Abstract
Binding of the human immunodeficiency virus type 1 (HIV-1) Gag protein precursor, Pr55Gag, to membrane is an indispensable step in virus assembly. Previously, we reported that a matrix (MA) residue 6 substitution (6VR) imposed a virus assembly defect similar to that observed with myristylation-defective mutants, suggesting that the 6VR change impaired membrane binding. Intriguingly, the 6VR mutation had no effect on Gag myristylation. The defective phenotype imposed by 6VR was reversed by changes at other positions in MA, including residue 97. In this study, we use several biochemical methods to demonstrate that the residue 6 mutation, as well as additional substitutions in MA amino acids 7 and 8, reduce membrane binding without affecting N-terminal myristylation. This effect is observed in the context of Pr55Gag, a truncated Gag containing only MA and CA, and in MA itself. The membrane binding defect imposed by the 6VR mutation is reversed by second-site changes in MA residues 20 and 97, both of which, when present alone, increase membrane binding to levels greater than those for the wild type. Both reduced and enhanced membrane binding imposed by the MA substitutions depend upon the presence of the N-terminal myristate. The results support the myristyl switch model recently proposed for the regulation of Gag membrane binding, according to which membrane binding is determined by the degree of exposure or sequestration of the N-terminal myristate moiety. Alternatively, insertion of the myristate into the lipid bilayer might be a prerequisite event for the function of other distinct MA-encoded membrane binding domains.
The human immunodeficiency virus type 1 (HIV-1) Gag polyprotein precursor, like those of other retroviruses, is sufficient to promote the assembly and release of immature virus-like particles in the absence of other viral proteins (for reviews, see references 44 and 46). Retroviral Gag proteins contain several functional domains critical for efficient virus assembly and release (for a review, see reference 11). These domains, which drive Gag membrane binding, Gag-Gag interactions, and late budding functions, have been referred to as M, I, and L, respectively (35). During or immediately after virus budding, Pr55Gag is cleaved by the viral protease (PR) to produce the mature Gag proteins p17 (MA), p24 (CA), p7 (NC), and p6; this proteolytic processing triggers a major conformational and morphological change in the virus particle, leading to core condensation and virion maturation.
HIV-1 MA has been implicated in both early and late phases of the virus life cycle. Based on the analysis of MA mutants which display defects in virus production, MA has been demonstrated to play a major role in the binding of Pr55Gag to membrane. The observation that large deletions (10, 17) and single amino acid changes (16) in MA cause a redirection of assembly to cytoplasmic compartments indicates that MA plays a role not only in promoting membrane binding in general but also in the targeting of Gag to the plasma membrane in particular. Deletions (9, 48, 52) and single amino acid substitutions (13, 15, 33) in MA can block HIV-1 envelope glycoprotein incorporation into virus particles, and HIV-1 MA has been implicated in early events postinfection (for a review, see reference 11).
During Pr55Gag synthesis, the N-terminal Gly residue of the MA domain is modified by the covalent attachment of a myristic acid moiety. The importance of N-terminal myristylation in Gag membrane binding and virus assembly has been definitively established (4, 8, 19, 23, 39, 41, 43, 49); however, the contribution of other MA domains to membrane binding is less well defined. A highly basic sequence located between HIV-1 MA residues 17 and 31 has been implicated in Gag membrane binding. It has been proposed that basic residues within this domain and elsewhere in MA form a positively charged surface which interacts with negatively charged phospholipids on the inner face of the lipid bilayer (21, 29, 54, 55). Data in support of a direct role for MA basic residues in membrane binding are mixed. A contribution of the basic domain is supported by mutational (12, 53, 54) and structural (for a review, see reference 7) data. In contrast, other studies observed that a basic domain deletion actually increased virus production (52), and large deletion mutants lacking all or most of MA but possessing a myristylation site are competent to produce virus particles in some studies (26, 38, 48).
It has been suggested, based on its relatively low hydrophobicity compared with other (longer) fatty acids, that myristate is used to promote the membrane binding of proteins which must interact with membranes in a reversible manner (45). In the case of the retinal rod calcium sensor protein recoverin, calcium binding causes a conformational change that exposes the myristate moiety, which in the absence of calcium is sequestered within the protein (2). According to this myristyl switch model, the calcium-triggered exposure of myristate induces the binding of recoverin to membrane (2). Based on the observation that Pr55Gag binds membrane more tightly than MA itself, it has been hypothesized that cleavage of Pr55Gag might cause a refolding of the MA domain, a sequestration of the N-terminal myristate, and a reduction in membrane binding potential (55). This model is supported by the findings that deletion of portions of MA which may be responsible for sequestration of myristate increased membrane binding (42, 55).
We previously reported that mutations in the N terminus of HIV-1 MA induced defects in virus assembly and release (16). Substitutions in the N-terminal five residues blocked or impaired Gag myristylation (32, 33); intriguingly, however, mutation of residue 6 from Val to Arg (6VR) caused virus assembly and release defects without affecting Gag myristylation (33). Because of the phenotypic similarities between the 6VR mutation and those affecting N-terminal myristylation (i.e., defective Gag processing and reduced virus production), we postulated that the 6VR substitution may impair binding of Gag to membrane (11, 33). Similar phenotypes were reported for N-terminal mutations in the MA of Moloney murine leukemia virus (20) and simian immunodeficiency virus (18). In the latter case, it was proposed that the effect of the N-terminal MA mutations was attributable to decreased hydrophobicity.
In our previous studies, we observed that the 6VR mutant, which replicated with markedly delayed kinetics in T cells (16) reverted in culture (33). The revertant viruses maintained the 6VR mutation but acquired second-site changes at several downstream positions in MA, including residue 97 (97KE). A 6VR/97KE double mutant displayed a complete reversal of the impaired virus production and delayed replication kinetics observed with 6VR.
Our goals in this study were severalfold: (i) to determine whether the domain of HIV-1 MA immediately downstream of the N-myristyl transferase recognition sequence (Gly-X-X-X-Ser/Thr) (45) is involved in Gag membrane binding, (ii) to biochemically characterize the basis for the ability of downstream MA mutations to overcome the virus assembly and replication defects imposed by the 6VR mutation, and (iii) to assess the role of Gag sequences downstream of MA on the membrane binding ability of wild-type and MA-mutant Gag by using assays which clearly distinguish membrane-bound Gag from assembled or aggregated complexes. Using three biochemical assays, we demonstrate that MA residues 6 to 8 play a critical role in membrane binding. This function is evident in the context of Pr55Gag, a truncated Gag containing only MA and CA, and MA itself. Mutations in residues 20 and 97 increase binding of Gag to membrane and as a result can reverse the membrane binding defect caused by the 6VR substitution. The contribution of MA residue 6 and 20 to membrane binding requires N-terminal myristylation.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa cells were maintained as previously described (14). Transfection of HeLa cells was performed by the calcium phosphate precipitation method as reported previously (16).
Plasmids, mutagenesis, and DNA cloning.
Construction of derivatives of the HIV-1 molecular clone pNL4-3 (1) containing MA amino acid changes 1GA, 4AD, 5SI, 6VR, 20LK, 97KE, and 6VR/97KE has been described previously (16, 33). The pNL4-3 derivative pNL4-3/PR−, which contains a PR active site mutation (Asp→Asn change at PR amino acid 25), has been described previously (22). PR− versions of MA mutant molecular clones were constructed by cloning the PR-containing SphI-EcoRI fragment (nucleotides 1443 to 5743) from pNL4-3/PR− into MA-mutant pNL4-3 derivatives.
Substitutions at MA amino acids 7 and 8 (7LV, 7LE, 8SA, and 8SE) and double mutations 1GA/6VR, 1GA/20LK, and 6VR/20LK were introduced into pNL4-3 by oligonucleotide-directed mutagenesis by using methods detailed previously (16). The molecular clone expressing p17 (MA) (pNL4-3/MAstop) was constructed by introducing stop codons at CA amino acid positions 1 and 2 by the same strategy used for MA mutagenesis (16). Introduction of 1GA and 6VR changes into pNL4-3/MAstop was performed as described above by using a 1.6-kbp StuI-SphI fragment of pNL4-3/MAstop subcloned into M13mp19 as a template for mutagenesis.
To construct the plasmid pNL4-3/p41stop, which expresses p41 (MA-CA), a stop codon was introduced at residue 1 of the p2 spacer peptide. Oligonucleotide-directed mutagenesis was performed by using an M13mp18 subclone harboring the SphI-PstI (1.4 kbp) fragment from pNL4-3 as a template and oligonucleotide 5′-AAGAGTTTTGTAAGAAGCAATGA-3′. After mutagenesis, the M13mp18-derived SphI-PstI fragment containing the stop codon was introduced into pNL4-3 and sequenced in its entirety. The same fragment was cloned into pNL4-3/1GA and pNL4-3/6VR to generate p41stop versions of these MA mutants.
Metabolic labeling and immunoprecipitation.
Metabolic labeling of transfected HeLa cells with either [35S]Cys, [35S]Met, or [3H]myristic acid was performed as previously described (14, 15, 33). Preparation of cell lysates, pelleting of labeled virions in the ultracentrifuge, and immunoprecipitation of cell- and virion-associated proteins with sera from patients with AIDS (National Institutes of Health AIDS Research and Reference Reagent Program, catalog no. 1983 and 1984) have been detailed previously (15, 50). For immunoprecipitation of labeled p17 (MA) from fractions of equilibrium flotation centrifugation assays (see below), 1.2 ml of fractionated samples was mixed with 0.3 ml of 5× triton lysis buffer (250 mM Tris-HCl [pH 7.5], 2.5% Triton X-100, 1.5 M NaCl, 50 mM iodoacetoamide, 1 mM phenylmethylsulfonyl fluoride, and 1 mg of leupeptin/ml), and precleared with recombinant protein G-agarose (GIBCO BRL, Gaithersburg, Md.). Subsequently, precleared samples were subjected to immunoprecipitation with mouse monoclonal anti-HIV-1 p17 antibody (Advanced Biotechnologies, Inc., Columbia, Md.) and immobilized on protein G-agarose, and precipitated proteins were analyzed as previously described (50).
Western blotting.
Western blotting of fractionated samples was performed as previously described (25). The following primary antibodies were used: sera from patients with AIDS, rabbit anti-gp41 serum (Fitzgerald Industries International, Inc., Concord, Mass.), and mouse monoclonal anticalnexin antibody (Affinity Bioreagent, Inc., Golden, Colo.). Horseradish peroxidase-conjugated antihuman immunoglobulin (Ig), antirabbit Ig, and antimouse Ig (all obtained from Amersham) were used as secondary antibodies. Quantitation of Western blotting data was performed by densitometry scanning.
Membrane binding analyses.
Cell fractionation and equilibrium sucrose density gradient centrifugation assays were performed as detailed previously (25) but with some modifications. Briefly, transfected HeLa cells were rinsed with ice-cold phosphate-buffered saline and collected by scraping 2 days posttransfection. Cells were washed once with 10 mM Tris-HCl (pH 7.5) containing 1 mM EDTA and 1 mM EGTA and suspended in 10 mM Tris-HCl containing 1 mM EDTA, 6% (wt/vol) sucrose, and Complete protease inhibitor cocktail (Boehringer Mannheim). Cell suspensions were subjected to sonication (15 s, twice) in ice water to achieve disruption of more than 90% of cells. After low-speed centrifugation, postnuclear supernatants were not adjusted (no salt) or adjusted to 1 M NaCl (high salt) and centrifuged at 100,000 × g for 1 h in a Beckman SW55Ti rotor. The resulting pellet and supernatant fractions were subjected to Western blotting as described above. For equilibrium sucrose density gradient centrifugation, postnuclear supernatants adjusted to 1 M NaCl as described above were loaded onto gradients composed of 20, 30, 40, 50, and 60% (wt/vol) sucrose in 10 mM Tris-HCl (pH 7.5) containing 1 mM EDTA (TE) and centrifuged at 100,000 × g for 16 h at 4°C in a Beckman SW55Ti rotor. Eleven fractions (480 μl each) were collected from the top of the gradients and were subjected to Western blotting analysis.
Our methods for equilibrium flotation centrifugation were modified from those reported by Spearman et al. (42). HeLa cells collected and washed as described above were resuspended in 10 mM Tris-HCl containing 1 mM EDTA, 10% (wt/vol) sucrose, and Complete protease inhibitor cocktail. Postnuclear supernatants were obtained after sonication of cell suspensions as described above. Then, 250 μl of postnuclear supernatants was mixed with 1.25 ml of 85.5% (wt/vol) sucrose in TE and placed on the bottom of a centrifuge tube. On top of this postnuclear-supernatant-containing 73% (wt/vol) sucrose mixture was layered 7 ml of 65% (wt/vol) sucrose in TE and 3.25 ml of 10% (wt/vol) sucrose in TE. The gradients were centrifuged at 100,000 × g for 18 h at 4°C in a Beckman SW41 rotor. Ten 1.2-ml fractions were collected from the top of the centrifuge tube for Western blotting or radioimmunoprecipitation as described above. For analysis of p17 (MA), postnuclear supernatants were obtained from [35S]Cys-labeled cells, and fractions were immunoprecipitated with anti-p17 antibody as described above.
RESULTS
Mutations near the N terminus of MA impair virus assembly and release without affecting myristylation.
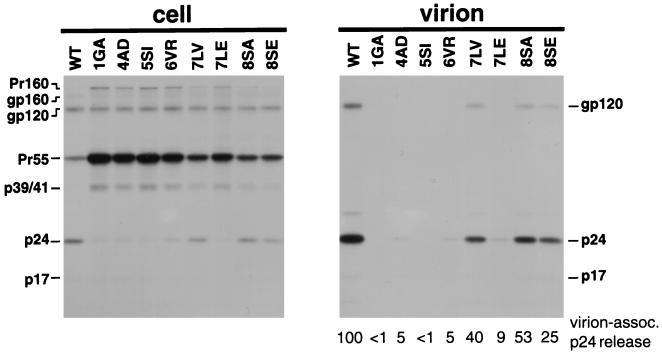
We previously reported that the Val-to-Arg substitution at MA amino acid 6 (6VR) markedly reduced virus assembly and release without affecting Gag myristylation (33). This finding suggested an unidentified role for residue 6 in the late stages of the virus life cycle. Since single amino acid changes at residues 9 (Gly→Glu), 10 (Gly→Arg), and 12 (Leu→Glu) had no effect on virus particle production (15, 16, 33), we sought to map the putative N-terminal assembly domain by determining whether mutation of other residues immediately following the N-myristyl transferase recognition sequence would affect virus particle production. Accordingly, we introduced additional single amino acid mutations in this region as follows: Leu→Val and Leu→Glu at amino acid 7 (7LV and 7LE, respectively) and Ser→Ala and Ser→Glu at amino acid 8 (8SA and 8SE, respectively) (Fig. 1). Following site-directed mutagenesis, these changes were introduced into the infectious molecular clone pNL4-3 (1). HeLa cells were transfected with wild-type or MA N-terminal mutant molecular clones and were metabolically labeled with [35S]Cys. Cell and virion lysates were prepared and subjected to immunoprecipitation analysis (Fig. 2). Consistent with our previous findings (16, 33), the 1GA, 4AD, 5SI, and 6VR mutations caused a marked reduction in virion release (Fig. 2, right panel). These mutations also reduced the efficiency of processing of cell-associated Gag and Gag-Pol precursors, as evidenced by the accumulation of Pr55Gag and Pr160Gag-Pol (Fig. 2, left panel). In addition, all the mutants with amino acid changes at residues 7 and 8 showed significantly reduced virus production (Fig. 2, right panel). Accumulation of Gag and Gag-Pol precursors in cell-associated material was also observed with the residue 7 and 8 mutants (Fig. 2, left panel), indicating a reduced efficiency of Gag precursor processing. These results suggest that MA amino acid residues 6 to 8 play a role in virus assembly and release.
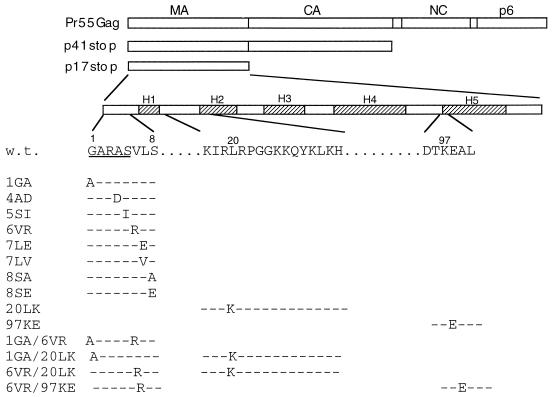
FIG. 1.
HIV-1 MA mutations analyzed for their effect on Gag membrane binding. At the top is indicated the linear organization of the Gag precursor Pr55Gag showing MA, CA, NC, and p6 domains. MA mutations were analyzed in the contexts of full-length Pr55Gag, a truncated Gag containing MA-CA (p41stop), and MA alone (MAstop). The locations of the five major α-helices in MA (H1 to H5) are indicated; the N terminus of MA and the sequences surrounding residues 20 and 97 are enlarged. The positions of the mutations are indicated. Dashes denote sequence identity with wild type (w.t.). The myristylation consensus sequence is underlined.
FIG. 2.
Effect of N-terminal MA mutations on Gag processing and virus production. HeLa cells transfected with wild-type (WT) pNL4-3 or derivatives containing the indicated MA mutations were metabolically labeled with [35S]Cys. Virions were pelleted in an ultracentrifuge. Cell (left panel)- and virion (right panel)-associated material was immunoprecipitated with sera from patients with AIDS (Materials and Methods). The relative levels of virion-associated p24 (normalized for cell-associated gp120) are indicated under each lane of the virion panel. The positions of the Pr160Gag-Pol (Pr160) and Pr55Gag (Pr55) precursors, the Env precursor gp160, the mature surface Env glycoprotein gp120, the Gag processing intermediates p41 and p39, and p24 (CA) and p17 (MA) are shown.
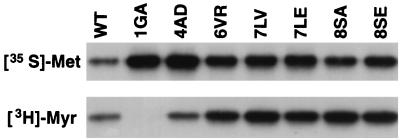
Mutations which block or impair HIV-1 Gag myristylation have been reported to disrupt virus assembly and reduce the efficiency of Gag processing (4, 16, 19, 27, 34). We previously demonstrated that the 6VR mutation did not impair Gag myristylation (33). However, since mutations at amino acids 7 and 8 are located close to the myristylation signal, we next examined whether these changes affected Gag myristylation. To eliminate differences in Pr55Gag levels resulting from differential rates of Gag processing observed with these mutants (Fig. 2), we analyzed the MA N-terminal mutants in the context of a PR− molecular clone (Materials and Methods). HeLa cells transfected with these mutants were metabolically labeled with [35S]Met or [3H]myristic acid, and cell-associated material was immunoprecipitated (Fig. 3). Consistent with our previous results (33), the 1GA and 4AD mutants showed no myristylation (1GA) or significantly impaired myristylation (4AD), whereas 6VR showed a wild-type level of 3H incorporation (Fig. 3). These results confirm that, unlike the 1GA and 4AD mutations, the 6VR change does not affect attachment of the N-terminal myristic acid moiety. Gag proteins with single amino acid changes at residues 7 and 8 also showed levels of myristylation at least as high as wild-type levels (Fig. 3). These results indicate that the defects in virus assembly and release caused by the mutations in MA residues 6 to 8 are not the result of disrupted Gag myristylation.
FIG. 3.
Effect of N-terminal MA mutations on Gag myristylation. HeLa cells were transfected with pNL4-3/PR− or derivatives containing the indicated MA mutations. Cells were metabolically labeled with either [35S]Met (top panel) or [3H]Myr (lower panel). Cell lysates were prepared and immunoprecipitated with sera from patients with AIDS. The Pr55Gag band is shown.
The N-terminal MA mutants display impaired membrane binding by several biochemical assays.
Although mutations in MA residues 6 to 8 do not impair myristylation (Fig. 3), the possibility remained that these changes might disrupt membrane binding. Thus, we next determined the membrane binding ability of Gag proteins with single amino acid changes at residues 6, 7, and 8, using several biochemical procedures.
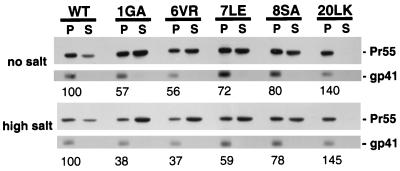
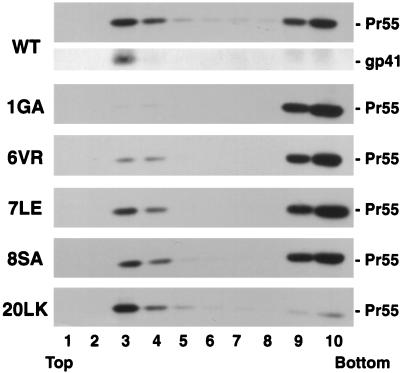
First, we performed cell fractionation assays commonly used to assess membrane binding (Materials and Methods) (Fig. 4). Approximately 70% of wild-type Pr55Gag was distributed in the pellet fraction under both no-salt and high-salt conditions. The 1GA mutant showed a marked decrease in pelleted Gag compared to that of the wild type. As we demonstrated previously (25), the 20LK change increased the percentage of Pr55Gag in the pellet fraction in the presence or absence of high-salt conditions. The 6VR, 7LE, and 8SA mutations decreased the amount of pelleted Gag, suggesting that Gag membrane binding is impaired by these changes. The transmembrane Env glycoprotein gp41, which is found almost exclusively in the pellet, serves as a fractionation control.
FIG. 4.
Cell fractionation of MA-mutant Pr55Gag. HeLa cells were transfected with pNL4-3/PR− or derivatives containing the indicated MA mutations. Postnuclear supernatants were treated with no salt (top panel) or high salt (1 M NaCl; lower panel) and fractionated (Materials and Methods). Pr55Gag and the transmembrane Env glycoprotein gp41 were detected by Western blotting. The percentage of Pr55Gag in the pellet fraction, normalized to the wild-type (WT) value, is indicated under each lane. Approximately 70% of wild-type Pr55Gag was detected in the pellet under either no- or high-salt conditions.
In the cell fractionation approach described above, the pellets likely contain non-membrane-bound material such as large protein complexes (28), misfolded protein aggregates, and cytoskeleton-bound protein in addition to authentic membrane-bound material. Thus, we cannot definitely assess the extent of Gag membrane binding by using this technique. To more accurately separate membrane-bound Gag from pelletable but non-membrane-bound material, we next examined Gag distribution by equilibrium sucrose density gradient centrifugation (data not shown). Postnuclear supernatants of transfected HeLa cells, prepared as in Fig. 4, were layered onto 20 to 60% (wt/vol) sucrose gradients, which were spun at 100,000 × g for 16 h. Under high-salt conditions, most wild-type Pr55Gag was recovered in two peaks: the lighter peak, sedimenting to fractions 1 to 3 (1.06 to 1.10 g/cm3 density), and the heavier peak, sedimenting to fractions 6 to 9 (1.15 to 1.20 g/cm3 density). An internal marker for cellular membranes, gp41, partially overlapped with the denser Pr55Gag peak. The 1GA mutant displayed increased Pr55Gag in fractions 1 to 3 and reduced Gag in fractions 6 to 9 compared to the wild type. These observations suggested that membrane-bound Pr55Gag was recovered mainly from fractions 6 to 9, whereas cytosolic Gag was distributed primarily in fractions 1 to 3. The 6VR mutant showed a distribution similar to 1GA: a decreased amount of Pr55Gag in membrane-containing fractions and an increased amount in cytosolic fractions compared to the wild type.
The fractionation and sucrose gradient results presented above suggest that mutations in MA residues 6 to 8 decrease the binding of Pr55Gag to membrane. However, we considered it necessary to assess the membrane binding of Pr55Gag in an assay that can definitively distinguish membrane-bound Gag from non-membrane-bound complexes. To this end, we analyzed the distribution of Gag, using equilibrium flotation centrifugation. This approach has been used successfully to study membrane binding of the vesicular stomatitis virus M protein (3, 5, 6) and has also been applied to the analysis of HIV-1 MA membrane binding (42). Postnuclear supernatants from transfected HeLa cell homogenates were adjusted to 73% (wt/vol) sucrose, placed at the bottom of centrifuge tubes, overlaid with 65% (wt/vol) and 10% (wt/vol) sucrose, and centrifuged for 18 h at 100,000 × g (Materials and Methods). Fractions were collected from the top of the gradient and were analyzed by Western blotting with sera from patients with AIDS or with anti-gp41 antibody. As shown in Fig. 5, gp41 was recovered almost exclusively in fractions 3 and 4, which correspond to the boundary between 10 and 65% sucrose. The flotation of gp41 to fractions 3 and 4 was eliminated by treatment with 1% Triton X-100 prior to ultracentrifugation (data not shown). Calnexin, an endoplasmic reticulum-resident transmembrane protein (47), was also recovered predominantly in these fractions (data not shown). Consistent with membrane flotation data obtained by other investigators (3, 5, 6, 42), these results suggest that most of the plasma membrane, as well as other cellular membranes, float to the 10%-65% sucrose interface. Under these conditions, 35% of wt Pr55Gag was detected in fractions 3 and 4 (Fig. 5). The flotation of Pr55Gag was largely abolished when postnuclear supernatants were treated with 1% Triton X-100 (data not shown), indicating that detergent-resistant protein complexes that are not membrane bound (28) could not float. The percentage of 1GA Pr55Gag that was recovered in fractions 3 and 4, normalized for the wild type, was reduced to 2%, while the remainder stayed in the bottom fractions (i.e., 0.8% of total 1GA Pr55Gag was present in fractions 3 and 4). The 20LK mutant displayed an increase, relative to wild type, in the percentage of Gag present in the floated fractions. 6VR Pr55Gag showed a clear reduction in membrane binding, with a distribution intermediate between wild type and 1GA. The amount of 7LE and 8SA Pr55Gag in the floated fractions was also reduced relative to wild type. The 6VR, 7LE, 8SA, and 20LK mutants showed 5.5%, 23.5%, 19.9%, and 89.2%, respectively, of Pr55Gag in membrane-containing fractions (3 and 4). Similar data were obtained for wild-type, 1GA, and 6VR by flotation assays performed in the presence of high salt (1 M NaCl) (data not shown). These results, together with those obtained by the cell fractionation and sucrose gradient methods, suggest that the N-terminal amino acids immediately following the MA myristylation recognition signal are involved in Gag membrane binding.
FIG. 5.
Analysis of MA mutants by membrane flotation centrifugation. HeLa cells were transfected with pNL4-3/PR− or derivatives containing the indicated MA mutations. Postnuclear supernatants were prepared and subjected to membrane flotation centrifugation (Materials and Methods), during which membrane-bound material floats to the interface between 10 and 65% sucrose (fractions 3 and 4). The transmembrane Env glycoprotein gp41, which is found almost exclusively in fractions 3 and 4, serves as a control membrane-bound protein. Pr55Gag and gp41 were detected by Western blotting.
N-terminal MA mutations reduce membrane binding independently of the Gag NC domain.
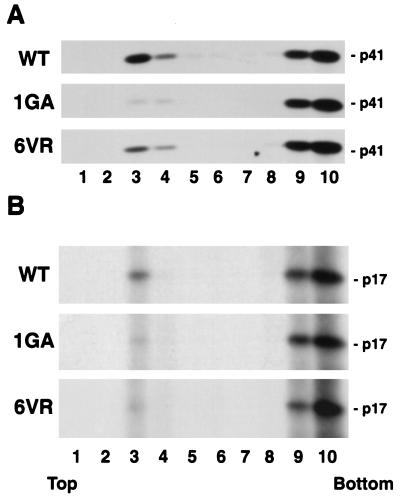
It has been suggested that a sequence located near the N terminus of NC, which has been referred to as the interaction, or I, domain (35), is involved in enhancing the binding of Pr55Gag to membrane (37, 40). It seemed possible that the impaired membrane binding imposed by mutations in MA residues 6 to 8 could result from an indirect effect involving the NC domain, perhaps via a global change in Pr55Gag conformation which might destabilize the association of Gag with membrane. To examine this possibility, we introduced a stop codon immediately after the CA domain in pNL4-3 to generate the molecular clone pNL4-3/p41stop (Fig. 1). We then introduced several of the MA mutations described above into pNL4-3/p41stop; this enabled us to analyze the effects of these mutations on membrane binding in the context of a p41 (MA-CA) Gag protein. Membrane flotation analysis (Fig. 6A) indicated that 25% of wild-type p41 was recovered in fractions 3 and 4, whereas only 0.6% of 1GA p41 and 4.5% of 6VR p41 was detected in the membrane-containing fractions. These results indicate that the effect of the MA N-terminal mutations on membrane binding is independent of Gag sequences C terminal to CA, including the NC domain.
FIG. 6.
Membrane flotation centrifugation with p41 (MA-CA) and p17 (MA). HeLa cells were transfected with pNL4-3/p41stop (which expresses a truncated Gag protein lacking sequences C-terminal to CA; panel A) or pNL4-3/MAstop (which expresses MA; panel B) or derivatives containing the 1GA or 6VR MA mutations. Postnuclear supernatants were prepared and subjected to membrane flotation centrifugation; p41 (MA-CA) was detected by Western blotting, and p17 (MA) was detected by immunoprecipitation (Materials and Methods).
To directly compare membrane binding of Pr55Gag and p41 (MA-CA), we performed the following experiment. Cells expressing wild-type Pr55Gag or p41 were mixed and disrupted by sonication, and postnuclear supernatants were subjected to membrane flotation centrifugation. We then assessed the levels of Pr55Gag and p41 in each fraction by Western blotting. This approach enabled us to eliminate any sample-to-sample variability in the assay. The results indicated that p41 (MA-CA) membrane binding was decreased from that of Pr55Gag; the percentages of Pr55Gag and p41 (MA-CA) present in membrane-containing fractions were approximately 32 and 14%, respectively. Similar results were obtained when postnuclear supernatants were prepared separately from Pr55Gag- and p41 (MA-CA)-expressing cells; in this case, the percentages of Pr55Gag and p41 (MA-CA) present in membrane-containing fractions were 32 and 23%, respectively.
The 6VR mutation reduces membrane binding of p17 (MA).
It has been proposed that following Pr55Gag cleavage by the viral PR, MA undergoes a conformation change which results in the masking or sequestration of the myristic acid moiety (55). p17 (MA) thus displays a significantly reduced affinity for membrane compared with Pr55Gag (42, 55). To determine whether the 6VR change influences membrane binding in the context of p17 (MA), we introduced a stop codon immediately after the MA coding region in pNL4-3 to generate pNL4-3/MAstop (Fig. 1). We then constructed 1GA and 6VR mutant derivatives of this molecular clone. Since it has been reported that MA forms complexes that might pellet in a fractionation assay even if not bound to membrane (31, 42), we performed membrane flotation centrifugation to directly assess the effect of the 1GA and 6VR mutations on membrane binding (Fig. 6B). Approximately 9% of wild-type p17 (MA) was detected in the membrane-containing fractions. We observed that both 1GA and 6VR mutations caused marked reductions in the amount of p17 (MA) present in the membrane fractions, indicating that both mutations reduce membrane binding in the context of p17 (MA).
The 6VR-induced membrane binding defect can be reversed by second-site changes in MA.
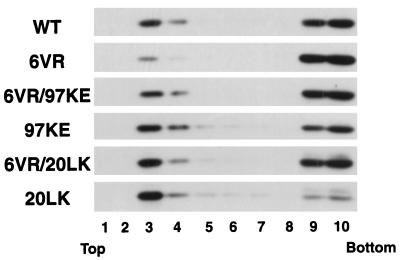
We previously isolated and characterized a set of viral revertants obtained by passaging the 6VR mutant in a T-cell line (33). One of these revertants contained a second-site change at MA residue 97 (97KE) which fully reversed the defects in virus replication and assembly caused by 6VR. These results, together with the findings described above, suggest that the 97KE change may reverse the 6VR phenotype by increasing Gag membrane binding. To examine whether the 97KE substitution increases Pr55Gag membrane binding in the presence or absence of the 6VR change, we transfected HeLa cells with pNL4-3/PR− molecular clones containing the 97KE or 6VR/97KE mutations and performed membrane flotation centrifugation (Fig. 7). Relative to wild type, the 97KE mutant showed an increase in membrane-bound Pr55Gag. Similarly, the 6VR/97KE double mutant showed an increased amount of floated Pr55Gag compared to the 6VR single mutant. These results indicate that the 97KE substitution increases Gag membrane binding both in the presence and absence of the 6VR change, thereby elucidating the biochemical mechanism by which the 97KE change reverses the 6VR-imposed defects in virus replication and assembly.
FIG. 7.
Opposing effect of MA mutations on membrane binding. HeLa cells were transfected with pNL4-3/PR− or derivatives containing the indicated MA mutations. Postnuclear supernatants were prepared and subjected to membrane flotation centrifugation (Materials and Methods). The amount of Gag in each fraction was determined by Western blotting.
We also investigated whether another mutation (20LK), which we demonstrated to increase membrane binding (Fig. 5) (25), could reverse the virus assembly and release defect imposed by 6VR. A derivative of pNL4-3/PR− was constructed which expressed a 6VR/20LK double mutant and was analyzed by membrane flotation centrifugation (Fig. 7). The percentage of 6VR/20LK Pr55Gag found in fractions 3 and 4 was significantly increased compared to that of 6VR Pr55Gag, indicating that the 6VR-imposed defect in membrane binding is reversed by the 20LK change. These results demonstrate a functional relationship between the MA N-terminal domain and downstream sequences in promoting efficient binding of Gag to membrane.
The effect of the 6VR and 20LK changes on membrane binding is dependent on Gag myristylation.
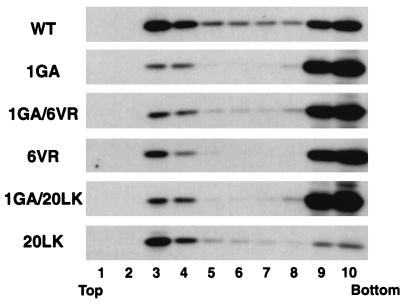
Although the mutations in residues 6 to 8 do not impair the covalent attachment of myristic acid to the N-terminal Gly of MA (Fig. 3), these changes could affect the ability of the myristate group to function effectively in membrane binding. Alternatively, the N-terminal domain could promote membrane binding in a manner independent of the myristate moiety. To explore these possibilities, we investigated whether the 6VR change would decrease membrane binding in the absence of N-terminal myristylation. We constructed a 1GA/6VR double mutant and compared its membrane binding properties with those of 1GA in membrane flotation assays (Fig. 8). As we showed above, even in the absence of myristylation, a small amount of Pr55Gag is recovered in the floated fractions (Fig. 5 and 8; 1GA). Analysis in the same experiment indicated that the amount of 1GA/6VR Gag in membrane-containing fractions was not less than that observed with 1GA Gag (Fig. 8), suggesting that the 6VR phenotype is dependent upon N-terminal myristylation.
FIG. 8.
Membrane flotation centrifugation of 1GA/6VR and 1GA/20LK double mutants. HeLa cells were transfected with pNL4-3/PR− or derivatives containing the indicated MA mutations. Postnuclear supernatants were prepared and subjected to membrane flotation centrifugation (Materials and Methods). The amount of Gag in each fraction was determined by Western blotting.
We also investigated whether the increase in membrane binding induced by the 20LK change was similarly dependent upon myristylation. We constructed a 1GA/20LK double mutant and compared its Gag membrane binding ability with that of 1GA. 1GA and 1GA/20LK Pr55Gag showed similar distributions in membrane flotation assays (Fig. 8). These results suggest that N-terminal Gag myristylation is prerequisite for the 20LK-induced enhancement of Gag membrane binding.
DISCUSSION
HIV-1 MA has been shown to contain functional domains involved in several steps in virus particle assembly and release, including membrane binding of Pr55Gag (for a review, see reference 11). In this study, we determined that a sequence near the N terminus of MA (residues 6 to 8) plays a critical role in HIV-1 Gag membrane binding. Single amino acid changes in this region impaired virus production and reduced the efficiency of Pr55Gag processing (Fig. 2). Despite wild-type levels of N-myristylation (Fig. 3), these mutants displayed significantly impaired Pr55Gag membrane binding, as determined by cell fractionation, equilibrium sucrose density gradient, and equilibrium flotation centrifugation assays (Fig. 4 and 5). This N-terminal MA sequence also reduced membrane binding of both p41 (MA-CA) (Fig. 6A) and p17 (MA) (Fig. 6B). Analysis of double mutants showed that decreased membrane binding imposed by amino acid changes at residue 6 (6VR) was reversed by second-site amino acid changes in other regions of MA (i.e., residues 20 and 97). Introduced singly into Pr55Gag, the amino acid 20 (20LK) and 97 (97KE) changes resulted in a mutant Pr55Gag which bound membrane more efficiently than did the wild type (Fig. 7). Finally, the ability of 6VR to decrease and 20LK to increase membrane binding was not observed in the absence of N-terminal Gag myristylation (Fig. 8).
MA residues 6 to 8 may contribute to Gag membrane binding by several possible mechanisms. (i) The somewhat hydrophobic nature of the MA N terminus may promote a direct interaction with the lipid bilayer. Although results obtained with a mutation near the N terminus of the simian immunodeficiency virus MA support this hypothesis (18), we observed membrane binding defects with the 8SA mutation, which increases local hydrophobicity. Furthermore, the conservative 7LV change also impaired virus assembly and release, and substitution of the residue 9 Gly for Glu had no significant effect on virus assembly or replication kinetics (16). These findings argue that the overall hydrophobicity of the MA N terminus is insufficient to account for its role in membrane binding. The 7LE mutation may induce defects in addition to the impaired membrane binding, since its impact on virus release is as severe as that of 6VR (Fig. 2), yet it has a less marked effect on membrane binding (Fig. 5). (ii) In accordance with the myristyl switch model (see introduction), mutations in MA residues 6 to 8 could prevent the exposure of the myristate group, whereas substitutions at amino acids 20 and 97 could increase myristate exposure. Our observation that the residue 20 mutation has no effect on membrane binding in the absence of MA myristylation (i.e., 1GA/20LK; Fig. 8) supports this hypothesis. We also observed that the 6VR mutation failed to decrease membrane binding of Gag in the absence of the N-terminal myristate (i.e., 1GA/6VR; Fig. 8). (iii) MA residues 6 to 8 could interact directly with the lipid bilayer or with a component of the membrane. This model can be rationalized with the 1GA/6VR data by considering that the binding of this N-terminal domain might occur only after initial interaction of the myristate moiety with the bilayer. It is interesting to consider the possibility that Gag might interact with a proteinaceous component of the plasma membrane (i.e., a Gag receptor). The interaction of other myristylated proteins, for example v-Src and MARCKS, is thought to involve protein-protein contacts (30). Although the existence of a Gag receptor has been postulated (51), to date no such molecule has been identified.
We urge caution in interpreting the impact of MA mutations on membrane binding based only on their effects on myristate exposure. Although myristate might be a prerequisite for efficient membrane binding, a variety of considerations argue against the myristate moiety being the only factor involved in the binding of authentic HIV-1 Gag to membrane. (i) It has been proposed that the Gibbs free energy of binding (ΔGu) contributed by the myristate group, which has been estimated for small peptides at approximately 8 kcal/mol, is insufficient for stable attachment of a myristylated protein to cellular membrane (36). (ii) The N-terminal 31 amino acid sequence of HIV-1 MA, which includes the highly basic domain, confers membrane binding ability upon heterologous proteins; some binding occurs even in the absence of myristylation (54). (iii) In this study, we observed a reduced but still detectable amount of 1GA Gag in membrane fractions in membrane flotation assays, implying that nonmyristylated Gag still retains some ability to bind membrane. However, we cannot rule out the possibility that the 1GA Gag recovered in membrane-containing fractions might represent a population of molecules which does not bind membrane directly but, rather, interacts with other membrane-bound components. (iv) In the three-dimensional structure of HIV-1 MA (21, 29), a number of basic amino acids, including several in the highly basic domain, are clustered on one side of MA such that they could contribute membrane binding energy by interacting with negatively charged phospholipids on the inner face of the lipid bilayer. The orientation of basic residues on the predicted membrane binding face appears to be a highly conserved feature of retroviral MA protein structure (for review, see reference 7). (v) The MA domains of several retroviruses, including Rous sarcoma virus and equine infectious anemia virus, are not myristylated yet possess the ability to direct Gag binding to membrane.
In this study, we utilized three different methods to examine Gag membrane binding. Although these assays all demonstrated that the N-terminal MA mutants display reduced membrane binding compared with that of the wild type, only the membrane flotation assay can separate membrane-bound material from non-membrane-bound protein complexes such as the recently described detergent-resistant Gag complex (28). This point is illustrated clearly by comparing the effect of the 1GA mutation by the three methods; in fractionation assays and sucrose gradient analyses the effect of the 1GA mutation was fairly modest, while in membrane flotation assays its impact was profound (Fig. 4 and 5). These observations suggest that much of the Gag observed in the pellet in fractionation assays and in membrane-containing fractions in the sucrose gradient analyses represents non-membrane-bound Gag complexes. This supposition is supported by the finding that treatment of postnuclear supernatants with Triton X-100 before fractionation shifted the Env glycoproteins to the supernatant fraction but had little effect on the distribution of Pr55Gag. In contrast, the same treatment before membrane flotation centrifugation largely abolished the migration of both Gag and gp41 to membrane-containing fractions (32). Taken together, these results are consistent with the notion that a significant amount of Gag multimerization precedes the binding of Gag to membrane. Although the contribution of NC to membrane binding observed in our study was considerably smaller than reported previously (40), the reduced membrane binding observed with p41 (MA-CA) versus Pr55Gag suggests that NC sequences enhance membrane binding. This effect may be mediated by the ability of NC to promote Gag-Gag interactions (for review, see reference 11). By carefully defining the advantages and limitations of each assay, we can utilize combinations of cell fractionation, sucrose gradient, and membrane flotation assays to gain additional insights into membrane binding and virus assembly.
Our studies suggest that the affinity of MA for membrane must be precisely balanced to enable Gag to function appropriately in early and late phases of the HIV-1 life cycle. The 20LK and 97KE changes, both of which increase Gag membrane binding (25) (Fig. 5 and 7), cause an early postentry defect in virus infectivity (24, 25). Decreased membrane binding reduces the efficiency of virus assembly and release, as observed in this and previous studies, whereas increased membrane binding may cause retention of MA at the membrane and consequently destabilization of the core/preintegration complex early postentry (25). Natural emergence of the 97KE substitution as a compensatory second-site change during replication of the 6VR mutant (33) is consistent with this hypothesis. Efforts are currently under way to further elucidate the role of membrane binding in HIV-1 replication.
ACKNOWLEDGMENTS
We thank R. Kiernan, T. Murakami, M. Martin, and R. Willey for helpful suggestions and critical review of the manuscript. Sera from patients with AIDS were obtained through the NIH AIDS Research Reference and Reagent Program (from L. Vujcic).
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames J B, Ishima R, Tanaka T, Gordon J I, Stryer L, Ikura M. Molecular mechanics of calcium-myristoyl switches. Nature. 1997;389:198–202. doi: 10.1038/38310. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann J E, Fusco P J. The M protein of vesicular stomatitis virus associates specifically with the basolateral membranes of polarized epithelial cells independently of the G protein. J Cell Biol. 1988;107:1707–1715. doi: 10.1083/jcb.107.5.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong L D, Rose J K. Interactions of normal and mutant vesicular stomatitis virus matrix proteins with the plasma membrane and nucleocapsids. J Virol. 1994;68:441–447. doi: 10.1128/jvi.68.1.441-447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong L D, Rose J K. Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J Virol. 1993;67:407–414. doi: 10.1128/jvi.67.1.407-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conte M R, Matthews S. Retroviral matrix proteins: a structural perspective. Virology. 1998;246:191–198. doi: 10.1006/viro.1998.9206. [DOI] [PubMed] [Google Scholar]
- 8.Copeland N G, Jenkins N A, Nexo B, Schultz A M, Rein A, Mikkelsen T, Jorgensen P. Poorly expressed endogenous ecotropic provirus of DBA/2 mice encodes a mutant Pr65gag protein that is not myristylated. J Virol. 1988;62:479–487. doi: 10.1128/jvi.62.2.479-487.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorfman T, Mammano F, Haseltine W A, Gottlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Facke M, Janetzko A, Shoeman R L, Krausslich H G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed E O. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 12.Freed E O, Englund G, Martin M A. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed E O, Martin M A. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J Virol. 1994;68:2503–2512. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed E O, Martin M A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallina A, Mantoan G, Rindi G, Milanesi G. Influence of MA internal sequences, but not of the myristylated N-terminus sequence, on the budding site of HIV-1 Gag protein. Biochem Biophys Res Commun. 1994;204:1031–1038. doi: 10.1006/bbrc.1994.2566. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez S A, Affranchino J L. Substitution of leucine 8 in the simian immunodeficiency virus matrix protein impairs particle formation without affecting N-myristylation of the Gag precursor. Virology. 1998;240:27–35. doi: 10.1006/viro.1997.8919. [DOI] [PubMed] [Google Scholar]
- 19.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granowitz C, Goff S P. Substitution mutations affecting a small region of the Moloney murine leukemia virus MA gag protein block assembly and release of virion particles. Virology. 1994;205:336–344. doi: 10.1006/viro.1994.1650. [DOI] [PubMed] [Google Scholar]
- 21.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen E C, Kjeldgaard N O, Pedersen F S, Jorgensen P. A nucleotide substitution in the gag N terminus of the endogenous ecotropic DBA/2 virus prevents Pr65gag myristylation and virus replication. J Virol. 1988;62:3217–3223. doi: 10.1128/jvi.62.9.3217-3223.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiernan, R. E., and E. O. Freed. Unpublished data.
- 25.Kiernan R E, Ono A, Englund G, Freed E O. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y M, Tian C J, Yu X F. A bipartite membrane-binding signal in the human immunodeficiency virus type 1 matrix protein is required for the proteolytic processing of gag precursors in a cell type-dependent manner. J Virol. 1998;72:9061–9068. doi: 10.1128/jvi.72.11.9061-9068.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y M, Yu X F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 29.Massiah M A, Starich M R, Paschall C, Summers M F, Christensen A M, Sundquist W I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 30.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 31.Morikawa Y, Zhang W H, Hockley D J, Nermut M V, Jones I M. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J Virol. 1998;72:7659–7663. doi: 10.1128/jvi.72.9.7659-7663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono, A., and E. O. Freed. Unpublished data.
- 33.Ono A, Huang M, Freed E O. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J Virol. 1997;71:4409–4418. doi: 10.1128/jvi.71.6.4409-4418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal R, Reitz M S, Jr, Tschachler E, Gallo R C, Sarngadharan M G, Veronese F D. Myristoylation of gag proteins of HIV-1 plays an important role in virus assembly. AIDS Res Hum Retroviruses. 1990;6:721–730. doi: 10.1089/aid.1990.6.721. [DOI] [PubMed] [Google Scholar]
- 35.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peitzsch R M, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 37.Platt E J, Haffar O K. Characterization of human immunodeficiency virus type 1 Pr55gag membrane association in a cell-free system: requirement for a C-terminal domain. Proc Natl Acad Sci USA. 1994;91:4594–4598. doi: 10.1073/pnas.91.10.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reil H, Bukovsky A A, Gelderblom H R, Gottlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rein A, McClure M R, Rice N R, Luftig R B, Schultz A M. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultz A M, Rein A. Unmyristylated Moloney murine leukemia virus Pr65gag is excluded from virus assembly and maturation events. J Virol. 1989;63:2370–2373. doi: 10.1128/jvi.63.5.2370-2373.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spearman P, Wang J J, Vander Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 45.Towler D A, Gordon J I, Adams S P, Glaser L. The biology and enzymology of eukaryotic protein acylation. Annu Rev Biochem. 1988;57:69–99. doi: 10.1146/annurev.bi.57.070188.000441. [DOI] [PubMed] [Google Scholar]
- 46.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–69. [PubMed] [Google Scholar]
- 47.Wada I, Rindress D, Cameron P H, Ou W J, Doherty J J D, Louvard D, Bell A W, Dignard D, Thomas D Y, Bergeron J J. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- 48.Wang C T, Zhang Y, McDermott J, Barklis E. Conditional infectivity of a human immunodeficiency virus matrix domain deletion mutant. J Virol. 1993;67:7067–7076. doi: 10.1128/jvi.67.12.7067-7076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weaver T A, Panganiban A T. N myristoylation of the spleen necrosis virus matrix protein is required for correct association of the Gag polyprotein with intracellular membranes and for particle formation. J Virol. 1990;64:3995–4001. doi: 10.1128/jvi.64.8.3995-4001.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willey R L, Bonifacino J S, Potts B J, Martin M A, Klausner R D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 52.Yu X, Yuan X, Matsuda Z, Lee T H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan X, Yu X, Lee T H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]