Abstract
The Epstein-Barr virus BMLF1 gene product EB2 has been shown to efficiently transform immortalized Rat1 and NIH 3T3 cells, to bind RNA, and to shuttle from the nucleus to the cytoplasm. In transient-expression assays EB2 seems to affect mRNA nuclear export of intronless RNAs and pre-mRNA 3′ processing, but no direct proof of EB2 being involved in RNA processing and transport has been provided, and no specific functional domain of EB2 has been mapped. Here we significantly extend these findings and directly demonstrate that (i) EB2 inhibits the cytoplasmic accumulation of mRNAs, but only if they are generated from precursors containing weak (cryptic) 5′ splice sites, (ii) EB2 has no effect on the cytoplasmic accumulation of mRNA generated from precursors containing constitutive splice sites, and (iii) EB2 has no effect on the 3′ processing of precursor RNAs containing canonical and noncanonical cleavage-polyadenylation signals. We also show that in the presence of EB2, intron-containing and intronless RNAs accumulate in the cytoplasm. EB2 contains an Arg-X-Pro tripeptide repeated eight times, similar to that described as an RNA-binding domain in the herpes simplex virus type 1 protein US11. As glutathione S-transferase fusion proteins, both EB2 and the Arg-X-Pro repeat bound RNA in vitro. However, by using EB2 deletion mutants, we demonstrated that the effect of EB2 on splicing and RNA transport requires the C-terminal half of the protein but not the Arg-X-Pro repeat.
The Epstein-Barr virus (EBV) is a human gammaherpesvirus which is associated with several malignancies, such as Burkitt’s lymphoma, nasopharyngeal carcinoma, Hodgkin’s disease, gastric carcinoma, breast carcinoma, and B- and T-cell lymphomas. Although it is still unclear if and how EBV contributes to the emergence of such malignancies, it is well documented that several EBV gene products act coordinately to induce the permanent proliferation of quiescent B lymphocytes in vitro (reviewed in reference 16).
Although the functions of many EBV gene products have been partially characterized, very little is known for certain others. Among these, the early nuclear protein EB2 (9), which is also known as BMLF1 or SM, was originally described as a promiscuous transcription factor because it activates the transient expression of the chloramphenicol acetyltransferase (CAT) gene placed under the control of many different promoters (18). However, the increase in CAT enzyme expression did not always correlate with an increase in the amount of specifically initiated CAT RNAs (5, 10, 14). Moreover, it was shown that the effect of EB2 on gene expression in transient transfections was not promoter dependent but was reporter gene dependent (13). EB2 has also been shown to efficiently induce the growth of Rat1 and NIH 3T3 cells at a low density under soft agar (11). From these results, it was concluded that EB2 is a nuclear protein that modulates gene expression nontranscriptionally by mechanisms not yet known, and that might explain how EB2 induces the transformation of immortalized cells. Subsequently, it has been demonstrated in transfection assays that (i) EB2 inhibited the expression of intron-containing genes and activated the expression of intronless genes (mainly CAT gene-based constructs) (28), (ii) EB2 could affect the 3′ processing of the EBV DNA polymerase pre-mRNA (15), and (iii) EB2 has properties of an RNA export protein and increases the cytoplasmic accumulation of intronless EBV replication gene mRNA but not that of intron-containing EBV replication gene mRNA (33). Taken altogether, the published results point to EB2 as a viral protein involved in the regulation of pre-mRNA processing and transport. However, since the effects of EB2 are seen both on EBV-derived reporter RNAs and on CAT-containing reporter RNAs, it is not specific to EBV RNAs. Moreover, no experiment was done to directly assess if EB2 affects pre-mRNA processing, mRNA stability, and/or mRNA nucleocytoplasmic transport and which mechanisms and domains of EB2 are required for its effects on mRNA processing and transport.
In this report, we significantly extend the results of previous studies and directly demonstrate that EB2 acts nontranscriptionally by inhibiting the cytoplasmic accumulation of polyadenylated RNAs when they are generated by the use of weak (cryptic) 5′ splice sites but not by the use of constitutive 5′ splice sites carried by RNA precursors initiated at the same promoter. In both cases, EB2 also induces the cytoplasmic accumulation of unspliced RNAs. These results suggest that EB2 affects RNA splicing and nuclear export. We also directly show that EB2 has no effect on the 3′ processing of precursor RNAs containing canonical and noncanonical cleavage-polyadenylation signals (CPS). EB2 contains an Arg-X-Pro tripeptide repeat similar to that described as an RNA-binding domain in the herpes simplex virus type 1 (HSV-1) US11 protein (27, 31). As glutathione S-transferase fusion proteins, both EB2 and the Arg-X-Pro repeat bound RNA in vitro. However, by using EB2 deletion mutants spanning the entire EB2 protein, we demonstrate that the effect of EB2 on splicing and on RNA nuclear export does not require the Arg-X-Pro repeat but the C-terminal half of the protein.
MATERIALS AND METHODS
Plasmid vectors. (i) Reporter plasmids.
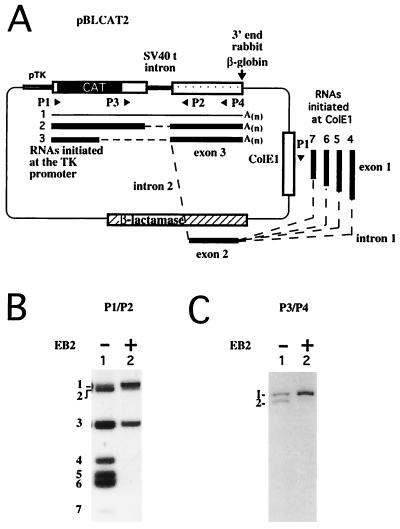
Plasmid pBLCAT2 (19) and plasmid pSV2β (5) are briefly described in Fig. 1A. The vector Paac, derived from pBluescript KSII+ (Stratagene), contains the cytomegalovirus (CMV) immediate-early promoter cloned in the NotI restriction site. Downstream of the polylinker, the early simian virus 40 (SV40) cleavage and polyadenylation sequence was inserted in the HincIII and KpnI restriction sites.
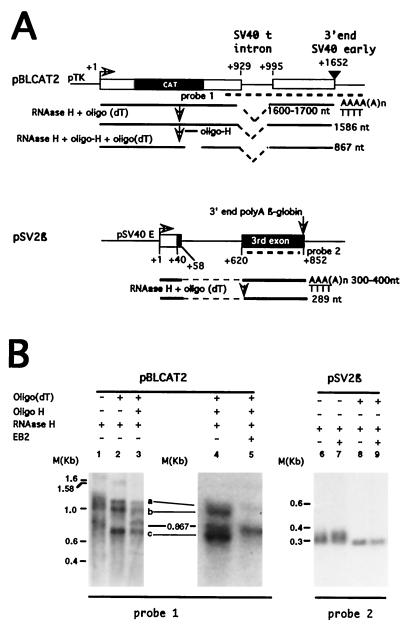
FIG. 1.
EB2 inhibits the expression of differentially spliced polyadenylated RNAs expressed from pBLCAT2. (A) Diagrams of the expected processed RNAs generated from plasmids pBLCAT2 and pSV2β. The genetic contents of the pre-mRNAs initiated at the tk promoter (pTK) in pBLCAT2 and at the early SV40 promoter in pSV2β (pSV40E) are indicated. The CAT ORF in pBLCAT2 and the rabbit β-globin sequences in pSV2β are shaded. DNA probes 1 and 2 were used for the Northern blot analysis shown in panel B. The deadenylation and cleavage of the expected processed RNAs by RNase H in the presence of oligo(dT) or oligo(dT) plus oligo(H) and the lengths of the expected RNase H-treated RNA products are also indicated. (B) Northern blot analysis of the RNAs transiently expressed in HeLa cells from plasmids pBLCAT2 (lanes 1 to 5) and pSV2β (lanes 6 to 9). The plasmids transfected, the proteins expressed, and the RNase H treatments are indicated above the panels. The 32P-labeled DNA probes used are indicated below the panels. M(Kb), molecular size markers (number of kilobases).
Plasmid Paac-ALT1 (see Fig. 3B) contains an 861-bp DNA fragment amplified by PCR from pBLCAT2 sequences located between positions 2834 and 3689 (coordinate 1 being the sequence 5′-TCGC, located 650 bp upstream of the CAT ATG) with primers 5′-CGCGGATCCCCGGATACCTGTCCGC and 5′-CCGGAATTCGACTGGATGGAGGCGG, generating a double-stranded DNA fragment that was further cut by BamHI and EcoRI and ligated into Paac digested with BamHI and EcoRI.
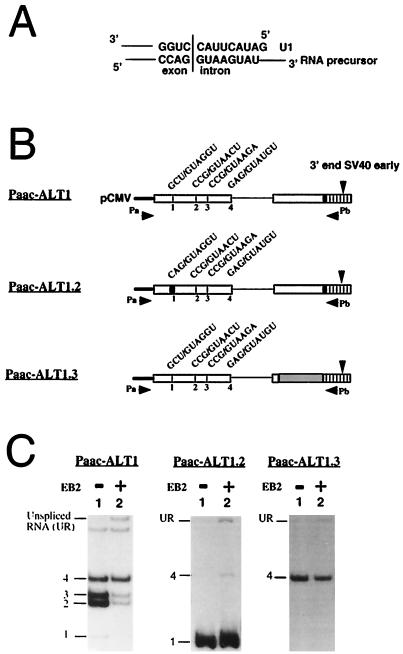
FIG. 3.
The effect of EB2 on alternative splicing is dependent on cis determinants of 5′ splice site selection. (A) Representation of a 5′ splice site sequence hybridizing perfectly to the 5′ end of U1 snRNA. (B) Diagram of the minigenes transfected into HeLa cells and the primers used for RT-PCR analysis of the cytoplasmic RNAs expressed. The alternative 5′ splice sites are indicated by numbers 1 to 4, and their sequences are presented over the diagram of each construction. Twenty PCR cycles were done in the RT-PCR. (C) Splicing patterns of Paac-ALT1, Paac-ALT1.2, and Paac-ALT1.3 minigenes upon cotransfection into HeLa cells with a vector expressing (lanes 2) or not expressing (lanes 1) EB2.
Plasmid Paac-ALT1.2 (see Fig. 3B) is a Paac-ALT1 derivate in which the first intron-exon boundary has the rabbit β-globin exonic sequence CAG. This was done by amplifying a DNA fragment from Paac-ALT1 by PCR with primer 5′-CGCGGATCCCCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCAATGCTCAGCAGGTAGGTATCTCAGTTCGGTGT AGG and primer 2. The PCR-amplified fragments were further cut by BamHI and EcoRI and ligated into Paac cut with BamHI and EcoRI.
Plasmid Paac-ALT1.3 (see Fig. 3B) is a Paac-ALT1 derivate in which the 175 bp of the second exon sequence were replaced by 175 bp of the rabbit β-globin second exon sequence, except for the intron-exon boundary. This was done by amplifying the β-globin second exon sequence by PCR with single-stranded oligonucleotides 5′-CGTTCATCCATAGTTGCCGCTGGTTGTCTACCCATG and 5′-CCGGAATTCGCTTAGCAAAGGTGCCTTTGAGG. The PCR product obtained was used as an oligonucleotide to amplify a DNA fragment from plasmid Paac-ALT1 with oligonucleotide 1 by PCR. The DNA fragment amplified was then digested with BamHI and EcoRI and ligated into Paac cut with BamHI and EcoRI. All the constructions were sequenced.
The plasmids pUCβ128SV and pUCβΔ128SV (7) were kindly provided by A. Krainer. Briefly, the plasmid pUCβ128SV contains the human β-globin gene cloned under the control of the SV40 early promoter in the plasmid pUC19. The plasmid pUCβΔ128SV contains a β-thalassemia allele in which the G at position 1 of intron 1 is mutated to an A, unmasking three cryptic 5′ splice sites (see Fig. 4).
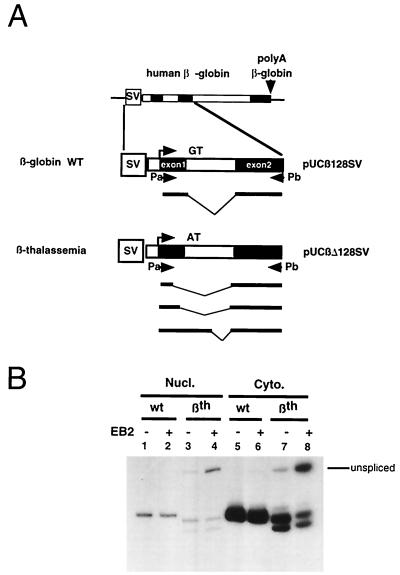
FIG. 4.
Effect of EB2 on alternative splicing versus constitutive splicing. (A) Diagrams of the human β-globin gene and the β-thalassemia gene and primers used for RT-PCR analysis. The expected splicing products are indicated below the wt β-globin gene and the βthal gene. Twenty PCR cycles were done in the RT-PCR. (B) Patterns of splicing of the wt β-globin gene (lanes 1, 2, 5, and 6) and the βthal gene (lanes 3, 4, 7, and 8), upon cotransfection with a vector not expressing (lanes 1 and 6) or expressing EB2 (lanes 2 and 7), in the nucleus (lanes 1 and 2) and the cytoplasm (lanes 6 and 7).
The plasmid I22, containing the PvuII-EcoRI fragment of the β-globin gene under the control of the CMV promoter, is essentially the same as plasmid pIM124 (22), except that the CUAGCUAGCUAG sequence, including three stop codons located downstream of exon 3 of the β-globin gene, was deleted.
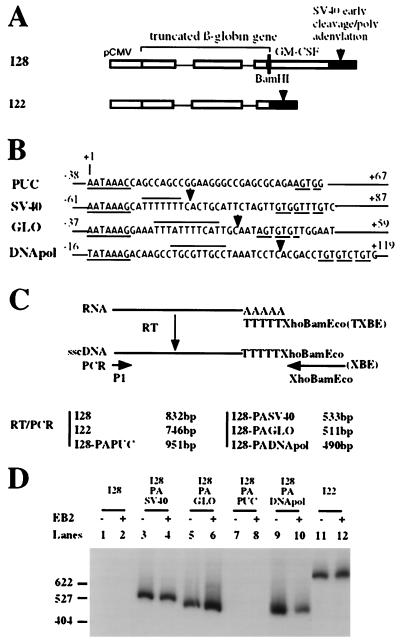
Plasmid I28 contains the rabbit β-globin gene with the third exon truncated, followed by the granulocyte-macrophage colony-stimulating factor (GM-CSF) RNA unstability sequences (22). The I28 precursor RNA initiated at the CMV promoter contains the β-globin sequences, followed by the GM-CSF RNA unstability sequences and the early SV40 CPS. As such this RNA is unstable but can be stabilized by inserting a functional CPS between the β-globin sequences and the GM-CSF instability sequences. Plasmid I28-PAPUC has a pUC18 cryptic CPS, plasmids I28-PASV40 and I28-PAGLO have the canonical SV40 early and β-globin CPS, and I28-PADNApol has the EBV DNA polymerase noncanonical CPS.
(ii) Expression plasmids.
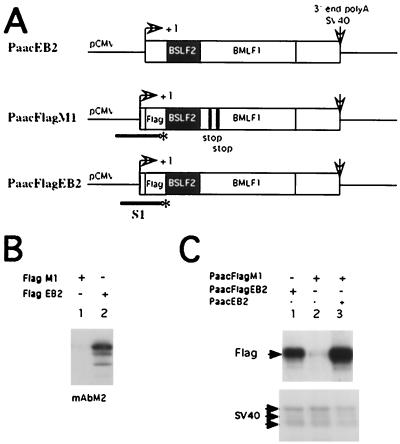
The EB2-expressing plasmid PaacEB2 was constructed by subcloning the BSLF2-BMLF1 intronless cDNA pcdM80 (5) into the XbaI-HindIII sites in Paac. The EB2 protein was tagged with the Flag epitope detected by monoclonal antibody M2, to generate plasmid PaacFlagEB2. From PaacFlagEB2, several deletion mutants were generated (see Fig. 9A). Firstly, XhoI sites were introduced at various positions in the BSLF2-BMLF1 cDNA by the PCR method described previously (22a). Deletion mutants were then generated by ligating different DNA fragments generated by XhoI digestion.
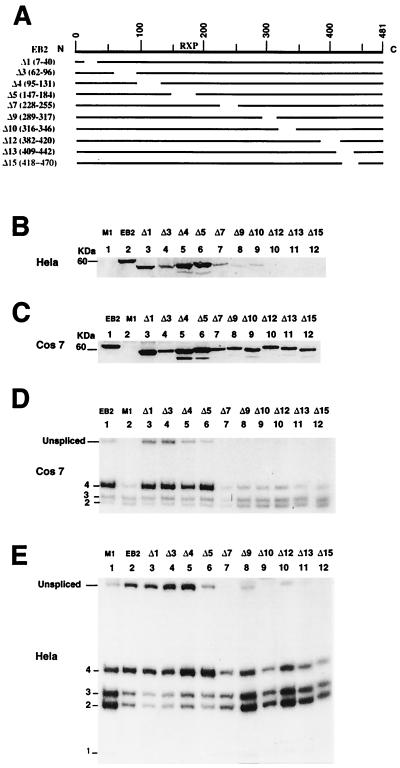
FIG. 9.
The C-terminal domain of EB2 is essential for its function, but the Arg-X-Pro repeat is not. (A) Diagrams of the EB2 deletion mutants generated. The names of the mutants and the amino acids deleted are indicated on the left side of the panel. The approximate location of the Arg-X-Pro repeat (RXP) is indicated above the diagram. (B and C) Expression of EB2 and EB2 mutants in HeLa and Cos7 cells evaluated by Western blotting and a polyclonal antibody against EB2. M1 is an EB2 mutant in which two stop codons have been inserted downstream of the BMLF1 AUG and which does not express the EB2 protein. (D and E) Effects of EB2 and EB2 mutants on the splicing pattern of the Paac-ALT1 minigene in Cos7 and HeLa cells. Twenty PCR cycles were done in the RT-PCR.
Plasmid PaacFlagM1 was generated from PaacFlagEB2 after the insertion of two stop codons downstream of the BMLF1 AUG, and EB2 is not expressed from PaacFlagM1, in contrast to PaacFlagEB2 (see Fig. 7B, lane 1 and lane 2). Plasmid PaacFlagM1 was included in each transfection labeled “minus EB2.”
FIG. 7.
EB2 does not affect cleavage-polyadenylation. (A) Diagram of the reporter gene used in the assay. (B) Sequences of the CPS inserted in I28 to generate I28-PAPUC, I28-PASV40, I28-PAGLO, and I28-PADNApol. The essential sequence elements for cleavage-polyadenylation are underlined or overlined. Cleavage sites are indicated by arrows. (C) Schematic representation of the RT-PCR assay and the expected size of the RT-PCR product generated. Twenty PCR cycles were done in the RT-PCR. sscDNA, single-stranded cDNA. (D) Semiquantitation by RT-PCR of the cytoplasmic RNAs expressed in HeLa cells transfected as indicated above the panel. Double-stranded DNA size markers are indicated on the left.
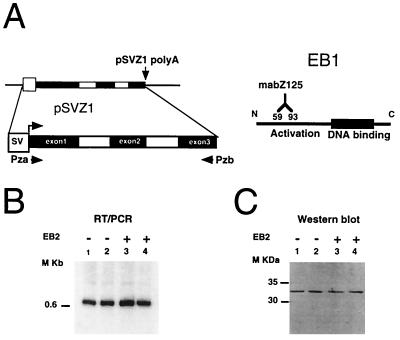
Plasmid pSVZ1 (see Fig. 5A) has been described elsewhere (9).
FIG. 5.
EB2 has no effect on constitutive splicing of an EBV immediate-early pre-mRNA. (A) Diagrams of the BZLF1 reporter gene and the primers used for RT-PCR. Twenty PCR cycles were done in the RT-PCR. The BZLF1 gene product EB1 is also schematically represented, with the epitope stained by the monoclonal antibody mabZ125. EB2 has no effect on the expression of the BZLF1 gene transfected into HeLa cells, and this is seen both at the level of the correctly processed polyadenylated RNAs by RT-PCR (B) and at the level of the EB1 protein, expressed by Western blotting and staining with mabZ125 (C).
Transfections and immunoblotting.
The plasmids used for transfection were prepared by the alkaline lysis method and purified through two CsCl gradients. The DNAs were in the same topological state as assayed by agarose gel electrophoresis. HeLa cells were grown at 37°C in Dulbecco modified Eagle medium (Gibco) supplemented with 10% fetal calf serum and were seeded at 8 × 105 cells per 100-mm-diameter petri dish 10 h prior to transfection. Transfections were performed by the calcium precipitate method as described in reference 5. Transfected cells were collected 48 h after transfection. Immunoblots were performed and stained with the anti-EB2 antibody as described previously (32).
RNA analysis. (i) RNA extraction.
Transfected cells were washed with phosphate-buffered saline, and cytoplasmic RNAs were extracted as described previously (5). Cells were harvested and lysed with Nonidet P-40. The nuclei were pelleted and kept for further nuclear RNA extractions. Cytoplasmic RNAs were extracted with phenol-chloroform from the supernatant, further treated with RNase-free DNase, and ethanol precipitated.
Nuclear RNAs were extracted from the nuclear pellet by using the Dynabeads kit designed for the purification of poly(A)+ RNA, according to the manufacturer’s instructions (Dynal). RNA were further treated with RNase-free DNase and ethanol precipitated.
(ii) Northern blots and quantitative S1 nuclease mapping.
Cytoplasmic RNAs (20 μg) were size separated on a formamide gel and transferred to a nitrocellulose membrane (Schleicher & Schuell). The immobilized RNAs were hybridized for 18 h at 42°C with 32P-labeled DNA fragments in a solution containing 50% formamide, 1% sodium dodecyl sulfate (SDS), 10% dextran sulfate, 1 M NaCl, and 150 mg of herring sperm DNA per ml. The filters were washed twice with 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) at room temperature, twice with 0.5× SSPE–0.1% SDS at 65°C, and once with 0.1× SSPE–0.1% SDS at 65°C.
When indicated, RNase H treatment was performed as follows. After heat denaturation, cytoplasmic RNAs were incubated with oligo(dT) with or without oligo(H) (Fig. 1A) for 10 min at room temperature. After the addition of 50 mM KCl and another incubation of 10 min, the hybrids were digested with 1 U of RNase H for 30 min at 37°C in 20 mM Tris-HCl (pH 7)–28 mM MgCl2. RNAs were extracted with phenol-chloroform and ethanol precipitated. Oligo(H) is located on the pBLCAT2 sequence between positions 1229 and 1258 according to the numbering described above.
Quantitative S1 nuclease mapping was performed essentially as described in reference 5.
(iii) Reverse transcription (RT)-PCR.
Cytoplasmic RNAs (5 μg) were reverse transcribed in a final volume of 20 μl with oligo(dT) or oligo(dT) XBE (see Fig. 7C) as described by the manufacturer (Promega). Labeled PCR was performed by using 2 μl of cDNA and 1 μCi of [32P]dCTP. For each PCR experiment, the amounts of amplified DNA fragments were compared at 15, 20, and 25 cycles, in the absence and presence of EB2. In each experiment described in this report, the amount of amplified DNA fragments increased with the number of cycles, but the relative amount of PCR-amplified DNA fragments in the absence and presence of EB2 was comparable at each number of cycles used.
RNA initiated at the thymidine kinase (tk) and ColE1 promoters in pBLCAT2 were detected with the forward primer P1 (5′-AACACCGACCGACCCTGC) located between bp 541 and 559 in pBLCAT2. The primer P1 sequence was also partially found in the ColE1 region in pBLCAT2 between positions 2818 and 2828 (5′-CCGACCGACCCTGC). The reverse primer P2 (5′-TTAAAGGCATTCCACCACTGC) is located downstream from the small t intron between positions 1571 and 1592.
For plasmid pSV2β, to detect RNA initiated at the SV40 promoter, primer pSV2β-P2 (5′-AGTGAGGAGGCTTTTTTGG) hybridizing in the SV40 promoter downstream of the +1 nucleotide and primer pSV2β-P3 (5′-GTGAGTCAGGGTATTGGCC) in the third exon of the rabbit β-globin gene were used. To detect RNA initiated at the ColE1 promoter, primers pBLCAT2-P1 and pSV2β-P3 were used.
For plasmids pUCβ128SV and pUCβΔ128SV, the forward primer Pa (5′-CATTTGCTTCTGACACAACTG) located in the first exon of the human β-globin gene and the reverse primer Pb (5′-GTGCAGCTCACTCAGTGTGGC) located in the second exon of the human β-globin gene were used to detect RNA initiated at the SV40 promoter.
To detect the RNAs specifically initiated at the CMV promoter in plasmids Paac-ALT1, Paac-ALT2, Paac-ALT1.2, and Paac-ALT1.3, the forward primer Pa (5′-CAGATCTCTAGAACTAGTGG) located in the polylinker region upstream from the first exon and the reverse primer Pb (5′-GGTATCGATAAGCTTGATATC) located in the SV40 sequences were used.
To detect BZLF1 RNAs initiated at the SV40 early promoter, primer Pza (5′-GTAAAATTTACACCTGACCC) located at the 5′ end of the BZLF1 RNA and primer Pzb (5′-TCCTCGTGTAAAACATCTGG) located immediately upstream of the BZLF1 CPS were used. The RT-PCR product expected from correctly spliced BZLF1 RNA is 691 bp.
To detect the RNAs initiated at the CMV promoter in plasmids I22 and I28 and derivates, the forward primer P1 (5′-CGACTGCTGCTTACACTTGC) located in the first exon of the rabbit β-globin gene and the reverse primer XBE were used (see Fig. 7C).
EMSAs.
Different double-stranded DNA fragments were produced by amplifying different portions of the Paac-ALT1 reporter gene by PCR. This was done by using a 5′ primer carrying the T7 promoter from which RNAs would be initiated at the Paac-ALT1 +1 nucleotide and 3′ primers located at different positions in the Paac-ALT1 gene. RNA probes were transcribed, 32P labeled in vitro with the T7 RNA polymerase, purified on denaturing gels, and used in electrophoretic mobility shift assays (EMSAs). The GST, GST–Arg-X-Pro, and GST-EB2 proteins were produced and purified by standard techniques. The RNA-protein interactions were done as described previously (12). The RNA-protein complexes were separated by electrophoresis in a nondenaturing acrylamide gel and visualized by autoradiography as described previously (12).
RESULTS
EB2 inhibits the expression of differentially spliced RNAs transcribed from pBLCAT2.
It has been suggested that EB2 affects the processing and transport of RNAs in transient-expression assays, and this has been observed both on EBV-derived reporter RNAs and on CAT-containing reporter RNAs. However, experiments directly proving that the processing of these reporter RNAs was correct and affected by EB2 have not yet been performed.
In order to study the effect of EB2 on the processing of the pre-mRNAs expressed from the CAT reporter RNA transcribed from pBLCAT2 (Fig. 1A), we first determined by Northern blotting if the cytoplasmic CAT RNAs expressed in HeLa cells transfected with pBLCAT2 were correctly cleaved and polyadenylated. We thus compared the sizes of the CAT RNAs before and after the removal of the poly(A) tail by treatment with RNase H in the absence or presence of oligo(dT) (Fig. 1A). The correctly 3′-processed CAT RNAs generated from a pre-mRNA initiated at the tk promoter in pBLCAT2 should be close to 1.7 kb but should be 1.58 kb when the poly(A) tail is degraded. This was what we observed (Fig. 1B, lanes 1 and 2), demonstrating that the pre-mRNAs initiated at the tk promoter were correctly cleaved and polyadenylated. Surprisingly, several RNAs smaller than 1.58 kb (seen as a smear in Fig. 1B, lane 1) appeared as homogeneous species on the Northern blot after the removal of the poly(A) tail, ranging from about 0.7 to over 1 kb, and are called here a, b, and c (Fig. 1B, lane 2).
For a better evaluation of the size of the poly(A) tail, we compared the sizes of the CAT RNAs after treatment with RNase H in the presence of oligo(dT) and of oligo(H) located upstream of the small t intron (Fig. 1A). Interestingly, when the cytoplasmic RNAs were hybridized with both oligo(dT) and oligo(H) and digested with RNase H, the sizes of RNAs a, b, and c were unaffected (Fig. 1B, lane 3). However, as expected, the 1.58-kb CAT RNA was cleaved with RNase H into two fragments (Fig. 1A): the fragment that was recognized by probe 1 was 867 nucleotides (nt) (Fig. 1B, lane 3). Moreover, when EB2 was expressed, the small RNA species either decreased in quantity (species a) or disappeared (species b and c), whereas the amount of the 867-nt RNA fragment was not detectably affected (Fig. 1B, lanes 4 and 5).
Plasmid pSV2β (Fig. 1A) was added to the transfections presented in lanes 4 and 5 as an internal control of transfection efficiency. The correctly processed RNAs generated from pre-mRNAs initiated at the SV40 promoter in pSV2β should appear on a Northern blot incubated with the 32P-labeled DNA probe, called probe 2 (Fig. 1A), as 300 to 400 nt long but should be 289 nt when the poly(A) tail is degraded (Fig. 1A). This is what we observed (Fig. 1B, lanes 6 to 9). The amount of pSV2β RNAs detected demonstrated that the transfections were comparable and that the pSV2β pre-mRNAs were correctly processed.
The results presented above suggested strongly that differentially spliced small polyadenylated RNAs were generated from RNA precursors transcribed from pBLCAT2 and that EB2 inhibited their expression. However, EB2 had no apparent effect on the expression of polyadenylated RNAs generated from RNA precursors initiated at the tk promoter in pBLCAT2 or at the SV40 promoter in pSV2β.
EB2 inhibits the use of competing weak 5′ splice sites.
In order to precisely characterize the small polyadenylated RNAs a, b, and c, we PCR amplified an oligo(dT)-primed cDNA made by RT of total cytoplasmic RNAs isolated from the transfected cells. HeLa cells were transfected with a pBLCAT2 derivate, pBLCAT2β, in which we replaced the SV40 CPS with that of the rabbit β-globin gene. This was done to show that the inhibition of the expression by EB2 of RNAs a, b, and c was independent of the 3′ processing signal present in the RNA precursor. The PCR primers P1 and P2 were used, with primer P1, hybridizing both close to the 5′ end of the tk precursor RNA and in the ColE1 region, and primer P2, hybridizing downstream of the 3′ splice site of the SV40 small t intron (Fig. 2A). The PCR products were labeled with [α-32P]dCTP and separated by electrophoresis on a nondenaturing acrylamide gel. As shown in Fig. 2B, lane 1, several PCR products numbered 1 to 7 were obtained and sequenced. Similar results were obtained with pBLCAT2 (data not shown). Their genetic contents (Fig. 2A) demonstrate that two RNA precursors are transcribed from pBLCAT2β.
FIG. 2.
EB2 inhibits the expression of polyadenylated RNAs generated by utilization of alternative cryptic 5′ splice sites. (A) Diagrams of the processed RNAs generated from RNA precursors initiated at the tk promoter and at a cryptic promoter in the vector, after transfection of pBLCAT2β into HeLa cells, and primers used for RT-PCR analysis. (B) 32P-labeled RT-PCR products obtained with primers P1 and P2. (C) 32P-labeled RT-PCR products obtained with primers P3 and P4. Twenty PCR cycles were done in the RT-PCR.
One RNA precursor is initiated at promoter tk from which three polyadenylated RNAs are generated: poly(A) RNA 1 is the unspliced precursor, poly(A) RNA 2 has the small t intron excised, and poly(A) RNA 3 has a longer intron excised, by the usage of a cryptic 5′ splice site located in the CAT open reading frame (ORF). Poly(A) RNA 3 has been identified as the small RNA species a in Fig. 1B, lane 3 (data not shown).
The other RNA precursor is initiated upstream of ColE1 (the promoter has not been precisely mapped), from which four polyadenylated RNAs, poly(A) RNAs 4, 5, 6, and 7, are generated by the excision of two introns. Intron 1 is excised by the alternative usage of four 5′ splice sites and a 3′ splice site located in the β-lactamase gene. Intron 2 is excised by the usage of a 5′ splice site located in the β-lactamase gene and the SV40 small t intron 3′ splice site. The poly(A) RNAs 4, 5, 6, and 7 were identified as part of RNA species b and c in Fig. 1B, lane 3 (data not shown).
We next evaluated the effect of EB2 on the expression of the pBLCAT2β poly(A) RNAs 1 to 7 by RT-PCR. In the presence of EB2, we observed a complete disappearance of cytoplasmic poly(A) RNAs 4, 5, 6, and 7 and a net decrease in poly(A) RNAs 3 and 2, while the cytoplasmic accumulation of unspliced poly(A) RNA 1 increased (Fig. 2B, lane 2). The absolute amount of PCR-amplified DNA fragments increased at 15, 20, and 25 PCR cycles, but the relative abundance of the PCR-amplified DNA fragments in the presence or absence of EB2 was comparable at 15, 20, and 25 PCR cycles (data not shown), demonstrating that our RT-PCR assay was quantitative. Such a quantitation was also done for all of the RT-PCRs shown here. By using PCR primers P3 and P4 flanking the small t intron, we confirmed that the fraction of the cytoplasmic RNAs initiated at the tk promoter was unspliced (Fig. 2C, lane 1) and that in the presence of EB2 the amount of unspliced cytoplasmic RNAs increased (Fig. 2C, lane 2). Alternative splicing often occurs at competing weak 5′ splice sites (for a review see reference 8). Our results strongly suggest that EB2 inhibits the utilization of competing weak 5′ splice sites in the RNA precursors initiated at a cryptic promoter in the vector sequences, independently of the 3′ processing signal present in the precursor RNA. However, EB2 has no effect on the constitutive splicing of the RNA precursor initiated at the SV40 early promoter in pSV2β (Fig. 1B, lanes 6 to 9).
The effect of EB2 on splicing is dependent on cis determinants of 5′ splice site selection.
We next wanted to introduce modifications in the sequence of the ColE1 RNA precursor, to transform alternative 5′ splicing to constitutive 5′ splicing. We could therefore compare the effect of EB2 on the processing of RNA precursors initiated at the same promoter but containing either competing weak 5′ splice sites or a constitutive 5′ splice site. This should be done without affecting the ColE1 and β-lactamase functions. To achieve this, we first constructed a minigene called Paac-ALT1, from which an RNA precursor containing the vector’s four 5′ splice sites, intron 1, part of the β-lactamase exon 2, and the SV40 early CPS was transcribed under the control of the CMV promoter (Fig. 3B). When this plasmid was transiently transfected into HeLa cells, all four 5′ splice sites were used (Fig. 3C, left panel, lane 1). However, EB2 inhibited the utilization of 5′ splice sites 1, 2, and 3 but not that of the proximal 5′ splice site 4, while unspliced RNAs appeared in the cytoplasm (Fig. 3C, left panel, lane 2).
We first tried to increase the efficiency of the utilization of splice site 1 in Paac-ALT1 by changing the exon-intron boundary sequence CGC(U/G)UAGGUAU to GCA(G/G)UAGGUAU in the RNA precursor (Fig. 3B), which hybridized more extensively to the 5′ end of U1 (Fig. 3A). Upon the transfection of plasmid Paac-ALT1.2 in HeLa cells, we observed that alternative splicing reverted to constitutive splicing by the unique usage of the 5′ splice site 1 (Fig. 3C, center panel, lane 1). EB2 did not inhibit the constitutive usage of splice site 1, but the usage of 5′ splice site 4 was weakly increased, and unspliced RNAs were also detected in the cytoplasms of transfected cells (Fig. 3C, center panel, lane 2).
Among several competing weak 5′ splice sites, the exclusive use of the proximal site could be induced by downstream exonic splicing enhancers and factors bound to them (for a review, see reference 8). We therefore generated plasmid Paac-ALT1.3 by changing the β-lactamase second exon in Paac-ALT1 to the rabbit β-globin second exon, except that the sequence CA(G/T)TGC at the intron-exon boundary was unchanged (Fig. 3B). When this construction was transfected into HeLa cells, the only RNAs that accumulated in the cytoplasm were those generated by the usage of splice site 4, proximal to the second exon (Fig. 3C, right panel, lane 1). EB2 had no effect on the usage of splice site 4, and again unspliced RNAs were detected in the cytoplasms of transfected cells (Fig. 3C, right panel, lane 2). The results described above confirmed that EB2 inhibits the use of competing weak 5′ splice sites but has no effect on constitutive splicing in HeLa cells.
Effect of EB2 on constitutive splicing versus alternative splicing.
In order to further study the effect of EB2 on alternative splicing versus constitutive splicing, we also used a more physiological assay: the wild-type (wt) human β-globin gene and a β-thalassemic allele (βthal) (35), containing a G-to-A transition at position 1 of intron 1 (IVS1) which causes the activation of three cryptic 5′ splice sites, otherwise completely silent in the wt gene (Fig. 4A). Plasmid pUCβ128SV containing the wt human β-globin gene activated by the SV40 enhancer and the βthal counterpart pUCβΔ128SV were transiently transfected in HeLa cells, in the presence or absence of an EB2-expressing vector. As evaluated by RT-PCR, EB2 had no effect on the cytoplasmic accumulation of correctly processed wt β-globin RNAs (Fig. 4B, lanes 5 and 6), although EB2 detectably increased the cytoplasmic accumulation of unspliced wt β-globin RNA. However, when the βthal gene was transfected into HeLa cells, the three cryptic 5′ splice sites were used alternatively and EB2 strongly decreased their utilization (Fig. 4B, lanes 7 and 8). It should be noted that the inhibition of alternative splicing was followed by an increase in the amount of unspliced RNAs and that this was seen in the cytoplasm (Fig. 4B, lane 8) and in the nuclear fraction (Fig. 4B, lane 4). Moreover, EB2 did not induce the nuclear accumulation of a particular species of RNA as illustrated by the nucleocytoplasmic repartition of wt β-globin and βthal transcripts (Fig. 4B, lanes 1 to 8). It can be concluded from the experiments described above that EB2 inhibits the alternative utilization of cryptic 5′ splice sites.
However, EB2 had no effect on constitutive splicing, but since this was only seen for the splicing of RNA precursors containing β-globin splice sites, we also used plasmid pSVZ1 carrying the EBV gene BZLF1, from which a pre-mRNA containing three constitutive exons was transcribed under the control of the SV40 early promoter (Fig. 5A). Upon the transfection of pSVZ1 into HeLa cells, we observed by RT-PCR that correctly processed EB1 mRNAs accumulated in the cytoplasms of transfected cells (Fig. 5B, lanes 1 and 2) and that EB2 had no effect on the cytoplasmic accumulation of these RNAs (Fig. 5B, lanes 3 and 4). We also examined the amount of EB1 protein accumulating in transfected HeLa cells by immunoblotting. As shown in Fig. 5C, EB2 also had no inhibitory effect on the amount of EB1 protein expressed in HeLa cells.
EB2 increases the cytoplasmic accumulation of its own intronless RNA.
EB2 has no effect on the cytoplasmic accumulation of mRNAs generated from precursors containing constitutive introns (this report), including the EBV BBLF2/3 mRNA (33). However, EB2 increases the cytoplasmic accumulation of intronless EBV RNAs (33) and CAT reporter RNAs (28). Surprisingly it has been reported that the cytoplasmic accumulation of the BBLF2/3 RNA generated from an intronless cDNA was also EB2 independent (33). In order to test if this is a general property of EB2, we evaluated, by quantitative S1 nuclease analysis, the effect of EB2 on the cytoplasmic accumulation of its own intronless RNA transcribed from a cDNA. The 32P-labeled 5′ end of the single-stranded S1 probe is located in the Flag sequence and allows the detection of RNAs initiated at the CMV promoter in plasmids PaacFlagM1 and PaacFlagEB2 but not the RNAs expressed from PaacEB2 (Fig. 6A). The cytoplasmic accumulation of EB2 mRNAs (Fig. 6C, upper panel, lane 1) and protein (Fig. 6B, lane 2) occurred in the cytoplasms of HeLa cells transfected with plasmid PaacFlagEB2, but when the BMLF1 ORF was interrupted by two stop codons (Fig. 6A, the diagram of PaacFlagM1), no EB2 protein (Fig. 6B, lane 1) and very few EB2 mRNAs (Fig. 6C, upper panel, lane 2) were detected in the cytoplasms of transfected HeLa cells. When PaacFlagM1 was cotransfected with the EB2 expression vector PaacEB2, EB2 strongly increased the cytoplasmic accumulation of M1 mRNAs (Fig. 6C, upper panel, lane 3). Plasmid pSV2β was added to each transfection as an internal control, and equal amounts of specifically initiated RNAs were detected, demonstrating that the transfections were comparable and that EB2 had no effect on the cytoplasmic accumulation of pSV2β mRNAs (Fig. 6C, lower panel). These results also demonstrate that contrary to BBLF2/3 intronless mRNAs (33), the cytoplasmic accumulation of BMLF1 intronless mRNAs is EB2 dependent.
FIG. 6.
EB2 increases the cytoplasmic accumulation of its own RNA. (A) Diagrams of the reporter genes and the single-stranded S1 probe used. (B) Visualization of the Flag EB2 (mAbM2) protein expressed in HeLa cells transfected as indicated in lanes 1 and 2. (C) Quantitative S1 nuclease analysis of the cytoplasmic RNAs expressed in HeLa cells transfected as indicated above the upper panel. The specific protected bands are indicated by Flag (upper panel), for the RNAs initiated at the CMV promoter in PaacFlagM1 and PaacFlag-EB2, and by SV40 for the internal control (lower panel).
EB2 does not affect 3′ end processing.
In the presence of EB2, there is a strong decrease in the cytoplasmic accumulation of polyadenylated RNAs generated from precursors containing weak 5′ splice sites and a strong increase in the cytoplasmic accumulation of BMLF1 intronless RNAs. However, it has also been proposed, but not proved, that EB2 could increase 3′-end processing of EBV B95-8 DNA polymerase intronless pre-mRNAs (15) and therefore could affect cleavage-polyadenylation. We therefore directly determined if EB2 had an effect on cleavage-polyadenylation at canonical and noncanonical signals. To do so we used plasmid I28 from which a rabbit β-globin RNA precursor containing the GM-CSF instability sequences is initiated at the CMV promoter (Fig. 7A). In this assay, the GM-CSF instability sequences could be excluded from the β-globin RNA precursor by inserting functional canonical CPS between the β-globin sequences and the GM-CSF instability sequences (Fig. 7B). Noncanonical CPS could also be tested in this assay, like one found in pUC18 (Fig. 7B) or the EBV DNA polymerase CPS (Fig. 7B). Upon the transfection of plasmid I28 and derivates into HeLa cells, the cytoplasmic polyadenylated β-globin RNAs were quantitated by RT-PCR (Fig. 7C) (see Materials and Methods) and compared to stable cytoplasmic polyadenylated β-globin RNAs expressed from plasmid I22 devoid of GM-CSF instability sequences (Fig. 7A). As shown in Fig. 7D and compared to cytoplasmic β-globin RNAs accumulating upon transfection of plasmid I22 (lane 11), no β-globin RNAs expressed from plasmid I28 (lane 1) or I28-PAPUC (lane 7) were detected in the cytoplasms of transfected cells, and EB2 had no effect on the cytoplasmic accumulation of I28 (lane 2) or I28-PAPUC (lane 8) mRNAs. However, the GM-CSF instability sequences were excluded from the rabbit β-globin precursor as efficiently by the SV40 early CPS (Fig. 7D, lane 3), the rabbit β-globin CPS (lane 6), or the EBV DNA polymerase noncanonical CPS (lane 9). Moreover, EB2 had no mandatory effect on the efficiency of cleavage-polyadenylation at canonical CPS (Fig. 7D, lanes 4, 6, and 12), while it decreased slightly the cleavage-polyadenylation at the noncanonical DNA polymerase CPS (lanes 9 and 10). Similar results were obtained by Northern blotting (data not shown). These results directly demonstrate that EB2 has no effect on cleavage-polyadenylation and that the EBV DNA polymerase CPS is as efficient as the SV40 and β-globin CPS in our assay.
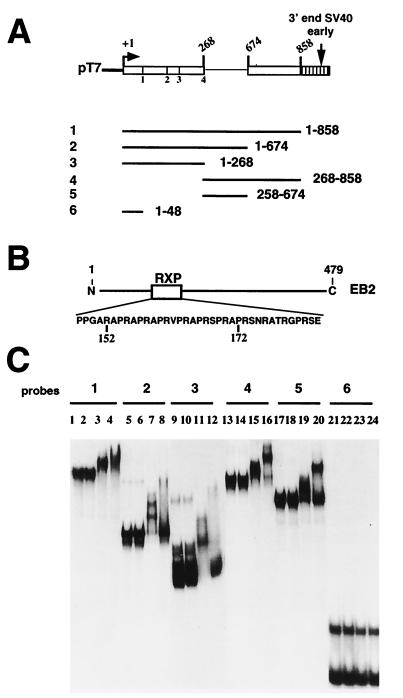
Both EB2 and the EB2 Arg-X-Pro repeat bind RNA in vitro.
Since EB2 appears to inhibit the use of competing weak (cryptic) 5′ splice sites and induce the cytoplasmic accumulation of intron-containing and intronless RNAs, we then determined if EB2 can bind directly to RNA. In EB2 there is an Arg-X-Pro tripeptide repeat located between amino acids 152 and 172 (Fig. 8B), similar to that in the HSV-1 US11 protein, which has been described as an RNA-binding domain (27, 31). We therefore purified a GST protein, a GST-EB2 fusion protein, and a GST–Arg-X-Pro repeat fusion protein (data not shown) and asked whether these proteins could bind in vitro to 32P-labeled RNAs transcribed in vitro from the Paac-ALT1 reporter gene and containing different sequences (Fig. 8A). As shown in Fig. 8C, the GST protein did not interact with any of the RNA probes used (lanes 2, 6, 10, 14, 18, and 22), and both the GST-EB2 and the GST–Arg-X-Pro repeat fusion proteins did not bind to RNA probe 6, which contains 48 nt of the Paac-ALT1 exon sequence located upstream of 5′ splice site 1, but is devoid of the 5′ splice site 1 sequences (lanes 23 and 24). The GST–Arg-X-Pro repeat fusion protein bound to RNA probes 1 to 5 (Fig. 8C, lanes 3, 7, 11, 15, and 19). However, on the shorter RNA probes 2, 3, and 5 (lanes 7, 11, and 19), several RNA–GST–Arg-X-Pro complexes could be separated, suggesting that more than one molecule of GST–Arg-X-Pro bound per molecule of RNA. The GST-EB2 fusion protein bound to RNA probes 1 to 5 (Fig. 8C, lanes 4, 8, 12, 16, and 20) but did not bind efficiently to RNA probes 2, 3, and 5, which lack the second exon sequence (lanes 8, 12, and 20), suggesting that contrary to GST–Arg-X-Pro, the GST-EB2 protein bound Paac-ALT1 RNA containing the second exon sequence more efficiently.
FIG. 8.
EB2 and the EB2 Arg-X-Pro repeat bind to RNA in vitro as GST fusion proteins. (A) Schematic representations of the RNA probes generated from the Paac-ALT1 minigene. (B) Diagram of the EB2 protein and of the Arg-X-Pro (RXP) polypeptide, which were fused to GST. (C) EMSAs were performed with probes 1 to 6. The 32P-labeled RNA probes used were loaded alone in lanes 1, 5, 9, 13, 17, and 21 or were loaded after incubation with the GST protein (lanes 2, 6, 10, 14, 18, and 22), the GST–Arg-X-Pro protein (lanes 3, 7, 11, 15, 19, and 23), or the GST-EB2 protein (lanes 4, 8, 12, 16, 20, and 24).
The EB2 Arg-X-Pro repeat is dispensable for the effect of EB2 on alternative splicing and unspliced RNA export.
In order to evaluate if the Arg-X-Pro tripeptide repeat was a functional domain in EB2, we generated a series of deletions in the EB2-encoding ORF BMLF1 (Fig. 9A) and first examined if the EB2 deletion mutants were equally produced in HeLa cells. As shown in Fig. 9B, among the EB2 proteins transfected, N-terminal mutants Δ1, Δ3, Δ4, and Δ5 were expressed at a level comparable to the wt EB2 protein, while the other mutated EB2 proteins were either poorly expressed (Δ7, Δ9, and Δ10) or not detectable (Δ12, Δ13, and Δ15). We also transfected the EB2 mutants into Cos7 cells, and in these cells we observed that all the mutated proteins were expressed at levels similar to those of the wt EB2 protein (Fig. 9C). We next examined, by RT-PCR, the effect of EB2 and EB2 deletion mutants on the RNAs generated from Paac-ALT1 by the usage of alternative 5′ splice sites. Upon the transfection of Paac-ALT1 in Cos7 cells, mainly RNA species 2, 3, and 4 were detected in the cytoplasmic fraction (Fig. 9D, lane 2), but surprisingly and contrary to what was observed in HeLa cells, EB2 did not inhibit the usage of the competing weak 5′ splice sites 1, 2, and 3 but favored the cytoplasmic accumulation of RNAs generated by the use of the proximal 5′ splice site 4 (Fig. 9D, lane 1). Among the mutants tested, mutants Δ7 to Δ15 (Fig. 9D, lanes 7 to 12) were inactive with respect to the effect of the wt protein (Fig. 9D; compare lanes 1 and 2), although the mutant Δ5, lacking the Arg-X-Pro repeat, was as active as the wt protein (Fig. 9D; compare lanes 1 and 6). The mutant protein Δ5 was also expressed in HeLa cells at a level comparable to that of the EB2 wt protein (Fig. 9B; compare lanes 2 and 6), and the deletion of the Arg-X-Pro domain did not affect the activity of the mutated protein on the alternative splicing and cytoplasmic transport of unspliced RNAs (Fig. 9E; compare lanes 1 and 6). It should be noted that unspliced RNAs also appeared in the cytoplasms of Cos7 cells (Fig. 9D, lanes 1 and 3 to 6) only when active EB2 proteins were expressed and that the mutants inactive in Cos7 cells were also those poorly expressed in HeLa cells (Fig. 9B, lanes 7 to 12). The poor expression of the EB2 mutated proteins Δ7, Δ9, Δ10, Δ12, Δ13, and Δ15 in HeLa cells is due to the inactivation of EB2 functions, since the cytoplasmic accumulation of EB2 intronless mRNAs and by extension of the EB2 protein are EB2 dependent (Fig. 6C).
DISCUSSION
Our findings directly demonstrate that the EBV protein EB2 acts by nontranscriptional mechanisms. Our findings also strongly suggest that EB2 binds RNA with no apparent specificity, inhibits the cytoplasmic accumulation of RNAs generated by use of competing weak (cryptic) 5′ splice sites, and induces the cytoplasmic accumulation of both intronless and intron-containing RNAs.
Our initial observation involved transiently transfecting plasmid pBLCAT2 into HeLa cells and observing by Northern blotting that the RNAs initiated at the tk promoter and processed at the expected constitutive signals were less abundant than several differentially spliced smaller polyadenylated RNAs. By RT-PCR, we determined the genetic contents of these small RNAs. We found that they were generated from an RNA precursor initiated in the vector sequences, by the alternative usage of 5′ splice sites. Their cytoplasmic accumulation decreased strongly in the presence of EB2. Furthermore, we found by RT-PCR that the cryptic promoter in the vector sequences was located about 300 bp upstream of primer P1 (Fig. 2A) (data not shown). Our RT-PCR quantitation completely corroborated our Northern blotting quantitation, except for the two CAT-containing RNAs 1 and 2, which were separated as RT-PCR products (Fig. 2C) but were indistinguishable on the Northern blot (Fig. 1B, lanes 1 to 5). However, our results demonstrated that the effect of EB2 on the alternative use of 5′ splice sites could be reliably quantitated under our RT-PCR conditions and was independent of the 3′ processing signal present in the RNA precursors.
Several pBLCAT2 derivatives have been generated to prevent the background transcription of RNAs initiated at cryptic, although not identified, promoters within the vector, and these pBLCAT2 derivatives indeed had a lower CAT activity background level (3). However, our results demonstrate that none of the processed RNAs generated from RNAs initiated at the cryptic ColE1 promoter contain the CAT coding sequence and therefore cannot generate a high background level of unspecific CAT activity. Moreover, they also show that most of the transient-expression assays that have been published have been done with competing promoters and that composite plasmid-gene reporter RNAs could have been generated, obscuring the conclusions made.
All the spliced RNAs accumulating in the cytoplasm of HeLa cells transfected with pBLCAT2 or pBLCAT2β were generated by alternative 5′ splicing, and their accumulation in the cytoplasm was inhibited, although to different extents, by EB2 (Fig. 2A and B). We never observed an effect of EB2 on the cytoplasmic accumulation of polyadenylated RNAs generated by the use of constitutive splice sites in the pSV2β RNA precursor (Fig. 1B), the human β-globin pre-mRNA (Fig. 4B), or the BZLF1 pre-mRNA (Fig. 5B). However, when human β-globin pre-mRNA constitutive splicing was transformed into alternative splicing by a β-thalassemia mutation, EB2 inhibited the cytoplasmic accumulation of the three polyadenylated RNAs generated by the alternative use of competing cryptic 5′ splice sites (Fig. 4B). Similarly, from the minigene Paac-ALT1, several polyadenylated RNAs were generated by the alternative usage of competing weak 5′ splice sites, and EB2 inhibited differentially their cytoplasmic accumulation (Fig. 3C). However, when alternative splicing (Fig. 3C, left panel) was transformed to constitutive splicing (Fig. 3C, Paac-ALT1.2 and Paac-ALT1.3), EB2 had no effect on the usage of the constitutive 5′ splice sites. In the experiments described above, the transcription of the RNA precursors carrying either constitutive splice sites or alternative splice sites was initiated at the same promoter, demonstrating that the effect of EB2 was not transcriptional. We also used a β-globin construct from which an RNA precursor containing a duplication of a constitutive β-globin 5′ splice site was transcribed (construction 5′-D16 [26]). In HeLa cells, the distal 5′ splice site was much more efficiently used than the proximal 5′ splice site, and a cryptic splice site located upstream of the distal 5′ splice site was poorly used (39 and data not shown). We observed that EB2 had no effect on the alternative usage of the duplicated constitutive 5′ splice sites but inhibited the cytoplasmic accumulation of RNAs generated by the use of the cryptic 5′ splice site (data not shown). One puzzling result is that in Cos7 cells, although EB2 induced the cytoplasmic accumulation of unspliced RNAs, it favored the cytoplasmic accumulation of RNAs generated by the use of the proximal 5′ splice site 4 (Fig. 9C, lane 1). This observation reinforces the idea that EB2 is a regulator of splicing and might reflect the fact that Cos1 cells and HeLa cells differ in their RNA processing machinery, i.e., in SR proteins. Since EB2 and mutated EB2 proteins are expressed from intronless RNAs, our results also strongly suggest that intronless RNAs are more stable in Cos1 cells than in HeLa cells. It is not clear at the moment if EB2 selectively affects the use of weak 5′ splice sites or increases the RNA export of poor RNA substrates for splicing.
Indeed, one striking observation is that in the presence of EB2, the decrease in the cytoplasmic accumulation of poor RNA substrates for splicing was always accompanied by an increase in the cytoplasmic accumulation of unspliced RNAs. This was, however, also seen with RNA templates containing constitutive splice sites (Fig. 4B, lanes 5 and 6), but when poor RNA templates for splicing were used, the cytoplasmic accumulation of unspliced RNAs was more efficient (Fig. 4; compare lanes 6 and 8) (Fig. 3; compare lanes 1 and 2 for Paac-ALT1 with lanes 1 and 2 for Paac-ALT1.2). Taken together, our results suggest that EB2 could induce, directly or indirectly and in a cell- and template-specific manner, the nuclear export of intron-containing and intronless RNAs. This EBV property is relevant to EBV biology, since there are EBV early intronless RNAs and also several early and late EBV intron-containing RNAs that accumulate in the cytoplasm of infected cells during the productive cycle, like the BMLF1 mRNA (5 and references therein), the BZLF1-BRLF1 bicistronic mRNAs (20), the BLLF2 mRNA (1), and the late BNLF1 mRNA (4), and these intron-containing RNAs are likely to be poor substrates for splicing.
The mechanisms by which EB2 exerts such functions are unclear. EB2 binds to pUC18-derived RNA in vitro and to EBV RNAs (28, 33), and there is no RNA recognition motif in EB2, characteristic of RNA-binding proteins involved in spliceosome assembly and function or in RNA nucleocytoplasmic transport (for reviews see references 17, 23, and 34). However, one domain of EB2 has homology with an RNA-binding domain identified in the HSV-1 US11 protein (27, 31), which contains a repeat of the tripeptide sequence Arg-X-Pro, and is located in EB2 between amino acids 152 and 172 (Fig. 8B). Nevertheless, we found that there is no specific binding of a GST–Arg-X-Pro fusion protein to the Paac-ALT1 RNA precursor. Moreover, the deletion of this domain in EB2 (Fig. 9A) did not impair its effect on alternative splicing and on the nuclear export of intron-containing RNAs (Fig. 9D and E, lanes 6). It is noteworthy that the GST-EB2 fusion protein bound to the Paac-ALT1 RNA precursor sequences with some specificity, since binding was more efficient when the second exon sequences were part of the RNA probe used in the mobility shift assay (Fig. 8C). In addition, the GST-EB2 protein did not bind to the smaller 48-nt RNA probe 6, which lacks a 5′ splice site. There is also a putative nuclear export signal (NES) in EB2 (36), located between amino acids 226 and 237 and homologous to the human immunodeficiency virus type 1 Rev NES (36). This sequence is deleted in mutant Δ7 (Fig. 9A), and this EB2 mutant is inactive in unspliced RNA export in Cos7 cells (Fig. 9D, lane 7). We have not yet precisely characterized the domain(s) of EB2 involved in RNA binding or specifically mutated the NES sequence to examine the effect of this mutation on the functions of EB2 in transient-expression assays.
EB2 also has some functional homologies to SR proteins that affect, when overexpressed, alternative splicing in a cell- and template-dependent manner (for a review see reference 17). Indeed, EB2 favored the cytoplasmic accumulation of RNAs generated by the use of the proximal 5′ splice site 4 (Fig. 7D and 3C). Although there is no RNA-binding domain in EB2 characteristic of SR proteins, scattered RS and SR dipeptides are found in EB2, like those found in the splicing regulators SRp160 (2) and Sip1 (38), which also do not contain RNA-binding domains. Moreover, EB2 colocalizes with the SR protein SC35 and shuttles between the nucleus and the cytoplasm (33), and a specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm (6). It is not clear at present if EB2 actively transports RNAs in the cytoplasm or is cotransported with RNAs into the cytoplasm where it might perform important functions.
Another unanswered question is the role of EB2 in the EBV replicative cycle. Other herpesviruses contain genes in their genomes from which proteins with homology to EB2 are expressed. The herpesvirus saimiri ORF 57 protein, p52, which is highly homologous to EB2 (24), has been shown to repress the expression of target genes containing an intron, but no direct study on splicing was done (37). The HSV-1 ICP27 protein has limited amino acid homology to EB2 and has been implicated in different steps of pre-mRNA processing, such as 3′-end processing (30), selection of poly(A) site usage (21), and inhibition of splicing (30) and more recently in the nucleocytoplasmic transport of some viral intronless RNAs (25, 29). EB2 also shuttles from the nucleus to the cytoplasm, and it has been suggested that the function of EB2 would be to increase the nuclear export of intronless EBV replication gene mRNAs, which will otherwise be sequestered and degraded in the nucleus (33). EB2 could therefore be essential for the efficient replication of the viral genome, although that has not been shown in infected cells. It could also be that EB2 affects the expression of highly spliced EBNA or LMP RNAs, but that point has not been addressed yet. However, the EB2-associated increased cytoplasmic accumulation of intronless RNAs is not specific to EBV RNAs, since it is also seen with CAT-containing reporter RNAs (28) and when the intronless RNA is reduced to only the bacterial CAT coding sequences (data not shown). Moreover, we also show that EB2 increases the cytoplasmic accumulation of intron-containing RNAs and that there are intron-containing RNAs that accumulate in the cytoplasm of EBV-infected cells in which a productive cycle is taking place (4, 5, 20). Finally, EB2 has been shown to induce the growth of Rat1 and NIH 3T3 cells at a low density under soft agar (11). It is therefore interesting to see if the effect of EB2 on the processing and/or transport of cellular RNAs is linked to cell transformation.
Although our results and the results of others point to the fact that EB2 is an RNA-binding protein affecting RNA splicing and transport in transient-expression assays, the function of EB2 in the EBV productive cycle is not yet known. This function has now to be directly tested by characterizing specific RNA and protein targets for EB2 and by producing a recombinant EBV expressing a conditional EB2 protein, in order to quantify and characterize EBV RNAs expressed from EBV early genes in infected cells expressing, or not expressing, a functional EB2 protein.
ACKNOWLEDGMENTS
M.B. and F.H. contributed equally to this work.
We thank H. Gruffat, E. Manet, and I. Mikaelian (U412 INSERM, Lyon, France) and Angela Krämer (Département de Biologie, Université de Genève, Geneva, Switzerland) for constructive discussions and Conrad B. Bluink for critical reading of the manuscript. The human β-globin and the β-thalassemia and pCDNA3-D16 constructs were kindly provided, respectively, by A. Krainer (Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.) and Jane Wu (Washington University School of Medicine, St. Louis, Mo.).
This work was supported by INSERM and by ARC (contract 9439 to A.S.).
REFERENCES
- 1.Baer R, Bankier A, Biggin M, Deininger P, Farrell P, Gibson T, Hatfull G, Hudson G, Satchwell S, Deguin C, Tuffnell P, Barrell B. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe B J, Issner R, Sharp P A. A co-activator of pre-mRNA splicing. Genes Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshart M, Klüppel M, Schmidt A, Schütz G, Lucckow B. Reporter constructs with low background activity utilizing the cat gene. Gene. 1992;110:129–130. doi: 10.1016/0378-1119(92)90456-y. [DOI] [PubMed] [Google Scholar]
- 4.Boss H, Berger R, Kuklik R C, Ifner T, Mueller L N. Enhancement of Epstein-Barr virus membrane protein (LMP) expression by serum, TPA, or n-butyrate in latently infected cells. Virology. 1987;159:161–165. doi: 10.1016/0042-6822(87)90360-6. [DOI] [PubMed] [Google Scholar]
- 5.Buisson M, Manet E, Trescol-Biemont M-C, Gruffat H, Durand B, Sergeant A. The Epstein-Barr virus (EBV) early protein EB2 is a posttranscriptional activator expressed under the control of EBV transcription factors EB1 and R. J Virol. 1989;63:5276–5284. doi: 10.1128/jvi.63.12.5276-5284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caceres J F, Screaton G R, Krainer A. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caceres J F, Stamm S, Helfman D H, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 8.Chabot B. Directing alternative splicing. Trends Genet. 1996;12:472–477. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- 9.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV) encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook I D, Shanahan F, Farrell P J. Epstein Barr virus SM protein. Virology. 1994;205:217–227. doi: 10.1006/viro.1994.1637. [DOI] [PubMed] [Google Scholar]
- 11.Corbo L, Le Roux F, Sergeant A. The EBV early gene product EB2 transforms rodent cells through a signalling pathway involving c-Myc. Oncogene. 1994;9:3299–3304. [PubMed] [Google Scholar]
- 12.Derek A M, Mikaelian I, Zemmel R W, Green S M, Lowe A D, Kimura T, Singh M, Butler J G, Gait M J, Karn J. Co-operative Rev binding to stem I of the Rev-response element modulates human immunodeficiency virus type-1 late gene expression. J Mol Biol. 1994;241:193–207. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- 13.Kenney S, Kamine J, Holley-Guthrie E, Mar E C, Lin J C, Markovitz D, Pagano J. The Epstein-Barr virus immediate-early gene product, BMLF1, acts in trans by a posttranscriptional mechanism which is reporter gene dependent. J Virol. 1989;63:3870–3877. doi: 10.1128/jvi.63.9.3870-3877.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenney S, Kamine J, Markovitz D, Fenrick R, Pagano J. An Epstein-Barr virus immediate-early gene product trans-activates gene expression from the human immunodeficiency virus long terminal repeat. Proc Natl Acad Sci USA. 1988;85:1652–1656. doi: 10.1073/pnas.85.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Key S C S, Yoshizaki T, Pagano J S. The Epstein-Barr virus (EBV) SM protein enhances pre-mRNA processing of the EBV DNA polymerase transcript. J Virol. 1998;72:8485–8492. doi: 10.1128/jvi.72.11.8485-8492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D, Howley P, et al., editors. Fields virology. Philadelphia, Pa: Raven Publishers; 1996. pp. 2343–2395. [Google Scholar]
- 17.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 18.Lieberman P M, O’Hare P, Hayward G S, Hayward S D. Promiscuous trans activation of gene expression by an Epstein-Barr virus-encoded early nuclear protein. J Virol. 1986;60:140–148. doi: 10.1128/jvi.60.1.140-148.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luckow B, Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manet E, Gruffat H, Trescol-Biemont M-C, Moreno N, Chambard P, Giot J-F, Sergeant A. Epstein-Barr virus bicistronic mRNA generated by facultative splicing code for two transcriptional transactivators. EMBO J. 1989;8:1819–1826. doi: 10.1002/j.1460-2075.1989.tb03576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGregor F, Phelan A, Dunlop J, Clements J B. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikaelian I, Krieg M, Gait M J, Karn J. Interaction of INS (CRS) elements and the splicing machinery regulates the production of Rev-responsive mRNAs. J Mol Biol. 1996;257:246–264. doi: 10.1006/jmbi.1996.0160. [DOI] [PubMed] [Google Scholar]
- 22a.Mikaelian I, Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagai K, Oubridge C, Ito N, Avis J, Evans P. The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- 24.Nicholas J, Gompels U A, Craxton M A, Honess R W. Conservation of sequence and function between the product of the 52-kilodalton immediate-early gene of herpesvirus saimiri and the BMLF1-encoded transcriptional effector (EB2) of Epstein-Barr virus. J Virol. 1988;62:3250–3257. doi: 10.1128/jvi.62.9.3250-3257.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phelan A, Clements B. Herpes simplex virus type 1 immediate-early protein IE63 shuttles between nuclear compartments and the cytoplasm. J Gen Virol. 1997;78:3327–3331. doi: 10.1099/0022-1317-78-12-3327. [DOI] [PubMed] [Google Scholar]
- 26.Reed R, Maniatis T. A role for exon sequence and splice-site proximity in splice-site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 27.Roller R J, Roizman B. The herpes simplex virus US11 open reading frame encodes a sequence-specific RNA-binding protein. J Virol. 1990;64:3463–3470. doi: 10.1128/jvi.64.7.3463-3470.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruvolo V, Wang E, Boyle S, Swaminathan S. The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc Natl Acad Sci USA. 1998;95:8852–8857. doi: 10.1073/pnas.95.15.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandri-Goldin R M, Mendoza G E. A herpes virus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 31.Scharer-Uthurralt N, Evard M, Kindbeiter K, Madjar J J, Diaz J J. Distinct domains of HSV1 US11 protein mediate post-transcriptional transactivation of HTLV-1 envelope glycoprotein gene expression and specific binding to the Rex responsive element. J Gen Virol. 1998;79:1593–1602. doi: 10.1099/0022-1317-79-7-1593. [DOI] [PubMed] [Google Scholar]
- 32.Segouffin C, Gruffat H, Sergeant A. Repression by RAZ of Epstein-Barr virus bZIP transcription factor EB1 is dimerization independent. J Gen Virol. 1996;77:1529–1536. doi: 10.1099/0022-1317-77-7-1529. [DOI] [PubMed] [Google Scholar]
- 33.Semmes O J, Chen L, Sarisky R T, Gao Z, Zhong L, Hayward S D. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J Virol. 1998;72:9526–9534. doi: 10.1128/jvi.72.12.9526-9534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 35.Treisman R, Orkin S H, Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983;302:591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- 36.Wen W, Meinkoth J L, Tsien R Y, Taylor S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 37.Whitehouse A, Cooper M, Meredith D M. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J Virol. 1998;72:857–861. doi: 10.1128/jvi.72.1.857-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W-J, Wu J Y. Sip1, a novel RS domain-containing protein essential for pre-mRNA splicing. Mol Cell Biol. 1998;18:676–684. doi: 10.1128/mcb.18.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W-J, Wu J Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]