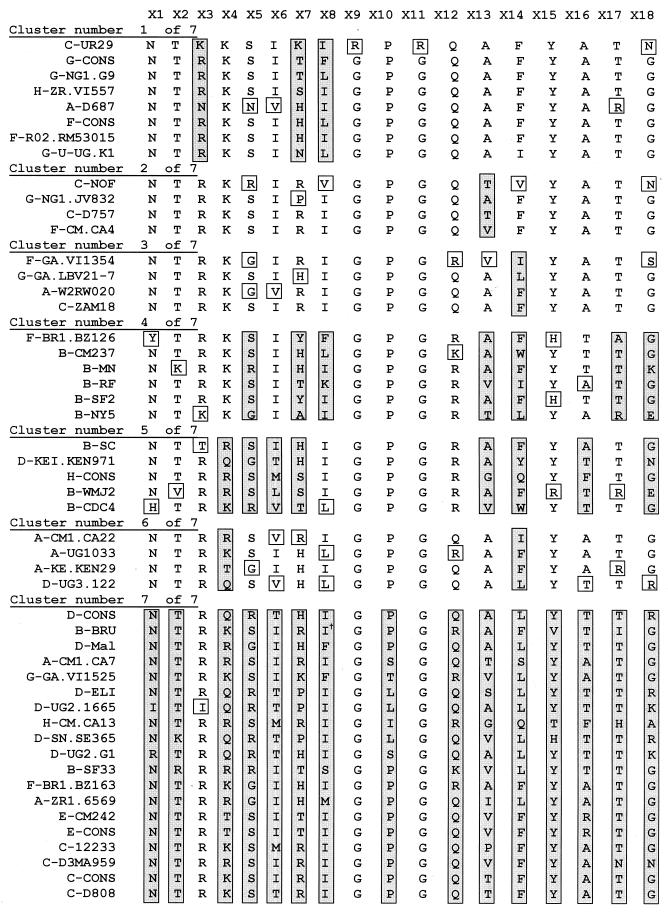
The amino acid sequences of the 50 V3 peptides, grouped by clusters, are shown in Fig. 5. The peptides studied all overlapped at 19 positions. The aligned sequences for the peptides at 18 of these positions are shown in Fig. 5. The amino acids immediately preceding the position designated as X1 in Fig. 5 are not shown although they were present in all peptides; this is because, in several cases, the appropriate amino acid from the virus sequence was replaced with a cysteine by the manufacturer. The degree of amino acid variation at each position within each cluster is noted, and among the peptides within each individual cluster studied, most amino acids are invariant or are replaced by only a single alternative amino acid (depicted in an open box). Those residues shown in shaded boxes reflect amino acids at positions in which maximal variation occurs within that cluster. For example, cluster 7 (Fig. 4 and 5) shows the maximum variation, with 15 of 18 positions containing multiple amino acids. In fact, this cluster is the least reactive cluster (see below), and the members of this cluster will probably be reclassified into additional clusters when additional MAbs become available to study their reactivities. In contrast, the sequences of amino acids in peptides that belong to each of the other six clusters display distinct patterns. For example, clusters 2 and 3 show considerable stability, with only a single position out of 18 displaying heterogeneity. These two clusters differ from one another in the single position in each which is heterogeneous: position 13 in cluster 2 and position 14 in cluster 3. Similarly, clusters 6 and 1 are quite stable, with 2 and 3 of the 18 positions being heterogeneous, respectively; in cluster 6, the variation appears at positions 4 and 14, while in cluster 1, the variation occurs at positions 3, 7, and 8. Finally, clusters 4 and 5 are heterogeneous at 7 and 8 of the 18 positions, respectively.
FIG. 5.
Sequences at 18 positions in V3 of 50 peptides classified into mathematically defined clusters. The first letter of each peptide name designates the clade of the virus from which that peptide was derived. The symbol † near the second isoleucine (I) in the sequence of B-BRU (cluster 7) shows the position of a QR insert. Open and shaded boxes are defined in the text.
Inspection of the sequences of the 50 peptides listed in Fig. 5 shows that those peptides which are most similar in structure often belong to the same peptide cluster. For example, peptides F-CONS and F-R02.RM53015 differ from one another at only one position (position 8, where the amino acids L and I, respectively, are found). Not surprisingly, these two peptides react quite similarly with the 21 MAbs (Fig. 1); this yields similar profiles of activity, which, by definition, results in these peptides clustering together. However, nonconservative changes at particularly important positions, such as position 7 (54), change peptide reactivities sufficiently to alter the cluster into which each peptide falls (e.g., compare peptide H-ZR.VI557 [cluster 1] to C-ZAM18 [cluster 3]).
Peptides in all of the clusters have a net positive charge. The only notable distinction between the clusters due to charged amino acids is found in clusters 4 and 5, in which arginine predominates at position 12, located at the tip of the V3 loop, whereas in all other peptide clusters glutamine predominates at this position.
Depiction of the reactivities of the members of each MAb cluster with the members of each peptide cluster.
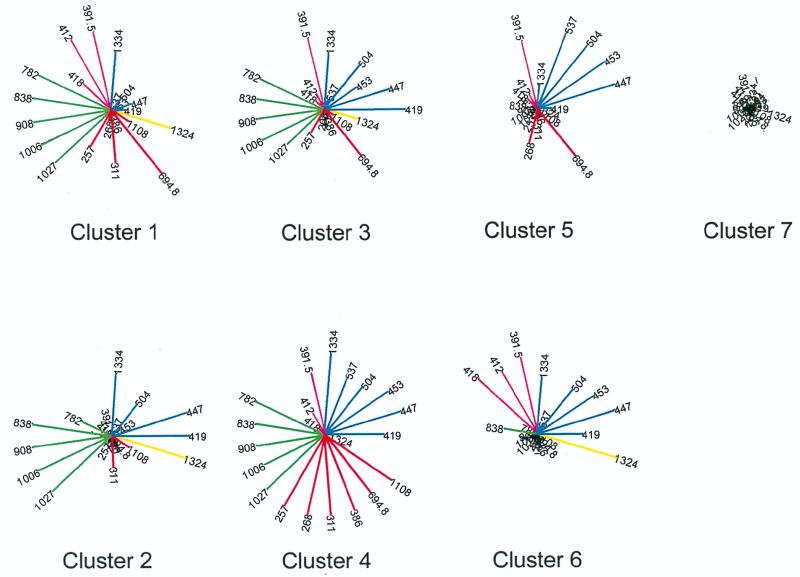
To more easily visualize the relationships between the MAb clusters and the peptide clusters, the median of the log specific reactivity of all peptides in each of the seven peptide clusters with each MAb is shown in star plots in Fig. 6. Each spoke represents the median LSR of the peptides in the designated cluster with each MAb. The length of each line is proportional to the LSR, and the color of each spoke corresponds to the MAb cluster to which the MAb belongs. This graphic representation shows clearly that cluster 7 peptides were the least reactive while the peptides of cluster 4 reacted with the maximum number of MAbs. Each peptide cluster displays a characteristic and distinct pattern of reactivity. For example, MAb 1324, from the clade E-infected patient, reacts well with peptides of clusters 1, 2, 3, and 6 but not with peptides of cluster 4, 5, or 7. Similarly, the members of MAb cluster III (selected with V3RF) react best with peptides of clusters 1 through 4 but poorly or not at all with members of peptide clusters 5, 6, and 7. The distinct patterns suggest that immunologically related families of HIV exist and can be identified on the basis of their reactivities with monoclonal reagents.
FIG. 6.
Star plots of the median LSRs of members of each peptide cluster with each of the 21 MAbs tested. Each of the seven peptide clusters (clusters 1 through 7) is shown individually. The length of each spoke represents the median reactivity of all of the peptides in that cluster with the designated MAb. The cluster to which each MAb belongs is denoted by color: yellow for MAb cluster I, red for cluster II, green for cluster III, pink for cluster IV, and blue for cluster V.
DISCUSSION
In order to establish a method for identifying groups of HIVs based on antigenic relatedness (designated here as immunotypes), the immunochemical cross-reactivity of the highly variable V3 region of HIV gp120 was studied by using 21 human anti-V3 MAbs and 56 V3 peptides derived from the V3 sequences of isolates from groups M (clades A through H) and O. The immunochemical data were analyzed by a multivariate statistical approach in order to identify immunotypes. To determine the validity of this mathematical approach, the characteristics of the five mathematically identified groups into which the 21 MAbs were divided were studied in light of their known immunochemical properties (16, 17, 20). The five mathematically defined groups of MAbs each displayed distinct immunologic characteristics based on their specificities for distinct regions within V3 (Fig. 3).
With the general concordance between the mathematical and immunochemical groupings of the MAbs providing a validation of the mathematical method, we approached our primary objective: determining whether immunologically related clusters of V3 peptides could be identified and, if so, what characterized the members of each cluster. The analysis disclosed that the 50 peptides from group M (clades A through H) studied could be divided into approximately seven peptide clusters (Fig. 4). Further subdivision, especially of the least reactive cluster (cluster 7), can be anticipated when additional MAbs with different specificities within V3 become available and are included in the analysis. Nonetheless, each of the seven clusters now defined contains peptides from more than one clade, a finding which is consistent with previously published data revealing the independence of genotypic and immunologic classifications (29, 36, 44, 47, 53). Further analysis of the amino acid sequences of the peptides within each of the seven peptide clusters revealed that each mathematically defined cluster bore a unique signature sequence (Fig. 5). Thus, the method of multivariate analysis of the immunochemical reactivities of the peptides with the MAbs was validated by identifying unique patterns within the primary structures of the peptides in each cluster.
Various mathematical methods have been used previously to identify relatedness among diverse molecules and organisms. Such analyses have been used to identify major histocompatibility complex class II antigens (5), dopamine, norepinephrine, and serotonin receptors in the brain (31), distinct categories of rhinoviruses (2), and serotypes of polio (10a). Several forms of mathematical cluster analysis have also been used to analyze data for HIV in attempts to identify HIV immunotypes (4, 29, 44); these studies are revealing because viral antigens were analyzed with polyclonal serum antibodies from subjects infected with one of several HIV clades. However, none of these studies used the combination of MAbs, viral antigens from multiple clades, and multivariate statistical methods that was used here.
The definition and description of HIV immunotypes have been slow to develop. Data showing that human monoclonal and polyclonal antibodies display broad cross-clade immunochemical reactivity with viral peptides and proteins provided evidence that immunotypes might exist (3, 8, 34, 36). Similar conclusions emerged from the description of functionally active antibodies that display cross-clade neutralization of primary isolates (29, 36, 44, 45, 53), and data presented by Mascola et al. suggest that clade E viruses constitute an immunologically distinct group within group M (33). Generally, the patterns of cross-clade reactivity were scrutinized to determine if sera from patients infected with a virus from one clade preferentially reacted with peptides, proteins, or virions from a virus from that same clade. The consensus derived from these studies, as well as the study described above, is clear: in general, patterns of immunologic reactivity do not correspond to genotypic classifications. More complex analyses of the data matrices were performed in only three of these previously published studies and on the data presented above. In analyses performed by Kostrikis et al., immunotypes were not delineated from studies of primary isolate neutralization with polyclonal sera; this was either because of the type of mathematical analysis performed or because of the noniterative nature of their data (29). However, Nyambi et al. (44), using primary isolates and HIV-positive sera, identified eight “neutralization clusters.” Plantier et al. identified five to nine serologic groups based on data from ELISAs performed with human HIV-positive sera and V3 peptides (47), while the study presented above, unique in its use of MAbs to identify immunotypes, defined approximately seven immunotypes. Thus, in three of four studies, immunologically defined groups have been identified. The fact that fewer than 10 such groups were found in each of these studies suggests that the number of existing immunotypes may be rather small.
The need to identify HIV immunotypes is predicated on the fact that vaccines against immunologically diverse organisms are composed of components which are representative of the various serotypes that make up a particular family of organisms. Vaccines for polio, influenza, and Streptococcus pneumoniae serve as examples of this precept. HIV is also an immunologically diverse group of organisms, and it is therefore unlikely that any single form of virus or product from any single form of HIV will induce immunity to all forms of HIV. As with the vaccines against other immunologically diverse organisms, such as those mentioned above, a polyvalent HIV vaccine will undoubtedly be required. It is not known, however, how many or which HIVs would be best to incorporate into a vaccine, even one intended for use in only a limited geographic area. Moreover, it may not be necessary to incorporate representatives of all HIV immunotypes into a vaccine, even one designed for global use, if other precedents apply (such as the S. pneumoniae vaccine, in which not all serotypes of the organism are used).
Polyvalent HIV vaccines are now being designed. Such vaccines could be constructed by incorporating viruses from different HIV genotypes. Indeed, initial tests of this approach are being conducted with a candidate vaccine incorporating gp120 molecules from clade B and E viruses (1). It appears that the viruses upon which this bivalent vaccine is based might represent two different immunotypes (33). Generally, however, HIV genotypes do not correspond to HIV immunotypes, and therefore the choice of vaccine components based solely on genetically defined clades may be inappropriate.
Choosing representatives of the various immunotypes to incorporate into a polyvalent vaccine is, as noted above, an approach which has proven useful for prevention of other diseases. However, the seven immunotypes found in this study may eventually differ both in number and in nature from clusters defined immunochemically with sera or with additional MAbs derived from subjects infected with other clades. Similarly, the number and nature of clusters may eventually differ if other envelope epitopes are examined immunochemically, if analyses are performed with gp120 molecules or intact virions, or if results based on antibody function, such as neutralization, are used for analysis in place of immunochemical reactivity. Indeed, it is probable that epitopes other than V3, and functional assays, rather than immunochemical assays, will provide better targets and tools for the definition of relevant immunotypes. The work described above establishes methods for identifying these potentially more-relevant immunotypes and demonstrates that immunotypes can be defined even for highly variable regions of the HIV envelope. Indeed, in ongoing work in our laboratory, several additional epitopes and antibody-mediated functions are being investigated to define immunotypes (see reference 43) and comparisons between the immunotypes identified by these different techniques will be performed to determine which are most relevant to protective immune responses. These ongoing studies, utilizing the analytic techniques described above, should identify relevant HIV immunotypes. In the meantime, since these and other studies (12, 16, 17, 36, 44, 53) have established the ability of the human immune system to produce extensive cross-clade immunity, there is every reason to believe that an appropriately constructed and delivered polyvalent HIV vaccine will induce a broad protective response. To construct such a vaccine, it is critical to understand, as completely as possible, the antigenic structure of HIV, to establish and identify immunologic classification for HIV, and to choose rationally among the HIV immunotypes the minimum number and types of viruses that will induce the broadest protective responses.
ACKNOWLEDGMENTS
We thank Lawrence Loomis-Price of the H. M. Jackson Foundation for the synthesis of the majority of the V3 peptides and Dale Lawrence of the Division of AIDS at NIH for many valuable, in-depth discussions concerning the current and past uses of cluster analysis in the study of epitopes.
This work was supported by NIH grants AI 32424, AI 07382, AI 36085, and AI 44302, by the VA Research Center for AIDS and HIV Infection, and by research funds from the Department of Veterans Affairs.
REFERENCES
- 1.Altman, L. K. 4 June 1998. FDA authorizes first full testing for HIV vaccine, p. A1. New York Times, New York, N.Y. [PubMed]
- 2.Andries K, Dewindt B, Snoeks J, Wouters L, Moereels J, Lewi P J, Janssen P A J. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogencity. J Virol. 1990;64:1117–1123. doi: 10.1128/jvi.64.3.1117-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baillou A, Brand D, Denis F, M’Boup S, Chout R, Goudeau A, Barin F. High antigenic cross-reactivity of the V3 consensus sequences of HIV-1 gp120. AIDS Res Hum Retroviruses. 1993;9:1209–1215. doi: 10.1089/aid.1993.9.1209. [DOI] [PubMed] [Google Scholar]
- 4.Barin F, Lahbabi Y, Buzelay L, LeJeune B, Baillou-Beaufils A, Denis F, Mathiot C, M’Boup S, Vithayasai V, Dietrich U, Goudeau A. Diversity of antibody binding to V3 peptides representing consensus sequences of HIV type 1 genotypes A to E: an approach for HIV type 1 serological subtyping. AIDS Res Hum Retroviruses. 1996;12:1279–1289. doi: 10.1089/aid.1996.12.1279. [DOI] [PubMed] [Google Scholar]
- 5.Bodmer J, Pickbourne P, Bodmer W, Batchelor R, Dewar P, Dick H, Entwistle C, Festenstein H, Gelsthorpe K, Joysey V, Mackintosh P, Lawler S, Morris P, Pegrum G D, Harris R, Taylor M. Serological identification of Ia antigens: report of a British region Ia workshop. Tissue Antigens. 1976;8:359–371. doi: 10.1111/j.1399-0039.1976.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 6.Burton D R, Pyati J, Koduri R, Sharp S J, Thornton G B, Parren P W H I, Sawyer L S W, Hendry R M, Dunlop N, Nara P L, Lamacchia M, Garratty E, Stiehm E R, Bryson Y J, Cao Y, Moore J P, Ho D D, Barbas C F., III Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 7.Cecilia D, KewalRamani V N, O’Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman D R, Zolla-Pazner S. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J Virol. 1998;72:6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheingsong-Popov R, Callow D, Beddows S, Shaunak S, Wasi C, Kaleebu P, Gilks C, Petrascu I V, Garaev M M, Watts D M, Constantine N T, Weber J N. Geographic diversity of human immunodeficiency virus type 1: serologic reactivity to Env epitopes and relationship to neutralization. J Infect Dis. 1992;165:256–261. doi: 10.1093/infdis/165.2.256. [DOI] [PubMed] [Google Scholar]
- 9.Conley A J, Gorny M K, Kessler II J A, Boots L J, Lineberger D, Emini E A, Ossorio M, Koenig S, Williams C, Zolla-Pazner S. Neutralization of primary HIV-1 virus isolates by the broadly-reactive anti-V3 monoclonal antibody, 447-52D. J Virol. 1994;68:6994–7000. doi: 10.1128/jvi.68.11.6994-7000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conley A J, Kessler J A, Boots L J, Tung J-S, Arnold B A, Keller P M, Shaw A R, Emini E A. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc Natl Acad Sci USA. 1994;91:3348–3352. doi: 10.1073/pnas.91.8.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Davies, D. Personal communication.
- 11.Expert Group of the Joint United Nations Programme On HIV/AIDS. HIV-1 subtypes for epidemiology, pathogenicity, vaccines, and diagnostics. AIDS. 1997;11:17–36. [PubMed] [Google Scholar]
- 12.Ferrari G, Humphrey W, McElrath M J, Excler J L, Duliege A M, Clements M L, Corey L C, Bolognesi D P, Weinhold K J. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci USA. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouts T R, Binley J M, Trkola A, Robinson J E, Moore J P. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorny M K, Conley A J, Karwowska S, Buchbinder A, Xu J-Y, Emini E A, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorny M K, Gianakakos V, Sharpe S, Zolla-Pazner S. Generation of human monoclonal antibodies to HIV. Proc Natl Acad Sci USA. 1989;86:1624–1628. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorny M K, Mascola J R, Israel Z R, VanCott T C, Williams C, Balfe P, Hioe C, Brodine S, Burda S, Zolla-Pazner S. A human monoclonal antibody specific for the V3 loop of HIV type 1 clade E cross-reacts with other HIV type 1 clades. AIDS Res Hum Retroviruses. 1998;14:213–221. doi: 10.1089/aid.1998.14.213. [DOI] [PubMed] [Google Scholar]
- 17.Gorny M K, VanCott T C, Hioe C, Israel Z R, Michael N L, Conley A J, Williams C, Kessler II J A, Chigurupati P, Burda S, Zolla-Pazner S. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and inter-clade cross-reactivity. J Immunol. 1997;159:5114–5122. [PubMed] [Google Scholar]
- 18.Gorny, M. K., T. C. VanCott, C. Williams, and S. Zolla-Pazner. Effects of oligomerization on the epitopes of HIV-1 envelope glycoproteins. Submitted for publication. [DOI] [PubMed]
- 19.Gorny M K, Xu J-Y, Gianakakos V, Karwowska S, Williams C, Sheppard H W, Hanson C V, Zolla-Pazner S. Production of site-selected neutralizing human monoclonal antibodies against the third variable domain of the HIV-1 envelope glycoprotein. Proc Natl Acad Sci USA. 1991;88:3238–3242. doi: 10.1073/pnas.88.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorny M K, Xu J-Y, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150:635–643. [PubMed] [Google Scholar]
- 21.Goudsmit J, Debouck C, Meloen R H, Smit L, Bakker M, Asher D M, Wolff A V, Gibbs C J, Jr, Carleton Gajdusek D. Human immunodeficiency virus type 1 neutralization epitope with conserved architecture elicits early type-specific antibodies in experimentally infected chimpanzees. Proc Natl Acad Sci USA. 1988;85:4478–4482. doi: 10.1073/pnas.85.12.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartigan J. Clustering algorithms. J. New York, N.Y: Wiley and Sons; 1975. [Google Scholar]
- 23.Hioe C E, Xu S, Chigurupati P, Burda S, Williams C, Gorny M K, Zolla-Pazner S. Neutralization of HIV-1 primary isolates by polyclonal and monoclonal human antibodies. Int Immunol. 1997;9:1281–1290. doi: 10.1093/intimm/9.9.1281. [DOI] [PubMed] [Google Scholar]
- 24.Janssens W, Heyndrickx L, Fransen K, Mitte J, Peeters M, Nkengasong J N, Ndumbe P M, Delaporte E, Perret J-L, Atende C, Piot P, van der Groen G. Genetic and phylogenetic analysis of env subtypes G and H in Central Africa. AIDS Res Hum Retroviruses. 1994;10:877–879. doi: 10.1089/aid.1994.10.877. [DOI] [PubMed] [Google Scholar]
- 25.Janssens W, Nkengasong J N, Heyndrickx L, Fransen K, Ndumbe P M, Delaporte E, Peeters M, Perret J-L, Ndoumou A, Atende C, Piot P, van der Groen G. Further evidence of the presence of genetically aberrant HIV-1 strains in Cameroon and Gabon. AIDS. 1994;8:1012–1013. doi: 10.1097/00002030-199407000-00022. [DOI] [PubMed] [Google Scholar]
- 26.Keller P A, Arnold B A, Shaw A R, Tolman R L, Middlesworth F V, Bondy S, Rusiecki V K, Koenig S, Zolla-Pazner S, Conrad P, Emini E A, Conley A J. Identification of HIV vaccine candidate peptides by screening random phage epitope libraries. Virology. 1992;169:893–897. doi: 10.1006/viro.1993.1179. [DOI] [PubMed] [Google Scholar]
- 27.Kessler J A, II, McKenna P M, Emini E A, Chan C P, Patel M D, Gupta S K, Mark III G E, Barbas III C F, Burton D R, Conley A J. Recombinant human monoclonal antibody IgG1b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res Hum Retroviruses. 1997;13:575–582. doi: 10.1089/aid.1997.13.575. [DOI] [PubMed] [Google Scholar]
- 28.Korber B T M, MacInnes K, Smith R F, Myers G. Mutational trends in V3 loop protein sequences observed in different genetic lineages of human immunodeficiency virus type 1. J Virol. 1994;68:6730–6744. doi: 10.1128/jvi.68.10.6730-6744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kostrikis L G, Cao Y, Ngai H, Moore J P, Ho D D. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J Virol. 1996;70:445–458. doi: 10.1128/jvi.70.1.445-458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kostrikis L G, Bagdades E, Cao Y, Zhang L, Dimitriou D, Ho D D. Genetic analysis of human immunodeficiency virus type 1 strains from patients in Cyprus: identification of a new subtype designated subtype I. J Virol. 1995;69:6122–6130. doi: 10.1128/jvi.69.10.6122-6130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewi P J. Spectral mapping, a technique for classifying biological activity profiles of chemical compounds. Drug Res. 1976;26:1295–1300. [PubMed] [Google Scholar]
- 32.Louwagie J, McCutchan F E, Peeters M, Brennan T P, Sanders-Buell E, Eddy G A, van der Groen G, Fransen K, Gershy-Damet G-M, Deleys R, Burke D S. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence for multiple genotypes. AIDS. 1993;7:769–780. doi: 10.1097/00002030-199306000-00003. [DOI] [PubMed] [Google Scholar]
- 33.Mascola J R, Louwagie J, McCutchan F E, Fischer C L, Hegerich P A, Wagner K F, Fowler A K, McNeil J G, Burke D S. Two antigenically distinct subtypes of HIV-1: viral genotype predicts neutralization immunotype. J Infect Dis. 1994;169:48–54. doi: 10.1093/infdis/169.1.48. [DOI] [PubMed] [Google Scholar]
- 34.Mauclere P, Damond F, Apetrel C, Loussert-Ajaka I, Souquiere S, Buzelay L, Dalbon P, Jolivet M, Lobe M M, Brun-Vezinet F, Simon F, Barin F. Synthetic peptide ELISAs for detection of and discrimination between group M and group O HIV type 1 infection. AIDS Res Hum Retroviruses. 1997;13:987–993. doi: 10.1089/aid.1997.13.987. [DOI] [PubMed] [Google Scholar]
- 35.Milligan G W, Cooper M C. An examination of procedures for determining the number of clusters in a data set. Psychometrika. 1985;50:159–179. [Google Scholar]
- 36.Moore J P, Cao Y, Leu J, Qin L, Korber B, Ho D D. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J Virol. 1996;70:427–444. doi: 10.1128/jvi.70.1.427-444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore J P, Trkola A, Korber B, Boots L J, Kessler II J A, McCutchan F E, Mascola J, Ho D D, Robinson J, Conley A J. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol. 1995;69:122–130. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgado M G, Sabino E C, Shpaer E G, Bongertz V, Brigido L, Guimaraes M D C, Castilho E A, Galvao-Castro B, Mullins J I, Hendry R M, Mayer A. V3 region polymorphisms in HIV-1 from Brazil: prevalence of subtype B strains divergent from North American/European prototype and detection of subtype F. AIDS Res Hum Retroviruses. 1994;10:569–576. doi: 10.1089/aid.1994.10.569. [DOI] [PubMed] [Google Scholar]
- 39.Myers G, Korber B, Foley B, Smith R F, Jeang K-T, Mellors J W, Wain-Hobson A. Human retroviruses and AIDS. Los Alamos, N.Mex: Theoretical Biology and Biophysics, Los Alamos National Laboratories; 1996. [Google Scholar]
- 40.Myers G, MacInnes K, Korber B. The emergence of simian/human immunodeficiency viruses. AIDS Res Hum Retroviruses. 1992;8:373–386. doi: 10.1089/aid.1992.8.373. [DOI] [PubMed] [Google Scholar]
- 41.Myers G, Pavlakis G N. Evolutionary potential of complex retroviruses. In: Levy J A, editor. The retroviruses. New York, N.Y: Plenum Press; 1992. pp. 51–104. [Google Scholar]
- 42.Nkengasong J N, Janssens W, Heyndrickx L, Fransen K, Ndumbe P M, Motte J, Leonaers A, Ngolle M, Ayuk J, Piot P, van der Groen G. Genetic subtypes of HIV-1 in Cameroon. AIDS. 1994;8:1405–1412. doi: 10.1097/00002030-199410000-00006. [DOI] [PubMed] [Google Scholar]
- 43.Nyambi P N, Gorny M K, Bastiani L, van der Groen G, Williams C, Zolla-Pazner S. Mapping of epitopes exposed on intact HIV-1 virions: a new strategy for studying the immunologic relatedness of HIV-1. J Virol. 1998;72:9384–9391. doi: 10.1128/jvi.72.11.9384-9391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nyambi P N, Nkengasong J, Lewi P, Andries K, Janssens W, Fransen K, Heyndrickx L, Piot P, van der Groen G. Multivariate analysis of human immunodeficiency virus type 1 neutralization data. J Virol. 1996;70:6235–6243. doi: 10.1128/jvi.70.9.6235-6243.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nyambi P N, Nkengasong J, Peeters M, Simon F, Eberle J, Janssens W, Fransen K, Willems B, Vereecken K, Heyndrickx L, Piot P, van der Groen G. Reduced capacity of antibodies from patients infected with human immunodeficiency virus type-1 (HIV-1) group O to neutralize primary isolates of HIV-1 group M viruses. J Infect Dis. 1995;172:1228–1237. doi: 10.1093/infdis/172.5.1228. [DOI] [PubMed] [Google Scholar]
- 46.Palker T J, Clark M E, Langlois A L, Matthews T J, Weinhold K J, Randall R R, Bolognesi D P, Haynes G. Type-specific neutralization of HIV with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci USA. 1988;85:1758–1762. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plantier J-C, Le Pogam S, Poisson F, Buzelay L, Lejeune B, Barin F. Extent of antigenic diversity in the V3 region of the surface glycoprotein, gp120, of human immunodeficiency virus type 1 group M and consequences for serotyping. J Virol. 1998;72:677–683. doi: 10.1128/jvi.72.1.677-683.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starcich B R, Hahn B H, Shaw G M, McNeely P D, Modrow S, Wolf H, Parks E S, Parks W P, Josephs S F, Gallo R C, Wong-Staal F. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 49.Trkola A, Pomales A B, Yuan H, Korber B, Maddon P J, Allaway G P, Katinger H, Barbas III C F, Burton D R, Ho D D, Moore J P. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.VanCott T C, Bethke F R, Burke D S, Redfield R R, Birx D L. Lack of induction of antibodies specific for conserved, discontinuous epitopes of HIV-1 envelope glycoprotein by candidate AIDS vaccines. J Immunol. 1995;155:4100–4110. [PubMed] [Google Scholar]
- 52.VanCott T C, Veit S C D, Kalyanaraman V, Earl P, Birx D L. Characterization of a soluble oligomeric HIV-1 gp160 protein as a potential immunogen. J Immunol Methods. 1995;183:103–117. doi: 10.1016/0022-1759(95)00038-c. [DOI] [PubMed] [Google Scholar]
- 53.Weber J, Fenyö E-M, Beddows S, Kaleebu P, Björndal Å the WHO Network for HIV Isolation and Characterization. Neutralization serotypes of human immunodeficiency virus type 1 field isolates are not predicted by genetic subtype. J Virol. 1996;70:7827–7832. doi: 10.1128/jvi.70.11.7827-7832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolfs T F W, Szart G, Bakker M, Valk M, Kuiken C L, Goudsmit J. Naturally occurring mutations within HIV-1 V3 genomic RNA lead to antigenic variation dependent on a single amino acid substitution. Virology. 1992;185:195–205. doi: 10.1016/0042-6822(91)90767-6. [DOI] [PubMed] [Google Scholar]
- 55.Zolla-Pazner S, O’Leary J, Burda S, Gorny M K, Kim M, Mascola J, McCutchan F. Serotyping of primary human immunodeficiency virus type 1 isolates from diverse geographic locations by flow cytometry. J Virol. 1995;69:3807–3815. doi: 10.1128/jvi.69.6.3807-3815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]