Abstract
Flowering Chinese cabbage (Brassica campestris L. ssp. Chinensis var. utilis Tsen et Lee) is a widely consumed vegetable in southern China with significant economic value. Developing product organs in the flowering Chinese cabbage involves two key processes: bolting and flowering. Nuclear factor Y (NF-Y) is a heterotrimeric transcription factor known for its crucial role in various plant developmental processes. However, there is limited information available on the involvement of this gene family during flowering during Chinese cabbage development. In this study, 49 BcNF-Y genes were identified and characterized along with their physicochemical properties, gene structure, chromosomal location, collinearity, and expression patterns. We also conducted subcellular localization, yeast two-hybrid, and transcriptional activity assays on selected BcNF-Y genes. The findings of this study revealed enhanced expression levels of specific BcNF-Y genes during the stalk development and flowering stages in flowering Chinese cabbage. Notably, BcNF-YA8, BcNF-YB14, BcNF-YB20, and BcNF-YC5 interacted with BcRGA1, a negative regulator of GA signaling, indicating their potential involvement in GA-mediated stalk development. This study provides valuable insights into the role of BcNF-Y genes in flowering Chinese cabbage development and suggests that they are potential candidates for further investigating the key regulators of cabbage bolting and flowering.
Keywords: flowering Chinese cabbage, NF-Y gene family, gibberellin, bolting
1. Introduction
Flowering Chinese cabbage (Brassica campestris L. ssp. Chinensis var. utilis Tsen et Lee) is a highly cultivated and productive vegetable in southern China. It belongs to the Chinese cabbage subspecies of Brassica species in the Brassicaceae family. The main product organ is the flowering stalk. The formation of the flowering stalk involves two simultaneous processes: bolting, which is characterized by stem elongation and thickening, and flowering. Gibberellin (GA) is a crucial factor influencing bolting in flowering Chinese cabbage. Treatment with exogenous gibberellin A3 (GA3) has been shown to induce the up-regulation of BcSOC1 and genes encoding cell wall structural proteins (BcEXPA11, BcXTH3) in flowering Chinese cabbage, thereby facilitating early bolting and flowering processes. Moreover, low temperatures trigger GA accumulation at the stem tip, accelerating growth and flowering. Conversely, using a GA synthesis inhibitor (PAC) suppresses the initiation of bolting and flowering [1,2,3,4]. BcSOC1, a key regulator in developing the flowering stalk, promotes early flowering and bolting when overexpressed. This is probably due to the upregulation of genes encoding cell wall structural proteins. In contrast, the knockdown of BcSOC1 leads to significant phenotypic differences in plants, underscoring its importance in flowering Chinese cabbage [5]. Loss-of-function mutations in BcRGL1, a DELLA protein, affect the expression of various genes in flowering Chinese cabbage. These genes include the GA regulatory gene BcGASA6, flower-related genes BcSOC1 and BcLFY, and genes encoding cell wall structural proteins BcEXPA11 and BraXTH3. These mutations promote flowering and bolting. Conversely, overexpression of BcRGL1 suppressed both bolting and flowering in flowering Chinese cabbage [6].
The nuclear factor Y (NF-Y) family, known as the CCAAT-binding factor (CBF) or heme-activating protein, is a conserved heterotrimeric transcription factor complex in eukaryotes. It consists of three subfamilies: NF-YA, NF-YB, and NF-YC. The NF-Y complex is formed by associating three members from different subfamilies [7,8,9]. NF-Y has been extensively studied and implicated in various biological processes, including plant stress responses, hormone signaling pathways, and seed development, among others [10,11,12,13,14,15,16,17,18]. For example, in rice, the complex formed by NF-YB1, NF-YC12, and bHLH144 binds to the Wx promoter, activating Wx transcription and regulating seed quality [19]. In poplar, PdNF-YB21 promotes root development by enhancing the expression of PdNCED3, a key gene involved in ABA synthesis, increasing ABA levels in the roots and promoting growth under drought conditions [12]. NF-YCs bind to the promoter of BR6ox2 (a gene involved in BR biosynthesis) to inhibit BR biosynthesis. Furthermore, they maintain the stability of BIN2, a key inhibitor of the BR signaling pathway, thus influencing the photoregulation of hypocotyl elongation in Arabidopsis [20].
In recent years, the involvement of NF-Y family members in flowering and gibberellin signaling has gained significant attention. Studies have revealed that NF-Ys form a complex with CONSTANS to recognize key regulatory motifs in the promoter region of FT genes, thereby affecting flowering time [21,22,23,24,25,26,27]. In the photoperiodic and GA pathways, GA-mediated degradation of DELLA proteins releases NF-Y from the DELLA-NF-Y complex. This enables NF-Y to bind to NFYBE, a critical motif in the SOC1 promoter, thereby promoting SOC1 expression and accelerating flowering in Arabidopsis [28]. In Arabidopsis, the NF-YC-RGL2 module regulates seed germination by binding to a distinct CCAAT motif within the ABI5 promoter, a key gene in the ABA signaling pathway. It also co-regulates a group of genes responsive to GA and ABA [29]. In roses, RhNF-YC9 regulates the petal expansion rate by influencing GA accumulation and expression of cell proliferation-related genes [30]. Moreover, DELLA proteins in Arabidopsis suppress SOC1 expression by weakening the binding of NF-YCs to SOC1 through BRM. This promotes the binding of BRM to SOC1, collectively inhibiting flowering [31]. These findings highlight the crucial role of the NF-Y gene family in plant flowering and the GA pathway. Nevertheless, our understanding of the essential characteristics and functional significance of NF-Y transcription factors in the flowering Chinese cabbage is still limited. Therefore, this study aimed to investigate the role of the NF-Y gene family in the product organ development process of flowering Chinese cabbage.
Using bioinformatics methods, 49 BcNF-Y genes were identified in flowering Chinese cabbage, with 17 BcNF-A, 20 BcNF-YB, and 12 BcNF-YC. Comprehensive analyses were conducted to investigate various aspects of these genes, including their physicochemical properties, gene structure, motif composition, and conserved structural domains. Phylogenetic tree analyses were conducted using data from flowering Chinese cabbage, rice, and Arabidopsis to examine their evolutionary relationship. Additionally, collinearity analysis between flowering Chinese cabbage and Arabidopsis was performed to determine the potential roles of these genes. The expression patterns of NF-Y gene family members were examined in different developmental stages of flowering Chinese cabbage tissues and under GA treatment. Notably, several NF-Y family members that interact with DELLAs have been identified and further characterized for their subcellular localization and transcriptional activity. This study offers valuable insights into the involvement of the NF-Y gene family in the development of flowering Chinese cabbage. Further, it provides a good set of candidate genes for future investigations that can be explored in the future to identify key regulators of flowering Chinese cabbage development.
2. Results
2.1. Identification of BcNF-Y Family Members
A comprehensive analysis of BcNF-Y genes was performed in the flowering Chinese cabbage genome. This resulted in identifying 49 non-redundant BcNF-Y genes (17 BcNF-YAs, 20 BcNF-YBs, and 12 BcNF-YCs). These genes were named BcNF-YA1 to BcNF-YA17 (NF-YA subfamily), BcNF-YB1 to BcNF-YB20 (NF-YB subfamily), and BcNF-YC1 to BcNF-YC12 (NF-YC subfamily). Physicochemical analysis of the encoded proteins revealed significant variations in protein length. The length ranges from 102 (BcNF-YB2) to 1374 amino acids (BcNF-YA12). Additionally, the predicted molecular weight of the proteins varied, the lowest being 11670.12 Da (BcNF-YB11) and the highest being 155711.3 Da (BcNF-YA5). The theoretical isoelectric point (PI) of the proteins ranged from 4.27 (BcNF-YB8) to 10.11 (BcNF-YB19) (Table 1).
Table 1.
Information of the BcNF-Y genes family in flowering Chinese cabbage.
| Gene Name | CDS (bp) | Length (AA) | pI | MW (Da) | Homologs in Arabidopsis |
|---|---|---|---|---|---|
| BcNF-YA1 | 2430 | 809 | 9.36 | 34,401.98 | AtNF-YA6 |
| BcNF-YA2 | 945 | 314 | 9.87 | 13,514.28 | AtNF-YA2 |
| BcNF-YA3 | 720 | 239 | 9.54 | 28,766 | AtNF-YA1 |
| BcNF-YA4 | 963 | 320 | 9.99 | 20,085.81 | AtNF-YA3 |
| BcNF-YA5 | 1653 | 550 | 7.16 | 155,711.3 | AtNF-YA1 |
| BcNF-YA6 | 846 | 281 | 9.74 | 26,426.97 | AtNF-YA2 |
| BcNF-YA7 | 927 | 308 | 6.87 | 44,034.04 | AtNF-YA9 |
| BcNF-YA8 | 360 | 119 | 9.51 | 24,406.47 | AtNF-YA4 |
| BcNF-YA9 | 696 | 231 | 9.61 | 27,110.81 | AtNF-YA5 |
| BcNF-YA10 | 450 | 149 | 6.99 | 88,896.45 | AtNF-YA9 |
| BcNF-YA11 | 783 | 260 | 9.04 | 35,419.5 | AtNF-YA5/6 |
| BcNF-YA12 | 4125 | 1374 | 6.32 | 26,437.07 | AtNF-YA3 |
| BcNF-YA13 | 540 | 179 | 8.99 | 34,091.57 | AtNF-YA3 |
| BcNF-YA14 | 717 | 238 | 9.68 | 30,882.8 | AtNF-YA5 |
| BcNF-YA15 | 645 | 214 | 6.36 | 61,377.49 | AtNF-YA7 |
| BcNF-YA16 | 1188 | 395 | 9.95 | 17,017.99 | AtNF-YA8 |
| BcNF-YA17 | 711 | 236 | 9.9 | 25,671.99 | AtNF-YA10 |
| BcNF-YB1 | 414 | 137 | 7.07 | 14,745.38 | AtNF-YB3 |
| BcNF-YB2 | 309 | 102 | 6.91 | 25,881.06 | AtNF-YB12 |
| BcNF-YB3 | 372 | 123 | 5.06 | 23,121.14 | AtNF-YB2 |
| BcNF-YB4 | 804 | 267 | 7.76 | 13,714.15 | AtNF-YB8/10 |
| BcNF-YB5 | 438 | 145 | 5.1 | 25,587.71 | AtNF-YB3 |
| BcNF-YB6 | 1497 | 498 | 8.84 | 19,813.36 | AtNF-YB7 |
| BcNF-YB7 | 684 | 227 | 5.04 | 15,948.19 | AtNF-YB8/10 |
| BcNF-YB8 | 468 | 155 | 4.27 | 24,485.01 | AtNF-YB5 |
| BcNF-YB9 | 357 | 118 | 5.96 | 25,267.27 | AtNF-YB1 |
| BcNF-YB10 | 408 | 135 | 8.92 | 13,174.68 | AtNF-YB3 |
| BcNF-YB11 | 369 | 122 | 4.8 | 11,670.12 | AtNF-YB4 |
| BcNF-YB12 | 681 | 226 | 5.01 | 56,994.71 | AtNF-YB2 |
| BcNF-YB13 | 693 | 230 | 7.07 | 15,677.32 | AtNF-YB9 |
| BcNF-YB14 | 666 | 221 | 8.8 | 29,546.27 | AtNF-YB11 |
| BcNF-YB15 | 510 | 169 | 9.61 | 25,962.1 | AtNF-YB13 |
| BcNF-YB16 | 456 | 151 | 6.91 | 14,458.08 | AtNF-YB7 |
| BcNF-YB17 | 363 | 120 | 5.21 | 13,011.72 | AtNF-YB2 |
| BcNF-YB18 | 648 | 215 | 6.65 | 17,694.84 | AtNF-YB9 |
| BcNF-YB19 | 969 | 322 | 10.11 | 12,238.18 | AtNF-YB9 |
| BcNF-YB20 | 552 | 183 | 5.34 | 29,312.26 | AtNF-YB8/10 |
| BcNF-YC1 | 717 | 238 | 7.12 | 63,811.16 | AtNF-YC11 |
| BcNF-YC2 | 2070 | 689 | 5.7 | 35,706.48 | AtNF-YC11 |
| BcNF-YC3 | 525 | 174 | 6.22 | 23,990.09 | AtNF-YC10 |
| BcNF-YC4 | 693 | 230 | 5.72 | 12,891.28 | AtNF-YC9 |
| BcNF-YC5 | 636 | 211 | 6.7 | 17,457.36 | AtNF-YC1 |
| BcNF-YC6 | 813 | 270 | 4.74 | 19,047.31 | AtNF-YC4 |
| BcNF-YC7 | 330 | 109 | 8.49 | 17,259.75 | AtNF-YC13 |
| BcNF-YC8 | 417 | 138 | 8.9 | 26,409.45 | AtNF-YC2 |
| BcNF-YC9 | 1734 | 577 | 6.93 | 75,108.26 | AtNF-YC9 |
| BcNF-YC10 | 687 | 228 | 6.13 | 12,824.61 | AtNF-YC9 |
| BcNF-YC11 | 339 | 112 | 5.1 | 25,421.53 | AtNF-YC10 |
| BcNF-YC12 | 459 | 152 | 9.1 | 19,965.36 | AtNF-YC11 |
2.2. Analysis of Motifs and Conserved Domains in BcNF-Y Family Members
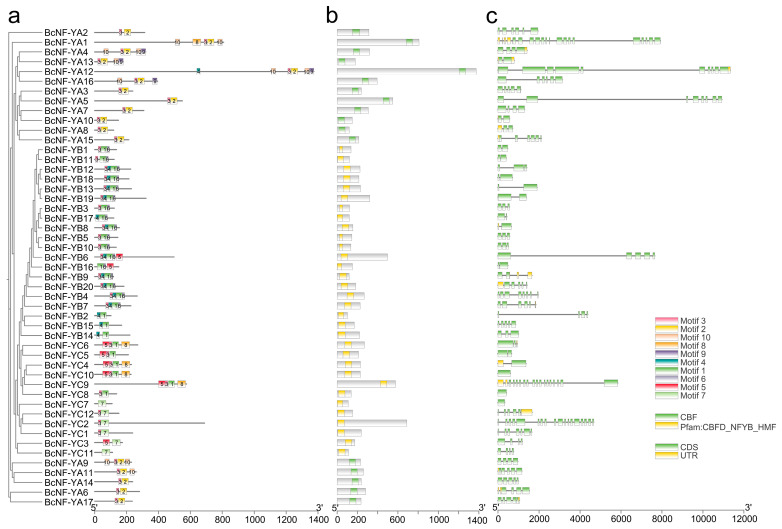
The MEME v5.4.1 software was used to identify motifs and analyze the motif composition of BcNF-Y proteins. The analysis revealed that members within the same subfamily exhibited similar conserved motifs (Figure 1a). In the BcNF-Y family, the BcNF-YA subfamily possessed a unique CBF domain, while the BcNF-YB and BcNF-YC subfamilies shared the CBFD_NFYB_HMF domain (Figure 1b). The presence of the same domain in BcNF-YB and BcNF-YC suggests functional similarities between these subfamilies. All members of the BcNF-YA subfamily contained motifs 2 and 3, with motif 2 exclusively present in the BcNF-YA subfamily, suggesting its potential role as a constituent of the CBF domain (Figure 1a). The BcNF-YB subfamily consistently harbored motif 1, except for BcNF-YB2, BcNF-YB14, and BcNF-YB15, which also contain motif 6. In contrast, the BcNF-YC subfamily exhibited a unique motif, motif 7 or 1 (Figure 1a and Figure S1).
Figure 1.
Schematic representation of protein and gene structure. (a) Distribution of conserved motifs in BcNF-Y proteins; different colors represent the 10 conserved domains identified. (b) Each domain is visually depicted as a colored box. (c) Gene structure of BcNF-Y genes.
Notably, significant differences in the number of introns were observed among the members of the BcNF-Y family in flowering Chinese cabbage. Moreover, while BcNF-YC7 and BcNF-YC8 lacked introns, BcNF-YA1 had the highest number of introns (n, 19). Most members had a substantial number of introns (Figure 1c, Table S1). These findings suggest that the NF-Y family may exhibit diverse regulatory patterns and functional roles in flowering Chinese cabbage.
2.3. Phylogenetic Analysis and Chromosomal Distribution
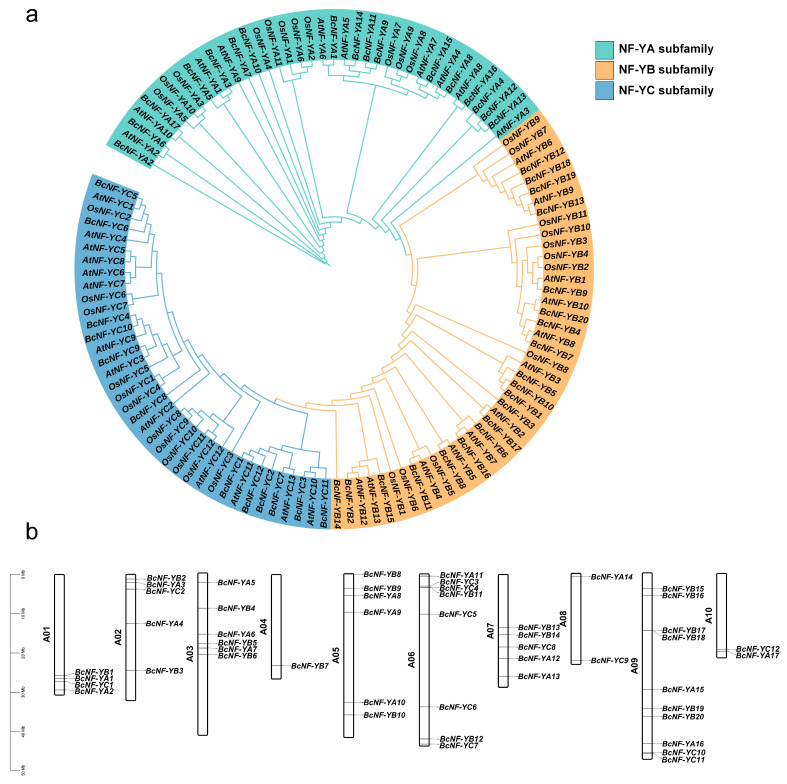
To classify the BcNF-Y family members into distinct subfamilies, a neighbor-joining phylogenetic tree was constructed using the rice and Arabidopsis NF-Y protein sequences (Table S2). The phylogenetic analysis helped us predict the function of BcNF-Y by examining the relationship between NY-F genes from different species. The results showed three BcNF-Y subfamilies. NF-YA exhibited noticeable differences from the other two subfamilies, while NF-YB and NF-YC showed closer evolutionary relationships (Figure 2a).
Figure 2.
Phylogenetic analysis of NF-Y proteins in Brassica campestris, rice, Arabidopsis, and chromosomal localization of BcNF-Y. (a) Unrooted neighbor-joining phylogenetic tree of NF-Y in flowering Chinese cabbage, Arabidopsis, and rice. The phylogenetic tree is divided into three subfamilies, and different colors are used to distinguish members of each subfamily. (b) Chromosomal distribution of 49 BcNF-Y genes in flowering Chinese cabbage.
The chromosomal localization of the 49 BcNF-Y genes in flowering Chinese cabbage was visualized using TBtools software (V1.0987). Based on their positions on chromosomes 1–10 (from top to bottom) and their classification into subfamilies, they were designated as follows: NF-YA subfamily (BcNF-YA1 to BcNF-YA17), NF-YB subfamily (BcNF-YB1 to BcNF-YB20), and NF-YC subfamily (BcNF-YC1 to BcNF-YC12) (Figure 2b). The findings revealed an uneven distribution of BcNF-Y genes on the chromosomes, with chromosome A04 containing only one BcNF-Y gene and chromosome A09 harboring 10 BcNF-Y genes.
2.4. Gene Duplication and Collinearity Analysis
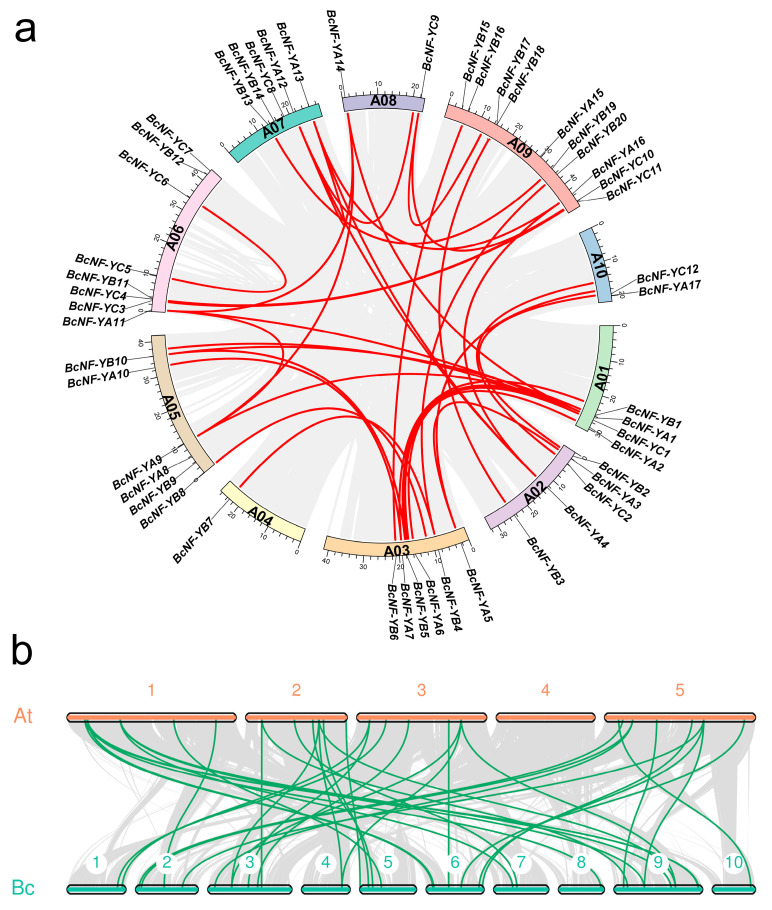
Analyzing the collinearity between genes provides valuable insights into gene family expansion and duplication events. In the case of BcNF-Y genes, collinearity analysis revealed 36 duplicated events within the gene family, with chromosome 3 showing the highest number of duplication events (Figure 3a). Among these duplicates, only BcNF-YC5 and BcNF-YC6 occurred on the same chromosome, while the rest occurred on different chromosomes (Figure 3a). The expansion of the BcNF-Y gene family is primarily attributed to segmental duplications.
Figure 3.
Gene duplication and collinearity analysis in Brassica campestris. (a) Duplication analysis of BcNF-Y genes on different chromosomes. The red lines connect BcNF-Y genes with a collinearity relationship. (b) Collinearity analysis of NF-Ys from Brassica campestris and Arabidopsis. The green lines indicate the presence of a collinearity relationship between NF-Y genes.
Collinearity between genes often indicates the presence of homologous sequences that may share similar functions. By examining the collinearity between BcNF-Y and AtNF-Y genes and performing BLASTP and phylogenetic comparisons, the homologous genes of BcNF-Y in Arabidopsis were identified. The results showed that all 43 BcNF-Y genes have homologs in Arabidopsis. Specifically, BcNF-YB4, BcNF-YB7, and BcNF-YB20 correspond to two homologous genes (AtNF-YB8/10) in Arabidopsis, while BcNF-YA11 corresponds to two homologous genes (AtNF-YA5/6) (Figure 3b, Table 1). Understanding the homology between BcNF-Y and AtNF-Y contributes to better comprehending the function of BcNF-Y genes, considering the extensive studies conducted on Arabidopsis AtNF-Y genes.
2.5. Analysis of BcNF-Y Gene Expression in Different Periods and Tissues
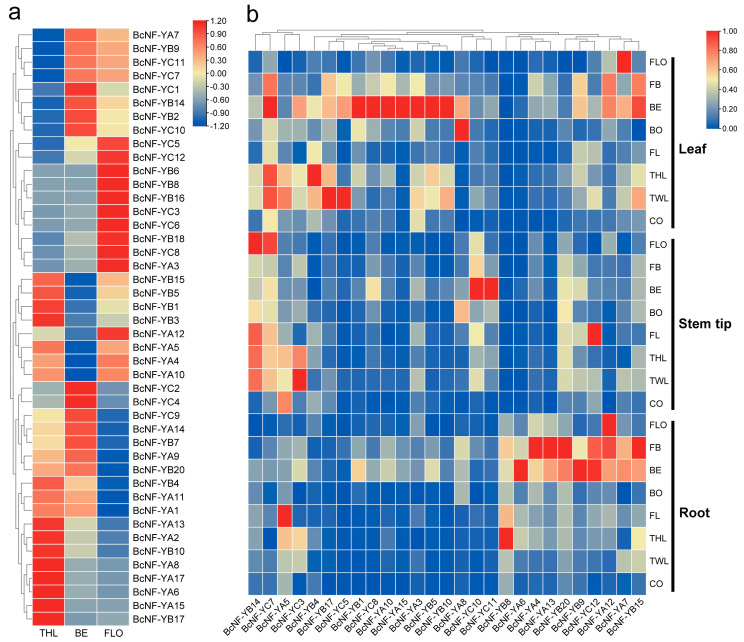
To explore the expression patterns of BcNF-Y genes throughout the growth and development of flowering Chinese cabbage, transcriptome analysis was conducted using data from various developmental stages, including three leaf, bud emergence, and flowering stages [2]. The results showed diverse expression patterns of BcNF-Y genes during the developmental stages of flowering Chinese cabbage (Figure 4a). Among the 18 BcNF-Y genes analyzed (11 BcNF-YA and 7 BcNF-YB), their expression levels were upregulated during the three-leaf stage. Additionally, 15 BcNF-Y genes (3 BcNF-YA, 5 BcNF-YB, and 7 BcNF-YC) showed increased expression levels during the bud emergence stage, while 11 BcNF-Y genes (2 BcNF-YA, 4 BcNF-YB, and 5 BcNF-YC) exhibited elevated expression levels during the flowering stage (Figure 4a). Specifically, BcNF-YA genes exhibited high expression during the seedling stage, while BcNF-YC genes demonstrated higher expression levels, specifically during flower stalk development. These findings indicate that BcNF-YA may play a more prominent role during the seedling stage, while BcNF-YC appears to be closely associated with stalk and flower development.
Figure 4.
Tissue-specific expressions of BcNF-Y genes in flowering Chinese cabbage. (a) Heatmap construction based on the transcripts per million (TPM) values. (b) Heatmap of BcNF-Y gene expression patterns in different periods and tissues. CO, cotyledon stage; TWL, two-leaf stage; THL, three-leaf stage; FL, four-leaf stage; BO, bolting stage; BE, bud emergence stage; FB, fast bolting stage; FLO, flowering stage.
Differential gene expression across different developmental stages and in different tissues often indicates their distinct functions. To gain further insight into the role of NF-Y in the growth and development of flowering Chinese cabbage and identify potential BcNF-Y genes associated with bolting and flowering, qRT-PCR analysis was performed on 27 BcNF-Y genes across eight stages (cotyledon, first leaf, second leaf, third leaf, fourth leaf, bolting, bud emergence, rapid bolting, and flowering) and three tissues (root, stem tip, and leaf). The findings revealed that during bud emergence and fast bolting, 18 genes were upregulated in specific tissues (Figure 4b). In the bud emergence, two BcNF-Y genes exhibited elevated expression in the stem apex, while seven genes showed increased expression in the leaves. Additionally, nine genes were upregulated in the root during bud emergence or rapid bolting stage. Notably, BcNF-YA showed higher expression in the leaves and roots than in the stem apex. Among the genes highly expressed in the stem apex, five of the six BcNF-Y genes belonged to the BcNF-YC subfamily, whereas the BcNF-YB subfamily members exhibited no specific expression trends (Figure 4b). These findings indicate that the BcNF-YC subfamily may play a crucial role in the bolting process of flowering Chinese cabbage, while BcNF-YA may primarily contribute to leaf and root development.
Overall, the findings indicate that a significant number of BcNF-Y genes were highly expressed during the critical stages of flowering and bolting, suggesting their key roles in the development of flowering Chinese cabbage.
2.6. BcNF-Y Gene Expression Analysis in Flowering Chinese Cabbage under Different Exogenous Hormone Spraying Conditions
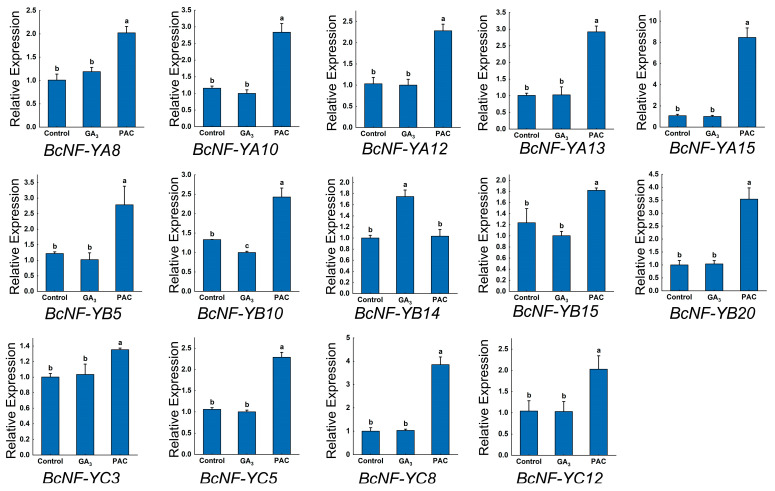
To investigate the influence of the NF-Y gene family on GA-mediated bolting of flowering Chinese cabbage, we analyzed the transcript levels of NF-Y gene family members following exogenous spraying with GA3 and PAC (a GA inhibitor). After PAC treatment, substantial upregulation in transcript levels was observed for all members of the NF-YA and NF-YC subfamilies. Conversely, no discernible response to the GA treatment was observed among the members (Figure 5).
Figure 5.
Effects of GA3 and PAC treatments on BcNF-Ys expression in flowering Chinese cabbage. Data represent the mean ± standard error for three biological experiments. Different letters (a, b, and c) indicate significant differences (p < 0.05).
NF-YB subfamily members exhibited distinct expression patterns (Figure 5). BcNF-YB5, BcNF-YB15, and BcNF-YB20 exhibited significantly elevated transcript levels after PAC treatment, whereas their expression levels were unaffected by GA treatment. However, BcNF-YB14 showed the opposite trend, with significantly increased expression after GA treatment and no change in expression after PAC treatment (Figure 5). BcNF-YB10 responded to GA3 and PAC treatments, with GA3 treatment significantly reducing its expression levels and PAC treatment significantly increasing its transcript levels (Figure 5).
2.7. Interactions of DELLAs with NF-Y Subunits and Inter-Subunit Interactions
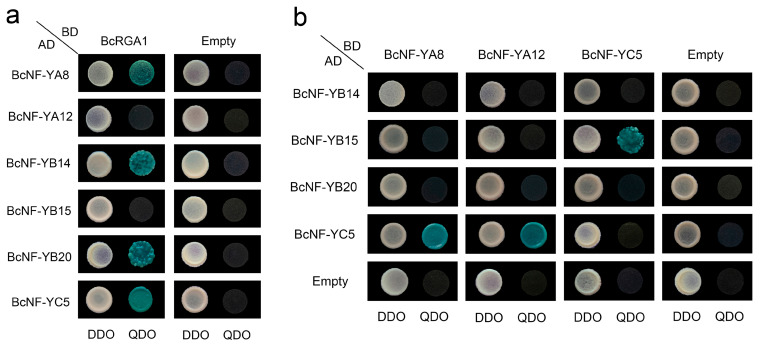
DELLA proteins have crucial regulatory functions in the GA signaling pathway and are essential for plant development. To determine the interactions between the BcNF-Y and BcDELLA proteins in flowering Chinese cabbage, yeast two-hybrid experiments were conducted. The experiment revealed that BcNF-YA8, BcNF-YB14, BcNF-YB20, and BcNF-YC5 interacted with BcRGA1, while BcNF-YA12 and BcNF-YB15 did not interact with BcRGA1 (Figure 6a).
Figure 6.
Protein interaction determination using yeast two-hybrid. (a) Yeast two-hybrid assay was employed to investigate protein interactions between BcNF-Ys and BcRGA1. (b) Interactions among NF-Y subfamily members at the protein. DDO and QDO represent SD/-Trp-Leu medium and SD/-Trp-His-Leu-Ade medium, respectively. Positive bacteria were stained using X-α-Gal. The combinations containing AD-empty or BD-empty were used as negative controls.
To determine the interactions among BcNF-Y subfamily members in flowering Chinese cabbage, yeast two-hybrid experiments were conducted. The results showed that BcNF-YA8 and BcNF-YA12 did not interact with BcNF-YB14, BcNF-YB15, or BcNF-YB20 (Figure 6b). However, BcNF-YC5, BcNF-YC5, and BcNF-YC5 interacted with BcNF-YA8, BcNF-YA12, and BcNF-YB15, respectively, suggesting their potential involvement in complex formation in flowering Chinese cabbage.
2.8. Subcellular Localization and Transcriptional Activation Analysis of NF-Y Genes
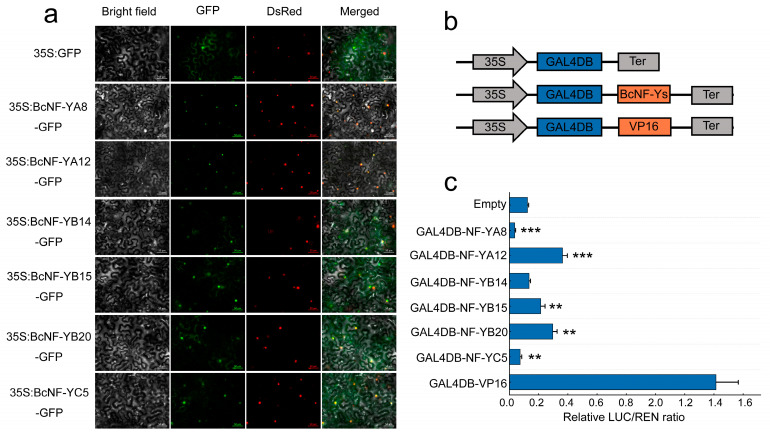
To further elucidate the functions of the BcNF-Y family, selected NF-Y genes were analyzed for their subcellular localization. The results show that BcNF-YA8-GFP and BcNF-YA12-GFP fluorescence was exclusively observed in the nucleus, indicating their localization within the nucleus (Figure 7a). In contrast, the BcNF-YB14-GFP, BcNF-YB20-GFP, BcNF-YC5-GFP, and BcNF-YB15-GFP fusion proteins exhibited fluorescence in the cell membrane and nucleus, indicating that these six NF-Y gene family members were localized in the cell membrane and nucleus of flowering Chinese cabbage (Figure 7a).
Figure 7.
Subcellular localization and transcriptional activation analysis. (a) Localization of BcNF-Ys within the cells of Nicotiana benthamiana was examined. (b) Schematic illustration for transcriptional activity assay. (c) The relative luciferase (LUC) activities observed in Nicotiana benthamiana leaves indicate the transcriptional activation potential of BcNF-Y proteins. ‘**’ indicates p-value < 0.01, and ‘***’ indicates p-value < 0.001, by one-way ANOVA analysis with Tukey’s HSD test.
Selected members were subjected to transcriptional activity analysis to assess the transcriptional activation abilities of BcNF-Y gene family members. The coding sequence (CDS) of BcNF-Y was fused to the GAL4 DNA-binding domain (GAL4DB) to generate an effector construct, while an empty vector and a VP16 served as the negative and positive controls, respectively (Figure 7b). The results showed that GAL4DB-BcNF-YB15, GAL4DB-BcNF-YB20, and GAL4DB-BcNF-YA12 exhibited significantly enhanced relative luciferase activity than in the negative control, indicating that GAL4DB-BcNF-YB15, GAL4DB-BcNF-YB20, and GAL4DB-BcNF-YA12 may function as transcriptional activators in flowering Chinese cabbage (Figure 7c). Conversely, GAL4DB-BcNF-YA8 and GAL4DB-BcNF-YC5 exhibited significantly decreased relative luciferase activities, suggesting that GAL4DB-BcNF-YA8 and GAL4DB-BcNF-YC5 may function as transcriptional repressors (Figure 7c). GAL4DB-BcNF-YB14 exhibited no significant difference in relative luciferase activity, indicating that the full-length BcNF-YB14 protein cannot regulate transcription.
3. Discussion
The NF-Y transcription factor family plays a crucial role in various plant growth and development stages, and extensive research has been conducted on various plant species [32,33,34,35]. In this study, we identified 49 BcNF-Y genes, consisting of 17 BcNF-A, 20 BcNF-YB, and 12 BcNF-YC genes. Furthermore, investigation of the expression patterns of these BcNF-Y genes across distinct developmental stages and tissues of flowering Chinese cabbage provided insight into their potential roles in plant development. Additionally, specific NF-Y proteins that interacted with BcRGA1 were identified, and subsequent subcellular localization and transcriptional activation assays were used to elucidate their functional properties. These comprehensive findings of this study lay the groundwork for further functional validation of BcNF-Y genes and the identification of key regulators involved in developing flower stalks in flowering Chinese cabbage.
Compared to Arabidopsis, a closely related cruciferous family member, flowering Chinese cabbage exhibited an expansion of the NF-Y gene family with the addition of 13 members [36]. The identification of fragment duplication events supports this expansion. Each subfamily exhibited distinct structural features, conserved domains, and motif compositions, indicating the evolutionary conservation of NF-Y genes in flowering Chinese cabbage. Introns play a critical role in splicing processes [37]. Alternative splicing enables a single gene to generate different mature or messenger RNAs, thereby expanding the proteome of an organism [38]. Furthermore, the number of introns in BcNF-Y family members was higher than in other plant species, such as alfalfa, castor, and peach. This suggests a potentially significant regulatory pattern and functional role for BcNF-Y genes in flowering Chinese cabbage [34,35,39].
NF-Y factors typically form heterotrimeric complexes that recognize and bind to CCAAT sequences to regulate transcription [40,41]. In the case of NF-YB and NF-YC, they form dimers in the cytoplasm and then translocate to the nucleus, where they recruit NF-YA to assemble a functional heterotrimeric complex [9]. There was no interaction between BcNF-YA and BcNF-YB in the yeast two-hybrid system, which is consistent with observations in Arabidopsis [42]. The complexity of NF-Y subunit interactions allows for better targeting of CCAAT boxes and fine-tuning of gene regulation [36].
In flowering Chinese cabbage, 18 BcNF-Y genes were found to be significantly upregulated during the bud emergence and fast-bolting stages, indicating their potential involvement in flowering stalk development. In Arabidopsis, AtNF-YC3, AtNF-YC4, and AtNF-YC9 are important factors in the CO-mediated flowering pathway and can redundantly regulate GA- and ABA-mediated seed germination [29]. Similarly, AtNF-YA2, AtNF-YB2, and AtNF-YC9 can bind to NFYBE, leading to the activation of SOC1 transcription and regulation of flowering in Arabidopsis [28]. Moreover, silencing the rose homologs of AtNF-YC9 and RhNF-YC9 resulted in reduced GA accumulation and significant downregulation of genes involved in cell expansion, inhibiting petal expansion [30]. In flowering Chinese cabbage, BcNF-YC10 is a homolog of AtNF-YC9 and exhibits significant upregulation in the stem tip during the bud emergence stage. Based on this observation, we hypothesized that BcNF-YC10 is an important candidate gene involved in developing flowering Chinese cabbage flower stalks. In Arabidopsis, overexpression of AtNF-YA2 leads to a delay in flowering, and miR169 regulates the stress-mediated flowering process and lateral root development by regulating AtNF-YA2 [43]. In flowering Chinese cabbage, BcNF-YA6 is a homolog of AtNF-YA2, exhibiting significantly high expression in roots at the bud emergence stage. This suggests that BcNF-YA6 may play a functional role comparable to AtNF-YA2. Therefore, we hypothesized that BcNF-YA6 is crucial in flowering and root development in flowering Chinese cabbage. Further studies on the overexpression and knockdown of BcNF-Ys could help elucidate their roles in flowering Chinese cabbage bolting and flowering.
GA is a well-known regulator of flowering and bolting in flowering Chinese cabbage. Treating with exogenous GA3 can help accelerate bolting and flowering processes while promoting stem elongation [44]. Although most BcNF-Y family members showed increased expression following treatment with PAC (a GA synthesis inhibitor), their responses to GA treatment were insignificant. However, BcNF-YB14 responded to GA and PAC, with its expression significantly upregulated in the leaves during the bolting stage. This suggests that BcNF-YB14 may be involved in GA-mediated bolting in flowering Chinese cabbage plants. In flowering Chinese cabbage, DELLA proteins function as negative regulators of the GA signaling pathway, influencing the bolting and flowering processes. One such DELLA protein in flowering Chinese cabbage is BcRGA1 [4]. Using yeast two-hybrid analysis, the interactions between BcRGA1 (a DELLA protein) and specific BcNF-Y proteins, including BcNF-YA8, BcNF-YB14, BcNF-YB20, and BcNF-YC5 were identified. During the bolting stage, BcNF-YA8 was significantly expressed in the leaves, while BcNF-YB14 was highly expressed in stem tips during the flowering stage, and BcNF-YB20 was highly expressed in roots during the fast bolting stage. Transcriptional activity analysis revealed that BcNF-YA8, BcNF-YB20, and BcNF-YC5 have regulatory roles in downstream transcription. However, BcNF-YB14 did not exhibit transcriptional regulation, possibly because of the presence of a transcriptional repressor domain in the full-length protein. These findings suggest that these four identified BcNF-Y genes likely participate in GA-mediated flowering and bolting processes in flowering Chinese cabbage.
Transcriptional regulation mediated by NF-Y involves a complex mechanism. In Arabidopsis, CO can substitute NF-YA to form a CO/NF-YB/NF-YC trimer, subsequently binding to and regulating the FT promoter [45]. The entire heterotrimeric complex can recruit additional transcription factors, thereby modulating its binding affinity for CCAAT boxes [46,47]. Moreover, a pair of AtNF-Y protein complexes (NFY and NF-CO) are in close proximity to each other and simultaneously bind to two CCAAT boxes located >5.3 kb apart on the FT promoter, regulating flowering [25,45]. The transcriptional regulatory effects of NF-Y on downstream genes primarily rely on intact NF-Y complexes. However, the identification of intact and active NF-Y complexes remains challenging. Nevertheless, we did not identify any candidate combinations of the NF-Y complexes. Advancements in bioinformatics and more accurate protein interaction predictions can expedite the exploration of NF-Y complex compositions and facilitate functional studies of NF-Y complexes.
4. Materials and Methods
4.1. Identification of the BcNF-Y Gene Family
AtNF-Ys proteins sequence of Arabidopsis were derived from a previous report [36]. The protein sequences of AtNF-Ys were used as queries and compared to the Brassica campestris genome using BLASTP with an e-value cutoff of 1 × 10−5 and an identity threshold of >40%. On the other hand, the hidden Markov model (HMM) profile for PF02045 and PF00808 were downloaded from the Pfam database (https://www.ebi.ac.uk/interpro/, accessed on 7 October 2022), which was used to conduct another search with HMMER v3.3.1 (e-value 1 × 10−5). The non-redundant protein sequences obtained from both methods were further analyzed using the NCBI Conserved Domain (CD) tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 7 October 2022) and the SMART database (https://smart.embl.de/, accessed on 7 October 2022) for confirmation.
4.2. Analysis of Gene Structure, Domains, and Conserved Motifs
Conserved motifs were identified using the online program MEME v5.4.1 with the find motifs parameter set to 10, while other parameters were kept as default (https://meme-suite.org/meme/; accessed on 7 October 2022). The SMART database (https://smart.embl.de/, accessed on 7 October 2022) was used to identify conserved domains with the default parameters. TBtools software (V1.0987) was used to conduct the structural visualization analysis [48].
4.3. Phylogenetic Analysis
NF-Y gene sequences from Arabidopsis (AtNF-Ys) and rice (OsNF-Ys) were obtained from previous studies [36,49]. To align all NF-Y, we used the L-INS-I method of the MAFFT sequence alignment program [50]. A neighbor-joining phylogenetic tree was then constructed using MEGA11, and the reliability of the tree was assessed using a bootstrap test with 1000 replicates [51].
4.4. Chromosomal Locations, Synteny Analysis
The chromosomal location of BcNF-Y genes was determined using TBtools software (V1.0987). To analyze the synteny of NF-Y genes, we employed MCScanX with the default parameters in TBtools (V1.0987) to perform synteny analysis of NF-Y genes was performed in Arabidopsis, rice, Brassica campestris, and within Brassica campestris [52]. Homologous genes between Brassica campestris and Arabidopsis thaliana were identified, and synteny analysis was conducted. The results of the analysis were visualized using TBtools.
4.5. Plant Materials and Treatments
The “youIv501” variety of flowering Chinese cabbage plants was cultivated in a daylight greenhouse located at the Department of Facility Horticulture, South China Agricultural University, using a substrate potting technique. To initiate the experiment, the seeds were sterilized and placed on Petri dishes containing a moist filter for 1 d. Following germination, the seeds were transferred to cavity trays and then transplanted into seedling pots filled with a substrate mixture composed of peat, vermiculite, and perlite (in a ratio of 3:1:1) when the seedlings reached the three-leaf stage. Tissue samples were collected at various seedling developmental stages of the seedlings, including the cotyledon, two-leaf, three-leaf, four-leaf, bolting, bud emergence, fast bolting, and flowering stages. Stem tips, roots, and leaf tissues were collected at each stage. For the exogenous hormone treatment, GA3 (200 mg/L) and PAC (GA synthesis inhibitor) (10 mg/L) were sprayed onto the seedlings at the three-leaf stage. Stem tip samples were collected 12 h after the hormone treatment. Each treatment was replicated three times with 20 seedlings per treatment. All collected samples were stored at −80 °C and later used for RNA extraction.
4.6. RNA Extraction and qRT-PCR
Total RNA was extracted from the sample using the Eastep® Super Total RNA Extraction Kit, and genomic DNA was removed. The cDNAs were prepared using Hiscript QRT SuperMix (Vazyme, Nanjing, China). The real-time PCR (qRT-PCR) were performed using the ChamQ SYBR Color qPCR Master Mix (Vazyme, Nanjing, China). Primers were designed using the NCBI BLAST tool with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal reference gene. The relative expression levels were determined using the 2−∆∆Ct method. qRT-PCR was conducted following the previously described protocol [44]. Table S3 lists the primers used for the qRT-PCR analyses.
4.7. Yeast Two-Hybrid Assay
Full-length coding sequences (CDS) of BcNF-Ys and BcRGA1 genes were cloned into pGADT7 and pGBKT7 vectors, respectively. The individual vectors were co-transformed into yeast cells using the Y2HGold Competent Cells (WEIDI, Shanghai, China). The co-transformed yeast cells were then plated on a selective medium lacking leucine and tryptophan (SD/-Leu/-Trp) to ensure the presence of both plasmids. The yeast transformants were then plated on selective media lacking leucine, tryptophan, histidine, and adenine (SD/-Trp/-Leu/-His/-Ade) to assess protein interactions. X-α-Gal medium was used to visualize and confirm the interactions, enabling the identification of blue and white colonies. Table S4 shows all yeast two-hybrid primers.
4.8. DLR Assay
The transient transcriptional activity assay was conducted on Nicotiana benthamiana leaves using a previously described method [53,54]. The full-length coding sequences (CDS) of BcNF-Ys genes were cloned into the GAL4DB vector to generate the GAL4DB-BcNF-Ys vector. For the experimental setup, GAL4DB-VP16 was used as the positive control, GAL4DB-Empty as the negative control, and GAL4DB-BcNF-Ys as the test vector. Agrobacterium tumefaciens strains harboring different plasmids were infiltrated into tobacco leaves, followed by a 48 h incubation period. Determination the luciferase/Renilla luciferase (Luc/Ren) ratio, enabling the analysis of transcription-al activity [6]. Table S5 lists all DLR assay primers.
4.9. Subcellular Localization
The full-length CDS sequence of BcNF-Ys without the stop codon was cloned into the pSUPER1300 vector. The pSUPER1300 empty vector was used as a control. The recombinant vectors and a localization signal (DsRed) were introduced into Agrobacterium tumefaciens GV3101 and co-injected into tobacco leaves. After two days, the fluorescence signal was observed using laser scanning confocal microscopy (Axioimager.D2). Table S6 lists all subcellular localization primers.
Supplementary Materials
The supporting information can be downloaded from: https://www.mdpi.com/article/10.3390/ijms241511898/s1.
Author Contributions
Formal analysis, Z.J. and W.L.; investigation, Z.J., Y.W. (Yuting Wang), Y.W. (Yudan Wang) and X.O.; writing—original draft preparation, Z.J.; writing—review and editing, Y.W. (Yuting Wang) and X.L.; methodology, R.C., S.S. and W.S.; visualization, Z.J. and W.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All important data are included in the article and supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was funded by the National Natural Science Foundation of China (32072656, 31972481) and the China Agriculture Research System.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Song S., Lei Y., Huang X., Su W., Chen R., Hao Y. Crosstalk of Cold and Gibberellin Effects on Bolting and Flowering in Flowering Chinese Cabbage. J. Integr. Agric. 2019;18:992–1000. doi: 10.1016/S2095-3119(18)62063-5. [DOI] [Google Scholar]
- 2.Huang X., Lei Y., Guan H., Hao Y., Liu H., Sun G., Chen R., Song S. Transcriptomic Analysis of the Regulation of Stalk Development in Flowering Chinese Cabbage (Brassica campestris) by RNA Sequencing. Sci. Rep. 2017;7:15517. doi: 10.1038/s41598-017-15699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kou E., Huang X., Zhu Y., Su W., Liu H., Sun G., Chen R., Hao Y., Song S. Crosstalk between Auxin and Gibberellin during Stalk Elongation in Flowering Chinese Cabbage. Sci. Rep. 2021;11:3976. doi: 10.1038/s41598-021-83519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan H., Huang X., Zhu Y., Xie B., Liu H., Song S., Hao Y., Chen R. Identification of DELLA Genes and Key Stage for GA Sensitivity in Bolting and Flowering of Flowering Chinese Cabbage. Int. J. Mol. Sci. 2021;22:12092. doi: 10.3390/ijms222212092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Huang X., Huang X., Su W., Hao Y., Liu H., Chen R., Song S. BcSOC1 Promotes Bolting and Stem Elongation in Flowering Chinese Cabbage. Int. J. Mol. Sci. 2022;23:3459. doi: 10.3390/ijms23073459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Song S., Hao Y., Chen C., Ou X., He B., Zhang J., Jiang Z., Li C., Zhang S., et al. Role of BraRGL1 in Regulation of Brassica rapa Bolting and Flowering. Hortic. Res. 2023:uhad119. doi: 10.1093/hr/uhad119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bucher P. Weight Matrix Descriptions of Four Eukaryotic RNA Polymerase II Promoter Elements Derived from 502 Unrelated Promoter Sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 8.McNabb D.S., Xing Y., Guarente L. Cloning of Yeast HAP5: A Novel Subunit of a Heterotrimeric Complex Required for CCAAT Binding. Genes Dev. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- 9.Myers Z.A. NUCLEAR FACTOR-Y: Still Complex after All These Years? Curr. Opin. Plant Biol. 2018;45:96–102. doi: 10.1016/j.pbi.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Wang J., Li G., Li C., Zhang C., Cui L., Ai G., Wang X., Larkin R.M., Ye Z., Zhang J., et al. NF-Y Plays Essential Roles in Flavonoid Biosynthesis by Modulating Histone Modifications in Tomato. New Phytol. 2020;229:3237–3252. doi: 10.1111/nph.17112. [DOI] [PubMed] [Google Scholar]
- 11.Ke X., Xiao H., Peng Y., Wang J., Lv Q., Wang X. Phosphoenolpyruvate Reallocation Links Nitrogen Fixation Rates to Root Nodule Energy State. Science. 2022;378:971–977. doi: 10.1126/science.abq8591. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y., Zhang Y., Wang X., Han X., An Y., Lin S., Shen C., Wen J., Liu C., Yin W., et al. Root-specific NF-Y Family Transcription Factor, PdNF-YB21, Positively Regulates Root Growth and Drought Resistance by Abscisic Acid-mediated Indoylacetic Acid Transport in Populus. New Phytol. 2020;227:407–426. doi: 10.1111/nph.16524. [DOI] [PubMed] [Google Scholar]
- 13.Niu B., Zhang Z., Zhang J., Zhou Y., Chen C. The Rice LEC1-Like Transcription Factor OsNF-YB9 Interacts with SPK, an Endosperm-specific Sucrose Synthase Protein Kinase, and Functions in Seed Development. Plant J. 2021;106:1233–1246. doi: 10.1111/tpj.15230. [DOI] [PubMed] [Google Scholar]
- 14.Swain S. The Multifaceted Roles of NUCLEAR FACTOR-Y in Arabidopsis thaliana Development and Stress Responses. Biophys. Acta. 2016;1860:636–644. doi: 10.1016/j.bbagrm.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Li W.-X., Oono Y., Zhu J., He X.-J., Wu J.-M., Iida K., Lu X.-Y., Cui X., Jin H., Zhu J.-K. The Arabidopsis NFYA5 Transcription Factor Is Regulated Transcriptionally and Posttranscriptionally to Promote Drought Resistance. Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato H., Mizoi J., Tanaka H., Maruyama K., Qin F., Osakabe Y., Morimoto K., Ohori T., Kusakabe K., Nagata M., et al. Arabidopsis DPB3-1, a DREB2A Interactor, Specifically Enhances Heat Stress-Induced Gene Expression by Forming a Heat Stress-Specific Transcriptional Complex with NF-Y Subunits. Plant Cell. 2014;26:4954–4973. doi: 10.1105/tpc.114.132928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu T., Liu Y., Fu J., Ma J., Fang Z., Chen J., Zheng L., Lu Z., Zhou Y., Chen M., et al. The NF−Y−PYR Module Integrates the Abscisic Acid Signal Pathway to Regulate Plant Stress Tolerance. Plant Biotechnol. J. 2021;19:2589–2605. doi: 10.1111/pbi.13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H., Ye T., Zhong B., Liu X., Jin R., Chan Z. AtHAP 5A Modulates Freezing Stress Resistance in Arabidopsis Through Binding to CCAAT Motif of AtXTH21. New Phytol. 2014;203:554–567. doi: 10.1111/nph.12812. [DOI] [PubMed] [Google Scholar]
- 19.Bello B.K., Hou Y., Zhao J., Jiao G., Wu Y., Li Z., Wang Y., Tong X., Wang W., Yuan W., et al. NF-YB1-YC12-BHLH144 Complex Directly Activates Wx to Regulate Grain Quality in Rice (Oryza sativa L.) Plant Biotechnol. J. 2019;17:1222–1235. doi: 10.1111/pbi.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W., Tang Y., Hu Y., Yang Y., Cai J., Liu H., Zhang C., Liu X., Hou X. Arabidopsis NF-YCs Play Dual Roles in Repressing Brassinosteroid Biosynthesis and Signaling during Light-Regulated Hypocotyl Elongation. Plant Cell. 2021;33:2360–2374. doi: 10.1093/plcell/koab112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., Coupland G. CONSTANS and the CCAAT Box Binding Complex Share a Functionally Important Domain and Interact to Regulate Flowering of Arabidopsis. Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumimoto R.W., Adam L., Hymus G.J., Repetti P.P., Reuber T.L., Marion C.M., Hempel F.D., Ratcliffe O.J. The Nuclear Factor Y Subunits NF-YB2 and NF-YB3 Play Additive Roles in the Promotion of Flowering by Inductive Long-Day Photoperiods in Arabidopsis. Planta. 2008;228:709–723. doi: 10.1007/s00425-008-0773-6. [DOI] [PubMed] [Google Scholar]
- 23.Kumimoto R.W., Zhang Y., Siefers N., Holt B.F. NF-YC3, NF-YC4 and NF-YC9 Are Required for CONSTANS-Mediated, Photoperiod-Dependent Flowering in Arabidopsis thaliana. Plant J. 2010;63:379–391. doi: 10.1111/j.1365-313X.2010.04247.x. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Naim O., Eshed R., Parnis A., Teper-Bamnolker P., Shalit A., Coupland G., Samach A., Lifschitz E. The CCAAT Binding Factor Can Mediate Interactions between CONSTANS-Like Proteins and DNA. Plant J. 2006;46:462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- 25.Cao S., Kumimoto R.W., Gnesutta N., Calogero A.M., Mantovani R., Holt B.F., Distal A. A Distal CCAAT /NUCLEAR FACTOR Y Complex Promotes Chromatin Looping at the FLOWERING LOCUS T Promoter and Regulates the Timing of Flowering in Arabidopsis. Plant Cell. 2014;26:1009–1017. doi: 10.1105/tpc.113.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siriwardana C.L., Gnesutta N., Kumimoto R.W., Jones D.S., Myers Z.A., Mantovani R., Holt B.F. NUCLEAR FACTOR Y, Subunit A (NF-YA) Proteins Positively Regulate Flowering and Act through FLOWERING LOCUS T. PLOS Genet. 2016;12:e1006496. doi: 10.1371/journal.pgen.1006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiwari S.B., Shen Y., Chang H., Hou Y., Harris A., Ma S.F., McPartland M., Hymus G.J., Adam L., Marion C., et al. The Flowering Time Regulator CONSTANS Is Recruited to the FLOWERING LOCUS T Promoter via a Unique Cis-Element. New Phytol. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- 28.Hou X., Zhou J., Liu C., Liu L., Shen L., Yu H. Nuclear Factor Y-Mediated H3K27me3 Demethylation of the SOC1 Locus Orchestrates Flowering Responses of Arabidopsis. Nat. Commun. 2014;5:4601. doi: 10.1038/ncomms5601. [DOI] [PubMed] [Google Scholar]
- 29.Liu X., Hu P., Huang M., Tang Y., Li Y., Li L., Hou X. The NF-YC–RGL2 Module Integrates GA and ABA Signalling to Regulate Seed Germination in Arabidopsis. Nat. Commun. 2016;7:12768. doi: 10.1038/ncomms12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C., Hussain N., Wang Y., Li M., Liu L., Qin M., Ma N., Gao J., Sun X. An Ethylene-Inhibited NF-YC Transcription Factor RhNF-YC9 Regulates Petal Expansion in Rose. Hortic. Plant J. 2020;6:419–427. doi: 10.1016/j.hpj.2020.11.007. [DOI] [Google Scholar]
- 31.Zhang C., Jian M., Li W., Yao X., Tan C., Qian Q., Hu Y., Xu L., Hou X. Gibberellin Signaling Modulates Flowering via the DELLA-BRAHMA-NF-YC Module in Arabidopsis. Plant Cell. 2023:koad166. doi: 10.1093/plcell/koad166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren C., Zhang Z., Wang Y., Li S., Liang Z. Genome-Wide Identification and Characterization of the NF-Y Gene Family in Grape (Vitis vinifera L.) BMC Genom. 2016;17:605. doi: 10.1186/s12864-016-2989-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lv M., Cao H., Wang X., Zhang K., Si H., Zang J., Xing J., Dong J. Identification and Expression Analysis of Maize NF-YA Subunit Genes. PeerJ. 2022;10:e14306. doi: 10.7717/peerj.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M., Li G., Liu W., Dong X., Zhang A. Genome-Wide Analysis of the NF-Y Gene Family in Peach (Prunus persica L.) BMC Genom. 2019;20:612. doi: 10.1186/s12864-019-5968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Xu W., Chen Z., Han B., Haque M.E., Liu A. Gene Structure, Expression Pattern and Interaction of Nuclear Factor-Y Family in Castor Bean (Ricinus communis) Planta. 2018;247:559–572. doi: 10.1007/s00425-017-2809-2. [DOI] [PubMed] [Google Scholar]
- 36.Siefers N., Dang K.K., Kumimoto R.W., Bynum W.E., Tayrose G., Holt B.F. Tissue-Specific Expression Patterns of Arabidopsis NF-Y Transcription Factors Suggest Potential for Extensive Combinatorial Complexity. Plant Physiol. 2009;149:625–641. doi: 10.1104/pp.108.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrillo E. Do Not Panic: An Intron-Centric Guide to Alternative Splicing. Plant Cell. 2023;35:1752–1761. doi: 10.1093/plcell/koad009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keren H., Lev-Maor G., Ast G. Alternative Splicing and Evolution: Diversification, Exon Definition and Function. Nat. Rev. Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 39.An Y., Suo X., Niu Q., Yin S., Chen L. Genome-Wide Identification and Analysis of the NF-Y Transcription Factor Family Reveal Its Potential Roles in Salt Stress in Alfalfa (Medicago sativa L.) Int. J. Mol. Sci. 2022;23:6426. doi: 10.3390/ijms23126426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baudin M., Laloum T., Lepage A., Rípodas C., Ariel F., Frances L., Crespi M., Gamas P., Blanco F.A., Zanetti M.E., et al. A Phylogenetically Conserved Group of Nuclear Factor-Y Transcription Factors Interact to Control Nodulation in Legumes. Plant Physiol. 2015;169:2761–2773. doi: 10.1104/pp.15.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H., Fischer R.L., Goldberg R.B., Harada J.J. Arabidopsis LEAFY COTYLEDON1 Represents a Functionally Specialized Subunit of the CCAAT Binding Transcription Factor. Proc. Natl. Acad. Sci. USA. 2003;100:2152–2156. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hackenberg D., Wu Y., Voigt A., Adams R., Schramm P., Grimm B. Studies on Differential Nuclear Translocation Mechanism and Assembly of the Three Subunits of the Arabidopsis Thaliana Transcription Factor NF-Y. Mol. Plant. 2012;5:876–888. doi: 10.1093/mp/ssr107. [DOI] [PubMed] [Google Scholar]
- 43.Sorin C., Declerck M., Christ A., Blein T., Ma L., Lelandais-Brière C., Njo M.F., Beeckman T., Crespi M., Hartmann C. A MiR169 Isoform Regulates Specific NF—YA Targets and Root Architecture in a Rabidopsis. New Phytol. 2014;202:1197–1211. doi: 10.1111/nph.12735. [DOI] [PubMed] [Google Scholar]
- 44.Ou X., Wang Y., Zhang J., Xie Z., He B., Jiang Z., Wang Y., Su W., Song S., Hao Y., et al. Identification of BcARR Genes and CTK Effects on Stalk Development of Flowering Chinese Cabbage. Int. J. Mol. Sci. 2022;23:7412. doi: 10.3390/ijms23137412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gnesutta N., Kumimoto R.W., Swain S., Chiara M., Siriwardana C., Horner D.S., Holt B.F., Mantovani R. CONSTANS Imparts DNA Sequence Specificity to the Histone Fold NF-YB/NF-YC Dimer. Plant Cell. 2017;29:1516–1532. doi: 10.1105/tpc.16.00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang M., Hu Y., Liu X., Li Y., Hou X. Arabidopsis LEAFY COTYLEDON1 Mediates Postembryonic Development via Interacting with PHYTOCHROME-INTERACTING FACTOR4. Plant Cell. 2015;27:3099–3111. doi: 10.1105/tpc.15.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J.-X., Howell S.H. BZIP28 and NF-Y Transcription Factors Are Activated by ER Stress and Assemble into a Transcriptional Complex to Regulate Stress Response Genes in Arabidopsis. Plant Cell. 2010;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 49.Thirumurugan T., Ito Y., Kubo T., Serizawa A., Kurata N. Identification, Characterization and Interaction of HAP Family Genes in Rice. Mol. Genet. Genom. 2008;279:279–289. doi: 10.1007/s00438-007-0312-3. [DOI] [PubMed] [Google Scholar]
- 50.Katoh K., Standley D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K., Stecher G., Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.-H., Jin H., Marler B., Guo H., et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Y., Zhao X., Aiwaili P., Mu X., Zhao M., Zhao J., Cheng L., Ma C., Gao J., Hong B. A Zinc Finger Protein BBX19 Interacts with ABF3 to Affect Drought Tolerance Negatively in Chrysanthemum. Plant J. 2020;103:1783–1795. doi: 10.1111/tpj.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. Repression Domains of Class II ERF Transcriptional Repressors Share an Essential Motif for Active Repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All important data are included in the article and supplementary material.