Abstract
Utilizing genetic modules of simple retroviruses, we have developed a novel generation of gene transfer vectors with improved therapeutic potential. In the 5′ untranslated “leader” sequences, all AUG codons which may aberrantly initiate translation and all viral coding sequences were removed. Thus, the probability of expressing unwanted peptides and the potential for homologous recombination with retroviral genes were largely reduced, and the cloning capacity was increased. The transgene was inserted to replace the viral gag sequences, and a new minimal splice acceptor was introduced, resulting in increased expression with all genes tested (those coding for human multidrug resistance 1 and enhanced green fluorescent protein, as well as the lacZ gene). These vectors may represent attractive tools for human gene therapy, because they increase the efficiency of transgene expression and may also increase safety in medical applications.
For reasons of efficiency and safety, vectors based on simple retroviruses such as murine leukemia viruses represent the “gold standard” for stable gene transfer in mammalian cells. Therefore, they have found widespread use not only in molecular and cell biology, but also in human gene therapy (4, 20, 21). Retroviruses replicate via a genomic RNA intermediate that is incorporated as a dimer in retroviral particles, transferred to target cells, reverse transcribed into proviral DNA, and then stably inserted into chromosomes (11). The packaging and dimerization domain (Ψ) required for the incorporation of genomic RNA in viral particles is found in the 5′ untranslated region (leader). Ψ sequences generate a complex structure recognized by nucleocapsid Gag proteins in the assembly of the retroviral particle (7). Ψ is located in between the retroviral splice donor and the viral gag-pol fusion gene. Inclusion of the 400-bp 5′ coding sequences of gag may elevate infectious vector titers harvested from safety-modified retroviral packaging cells releasing replication-incompetent vectors (2, 6). Vectors containing this fragment were denoted gag+ (G+) vectors.
A second and independent function of the leader is to direct translation of genes expressed from the promoter located in the U3 region of the long terminal repeat (LTR). Gag-Pol proteins are translated from the unspliced leader, including Ψ, while Env is translated from a spliced subgenomic RNA (11). Splicing efficiency is controlled by the interaction of the splice donor with the branch site and the splice acceptor located upstream of env. While the spliced RNA is translated by a cap-dependent ribosomal scanning mechanism, the unspliced genomic message may also be translated by cap-independent internal ribosome entry (8), followed by scanning for an appropriate AUG start codon. In either case, the translation efficiency of the gene of interest is dependent on the context of its start codon (homology to Kozak consensus, no aberrant start codon located upstream, and no interference with RNA folding). Thus, a conflict arises between the optimal structure for high-titer packaging (long leader containing the folded Ψ region) and the requirements for optimal gene translation. This dual and partially conflicting function of the leader has impeded the development of efficient vectors that fulfill the rigorous safety criteria relevant to human gene therapy.
All vectors used so far in somatic gene transfer contain the long G+ leader comprising at least 1,000 bp. A widely used version is the LX vector (19). An alternative vector architecture that has attracted great attention is MFG (26). This is a G+ vector with additional 400 bp of the 3′ end of the retroviral pol gene (P+), including the retroviral branch site and splice acceptor for the env message. (We refer to this assembly as G+P+.) Here, a proportion of the vector message is spliced to a subgenomic RNA, leading to enhanced expression in the case of some, but not all, of the transgenes of interest (17). The G+ fragment contains a weaker splice acceptor (2, 6), which can give rise to a subgenomic RNA with improved translation efficacy (Fig. 1).
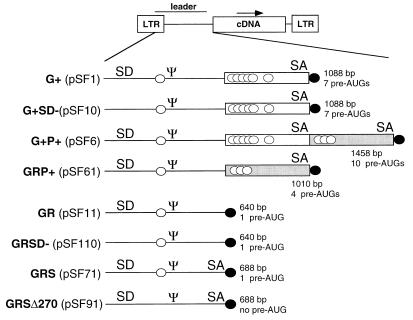
FIG. 1.
Design of leader sequences. A schematic representation of the 5′ untranslated sequences used in retroviral vectors containing viral gene fragments (first set) and the novel design analyzed in this study (second set) is shown. Ψ, packaging signal; SD, splice donor; SA, splice acceptor. Open boxes indicate the viral gag gene fragment (G+), and shaded boxes indicate the viral pol gene fragment (P+). Potential AUG start codons preceding the cDNA are indicated by open circles, and the AUG of the inserted cDNA is indicated by a solid circle. G+SD−, G+ vector with destroyed splice donor; GRP+, gag replacement with P+ present; GR, gag replacement; GRSD−, GR vector with destroyed splice donor; GRS, gag replacement with SA oligonucleotide. GRSΔ270, GRS leader with destroyed AUG at position +270.
Both G+ and G+P+ vectors still express significant amounts of the unspliced message (17), which are also subject to ribosome scanning. Therefore, it was disturbing to note a number of potential AUG start codons in the unspliced leader of G+ or G+P+ vectors. While the start codon of Gag had been mutated in both vectors, 7 additional AUGs remain in the leader of G+ and even 10 AUGs remain in G+P+ vectors (Fig. 1 and Table 1). These aberrant AUGs may initiate translation of polypeptides with unknown functions. This creates a potential hazard in terms of toxicity or immunogenicity in human gene therapy.
TABLE 1.
Potential aberrant AUGs and encoded peptides in conventional vectors
| AUG positiona | Kozak match [(A/G)CC-AUG-G] | Length of ORFb | Peptide sequence |
|---|---|---|---|
| 270 | CUA-AUG-U (poor) | 10 | MFAPASVLVG |
| 680 | AAG-AUG-U (poor) | 10 | MSSGSLTTSR |
| 713 | UAG-AUG-U (poor) | 65 | MSRRDDGLPSALQNGQPLTSDGRETAPLTETSSPRLRSRSFHLARMDTQTRSPTS |
| 729 | ACG-AUG-G (good) | 12 | MGYLLLCRMANL |
| 753 | AGA-AUG-G (partial) | 4 | MANL |
| 774 | CGG-AUG-G (partial) | 7 | MAARRHL |
| 848 | CGC-AUG-G (good) | 10 | MDTQTRSPTS |
| 1077c | CAU-AUG-A (poor) | 11 | MRSYMGHPRPL |
| 1089c | UAU-AUG-G (partial) | 7 | MGHPRPL |
| 1130c | GAC-AUG-A (partial) | >110 | MTRVTNSPSLQAHLQALYLVGHEVWRPLAAAYQEQLDRPVVPHPYRVGDTVWVRRHQTKNLEPRWKGPYTVLLTTPTALKVDGIAAWIHAAHVKAADPGGGPSSRLRPP (continued in inserted cDNA) |
The second safety concern associated with remnants of viral genes in vectors designed for medical purposes is the potential for homologous recombination, or template switching during reverse transcription, with related viral sequences endogenous to the packaging cell or to the target cell. This may promote the generation of novel retroviruses, including replication-competent forms, with potential pathogenicity (1, 22). Therefore, there is a great demand for the design of vectors that avoid these hazards and that simultaneously show efficient and specific translation of the transgenes of choice for complete penetrance of the desired phenotype. Here, we describe the development of a novel type of retroviral leader that removes all recombinogenic viral coding sequences and all aberrant AUG initiation codons and generates an artificial intron in order to exclude translational silencing (18), thus improving transgene expression.
MATERIALS AND METHODS
Vector constructions.
All vectors used here are based on pSF1 (GenBank accession no. AJ224005) with the 3′ LTR of spleen focus-forming virus (SFFVp) and the leader of the murine embryonic stem cell virus (MESV), which is based on dl587rev (12, 15). The variations of the leader sequence analyzed in this study are schematically shown in Fig. 1.
(i) G+ vectors.
The G+ vectors contain remnants of the gag gene and thus represent the leader found in pSF1. pSF1m with the human multidrug resistance 1 (MDR1) cDNA, pSF1L with a lacZ-Neor fusion gene, and pSF1γ with the eGFP cDNA have been described previously (14, 15). For construction of pSF1K, containing a lacZ-Neor fusion gene with a Kozak-matching start codon, PCR was performed with pSF1L as template, forward primer 5′-GCG-GCC-GCC-ATG-TCG-TTT-ACT-TTG-ACC-3′, and reverse primer 5′-CCC-CAG-CGA-CCA-GAT-GAT-CAC-ACT-CGG-G-3′. Thirty cycles (1 min at 95°C, 1 min at 55°C, and 5 min at 72°C) were run by using 2.5 U of Pfu DNA polymerase (Stratagene GmbH, Heidelberg, Germany). The PCR product was subcloned, digested (NotI and BclI), and ligated into pSF1 (opened with NotI and BamHI) together with the BclI-BamHI fragment of pSF1L.
(ii) G+SD− vectors.
The G+SD− vectors are splice donor-defective versions of G+ vectors. The basic plasmid of G+SD− vectors was designated pSF10. The splice donor was destroyed by site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene GmbH) according to the manufacturer’s instructions with pSF1 as a template, forward primer 5′-ACC-GAC-CCC-CCC-GCC-GGG-AAC-TAA-GCT-GGC-CAG-CGG-TCG-TT-3′, and reverse primer 5′-AAC-GAC-CGC-TGG-CCA-GCT-TAG-TTC-CCG-GCG-GGG-GGG-TCG- GT-3′ at an annealing temperature of 55°C.
(iii) G+P+ vectors.
The G+P+ vectors are similar to MFG vectors and contain remnants of both gag and pol. The basic plasmid was designated pSF6. The P+ fragment was generated by PCR with Moloney murine leukemia virus cDNA as a template, forward primer 5′-CCG-CTC-GAG-CAT-ATG-AGA-TCT-TAT-ATG-GGG-CAC-C-3′, and reverse primer 5′-ATA-GTT-TTA-GCG-GCC-GCA-GTC-TAG-AGG-ATG-GTC-CAC-CCC-CGG-3′. Thirty cycles (40 s at 95°C, 50 s at 55°C, and 1 min at 72°C) were run with 2.5 U of Pfu DNA polymerase. The PCR product was subcloned, digested (XhoI and NotI), and ligated into pSF1 (opened with SalI and NotI).
(iv) GRP+ vectors.
The GRP+ vectors contain the P+ fragment inserted in a gag replacement (GR [described below]) vector and are similar to the vectors reported by Kim et al. (16). The basic vector was designated pSF61. PCR was performed with pSF1 as a template, forward primer 5′-GTG-CCG-GCA-TCT-AAT-GTT-TGC-GCC-TGC-G-3′, and reverse primer 5′-GTC-CGC-TCG-AGC-TAA-TTT-TCA-GAC-AAA-TAC-AGA-AAC-ACA-GTC-3′. Thirty cycles (30 s at 95°C, 40 s at 55°C, and 2 min at 72°C) were run with 2.5 U of Pfu DNA polymerase. The PCR product was subcloned, digested (BglII and XhoI), and ligated into pSF1 (deleted between BglII and NotI) together with the P+ fragment (digested with SalI and NotI) as described above.
(v) GR vectors.
GR vectors were created by having the transgene coding sequences inserted to substitute for gag. The basic vector was designated pSF11. PCR was used to delete gag sequences from pSF1 by using forward primer 5′-GTG-CCG-GCA-TCT-AAT-GTT-TGC-GCC-TGC-G-3′ and reverse primer 5′-ATA-GTT-TAG-CGG-CCG-CTA-ATT-TTC-AGA-CAA-ATA-CAG-AAA-CAC-AGT-C-3′. Thirty cycles (30 s at 95°C, 40 s at 55°C, and 2 min at 72°C) were run with 2.5 U of Pfu DNA polymerase. The PCR product was subcloned, digested (BglII and NotI), and ligated into pSF1 (opened with BglII and NotI). Thus, these vectors contain the entire untranslated region 5′ of the gag reading frame as derived from MESV.
(vi) GRSD− vectors.
GRSD-vectors are splice donor-defective versions of GR vectors. The basic plasmid was designated pSF110. Site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene GmbH), used according to the manufacturer’s instructions, was performed with pSF11 as a template, forward primer 5′-ACC-GAC-CCC-CCC-GCC-GGG-AAC-TAA-GCT-GGC-CAG-CGG-TCG-TT-3′, and reverse primer 5′-AAC-GAC-CGC-TGG-CCA-GCT-TAG-TTC-CCG-GCG-GGG-GGG-TCG-GT-3′ at an annealing temperature of 55°C.
(vii) GRS vectors.
GRS vectors contain a minimal splice acceptor oligonucleotide (SAO) inserted in a GR vector 5′ of the multiple cloning site for the transgene. The basic plasmid was designated pSF71. The SAO was obtained by annealing oligonucleotides 5′-TCG-ACA-AAG-TTA-AGT-AAT-AGT-CCC-TCT-CTC-CAA-GCT-CAC-TTA-CAG-GC-3′ and 5′-GGC-CGC-CTG-TAA-GTG-AGC-TTG-GAG-AGA-GGG-ACT-ATT-ACT-TAA-CTT-TG-3′. The annealed SAO was ligated with the PCR product generated for the GRP+ vector (see above) and cloned into pSF1 opened with BglII and NotI.
GRSΔ270 vectors.
The GRSΔ270 vectors are GRS vectors devoid of the residual aberrant AUG found in the leader at position 270. The basic vector was assigned pSF91. Site-directed mutagenesis was performed with pSF71 as template, forward primer 5′-GTG-CCG-GCA-TCT-AGT-GTT-TGC-GCC-TGC-G-3′, and reverse primer 5′-CGC-AGG-CGC-AAA-CAC-TAG-ATG-CCG-GCA-C-3′ at an annealing temperature of 55°C.
All basic vector plasmids were sequenced to confirm the correctness of the leader sequences. For construction of the eGFP cDNA-containing vectors, pSF6, pSF10, pSF11, pSF61, pSF71, and pSF91 were digested (NotI and EcoRI) and the eGFP cDNA excised from pSF1γ was inserted, resulting in the vectors pSF1γ, pSF11γ, pSF71γ, pSF91γ, pSF6γ, and pSF10γ.
For construction of the MDR1 cDNA-containing vectors pSF6m, pSF11m, pSF61m, and pSF71m, the corresponding basic vectors were digested (NotI and BamHI) and the MDR1 cDNA from pSF1m was inserted.
For construction of the Kozak-lacZ-Neor-containing vectors pSF6K, pSF11K, pSF61K, and pSF71K, the cDNA from pSF1K was inserted into the corresponding basic vectors by using NotI and BamHI sites.
To construct the Neor-containing vectors pSF11N, pSF110N, and pSF91N, the Neor cDNA from pSF1N (15) was introduced between restriction sites for NotI and BamHI of the corresponding basic vectors.
Cell culture and vector production.
Retroviral packaging cells GP&env86 (ecotropic) and GP&envAM12 (amphotropic) or PG13 (gibbon ape leukemia virus envelope) were used as previously described (13–15). For the establishment of the MDR1 and lacZ-Neor producer cells, 10 μg of the corresponding retroviral vector plasmid DNA was electroporated in GP&env 86 cells; the supernatant was used to transduce GP&envAm12 cells. A similar protocol was used to establish PG13 cells transduced with Neor vectors. Selection of packaging cells was performed with colchicine (Sigma-Aldrich GmbH, Deisenhofen, Germany) at 40 ng/ml for the MDR1 constructs and with G418 at 0.4 mg/ml (Life Technologies, Paisley, Scotland) for the lacZ-Neor and Neor vectors. The surviving producers were pooled (MDR1, lacZ-Neor) or kept as individual clones (Neor). For the establishment of the eGFP producer cells, 20 μg of the corresponding retroviral vector plasmid DNA was coelectroporated in GP&envAM12 cells together with 2 μg of plasmid DNA coding for Neor. Surviving producers were pooled. Maintenance and retroviral transduction of target cells K562 and CEM were performed as described previously (3, 13–15); the multiplicity of infection (MOI) was kept below 1, resulting in single-copy integrations. Mass cultures of K562 cells transduced with MDR1 vectors were established with 20 ng of colchicine per ml and selected in increasing amounts of colchicine in soft agar colony assays at three different cell densities, each plated in triplicates. Retroviral supernatant titration on HT1080 target cells was performed in triplicates by endpoint dilution of cell-free supernatants in the presence of 4 μg of Polybrene/ml. Twenty-four hours after transduction, cells were exposed to G418 at 0.4 mg/ml, and the surviving clones were counted microscopically.
Flow cytometry.
Flow cytometry was performed with a FACScalibur (Becton Dickinson) with CellQuest (Becton Dickinson) software. Expression of eGFP was measured 4 days after transduction. The cells were morphologically gated on the living population, and the mean fluorescence of the transduced cells was determined.
β-Galactosidase activity.
The β-galactosidase activity was determined with the Luminescent β-gal Genetic Reporter System II (Clontech, Palo Alto, Calif.) according to the manufacturer’s instructions on the LB 952 T/16 luminometer (Berthold GmbH, Wildbad, Germany). Protein extracts were prepared, and the protein concentration was measured as previously described (14, 15).
RT-PCR.
RNA was isolated from untransduced K562 and K562 transduced with SF1m or SF71m by using the High Pure RNA isolation kit (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer’s instructions. Reverse transcription (RT)-PCR was performed with the Titan One Tube RT-PCR kit (Boehringer Mannheim) according to the manufacturer’s instructions, with a 55°C annealing temperature. The oligonucleotides used were 5′-TCT-CCT-CAG-ATT-GAT-TGA-CTG-C-3′ (forward primer) and 5′-TTC-CTG-CAT-TTG-CAA-AGA-TAT-C-3′ (reverse primer).
Nucleotide sequence accession number.
Leader sequences of pSF11, pSF71, and pSF91 are available in the EMBL, GenBank, and DDBJ databases under accession no. AJ132035 (pSF11), AJ132036 (pSF71), and AJ132037 (pSF91).
RESULTS
Increased expression of drug resistance genes by using improved 5′ untranslated sequences.
To develop improved leader sequences fulfilling stringent criteria of safety and efficiency, we examined the evolutionary architecture of murine leukemia viruses and inserted the transgene of interest to exactly replace gag-pol in frame (Fig. 1). This design removes all residual viral coding sequences, but retains the entire untranslated region upstream of gag-pol. Thus, these vectors mimic the original translational control of Gag, which in simple retroviruses is synthesized at far greater levels than Env (11). This architecture is different from former versions of gag-deleted vectors which used the PstI site 58 bp upstream of the gag initiation codon (2, 6, 16). We analyzed the potential of this GR leader in the context of a retroviral vector developed for somatic gene therapy. This vector expresses the MDR1 gene under transcriptional control of the LTR of SFFVp, with leader sequences based on MESV/dl587rev (3, 13). This vector mediates dominant multidrug resistance in human hematopoietic progenitor cells (13, 14).
K562 cells were transduced at an MOI of <1 and selected as uncloned mass cultures with 20 ng of colchicine/ml, resulting in the death of untransduced cells and survival of about 50% of transduced cells (3, 13). The low MOI, the high transcriptional activity of the vectors in K562 cells, and the moderate selection level ensure that the majority of the cells contributing to the mass cultures contain single-copy integrations of the vector (3, 14). Thus, measurements of expression levels reflect the functional strength of the vector rather than differences in gene dosage. To monitor the levels of expression of MDR1 provided by the vectors, K562 mass cultures were replated in increasing doses of colchicine. In this sensitive biological assay, cloning efficiency is strictly dependent on the expression levels of the membrane-located drug efflux pump encoded by MDR1 (3, 13, 14).
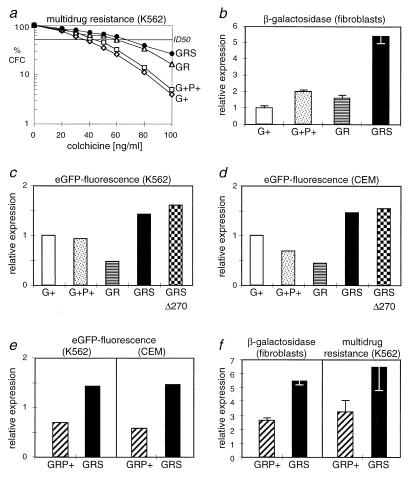
As shown in Fig. 2a, chemoprotection of transduced hematopoietic cells by MDR1 expression from the original construct containing a G+ leader was still limiting when cells were challenged with intensified drug doses. The G+P+ vector led to slightly elevated drug resistance. In contrast, cells transduced with the novel GR vectors had significantly higher cloning efficiencies, indicating that the translation efficiency of MDR1 was improved (Fig. 2a). Expression could be further enhanced by introducing an artificial SAO in front of MDR1. The SAO contains the minimal branch site (partially degenerated), pyrimidine tract, and splice acceptor sequences deduced from SFFVp (28). This novel leader assembly containing the SAO was named GRS (gag replacement with splice) (Fig. 1). With this leader, at high doses of colchicine, 6.5-fold-increased cloning efficiencies were obtained compared to those in cells transduced with the G+ vector (Fig. 2a). Compared to G+ vectors, the elevated cloning efficiencies obtained with vectors containing leaders GR and GRS were highly significant (P < 0.001 according to Student’s t test). In cells transduced with the GRS vector, the 50% inhibitory dose (ID50) of colchicine was almost doubled (68 ng/ml for the GRS vector versus 36 ng/ml for the G+ vector versus 3 ng/ml for untransduced cells).
FIG. 2.
The novel GRS leader (gag replacement with splice) increases expression from retroviral vectors. (a) The expression of multidrug resistance in K562 cells transduced with MDR1 vectors is improved with GR leader sequences. Uncloned mass cultures of cells transduced with the vectors as described in Materials and Methods were plated in colony assays containing increasing doses of the cytotoxic agent colchicine, which is recognized by the MDR1-encoded drug efflux pump. The surviving colonies were microscopically counted. Data are expressed as relative cloning efficiency of colony-forming cells (CFC) with respect to the unselected control. The novel leaders GR and, even better, GRS significantly increase the ID50 of the drug, which is 3 ng/ml for untransduced cells. Shown are mean values of two separate experiments, with each culture plated for nine determinations. Most standard deviations were below 15%; only at doses of >60 ng of colchicine/ml were standard deviations up to 26%. Differences between vectors GRS and G+ are highly significant (P < 0.001). Regression analysis revealed the linearity of the curves (r values of >0.995). The following monocistronic vectors described in Materials and Methods were used: SF1m (G+), SF6m (G+P+), SF11m (GR), and SF71m (GRS). (b) The GRS leader improves expression of β-galactosidase from vectors containing the lacZ-Neor fusion gene. Transduced NIH-3T3-based fibroblasts were selected as a mass culture with G418 at 400 μg/ml, and β-galactosidase activity was determined in cell extracts, as described in Materials and Methods. Data are expressed as relative protein expression with respect to cells transduced with SF1K containing the G+ leader. Standard deviations of three experiments were below 20%. The following monocistronic vectors described in Materials and Methods were used: SF1K (G+), SF6K (G+P+), SF11K (GR), and SF71K (GRS). (c and d) Increased expression of eGFP with the GRS leader in human hematopoietic (c) and lymphoid (d) cells. The destruction of the final residual AUG at +270 slightly increases expression from the GRS leader (GRSΔ270). Data represent the mean fluorescence of unselected transduced cells measured by flow cytometry as described in Materials and Methods, expressed as fold increase with respect to cells transduced with SF1γ containing the G+ leader. A total of 200,000 cells were measured per experiment. The data were reproduced in two further sets of experiments, with standard deviations of <11%. Differences between GRS vectors and vectors G+, G+P+, and GR were significant (P < 0.005). The following monocistronic vectors described in Materials and Methods were used: SF1γ (G+), SF6γ (G+P+), SF11γ (GR), SF71γ (GRS), and SF91γ (GRSΔ270). (e and f) The splice acceptor oligonucleotide contained in the GRS leader is superior to the pol gene fragment (P+) for protein expression with all genes tested. (e) Increase in mean fluorescence with respect to the G+ vector. Data were accumulated and expressed as described for panels c and d. The following monocistronic vectors described in Materials and Methods were used: SF1γ (G+), SF61γ (GRP+), and SF71γ (GRS). (f) The GRS vector is superior to the GRP+ vector in expression of MDR1. Data are expressed with respect to those achieved with the G+ vector, shown for cloning efficiency in the presence of 100 ng of colchicine/ml. Mean values of nine determinations with error bars representing standard deviations are presented, showing significant differences between vectors GRS and GRP+ (P < 0.005). Data were reproduced in a second experiment. The following monocistronic vectors described in Materials and Methods were used: SF1m (G+), SF61m (GRP+), and SF71m (GRS). Assay procedures were as described for panel a.
The efficiency of the novel leader GRS is not dependent on the transgene.
To analyze the function of the leaders GR and GRS with other transgenes, we introduced the cytoplasmic marker lacZ-Neor genes or the gene coding for enhanced green fluorescent protein (eGFP) (15). Again, all transductions were performed at a limited MOI of <1 to ensure single-copy integrations in the majority of the cells analyzed, independent of the vector used. Vectors with the GRS leader mediated the strongest expression of the lacZ-Neor fusion gene. In transduced murine fibroblasts, β-galactosidase levels were about five and three times, respectively, higher than those expressed from vectors G+ and G+P+ (Fig. 2b). In cells transduced with the GR vector lacking SAO, the efficiency of expression was similar to that in the constructs containing viral gene fragments. Equivalent data were obtained with K562 (data not shown).
The eGFP vectors were analyzed in K562 as well as in CEM T lymphoblasts. In both cells, fluorescence intensities obtained with GR vectors were lower than those achieved with the G+ or G+P+ constructs. This indicated that efficient splicing of the 5′ untranslated region was important for high expression of eGFP. Accordingly, presence of the SAO in the GRS vector mediated significantly enhanced expression levels, which, as with MDR1 and lacZ-Neor, were significantly superior to those achieved with the G+ or G+P+ leader (Fig. 2c and d).
Mutation of the final aberrant start codon increases safety and efficiency.
To further increase the safety of the GRS vectors, we also mutated the final aberrant AUG at position +270 of the leader (Fig. 1 and Table 1). Vectors containing this leader (GRSΔ270) reproducibly mediated slightly, although not significantly, increased expression of eGFP (Fig. 2c and d). This suggested that this AUG with a poor Kozak match (Table 1) may be utilized with a low probability of initiation of translation.
The artificial 5′ intron is important for high transgene expression.
We further addressed the importance of the splice sites and the intronic sequences for vector expression. In vectors expressing either MDR1 or eGFP, the GRS leader mediated up to 10-fold-stronger drug resistance or mean fluorescence, respectively, than vectors containing the internal ribosomal entry site (IRES) from encephalomyocarditis virus for translational control of the transgene or the U3 region of the LTR as internal promoter (data not shown). Destruction of the retroviral splice donor (leader G+SD− [Fig. 1]) reduced eGFP expression to 45% (K562) or 11% (CEM) of the levels achieved with the original G+ vector (data not shown). Compared to this splice donor-defective leader, the GRS vector thus mediated 7-fold (K562)- or 31-fold (CEM)-higher eGFP expression. Interestingly, the minimal SAO contained in GRS vectors mediated better expression than the larger P+ fragment, which contains three aberrant AUGs (GRP+ vectors [Fig. 1]). This was seen with all genes (eGFP, lacZ-Neor, and MDR1) and suggested improved splicing efficiency and/or better expression from the unspliced message (Fig. 2e and f). The SAO also improved expression from G+ vectors, most likely by increasing splicing efficiency (data not shown).
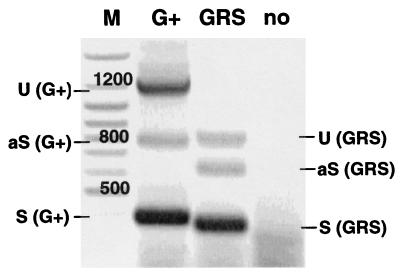
Alternative splicing of the intron present in GRS vectors was verified by using RT-PCR. RNA was harvested from multidrug-resistant K562 mass cultures transduced with vectors containing leader G+ or GRS as well as from untransduced K562 cells (negative control). Subgenomic products of the expected sizes were obtained for both leaders, as shown in Fig. 3. In the GRS vector, the genomic RNA appeared to be reduced in favor of the major splice product. The major splice product of the GRS vector was isolated, subcloned, and sequenced. Thus, the splice sites of the GRS leader were determined as indicated in Fig. 4.
FIG. 3.
Efficient splicing of 5′ untranslated leader sequences expressed from GRS vectors. RT-PCR was performed with RNA extracted from mass cultures of K562 cells transduced with SF1m (G+ leader) and SF71m (GRS leader) using primers annealing upstream of the retroviral splice donor and downstream of the start codon of the MDR1 cDNA. Sizes of the unspliced (U) RNA are 1,214 and 833 bp for SF1m (G+) and SF71m (GRS), respectively. The sizes of the spliced (S) RNA are 376 bp and 364 bp for SF1m (G+) and SF71m (GRS), respectively. The identity of the spliced RNA was confirmed by sequencing of subcloned RT-PCR products (Fig. 4). We also observed previously unknown, minor alternative splice (aS) products of both leader sequences whose origins remain to be determined. M, molecular weight marker; no, untransduced cells.
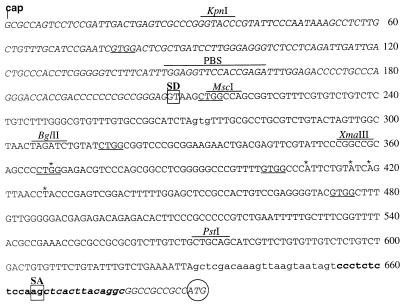
FIG. 4.
Sequence of the GRSΔ270 leader (gag replacement with splice) based on MESV, starting from the cap site and ending with the ATG of the cDNA (circled). The primer binding site (PBS) and recognition sites for endonucleases KpnI, MscI, BglII, XmaIII, and PstI are indicated above the sequence. The splice donor (SD) and splice acceptor (SA) are boxed. The mutated AUG at position 270 and the SAO are shown in lowercase, and the stretch of the SAO with continuous sequence homology to pol is shown in boldface. Sequences contained in the major splice product shown in Fig. 3 are shown in italics. Putative non-AUG start codons are underlined, including the partially degenerated start codon for the glyco-Gag at position 366 (see Discussion). The sequence was obtained from plasmid pSF91 and by subcloning and repetitive sequencing of the major splice product of the RT-PCR shown in Fig. 3. Asterisks delineate sequence deviations (three A-G switches and one T insertion) compared to the published sequence of dl587rev (12). These are present in MESV and all derived leader variants analyzed in this study, including pSF1 (GenBank accession no. AJ224005).
The novel leader design does not interfere with high vector titers.
Finally, we analyzed whether the deletion of the G+ sequences and the introduction of the stronger intron affected vector titers. In safety-modified retroviral packaging cells, monocistronic G+ vectors containing the LTR of SFFVp and the leader of MESV are produced at normal titers of between 106 and 107 infectious particles/ml of tissue culture supernatant (15). In the process of our experiments with vectors containing MDR1 or eGFP, no profound differences in transfer efficiencies were observed with leader sequences G+, G+P+, and GR, whereas GRS vectors had slightly lower titers. This was confirmed with endpoint dilution of cell-free supernatants from single clones stably transduced with Neor vectors (Table 2). Titers of GR vectors were not reduced but rather were slightly elevated compared with those of the corresponding G+ vectors. Titers of GRS vectors were on average fourfold lower than those of GR vectors, which can be explained by reduced expression of unspliced genomic message. However, mean titers of GRS vectors were reduced only 25% compared with G+ vectors. Interestingly, strongly reduced titers of GR vectors were only observed when the splice donor was destroyed (see Discussion).
TABLE 2.
Splice sites are more important than viral coding sequences for vector titersa
| Vector | Gag+ | Splice donor | Splice acceptor oligonucleotide | Titer (105)/mlb |
|---|---|---|---|---|
| G+ (SF1N) | Yes | Yes | No | 2.4 ± 0.7 |
| GR (SF11N) | No | Yes | No | 7.2 ± 1.1 |
| GRΔSD (SF110N) | No | No | No | 0.2 ± 0.09 |
| GRS (SF91N) | No | Yes | Yes | 1.8 ± 0.8 |
Titration of cell-free supernatant was performed in limiting dilution on HT1080 target cells seeded in triplicates. HT1080 cells were selected at 0.4 mg of G418 per ml, and clones were counted after 14 days. In this experimental setting, titers of >106/ml are rarely observed. An internal standard is provided by the gag+ (G+) vector SF1N.
Values represent the mean titer ± standard deviation of 24 clones of transduced PG13 cells for each vector.
DISCUSSION
We have presented a new design of retroviral vectors completely lacking viral coding sequences and aberrant AUG start codons for initiation of translation. These vectors combine genetic elements of simple retroviruses that have been evolutionarily selected for high-level gene expression in vivo. The assembly of these modules followed guidelines for efficiency and safety which are relevant to human gene therapy. The novel GRS leader with the minimal SAO creates an efficacious artificial intron that is compatible with sufficient vector titers. This unique design represents an important improvement compared to vectors with large remnants of gag (G+ leader, equivalent to LX described in reference 19) gag and pol (G+P+ leader, equivalent to MFG described in references 17 and 26), or pol only (GRP+, equivalent to the MFG variants described in reference 16). Three features of GRS vectors support this conclusion: increased specificity of translation, high efficiency of vector expression, and reduced likelihood for viral recombination.
Specificity of translation.
The vectors presented in this study are the first designed to largely abrogate the expression of aberrant proteins or peptides that may be toxic or immunogenic. Six aberrant AUGs residing in the G+ fragment were eliminated by replacement with the transgene, and another three were eliminated by the design of the SAO instead of utilizing the P+ fragment to generate a splice-competent leader. Mutagenesis of the final residual aberrant AUG upstream of the transgene slightly increased the expression of the protein of interest.
Initiation of translation from non-AUG start codons is a very rare event in mammalian cells; it requires the presence of a purine at position −3 and the G at +4 from a non-AUG triplet such as GUG, ACG, or CUG and also appropriate sequences in the second codon (9). The only known functional non-AUG start codon in murine leukemia viruses is the Kozak-matching ACC-CUG-G that can give rise to the expression of the glycosylated Gag (25). This is partially degenerated in the MESV-based sequences used for construction of our vectors (Fig. 4), in contrast to vectors with leaders derived from Moloney murine leukemia or sarcoma virus (16, 19, 26). However, another CUG and a GUG still match the Kozak consensus (G residue at +4, purine at −3), while a partial match (absence of the purine at −3) is fulfilled by one further CUG and three GUGs (Fig. 4). Putative ACG initiation codons are not found. The spliced leader contains only one putative non-AUG initiation codon (Fig. 4).
It remains to be seen whether these non-AUG codons also have to be mutated to completely abrogate aberrant initiation of translation. If so, this issue is at least equally relevant for other vectors, especially for those containing large gene fragments upstream of the cDNA of interest.
High efficiency of expression.
GRS vectors mediate increased expression of transferred genes with every cDNA (MDR1, lacZ-Neor, eGFP) and in any cellular background examined (murine fibroblasts, human myeloid, and lymphoid cells). More variable results were observed with leaders G+, G+P+, GR, and GRP+. The increased efficiency was achieved by exploiting two additive posttranscriptional mechanisms of gene regulation. First, the introduction of an intron in the 5′ untranslated sequences may prevent translational silencing of eukaryotic genes (18). This interpretation is supported by the data showing that the GRS leader with the artificial intron gave much higher levels of expression than splice-defective vectors and vectors utilizing IRES sequences or internal promoters for translational control of the transgene. Second, the optimized specificity of translation (described above) may add to the increased efficiency, since no transcripts are lost by recruitment of aberrant reading frames.
Optimal posttranscriptional processing is of considerable relevance when vector transcription may be impaired, as in primitive hematopoietic cells (3, 4, 13). The vectors presented in this study were based on FMEV (a Friend mink cell focus-forming–MESV hybrid vector), which provides the strongest transcriptional activity known for retroviral vector expression in hematopoietic cells (3–5, 13, 15). The combination with the GRS leader allows very efficient levels of gene expression in hematopoietic cells. For the transfer of genes such as MDR1, which mediate cancer drug resistance for in vivo selection of primitive hematopoietic cells, high-level gene expression is of outstanding importance to increase the numbers of protected cells, to suppress accumulation of sublethal damage in surviving cells, and thus to improve their proliferative capacity and differentiation potential (4, 13). High levels of vector expression will also improve safety and efficacy in other gene therapy strategies, e.g., in those utilizing transfer and expression of marker genes for the enrichment of transduced cells in vitro (15, 24) or suicide genes for the negative selection of transduced cells in vivo (10).
Reduced likelihood for retroviral recombination.
Deletion of the gag gene and replacement of the pol fragment with the minimal SAO not only increases cloning efficiency. More significantly, it also reduces the potential for recombination with naturally occurring retroviruses, which show a high degree of sequence conservation in pol (11). While more conventional retroviral vectors contain 400 bp (G+ and GRP+) or even 800 bp (G+P+) of viral coding sequences, the SAO used in this study shows only a small remnant of 26 bp in the pyrimidine tract and the splice acceptor consensus that are homologous to pol of Moloney murine leukemia virus (Fig. 4). However, we cannot completely rule out that these sequences may occasionally trigger recombination with viral coding sequences, as shown for other stretches with limited homology (23). If evidence is obtained that the SAO is involved in recombination with viral coding sequences, the residual homology can be abolished by sequence wobbling or by designing a nonretroviral SAO.
Sufficient vector titers.
Our data are in agreement with those reported by Kim et al. (16) indicating that G+ sequences are not required for high-titer packaging. This was demonstrated with statistically sufficient numbers of stable producer clones (Table 2) rather than using transiently transfected mass cultures (16). In addition, our data offer a novel interpretation of those presented by Bender et al. (6), who compared titers of G+ vectors containing an intact splice donor with those of gag-deleted vectors containing a mutated splice donor. In a synopsis of the available data, we suggest that, in addition to the Ψ domain, the presence of the splice donor is a major prerequisite for high titers. This may indicate a function of the splice donor in RNA export or in the folding of the Ψ domain. Thus we conclude that splice sites play more crucial roles for vector titers than the presence of viral coding sequences. The observations of slightly (25% when compared with G+ vectors) reduced titers of GRS vectors with Ψ located in a strong intron is likely to reflect reduced expression of the genomic message in packaging cells. This may be compensated for by increasing vector transcription in packaging cells, by utilizing cells with more potent packaging functions or simply by screening a greater number of producer clones. Using improved, fibronectin-assisted protocols such as those described by Schilz et al. (27), the titers obtained with GRS vectors are sufficient for transducing primitive human CD34+/38− cells, including the population of SCID-repopulating cells. Thus, moderate titer reductions represent a minor drawback for therapeutic applications of retroviral vectors ex vivo.
In summary, the novel vectors presented in this study show increased efficiency of retroviral gene expression and are expected to increase the safety of somatic gene therapy. The optimization of the leader is independent of the specific promoter, transgene, viral backbone, or target cell used. Therefore, among the attempts to identify appropriate promoters or sequences that prevent epigenetic silencing of transgenes, the development described here adds an important component to the design of therapeutic vectors.
ACKNOWLEDGMENTS
We thank Joachim Nowock and Carol Stocking for critical review of the manuscript.
W.O. and C.B. were supported by a grant of the Bundesministerium für Bildung und Forschung (O1KV9530/9811), and M.H. was supported by a grant of the José Carreras Leukemia Fund. The Heinrich-Pette-Institute is financially supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit, Germany.
REFERENCES
- 1.Anderson W F, McGarrity G J, Moen R C. Report to the NIH recombinant DNA advisory committee on murine replication competent retrovirus (RCR) assay. Hum Gene Ther. 1993;4:311–321. doi: 10.1089/hum.1993.4.3-311. [DOI] [PubMed] [Google Scholar]
- 2.Armentano D, Yu S-F, Kantoff P W, von Rüden T, Anderson W F, Gilboa E. Effect of internal viral sequences on the utility of retroviral vectors. J Virol. 1987;61:1647–1650. doi: 10.1128/jvi.61.5.1647-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baum C, Hegewisch-Becker S, Eckert H-G, Stocking C, Ostertag W. Novel retroviral vectors for efficient expression of the multidrug resistance (mdr-1) gene in early hematopoietic cells. J Virol. 1995;69:7541–7547. doi: 10.1128/jvi.69.12.7541-7547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baum C, Stocking C, Wagener T, Eckert H-G, Ostertag W. Gene transfer and transgene expression in hematopoietic cells. In: Strauss M, Barranger J A, editors. Concepts in gene therapy. Berlin, Germany: De Gruyter; 1997. pp. 233–266. [Google Scholar]
- 5.Baum C, Itoh K, Meyer J, Laker C, Ito Y, Ostertag W. The potent enhancer activity of the polycythemic strain of spleen focus-forming virus in hematopoietic cells is governed by a binding site for Sp1 in the upstream control region and by a unique enhancer core motif, creating an exclusive target for PEBP/CBF. J Virol. 1997;71:6323–6331. doi: 10.1128/jvi.71.9.6323-6331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 8.Berlioz C, Darlix J-L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeck R, Kolakofsky D. Positions +5 and +6 can be major determinants of the efficiency of non-AUG initiation codons for protein synthesis. EMBO J. 1994;13:3608–3617. doi: 10.1002/j.1460-2075.1994.tb06668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonini C, Ferrari G, Verzeletti S, Servida P, Zappone E, Ruggieri L, Ponzoni M, Rossini S, Mavilio F, Traversari C, Bordignon C. HSV-TK gene transfer into donor lymphocytes for control of allogeneic graft-versus-leukemia. Science. 1997;276:1719–1724. doi: 10.1126/science.276.5319.1719. [DOI] [PubMed] [Google Scholar]
- 11.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 763–844. [Google Scholar]
- 12.Colicelli J, Goff S P. Isolation of a recombinant murine leukemia virus utilizing a new primer tRNA. J Virol. 1986;57:37–45. doi: 10.1128/jvi.57.1.37-45.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert H-G, Stockschläder M, Just U, Hegewisch-Becker S, Grez M, Zander A, Ostertag W, Baum C. High-dose multidrug resistance in primary human hematopoietic progenitor cells transduced with optimized retroviral vectors. Blood. 1996;88:3407–3415. [PubMed] [Google Scholar]
- 14.Hildinger M, Fehse B, Hegewisch-Becker S, John J, Rafferty J A, Ostertag W, Baum C. Dominant selection of hematopoietic progenitor cells with retroviral mdr1-coexpression vectors. Hum Gene Ther. 1998;9:33–42. doi: 10.1089/hum.1998.9.1-33. [DOI] [PubMed] [Google Scholar]
- 15.Hildinger M, Eckert H G, Schilz A J, John J, Ostertag W, Baum C. FMEV vectors: both retroviral long terminal repeat and leader are important for high expression in transduced hematopoietic cells. Gene Ther. 1998;5:1575–1579. doi: 10.1038/sj.gt.3300759. [DOI] [PubMed] [Google Scholar]
- 16.Kim S H, Yu S S, Park J S, Robbins P D, An C S, Kim S. Construction of retroviral vectors with improved safety, gene expression, and versatility. J Virol. 1998;72:994–1004. doi: 10.1128/jvi.72.2.994-1004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krall W J, Skelton D C, Yu X J, Riviére I, Lehn P, Mulligan R C, Kohn D B. Increased levels of spliced RNA account for augmented expression from the MFG retroviral vector in hematopoietic cells. Gene Ther. 1995;3:37–48. [PubMed] [Google Scholar]
- 18.Matsumoto K, Montzaka Wassarman K, Wolffe A P. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller A D, Rosman D J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 20.Miller A D, Miller D G, Garcia J V, Lynch C M. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 1993;217:581–599. doi: 10.1016/0076-6879(93)17090-r. [DOI] [PubMed] [Google Scholar]
- 21.Mulligan R C. The basic science of gene therapy. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 22.Munk C, Lohler J, Prassolov V, Just U, Stockschlader M, Stocking C. Amphotropic murine leukemia viruses induce spongiform encephalomyelopathy. Proc Natl Acad Sci USA. 1997;94:5837–5842. doi: 10.1073/pnas.94.11.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otto E, Jones-Trower A, Vanin E F, Stambaugh K, Mueller S N, Anderson W F, McGarrity G J. Characterization of a replication-competent retrovirus resulting from recombination of packaging and vector sequences. Hum Gene Ther. 1994;5:567. doi: 10.1089/hum.1994.5.5-567. [DOI] [PubMed] [Google Scholar]
- 24.Pawliuk R, Eaves C J, Humphries R K. Sustained high-level reconstitution of the hematopoietic system by preselected hematopoietic cells expressing a transduced cell-surface antigen. Hum Gene Ther. 1997;8:1595–1604. doi: 10.1089/hum.1997.8.13-1595. [DOI] [PubMed] [Google Scholar]
- 25.Portis J L, Spangrude G J, McAtee F J. Identification of a sequence in the unique 5′ open reading frame of the gene encoding glycosylated Gag which influences the incubation period of neurodegenerative disease induced by a murine retrovirus. J Virol. 1994;68:3879–3887. doi: 10.1128/jvi.68.6.3879-3887.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riviére I, Brose K, Mulligan R C. Effects of retroviral vector design on expression of human adenosine deaminase in murine bone marrow transplant recipients engrafted with genetically modified cells. Proc Natl Acad Sci USA. 1995;92:6733–6737. doi: 10.1073/pnas.92.15.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schilz A J, Brouns G, Knobeta H, Ottmann O G, Hoelzer D, Fauser A A, Thrasher A J, Grez M. High efficiency gene transfer to human hematopoietic SCID-repopulating cells under serum-free conditions. Blood. 1998;92:3163–3171. [PubMed] [Google Scholar]
- 28.Wolff L, Kaminchick J, Hankins W D, Ruscetti S K. Sequence comparison of the anemia- and polycythemia-inducing strains of Friend spleen focus-forming virus. J Virol. 1985;53:570–578. doi: 10.1128/jvi.53.2.570-578.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]