Abstract
Many stages of the herpes simplex virus maturation pathway have not yet been defined. In particular, little is known about the assembly of the virion tegument compartment and its subsequent incorporation into maturing virus particles. Here we describe the construction of a herpes simplex virus type 1 (HSV-1) recombinant in which we have replaced the gene encoding a major tegument protein, VP22, with a gene expressing a green fluorescent protein (GFP)-VP22 fusion protein (GFP-22). We show that this virus has growth properties identical to those of the parental virus and that newly synthesized GFP-22 is detectable in live cells as early as 3 h postinfection. Moreover, we show that GFP-22 is incorporated into the HSV-1 virion as efficiently as VP22, resulting in particles which are visible by fluorescence microscopy. Consequently, we have used time lapse confocal microscopy to monitor GFP-22 in live-cell infection, and we present time lapse animations of GFP-22 localization throughout the virus life cycle. These animations demonstrate that GFP-22 is present in a diffuse cytoplasmic location when it is initially expressed but evolves into particulate material which travels through an exclusively cytoplasmic pathway to the cell periphery. In this way, we have for the first time visualized the trafficking of a herpesvirus structural component within live, infected cells.
The herpesvirus particle is made up of four concentric compartments, namely, the DNA core, the capsid, the tegument, and the envelope (4, 24). The tegument is the least understood compartment of the virion in relation to the functions of its individual constituents, its role in virus entry, and its mechanism of assembly and incorporation into the maturing virion. In particular, the site of tegument assembly within the cell remains to be determined. While it is well established that during virus replication genomic DNA is packaged into assembling capsids within the nucleus (25), the subsequent sites of tegument assembly, capsid envelopment, and virion maturation are somewhat controversial. Early electron microscopy studies showing the thickening of nuclear membranes next to assembled capsids (17, 18, 27) were interpreted to mean that the virion envelope was acquired at the inner nuclear membrane and that maturation of envelope glycoproteins occurred as vesicles containing these virions moved through the secretory pathway. Nonetheless, this model has always been complicated by the presence of numerous unenveloped capsids in the cytoplasm of infected cells (33), a feature which has previously been suggested to reflect a “dead-end” pathway of terminal de-envelopment (2). However, several recent reports on the localization and processing of virus glycoproteins provide evidence that these naked capsids may be on the true pathway of virus assembly, whereby virions budding through the nuclear membrane would proceed through a further stage of de-envelopment at the outer nuclear membrane and acquire their final envelope downstream in the secretory pathway (1, 3, 29, 33). It is therefore likely that the identification of the cellular site of tegument assembly will help to address the issues surrounding the herpesvirus maturation pathway(s).
The herpes simplex virus type 1 (HSV-1) protein VP22 is a major tegument component of the virus particle (11, 13, 28). While the exact role of VP22 during virus infection remains unclear, we have shown that VP22 exhibits several fascinating properties. During expression in tissue culture cells by either transient transfection or virus infection, VP22 exhibits the property of intercellular spread, which is so efficient that an individual cell expressing the protein can deliver it to as many as 200 surrounding cells (8). Furthermore, we have recently shown that in cells which actively synthesize VP22, during either transient transfection or virus infection, the protein reorganizes and stabilizes the cellular microtubule network, and as such VP22 is the first animal virus-encoded protein shown to possess the properties of a cellular microtubule-associating protein (9). Thus, not only is VP22 a major structural component of the virus particle, but it also exhibits several interesting cellular interactions which may be important to the virus replication cycle.
We have previously shown that a green fluorescent protein (GFP)-VP22 fusion protein (GFP-22) is competent for both intercellular movement and interaction with microtubules (8), suggesting that the addition of GFP onto the VP22 open reading frame has no effect on VP22 activities within the cell. In this report we describe the construction of an HSV-1 recombinant in which we have exchanged the single copy of the VP22 open reading frame in the HSV-1 genome, gene UL49 (10), with a gene encoding GFP-22. Surprisingly, this virus is fully viable and exhibits growth kinetics similar to those of its parental virus. Moreover, GFP-22 is incorporated into the virus particle with the same efficiency as VP22. The presence of GFP-22 in the virion results in fluorescent particles which are readily visualized with a light microscope. Furthermore, we show that newly synthesized GFP-22 is detectable as early as 3 h after infection at a high multiplicity, allowing the direct visualization of GFP-22 within live cells. As a consequence of such sensitive detection of GFP-22 throughout infection, we have been able to use time lapse confocal microscopy to monitor the trafficking of GFP-22 within individual cells, at both high and low multiplicities of infection, the results of which we present as time lapse animations. Thus, we have generated a reagent which will enable the visualization of several aspects of HSV-1 infection in live cells, including virus entry, assembly, trafficking, and egress.
MATERIALS AND METHODS
Cells and virus infections.
Vero cells, COS-1 cells, and BHK-1 cells were maintained in Dulbecco’s modified minimal essential medium containing 10% newborn calf serum. The parental virus used in this study was HSV-1 strain 17.
Virions were purified from extracellular virus released into the infected cell medium. Approximately 5 × 108 BHK cells were infected at a multiplicity of 0.05. Four days later, the infected cells were harvested into the cell medium and centrifuged for 30 min at 3,000 rpm in a Sorvall GS3 rotor. The supernatant was removed and centrifuged for a further 90 min at 10 K to pellet the extracellular virus. The resulting crude virus pellet was resuspended in 1 ml of phosphate-buffered saline (PBS) and laid onto a 12-ml 5 to 15% Ficoll gradient, which was centrifuged at 12,000 rpm in a Sorvall TH641 rotor for 2 h. The virion band was harvested through the side of the tube, diluted in 10 ml of PBS, and pelleted for 1 h at 25,000 rpm in a Sorvall TH641 rotor. Following resuspension in 0.5 ml of PBS, the virion stock was placed at −70°C.
Infectious virus DNA was also produced from extracellular virus. BHK cells were infected as described above, and the crude extracellular virus pellet was resuspended in 5 ml of 10 mM Tris-HCl (pH 7.6)–1 mM EDTA–1% sodium dodecyl sulfate (SDS)–50 μg of proteinase K/ml. After gentle mixing the suspension was incubated at 50°C for 4 h, extracted twice with phenol and twice with phenol-chloroform, and ethanol precipitated.
Construction of GFP-22 virus.
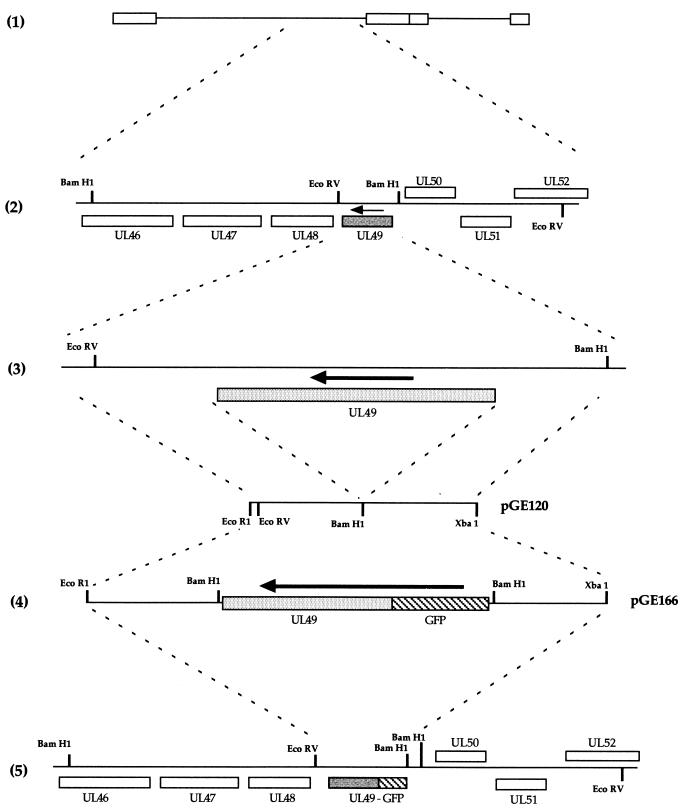
The construction of the GFP-22-expressing virus is detailed in Fig. 1. The two 400-bp flanking sequences of the HSV-1 UL49 gene (Fig. 1, diagram 3) were amplified together by PCR from purified genomic DNA to construct a single 800-bp fragment incorporating an EcoRI site at one end, an XbaI site at the other, and a BamHI site engineered in place of the UL49 gene (Fig. 1, diagram 3). This was inserted into plasmid pSP72 (Promega) as an EcoRI/XbaI fragment to produce plasmid pGE120 (Fig. 1, diagram 3). A GFP-UL49 cassette contained on a BamHI fragment was then inserted into the BamHI site of pGE120 to produce plasmid pGE166 (Fig. 1, diagram 4), which consisted of GFP-UL49 surrounded by the UL49 flanking sequences and hence driven by the UL49 promoter.
FIG. 1.
Construction of the GFP-22-expressing virus. (1) Schematic representation of the HSV-1 genome. (2) The region of the genome which contains the UL49 gene (shaded). The arrow indicates the direction of transcription. (3) The EcoRV/BamHI fragment which contains the UL49 gene and its flanking sequences. The 400-bp sequences on either side of the UL49 gene were amplified by PCR with EcoRI and XbaI sites at either end, and a BamHI site in place of the UL49 gene (4). The EcoRI/XbaI fragment was inserted into EcoRI/XbaI-digested pSP72 to make pGE120. A GFP-VP22 BamHI cassette was inserted into the BamHI site of pGE120 to make pGE166. (5) The structure of the genome resulting from recombination of plasmid pGE166 with HSV-1 DNA.
Equal amounts (2 μg) of plasmid pGE166 and infectious HSV-1 strain 17 DNA were transfected into 106 COS-1 cells grown in a 60-mm-diameter dish by using the calcium phosphate precipitation technique modified with BES [N,N-bis(2-hydroxyl)-2-aminoethanesulfonic acid]-buffered saline in place of HEPES-buffered saline. Four days later, the infected cells were harvested into the cell medium and subjected three times to freeze-thawing, and the resulting virus was titrated on Vero cells. Around 6,000 plaques were then plated onto Vero cells and screened for possible recombinants by GFP fluorescence.
Virus genomic DNA screening.
Virus DNA for restriction digestion was purified from 5 × 107 infected BHK cells. Infected cell pellets were washed in cold PBS and resuspended in 5 ml of 10 mM Tris-HCl (pH 7.5)–2 mM MgCl2–10 mM NaCl–0.5% Nonidet P-40. After vortexing, the cells were left on ice for 5 min and vortexed again. Nuclei were pelleted out at 2,000 rpm in a benchtop centrifuge, and 0.5% SDS and 10 mM EDTA were added to the supernatant. This was extracted twice with phenol-chloroform and once with chloroform and then was ethanol precipitated. After pelleting, the nucleic acid was resuspended in 100 ml of H2O and digested overnight with EcoRV in the presence of RNase A. Electrophoresis was carried out in a 0.8% agarose gel for 8 h at 100 V, and the gel was transferred to a nylon membrane by standard procedures. The Southern blots were then hybridized with a 32P-labelled DNA probe synthesized by the random priming of fragments specific for either UL49 or GFP.
SDS-PAGE and Western blot analysis.
Solubilized proteins were subjected to SDS-polyacrylamide gel electrophoresis (PAGE), and the gels were either stained with Coomassie blue or transferred to nitrocellulose filters and reacted with the appropriate primary antibody. A horseradish peroxidase-linked secondary conjugate was used, and reactive bands were visualized with the enhanced chemiluminescence (ECL) detection reagents (Amersham).
The polyclonal anti-VP22 antibody, AGV30, has been described previously (8). Monoclonal antibodies against α tubulin and acetylated tubulin were obtained from Sigma. The monoclonal anti-GFP antibody was obtained from Clontech. Antibodies against IE110 (11060), thymidine kinase (TK), and VP16 (LP1) were kindly provided by Roger Everett, David Gower, and Tony Minson, respectively.
Live-cell microscopy and time lapse analysis.
Cells for short-term live analysis of GFP expression were plated onto 24-mm coverslips placed in six-well trays (Costar). At the specified time of analysis, the coverslip was transferred to a 35-mm Attofluor cell chamber (Molecular Probes), 2 ml of fresh medium was added, and the cells were examined with a Zeiss LSM 410 inverted confocal microscope. Resulting images were processed by using Adobe Photoshop software.
Cells for long-term time lapse analysis were plated onto 42-mm coverslips contained in 60-mm dishes. Prior to analysis, the coverslip was transferred to a Bachhoffer POC-chamber (obtained from Carl Zeiss) in open cultivation mode. This chamber was placed on a Saur heated frame (obtained from Carl Zeiss) on the microscope and covered with a Perspex lid through which a constant supply of 5% CO2 was fed. XYZT software from Zeiss was used to collect a Z series of images for each point in the time series, and these were then merged to produce an individual Z image for each time point. Animation of the time series was carried out by using NIH-Image software, and each series was saved as a Quicktime video. The animations accompanying Fig. 7 and 8 are located at http://mc11.mcri.ac.uk/mov/figure7.mov and http://mc11.mcri.ac.uk/mov/figure8.mov.
FIG. 7.
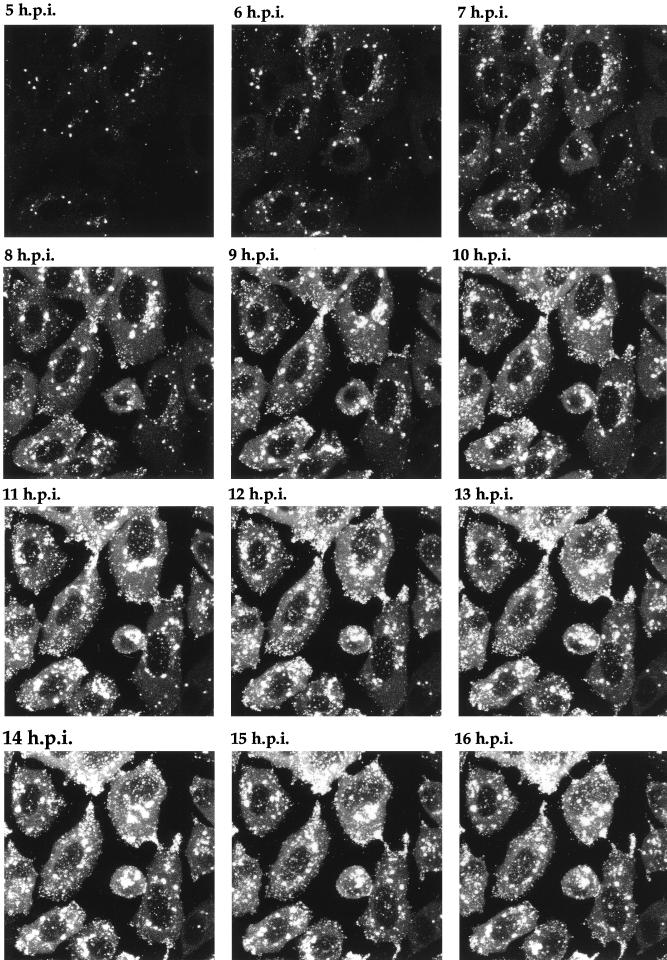
Time lapse analysis of GFP-22 trafficking in a high-multiplicity 166v infection. Vero cells were infected with 166v at 10 PFU per cell and were transferred to the heated chamber 5 h postinfection (5 h.p.i.). A single field was chosen for analysis, and images were collected every 5 min for a further 15 h. A time point representing each hour is shown, and the corresponding animation can be found at http://mc11.mcri.ac.uk/mov/figure7.mov.
FIG. 8.
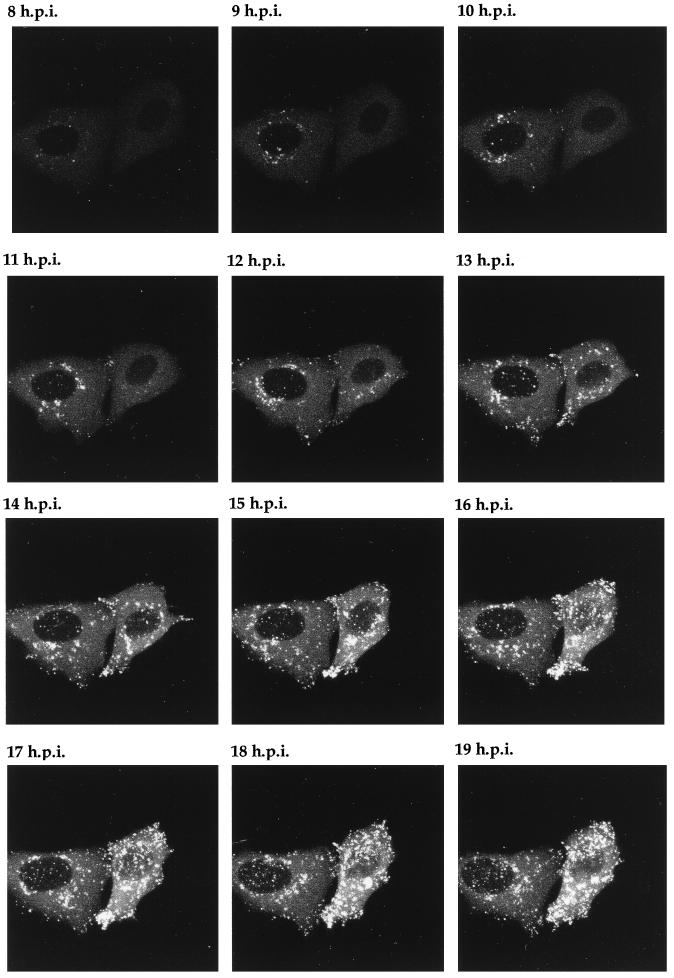
Time lapse analysis of GFP-22 trafficking in a low-multiplicity 166v infection. Vero cells infected with 166v at 0.02 PFU per cell were transferred to the heated chamber 8 h postinfection (8 h.p.i.). A single field of cells was chosen for further analysis, and images were collected every 5 min for a further 14 h. A time point representing each hour is shown, and the corresponding animation can be found at http://mc11.mcri.ac.uk/mov/figure8.mov.
RESULTS
Construction of an HSV-1 recombinant virus expressing GFP-22.
The HSV-1 structural protein VP22 is encoded by the UL49 gene (10) located in the Bam F restriction fragment of the unique long region of the genome (Fig. 1, diagrams 1 and 2). UL49 was replaced with the gene encoding GFP-22, as described in detail in Materials and Methods. Briefly, plasmid pGE166, consisting of the GFP-22 open reading frame surrounded by UL49 flanking sequences (Fig. 1, diagram 4) was cotransfected into COS-1 cells together with purified HSV-1 strain 17 genomic DNA and incubated for 4 days until cytopathic effect was present in all cells. Virus was harvested, and around 6,000 plaques were plated onto Vero cells and screened by using GFP fluorescence to identify potential recombinants. Two green plaques were detected and plaque purified a further two times. One of these viruses was chosen for further analysis and was designated 166v (Fig. 1, diagram 5).
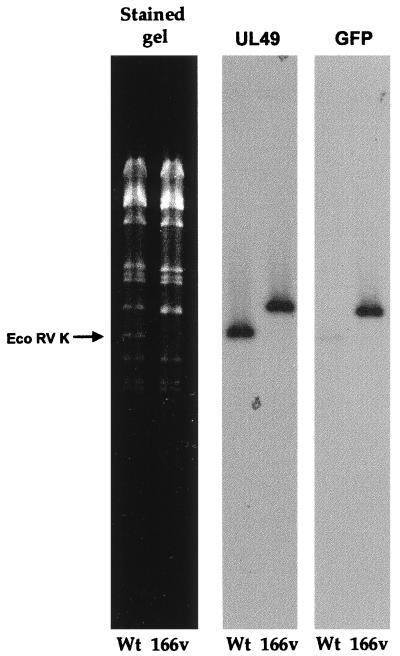
To ensure that recombination had taken place in the correct location on the genome and that the endogenous copy of the VP22 gene had been replaced by the GFP-22 gene, genomic DNA was purified from both the wild-type (WT) S17 virus and the 166v virus and was subjected to restriction digestion with EcoRV (Fig. 2). Incorporation of GFP-22 into the genome should result in an increase in the size of the EcoRV K fragment of the genome from 5.55 to 6.3 kb (Fig. 1; compare diagrams 2 and 5). The restriction pattern of EcoRV-digested virus DNA shows the loss of the 5.55-kb K fragment in the recombinant virus and the appearance of a larger fragment of 6.3 kb (Fig. 2, stained gel). Southern blotting carried out on the gel by using both a UL49 probe and a GFP probe (Fig. 2) indicated that this new larger fragment hybridized with both sequences, confirming the presence of the GFP-22 gene in the EcoRV K fragment.
FIG. 2.
Southern blotting of virus DNA confirms the presence of the GFP-22 cassette in the HSV-1 genome. Viral DNA purified from cells infected with either S17 or 166v was digested overnight with EcoRV and electrophoresed in a 0.8% agarose gel containing ethidium bromide. The gel was photographed (stained gel), transferred to a nylon membrane, and hybridized with a probe specific for the UL49 gene. The same membrane was stripped and rehybridized with a probe specific for GFP. Arrow points to the WT EcoRV K fragment.
166v has the same growth characteristics as WT HSV-1.
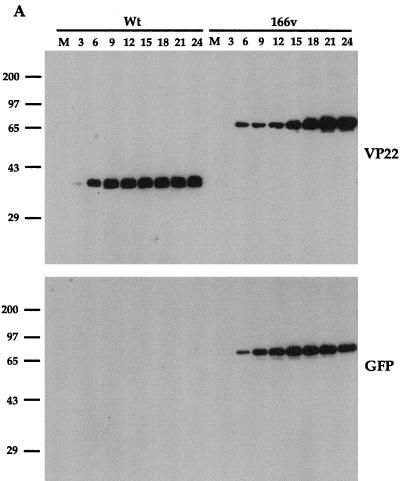
To ascertain the growth properties of the GFP-22 virus, a time course of infection was carried out for both WT and 166v viruses. Vero cells were infected at a multiplicity of 10 and harvested every 3 h postinfection up to 24 h. Total-cell lysates were analyzed by SDS-PAGE and Western blotting, and the kinetics of synthesis of several virus proteins were assessed (Fig. 3). Western blotting with both the anti-VP22 polyclonal antibody AGV30 and the anti-GFP monoclonal antibody (Fig. 3A) indicated that 166v synthesized a GFP-22 fusion protein of the correct size, 65 kDa, confirming the loss of the endogenous VP22 gene from the 166v virus. Moreover, the GFP-22 fusion protein remained intact throughout the course of the infection, as judged by both the VP22 and the GFP Western blotting (Fig. 3A). In addition, the overall kinetics of VP22 and GFP-22 synthesis were very similar, with both VP22 and GFP-22 first being detected 6 h postinfection (Fig. 3A). The kinetics of synthesis of three other virus proteins representing different classes of genes, namely, the immediate-early gene IE110, the early gene TK, and the late gene VP16, were identical for both the viruses (Fig. 3B). These results suggest that the 166v virus enters cells and initiates virus replication as efficiently as the WT virus.
FIG. 3.
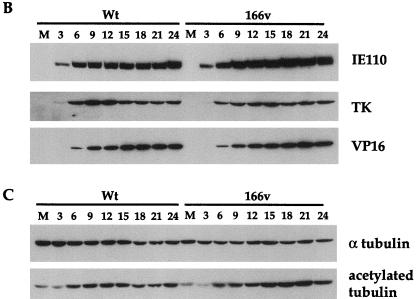
The GFP-22 virus replicates as efficiently as the WT virus. Vero cells infected with either the WT virus or 166v at 10 PFU per cell were harvested every 3 h after infection, up to 24 h, as indicated above the gels. Equal amounts of total cell lysates were analyzed by SDS-PAGE followed by Western blotting with antibodies against VP22 and GFP (A); IE110, TK, and VP16 (B); or α tubulin and acetylated tubulin (C).
We have recently shown that a feature of VP22 in both transfection and virus infection is its ability to stabilize the cellular microtubule network (9). One consequence of such microtubule stability is an increase in the level of acetylation present on α tubulin (21), a modification which is detectable with an antibody specific for the acetylated form of α tubulin. In agreement with our previous results, Western blotting of the same time course samples used above with antibodies specific for both α tubulin and acetylated tubulin (Fig. 3C) showed an increase in acetylated-tubulin levels in the WT virus infection in comparison to overall α tubulin levels, correlating with the synthesis of VP22 (Fig. 3C). Moreover, the 166v virus was also able to induce the acetylation of α tubulin as efficiently as the parental virus (Fig. 3C). Therefore, it appears that 166v virus infection increases the stability of cellular microtubules as efficiently as WT virus infection.
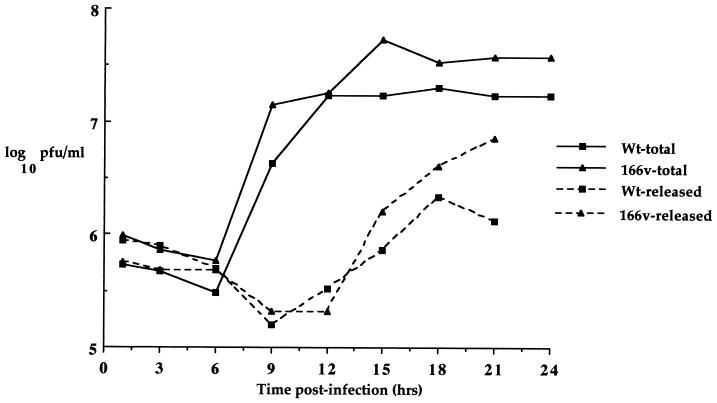
While these results demonstrated that virus gene expression was similar in 166v- and WT virus-infected cells, it was possible, since VP22 is a structural component of the virus particle, that the virus could be in some way restricted for assembly and/or release from the cell. To assess the rate of virus assembly and egress, one-step growth curves were carried out for both the 166v and the WT virus (Fig. 4). Vero cells were infected at a multiplicity of 10 and harvested every 3 h for 24 h, and both total-virus yield and extracellular-virus yield were calculated for both viruses. The results show that the growth curves for total and released virus from WT virus and 166v infections were very similar (Fig. 4), implying that the rates of both virus assembly and virus egress from the cell were the same for the two viruses.
FIG. 4.
One-step growth curves for WT and 166v viruses. Vero cells infected with either the WT virus or 166v at 10 PFU per cell were harvested every 3 h after infection up to 24 h and were titrated onto Vero cells. Total, virus yield from intra- and extracellular virus. Released, virus yield from extracellular virus.
GFP-22 is efficiently incorporated into 166v virus particles.
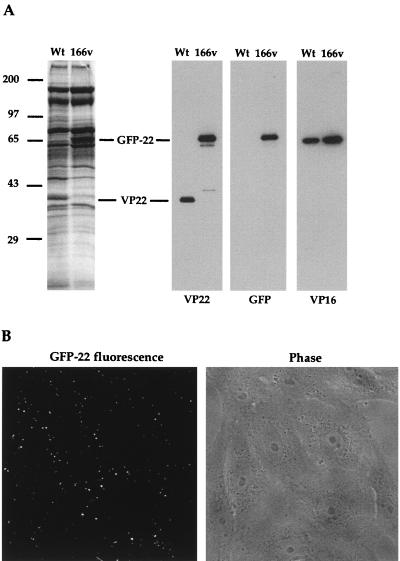
VP22 is a major component of the tegument of the virus particle. However, it has not yet been determined if VP22 is essential for virus assembly, and it was therefore of interest to determine if GFP-22 was incorporated into virus particles produced during 166v infection. Virions of both the 166v and the parental virus were purified by pelleting extracellular virus released into infected cell medium and purifying the particles on a 5 to 15% Ficoll gradient. Approximately equivalent numbers of particles were analyzed by SDS-PAGE followed either by Coomassie blue staining or by Western blotting (Fig. 5A). The total virion protein profiles showed that the 38-kDa VP22 species present in the WT profile had been lost from the 166v profile and that a new, 65-kDa species, the correct size for GFP-22, was present (Fig. 5A, left panel). Western blotting with both the anti-VP22 and anti-GFP antibodies confirmed that this new virion component represented the GFP-22 fusion protein (Fig. 5A, right panels). It is noteworthy that, in spite of the fact that GFP-22 is almost twice the size of VP22, the levels of GFP-22 and VP22 incorporated into their respective particles were roughly equivalent when compared to a VP16 loading control (Fig. 5A, right panels).
FIG. 5.
Incorporation of GFP-22 into virus particles. (A) Purified WT and 166v virions were solubilized and analyzed by SDS-PAGE on a 9% acrylamide gel, followed by either Coomassie blue staining (left) or Western blotting with antibodies against VP22, GFP, or VP16 (right). VP22 and GFP-22 species are indicated. (B) 166v virions are fluorescent. Approximately 10 PFU of purified 166v virions per cell was laid onto a monolayer of Vero cells maintained at 4°C. Thirty minutes later the cells were examined live by both phase-contrast (Phase) and fluorescence (GFP-22 fluorescence) microscopy.
The inclusion of GFP-22 into the virions of 166v raised the possibility that these particles would be detectable by fluorescence. To determine if this was the case, gradient-purified virions were laid onto a coverslip of Vero cells maintained at 4°C at a multiplicity of 10, and the cells were examined 30 min later by fluorescence microscopy. A typical field, showing both GFP fluorescence and the phase image of the Vero cells, is shown in Fig. 5B. Strikingly, fluorescent particles were readily visualized on the outer surfaces of these cells (Fig. 5B, left panel).
Detection of newly synthesized GFP-22 in infected cells.
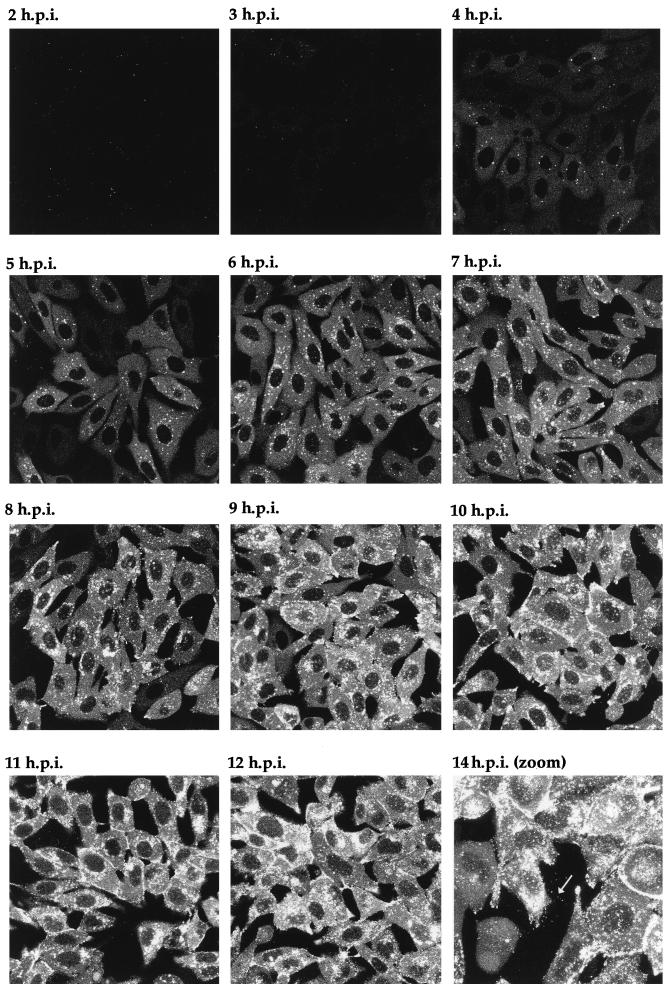
One of the major possibilities for a virus expressing a fluorescent structural protein would be the ability to localize that protein within the cell, to follow its trafficking as infection progresses, and ultimately to visualize the pathway of virus assembly in live cells. To determine the stage of infection at which newly synthesized GFP-22 could initially be detected by fluorescence, and to analyze its localization in live cells at various stages of infection, Vero cells were infected with the 166v virus at a multiplicity of 10, and the cells were examined for GFP fluorescence every hour over a period of 14 h. Representative images collected at each time point are shown in Fig. 6. Notably, GFP-22 was first observed in the majority of cells as early as 3 h postinfection, at which time the protein was localized in a diffuse cytoplasmic pattern (Fig. 6). By 4 h postinfection, GFP-22 was visible in most cells, in the same diffuse cytoplasmic location, but with a small number of additional intensely fluorescent spots situated throughout the cytoplasm (Fig. 6). Over the next 4 h the diffuse intensity of GFP-22 fluorescence in the cytoplasm increased, while a new form of particulate GFP-22 started to accumulate at the edge of the nucleus (Fig. 6). This material increased in intensity and localized to other regions of the cytoplasm, and by 10 h a similar form of GFP-22 was present at the peripheries of the cells (Fig. 6). At 14 h, fluorescent particles, presumably virions, were visible outside the cells (Fig. 6). After 18 h, the cells were intensely fluorescent, and it became difficult to discern any more information from them. These preliminary results suggest that throughout infection GFP-22 is concentrated primarily in the cytoplasm of the cell, where it exhibits distinctive patterns of localization, ranging from an initial diffuse pattern to an accumulation around the nucleus, followed by a cell periphery location.
FIG. 6.
Live-cell analysis of GFP-22 localization during a high-multiplicity infection of 166v. Vero cells were infected with 166v at 10 PFU per cell and were examined every hour up to 14 h postinfection (14 h.p.i.) for GFP-22 fluorescence. The same settings for the confocal microscope were used at each time point. Extracellular fluorescent particles can be seen in the image taken at 14 h postinfection (arrow).
Time lapse analysis of virus infection.
The ultimate elegance of a GFP-incorporating virus is the potential for monitoring virus infection in individual living cells. To determine the optimum conditions for the long-term maintenance of cells for confocal microscopy, so that time lapse analysis could be carried out, we examined a variety of growth conditions. The final conditions used were as follows. Cells were grown on a glass coverslip, which was then sealed into our microscope chamber at the required time. Standard Dulbecco’s minimal essential medium was added to the cells, and the entire chamber was maintained at 37°C by placing it on a heated platform seated on the microscope. With the provision of a constant supply of 5% CO2, the cells could be maintained on the microscope as long as 48 h. To generate time lapse data on cells infected with the GFP-22-encoding virus, Vero cells on a coverslip were infected at a multiplicity of 10 and placed in the chamber, and a single field was chosen for time lapse analysis beginning at 5 h postinfection. Images were collected every 5 min for a further 12 h to form the time series represented as both an animation (http://mc11.mcri.ac.uk/mov/figure7.mov) and the static images of hourly intervals shown in Fig. 7. At the first time point (5 h postinfection), the bright fluorescent spots observed previously (at 4 h postinfection) (Fig. 6) were also observed in the cytoplasm of these infected cells (Fig. 7). As the infection progressed over the next few hours, these spots were engulfed in the mass of particulate GFP-22 which formed at the edge of the nucleus (Fig. 7, 10 h postinfection). This same material then travelled through the cytoplasm from the edge of the nucleus to the cell periphery, and in particular to the extremities of the cells (Fig. 7, 14 h postinfection).
It was clear from the evolving localization patterns observed for GFP-22 over the 16 h of a high-multiplicity infection (Fig. 6 and 7) that after the initiation of VP22 synthesis its localization altered rapidly, and detailed trafficking of the protein may therefore be difficult to assess. Thus, to slow down the infection process and enable GFP-22 trafficking to be examined more precisely, Vero cells were infected at a low multiplicity so that approximately 1 cell in 50 was infected. Eight hours postinfection it was possible to detect individual cells which were expressing GFP-22 in its early diffuse cytoplasmic pattern. Two such cells were chosen for further analysis by time lapse confocal microscopy (Fig. 8), and images of these cells were collected every 5 min over a period of 15 h. The resulting time series of 150 images was animated to produce a time lapse analysis of infection progressing in these two cells (http://mc11.mcri.ac.uk/mov/figure8.mov), with hourly images presented as a gallery in Fig. 8. Between 8 and 12 h postinfection, the diffuse GFP-22 evolved into particulate GFP-22 in a manner similar to that seen during the high-multiplicity infection (Fig. 8), with the exception that none of the bright fluorescent spots which had been seen at the earlier stages of the high-multiplicity infection (Fig. 6, 4 and 5 h postinfection) were observed during this low-multiplicity infection. The particulate material localized initially around the nucleus (Fig. 8, e.g., 9 h postinfection), but over the next few hours it appeared in other regions of the cytoplasm (Fig. 8, e.g., 13 h postinfection). As more of this particulate material appeared in the cytoplasm of these two cells, it began to move to the periphery of the cell (Fig. 8, 15 h postinfection; seen clearly in the right-hand cell) and in particular to the apices of the cell. This trafficking continued for the rest of the time course. It can be noted from this time lapse, and in particular from the animation, that at the later stages of infection there was continued movement of the cell membranes and that projections containing a high concentration of GFP-22 were actively thrown out from the cytoplasm. Thus, we suggest that these processes may represent tracks along which virus egress takes place and that in the future it may be possible to visualize the progression of individual GFP-22-containing vesicles along these projections.
DISCUSSION
While certain features of the herpesvirus replication cycle are now broadly understood, the fine details of virion trafficking and maturation remain to be elucidated. One of the major obstacles to our full understanding of virus assembly and egress has been the inability to analyze and follow virus trafficking at the single-cell level. The development of GFP technology has recently begun to overcome problems associated with the study of protein trafficking in several aspects of cell biology, such as endoplasmic reticulum-to-Golgi apparatus transport (22, 26), the cytoskeleton (16, 20, 30), and cell division (6, 12). Furthermore, the use of time lapse analysis of cells expressing GFP fusion proteins has for the first time enabled the tracking of individual proteins over time at the subcellular level (15, 19, 32). In this report we present the first time lapse images of virus infection in live cells captured by the use of a herpesvirus incorporating a GFP-tagged protein into its structure. This virus produces fluorescent particles which can be readily resolved by fluorescence microscopy and can therefore be traced through the different stages of virus replication. Thus, we have generated an extremely powerful tool for the analysis of herpesvirus infection.
The protein to which we have fused GFP for incorporation into the virion is the major tegument protein VP22. Although we have recently identified several unusual properties of this protein (8, 9), the exact role of VP22 in virus infection is still unclear. We have previously shown that the fusion of GFP to VP22 has little effect on any of the properties of VP22 which we have identified by transient transfection. Strikingly, we show here that the addition of the 27-kDa GFP protein onto the VP22 open reading frame also has little effect on any aspect of virus infection, including efficiency of virus entry, the rate of virus assembly, and the rate of release from the cell. Moreover, the tegument of the virus particle appears flexible enough to accommodate the same number of GFP-22 molecules as VP22 molecules, in spite of the near-doubling in size of the protein, with no apparent increase in size of the individual particles, as judged by electron microscopy (data not shown), and with no obvious changes in the ratio of other tegument proteins within the virions. This suggests that the tegument compartment exhibits some flexibility in its structure and is capable of incorporating fusion proteins or, as previously shown, excess copies of at least some of the constituent proteins (14). Interestingly, it has recently been shown that the HSV-1 capsid can also accommodate a GFP-fusion protein in the form of GFP-VP26, which is located at the tips of the capsid hexons (5).
Our preliminary analysis of the 166v infection demonstrated that newly synthesized GFP-22 was detectable in live cells as early as 3 h postinfection. This sensitivity of GFP-22 fluorescence suggested that it may be possible, in combination with time lapse imaging of individual cells, to analyze the trafficking of GFP-22, and hence of HSV-1 virions, dynamically in live cells. We have therefore produced time lapse animations of 166v infections and have subsequently built up an overall picture of GFP-22 trafficking during infection. These animations show us that when GFP-22 is initially synthesized, it appears in a diffuse cytoplasmic pattern. However, as infection progresses, the GFP-22 molecules take on a more distinctive pattern, concentrated in particles to one side of the nucleus, in a pattern reminiscent of the Golgi apparatus. This material then travels towards the cell periphery and eventually appears extracellularly as individual fluorescent particles. Thus, we would suggest that this pathway reflects GFP-22 incorporation into assembling virions, followed by virion trafficking and egress from the cell. Moreover, further studies on the relative proportion of infected-cell VP22 which is incorporated into virions should help to address the relevance of this pathway.
Central to the debate concerning the herpesvirus maturation pathway(s) is the identification of the tegument assembly site within the cell. It has recently been demonstrated that at least two HSV-1 tegument proteins localize in the nucleus close to putative sites of capsid assembly, termed assemblons (31), an observation taken to mean that tegument proteins may assemble into the virus particle in the nucleus. However, we have previously shown by immunofluorescence studies that VP22 and another tegument protein, VP16, colocalize in the cytoplasm rather than the nucleus of both infected and transfected cells (7), suggesting that during coexpression these proteins are targeted to the cytoplasmic compartment. Moreover, it has been demonstrated that the tegument can assemble independently of the capsid (23). Our results presented here demonstrate that throughout the course of infection, GFP-22 appears almost entirely cytoplasmic. While we cannot exclude the possibility that GFP-22 travels through the nucleus so rapidly and efficiently that it is never detected there, or that GFP-22 fluorescence varies depending on its cellular location, the animations provide strong evidence that this tegument protein is incorporated into the virion at a stage downstream of capsid translocation through the nuclear envelope. Consequently, these results would suggest that the final envelope of the mature virion is acquired at a cytoplasmic location, further along the exocytotic pathway from the site of VP22 inclusion into the virion.
The development of a GFP-labelled herpesvirus and the demonstration that the incorporation of GFP-22 into the virus particle is in no way detrimental to virus replication open up a wide range of applications for such a reagent. Purified virions could be used to analyze virus entry, and in particular the fate of the tegument after the envelope has fused to the cell membrane. Moreover, detailed studies on the kinetics of virus egress could be conducted to assess the effect of virus infection on the exocytotic pathway. The GFP-22 virus could be combined with mutations in other genes to uncouple the individual steps of virus maturation as we now observe it in live cells. Thus, with the advent of GFP technology, it may now be possible to address many of the issues surrounding herpesvirus morphogenesis by the use of viruses incorporating GFP into the various compartments of the virion.
ACKNOWLEDGMENTS
We thank Tony Minson for antibody LP1, Roger Everett for antibody 11060, and David Gower for the anti-TK antibody.
This work was funded by Marie Curie Cancer Care.
REFERENCES
- 1.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campadelli-Fiume G, Farabegoli F, Di Gaeta S, Roizman B. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J Virol. 1991;65:1589–1595. doi: 10.1128/jvi.65.3.1589-1595.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung P, Banfield B W, Tufaro F. Brefeldin A arrests the maturation and egress of herpes simplex virus particles during infection. J Virol. 1991;65:1893–1904. doi: 10.1128/jvi.65.4.1893-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dargin D. The structure and assembly of herpes viruses. In: Harris J R, Horne R W, editors. Electron microscopy of proteins. 5. Virus structure. London, United Kingdom: Academic Press; 1986. pp. 359–437. [Google Scholar]
- 5.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellenberg J, Siggia E D, Moreira J E, Smith C L, Presley J F, Worman H J, Lippincott-Schwarz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott G, Mouzakitis G, O’Hare P. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott G, O’Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 9.Elliott G, O’Hare P. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott G D, Meredith D M. The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J Gen Virol. 1992;73:723–726. doi: 10.1099/0022-1317-73-3-723. [DOI] [PubMed] [Google Scholar]
- 11.Gibson W, Roizman B. Proteins specified by herpes simplex virus. X. Staining and radiolabeling properties of B capsid and virion properties in polyacrylamide gels. J Virol. 1974;13:155–165. doi: 10.1128/jvi.13.1.155-165.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanda T, Sullivan K, Wahl G M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 13.Knopf K W, Kaerner H C. Virus-specific basic phosophoproteins associated with herpes simplex virus type 1 (HSV-1) particles and the chromatin of HSV-1-infected cells. J Gen Virol. 1980;46:405–414. doi: 10.1099/0022-1317-46-2-405. [DOI] [PubMed] [Google Scholar]
- 14.Leslie J, Rixon F J, McLauchlan J. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology. 1996;220:60–68. doi: 10.1006/viro.1996.0286. [DOI] [PubMed] [Google Scholar]
- 15.Lippincott-Schwarz J, Cole N, Presley J. Unravelling Golgi membrane traffic with green fluorescent protein chimeras. Trends Cell Biol. 1998;8:16–20. doi: 10.1016/s0962-8924(97)01199-9. [DOI] [PubMed] [Google Scholar]
- 16.Ludin B, Matus A. GFP illuminates the cytoskeleton. Trends Cell Biol. 1998;8:72–77. [PubMed] [Google Scholar]
- 17.Morgan C, Rose H M, Holden M, Jones E P. Electron microscopic observations on the development of herpes simplex virus. J Exp Med. 1959;110:643–656. doi: 10.1084/jem.110.4.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nii S, Morgan C, Rose H M. Electron microscopy of herpes simplex virus. II. Sequence of development. J Virol. 1968;2:517–536. doi: 10.1128/jvi.2.5.517-536.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oatey P B, Van Weering D H, Dobson S, Gould G W, Tavare J M. GLUT4 vesicle dynamics in living 3T3 L1 adipocytes visualised with green-fluorescent protein. Biochem J. 1997;327:637–642. doi: 10.1042/bj3270637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson K, McIntosh J, Olmsted J. Analysis of MAP4 function in living cells using green fluorescent protein (GFP) chimeras. J Cell Biol. 1995;130:639–650. doi: 10.1083/jcb.130.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piperno G, LeDizet M, Chang X. Microtubules containing acetylated α-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Presley J F, Cole N B, Schroer T A, Hirschberg K, Zaal K J M, Lippincott-Schwarz J. ER-to-Golgi transport visualised in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 23.Rixon F J, Addison C, McLauchlan J. Assembly of enveloped tegument structures (L particles) can occur independently of virion maturation in herpes simplex virus type 1-infected cells. J Gen Virol. 1992;73:277–284. doi: 10.1099/0022-1317-73-2-277. [DOI] [PubMed] [Google Scholar]
- 24.Roizman B, Furlong D. The replication of herpesviruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. New York, N.Y: Plenum Press; 1974. pp. 229–403. [Google Scholar]
- 25.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, editors. Fundamental virology. New York, N.Y: Raven Press; 1991. pp. 849–895. [Google Scholar]
- 26.Scales S J, Pepperkok R, Kreis T. Visualisation of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz J, Roizman B. Concerning the egress of herpes simplex virus from infected cells. Electron microscope observations. Virology. 1969;38:42–49. doi: 10.1016/0042-6822(69)90126-3. [DOI] [PubMed] [Google Scholar]
- 28.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Genderen I L, Brandimarti R, Torrisi M R, Campadelli G, van Meer G. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology. 1994;200:831–836. doi: 10.1006/viro.1994.1252. [DOI] [PubMed] [Google Scholar]
- 30.Wacker I, Kaether C, Kromer A, Migala A, Almers W, Gerdes H H. Microtubule-dependent transport of secretory vesicles visualised in real time with a GFP-tagged secretory protein. J Cell Sci. 1997;110:1453–1463. doi: 10.1242/jcs.110.13.1453. [DOI] [PubMed] [Google Scholar]
- 31.Ward P L, Ogle W O, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus type 1. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weimer E A, Wenzel T, Deerinck T J, Ellisman M H, Subramani S. Visualisation of the peroxisomal compartment in living mammalian cells: dynamic behavior and association with microtubules. J Cell Biol. 1997;136:71–80. doi: 10.1083/jcb.136.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]