Abstract
Although most reports defining the human immunodeficiency virus type 1 (HIV-1) genomic RNA packaging signal have focused on the region downstream of the major 5′ splice site, others have suggested that sequences upstream of the splice site may also play an important role. In this study we have directly examined the role played by the HIV-1 TAR region in RNA packaging. For these experiments we used a proviral expression system that is largely independent of Tat for transcriptional activation. This allowed us to create constructs that efficiently expressed RNAs carrying mutations in TAR and to determine the ability of these RNAs to be packaged. Our results indicate that loss of sequences in TAR significantly reduce the ability of a viral RNA to be packaged. The requirement for TAR sequences in RNA packaging was further examined by using a series of missense mutations positioned throughout the entire TAR structure. TAR mutations previously shown to influence Tat transactivation, such as G31U in the upper loop region or UCU to AAG in the bulge (nucleotides [nt] 22 to 24), failed to have any effect on RNA packaging. Mutations which disrupted the portion of the TAR stem immediately below the bulge also had little effect. In contrast, dramatic effects on RNA packaging were observed with constructs containing mutations in the lower portion of the TAR stem. Point mutations which altered nt 5 to 9, 10 to 15, 44 to 49, or 50 to 54 all reduced RNA packaging 11- to 25-fold. However, compensatory double mutations which restored the stem structure were able to restore packaging. These results indicate that an intact lower stem structure, rather than a specific sequence, is required for RNA packaging. Our results also showed that RNA molecules retained within the nucleus cannot be packaged, unless they are transported to the cytoplasm by either Rev/Rev response element or the Mason-Pfizer monkey virus constitutive transport element.
While a detailed understanding of the mechanism for RNA packaging in human immunodeficiency virus (HIV) is lacking, it is widely believed that the HIV genome contains a packaging region that interacts with a viral factor to mediate the incorporation of full-length genomic RNA into virus particles (for a review, see reference 6). RNA mapping studies have shown that important determinants for RNA packaging map throughout the entire 5′-end region of the HIV type 1 (HIV-1) RNA, up to the start of gag (8, 37, 38, 43, 46, 48, 56, 63). However, most attention has focused on the region of viral RNA mapping between nucleotides 230 and 352 (4, 12, 32). This region has been called the HIV-1 Ψ region, in analogy to the packaging region of murine leukemia virus (MLV) RNA (44, 62).
The HIV-1 Ψ region forms four stem-loop structures termed SL1 through SL4 (4, 12, 32). SL1 contains the primary site of RNA dimer formation (14, 40, 45, 49, 50, 53, 59), and SL3 contains a specific binding site for HIV-1 nucleocapsid protein (NC) (5, 17, 42). Most data suggest that there are determinants in both SL1 and SL3 that are essential for efficient packaging. Since the major 5′ splice site is contained within SL2 (58) (which maps between SL1 and SL3), only unspliced RNA contains both the SL1 and SL3 determinants. This explains, at least in part, why only unspliced HIV RNA is efficiently packaged.
Despite the progress that has been made in defining the HIV-1 packaging signal, there is no unifying model to explain how this complex, multipartite signal is actually recognized by the assembling virus particle. The viral factor involved in this process appears to be NC (1, 7, 9, 25, 55, 64), but the details of the mechanism by which NC provides specificity remain elusive. For example, even though the three-dimensional structure of complex of NC and SL3 has recently been described (17), we do not understand why only two molecules of RNA are packaged into a particle that contains about 1,800 NC molecules and thus 1,800 SL3 binding sites. We also have little understanding of the role played by sequences mapping upstream of SL1.
The HIV-1 genome contains at least two other cis-acting sequence elements whose interactions with viral proteins have been well defined. The HIV-1 TAR element consists of a 57-nucleotide sequence that forms a stem-loop. It is found at both ends of HIV-1 genomic RNA, forming the first part of the “R” repeat present in all retroviral RNAs. The element regulates transcription through its interaction with the HIV-1 Tat protein and cellular factors (for reviews, see references 34 and 36). Parts of the TAR stem-loop not involved with the Tat interaction have also been shown to be essential for viral replication, though the reasons for their importance are much less well defined (31, 39). For example, elegant genetic studies have implicated the lower stem region of TAR as an essential element for viral infectivity (39).
Another RNA element in the HIV genome that is involved in the binding of a viral specific protein is the Rev response element (RRE). This 248-nucleotide element is located within the coding region of env and is present in full-length and singly spliced HIV RNAs. Rev binding is needed to mediate the export of the RRE-containing RNA from the nucleus to cytoplasm (for reviews, see references 26 and 54). An element functionally equivalent to the RRE, termed the constitutive transport element (CTE), has been identified in type D retroviruses (10, 65). The CTE mediates the nuclear export of intron containing RNA (20) by interacting exclusively with factors that are encoded by the cell.
The present study examines determinants of HIV-1 RNA packaging by introducing deletion and cluster mutations into the RNA and analyzing their effects on encapsidation. We have focused our effort mainly on the region mapping upstream of Ψ and, in particular, on the HIV TAR region. Our data strongly suggest that a distinct packaging determinant exists within the bottom part of the TAR stem. Our results suggest that it is the structure, rather than the sequence, of the TAR stem that is important. We also describe in this report other experiments that demonstrate that an RNA which is dependent on Rev for transport must interact with Rev (and presumably exit the nucleus) before it can be packaged.
MATERIALS AND METHODS
HIV-1 proviral strain and numbering system.
The HIV-1 proviral clone (pHR969) and its derivative plasmids are derived from HIV-1 HxB2 (GenBank accession no. K03455, M38432, and g327742). Sequences from pNL4-3 (GenBank accession no. M1992) and BH10 (GenBank accession no. M15654, K02008, K02009, and K02010) were also used. The HIV-1 numbering system used is derived from HxB2 and places nucleotide 1 at the start of R in the 5′ long terminal repeat (LTR).
Plasmids.
Plasmid pGAGPOL-RRE-r (pHR146) has been previously described (60). It contains the coding regions for the HIV-1 proteins Gag, Pol, Vif, and truncated Vpr, followed by the RRE and a rabbit β-globin intron and poly(A) site. Expression is directed by the simian virus 40 (SV40) late promoter.
pHR969 contains the full-length HIV-1 genome from the HIV-1 HxB2 proviral construct with the exception of a 330-bp in-frame deletion in pol making it noninfectious. The plasmid also contains cellular flanking sequences on each side of the viral genome as well as the SV40 origin of replication in the vector backbone.
pHR1256 was derived directly from pHR969. It has a deletion in the midportion of the genome corresponding to the central portion of the HIV-1 genome that is deleted in pHR146. Details of its construction are available on request.
pHR1486 was derived from sequences present in the vectors pHR1256 and RRE-r through a series of intermediate constructions. It contains a 220-bp SacI-BssHII fragment from the HIV-1 proviral clone pNL4-3 between the SacI site at position 28 and the BssHII site at position 248 in place of HxB2 sequences. Thus, it lacks the SacI site normally found at position 228 in HxB2.
pHR1573 was made by linearizing vector pHR1486 at the unique SacI site in TAR, repairing the 4-bp 3′ overhangs with T4 DNA polymerase and religation.
pHR1512, pHR1679, pHR1487, pHR1582, pHR1583, pHR1584, pHR1585, pHR1586, pHR1815, pHR1816, pHR1817, pHR1819, pHR1820, and pHR1821 are derivatives of pHR1486 which were made by PCR-based mutagenesis. Details of their construction are available on request.
pHR1152 (previously described [10]) was derived from pNL4-3. It produces a nonfunctional Rev protein due to a stop codon at amino acid 23 (TAT→TAA) in the first coding exon of rev. It also has the Mason-Pfizer monkey virus (MPMV) CTE (MPMV bp 8007 to 8240) inserted into a unique XhoI site at the start of the nef gene. Prior to insertion of the CTE, 150 bp were deleted from nef, moving the XhoI site just downstream of the nef start codon.
pCMVrev (pHR30), previously described as pRev1 (60), contains the HIV-1 Rev coding region under the control of simian cytomegalovirus immediate-early promoter-enhancer (35). pCMVtat (pHR136) is analogous to pCMVrev. It contains the Tat coding sequences from pCV1 (3) under the control of the simian cytomegalovirus immediate-early promoter-enhancer.
Plasmid pHR1290 was constructed by deleting the 2.3-kb BssHII-EcoRV fragment from the previously described pGem-11Zf(−)HIV (pHR243) (61) and religating the vector fragment. This plasmid was used for in vitro transcription to make the RNA standards for reverse transcription (RT)-PCR.
Cell culture and transfections.
Maintenance of and transfection CMT3-COS cells (23) have been previously described. In a standard packaging experiment, 15 μg of test plasmid and 10 μg of pCMVrev per 150-mm-diameter plate were transfected by using DEAE-dextran as previously described (28). In some experiments, plasmid pCMVtat was used for cotransfections as well (10 μg/150-mm-diameter plate).
In some instances, A5.6 cells were substituted for CMT3-COS cells to overcome the need for pCMVrev cotransfection. A5.6 cells are derived from CMT3-COS by stable transfection with pCMVrev and a hygromycin B resistance gene (pHR392). They constitutively express the HIV-1 Rev protein and are similar to the previously described cell line A5.9 (19).
Cellular and virion RNA isolation [total poly(A) selection].
Total poly(A)+ RNA was isolated from cells as previously described (27). To prepare virion RNA, virus particles were collected from the medium of transfected cells. To do this, medium was first cleared of cells and debris by centrifugation two times at 2,500 rpm for 15 min at 4°C in a tabletop centrifuge. The cleared medium was then subjected to ultracentrifugation through a 6-ml cushion of 20% sucrose for 1 h at 4°C and 27,000 rpm (100,000 × g), using a SW28 rotor. The pelleted virus particles were resuspended in 100 μl of stock buffer (200 mM NaCl, 200 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2). To prepare RNA, 3 ml of lysis buffer (200 mM NaCl, 200 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2, 2% sodium dodecyl sulfate, 200 μg of proteinase K [Sigma] per ml) together with 200 μg of yeast tRNA (GibcoBRL) as a carrier was added to the virions. RNA was then prepared by the procedure used to prepare cellular RNA.
DNase I treatment of RNA.
After ethanol precipitation, the RNA samples were resuspended in 20 mM Tris-HCl (pH 8.4)–2 mM MgCl2–125 mM KCl–40 U of rRNasin (Pharmacia)–20 U of DNase I (Boehringer Mannheim) and incubated at 37°C for 1 h. The RNA was then extracted with phenol-CHIASM (chloroform-isoamyl alcohol; 24:1) and reprecipitated.
In vitro transcription.
pHR1290 digested with EcoRI (20 U/μl; New England Biolabs) was used as a template to produce RNA in vitro for a standard curve. This plasmid produces RNA of the same sense as the HIV genome when transcription is driven by T7 RNA polymerase. The in vitro transcription reactions were performed according to instructions from Promega.
Oligonucleotides.
The sense-strand oligonucleotide used to detect HIV-1 gag-pol RNA was oligonucleotide 304, which hybridized to pol at positions 3167 to 3188 and had the sequence 5′-AGGGGTGCCCACACTAATGATG-3′. The antisense oligonucleotide used for this RNA was oligonucleotide 305, which hybridized to pol at positions 3664 to 3685 and had the sequence 5′-GGCTACATGAACTGCTACCAGG-3′.
To detect HIV-1 rev RNA, the sense-strand oligonucleotide used was oligonucleotide 298, which hybridized to the start of the first exon of rev at positions 5186 to 5206 and had the sequence 5′-ATGGCAGGAAGAAGCGGAGAC-3′. The antisense oligonucleotide used, 190, hybridized to the nef gene at positions 8137 to 8161) and had the sequence 5′-GCTGCTGTGTTGCTACTTGTGATTG-3′.
32P 5′-end labeling of oligonucleotides.
Oligonucleotides were labeled by using polynucleotide kinase and [γ-32P]ATP (6,000 Ci/mmol) as previously described (27). The specific activity of the labeled oligonucleotides was usually in the range 2 × 108 to 4 × 108 cpm/μg of oligonucleotide.
RT and cDNA synthesis.
The RT-PCR procedure of Arrigo et al. was followed, with some modifications (2). Both cellular and virion poly(A)-selected RNA was used as template to make cDNA for quantitative PCR. Each cellular RNA sample was a yield from a 150-mm-diameter cell culture plate; one such plate usually gave the yield of 4 to 8 μg of total poly(A)-selected cellular RNA. The cellular RNA samples were resuspended in 200 μl of diethylpyrocarbonate-treated H2O, and usually 1/200 (20 to 40 ng of RNA) was used per RT reaction. The virion RNA samples (from the medium of a 150-mm-diameter cell culture plate) were resuspended in 100 μl of H2O, and the yield from 1/10 of a plate was used per RT reaction. Each RT reaction was performed in a final volume of 50 μl containing 1 μl (40 U) of rRNasin, 10 μl of 5× RT buffer (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2, 50 mM dithiothreitol), 1 μl (100 ng/μl) of antisense oligonucleotide primer, 5 μl (1 μg/μl) of bovine serum albumin (New England Biolabs), 2.5 μl 10 mM each deoxynucleoside triphosphate, 5 μl (500 μg/ml) actinomycin D (Sigma), and 1 μl of M-Moloney murine leukemia virus reverse transcriptase (200 U/μl; GibcoBRL). To detect potential DNA contamination in the RNA samples, the same amounts of cellular and virion RNA samples were treated exactly the same way as the RT samples except that no reverse transcriptase was added to the tubes. Known amounts of serially diluted RNA standards which were made by in vitro transcription were also used as controls to measure the amounts of HIV-1-specific RNA in the test samples and to ensure the linearity of the RT-PCRs; they were treated the same way as other RT reactions. The reactions were carried out at 37°C for 2 h, after which 950 μl of Tris-EDTA buffer was added to each tube to give the volume of 1 ml. The cDNA was stored at −75°C until used for quantitative PCR.
Quantitative PCR.
For each quantitative PCR, 10 μl of cDNA (from the total volume of 1 ml) was used. To that was added 15 μl containing 15 ng of 32P-labeled 3′ oligonucleotide (specific activity, 2 × 108 to 4 × 108 cpm/μg), 15 ng of unlabeled 3′ oligonucleotide, 100 ng 5′ oligonucleotide, 2.5 μl of 10× modified PCR buffer (250 mM Tris pH 8.0, 50 mM MgCl2, 500 mM NaCl, 2.5 mM dATP, 2.5 mM dTTP, 2.5 mM dCTP, 2.5 mM dGTP, 1 μg of bovine serum albumin per μl), and 0.5 μl of Amplitaq DNA polymerase (5 U/μl; Perkin-Elmer), so that the final volume was 25 μl per reaction. The reactions were performed in 0.5-ml Eppendorf tubes; the reaction products were overlaid with 1 drop of mineral oil and placed in a DNA thermal cycler. The reactions were performed at 94°C for 1 min and 60°C for 2 min for 22 cycles.
Following the quantitative RT-PCRs, 5 μl of each sample was mixed with 4 μl of H2O and 1 μl of 10× restriction stop buffer; the mixture was loaded on a 2% agarose gel, which was subjected to electrophoresis in 0.5× Tris-acetate-EDTA buffer at 150 V (at 0.25-A setting) for about an hour. The agarose gel was overlaid with Saran Wrap, dried, and quantitated with a phosphorimager.
p24 enzyme-linked immunosorbent assay (ELISA) on HIV-1 virions.
The concentrated virus particles were diluted and assayed at the dilutions 1/62,500 and 1/312,500 to keep the test samples within the linear range of the assay. The assay was performed according to instructions from the manufacturer (Cellular Products Inc.). A molecular weight of 25,390 was used to convert the amount of p24 to moles.
Calculation of the virion RNA/virion p24 ratio.
The amount of virion RNA present in a sample was determined from the relative intensity of the RT-PCR product by using an RNA standard curve which was generated from in vitro-transcribed RNA. This amount was then multiplied by the appropriate dilution factor to calculate the total amount of virion RNA produced per plate. For virion p24, the value obtained in the ELISA by using the p24 standard curve was also multiplied by an appropriate dilution factor. A ratio of millimoles of virion RNA per mole of virion p24 was then calculated. For most experimental samples, a normalized value for this ratio was obtained by multiplying the value by 100 and dividing by the ratio obtained from the control transfection.
Calculation of the percentage of total RNA packaged.
Total virion RNA RI (relative intensity) units per plate were determined from the relative intensity of the RT-PCR product by multiplying the value obtained from the phosphorimager by the appropriate dilution factor for each test sample. To calculate the cellular plasmid-derived RNA RI units per plate, total cellular RNA was prepared and quantitated by optical density. Usually 20 ng of this RNA was tested by RT-PCR for plasmid-derived RNA content. The RI units obtained from this sample were then multiplied by the appropriate dilution factor to obtain the total RI units per plate. Since the yield of total RNA from each plate varied somewhat, it was assumed that the plate that gave the highest yield of total cellular RNA in each experiment had a yield closest to 100%, and the dilution factor was determined from this value. Yields usually did not vary more than twofold. Finally, the total RI units from both cellular and virion samples were added together and the percentage of the RNA which was found in the virions was calculated.
RESULTS
Packaging of RNA expressed from transient expression vectors.
We have previously described an SV40 late replacement vector, pGAGPOL-RRE-r, and its derivatives, that were used to study assembly interactions between the Pr55gag and Pr160gag-pol precursors (60, 61). In those studies we demonstrated that virus-like particles efficiently formed and budded when this plasmid and a plasmid that expressed Rev were transfected into appropriate cells. However, it was never determined whether the virus-like particles produced packaged viral RNA.
The HIV RNA produced by pGAGPOL-RRE-r lacks the first 228 nucleotides found in a normal HIV genome but retains sequences further downstream known to be involved in RNA packaging; in addition, sequences 3′ of the SalI site in the middle of the HIV-1 genome are replaced with a removable rabbit β-globin intron and poly(A) signal (Fig. 1). Thus, it was of interest to determine whether RNA produced by this plasmid could be packaged into particles, since this would shed light on the sequence requirements for RNA packaging by HIV.
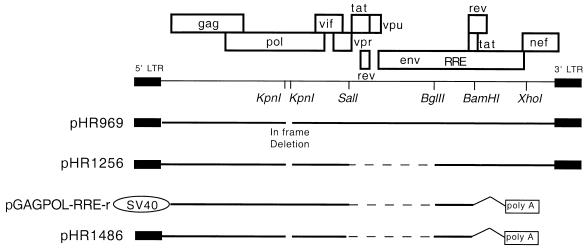
FIG. 1.
Schematic representation of HIV-1 sequences present in six plasmids used in this study. The upper portion shows a map of the entire HIV-1 genome, with a few key restriction enzyme sites marked. The two KpnI sites in pol were used to create a 330-bp in-frame deletion in the proviral clone pHR969 in order to make it noninfectious. Plasmids pHR1256 and pHR1486 were derived from pHR969 and hence contain the same deletion. These plasmids, as well as pGAGPOL-RRE-r, also contain a deletion in the midportion of the viral genome (between SalI and BglII) shown as a dashed line. Expression of pGAGPOL-RRE-r is directed by the SV40 late promoter rather than the 5′ LTR. pGAGPOL-RRE-r also lacks 228 bp at the 5′ end of the viral RNA. pGAGPOL-RRE and pHR1486 contains an intron and poly(A) signal derived from the rabbit β-globin gene.
In the first experiment, we compared the packaging efficiency of pGAGPOL-RRE-r to those of the proviral clone pHR969 and its derivative pHR1256. pHR969 is a full-length viral clone, derived from HxB2, that contains a small in-frame deletion in the RNase H domain of pol. This renders it noninfectious. The plasmid also contains an SV40 origin of replication which allows Tat-independent expression in cells containing SV40 T antigen (data not shown). pHR1256 is derived from pHR969 and was used for comparison since it shares a common deletion in the central portion of the genome with pGAGPOL-RRE-r but contains an intact 5′ LTR. Both pHR1256 and pGAGPOL-RRE-r lack a functional rev gene and require cotransfection with a Rev-expressing plasmid to obtain cytoplasmic RNA expression. All three plasmids are diagrammed in Fig. 1.
CMT3-COS cells were transfected with the various plasmids by using DEAE-dextran, and the cells and medium were quantitatively harvested at 65 h posttransfection. Both cellular and particle-derived RNA were prepared. Quantitative RT-PCR was then performed to determine the amount of RNA in each fraction (see Materials and Methods). The results of this RNA packaging analysis are presented in Fig. 2 and Table 1.
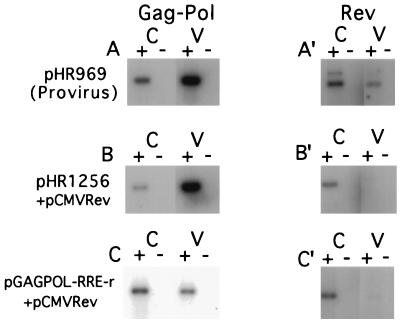
FIG. 2.
RT-PCR analysis of total poly(A)-selected cellular (C) and virion RNA (V) isolated from CMT3-COS cells transiently transfected with the indicated plasmids. (A to C) PCR products derived from the Gag-Pol RNA; (A′ to C′) PCR products derived from Rev-specific RNA. The reactions for all RNA samples were performed in the presence (+) and absence (−) of reverse transcriptase. Both primers used to detect the Gag-Pol RNA anneal in pol (primers 304 and 305), while the primers used to detect Rev RNA anneal in the first exon of rev and in nef (primers 190 and 298). The same 3′ primers were used for the RT reaction and the PCR. The upper band in panel A′ is likely derived from the Tev mRNA, which is not produced by the other plasmids. The RT-PCR products derived from the proviral clone pHR969 (A and A′) and pHR1256 + pCMVrev (B and B′) were exposed for 5 h on the phosphorimager; the products from pGAGPOL-RRE-r plus pCMVrev (C and C′) were exposed for 36 h. Quantitation of the data is given in Table 1.
TABLE 1.
RNA packaging measured as a percentage of total RNA produced (quantitative analysis of the data shown in Fig. 2)
| Fig. 2 panel, plasmid | RNA | Total intensity/plate
|
% RNA in virions | |
|---|---|---|---|---|
| Cellular RNAa | Virion RNAb | |||
| A, pHR969(provirus) | Gag-Pol | 7.10 × 108 | 1.33 × 108 | 15.7 |
| A′, pHR969(provirus) | Rev | 8.70 × 107 | 7.12 × 105 | 0.80 |
| B, pHR1256 | Gag-Pol | 1.68 × 108 | 1.20 × 108 | 41.7 |
| B′, pCMVrev | Rev | 3.62 × 107 | <10 | <0.00027 |
| C, pGAGPOL-RRE-r | Gag-Pol | 9.25 × 107 | 1.37 × 106 | 1.46 |
| C′, pCMVrev | Rev | 3.55 × 107 | 5.93 × 104 | 0.16 |
The measured intensities of the bands in the lanes marked C in Fig. 2 were multiplied by a correction factor of 357 as described in Materials and Methods.
The measured intensities of the bands in the lanes marked V in Fig. 2 were multiplied by a correction factor of 10 as described in Materials and Methods.
Figure 2 shows the PCR products obtained from the RNA derived from both cells and pelleted virus particles. Both Gag-Pol and Rev RNAs were analyzed by using specific primer pairs. The figure shows that in all cases, there was no signal when reverse transcriptase was omitted from the reaction mixture, indicating that the RNA preparations were free from contaminating DNA. Experiments treating the virus particles with micrococcal nuclease before RNA extraction yielded essentially the same results, indicating that the signal from the particles was derived exclusively from packaged RNA (data not shown). The intensity of each band in Fig. 2 was measured with a phosphorimager and corrected for the dilution factor (see Materials and Methods); the results are tabulated in Table 1.
The data in Table 1 demonstrate that in the case of the full-length proviral clone pHR969, nearly 16% of the Gag-Pol RNA produced was packaged into particles. For pHR1256 cotransfected with pCMVrev, this value reached nearly 42%. On the other hand, for pGAGPOL-RRE-r cotransfected with pCMVrev, only about 1.5% of the vector RNA was incorporated into particles. For the Rev mRNA, incorporation into virus particles was in all cases less than 1% of the total, whether it was derived from the proviral clone (Fig. 2A′) or pCMVrev (Fig. 2B′ and C′). It is interesting that in this experiment, as well as in several others (data not shown), Rev RNA produced from pCMVrev was packaged significantly less well than the Rev RNA produced from the proviral clone. One reason for this could be that a portion of the HIV packaging signal mapping upstream of the 5′ splice site (see below) is retained in the Rev mRNA that is derived from the provirus but is missing in Rev mRNA from pCMVRev and that this sequence promotes a low level of packaging.
In a second experiment using the same combinations of plasmids, we analyzed RNA packaging by measuring the amount of RNA found in virus particles in comparison to the amount of capsid protein (CA) p24. This analysis is shown in Table 2. In this experiment, both pHR969 and pHR1256 produced RNAs that were packaged at a level of between 1.6 and 1.9 mmol of RNA/mol of p24, or more than 100 times more efficiently than the RNA produced from pGAGPOL-RRE-r. Interestingly, this value was within twofold of the theoretical value of 1.06 mmol of RNA/mol of p24, which can be calculated by assuming that every viral particle contained the theoretical 2 molecules of RNA per 1,890 molecules of CA p24 (52). Our data thus clearly show that in comparison to pHR1256 and pHR969, pGAGPOL-RRE-r lacks a critical determinant for RNA packaging.
TABLE 2.
RNA packaging measured as a ratio of RNA to p24
| Plasmid(s) | pmol of particle p24/plate | fmol of particle RNA/plate | mmol of particle RNA/mol of particle p24 | Normalized RNA/p24 |
|---|---|---|---|---|
| pHR969 (provirus) | 9.1 | 14.3 | 1.6 | 100 |
| pHR1256 + pCMVrev | 3.9 | 7.6 | 1.9 | 119 |
| pGAGPOL-RRE-r + pCMVrev | 12.2 | 0.2 | 0.02 | 1.3 |
The 5′ TAR element is a determinant for RNA packaging.
pGAGPOL-RRE-r differs from pHR1256 in several ways. It lacks sequences from both the extreme 5′ and 3′ ends of the HIV genome, and RNA derived from it is spliced to remove an intron near its 3′ end (Fig. 1). To determine which of these differences led to the differential in RNA packaging between pGAGPOL-RRE-r and pHR1256, we made a series of pHR1256 derived constructs and tested them for the ability to produce packageable RNA. Our results showed that sequences from the 3′ end of the genome were not needed for packaging from these constructs and that splicing of the RNA did not prevent it from being packaged (data not shown). These results confirmed previous studies by others working with slightly different systems (13, 37, 46, 48). Thus, it seemed likely that sequences present in pHR1256 at the extreme 5′ end of the genome, but lacking in pGAGPOL-RRE-r, contained packaging determinants.
To determine whether sequences at the 5′ end of the genome played a role in RNA packaging, we constructed plasmids which had a series of deletions within the TAR element, as shown in Fig. 3A. For these experiments, A5.6 cells were used in place of CMT3-COS cells. A5.6 cells are a clone of CMT-COS created by stable transfection with pCMVrev and a plasmid that confers hygromycin resistance. These cells constitutively express the Rev protein at levels similar to those expressed by the previously described (19) A5.9 cell line. Using these cells allowed us to omit plasmid pCMVrev from the transfection.
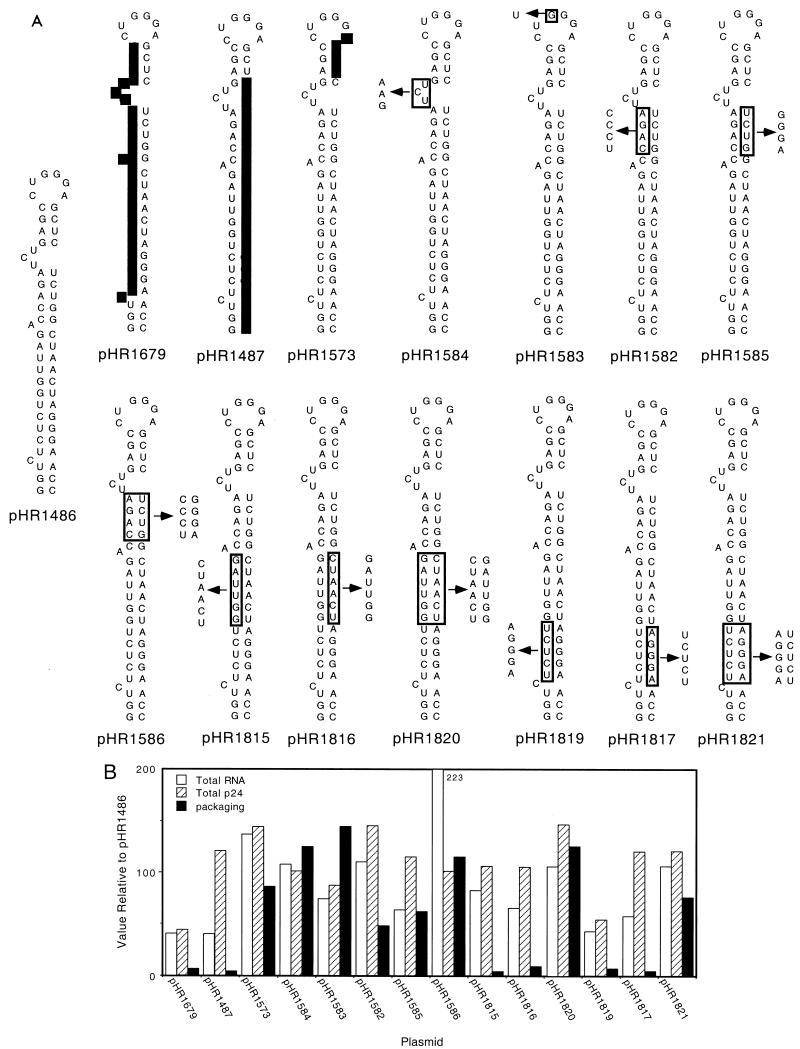
FIG. 3.
Packaging from plasmids expressing RNA with nucleotide changes in TAR. (A) Schematic representations of the TAR regions from 12 plasmids. Each mutant plasmid was derived from pHR1486. The changed nucleotides are boxed, and each arrow points to the new sequence. (B) Quantitation of RNA packaging. The rev-containing cell line A5.6 was transfected with either pHR1486 as a control or with one of the indicated plasmids. Total RNA, total p24, or particleRNA/particle 24 (packaging) was determined for each transfection as described in Materials and Methods. The measured values were normalized to the values obtained for the control, pHR1486, which were set to 100. The combined results from multiple experiments are shown.
Packaging efficiency for each construct was measured as a ratio of Gag-Pol RNA to CA p24 in pelleted particles. For comparison, these values were normalized to the ratio found in the pHR1486 control. pHR1486 produces an RNA which is identical to the RNA produced from pGAG-POL-RRE-r except for sequences at the 5′ end of the genome (Fig. 1). The results are shown in Fig. 3B. To assess possible differences in expression levels, which could complicate the interpretation of the results, the total p24 found in the medium and total plasmid-specific RNA were also measured in each case and normalized to the value for the pHR1486 control.
Figure 3B shows that the deletions in pHR1679 and pHR1487 seemed to have a slight (two- to threefold) effect on RNA stability, as judged by the amount of total RNA found in the cells compared to pHR1486. However, the effect of the deletions on RNA packaging was much greater. The deletion in pHR1573, on the other hand, had no effect on stability and led to an RNA that was efficiently packaged. The failure of pHR1679 and pHR1487 to produce packageable RNA suggested that the TAR stem was required for packaging.
The structure, but not the sequence, of the TAR stem is essential for RNA packaging.
To further evaluate the role of TAR in RNA packaging, we created a series of site-directed mutations throughout the TAR region (Fig. 3A). These mutations were made in the background of pHR1486, and the resulting plasmids were subjected to an RNA packaging analysis similar to the ones described above. The results are shown in Fig. 3B.
All of the constructs produced levels of particle p24 and total RNA that were within twofold of the control level, indicating that there was no gross effect of any of the mutations on RNA stability or translation. pHR1584 and pHR1583 produced RNA that was packaged at wild-type levels, indicating that the TAR bulge and loop are not determinants for RNA packaging even though they are binding sites for Tat and other factors required for transactivation. The mutations in pHR1582 and pHR1585 destroyed the upper part of the TAR stem but had only a modest (twofold) effect on packaging that was fully restored by the stem-restoring compensatory mutations in pHR1586.
Much more dramatic effects on RNA packaging were observed with mutations that destroyed the lower portions of the TAR stem. The mutations in pHR1815, pHR1816, pHR1819, and pHR1817 all reduced RNA packaging significantly (10- to 25-fold). Interestingly, the compensatory mutations in pHR1820 and pHR1821 which restored the TAR stem structure fully restored RNA packaging. These results allow us to conclude that the lower portion of the TAR stem must remain base paired for efficient RNA packaging. However, the actual sequence of the stem does not seem to matter.
Cytoplasmic localization of the Gag-Pol RNA, mediated by either Rev-RRE or the MPMV CTE, is necessary for RNA packaging.
It has been suggested that retroviral RNA packaging may utilize a special pool of unspliced Gag-Pol RNA that is not translated (reviewed in reference 6; 41). One way of separating the putative packaging pool from the pool of RNA that is to be translated could be selection of the RNA for packaging in the nucleus, before it has a chance to be transported by Rev and interact with the translational machinery. Consistent with this notion are several reports that demonstrate the presence of Gag precursors from various retroviruses in the nucleus (18, 24, 51, 57) and others that show that the HIV-1 Pr55gag and Pr160gag-pol precursors contain putative nuclear localization signals (11, 21, 22).
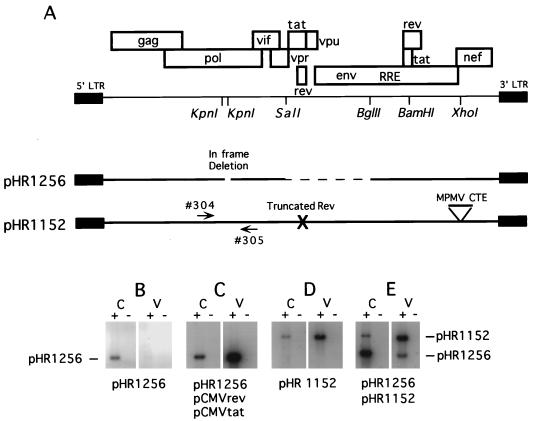
Transfection of cells with pHR1256 alone yields an nuclear form of Gag-Pol RNA that can be detected in total cellular RNA preparations despite the fact that no p24 is produced (Fig. 4B). To determine if this RNA could be packaged in the absence of Rev, we cotransfected pHR1256 together with pHR1152, which has been previously described as pRev(−)MPMV (10). pHR1152 lacks a functional rev gene but expresses Gag and Gag-Pol due to the presence of the MPMV CTE. Since pHR1256 has a 330-bp deletion in pol but pHR1152 does not, RNA produced from the two plasmids can be readily distinguished by using PCR primers which span the deleted region (Fig. 4A).
FIG. 4.
Lack of RNA packaging from a plasmid that expresses RNA which remains in the nucleus. (A) Schematic representation of the plasmids used in this study. pHR1256 contains a small in-frame deletion in pol as well as a larger deletion in the midportion of the genome. Because of the latter deletion, Rev coexpression is required for the nucleocytoplasmic transport of the Gag-Pol mRNA. pHR1152 is a full-length proviral clone with a mutated nonfunctional rev gene. The plasmid also contains the MPMV CTE inserted into the XhoI site of the nef gene. Gag-Pol expression from this plasmid is rev independent. Gag-Pol RNA from the two plasmids can be readily distinguished by using PCR primers 304 and 305 (arrows), which flank the KpnI sites in pol. (B) RT-PCR analysis of total poly(A)-selected cellular (C) and virion RNA (V) isolated from CMT3-COS cells transiently transfected with the indicated plasmids. The reactions for all RNA samples were performed in the presence (+) and the absence (−) of reverse transcriptase. Both primers used to detect the Gag-Pol RNA anneal in pol as shown in panel A. Quantitation of the data is given in Table 3.
Analysis of cellular and viral particle-derived RNA from the cotransfection of pHR1256 and pHR1152 is shown in Fig. 4E. As controls, cells were also transfected with pHR1256 alone (Fig. 4B), with pHR1256, pCMVrev and pCMVtat (Fig. 4C), or with pHR1152 alone (Fig. 4D). Quantitation of the results of this experiment is given in Table 3.
TABLE 3.
RNA packaging for an RNA remaining in the nucleus (measured as a percentage of total RNA produced)
| Plasmid(s)a | Total intensity/per plate
|
% Gag-Pol RNA in virions | |
|---|---|---|---|
| Cellular Gag-Pol RNAb | Virion-associated Gag-Pol RNAc | ||
| pHR1256 + pCMVrev + pCMVtat | 1.68 × 107 | 3.33 × 106 | 16.6 |
| pHR1152 | 8.72 × 105 | 1.05 × 105 | 10.7 |
| pHR1152 + pHR1256 |
1.46 × 106 | 1.52 × 105 | 9.4 |
| 1.74 × 107 | 3.46 × 104 | 0.2 | |
Values for pHR1256 alone were not determined since no virion particles were produced.
The measured intensities of the bands in the lanes marked C in Fig. 4B were multiplied by a correction factor of 360 as described in Materials and Methods.
The measured intensities of the bands in the lanes marked V in Fig. 4B were multiplied by a correction factor of 5 as described in Materials and Methods.
The control experiments demonstrated that RNA derived from pHR1256 could be packaged as efficiently as RNA derived from a proviral construct when Rev was supplied in trans from a second plasmid. About 17% of the pHR1256-specific RNA was packaged, compared to 11% of the RNA produced in the pHR1152 transfection. However, when pHR1256 and pHR1152 were cotransfected, RNA produced from pHR1152 was efficiently packaged, while RNA derived from pHR1256 was not. In this case, only 0.2% of the pHR1256 RNA was packaged, compared to 9.4% for the pHR1152-derived RNA. Since RNA from pHR1256 remains in the nucleus under these conditions (27), we can conclude that an RNA residing in the nucleus because of a block to its transport cannot be packaged.
DISCUSSION
Our results demonstrate that the HIV TAR element is required for efficient encapsidation of HIV RNA into virus particles. The region of TAR necessary for packaging maps to the lower portion of the TAR stem. It appears that the integrity of this stem is the important factor, as a series of compensatory double mutants, with changed stem sequence but unaltered two-dimensional structure, were still packaged. In contrast, mutants with changes in regions of TAR which are critically important for transactivation (the TAR bulge or loop) had little effect on packaging. These results are consistent with a previously published genetic study that demonstrated the necessity of base pairing in the lower portion of the TAR stem for optimal HIV replication (39). That study did not define the actual role played by the lower portion of the stem, although it did suggest that the lower stem structure might be necessary for transcription, RNA packaging, or reverse transcription. Other genetic studies have also suggested functions of TAR that go beyond its role in transactivation (15, 29–31). Furthermore, a recent report demonstrated decreased (about 3.5-fold) packaging in an HIV vector mutant containing a total deletion of TAR (48).
Previously published reports from other laboratories suggest that additional packaging determinants other than TAR are present in R-U5 (16, 47, 48, 63). The data suggest that at least two distinct stem loop structures exist in the R-U5 region and that both of these structures are needed for efficient encapsidation of HIV RNA. These results, together with our present data, and data showing the importance of regions mapping downstream of the primer binding site (SL1 to SL4) in RNA packaging, explain our previous observation that at least 1 kb of HIV RNA must be included in an HIV-1 vector for efficient packaging (8).
Mutational studies, such as the ones presented here, that measure incorporation of RNA into particles as an endpoint are subject to several general interpretations. Each identified packaging element could be a unique domain for the binding of a host or viral factor that somehow mediates the encapsidation of the RNA. Alternatively, all of the elements would not be involved in the binding to specific factors. Many would provide only the correct structural context for RNA folding, thus exposing the true packaging element(s) (i.e., the one[s] that binds the important host or viral factor[s]). It could also be that the TAR mutants somehow prevent the RNA from being delivered to the correct packaging compartment, though this seems less likely since none of the TAR mutants affect p24 production or RNA stability to any significant extent. We therefore cannot conclude that the TAR stem is necessary for the binding of a specific host or viral packaging factor. The stem structure might simply facilitate the formation of a correct structure elsewhere in the RNA. However, binding of NC to SL3 does not require other viral RNA sequences (5, 17, 42).
A recently published paper describes a novel model for the structure of the 5′ end of HIV RNA (33). The model is based primarily on electron microscopic data examining dimer RNA extracted from HIV-1 particles. In contrast to RNA extracted from other retroviruses, the results in this report show the dimer linkage region of HIV-1 RNA as a loop of about 325 nucleotides. The 5′ ends of the RNA are not free. It is thus proposed that the dimer linkage region consists of two parts: (i) an intramolecular base pairing of SL1 and (ii) an intramolecular base pairing of the R-U5 region and/or TAR which creates the circular loop between the 5′ end and SL1. If this model is correct, then the role of the bottom part of the TAR stem in packaging could be to help create this novel bipartite dimer linkage which may be necessary for efficient packaging.
We have also shown that HIV RNAs that are normally retained within the nucleus in the absence of Rev cannot be packaged into particles unless Rev is provided in trans or the CTE from MPMV is provided in cis. Since the CTE can fully substitute for the RRE to enable RNA packaging, it is unlikely that the RRE contains a direct packaging determinant as has been proposed in some earlier studies (56). Rather, Rev/RRE or the CTE is probably necessary to allow the RNA to exit from the nucleus so that it can be recognized by the packaging factors which presumably reside in the cytoplasm.
RNA export mediated by Rev/RRE or CTE may utilize export pathways different from the pathways that normally mediate the export of spliced cellular mRNA (see reference 26 for a review). It will therefore be of interest to determine if transport utilizing these elements is a critical factor in packaging or if RNA exported by the normal mRNA export pathway can also be packaged. Since all of the RNAs examined in the present study required Rev/RRE or CTE for export, it will be necessary to create other substrates before this question can be answered.
ACKNOWLEDGMENTS
We thank Joy Niesen for expert tissue culture assistance.
This work was supported by NIH grants AI34721 to M.-L.H. and AI38186 to D.R. and by the Charles H. Ross, Jr., and Myles H. Thaler Endowments at the University of Virginia.
REFERENCES
- 1.Aldovini A, Young R. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrigo S J, Weitsman S, Zack J A, Chen I S. Characterization and expression of novel singly spliced RNA species of human immunodeficiency virus type 1. J Virol. 1990;64:4585–4588. doi: 10.1128/jvi.64.9.4585-4588.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arya S K, Guo C, Josephs S F, Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III) Science. 1985;229:69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- 4.Baudin F, Marquet R, Isel C, Darlix J L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 5.Berglund J A, Charpentier B, Rosbash M. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 1997;25:1042–1049. doi: 10.1093/nar/25.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz R, Fisher J, Goff S P. RNA packaging. In: Krausslich H-G, editor. Morphogenesis and maturation of retroviruses. Berlin, Germany: Springer; 1996. pp. 177–218. [Google Scholar]
- 7.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz R D, Hammarskjold M L, Helga Maria C, Rekosh D, Goff S P. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 9.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjöld M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clever J L, Wong M L, Parslow T G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das A T, Klaver B, Berkhout B. The 5′ and 3′ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J Virol. 1998;72:9217–9223. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das A T, Klaver B, Klasens B I, van Wamel J L, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 18.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dundr M, Leno G H, Hammarskjöld M-L, Rekosh D, Helga-Maria C, Olson M O J. The roles of nuclear structure and function in the subcellular location of the HIV-1 Rev protein. J Cell Sci. 1995;108:2811–2823. doi: 10.1242/jcs.108.8.2811. [DOI] [PubMed] [Google Scholar]
- 20.Ernst R, Bray M, Rekosh D, Hammarskjöld M-L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freed E O, Englund G, Martin M A. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 23.Gerard R D, Gluzman Y. New host cell system for regulated simian virus 40 DNA replication. Mol Cell Biol. 1985;5:3231–3240. doi: 10.1128/mcb.5.11.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 25.Gorelick R J, Nigida S M, Bess J R, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammarskjöld M-L. Regulation of retroviral RNA export. Semin Cell Dev Biol. 1997;8:83–90. doi: 10.1006/scdb.1996.0127. [DOI] [PubMed] [Google Scholar]
- 27.Hammarskjöld M-L, Heimer J, Hammarskjöld B, Sangwan I, Albert L, Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammarskjöld M-L, Wang S-C, Klein G. High-level expression of the Epstein-Barr virus EBNA1 protein in CV1 cells and human lymphoid cells using a SV40 late replacement vector. Gene. 1986;43:41–50. doi: 10.1016/0378-1119(86)90006-5. [DOI] [PubMed] [Google Scholar]
- 29.Harrich D, Hsu C, Race E, Gaynor R B. Differential growth kinetics are exhibited by human immunodeficiency virus type 1 TAR mutants. J Virol. 1994;68:5899–5910. doi: 10.1128/jvi.68.9.5899-5910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrich D, Mavankal G, Mette S A, Gaynor R B. Human immunodeficiency virus type 1 TAR element revertant viruses define RNA structures required for efficient viral gene expression and replication. J Virol. 1995;69:4906–4913. doi: 10.1128/jvi.69.8.4906-4913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrich D, Ulich C, Gaynor R B. A critical role for the TAR element in promoting efficient human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:4017–4027. doi: 10.1128/jvi.70.6.4017-4027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison G P, Lever A M. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoglund S, Ohagen A, Goncalves J, Panganiban A T, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 34.Jeang K T. Tat, Tat-associated kinase, and transcription. J Biomed Sci. 1998;5:24–27. doi: 10.1007/BF02253352. [DOI] [PubMed] [Google Scholar]
- 35.Jeang K T, Rawlins D R, Rosenfeld P J, Shero J H, Kelly T J, Hayward G S. Multiple tandemly repeated binding sites for cellular nuclear factor 1 that surround the major immediate-early promoters of simian and human cytomegalovirus. J Virol. 1987;61:1559–1570. doi: 10.1128/jvi.61.5.1559-1570.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones K A. Taking a new TAK on tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 37.Kaye J F, Richardson J H, Lever A M. cis-acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J Virol. 1995;69:6588–6592. doi: 10.1128/jvi.69.10.6588-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim H-J, Lee K, O’Rear J J. A short sequence upstream of the 5′ major splice site is important for encapsidation of human immunodeficiency virus type 1 genomic RNA. J Virol. 1994;198:336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 39.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994;13:2650–2659. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 41.Levin J G, Grimley P M, Ramseur J M, Berezesky I K. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lochrie M A, Waugh S, Pratt J D, Clever J, Parslow T G, Polisky B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 1997;25:2902–2910. doi: 10.1093/nar/25.14.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mann R, Mulligan R C, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 45.Marquet R, Paillart J C, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mujeeb A, Clever J L, Billeci T M, James T L, Parslow T G. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat Struct Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 50.Muriaux D, Fosse P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV- 1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 51.Nash M A, Meyer M K, Decker G L, Arlinghaus R B. A subset of Pr65gag is nucleus associated in murine leukemia virus-infected cells. J Virol. 1993;67:1350–1356. doi: 10.1128/jvi.67.3.1350-1356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nermut M V, Hockley D J. Comparative morphlogy and structural classification of retroviruses. In: Krausslich H-G, editor. Morphogenesis and maturation of retroviruses. Berlin, Germany: Springer; 1996. pp. 1–24. [DOI] [PubMed] [Google Scholar]
- 53.Paillart J C, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 54.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 55.Poon D T, Li G, Aldovini A. Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J Virol. 1998;72:1983–1993. doi: 10.1128/jvi.72.3.1983-1993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richardson J H, Child L A, Lever A M. Packaging of human immunodeficiency virus type 1 RNA requires cis-acting sequences outside the 5′ leader region. J Virol. 1993;67:3997–4005. doi: 10.1128/jvi.67.7.3997-4005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schliephake A W, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. J Virol. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz S, Felber B K, Fenyo E M, Pavlakis G N. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J Virol. 1990;64:5448–5456. doi: 10.1128/jvi.64.11.5448-5456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4649. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith A J, Cho M I, Hammarskjöld M L, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith A J, Srinivasakumar N, Hammarskjöld M L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sorge J, Wright D, Erdman V D, Cutting A E. Amphotropic retrovirus vector system for human cell gene transfer. Mol Cell Biol. 1984;4:1730–1737. doi: 10.1128/mcb.4.9.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vicenzi E, Dimitrov D S, Engelman A, Migone T S, Purcell D F, Leonard J, Englund G, Martin M A. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J Virol. 1994;68:7879–7890. doi: 10.1128/jvi.68.12.7879-7890.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. . (Erratum, 71:5712, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]