Abstract
Hepatitis E virus (HEV) is the etiological agent for viral hepatitis type E, which is a major problem in the developing world. Because HEV cannot be cultured in vitro, very little information exists on the mechanisms of HEV gene expression and genome replication. HEV is a positive-strand RNA virus with three potential open reading frames (ORFs), one of which (ORF2) is postulated to encode the major viral capsid protein (pORF2). We earlier showed (S. Jameel, M. Zafrullah, M. H. Ozdener, and S. K. Panda, J. Virol. 70:207–216, 1996) pORF2 to be a ∼88-kDa glycoprotein, carrying N-linked glycans and a potential endoplasmic reticulum (ER)-directing signal at its N terminus. Treatment with the drugs brefeldin A and monensin suggest that the protein may accumulate within the ER. Based on mutational analysis, we demonstrate Asn-310 to be the major site of N-glycan addition. In COS-1 cell expression and in vitro translation experiments, we confirm the ER-translocating nature of the pORF2 N-terminal hydrophobic sequence and show that the protein is cotranslationally, but not posttranslationally, translocated across the ER membrane. Earlier, we had also demonstrated cell surface localization of a fraction of the COS-1 cell-expressed pORF2. Using glycosylation- and translocation-defective mutants of pORF2, we now show that while transit of pORF2 into the ER is necessary for its cell surface expression, glycosylation of the protein is not required for such localization. These results may offer clues to the mechanisms of gene expression and capsid assembly in HEV.
Hepatitis E virus (HEV) is a human pathogen prevalent in much of the developing world, where it causes sporadic and epidemic forms of acute viral hepatitis (3, 11, 26, 41). The disease has also been observed in parts of the world where hepatitis is not endemic (43), mostly in relation to travel to areas of endemicity but rarely without being associated with another known risk factor (32). The transmission of HEV is feco-oral, with only human-to-human transfer recognized so far (14). However, the recent discovery of a novel virus closely related to HEV in domestic swine (16) suggests possible zoonotic reservoirs as well.
The viral genome has been cloned from multiple geographically distinct isolates and shows a high degree of sequence conservation (1, 2, 8, 32, 36, 40). Based on sequence homology and genome organization, HEV was earlier classified as a hepevirus, a new genus in the family Calciviridae (17). It has also been suggested that HEV may be a nonenveloped alpha-like virus closely related to the rubella virus (13, 27). HEV is now classified, on an interim basis, in a separate family, the hepatitis E-like viruses (9). Phylogenetic grouping suggests that at least three closely related variants of HEV have been identified. These include the US-1 strain (32) and the swine HEV (16), the most variant Mexican strain (8), and another group comprising the highly homologous isolates from Burma (36), China (1, 2), India (24), and Pakistan (39).
The ∼7.5-kb positive-sense RNA genome contains three open reading frames (ORFs), designated ORF1, ORF2, and ORF3 (36). Though no experimental data are available, based on homology to other positive-sense RNA viruses, ORF1 is postulated to encode the HEV nonstructural polyprotein (13). Whether this polyprotein is processed is unknown, but it carries domains found in viral methyltransferases, papain-like cysteine proteases, RNA helicases, and RNA-dependent RNA polymerases (13). The remaining ORFs are found in the structural portion of the genome, with ORF2 postulated to encode the viral capsid protein (pORF2) and ORF3 postulated to code for a small protein (pORF3) whose function is as yet undefined (36).
In the absence of an in vitro culture system, little information is available on the mechanisms of HEV genome replication, protein expression, and processing. We have used a subgenomic expression strategy to study the properties and interactions of the HEV ORF2 and ORF3 proteins (10, 44). pORF3 expressed in animal cells was found to be a cytoskeleton-associated phosphoprotein which appears to be phosphorylated by the cellular mitogen-activated protein kinase. Similar expression analysis of pORF2 showed it to be an 88-kDa glycoprotein which is expressed intracellularly as well as on the cell surface and has the potential to form homodimers (10). Based on pulse-chase analysis, tunicamycin inhibition, and endoglycosidase sensitivity, we suggested that pORF2 is cotranslationally translocated via its N-terminal signal sequence into the endoplasmic reticulum (ER) where the signal is processed. We further suggested that the protein may be glycosylated in the ER at asparagine residues in one or more of the three possible N-linked glycosylation sites showing the Asn-X-Ser/Thr (N-X-S/T) sequence (10).
Using mutational analysis, we now conclusively demonstrate the presence of an ER-translocating signal sequence at the N terminus of pORF2. We also show that while all three asparagine residues in the N-X-S/T consensus are modified, Asn-310 is the major site of glycosylation. In vivo and in vitro assays suggest that pORF2 is cotranslationally translocated across the ER membrane. Localization studies suggest that while glycosylation of pORF2 is the result of its transit through the ER, it has no bearing on the cell surface expression of pORF2. These results and their implications for HEV capsid assembly are discussed.
MATERIALS AND METHODS
Vectors and mutagenesis.
The expression vector pSG-ORF2 has been described earlier (10). The ORF2[Δ2-34] mutant was generated by using an oligonucleotide reconstruction approach. Plasmid pSG-ORF2 was digested with BstEII and BamHI to obtain an ORF2 fragment lacking 127 bp at its 5′ end (A base in ATG start codon considered as position 1). The oligonucleotides used for reconstruction were 35F, CTAGATGGGCGGTTCCGGCGGTGGTTTCTGGG and 35R, GTCACCCCAGAAACCACCGCCGGAACCGCCTAC.
The annealed double-stranded oligonucleotide carried XbaI and BstEII sites at its 5′ and 3′ ends, respectively, and reconstructed ORF2 without the coding sequences for amino acids 2 to 34. The mutant gene was cloned in XbaI- and BamHI-digested plasmid pBSK(+) (Stratagene, GmbH) and verified by dideoxy sequencing. Subsequently, ORF2[Δ2-34] was cloned as a NotI-XhoI fragment in plasmid pMT3 (35) and from there as a PstI-BamHI fragment in plasmid pSGI (10). This resulted in plasmid pSG-ORF2[Δ2-34].
For site-directed mutagenesis, HEV ORF2 was cloned in plasmid pALTER-1 (Promega, Madison, Wis.) as a PstI-BamHI fragment, and mutagenesis was carried out by published methods (42). The oligonucleotides used for mutagenesis were as follows, with lowercase letters indicating changed bases for the Asn→Ala mutation at amino acid position 137, 310, or 562 in the ORF2 protein: 2-137, GATGTTGATAGGgcGTACTGCCG; 2-310, GGGGGTAAGGgcGCGAAACTCA; and 2-562, TGCAGTGGTGgcGTAATTATAAGG. All single-site mutants were verified by dideoxy sequencing by using primers specific for the ORF2 sequence. The dual- and triple-site mutants were reconstructed in the pALTER-1 background by using convenient restriction sites. The ORF2[137,310] mutant was generated by cloning a 1.3-kb EcoRI fragment from plasmid pALT-ORF2[310] into EcoRI-digested pALT-ORF2[137]. The other mutants were generated by cloning a 1-kb SpeI-BamHI fragment from plasmid pALT-ORF2[562] into SpeI-BamHI-digested pALT-ORF2[137] to yield ORF2[137,562], by cloning the fragment into SpeI-BamHI-digested pALT-ORF2[310] to yield ORF2[310,562], and by cloning the fragment into SpeI-BamHI-digested pALT-ORF2[137,310] to yield the triple mutant ORF2[137,310,562]. All the mutant ORF2 genes were moved as PstI-BamHI fragments into PstI-BamHI-digested expression vector pSGI.
Transfection and labeling of cultured cells.
COS-1 cells were maintained and transfected with lipofectin (GIBCO-BRL) as described earlier (10). Metabolic labeling of cells, treatment of cells with tunicamycin, preparation and immunoprecipitation of cell lysates, and endoglycosidase H digestions were also carried out as described earlier (10).
Treatment of cells with brefeldin A (BfA) and monensin were carried out essentially as described elsewhere (28). Briefly, 40 h posttransfection, cells were incubated in Dulbecco’s modified Eagle medium (DMEM) lacking cysteine and methionine for 1 h, followed by labeling each 60-mm-diameter plate of cells with 200 μCi of [35S]-Promix (Amersham)/ml for 30 min. After removal of the label, cells were further incubated in chase medium containing excess cysteine and methionine (complete DMEM) for another 3 h. BfA and monensin, when used, were present at the appropriate concentrations during the prelabeling, labeling, and chase periods. Steady-state labeling of transfected cells was done for 4 h as described earlier (10), with BfA present during the prelabeling and labeling periods.
Preparation of microsomes.
The preparation of microsomal membranes and protease protection of translocated pORF2 was carried out essentially as described previously (25). Forty hours posttransfection, COS-1 cells were labeled with [35S]promix for 4 h. The cells were washed twice with cold Tris-buffered saline (TBS; 50 mM Tris-HCl [pH 7.5], 150 mM NaCl), incubated in 0.1× TBS (0.45 ml per 60-mm-diameter plate) for 15 min on ice, and the swollen cells were disrupted by 20 strokes of Dounce homogenization. After the debris was removed at 2,500 rpm and 4°C for 20 min, the supernatant was made to 1× TBS by using 10× TBS. One milliliter of this supernatant was overlaid on a 2.7-ml cushion of 250 mM sucrose in TBS and centrifuged at 37,000 rpm and 4°C for 30 min in a SW60 rotor (Beckman). The pellet, comprising the microsomal fraction, was resuspended in 0.1 ml of TBS containing 2 mM tetracaine and 2 mM CaCl2 (per SW60 tube or two 60-mm-diameter plate of cells). For protease protection studies, the microsomal fraction was divided into three equal aliquots and treated as follows: (i) no additions, (ii) 25 μg of trypsin/ml, and (iii) 25 μg of trypsin/ml and 0.5% Nonidet P-40 (NP-40). After incubation on ice for 60 min, the reaction mixtures were adjusted to 40 μg of aprotinin/ml and 2 mM phenylmethylsulfonyl fluoride (PMSF) and incubated on ice for another 20 min. Subsequently, all reaction mixtures were adjusted to 1× radioimmunoprecipitation assay (RIPA) buffer and immunoprecipitated with antibodies to pORF2 as described earlier (10).
In vitro translation and translocation.
In vitro transcription and translation was carried out by using a coupled transcription-translation system (TNT coupled reticulocyte lysate system; Promega) according to the manufacturer’s instructions. In a 25-μl reaction mixture, 0.5 μg of circular plasmid DNA was transcribed with T7 RNA polymerase and translated by using reticulocyte lysate and 20 μCi of either [35S]-Promix (Amersham) [35S]methionine (BRIT, Mumbai, India). Canine pancreatic membranes (Promega), when present, were used at an optimal amount determined experimentally.
To assay for translocation of in vitro-synthesized proteins across microsomal membranes, trypsin protection was carried out essentially as described above. Following protein synthesis at 30°C for 90 min, the reaction mixtures were chilled on ice, and 20 μl of the sample loaded on a 1-ml cushion of 250 mM sucrose in TBS. The tubes were centrifuged at 11,400 rpm for 30 min in a refrigerated microfuge (Biofuge 17RS; Heraeus GmbH). The supernatant was discarded, and the pellet was resuspended in 40 μl of TBS containing 2 mM tetracaine and 2 mM CaCl2. The resuspended pellet was divided into three equal aliquots and treated with trypsin and NP-40 as described above for the COS-1 cell microsomal fraction. Following proteolysis, the reaction was terminated with aprotinin and PMSF, an equal volume of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading dye was added, and the samples were boiled and subjected to SDS-PAGE.
Alternatively, protease digestion was also carried out without prior separation of the canine pancreatic membranes after the translocation period (23). Following protein synthesis at 30°C for 90 min, in the absence or presence of membranes, the reaction mixtures were quenched with 100 μg of cycloheximide/ml and 200 μM methionine and kept at 30°C for another 60 min. The reactions were then chilled on ice, adjusted to 3 mM in tetracaine and 10 mM in CaCl2, and divided into three equal aliquots. These were treated on ice for 1 h as follows: (i) no additions, (ii) 250 μg of trypsin/ml, and (iii) 250 μg of trypsin/ml and 0.7% Triton X-100. The proteolytic reaction was terminated by transferring it, with a tip dipped in 200 mM PMSF, to an equal volume of boiling 2× SDS-PAGE loading dye. The samples were boiled and subjected to SDS-PAGE.
For posttranslational translocation, in vitro-coupled transcription-translation was carried out as described above at 30°C for 90 min in the absence or presence of canine pancreatic membranes. Cycloheximide was added to 100 μg/ml, and the reaction mixtures were incubated at 30°C for 15 min. To the reaction mixtures 1 or 2 μl of canine pancreatic membranes was added and incubated at 30°C for another 30 min. Trypsin protection was then carried out with either of the two methods described above for cotranslational translocation.
Immunofluorescence.
COS-1 cells were grown on autoclaved glass coverslips (18 mm diameter) placed individually inside the wells of a 12-well tissue culture plate and transfected with the appropriate plasmids. Forty hours posttransfection, cells were washed three times with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 30 min at room temperature, and again washed with PBS as earlier. The fixed cells were then incubated for 2 h at 37°C in a humid chamber with rabbit anti-pORF2 (1:200 dilution in 0.5% bovine serum albumin [BSA]–PBS), either in the absence or presence of 0.1% saponin. The antibody was earlier preadsorbed on fixed and untransfected COS-1 cells. Following three washes of 5 min each in PBS, the cells were incubated as described above for 1 h with anti-rabbit immunoglobulin G (IgG)–fluorescein isothiocyanate (FITC) conjugate (1:200 dilution in 0.5% BSA–PBS; Sigma). The cells were washed with PBS as described above, and the coverslips were mounted on glass slides with a drop of 20% glycerol. Viewing and photography were carried out on a fluorescence microscope (Wild Leitz, GmbH).
RESULTS
Domain analysis of pORF2.
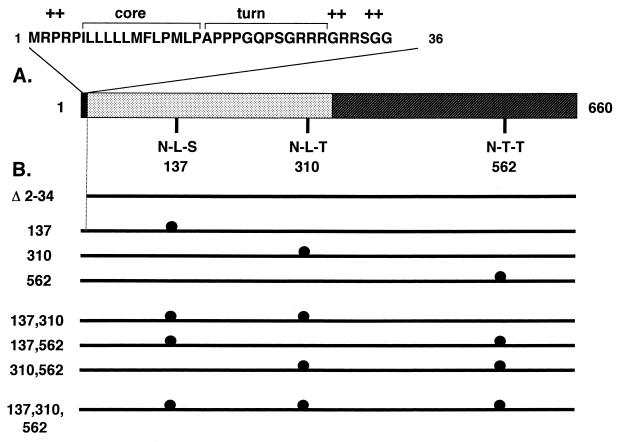
The 660-amino-acid protein encoded by ORF2 of HEV was analyzed for the presence of membrane-translocating signals and N-linked glycosylation sites. The hydropathy profile of pORF2 showed the presence of a highly hydrophobic stretch, rich in leucines, at its N terminus, followed by turn-inducing and positively charged amino acids. Such an N-terminal stretch makes up the export signal sequence generally found in proteins that are translocated across cellular membranes (31). A search for potential N-linked glycosylation sites with the consensus N-X-S/T (X, any amino acid except Pro) revealed the presence of three such sites within pORF2, Asn-137, Asn-310, and Asn-562. These features were found in the deduced amino acid sequences of ORF2 from all isolates of HEV sequenced so far and are shown in Fig. 1A. Various mutants used in this study are schematically illustrated in Fig. 1B, and the details of their construction are presented in Materials and Methods. Essentially, these included a deletion of the N-terminal putative signal sequence and Asn→Ala mutations at amino acid positions 137, 310, and 562, either alone or in different combinations.
FIG. 1.
ORF2 features and mutants. (A) The 660-amino-acid protein (pORF2) encoded by HEV ORF2 is shown schematically, with two major regions: the extreme N terminus, containing the putative signal sequence (filled), and the basic highly N-terminal half of the protein, containing about 10% arginine residues (dotted). The extreme N-terminal region is expanded to show three domains characteristic of eukaryotic signal sequences, the hydrophobic core domain, a region containing turn-inducing amino acids, and a positively charged domain (+). The positions of three potential N-linked glycosylation sites (N-X-S/T) are shown at amino acid residues 137, 310, and 562 in the primary sequence of pORF2. (B) Various mutants used in this study are shown schematically. The Δ2-34 mutant carries a 5′-end deletion in ORF2 corresponding to amino acids 2 to 34 in the polypeptide sequence. The glycosylation mutants with Asn→Ala changes at the indicated positions are shown (●). The nomenclature corresponds to the amino acid position(s) changed in that particular mutant.
pORF2 is glycosylated in the ER.
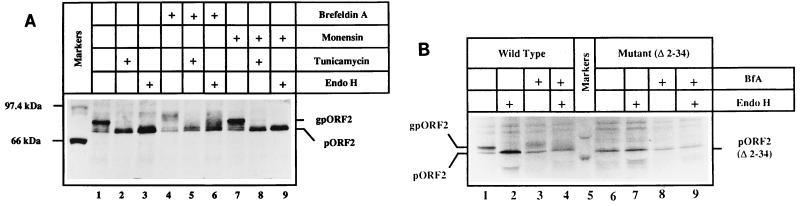
The complete susceptibility of glycosylated pORF2 (gpORF2) to endoglycosidase H (endo H) (10) suggested that it carried high-mannose residues, a modification that takes place within the ER (38). To further analyze this, we used the drugs brefeldin A and monensin. When the protein was transiently expressed in COS-1 cells, two forms, pORF2 and its glycosylated version, gpORF2, were observed, as earlier. The gpORF2 was susceptible to tunicamycin and endo H, both treatments reducing it to pORF2 (Fig. 2A, lanes 1 to 3). In a pulse-chase experiment, an alternatively modified and slower-migrating form of gpORF2 was observed upon treatment with 10 μg of BfA/ml (Fig. 2A, lanes 4 to 6). Some of this modified form of gpORF2 was endo H resistant, as apparent from the heterogeneous forms present above the pORF2 band. Such an effect was also observed with BfA concentrations as low as 1 μg/ml (data not shown). Used similarly, 5 μM monensin showed no effect on pORF2 glycosylation (Fig. 2A, lanes 7 to 9). This was also true with monensin concentrations as high as 20 μM (data not shown). The gpORF2 form, either in the absence of the drugs or in the presence of monensin, remained completely susceptible to tunicamycin and endo H treatments. These experiments confirmed our earlier observations and, as discussed later, suggested that the ER is the major site of intracellular accumulation of gpORF2.
FIG. 2.
Effects of brefeldin A and monensin on pORF2 expression. (A) COS-1 cells transfected with pSG-ORF2 were pulse-labeled with [35S]promix for 30 min, followed by a 3-h chase in complete DMEM. The cells were left untreated (lanes 1 and 3) or treated with 10 μg of brefeldin A (lanes 4 to 6)/ml, 5 μM monensin (lanes 7 to 9), or 10 μg of tunicamycin (lanes 2, 5, and 8)/ml during the prelabeling, pulse-labeling, and chase periods. Cell lysates were prepared, and the polypeptides were immunoprecipitated with a rabbit anti-pORF2 antiserum. Washed immunoprecipitates were subjected to mock treatment (lanes 1, 2, 4, 5, 7, and 8) or Endo H digestion (lanes 3, 6, and 9), and the polypeptides were analyzed on an SDS–7.5% polyacrylamide gel, followed by fluorography. Various treatments (+), the positions of gpORF2 and pORF2, and molecular size markers are indicated. (B) COS-1 cells transfected with pSG-ORF2 (lanes 1 to 4) or pSG-ORF2[Δ2-34] (lanes 6 to 9) were labeled with [35S]promix for 4 h in the absence (lanes 1, 2, 6, and 7) or presence (lanes 3, 4, 8, and 9) of 2.5 μg of brefeldin/ml. Following immunoprecipitation, the polypeptides in the precipitates were mock treated (lanes 1, 3, 5, and 7) or digested with Endo H (lanes 2, 4, 6, and 8) and analyzed on an SDS–7.5% polyacrylamide gel, followed by fluorography. The positions of gpORF2 and pORF2 are indicated. Only molecular size markers corresponding to 97.4 and 66 kDa are shown.
Treatment with BfA also resulted in significantly reduced levels of the nonglycosylated pORF2 (Fig. 2A, lanes 4 to 6). A labeling experiment was carried out in which steady-state expression levels of pORF2 or its N-terminal deletion mutant pORF2[Δ2-34] were estimated. As shown in Fig. 2B, steady-state levels of pORF2 and gpORF2 were significantly reduced in BfA-treated cells. Again, alternately modified and partially endo H-resistant forms of gpORF2 were observed (lanes 3 and 4). While BfA showed a generalized toxic effect on pORF2 expression, taken together with the pulse-chase results (Fig. 2A), it appeared to increase turnover of the ORF2 protein.
Asn-310 is the primary site of glycosylation.
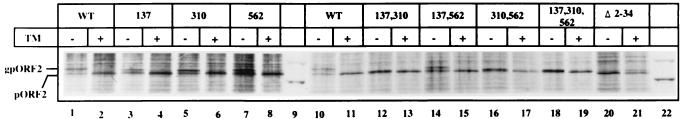
To determine which of the three potential N-linked glycosylation sites are modified in pORF2, we constructed a number of site-directed mutants in which one or more of the asparagine residue(s) in the N-X-S/T sequon were changed to alanine(s). COS-1 cells were transiently transfected with expression vectors containing either wild-type or mutant ORF2. The cells were metabolically labeled with [35S]methionine-cysteine in the absence or presence of tunicamycin, cell lysates were subjected to immunoprecipitation, and the proteins were analyzed by SDS-PAGE. The results are presented in Fig. 3. The single-site mutants ORF2[137], ORF2[310], and ORF2[562] all showed the presence of gpORF2 (lanes 3 to 8). The dual-site mutants ORF2[137,310] and [310,562], i.e., those that include Asn-310 along with either of the other two sites, were found to express pORF2, but the protein was not glycosylated (lanes 12 and 16). Alternatively, the protein may carry only minimal glycosylation, such that the nonglycosylated and glycosylated forms remain unresolvable on the gel. In contrast, the dual-site mutant ORF2[137,562], in which the Asn-310 residue was unchanged, expressed the pORF2 as well as gpORF2 forms of the protein (lane 14). However, the mobility of mutant gpORF2 was higher than that of wild-type gpORF2 (compare lanes 1 and 10 to lanes 3, 5, 7, and 14). As expected, the triple mutant ORF2[137,310,562] expressed only the nonglycosylated form of the protein (lane 18). Verification that the slower-moving forms in all the mutants were indeed gpORF2 was ascertained through tunicamycin inhibition experiments. These results strongly indicate that Asn-310 is the major, though not the only, site of glycosylation on the ORF2-encoded protein.
FIG. 3.
Expression and glycosylation of mutant pORF2 proteins. COS-1 cells transfected with pSG-ORF2 (lanes 1, 2, 10, and 11), pSG-ORF2[Asn→Ala mutants] (lanes 3 to 8 and 12 to 19), or pSG-ORF2[Δ2-34] (lanes 20 and 21) were labeled with [35S]promix for 4 h. Tunicamycin (10 μg/ml) was absent (−) or present (+) during the prelabeling and labeling periods as indicated. After immunoprecipitation, the polypeptides were analyzed on a SDS–7.5% polyacrylamide gel, followed by fluorography. The positions of gpORF2 and pORF2 are indicated. Only molecular size markers corresponding to 97.4 and 66 kDa are shown (lanes 9 and 22). WT, wild type; TM, tunicamycin.
pORF2 carries an N-terminal signal sequence.
To determine if the N-terminal region of pORF2 carried a functional signal sequence, a deletion mutant, ORF2[Δ2-34], was made in which sequences corresponding to residues 2 to 34 of pORF2 were deleted. This included the hydrophobic leucine-rich region as well as the charged arginine-rich region following it (Fig. 1A). This mutant ORF2 construct was also transfected into COS-1 cells and was found to express only the nonglycosylated form of the protein (Fig. 3, lanes 20 and 21). The presence of all three of the potential glycosylation sites in this deletion mutant indicated that the N-terminal region of pORF2 carried a signal sequence which directed the protein into the ER for glycosylation.
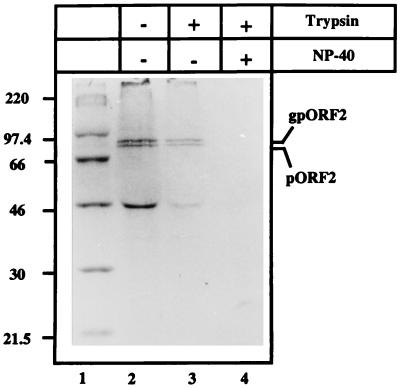
To further ascertain the ER-directing function of this signal sequence, in vivo and in vitro translocation assays were carried out. COS-1 cells transiently transfected with wild-type ORF2 or ORF2[Δ2-34] expression vectors were metabolically labeled with [35S]methionine-cysteine and subjected to subcellular fractionation. The microsomal fraction was prepared as described in Materials and Methods. The washed microsomes were then subjected to digestion with trypsin in the absence or presence of a nonionic detergent (NP-40). The wild-type pORF2/gpORF2 (Fig. 4, lane 2) was found in the microsomal fraction. The pORF2[Δ2-34] mutant was not found in the microsomal fraction but was present in the cytosolic fraction (data not shown). Further, wild-type pORF2/gpORF2 was resistant to trypsin digestion in the absence (lane 3) but not in the presence of NP-40 (lane 4). The protection from trypsin was, however, not quantitative. This can be explained by the protection of only fully translocated forms, while those in the process of being translocated would remain sensitive to trypsin. A cellular protein of ∼46 kDa was found to cross-react with anti-pORF2 (lanes 2 and 3). The possibility that this represented a cross-reacting polypeptide associated with the microsomal fraction is suggested by its presence in immunoprecipitates of mock-transfected cells as well (data not shown).
FIG. 4.
Microsomal localization of pORF2. COS-1 cells transfected with pSG-ORF2 were labeled with [35S]promix for 4 h and used to prepare the microsomal fraction as described in Materials and Methods. The microsomal fraction was then incubated in the absence (−) or presence (+) of 25 μg of trypsin/ml and/or 0.5% NP-40, on ice for 60 min. Following immunoprecipitation, the polypeptides were analyzed on an SDS–10% polyacrylamide gel, followed by fluorography. The positions of gpORF2 and pORF2 are indicated. Molecular size markers are shown (lane 1).
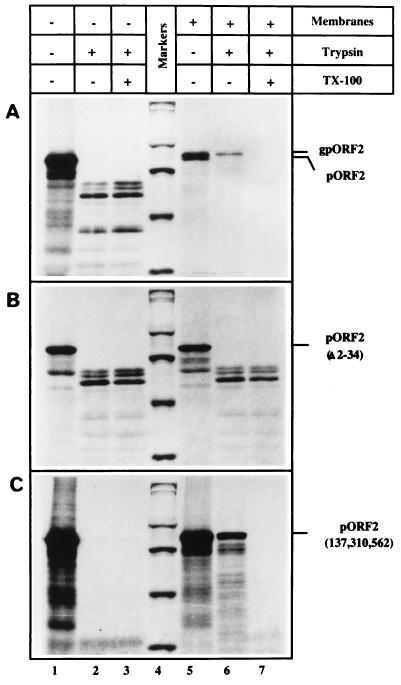
The translocation of pORF2, followed by trypsin resistance, was also carried out in vitro. By using a coupled transcription-translation system, pORF2/gpORF2 was synthesized in the absence or presence of added microsomal membranes. The wild-type protein (Fig. 5A), but not the Δ2-34 mutant protein (Fig. 5B), was found to be resistant to trypsin when synthesized in the presence of membranes. In the absence of membranes, both proteins were susceptible to trypsin. The triple Asn mutant protein, pORF2[137,310,562], was also resistant to trypsin only when synthesized in the presence of membranes (Fig. 5C). A few discrete bands smaller than the full-length polypeptide were observed in the trypsin-treated pORF2[Δ2-34] mutant synthesized in the presence of membranes (Fig. 5B, lanes 6 and 7). These appear identical to the bands observed when either pORF2 (Fig. 5A, lanes 2 and 3) or pORF2[Δ2-34] (Fig. 5B, lanes 2 and 3), synthesized in the absence of membranes, was treated with trypsin. This observation further reinforces the lack of pORF2[Δ2-34] translocation across microsomal membranes.
FIG. 5.
In vitro synthesis and translocation of pORF2 and its mutants. Coupled transcription-translation reactions were carried out by the TNT T7 system (Promega) programmed with plasmid pSG-ORF2 (A), pSG-ORF2[Δ2-34] (B), or pSG-ORF2[137,310,562] (C). Canine pancreatic membranes were absent (lanes 1 to 3) or present (lanes 4 to 7) during the in vitro reaction. Subsequently, each reaction mix was divided into three parts and either mock treated (lanes 1 and 5) or treated directly with 250 μg of trypsin/ml in the absence (lanes 2 and 6) or presence (lanes 3 and 7) of 0.7% Triton X-100. The protected polypeptides were analyzed on an SDS–7.5% polyacrylamide gel, followed by fluorography. Molecular size markers (lane 4) are shown as in Fig. 4. The positions of gpORF2, pORF2, pORF2[Δ2-34], and pORF2[137,310,562] are indicated.
Taken together, the results presented in Fig. 4 and 5 confirmed that pORF2 carrying the N-terminal signal sequence is cotranslationally translocated across the microsomal membrane.
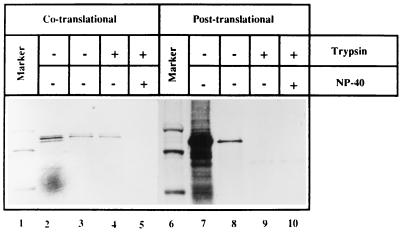
Experiments were carried out to distinguish between cotranslational and posttranslational translocation of pORF2. To assess any possible posttranslational translocation, the translation reaction was terminated with cycloheximide and in vitro-synthesized proteins were incubated with microsomal membranes. The membranes were pelleted through a cushion of 250 mM sucrose–TBS, and translocation across these membranes was assayed by resistance to trypsin. While pORF2 translated in the presence of membranes was protected from trypsin (Fig. 6, lanes 2 to 5), pORF2 synthesized in the absence of membranes was not protected from trypsin when translocation-competent microsomal membranes were added following translation (Fig. 6, lanes 7 to 10). Further, no gpORF2 band was observed when in vitro-translated pORF2 was incubated with the canine pancreatic membranes subsequent to its synthesis. These results showed that pORF2 can translocate across microsomal membranes in a cotranslational manner but not in a post-translational manner.
FIG. 6.
Co- and posttranslational translocation of pORF2. Plasmid pSG-ORF2 was subjected to in vitro-coupled transcription-translation. Following co- or posttranslational translocation of the in vitro-synthesized protein, as described in Materials and Methods, the membranes were pelleted down through a 250 mM sucrose–TBS cushion, resuspended in TBS, and divided into three equal aliquots. The aliquots were either mock treated (lanes 3 and 8) or treated with 25 μg trypsin/ml in the absence (lanes 4 and 9) or presence (lanes 5 and 10) of 0.5% NP-40. Input reactions, prior to membrane pelleting, are also shown (lanes 2 and 7). The positions of gpORF2 and pORF2 and the 97.4- and 66-kDa markers (lanes 1 and 6) are indicated.
pORF2 translocation, but not glycosylation, is required for its cell surface localization.
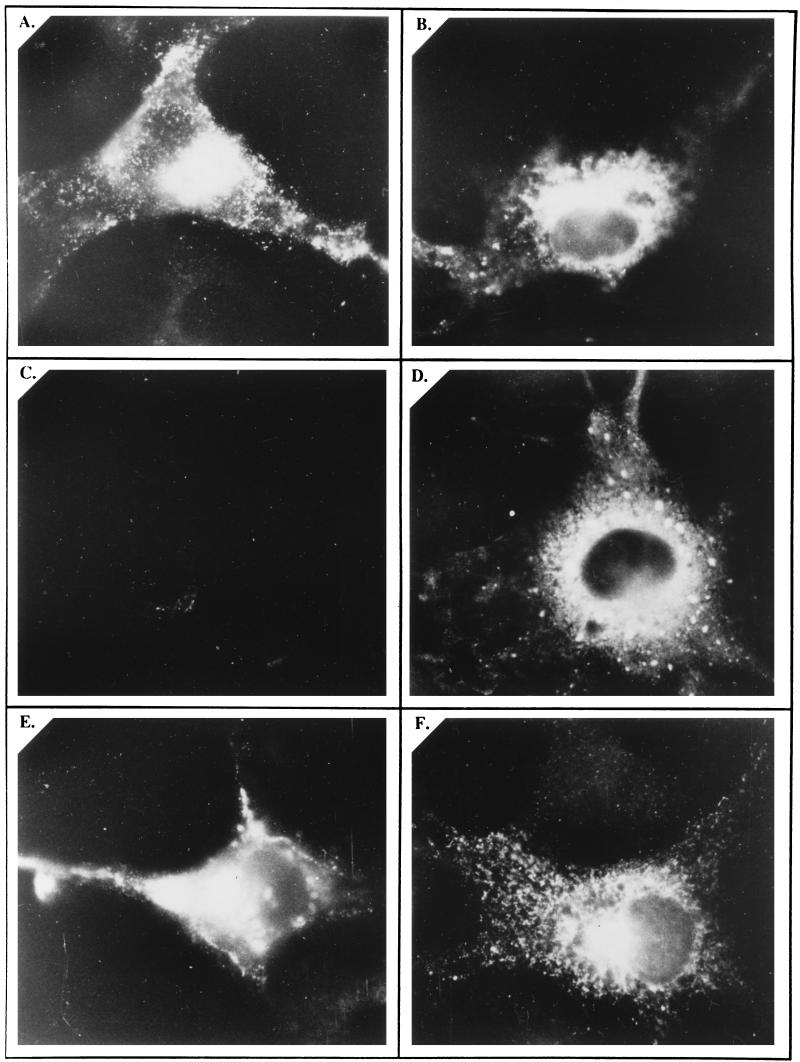
Previous results showed that a fraction of pORF2/gpORF2 is expressed on the cell surface. However, it was not clear whether glycosylation of pORF2 into its gpORF2 form was a prerequisite for such cell surface expression. Through indirect immunofluorescence, we analyzed the cell surface and intracellular localization of the wild-type and mutant forms of pORF2. While the wild-type (Fig. 7A and B) and the glycosylation-defective mutant pORF2[137,310,562] (Fig. 7E and F) proteins showed intracellular as well as cell surface localization, the signal sequence-deficient pORF2[Δ2-34] form of the protein localized intracellularly, but not to the cell surface (Fig. 7C and D). This result suggested that while pORF2 glycosylation is not required, its translocation to the ER and entry into the secretory network are prerequisites for its cell surface localization.
FIG. 7.
Cell surface and intracellular localization of pORF2 and its mutants. COS-1 cells transfected with pSG-ORF2 (A and B), pSG-ORF2[Δ2-34] (C and D), or pSG-ORF2[137,310,562] (E and F) were fixed with 4% paraformaldehyde–PBS and stained with rabbit anti-pORF2, followed by the goat anti-rabbit IgG-FITC conjugate. Antibody incubations were carried out in the absence (A, C, and E) or presence (B, D, and F) of 0.1% saponin for surface and intracellular staining, respectively. The stained cells were mounted in 20% glycerol and viewed and photographed with a fluorescence microscope. Representative views are presented.
DISCUSSION
Comparative analyses of homologous domains and consensus sequences in proteins provide good starting points for understanding structural and functional properties of new proteins. Analysis of the HEV putative major capsid protein, pORF2, suggested that this protein may go through the cellular secretory pathway, undergoing modifications en route. Earlier we showed that pORF2 carried N-linked glycans of the high-mannose type and that the nascent protein underwent processing subsequent to its synthesis (10). Here we provide additional details of pORF2 glycosylation and processing.
N-linked oligosaccharides are synthesized as a core unit of Glc3MangGlc-NAc2 attaching to asparagines at N-X-S/T sequences while the polypeptide chains are being translocated across the ER membrane (4). The glycosylation pattern of a glycoprotein reflects the cellular compartment through which it has passed during its synthesis and processing. Our earlier results suggested that pORF2 underwent glycosylation within the ER (10). Here we have analyzed the effects of the drugs brefeldin A and monensin on pORF2 glycosylation.
Brefeldin A is a fungal metabolite that disrupts intracellular membrane traffic at the ER-Golgi junction (20), resulting in a redistribution of Golgi enzymes into the ER. Since BfA collapses Golgi enzymes into the ER, it is not surprising to find some gpORF2 becoming endo H resistant, as observed in Fig. 2A and B. It was, however, surprising to find that wild-type gpORF2 was completely sensitive to endo H. This seems to indicate that the major site of intracellular accumulation of gpORF2 is the ER. Similar observations have been made in the case of rotavirus VP7 protein (19) and the spleen focus-forming virus glycoprotein, gp55 (40). Monensin, a metabolite of Streptomyces cinnamonensis, impairs Golgi function (21), and in our studies, did not interfere with the glycosylation of pORF2. This would be expected if most of the protein was not leaving the ER. Alternatively, it may move directly from the ER to the cell surface, as suggested for the hepatitis B virus M protein (34). The data presented here, though persuasive, are not sufficient for proving existence of the intracellular trafficking pathway of the HEV ORF2 protein. However, the observations do suggest that the HEV capsid, or at least a precursor, may assemble within the ER.
Tunicamycin and endoglycosidase digestion experiments carried out earlier showed that pORF2 was N-glycosylated (10). In its 660-amino-acid sequence, it contains three N-X-S/T motifs which can act as potential acceptor sites. These sites are conserved in the ORF2 sequences of all HEV isolates sequenced so far (1, 2, 8, 32, 36, 39), as well as in swine HEV (16). To analyze which one of these potential site(s) was glycosylated, we made site-directed mutants such that the asparagine residues at positions 137, 310, and 562, within the primary amino acid sequence of pORF2, were individually changed to alanines. The glycosylation pattern of these mutants suggested that Asn-310 was the primary site of modification though not the only one; the other two sites were also utilized. However, since double mutants carrying the Asn-310→Ala mutation along with either the Asn-137 or the Asn-562 mutation failed to produce gpORF2, it appears that Asn-310 is the major site of glycosylation. The inability of the mutant protein pORF2[137,310,562] to undergo glycosylation ruled out any more sites at which N-glycans can be added to the protein.
Nascent polypeptides crossing the ER membrane are cotranslationally N-glycosylated at the N-X-S/T motif by the oligosaccharide complex, which consists of several subunits (30) and lies close to the protein translocation channel. There appears to be structural specificity with respect to glycosylation efficiency in that the amino acids proline, aspartic acid, and glutamic acid are not tolerated at the X position (33). Further, statistical analysis of a wide range of N-glycosylated proteins suggests that proline residues either at the X position or next to the serine/threonine position greatly reduce the degree of glycosylation (5). In this context, it is interesting that a proline residue is present next to the 310N-L-T motif in the deduced pORF2 sequences from all except the Mexican isolate of HEV; yet, this appears to be the major glycosylation site within pORF2. The low overall amounts of N-glycans added to pORF2 may reflect this sequence constraint.
Secretory and plasma membrane proteins are targeted to the ER membrane via their signal peptides, which are usually located at the amino terminus of the nascent polypeptides (31). Signal sequences increase the specific interaction of a protein with its appropriate transport machinery. It is generally assumed that signal sequences have highly degenerate primary sequences but a common secondary structure or a similar distribution of charged and apolar residues (31). The signal peptide consists of three regions, an amino-terminal polar region of 2 to 40 amino acids, a central hydrophobic region of 7 to 20 amino acids, and a variable-length sequence containing turn-inducing amino acids, followed by the signal peptidase cleavage site (31). A hydropathy plot of pORF2 and close examination of its amino-terminal sequence revealed the presence of all these features. Taken together with our earlier pulse-chase studies, showing processing of the nascent pORF2 polypeptide (10), this suggested the presence of a functional ER-translocating signal sequence at the amino terminus of pORF2.
A deletion mutant of ORF2 was constructed in which sequences corresponding to amino acids 2 to 34 were deleted. This included all three regions of the putative signal sequence mentioned above. Expressed in COS-1 cells, the mutant protein showed no glycosylation, even though all three of the N-linked glycosylation sites were present.
The translocation of wild-type and mutant pORF2 polypeptides into microsomes was also studied by using resistance to trypsin as an assay for translocation. In transfected COS-1 cells a significant fraction of the wild-type protein was found within isolated microsomes. The N-terminally-deleted pORF2[Δ2-34] protein, however, was cytosolic in its subcellular localization. In an in vitro translation system, the wild-type but not the Δ2-34 mutant protein was sequestered within exogenously added microsomes. The glycosylation-null triple-Asn mutant protein, pORF2[137,310,562], which carried an intact N-terminal signal sequence, also translocated to the ER and was protected from trypsin digestion.
The in vitro synthesis of wild-type and mutant forms of pORF2, followed by a translocation assay with trypsin also provided some insight into the folding of this protein. Being rich in lysine and arginine residues, the protein has 59 possible trypsin digestion sites, rarely resulting in peptides 50 amino acids or longer. Yet, at least four distinct bands of about 40 to 60 kDa were observed upon tryptic digestion, suggesting extensive folding of the in vitro-synthesized polypeptide, resulting in the sequestration of a majority of trypsin-sensitive sites. The N-terminally-deleted protein lacking the first 34 amino acids, pORF2[Δ2-34], appeared to fold almost identically to the wild-type pORF2, as was apparent from the major tryptic fragments generated. Interestingly, when just three conserved residues involved in the N-linked glycosylation of pORF2 were changed, the mutant pORF2[137,310,562] became completely sensitive to trypsin in the absence of microsomal membranes. This observation is compatible with a drastically different folding of this mutant polypeptide.
Proteins are translocated across the ER membrane through two distinct pathways, one dependent upon the signal recognition particle (SRP) (29) and the other independent of it (6). The targeting route depends largely upon the hydrophobicity of the signal sequence present on the protein to be translocated (22). Proteins with a hydrophobicity index (according to Kyte-Doolittle) of +2.0 or less show SRP-independent translocation, while those with an index approaching +3.0 are SRP dependent (22). An analysis of the pORF2 signal sequence showed a hydrophobicity index of +3.0 for the core, suggesting an SRP-dependent mode of translocation. This is further supported by experiments showing cotranslational but not posttranslational translocation of pORF2 in the presence of exogenously added microsomal membranes. SRP-dependent translocation tends to be cotranslational (29), while the SRP-independent translocation is posttranslational (22). It will be of interest to determine if pORF2 interacts with components of the SRP and resident ER proteins.
The localization of wild-type and mutant ORF2 proteins at the cell surface was studied by indirect immunofluorescence of the transfected cells. Since both wild-type and the glycosylation-null triple-Asn mutant but not the signal-deleted mutant were found on the cell surface, the ER localization of pORF2 and its entry into the cellular secretory network are essential prerequisites for cell surface localization of pORF2/gpORF2. Glycosylation of pORF2 is not important for its appearance on the cell surface.
What is the functional significance of pORF2 glycosylation? It is not clear if the addition of N-glycans has any role to play in viral capsid assembly or virus-host cell interactions. The modification of pORF2 may occur simply because the protein has accessible N-linked glycosylation sites and is translocated to the ER, where the cellular machinery for such a modification is available. However, trypsin sensitivity studies on wild-type and mutant proteins, discussed above, suggest an important structural role for pORF2 glycosylation. Further, the pORF2 amino-terminal signal sequence and all three N-linked glycosylation sites are universally found in all isolates of human HEV as well as in the newly discovered swine HEV. Therefore, it is likely that pORF2 glycosylation has a functional significance as well. It is possible that all or part of the HEV capsid assembles within the ER. N-linked oligosaccharides are known to increase the solubility and stability of many proteins and thus to help in their proper folding (7). That would be critical for viral capsid assembly, since multiple identical subunits must come together to form that structure.
While glycosylation of surface proteins is essential for the assembly of enveloped viruses, little information exists about the function of carbohydrates on the capsid protein(s) of nonenveloped viruses. The rotavirus VP7 protein becomes resistant to endo H upon brefeldin A treatment (19), and a combination of tunicamycin and brefeldin A results in misfolding and interdisulfide bond aggregation of the lumenal VP7 protein (18). Further, while glycosylated VP7 was found to interact with protein disulfide isomerase, nonglycosylated VP7 did not. This result is taken to mean that the major function of carbohydrates on VP7 is to facilitate correct disulfide bond formation and protein folding (18). It has also been proposed that capsid glycosylation may render the virus more resistant to the gastric environment (18). Similar roles may be proposed for pORF2 glycosylation.
Following proper folding in the ER, N-glycans are also known to play a role in biosynthetic traffic beyond the ER (4). However, few studies have differentiated between the effects of N-glycans on protein folding in the ER and their role in subsequent transport to the plasma membrane. The requirements for glycosylation appear to be more stringent for membrane proteins than for secreted proteins. The cell surface expression of very few plasma membrane proteins, such as pORF2 described here, is unaltered by inhibition of glycosylation (4). However, we need to examine pORF2 transport kinetics quantitatively through pulse-chase and cell fractionation studies to be able to state this finding conclusively.
Another proposed role for N-glycans is sorting signals in polarized cells. For example, erythropoietin, an apically secreted protein in MDCK cells, is discharged equally from the apical and basolateral surfaces when its three N-glycosylation sites are removed by mutagenesis (12). Even in nonpolarized cells such as fibroblasts, the existence of two sorting pathways analogous to the apical and basolateral routes in MDCK cells is postulated (15). Because hepatocytes are polarized cells, newly synthesized HEV particles may exit the cell preferentially from one surface and N-glycosylation of pORF2 may be a determinant for that selectivity. Clearly, a number of questions are unanswered; these will be the focus of our future efforts.
While no in vitro culture system exists for HEV, a primary infectivity system with hepatocytes from experimentally infected nonhuman primates has recently become available (37). It will be of interest to develop comparative data by such a system for the expression and processing of HEV proteins.
ACKNOWLEDGMENTS
This work was supported by internal grants from the ICGEB.
We thank Satyajit Mayor for suggestions; Vir Chauhan, Seyed Hasnain, Vijay Kumar, and Sudhir Sopory for reviewing the manuscript; and Dipti Arora-Chugh for proofreading it.
REFERENCES
- 1.Aye T T, Uchida T, Ma X-Z, Iida F, Shikata T, Zhuang H, Win K M. Complete nucleotide sequence of a hepatitis E virus isolated from the Xinjiang epidemic (1986–1988) of China. Nucleic Acids Res. 1992;20:3512. doi: 10.1093/nar/20.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi S L, Purdy M A, McCaustland K A, Margolis H S, Bradley D W. The sequence of hepatitis E virus isolated directly from a single source during an outbreak in China. Virus Res. 1993;28:233–247. doi: 10.1016/0168-1702(93)90024-h. [DOI] [PubMed] [Google Scholar]
- 3.Bradley D W. Enterically-transmitted non-A, non-B hepatitis. Br Med Bull. 1990;46:442–461. doi: 10.1093/oxfordjournals.bmb.a072409. [DOI] [PubMed] [Google Scholar]
- 4.Fielder K, Simmons K. The role of N-glycans in the secretory pathway. Cell. 1995;81:309–312. doi: 10.1016/0092-8674(95)90380-1. [DOI] [PubMed] [Google Scholar]
- 5.Gavel Y, von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990;3:433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hann, B. C., and P. Walter. The signal recognition particle in S. cerevisiae. Cell 67:131–144. [DOI] [PubMed]
- 7.Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Cell Biol. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C C, Nguyen D, Fernandez J, Yun K Y, Fry K E, Bradley D W, Tam A W, Reyes G R. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV) Virology. 1992;191:550–558. doi: 10.1016/0042-6822(92)90230-m. [DOI] [PubMed] [Google Scholar]
- 9.International Committee on Taxonomy of Viruses. Eighth report, in press.
- 10.Jameel S, Zafrullah M, Ozdener M H, Panda S K. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J Virol. 1996;70:207–216. doi: 10.1128/jvi.70.1.207-216.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khuroo M S. Study of an epidemic of non-A, non-B hepatitis: possibility of another human hepatitis virus distinct from post-transfusion non-A, non-B type. Am J Med. 1980;68:818–823. doi: 10.1016/0002-9343(80)90200-4. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa Y, Sano Y, Ueda M, Higashio K, Narita H, Okano M, Matsumoto S I, Sasaki R. N-glycosylation of erythropoietin is critical for apical secretion by Madin-Darby canine kidney cells. Exp Cell Res. 1994;213:449–457. doi: 10.1006/excr.1994.1222. [DOI] [PubMed] [Google Scholar]
- 13.Koonin E V, Gorbalenya A E, Purdy M A, Rozanov M N, Reyes G R, Bradley D W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-stranded RNA plant and animal viruses. Proc Natl Acad Sci USA. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krawczynski K. Hepatitis E. Hepatology. 1993;17:932–941. [PubMed] [Google Scholar]
- 15.Matter K, Mellman I. Mechanisms of cell polarity: sorting and transport in epithelial cells. Curr Opin Cell Biol. 1994;6:545–554. doi: 10.1016/0955-0674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 16.Meng X-J, Purcell R H, Halbur P G, Lehman J R, Webb D M, Tsareva T S, Haynes J S, Thacker B J, Emerson S U. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller M J. Viral taxonomy. Clin Infect Dis. 1995;21:280. doi: 10.1093/clinids/21.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirazimi A, Svensson L. Carbohydrates facilitate correct disulfide bond formation and folding of rotavirus VP7. J Virol. 1998;72:3887–3892. doi: 10.1128/jvi.72.5.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirazimi A, von Bonsdorff C H, Svensson L. Effect of brefeldin A on rotavirus assembly and oligosaccharide processing. Virology. 1996;217:554–563. doi: 10.1006/viro.1996.0150. [DOI] [PubMed] [Google Scholar]
- 20.Misumi Y, Miki K, Takatsuki A, Tamura G, Ikehara Y. Novel blockage by brefeldin A of intracellular transport of secretory proteins in cultured rat hepatocytes. J Biol Chem. 1986;261:11398–11403. [PubMed] [Google Scholar]
- 21.Mollenhauer H H, Orr D J, Rowe L D. Alteration of intracellular traffic by monensin: mechanism, specificity and relationship to toxicity. Biochim Biophys Acta. 1990;1031:225–246. doi: 10.1016/0304-4157(90)90008-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng D T W, Brown J D, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostapchuk P, Hearing P, Ganem D. A dramatic shift in transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 1994;13:1048–1057. doi: 10.1002/j.1460-2075.1994.tb06353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panda S K, Nanda S K, Zafrullah M, Ansari I H, Ozdener M H, Jameel S. An Indian strain of hepatitis E virus (HEV): cloning, sequence, and expression of the structural region and antibody responses in sera from individuals from an area of high-level HEV endemicity. J Clin Microbiol. 1995;33:2653–2659. doi: 10.1128/jcm.33.10.2653-2659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prange R, Streeck R E. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell R H, Ticehurst J R. Enterically transmitted non-A, non-B hepatitis: epidemiology and clinical characteristics. In: Zuckerman A J, editor. Viral hepatitis and liver disease. New York, N.Y: Alan R. Liss, Inc.; 1988. pp. 131–137. [Google Scholar]
- 27.Purdy M A, Tam A W, Huang C C, Yarbough P O, Reyes G R. Hepatitis E virus: a non-enveloped member of the “alpha-like” RNA virus supergroup? Semin Virol. 1993;4:319–326. [Google Scholar]
- 28.Qiu Z, Tufaro F, Gillam S. Brefeldin A and Monensin arrest cell surface expression of membrane glycoproteins and release of rubella virus. J Gen Virol. 1995;76:855–863. doi: 10.1099/0022-1317-76-4-855. [DOI] [PubMed] [Google Scholar]
- 29.Rapoport T A, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 30.Reiss G, Hessen S, Gilmore R, Zufferey R, Aebi M. A specific screen for oligosaccharyltransferase mutations identifies the 9 kDa OST5 protein required for optimal activity in vivo and in vitro. EMBO J. 1997;16:1164–1172. doi: 10.1093/emboj/16.6.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 32.Schlauder G G, Dawson G J, Erker J C, Kwo P Y, Knigge M F, Smalley D L, Rosenblatt J E, Desai S M, Mushahwar I K. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J Gen Virol. 1998;79:447–456. doi: 10.1099/0022-1317-79-3-447. [DOI] [PubMed] [Google Scholar]
- 33.Shakin-Eshelman S H, Spitalnik S L, Kasturi L. The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J Biol Chem. 1996;271:6363–6366. doi: 10.1074/jbc.271.11.6363. [DOI] [PubMed] [Google Scholar]
- 34.Sheu S Y, Lo S J. Biogenesis of the hepatitis B viral middle (M) surface protein in a human hepatoma cell line: demonstration of an alternate secretion pathway. J Gen Virol. 1994;75:3031–3039. doi: 10.1099/0022-1317-75-11-3031. [DOI] [PubMed] [Google Scholar]
- 35.Swick A G, Janicot M, Cheval-Kastelic T, McLenithan J C, Lane M D. Promoter-cDNA-directed heterologous protein expression in Xenopus laevis oocytes. Proc Natl Acad Sci USA. 1992;89:1812–1816. doi: 10.1073/pnas.89.5.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tam A W, Smith M M, Guerra M E, Huang C C, Bradley D W, Fry K E, Reyes G R. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam A W, White R, Reed E, Short M, Zhang Y, Fuerst T R, Lanford R E. In vitro propagation and production of hepatitis E virus from in vivo-infected primary macaque hepatocytes. Virology. 1996;215:1–9. doi: 10.1006/viro.1996.0001. [DOI] [PubMed] [Google Scholar]
- 38.Tarentino A L, Trimble R B, Plummer T H. Enzymatic approaches for studying the structure, synthesis and processing of glycoproteins. Methods Cell Biol. 1989;32:111–139. doi: 10.1016/s0091-679x(08)61169-3. [DOI] [PubMed] [Google Scholar]
- 39.Tsarev S A, Emerson S U, Reyes G R, Tsareva T S, Letgers L J, Malik I A, Iqbal M, Purcell R H. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci USA. 1992;89:559–563. doi: 10.1073/pnas.89.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulmer J B, Palade G E. Effects of brefeldin A on the processing of viral envelope glycoproteins in murine erythroleukemia cells. J Biol Chem. 1991;266:9173–9179. [PubMed] [Google Scholar]
- 41.Wong D C, Purcell R H, Sreenivasan M A, Prasad S R, Pavri K M. Epidemic and endemic hepatitis in India: evidence for a non-A, non-B virus etiology. Lancet. 1980;2:876–879. doi: 10.1016/s0140-6736(80)92045-0. [DOI] [PubMed] [Google Scholar]
- 42.Yuckenberg P D, Witney F, Geisselsoder J, McClary J. Site-directed in vitro mutagenesis using uracil-containing DNA and phagemid vectors. In: McPherson M J, editor. Directed mutagenesis: a practical approach. Oxford, England: IRL Press; 1991. pp. 27–48. [Google Scholar]
- 43.Zaaijer H L, Kok M, Lelie P N, Timmerman R J, Chau K, van der Pal H J. Hepatitis E in The Netherlands: imported and endemic. Lancet. 1993;341:826. doi: 10.1016/0140-6736(93)90599-c. [DOI] [PubMed] [Google Scholar]
- 44.Zafrullah M, Ozdener M H, Panda S K, Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol. 1997;71:9045–9053. doi: 10.1128/jvi.71.12.9045-9053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]