Abstract
Infection with Kaposi’s sarcoma (KS)-associated herpesvirus (KSHV) or human herpesvirus 8 (HHV8) is common in certain parts of Africa, the Middle East, and the Mediterranean, but is rare elsewhere, except in AIDS patients. Nevertheless, HHV8 DNA is found consistently in nearly all classical, endemic, transplant and AIDS-associated KS lesions as well as in some rare AIDS-associated lymphomas. The concept that HHV8 genomes fall into several distinct subgroups has been confirmed and refined by PCR DNA sequence analysis of the ORF-K1 gene encoding a highly variable glycoprotein related to the immunoglobulin receptor family that maps at the extreme left-hand end of the HHV-8 genome. Among more than 60 different tumor samples from the United States, central Africa, Saudi Arabia, Taiwan, and New Zealand, amino acid substitutions were found at a total of 62% of the 289 amino acid positions. These variations defined four major subtypes and 13 distinct variants or clades similar to those found for the HIV ENV protein. The B and D subtype ORF-K1 proteins differ from the A and C subtypes by 30 and 24%, respectively, whereas A and C differ from each other by 15%. In all cases tested, multiple samples from the same patient were identical. Examples of the B subtype were found almost exclusively in KS patients from Africa or of African heritage, whereas the rare D subtypes were found only in KS patients of Pacific Island heritage. In contrast, C subtypes were found predominantly in classic KS and in iatrogenic and AIDS KS in the Middle East and Asia, whereas U.S. AIDS KS samples were primarily A1, A4, and C3 variants. We conclude that this unusually high diversity, in which 85% of the nucleotide changes lead to amino acid changes, reflects some unknown powerful biological selection process that has been acting preferentially on this early lytic cycle membrane signalling protein. Two distinct levels of ORF-K1 variability are recognizable. Subtype-specific variability indicative of long-term evolutionary divergence is both spread throughout the protein as well as concentrated within two 40-amino-acid extracellular domain variable regions (VR1 and VR2), whereas intratypic variability localizes predominantly within a single 25-amino-acid hypervariable Cys bridge loop and apparently represents much more recent changes that have occurred even within specific clades. In contrast, numerous extracellular domain glycosylation sites and Cys bridge residues as well as the ITAM motif in the cytoplasmic domain are fully conserved. Overall, we suggest that rather than being a newly acquired human pathogen, HHV8 is an ancient human virus that is preferentially transmitted in a familial fashion and is difficult to transmit horizontally in the absence of immunosuppression. The division into the four major HHV8 subgroups is probably the result of isolation and founder effects associated with the history of migration of modern human populations out of Africa over the past 35,000 to 60,000 years.
The recently discovered Kaposi’s sarcoma (KS)-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV8), is thought to be essential for the development of both classical and AIDS-associated forms of KS (5, 15) as well as being involved in AIDS-associated primary effusion lymphoma (PEL or body cavity-based lymphoma [BCBL]) (12) and multicentric Castleman’s Disease (62). HHV8 DNA is present in virtually all KS tumor samples and in the peripheral blood mononuclear cells in up to 50% of homosexual AIDS patients with KS (16, 19, 22, 46, 67). Serological evidence obtained by LANA immunofluorescent antibody assay indicates that infection is also widespread in those parts of southern Italy (5 to 20% seropositivity) and central and southern Africa (40 to 60% seropositivity) where endemic and classical KS have the highest incidence rates, reaching up to 1.0 and 10 per 100,000 person years, respectively, with a high preference for males (4, 25, 26, 34, 68). Similarly, KS patients and male homosexual AIDS patients, but not human immunodeficiency virus (HIV)-positive intravenous drug users and hemophiliacs, have extremely high seropositivity rates of 85 and 50%, respectively (25, 26). However, the seroprevalence in blood donors in the United Kingdom and United States may be no greater than 1% (68), which correlates with estimated classic KS incidence rates of as low as 0.014 and 0.165 per 100,000 person years in the United Kingdom and United States, respectively (4, 6).
HHV8 is a gamma-2 class herpesvirus that is distantly related to herpesvirus saimiri (HVS) and Epstein-Barr virus (EBV). Several reports have described three novel 10- to 13-kb segments of the HHV8 genome that encode divergent viral homologues of exogenously acquired cellular genes encoding interleukin-6 (IL6), dihydrofolate reductase (DHFR), MIP-IB, TS, MIP-IA, and BCL-2; several IRF-like genes; and FLIP, CYC-D, OX-2, and GCR, most of which have not been found previously in other HHVs (14, 45, 47–50, 59). The nearly complete primary nucleotide sequences of the 190-kb double-stranded DNA molecules of two HHV8 genomes, one derived from an AIDS BCBL cell line (59), and the other from a KS lesion (48), have been determined and were found to differ by only 0.4% from each other.
Because of the many questions that arise about the origin, distribution, transmissibility, and disease associations of HHV8, we have been interested in examining the levels of genetic variation and polymorphism in this virus. In initial studies, we carried out PCR sequencing on three small segments of the open reading frame 26 (ORF26) and ORF75 genes from 12 KS specimens from several different patients and from multiple distinct KS lesions from a single patient (70). All seven lesions tested from an AIDS patient with aggressive disseminated KS proved to be identical to one another, but the equivalent sequences from different patients varied significantly and fell into three distinct DNA patterns, referred to as A, B, and C subtypes. Overall nucleotide variation among the three groups reached between 1.0 and 1.5%, but less than 0.1% occurred within each group. In comparison, the average nucleotide difference levels between unrelated human beings is 0.3 to 0.4%. Compilation of existing data from the literature at that time over a very small 233-bp region of ORF26 (9, 17, 31, 46) implied that among U.S. AIDS-associated HHV8 samples, a large majority were very closely related or identical A subtype genomes, whereas a minor subpopulation were C subtype variants, and several samples from Africa appeared to represent a distinctive B subtype.
In the present study, we sought to take this molecular analysis further by examining a much larger group of samples within a highly variable region of the HHV8 genome. We and others have previously noted unexpectedly high levels of diversity in the gene encoding the ORF-K1 protein at the extreme left-hand side (LHS) of the genome (35, 38, 51). The ORF-K1 protein is a highly glycosylated cytoplasmic and membrane protein similar to the immunoglobulin receptor family that is expressed as an inducible early-lytic-cycle gene product in PEL cell lines (35, 38, 52). It is not related to any other known herpesvirus protein, but has the interesting properties of producing foci in DNA transfected primary cells and substituting for the saimiri transformation protein (STP) of HVS in T-cell transformation and tumorigenesis assays, and it also contains a functional immunoglobulin-receptor tyrosine-based activation motif (ITAM) (36, 38). Here we have analyzed the complete ORF-K1 coding regions of more than 60 different HHV8 DNA samples from a wide variety of classic and AIDS-associated KS lesions and PEL tumors or cell lines. The results revealed considerable amino acid polymorphism, particularly in the extracellular domain, giving ORF-K1 protein patterns that fall into four major subtypes, together with further subdivisions into 13 distinctive variants or clades. Moreover, individual clusters of related proteins closely correlate with the different geographical and ethnic backgrounds of the patients.
MATERIALS AND METHODS
KS tumor DNA samples.
Five KS DNA samples from East Coast U.S. AIDS patients were described previously (49). C282 and ASM70-80 were autopsy samples from patients with advanced disseminated KS from New York’s Sloan-Kettering/Memorial Hospital in 1984, whereas AKS1, AKS2 and AKS4 were biopsy samples obtained from AIDS patients in Baltimore, Md., and New York in 1995. The samples referred to as BKS8, BKS10, BKS11, BKS13, BKS14, BKS15, and BKS16 were all KS biopsies collected from unrelated AIDS patients in Galveston, Tex. EKS1, EKS2, and BKS12 were biopsy samples from classical KS patients in Baltimore and Galveston, respectively. BKS3 (CVU-14) and BKS4 (CVU-1) were direct biopsy and cultured cell supernatant samples obtained on different occasions from different lesions from a classical KS patient in Nashville, Tenn. 431KAP and 431NSC represent KS and adjacent normal skin biopsy DNA samples obtained from a male non-AIDS-associated KS patient from Zaire in 1984, whereas ST1 and ST2 represent KS biopsies from HIV-positive females in Uganda (49, 70). OKS3 and OKS4 were derived from paraffin-block sections of KS biopsies from male AIDS patients in Tanzania (kindly provided by Andrew Blauvelt, Dermatology Branch, National Cancer Institute, Bethesda, Md.). OKS7, OKS8, and OKS9 were from paraffin block sections of KS biopsies from AIDS patients of male black Haitian, female Hispanic Mexican, and female Hispanic Nicaraguan origin, respectively, seen at the University of Miami Medical Center, Miami, Fla., and provided by the AIDS Malignancy Bank. RKS1, RKS2, RKS3, RKS4, and RKS5 were all frozen KS biopsy samples from AIDS patients from Lusaka, Zambia. The TKS samples were all collected at the National Cheng Kung University Hospital, Taiwan, Republic of China (64). TKS1, TKS3, and TKS9 came from HIV-positive AIDS-associated KS patients, whereas TKS2, TKS5, TKS6, TKS7, and TKS10 were all from classic HIV-negative non-AIDS associated KS patients, and TKS11 represented an iatrogenic renal transplant KS patient. TKS10 differed from all of the other Taiwan samples by being from a Hwalien patient of indigenous ethnic background and probable South Pacific ancestry. SKS1, SKS2, SKS3, and SKS6 to SKS9 were iatrogenic KS samples from renal transplant patients from the King Faisal Hospital, Riyadh, Saudi Arabia (23). WKS1 came from an AIDS biopsy specimen from the University of North Carolina. Samples of KS specimens collected at the University of Auckland Hospital, Auckland, New Zealand, included ZKS1 and ZKS5 from HIV-positive male Caucasians, ZKS7 from a renal transplant recipient, ZKS3 and ZKS4 from two elderly Polynesian males with classic KS, and ZKS6 from an HIV-positive immigrant of Bushman ancestry from southern Africa. JKS15 and JKS20 represent archival KS paraffin block samples from an African-American AIDS patient and an HIV-negative African immigrant with endemic KS that were selected as the only two B subtype genomes detected within a collection of 15 specimens from the Johns Hopkins Department of Dermatology and AIDS Malignancy Clinic (11). Tissue DNA from paraffin block sections (OKS and JKS series) or OCT frozen sections (RKS series) were extracted by treatment with xylene, proteinase K, and Tween 20 or with proteinase K, sodium dodecyl sulfate, and phenol-chloroform-isoamyl alcohol respectively, followed by ethanol precipitation and resuspension in a small volume of distilled water.
PEL cell lines and BCBL tumor DNA samples.
A total of six different PEL cell lines were grown in RPMI medium plus 20% fetal calf serum. Total-cell DNA was extracted from washed pelleted cells by detergent lysis, proteinase K and RNase treatment, phenol extraction, and dialysis. HBL6 cells (24) were obtained as a gift from Patrick Moore (Columbia University, New York, N.Y.). BCBL1 cells were received from the AIDS Reagent Repository (30, 56), and the initial BC2 (13), BC3 (2), and BCP1 (8) cell cultures were obtained from the American Type Culture Collection. The JSC1 cell line was established from pleural effusion cells of an AIDS-associated BCBL patient at the Johns Hopkins Hospital Lymphoma Clinic and was a gift from Jennifer S. Cannon and Richard Ambinder (10). In addition, DNA was extracted directly from three other PEL tumor cell samples, namely, BCBL-R, obtained from tumor cells of an AIDS patient in Washington, D.C.; BKS1 (CVU-30), obtained from early-passage adherent cells from a pleural effusion sample from an AIDS KS patient in Nashville, Tenn.; and BKS6 (CVU-27) and BKS5 (CVU-19), representing filtered supernatant and uncultured pleural effusion cells, respectively, obtained on different occasions from an unusual AIDS-associated T-cell BCBL in Nashville. The latter two samples are collectively referred to here as BCBL-B.
HHV8 genomic DNA libraries and ORF-K1 phage and plasmid subclones.
Phage lambda clones derived from two BCBL-R tumor DNA genomic libraries in the λEMBL3 and λDASHII backgrounds were described previously (28, 51, 70). One clone (λD-S1), containing a 16-kb insert from the LHS genomic terminus equivalent to HHV8 (BC1) positions −1.8 to 14.5 kb, was isolated by hybridization with a PCR DNA probe representing part of the ORF6 (SSB) gene from the extreme left-hand terminus of λD3-80 (49). A 3.5-kb BamHI-BamHI plasmid genomic subclone (pDJA61) encompassing 1,600 bp of the proximal LHS terminal tandem repeat (TTR) sequences, plus all of ORF-K1 and part of ORF4 from λD-S1, was isolated and sequenced by primer walking procedures. Primers based on this sequence were then used to obtain the complete DNA sequence from genomic positions 20 to 1085 covering the whole of the ORF-K1 coding region for 71 KS and PEL or BCBL samples.
PCR amplification and sequencing primers.
The 870-bp ORF-K1 coding region from nucleotide positions 105 to 974 was usually amplified directly from HHV8-positive DNA samples as a 1,066-bp PCR product. BRL Taq polymerase (GIBCO BRL catalog no. 18038-042) was incubated with 20 to 100 ng of DNA template in a Techne PHC-3 thermocycler set at 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min over 35 cycles with the following outside primer pair (genomic nucleotide positions indicated in parentheses): LGH2089 (5′-GTTCTGCCAGGCATAGTC-3′ [21 to 38]) and LGH2088 (5′-AATAAGTATCCGACCTCAT-3′ [1085 to 1067]). Other commonly used additional internal ORF-K1 primers for either nested or direct PCR amplification and PCR sequencing of most or all subtypes were as follows: LGH2090 (5′-GAGTGATTTCAACGCCTTAC-3′ [193 to 212]), LGH2091 (5′-GAGTATTGTTGCAATACC-3′ [270 to 253]), LGH2505 (5′-CAACCTGTCTTACAAACC-3′ [401 to 418]), LGH2500 (5′-GTAACATGCTGACCACAAG-3′ [445 to 427]), LGH2507 (5′-CGTCTCGCCTGTCAAATC-3′ [589 to 606]), LGH2508 (5′-AGATACCACACATGGTT-3′ [840 to 864]), and LGH2092 (5′-GACACTCGTAGCTCTGAT-3′ [802 to 820]). Dideoxynucleotide double-stranded cycle sequencing (GIBCO BRL catalog no. 18196) with single 32P-labelled primers was then carried out with isolated agarose gel-purified DNA bands. The reaction products were fractionated on 7 M urea–6% polyacrylamide gels, and the autoradiographs were read manually. All sequencing was carried out with both complementary strands, and most analyses involved confirmation with redundant overlapping fragments from multiple independent PCR-amplified products. Several representative samples were also subjected to PCR sequencing over the 3′ end of the ORF4 gene by using primers LGH2094 (5′-GGTACTGACATTCAGG-3′ [966 to 982]) and LGH2095 (5′-TCACAAAAAGGAGGAGAAG-3′ [1540 to 1522]).
Phylogenetic analysis.
Deduced amino acid sequences were aligned manually with the program VisEd (version 1.2). The full alignment contained 63 ORF-K1 sequences. For each variant, there were 289 characters (amino acids plus gaps). Regions of ambiguous alignment (gap positions) were excluded from the phylogenetic analysis, and, consequently, 278 of the 289 amino acids were compared. All phylogenetic reconstructions were carried out by using the PHYLIP package, version 3.5C. Once the sequences were aligned, the amino acid divergence was calculated by using PRODIST, with the Dayhoff PAM matrix option. The branching pattern was estimated from the distance matrix by neighbor joining with the program NEIGHBOR. The SKS1 variant was used as an outgroup, but a similar topography was obtained by using other variants as the outgroup. Confidence levels for the branching pattern were estimated by a bootstrap resampling of the data. Bootstrap values were obtained from a consensus tree based on 1,000 randomly generated data sets by using SEQBOOT, PRODIST, CONDENSE, and NEIGHBOR.
Nucleotide sequence accession number.
The DNA sequence data for the prototype A1 (BCBL-R), A4 (BCBL-B), B (431KAP), C1 (ASM72), C3 (BC2), and D1 (TKS10), and D2 (ZKS3) subtype ORF-K1 genes are available from GenBank (accession no. AF133038 to AF133044). Other published ORF-K1 sequences in this analysis include those for HHV8 samples BC1 (U75678), KS-F (U93872), and BCBL-1 (U86667).
RESULTS
Unusually high levels of variability between the A, B, and C subtypes of the ORF-K1 protein.
Analysis of the DNA sequences encompassing the intact ORF-K1 gene coding region at the extreme LHS of the unique segment of HHV8 from each of the three prototype strains of the proposed A, B, and C subtypes revealed extraordinarily high levels of nucleotide variation (Fig. 1A and B). For example, a 1,066-bp fragment of cloned viral genomic DNA from BCBL-R (A) showed 8 and 16% nucleotide variations (not counting deletions) relative to the equivalent region of viral DNA from ASM72 (C) and 431KAP (B), respectively. This compares to only 0.4 and 1.4% nucleotide variations within an immediately adjacent 474-bp PCR product from the ORF4 gene region (not shown). Based on these data, the published PEL cell line ORF-K1 DNA sequences from the BC1 and BCBL-1 cell lines (35, 59) as well as the KS genome of Lee et al. (38) all represent A subtypes, whereas the KS genome of Neipel et al. (48) represents a C subtype. However, even within the A subgroup, the ORF-K1 genes from BCBL-R and BC1 display 16 nucleotide differences (1.5% variation), compared to only 0.2% variation in the ORF4 gene block. Lagunoff and Ganem (35) have also reported that the ORF-K1 coding region of BCBL1 shows 2% nucleotide differences from the equivalent region of BC1.
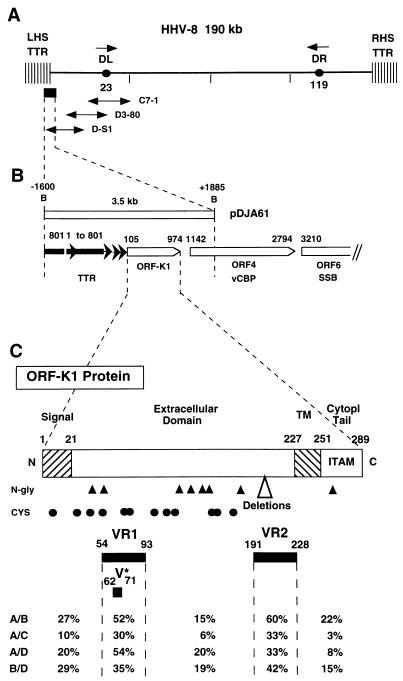
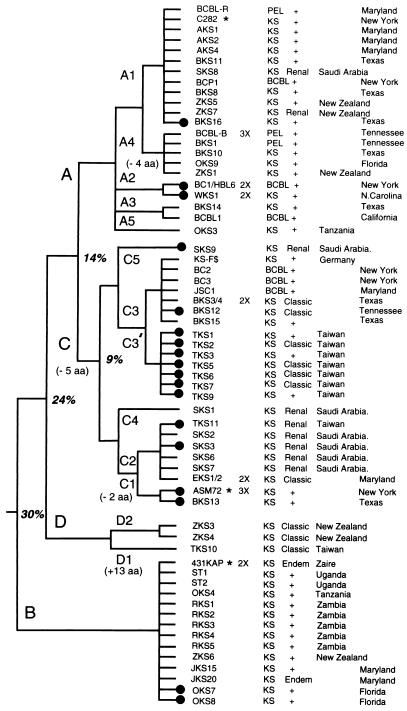
FIG. 1.
Organization of the left-hand end of the HHV8 genome encompassing the ORF-K1 gene. (A) Map of the genomic location of HHV8 (BCBL-R) lambda clone λD-S1 relative to other LHS clones and major features of the HHV8 DNA molecule. DL and DR, duplicated 1-kb ORI-like regions (left and right, respectively) at genomic coordinates 23 kb and 119 kb. (B) Organization and orientation of ORFs and terminal repeat unit sequences within an expanded area across the LHS TTR-unique region boundary encompassed within the 3.5-kb BamHI-BamHI fragment in plasmid subclone pDJA61 (genomic coordinates −1600 to +1885). Nucleotide positions of the initiator and terminator codons in the unique region are given above the ORFs (open arrows), and those of the TTR sequences are given above the different repeat unit fragments (solid arrows). (C) Predicted domain structure and key features of the highly variable 289-amino-acid ORF-K1 membrane receptor-like protein encoded between genomic nucleotide coordinates 105 and 974. Hatched bars denote signal peptide and transmembrane (TM) domains with amino acid boundaries indicated. Predicted N-glycosylation sites (NXS/NXT) (solid triangles) and the 12 conserved Cys residues (solid circles) are indicated. Cytopl, cytoplasmic. (Lower panel) Locations of highly variable VR1 and VR2 domains and the proposed hypervariable A subtype VR* domain and summary listing of major subtype amino acid difference values both within and outside of the VR blocks.
The HHV8 ORF-K1 gene is predicted to encode a 289-amino-acid polypeptide that has features of a membrane-bound and largely extracellular immunoglobulin receptor-like protein (Fig. 1C). Surprisingly, unlike the situation in the conserved ORF26 and ORF75 loci, a large majority (almost 85%) of the nucleotide variations observed in ORF-K1 also proved to lead to amino acid substitutions. For example, only 17 of the total of 135 nucleotides that differ between BCBL-R (A) and 431KAP (B) within the 870-bp ORF-K1 coding region represent silent (synonymous) mutations. Overall, the prototype A and C versions differ by a total of 39 amino acids (15%), and the A and B versions vary by 85 amino acids (29%). The differences between the A and C versions are concentrated primarily within two 40-amino-acid blocks, referred to as VR1 and VR2, extending from codons 54 to 92 and 199 to 227, with the latter including two small in-frame deletions. These two highly variable blocks extend even further in the B subtype, encompassing codons 1 to 92 surrounding VR1 (including the N-terminal signal peptide at positions 1 to 22), and codons 191 to 228 surrounding VR2, although there are no deletions relative to the A pattern. The putative transmembrane domain lying between positions 229 and 261 (Fig. 1C) is largely invariant among all three subtypes, but in contrast, although the A and C subtypes show few or no differences in the central region (positions 105 to 148) or in the C-terminal cytoplasmic tail (positions 262 to 289), the B subtype displays nearly 30% (12 of 38) amino acid differences within the cytoplasmic tail region.
Clade analysis of multiple HHV8 genomes based on ORF-K1 protein patterns.
To investigate whether the ORF-K1 protein patterns found within a much larger set of KS, PEL, and BCBL samples also all fell within the same three groups, or instead either represented a continuum or formed additional groups, we PCR amplified and sequenced the entire 1,066-bp ORF-K1 gene coding region encompassing genomic nucleotide positions 20 to 1085 from a total of 71 distinct HHV8-positive samples representing 63 different patients. The samples included 11 of the 12 analyzed previously in the ORF26 and ORF75 constant region (70), plus additional samples representing several BCBL cell lines (BC2, BC3, BCP1, BCBL1, and JSC1), an unusual T-cell like PEL tumor (BCBL-B), and KS samples derived primarily from the East Coast and southern United States, central and southern Africa, Saudi Arabia, Taiwan, and New Zealand.
A comparison of the complete or nearly complete 289-amino-acid ORF-K1 protein sequences from 63 different patients is presented in Fig. 2. With the exception of TKS10, ZKS3, and ZKS4, which formed a new D subtype, all samples were readily differentiated into three very distinctive groups corresponding to our previously assigned A, B, and C subtypes. Moreover, because of the high variability here, we were also able to further subdivide the ORF-K1 protein patterns into a total of 13 distinct variants based either on amino acid differences totalling 5% or greater or on the presence of common distinctive in-frame deletions within the VR2 domain. These variant subgroups have been assigned additional numerical descriptors (e.g., A1 to A5, C1 to C5, and D1 or D2). For the purposes of this analysis, we have reserved the term “clade” for several even narrower clusters of closely related genomes with obvious relatively recent common origins (such as those described below as A1′ or the C3′ cluster from Taiwan). A tabular diagram summarizing the geographic origins, subgroup assignments, and key clinical features of each of the 63 patients is presented in Fig. 3. The classification and clustering shown in Fig. 2 and 3 were generated by visual inspection, but subsequent computer-generated tree and radial dendrograms illustrating the overall phylogenetic distance and similar evolutionary branch relationships among the different subtypes and variants fully support these interpretations (Fig. 4).
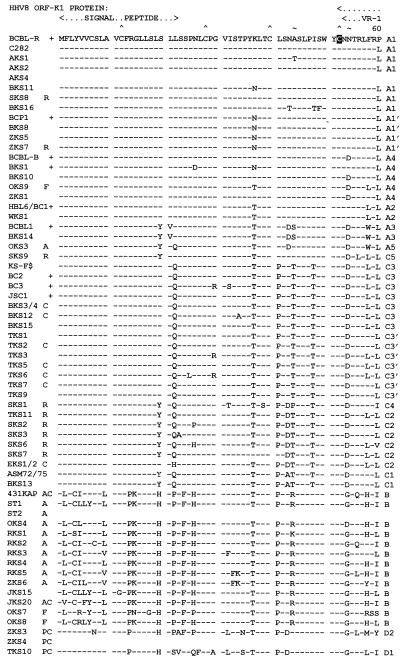
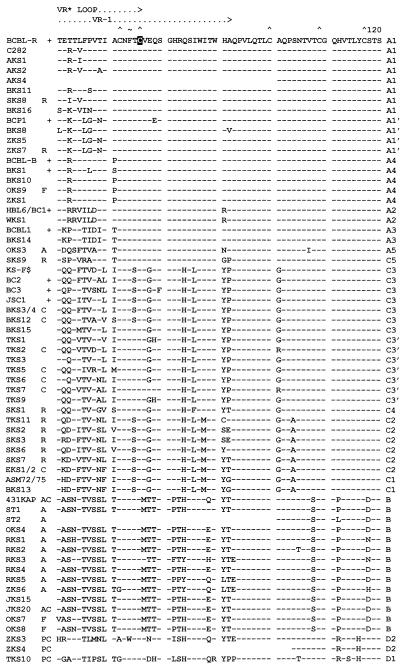
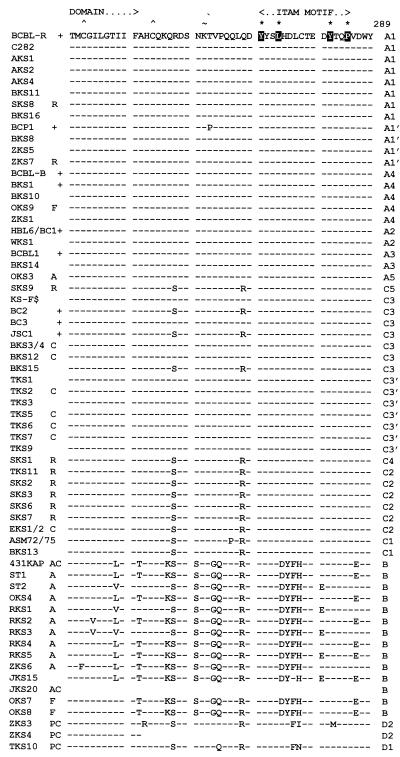
FIG. 2.
Amino acid alignment of the ORF-K1 proteins of 63 distinct HHV8 genomes. The complete amino acid sequence is given only for HHV8 (BCBL-R) on the top line, with amino acid identities indicated by dashes for the other genomes. Deletions in VR2 are indicated by gaps in parentheses, and subtype designations are given to the far right. The A1, A2, A3, B, and D2 subtype ORF-K1 proteins consist of 289 amino acids, whereas the A4 subtype has 285 amino acids, the C3 and C2 subtypes have 284 amino acids, the C1 subtype has 282 amino acids, and the single example of subtype D1 has 302 amino acids. The conserved Cys residues (∧) surrounding the predicted VR* loop and key amino acids in the predicted ITAMs (★) are highlighted. ∼, potential N-glycosylation site; +, BCBL or PEL tumor or cell line; C, classic or endemic non-HIV-associated KS; R, renal transplant-associated iatrogenic KS; A, patient lived in or was a direct immigrant from Africa; P, Pacific Islander; F, aggressive KS from Florida. Blanks indicate incomplete data for AKS4, ST1, and JKS20, for which insufficient original DNA was available. All samples designated as members of the A1, A4, A2, A3, C3, C2, C1, B, and D variants are grouped together. AKS, AIDS KS from Maryland; BKS, samples from Texas and Tennessee; OKS, AIDS KS samples from Tanzania or Florida; RKS, AIDS KS samples from Zambia; SKS, renal transplant KS from Saudi Arabia; TKS, samples from Taiwan; ZKS, samples from New Zealand. Note that the data for HBL6 (BCBL cell line) and WKS1 (KS) are identical to the published sequence for the BC1 cell line (59). Similarly, our results for the BCBL1 cell line and BKS14 KS are identical to each other and to published data for BCBL1 (35). Finally, the data for the BCBL-B cell line and for the BKS10 KS samples from different patients are also identical. KS-F shows data reported by Neipel et al. (48) for an AIDS KS sample from Germany.
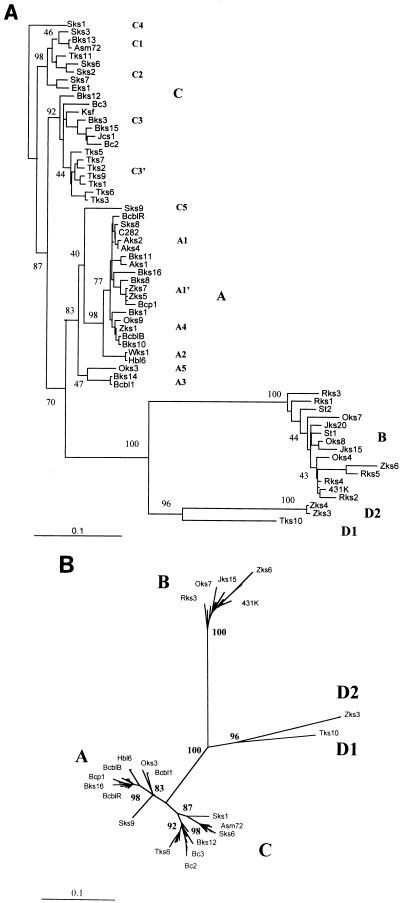
FIG. 3.
Summary of ORF-K1 subtype and clade patterns compared with disease type and geographic origins. The diagram includes a listing of the clinical characteristics and geographic source of each of the 63 individual HHV8 samples included in Fig. 2 together with the evolutionary relationships between the various ORF-K1 clades. ∗, prototype A, B, and C samples collected in 1984; +, AIDS-associated, HIV-positive patients; X, patients from which multiple independent samples were sequenced (all proved to be identical). KS, lesion biopsy, autopsy, or archival paraffin block samples; PEL, PEL tumor samples; BCBL, established lymphoblastoid cell lines; Classic and Endem, non-HIV-associated KS patients; Renal, iatrogenic renal transplant KS. Solid circles denote those genomes that have M rather than P alleles of the ORF-K15 gene (53). $, data from Neipel et al. (48).
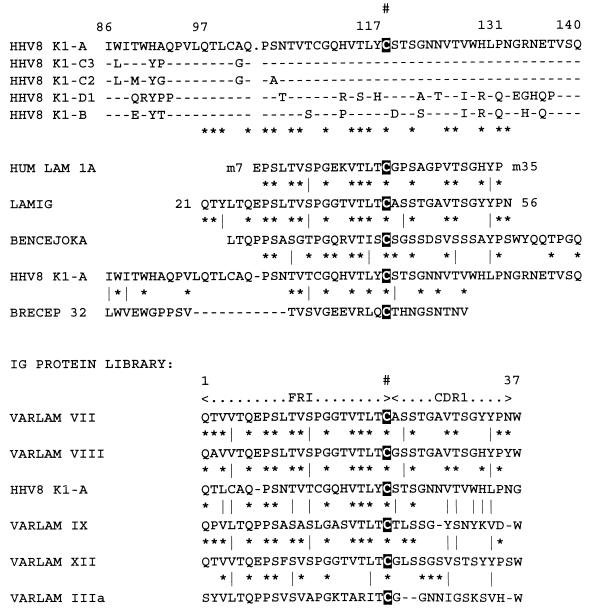
FIG. 4.
Predicted phylogenetic relationships among the 63 HHV8 ORF-K1 protein subtypes and variants examined here. The diagrams were generated by PHYLIP, PRODIST, and NEIGHBOR program analysis by Raphael Viscidi of the Department of Pediatrics, Johns Hopkins School of Medicine, based on estimated PAM distances and variances for each pair of the intact protein sequences. In-frame deletions and insertions in the VR2 block are not taken into account. (A and B) Linear (A) and radial (B) unrooted phylogenetic dendrograms with the SKS1 sequence as an outgroup. The length of each branch indicates genetic distance with the size scale for 0.1 (10% difference) indicated. Confidence levels (percent) from bootstrap analysis at major branch points are given. Not all samples have been labelled in the radial diagram because of space constraints.
Lack of variability among multiple samples from the same patient.
Considering the high degree of variability encountered among the ORF-K1 genes, it was important to establish whether any diversity occurs within a single infected host. We were able to address this question for six patients, but in all cases obtained identical sequence data for multiple samples derived at different times or from different lesions from the same infected individual. First, the ORF-K1 gene from the HBL6 cell line proved to have a sequence identical to that presented for BC1 by Russo et al. (59), which represents a separate cell line established at a different time from the same PEL patient (13, 24). Second, three separate KS lesions obtained at autopsy for our prototype C strain—one from skin (ASM72) and the other two from lung or lymph node lesions (ASM70 and ASM75)—were identical. Third, for our prototype B strain from a Ugandan endemic KS patient (1a), both a KS skin lesion biopsy sample (431KAP) and an adjacent control normal skin biopsy sample (431NSC) proved to have identical ORF-K1 sequences. Fourth, both a direct KS lesion biopsy (BKS3) and a separate cell culture-grown sample (BKS4) from another lesion, which was still HHV8 positive at second passage, proved to be identical. Fifth, two distinct KS biopsy specimens from different skin lesions biopsied at different times from the same patient with classic KS at Johns Hopkins Hospital (EKS1 and EKS2) were identical. Sixth, two pleural effusion samples (BKS5 and BKS6) obtained at different times (including one undergoing cell culture passaging) proved to be identical to an original T-cell-like PEL tumor sample from the same patient, referred to as BCBL-B. In addition, many individual samples were independently amplified and sequenced on both DNA strands across the same segment of the ORF-K1 gene on multiple occasions with different overlapping primer pairs without detecting any confirmed discrepancies. Therefore, although admittedly the procedures used would detect only the most abundant molecular type if mixtures existed, there is no evidence as yet that any patients were infected with multiple HHV8 isolates or that any of the variability is generated within a single infected individual.
Distinctive variants and clades within subtype A ORF-K1 proteins.
The 22 ORF-K1 protein sequences with A subgroup patterns fell into five distinct subgroups, referred to as A1, A2, A3, A4, and A5 variants, with the major branch containing the A1, A4, and A2 variants separating from the A3 and A5 branch with a bootstrap confidence level of 83% (Fig. 4A). Twelve genomes, including BCP1 and BCBL-R from PEL tumors, plus 4 KS genomes from New York and Maryland (C282, AKS1, AKS2, and AKS4), 3 KS samples from Texas and Tennessee (BKS8, BKS11, and BKS16), 2 from New Zealand (ZKS5 and ZKS7), and 1 from Saudi Arabia (SKS8) are all classified as having the A1 pattern. Although very similar, none were identical to one another, and almost all nucleotide changes gave amino acid substitutions. Even C282, AKS1, and AKS2, which showed no nucleotide differences at all within the 2,000 bp of constant region sequence analyzed previously (70), displayed 3, 8, and 5 nucleotide changes, respectively, in ORF-K1. The maximum variation found within the A1 subset was 10 of 289 amino acids (3%), and 7 of the 23 overall variant amino acid positions were concentrated over a 10-amino-acid stretch between codons 60 and 69 (referred to as part of the VR* domain [described later]). A distinctive cluster of four very closely related A1 genomes (BCP1, BKS8, ZKS5, and ZKS7) having a common LGVN feature at amino acid positions 66 to 69 is referred to as the A1′ clade.
Five more samples formed an A4 variant cluster (Fig. 3). Although overall the A4 patterns are very close to those of the A1 group (i.e., only six to eight amino acid differences from BCBL-R), all five contain a common 12-bp deletion encompassing codons 207 to 210 in the VR2 block. Most of them are also distinguishable from the A1 pattern at positions D-54, P-71, and Q-183. Two of the A4 DNAs, one from Tennessee (BCBL-B) and one from Texas (BKS10), were identical at the amino acid level, and one from New Zealand (ZKS1) differed by only one amino acid, whereas the others differed by three to five amino acids from each other and from the first three. Although, the dendrogram analysis (Fig. 4A) separated the A4 variant subset from the A1 subset with a bootstrap value of only 44, this does not take into account the characteristic four-amino-acid deletion.
The A2 pattern separates from the A1-plus-A4 branch on the dendrogram with a bootstrap value of 98. Identical A2 variants were found in both the HBL6 and BC1 cell lines, as well as in the WKS1 KS lesion. The A2 ORF-K1 pattern differs from the prototype A1 (BCBL-R) pattern by 14 amino acids (5%), with just a single additional synonymous nucleotide difference, and from all other A variants by between 7 and 8% of their amino acids (Fig. 3). Overall, 17 of the 19 A1, A1′, A2, and A4 samples in Table 1 came from AIDS patients, and the other 2 were from renal transplant patients (SKS8 and ZKS7). However, recent VR1 and VR2 data from three new samples of classic KS from Israel (not shown) reveals that they also cluster tightly into the A1′ clade, confirming the validity of this designation (9a).
TABLE 1.
Levels of variability among the ORF-K1 protein major subtypes and variantsa
| Subtype or variant | Total amino acid differences
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A4 | A2 | A3 | A5 | C5 | C3 | C4 | C2 | C1 | B | D1 | D2 | |
| A1 | 5+ | 15 | 19 | 19 | 26 | 40 | 33 | 35 | 39 | 85 | 75 | 73 | |
| A4 | 1.7+ | 20 | 17 | 23 | 26 | 40 | 32 | 33 | 35 | 84 | 71 | 75 | |
| A2 | 5.2 | 6.9 | 22 | 23 | 25 | 42 | 38 | 36 | 39 | 85 | 72 | 78 | |
| A3 | 6.6 | 5.9 | 7.6 | 17 | 23 | 39 | 30 | 30 | 33 | 83 | 69 | 73 | |
| A5 | 6.6 | 8.0 | 8.0 | 5.9 | 29 | 35 | 30 | 28 | 32 | 81 | 70 | 74 | |
| C5 | 9.0 | 9.0 | 8.6 | 8.0 | 10.0 | 36 | 32 | 32 | 36 | 86 | 76 | 72 | |
| C3 | 13.8 | 13.8 | 14.5 | 13.5 | 12.1 | 12.5 | 28 | 24 | 28 | 89 | 80 | 78 | |
| C4 | 11.4 | 11.1 | 13.1 | 10.4 | 10.4 | 11.1 | 9.7 | 15 | 17 | 82 | 71 | 70 | |
| C2 | 12.1 | 11.4 | 12.5 | 10.4 | 9.7 | 11.1 | 8.3 | 5.2 | 9+ | 84 | 72 | 73 | |
| C1 | 13.5 | 12.1 | 13.5 | 11.4 | 11.1 | 12.5 | 9.7 | 5.9 | 3.1+ | 84 | 77 | 75 | |
| B | 29.4 | 29.1 | 29.4 | 28.7 | 28.0 | 29.8 | 30.8 | 28.4 | 29.1 | 29.1 | 77 | 83 | |
| D1 | 26.0 | 24.6 | 24.9 | 23.9 | 24.2 | 26.3 | 27.7 | 24.6 | 24.9 | 26.6 | 26.6 | 60+ | |
| D2 | 25.3 | 26.0 | 27.0 | 25.3 | 25.6 | 24.9 | 27.0 | 24.2 | 25.3 | 26.0 | 28.7 | 20.8+ | |
| Total amino acids | 289 | 285 | 289 | 289 | 289 | 284 | 284 | 284 | 284 | 282 | 289 | 302 | 289 |
Summary showing numbers (top half) and percentages (bottom half) of pairwise amino acid differences between the prototype examples of each of the 13 ORF-K1 subtypes and variants recognized here. Deletions and insertions are not taken into account, but their contributions to variant classification designations are acknowledged by superscript plus signs where significant.
The second major branch of the A subtype patterns contains at least two variants, A3 and A5, that are distinguishable from the A1, A4, and A2 branch with a bootstrap value of 83. Again, the only two A3 variants found, one from the BCBL1 cell line, and the other from the BKS14 lesion, were identical to one another, but they differed from the A1, A4, and A2 amino acid patterns by 6 to 8%. Finally, OKS3, the only A subtype sample from Africa, showed a distinctive A5 pattern that also differed from each of the other A variants by between 6 and 8%. Although closest to A3 in the dendrogram (bootstrap value of 47), the A5 pattern also has several features more typical of B or C subtypes rather than A patterns (e.g., Q-22, Q-63, V-67, N-222, P-223, V-227, and P-228). Importantly, among an additional 11 subtype A ORF-K1 genes examined that are not included in Fig. 3 (because only VR1 and VR2 block data are available at present), there are several other distinctive sequence variants that fit best into the A3 category, especially among U.S. and European patients with classic KS. In addition, we and others have also now detected a relatively large subset of African samples that closely resemble the A5 ORF-K1 pattern in the VR1 and VR2 regions (1, 33a, 51a, 68a).
Subdivision of the C subtype ORF-K1 proteins into two major classes.
All subtype C ORF-K1 patterns differ from subtype A patterns by an average of 14 to 15% of their amino acids (bootstrap value of 70), and the majority (22 of 24) fall into one of two major branches consisting of either C1 and C2 variants or C3 and C4 variants (bootstrap value of 87). All C subtype ORF-K1 genes have the primary distinguishing characteristic of a 15-bp in-frame deletion of amino acids 201 to 205 in VR2, although the two C1 samples also contain a second 6-bp in-frame deletion of amino acids 213 and 214. Additional unique features that are common to nearly all C subtype patterns, but are not found in A or B patterns, are Q-22, T-44, T-48, G-78, L-86, G-101, A-165, and L-168.
The only two subtype C genomes that occurred among our original 12 samples (70) are referred to as the prototype C1 (ASM72) and C2 (EKS1) patterns. One additional U.S. sample (BKS13), one Taiwan sample (TKS11), and four Saudi Arabian samples (SKS2, SKS3, SKS6, and SKS7) turned out to be C1 or C2 variants, whereas six other U.S. samples, including those in three PEL cell lines (BC2, BC3, and JSC1), plus seven of the nine Taiwan KS samples and KS-F from Germany all fell into a large new C3 subgroup. The eight C1 and C2 patterns have six conserved amino acid variations at D-63, N-69, S (codon TCT)-75, M-88, A-104, and F-220 that distinguish them from all other ORF-K1 proteins, plus three at P-228, S-258, and R-269 that they share in common with B and D1 subtypes and three more at Y-20, D-43, and P-223 that they share in common only with A3 and A5 or A5 alone. Our rationale for distinguishing ASM72 and BKS13 as C1 patterns relative to EKS1, TKS11, SKS2, SKS3, SKS6, and SKS7 as C2 patterns is based primarily on the presence or absence of the 6-bp deletion. The two C1 samples also have three unique positions in common, A-43, H-62, and G-92, and differ from each other by only one amino acid. The dendrogram analysis also places SKS3 closer to C1 than to C2 when not taking into account the characteristic 6-bp deletion (bootstrap value of 46). Overall the eight C1 plus C2 variants display a total of 22 amino acid differences among them (8%) and are distinguished from the C3 group by a bootstrap value of 98.
The 14 examples of C3 variant ORF-K1 proteins are all different from one another, but show many common characteristics that make them a very distinctive branch (bootstrap value of 92), including the presence of Q-63, P-92, S-172, M-215, L-223, and Q-226 and the absence of the C1- and C2-specific changes listed above. Overall, there were a total of 38 variant amino acid positions within the group, with the 20 differences between BC2 and BC3 (7%) representing the extremes, but except for the clear separation of seven Taiwan samples into the C3′ clade (bootstrap value of 44), no other convincing clustering is discernible. Overall, the prototype C1-C2 pattern (ASM72) differs from the prototype C3 pattern (BC2) by nearly 10%, from A subtype patterns by 11 to 13.5% (plus the 5-amino-acid VR2 deletion), and from the B pattern by 29%, whereas C3 differs from the A subtypes by 12 to 14.5% (plus the deletion) and from the B pattern by 31% (Table 1).
Similarities and divergence among renal transplant samples from Saudi Arabia.
A high incidence of iatrogenic KS has been reported in studies of renal transplant patients of Middle Eastern and Jewish origin in both Saudi Arabia (54, 55) and Toronto (29). Foreman et al. (23) reported that four of five HHV8 genomes from the Saudi Arabian KS patients had in common an unusual ORF26 pattern that differs from those of five tested U.S. renal transplant KS samples that were mostly A subtypes. Seven of the same Saudi Arabian samples were sequenced here in the ORF-K1 region, and six proved to have C subtype patterns. Only SKS8 was different by being a typical A1 genome. In contrast, SKS2, SKS3, SKS6, and SKS7 all closely resemble our two other C2 patterns (EKS1 and TKS11), a subtype that has not yet been detected among our U.S. AIDS patient samples. Overall, 13 of the total of 20 KS samples that we have tested that were not AIDS associated had C-subtype ORF-K1 genes, including 4 from Taiwan and 6 from Saudi Arabia, plus 1 each from Texas, Tennessee, and Maryland. Furthermore, the C2 variant EKS1-EKS2 specimens from Maryland also came from an HIV-negative classic KS patient of Middle Eastern background.
Two Saudi Arabian samples, SKS1 and SKS9, proved to be unusual and have been assigned as the prototypes of novel C4 and C5 variants. Both include the characteristic C subtype 15-bp deletion, but SKS1 differs from all other C1, C2, and C3 genomes at between 5 and 7% of its amino acids and was excluded from the C1-C2 variant cluster on the dendrogram (with a bootstrap value of 98). SKS9 represents a special case that is otherwise closer to the A subtypes than to all other C subtypes by having 11 to 13% amino acid differences from the C1, C2, and C3 prototypes and 8 to 10% differences from each of the A subgroup ORF-K1 proteins (Table 1). The dendrogram analyses (Fig. 4) place SKS9 as a nomadic outlier of the A subgroup, but this does not take into account the characteristic C subtype VR2 deletion, which we consider to be an overriding factor leading to its classification as a C5 variant (Fig. 3). In fact, SKS9 more closely resembles A subtype patterns in the N-terminal half of the protein and C subtype patterns in the C-terminal half of the protein, although in neither case does it fit clearly into any of the current variant subgroups (Fig. 2). Therefore, SKS9 may either be an A/C chimera that arose originally by intertypic recombination within the ORF-K1 gene, or it could represent a true evolutionary intermediate.
African KS samples fall primarily into the ORF-K1 B subtype pattern.
Among the 14 different HHV8 KS samples sequenced that fell into the B subgroup, 9 came directly from Africa, including 1 endemic case of infection from 1984 in Zaire (431KAP), 2 from HIV-positive females in Uganda, 1 of 2 from HIV-positive patients in Tanzania, and all 5 from HIV-positive patients in Zambia. Among the other five B samples, at least four probably came indirectly from Africa. Two were from African immigrants to either New Zealand (ZKS6/AIDS) or the United States (JKS20 classic), whereas OKS7 and OKS8 came from AIDS KS patients of black Haitian and Hispanic Mexican backgrounds, respectively (both from Miami), and JKS15 came from an African-American AIDS patient in Baltimore. Only 1 of the 10 samples tested from Africa (OKS3=A5) did not have a B subtype ORF-K1 gene.
The B subtype ORF-K1 patterns show up to 30% overall amino acid differences from the A, C, and D subgroup patterns (Table 1), with numerous characteristic subtype-specific differences occurring essentially throughout the whole length of the protein (Fig. 2). Even within the N-terminal signal peptide region, 13 of 26 positions differ from BCBL-R (A1) and ASM72 (C1), and across the C-terminal cytoplasmic domain, 12 of 38 positions differ, and yet these domains are both virtually invariant between and within the A and C subtypes. Therefore, the divergence of B subtypes from a progenitor of the A and C subtypes probably represents a much older event that the separation of the A and C subtypes. There are no fewer than 40 totally invariant B-specific amino acid changes relative to the A and C patterns, with 22 others that are nearly invariant. All B subtype examples also differed significantly from one another, although no clear clustering or subgrouping was apparent even in the dendrogram analysis. A total of 58 residues show variations across the whole B subset, with the maximum range being the 26 amino acid (9%) differences between OKS7 and RKS5. RKS3 and RKS5 show significant differences from the other Zambian samples, whereas RKS5 and ZKS6 show some common unique features, such as F-33, K-34, Y-84, Q-89, E-93, and G-189. The three nonimmigrant American B samples have two unique positions in common (Y-5 and R-223), but they also differ significantly elsewhere. Only 431KAP and JKS20 represent endemic non-AIDS-associated KS samples, and no PEL samples from Africa have been examined as yet. Importantly, the B subtype ORF-K1 proteins are not only more diverged among themselves than are either the A or C subgroups, but their branch lengths in the dendrogram (Fig. 4) are also noticeably longer (especially ZKS6), supporting the idea that they may represent a more ancient and diverse group than the others.
Common features of the Taiwan C3′ clade and identification of a novel D subtype.
Interestingly, the seven C3 subtype Taiwan samples (four classic and three AIDS associated) all represent a distinctive subset with four common novel amino acid variations that are not found in any other C subtype genomes (represented by R-165, R-200, D-222, and the absence of S-65). However, they are also quite variable among themselves, with TKS2 and TKS9 having six common unique nucleotide variations at A-79, C-341, G-392, C-765, T-741, and C-742, although also differing at three positions, and with TKS5 being quite distinctive from all other HHV8 genomes tested, with unique nucleotide variations that include G-75, A-306, G-307, G-308, G-317, and A-734. Based on the obvious common lineage of this set of samples within the C3 subgroup, we refer to them as a distinct C3′ clade (bootstrap value of 44).
Among the other two samples from Taiwan, TKS11 came from a renal transplant recipient and proved to have a C2 subtype ORF-K1 gene, whereas TKS10, representing a classic non-AIDS-associated KS case in an aboriginal Hwalian patient from eastern Taiwan, proved to be the prototype of a novel D1 subtype genome. A major feature of TKS10 is a 39-bp duplication of amino acids 181 to 193 close to the VR2 region (Fig. 2). In addition, the TKS10 protein differed from all A and C variants by 24 to 28% and from the B prototype by 27% at the amino acid level (Table 1). The D1 ORF-K1 protein is more similar to B than to A and C in the VR1 block, but more similar to A and C than to B in VR2 and at the N and C termini. There are 18 common amino acid positions within B and D1 that differ from sites conserved in A and C, as well as 31 common amino acid positions between A, C, and D1 that differ from those in B subtypes (Fig. 2). On the other hand, TKS10 also has D-43 in common with the A3, A5, and C2 patterns; P-227 in common with the A5, C2, and B patterns; and both P-92 and R-200 in common only with many C3 patterns. The overall interpretation from both manual alignment (Fig. 2 and 3) and according to phylogenetic tree analysis (Fig. 4) is that the divergence of the D branch occurred at an intermediate level relative to the divergence of the A-plus-C branch from the B branch. Both the radial dendrogram (Fig. 4B) and the amino acid difference table (Table 1) suggest that the D branch point lies somewhat closer to the A-plus-C lineage than to the B lineage.
Additional D subtype genomes from the South Pacific.
After discovering that the unusual D subtype genome (TKS10) came from a classic KS patient who had a Pacific Island ethnic heritage rather than the Chinese heritage of the other Taiwan patients, we searched for additional archived diagnostic samples that might be representative of a Pacific branch of the virus. A total of six HHV8-positive KS lesion DNA samples from New Zealand were examined and proved to represent several different subtypes. Samples ZKS1 and ZKS5 from male Caucasian AIDS patients had typical A4 and A1 variant ORF-K1 genes, respectively, and the sample from a Caucasian renal transplant patient (ZKS7) was also an A1 subtype. In contrast, sample ZKS6 came from an HIV-positive recent immigrant from southern Africa and was a B subtype closely related to RKS5 from Zambia. Finally, the genes in two samples, referred to as ZKS3 and ZKS4, from HIV-negative elderly male Pacific Islanders with classic KS were dramatically different from all other ORF-K1 genes examined and have been provisionally designated as D2 variants within the same Pacific D subtype as TKS10.
The ZKS3 and ZKS4 ORF-K1 proteins differ by only one amino acid from each other, and although considerably different from the D1 pattern, they also display several features in common only with TKS10, such as L-32, D-43, S-83, Q-89, R-112, C-116, A-121, T-123, and G-133. They also have a number of other features in common only with both the B and D1 subtypes, such as P-14, H-20, G-54, I-126, R-128, Q-135, P-206, A-211, E-216, and F-277, but they do not have the 13-amino-acid duplication seen in TKS10 (Fig. 2). Overall, the D2 ORF-K1 proteins differ from the A and C prototypes by 25 to 27%, from B by 29%, and from D1 by nearly 21% at the amino acid level (Table 1). Although quantitatively the differences between the D1 and D2 ORF-K1 proteins are greater than those between the A and C subtype patterns, the dendrogram patterns clearly show them to be on a common distinctive branch (Fig. 4B), and it was deemed unwise to introduce an additional E designation until more samples of these subtypes have been found and evaluated. Furthermore, D1 and D2 proved to be virtually identical across the rest of their genomes (53).
DISCUSSION
Structural similarity between ORF-K1 and immunoglobulin family receptors.
The size and structure of the 289-amino-acid ORF-K1 protein show a distinct resemblance to those of a single-domain immunoglobulin family receptor with two extended loops (VR1 and VR2) added to the large extracellular domain. Intriguingly, ORF-K1 contains a 36-amino-acid block between positions 97 and 133 that has 50 to 55% amino acid identity to the extreme N terminus of the mature forms of the variable domains of certain immunoglobulin lambda light chains, in particular those referred to as subtypes VII, VIII, and XII (Fig. 5). This homology is not significantly affected by polymorphism of different ORF-K1 subtypes and includes conservation of the first Cys bridge residue of the immunoglobulin variable region (position C-117 in ORF-K1). Relative to classical immunoglobulin domain family proteins, VR1 is positioned between the N-terminal signal peptide and the beginning of the complementarity-determining region of the variable domain homology region, whereas VR2 lies adjacent to the single C-terminal transmembrane spanning (or TM) domain (see Fig. 1C). There are a total of 10 completely conserved Cys residues at positions 28, 40, 52, 76, 100, 109, 117, 154, 157, and 170 in the mature extracellular domain of ORF-K1, as well as seven predominantly conserved N-glycosylation motifs.
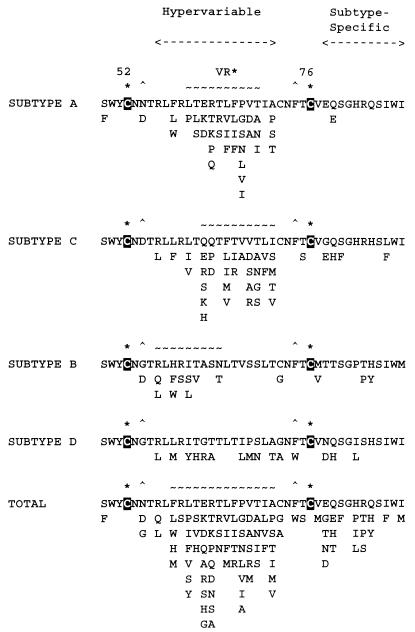
FIG. 5.
Homology between ORF-K1 and the variable region of immunoglobulin (IG) light chains. (Top panel) Typical variations seen within the major subtypes of ORF-K1 in the vicinity of the immunoglobulin homology. Dashes indicate matches to the ORF-K1 subtype A sequence (top line). The conserved Cys at position 117 in ORF-K1 is indicated in a black box. (Center panel) Amino acid alignments between the N termini of mature and unprocessed immunoglobulin lambda chains (positions 7 to 35 and 21 to 56, respectively), a Bence Jones protein, and a BCR protein with positions 86 to 140 of ORF-K1 subtype A. Dashes indicate spaces, asterisks denote identities to the ORF-K1A version, and vertical bars denote similarities. (Bottom panel) Homologies among different lambda chain variable region subtypes compared to the ORF-K1 (A) immunoglobulin homology region. The numbering used represents that of amino acid positions in the mature immunoglobulin forms lacking the signal peptide. FRI, flanking region I; CDR1, complementarity-determining region I. Other symbols are the same as for the center panel above.
The 38-amino-acid cytoplasmic tail domain in all ORF-K1 subtypes and variants also retains a highly conserved version of the ITAM found in components of T-cell receptor (TCR) and B-cell receptor (BCR) signalling complexes, as well as in Fc-like receptor proteins, including the CD3, CD8 and zeta subunits and surface immunoglobulin α and β (CD79) subunits (65, 66). Except for the unusual P at position 284, the double ITAM motifs of YYSLHDLCTEDYTQPV (A/C) and YYSLDYFHTEDYTQPV (B) between codons 270 to 285 near the extreme C terminus of ORF-K1 match better with the consensus YXXLX6–8YXXL sequence of a positively acting ITAM than they do with the LXYXXLX4–6LXYXXL sequence of a negative-acting ITAM found in major histocompatibility complex-recognizing receptors such as KIR (39). However, several functional SH2 tyrosine kinase target motifs (e.g., in the platelet-derived growth factor αβ receptors and ErbB2) that are known to bind to phospholipase C-γ also replace the YXXL motif with the same nonconsensus pattern found in the second ORF-K1 subregion, namely YXXPL (61). In cross-linked TCR and BCR immunoglobulin family receptors as well as in LMP2A of EBV, the ITAMs act as target sites for initial phosphorylation by SH2-SH3 class tyrosine kinases (such as SRC, LCK, and LYN, etc.) and subsequent binding to 2xSH2 class tyrosine kinases (such as SYK and ZAP70) that regulate T-cell or B-cell activation signals, including induction of interleukin 2 (3, 32). Recently, Lee et al. (38) have shown that insertion of the HHV8 ORF-K1 gene can functionally substitute for the STP oncogene of HVS by restoring the ability of an STP-deleted virus to both immortalize primary T cells and produce lymphoma in experimentally inoculated marmosets. Furthermore, they have also demonstrated that the C-terminal domain ITAM of ORF-K1 is functionally active in tyrosine protein kinase signaling via the SRC and SYK proteins in CD8/K1 chimeric proteins when introduced into COS cells (36). Therefore, ORF-K1 seems likely to play some critical, but as yet unknown, role involving signal transduction in HHV8 biology. The ORF-K1 protein is expressed as a stable and highly glycosylated membrane protein in DNA-transfected cells (36, 52), but, curiously, ORF-K1 mRNA is only expressed abundantly as an early phosphonoacetic acid-insensitive lytic cycle gene product in tetradecanoyl phorbol acetate- or butyrate-induced PEL cell lines (35, 52), and it is not yet known whether ORF-K1 is expressed at significant levels in the latent state or in KS lesions.
The extraordinarily high level of variability among ORF-K1 amino acid sequences can be separated into two distinct patterns. Throughout most of the protein, there are at most two to three alternative amino acids at any one variable position, and many of these changes are highly characteristic of a particular subtype or variant, evidently reflecting relatively ancient evolutionary divergence. On the other hand, a second level of variability occurs even among highly related genomes that cluster within specific variants or clades. The latter is particularly highly concentrated within the central VR* portion of VR1, where as many as 5 to 9 alternative amino acids are found at each of 12 of 15 consecutive residues between amino acid positions 57 and 71 (Fig. 6). Furthermore, this whole VR* block lies between two totally conserved NXS/T glycosylation sites as well as two adjacent completely conserved Cys residues at positions 52 and 76 that appear likely to mediate formation of a 23-amino-acid hypervariable loop with a disulfide bridge at the base. The high level of intratypic variability within the VR* loop contrasts dramatically with the region of VR1 immediately to the right between amino acids 78 and 88, in which the variability is almost exclusively subtype specific (Fig. 6). The VR* loop structure closely resembles the organization of the several hypervariable Cys-bridged loops (e.g., V3 and V4) in the extracellular domain of the gp160 ENV protein of HIV. All four HHV8 subtype ORF-K1 protein patterns have this VR* hypervariability within VR1, although it is most pronounced within the A and C subtypes and is much less evident among the B subtype samples (Fig. 6). In contrast, the VR2 region shows many fewer substitutions at individual positions than does VR1, but instead displays a number of distinctive short in-frame deletions or reiterations. Overall, the intratypic hypervariability in VR1 and VR2 has the effect of greatly exaggerating the apparent genetic divergence between the A and C subtypes, which are relatively well conserved elsewhere along the protein. However, the B and D subtypes have diverged from each other and from the A/C group virtually throughout the whole length of the protein.
FIG. 6.
Summary of amino acid substitutions found within the predicted Cys-bridged VR* loop segment within the extracellular domain of HHV8 ORF-K1. The diagram lists all alternative amino acids found within VR1 between amino acids 52 and 76 of the A, C, B, and D subtypes, together with an overall summary of all substitutions within this region among the 63 distinct HHV8 ORF-K1 genes studied here. ∗, conserved probable Cys-bridging residue; ^, conserved N-glycosylation site (NXS/T); ∼, potential N-glycosylation site.
Comparison of the levels of HHV8 ORF-K1 variation with strain variability in other herpesviruses.
The ORF-K1 gene shows very high levels of sequence divergence relative to those observed in other HHV8 genes. This is especially true at the protein level, where 4 major subtypes and at least 13 distinctive ORF-K1 variants have been defined. In six other HHV8 gene loci tested (53), similar subtype patterns can be discerned, but these involve maximally only 1 to 2% nucleotide changes, with only a small subset of the substitutions resulting in amino acid changes. In contrast, in ORF-K1, the 8 to 15% nucleotide variations between the A, B, C, and D subtypes translate into 15 to 30% amino acid differences. In fact, nearly 85% of the nucleotide changes observed in ORF-K1 are nonsynonymous (i.e., lead to amino acid changes), implying that a highly selective biological mechanism at the protein level is driving the phenomenon, rather than simple acquisition of random unselected mutations.
The only comparable strain variability situations detected previously in human herpesviruses involve the EBNA2 nuclear antigen in EBV, where the amino acid differences between the A and B subgroups in human EBV differ by 35%, compared to a 45% divergence level of both from the equivalent EBNA2 protein of the related baboon virus herpesvirus papio (18, 40). Although other latency genes in EBV (e.g., EBERs, EBNA1, and EBNA3abc) also show consistent A or B divergence patterns, the differences are much smaller, and known intrastrain variability within the A and B subtypes of EBNA1 and EBNA2 (69) appears to be much smaller than that within the ORF-K1 subtypes. It is also not clear as yet whether A and B differences can be clearly discerned within the lytic cycle genes of EBV. Interestingly, the in vitro transformation properties of the two EBNA2 subtypes differ somewhat (58, 60), and the EBV B subgroup predominates in Africa, whereas the A subgroup predominates elsewhere, although both are commonly present in AIDS patients.
Clear grouping into the predominantly African A and non-African B subgroups of HHV6 has also been demonstrated. These differences are most pronounced within the nuclear regulatory protein IE1, as well as in gpB and gpH, and appear to account for as much as 5% nucleotide variation overall across the whole genome (21, 27). Similarly, based on two variable loci within gpB and another in gpE of human cytomegalovirus (HCMV) there is an apparent dichotomy among HCMV isolates as well, although in this case, the two principal subtypes have become highly scrambled by recombination even within the gpB gene itself. The major sites of variability in HCMV gpB are largely limited to within the predicted N-terminal signal peptide and are close to an internal processing cleavage site, where they seem unlikely to influence either function or immune surveillance.
None of these other instances of herpesvirus variability have proved sufficient to permit clade analysis of the type presented here for HHV8, which is much more reminiscent of HIV ENV protein clade patterns (41, 43, 63). However, even the strain variability in HHV8 ORF-K1 does not approach the level described in the equivalent left-hand end of HVS, where the A, B, and C subtypes encode very different STP genes and the C subtype lacks the TIP gene altogether (44). Furthermore, the highly transforming C type of STP gene also displays considerable amino acid heterogeneity of up to 15% among different isolates (37). On the other hand, we have found that there are also two very different versions of the highly spliced ORF-K15 membrane protein gene at the right-hand end of the HHV8 genome (less than 30% identity only) that may represent a situation somewhat equivalent to that of the three different types of STP genes in HVS (51, 53).
Significance of ORF-K1 hypervariability.
Remarkably, the HHV8 ORF-K1 protein displays almost as much amino acid variability as do immunoglobulins and the HIV ENV proteins, and yet it is hardly conceivable that a herpesvirus has any similar special mechanisms to generate diversity at the high rate that those two proteins do. Since herpesvirus-infected cells are known to be more susceptible to the cell-mediated cytotoxicity arm than to antibody-mediated humoral immunity, the likelihood that the high variability of ORF-K1 simply represents a mechanism to evade recognition by neutralizing antibody appears somewhat implausible. Furthermore, unlike the ENV protein of HIV, there is no evidence at present that any variability is generated during infection of a single human host. Perhaps instead the variability represents biological selection for recognition of a polymorphic cell surface marker or for mimicking or avoiding a particular HLA repertoire that allows preferential virus entry or survival only in genetically appropriate host cells. Therefore, the apparently low infectivity and the far from universal distribution of HHV8 (outside of the AIDS epidemic) might result from an adaptation to preferential vertical or familial transmission, and the role of selective ORF-K1 mutation might only come into play during the earliest stages of horizontal transmission. Overall, the concept of the hypervariable ORF-K1 protein as an extracellular receptor protein that also participates in protein tyrosine kinase signaling and is expressed in the early lytic cycle, but perhaps not during latency, is quite puzzling and enigmatic at present.
The cladal pattern of HHV8 infection seen in AIDS patients in Maryland (for example), where six of the first seven samples tested were of the A1 subtype with variations concentrated within an 11-amino-acid subset of the VR* block, may result from both a very rare initial reactivation in a classic HHV8-positive individual, followed by occasional transmission to compatible recipients with accompanying selection for adaptive VR* mutations. The alternative option—that the VR* variability preceded the AIDS epidemic and that most AIDS KS represents reactivation of preexisting inapparent infections—seems less attractive. The hypothesis that much of the subtype A VR* variation has occurred within the AIDS epidemic may be testable in the future by tracing rates of genetic change within samples from prospective cohort studies, for example. However, it is also clear that AIDS KS does involve HHV8 viruses displaying several different genotypes, with the A1, A4, and C3 ORF-K1 variants being most common among the predominantly East Coast and southern U.S. samples that we have evaluated.
There is still debate about whether different HIV clades have significantly different biological properties, and this will be an interesting problem for HHV8 also. Luppi et al. (42) have stated that, in the absence of HIV infection, they find only A subtypes of HHV8 in classical KS samples in Italy, whereas C subtypes predominate in Italian nonmalignant lymphoid disorders. Unfortunately, those authors only examined a very narrow and relatively unrevealing 233-bp window within the ORF26 gene, which, as we demonstrate elsewhere, can be highly ambiguous for strain subtyping purposes (53). This is also the case among several claims for an association between HHV8 infection and multiple myeloma (57) or sarcoidosis (20), which both remain highly controversial at present. A biological distinction between A and C subtypes clearly does not apply to KS and PEL samples within U.S. AIDS patients. However, two of the three ORF-K1 B subtype genomes that we have detected so far among U.S. patients were from aggressive disseminated AIDS KS from Miami that also demonstrated unusual patches of lytic cycle HHV8 infection within the spindle cell regions (11). Perhaps there is a parallel here to the aggressiveness of childhood KS associated with B subtypes in central or southern Africa. Considering the high rates of seropositivity and evidence for frequent primary infection in young children in Zambia (34), compared to low overall seropositivity and seroconversion only after adolescence elsewhere (7), it may be plausible to suggest that a predominant variant present in parts of Africa can be transmitted readily from mother to infant or between young children, whereas the other subtypes may be restricted to having very low rates of postadolescent sexual transmission only. Obviously, an evaluation of whether these differences reflect viral subtype effects, rather than behavioral or environmental factors, or simply represent the large overall population load of the virus in that part of Africa may now be feasible.
Implications about HHV8 evolutionary history.
The most significant finding from our HHV8 ORF-K1 analysis is the close correlation observed between the clade patterns and the geographic or ethnic backgrounds of the infected patients. This is especially true with regard to the predominance of B subtypes in patients from central or southern Africa and in African emigrants, as well as for the C2 subtypes in Saudi Arabia and the C3′ clade in Taiwan. These results are not compatible with a single relatively recent cross-species transmission and as yet incomplete spread into present-day human populations (in which case there would be very little variation), but more likely attest to an ancient infection with very low penetrance and infrequent horizontal transmission of the virus, as well as to relatively rare multiple infections and consequent low recombination rates. In fact, the distribution patterns appear to be compatible with founder effects associated with the global spread of modern human populations in paleolithic times. In this scenario, the major European and northern Asian branches established 35,000 years ago (subtypes A and C) would be expected to be more similar to each other than to the modern African versions (subtype B), with an original divergence of the two major branches dating perhaps to 100,000 years ago, whereas the three D subtype samples encountered so far (all from classic KS patients of Pacific Island origin) might be representative of the earlier migration 60,000 years ago into southern Asia, Australia, and Oceania. Presumably the A and C variants in U.S. patients had their origins among immigrants from Europe and the Mediterranean, whereas the B variants in the United States are of the original African origin, perhaps via the West Indies.
An examination of additional samples from Asia and Oceania, western Africa, Scandinavian countries, and indigenous North and South American populations, etc., to determine how they relate to the four major subgroups identified so far, as well as studies to attempt to determine the rate at which HHV8 ORF-K1 divergence occurs, should also be very informative. At present, our data imply that little or no evolutionary change occurs within single infected individuals once either PEL tumors or KS lesions are established. However, since all of our sequence data were determined by PCR analysis of lesion tissue- or tumor-derived BCBL cell line DNA, we could be sampling only the most abundant or transformation-competent genome types, and if variation and selection occurred either during the earliest stages of infection or in virus in circulating lymphoid cells, we would be unlikely to have detected it here. Comparative analysis of viral DNA from multiple blood samples and from both early and late lesions from the same patient, as well as from known cases of spousal or partner transmission, will have to be carried out to try to address these issues.
Finally, the extensive ORF-K1 protein polymorphism detected here will likely provide important insights into the role of ORF-K1 in HHV8 biology as well as offer a unique opportunity for further study of HHV8 epidemiology. In addition, careful properly controlled sequence analysis of ORF-K1 variability also has the potential to resolve currently contentious issues of whether HHV8 DNA detected by some groups but not others by nested PCR with samples from multiple myeloma, sarcoid, saliva, and prostate, etc., represents contamination or valid low-level infection. Obviously, future efforts to consolidate our evolutionary models, as well as any attempts to make correlations between ORF-K1 subtypes and pathogenesis, will also have to take into account the potential complications of other polymorphic loci and the possibility of recombinant or chimeric genomes. There is already strong evidence for both of these complications in a minority of HHV8 genomes (51, 70), but an extensive evaluation of subtype patterns has revealed that most HHV8 samples show consistent subtype linkage patterns, even at conserved loci across the entire length of the genome (53).
ACKNOWLEDGMENTS
These studies were funded by Public Service grant R01 CA73585 to G.S.H. from the National Cancer Institute.
We thank Margit Lucskay for technical assistance and Sarah Heaggans for help with preparation of the manuscript. We also gratefully acknowledge receipt of discarded diagnostic KS samples from Andrew Blauvelt (National Institutes of Health), Jennifer Cannon and Richard Ambinder (JHU Oncology Center), Steve Harrington (University of Miami), D. Wade Gibson (JHU, Department of Pharmacology), and the AIDS Malignancy Bank.
REFERENCES
- 1.Alagouzoulou, L., J.-C. Zong, and G. S. Hayward. Unpublished data.
- 1a.Ambinder R F, Newman C, Hayward G S, Biggar R, Melbye M, Kestens L, Van Marck E, Piot P, Gigase P, Wright P B, Quinn T C. Lack of association of cytomegalovirus with endemic African Kaposi’s sarcoma. J Infect Dis. 1987;156:193–197. doi: 10.1093/infdis/156.1.193. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi’s sarcoma associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 3.Beaufils P, Choquet D, Mamoun R Z, Malissen B. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 1993;12:5105–5112. doi: 10.1002/j.1460-2075.1993.tb06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beral V. Epidemiology of Kaposi’s sarcoma. Cancer Surv. 1991;10:5–22. [PubMed] [Google Scholar]
- 5.Beral V, Peterman T A, Berkelman R C, Jaffe H W. Kaposi’s sarcoma among persons with AIDS: a sexually transmitted infection? Lancet. 1990;335:123–128. doi: 10.1016/0140-6736(90)90001-l. [DOI] [PubMed] [Google Scholar]
- 6.Biggar R J, Horm J, Fraumeni J, Jr, Greene M H, Goedert J J. Incidence of Kaposi’s sarcoma and mycosis fungoides in the United States including Puerto Rico, 1973–1981. J Natl Cancer Inst. 1984;73:89–94. [PubMed] [Google Scholar]
- 7.Blauvelt A, Sei S, Cook P M, Schulz T F, Jeang K-T. Human herpesvirus 8 infection occurs following adolescence in the United States. J Infect Dis. 1997;176:771–774. doi: 10.1086/517298. [DOI] [PubMed] [Google Scholar]
- 8.Boshoff C, Gao S-J, Healy L E, Matthews S, Thomas A J, Coignet L, Warnke R A, Strauchen J A, Matutes E, Kamel O W, Moore P S, Weiss R A, Chang Y. Establishing a KSHV positive cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood. 1998;91:1671–1679. [PubMed] [Google Scholar]
- 9.Boshoff C, Whitby D, Hatziioannou T, Fisher C, van der Walt J, Hatzakis A, Weiss R, Schulz T. Kaposi’s sarcoma-associated herpesvirus in HIV-negative Kaposi’s sarcoma. Lancet. 1995;345:1043–1044. doi: 10.1016/s0140-6736(95)90780-7. [DOI] [PubMed] [Google Scholar]
- 9a.Boshoff, C., J.-C. Zong, and G. S. Hayward. Unpublished data.
- 10.Cannon, J. S., A. C. Hawkins, C. A. Griffin, Q. Tao, M. Borowitz, G. S. Hayward, and R. F. Ambinder. Characterization of a new EBV+/HHV8+ primary effusion lymphoma-derived cell line. Submitted for publication.
- 11.Cannon, J. S., J. Nicholas, J. M. Orenstein, R. B. Mann, P. J. Browning, J. A. DiGiuseppe, E. Cesarman, G. S. Hayward, and R. F. Ambinder. Heterogeneity of viral IL6 expression in HHV-8-associated diseases. Submitted for publication. [DOI] [PubMed]
- 12.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 13.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two AIDS-related lymphoma cell lines containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 14.Cesarman E, Nador R G, Bai F, Bohenzky R A, Russo J J, Moore P S, Chang Y, Knowles D M. Kaposi’s sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 16.Chuck S, Grant R M, Katongole-Mbidde E, Conant M, Ganem D. Frequent presence of a novel herpesvirus genome in lesions of human immunodeficiency virus-negative Kaposi’s sarcoma. J Infect Dis. 1996;173:248–251. doi: 10.1093/infdis/173.1.248. [DOI] [PubMed] [Google Scholar]
- 17.Collandre H, Ferris S, Grau O, Montagnier L, Blanchard A. Kaposi’s sarcoma and new herpesvirus. Lancet. 1995;345:1043. doi: 10.1016/s0140-6736(95)90779-3. [DOI] [PubMed] [Google Scholar]
- 18.Dambaugh T, Hennessey K, Chamnankit L, Kieff E. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr virus nuclear antigen 2. Proc Natl Acad Sci USA. 1984;81:7632–7636. doi: 10.1073/pnas.81.23.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decker L L, Shankar P, Khan G, Freeman R B, Dezube B J, Lieberman J, Thorley-Lawson D A. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J Exp Med. 1996;184:283–288. doi: 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Alberti L, Piattelli A, Artese L, Favia G, Patel S, Saunders N, Porter S R, Scully C M, Ngul S-L, Teo C-G. Human herpesvirus 8 variants in sarcoid tissues. Lancet. 1997;350:1655–1659. doi: 10.1016/s0140-6736(97)10102-7. [DOI] [PubMed] [Google Scholar]
- 21.Di Luca D, Mirandola P, Ravaioli T, Bigoni B, Cassai E. Distribution of HHV-6 variants in human tissues. Infect Agents Dis. 1996;5:203–214. [PubMed] [Google Scholar]
- 22.Dupin N, Grandadam M, Calvez V, Gorin I, Aubin J T, Havard S, Lamy F, Leibowitch M, Huraux J M, Escande J P. Herpesvirus-like DNA sequences in patients with Mediterranean Kaposi’s sarcoma. Lancet. 1995;345:761–762. doi: 10.1016/s0140-6736(95)90642-8. [DOI] [PubMed] [Google Scholar]
- 23.Foreman K E, Alkan S, Krueger A E, Panella J R, Swinnen L J, Nickoloff B J. Geographically distinct HHV-8 DNA sequences in Saudi Arabian iatrogenic Kaposi’s sarcoma lesions. Am J Pathol. 1998;153:1001–1004. doi: 10.1016/S0002-9440(10)65642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaidano G, Cechova K, Chang Y, Moore P S, Knowles D M, Dalla-Favera R. Establishment of AIDS-related lymphoma cell lines from lymphomatous effusions. Leukemia. 1996;10:1237–1240. [PubMed] [Google Scholar]
- 25.Gao S-J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion to antibodies against Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 26.Gao S-J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi’s sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 27.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 28.Guo H-G, Browning P, Nicholas J, Hayward G S, Jiang Y W, Sadowska M, Tschachler E, Raffeld M, Columbini S, Gallo R C, Reitz M. Characterization of a chemokine receptor-related gene in human herpesvirus 8 and its expression in Kaposi’s sarcoma. Virology. 1997;228:371–378. doi: 10.1006/viro.1996.8386. [DOI] [PubMed] [Google Scholar]
- 29.Harwood A R, Osoba D, Hofstader S L, Goldstein M B, Cardella C, Holecek M J, Kunynetz R, Giammarco R A. Kaposi’s sarcoma in recipients of renal transplants. Am J Med. 1979;67:759–765. doi: 10.1016/0002-9343(79)90731-9. [DOI] [PubMed] [Google Scholar]
- 30.Herndier B G, Werner A, Arnstein P, Abbey N W, Demartis F, Cohen R L, Shuman M A, Levy J A. Characterization of a human Kaposi’s sarcoma cell line that induces angiogenic tumors in animals. AIDS. 1994;8:575–581. doi: 10.1097/00002030-199405000-00002. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y Q, Li J J, Kaplan M H, Poiesz B, Katabira E, Zhang W C, Feiner D, Friedman-Kien A E. Human herpesvirus-like nucleic acid in various forms of Kaposi’s sarcoma. Lancet. 1995;345:759–761. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- 32.Irving B A, Weiss A. The cytoplasmic domain of the T cell receptor zeta chain is sufficient to couple to receptor-associated signal transduction pathways. Cell. 1991;64:891–901. doi: 10.1016/0092-8674(91)90314-o. [DOI] [PubMed] [Google Scholar]
- 33.Kasolo F C, Mpabalwani E, Gompels U A. Infection with AIDS-related herpesviruses in human immunodeficiency virus-negative infants and endemic childhood Kaposi’s sarcoma in Africa. J Gen Virol. 1997;78:847–856. doi: 10.1099/0022-1317-78-4-847. [DOI] [PubMed] [Google Scholar]
- 33a.Kasolo, F., and U. Gompels. Personal communication.
- 34.Kedes D H, Oberskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 35.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) Virology. 1997;236:147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- 36.Lee H, Guo J, Li M, Choi J-K, DeMaria M, Rosenzweig M, Jung J U. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi’s sarcoma-associated herpesvirus. Mol Cell Biol. 1998;18:5219–5228. doi: 10.1128/mcb.18.9.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee H, Trimble J J, Yoon D-W, Regier D, Desrosiers R C, Jung J U. Genetic variation of herpesvirus saimiri subgroup A transforming protein and its association with cellular src. J Virol. 1997;71:3817–3825. doi: 10.1128/jvi.71.5.3817-3825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A, Desrosiers R C, Jung J U. Deregulation of cell growth by the K1 gene of Kaposi’s sarcoma-associated herpesvirus. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 39.Liebson P J. Signal transduction during natural killer cell activation: inside the mind of a killer. Immunity. 1997;6:655–661. doi: 10.1016/s1074-7613(00)80441-0. [DOI] [PubMed] [Google Scholar]
- 40.Ling P D, Ryon J J, Hayward S D. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louwagie J, Janssens W, Mascola J, Heyndrickx L, Hegerich P, van der Groen G, McCutchan F E, Burke D S. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J Virol. 1995;69:263–271. doi: 10.1128/jvi.69.1.263-271.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luppi M, Barozzi P, Marasca R, Ferrari M G, Torelli G. Human herpesvirus 8 strain variability in clinical conditions other than Kaposi’s sarcoma. J Virol. 1997;71:8082–8083. doi: 10.1128/jvi.71.10.8082-8083.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCutchan F E, Artenstein A W, Sanders-Buell E, Salminen M O, Carr J K, Mascola J R, Yu X-F, Nelson K E, Khamboonruang C, Schmitt D, Kieny M P, McNeil J G, Burke D S. Diversity of the envelope glycoprotein among human immunodeficiency virus type 1 isolates of clade E from Asia and Africa. J Virol. 1996;70:3331–3338. doi: 10.1128/jvi.70.6.3331-3338.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medveczky P, Szomolanyi E, Desrosiers R C, Mulder C. Classification of herpesvirus saimiri into three groups based on extreme variation in a DNA region required for oncogenicity. J Virol. 1984;52:938–944. doi: 10.1128/jvi.52.3.938-944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 46.Moore P S, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi’s sarcoma in patients with and those without HIV infection. N Engl J Med. 1995;332:1182–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 47.Moore P S, Gao S-J, Dominguez G, Cesarman E, Lungu O, Knowles D, Garber R, Pellett P E, McGeoch D J, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi’s sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo H-G, Reitz M S, Hayward G S. A single 13-kilobase divergent locus in Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains at least nine open reading frames homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H-G, Hayward G S, Reitz M S. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 51.Nicholas J, Zong J-C, Alcendor D J, Ciufo D M, Poole L J, Sarisky R T, Chiou C J, Zhang X, Wan X, Guo H-G, Reitz M S, Hayward G S. Novel organizational features, captured cellular genes and strain variability within the genome of KSHV/HHV8. J Natl Cancer Inst Monogr. 1998;23:79–88. doi: 10.1093/oxfordjournals.jncimonographs.a024179. [DOI] [PubMed] [Google Scholar]
- 51a.Pellet, P. Personal communication.
- 52.Poole, L. J., D. M. Ciufo, B. Chandran, and G. S. Hayward. Identification and expression of the immunoglobulin receptor-like glycoprotein encoded by the hypervariable ORF-K1 transforming gene of KSHV/HHV8. Submitted for publication.
- 53.Poole, L. J., J.-C. Zong, D. M. Ciufo, D. J. Alcendor, J. S. Cannon, R. Ambinder, J. Orenstein, M. S. Reitz, and G. S. Hayward. Comparison of genetic variability at multiple loci across the genomes of the major subgroups of Kaposi’s sarcoma associated herpesvirus (HHV8) reveals evidence for recombination and for two distinct types of ORF-K15 alleles at the right hand end. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 54.Qunibi W, Akhtar M, Sheth K, Ginn H E, Al-Furayh O, DeVol E B, Taher S. Kaposi’s sarcoma: the most common tumor after renal transplantation in Saudi Arabia. Am J Med. 1988;84:225–232. doi: 10.1016/0002-9343(88)90418-4. [DOI] [PubMed] [Google Scholar]
- 55.Qunibi W Y, Barri Y, Alfurayh O, Almeshari K, Khan B, Taher S, Sheth K. Kaposi’s sarcoma in renal transplant recipients: a report on 26 cases from a single institution. Transplant Proc. 1993;25:1402–1405. [PubMed] [Google Scholar]
- 56.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 57.Rettig M B, Ma H J, Vescio R A, Pold M, Schiller G, Belson D, Savage A, Nishikubo C, Wu C, Fraser J, Said J W, Berenson J R. Kaposi’s sarcoma-associated herpesvirus infection of bone marrow dendritic cells from multiple myeloma patients. Science. 1997;276:1851–1854. doi: 10.1126/science.276.5320.1851. [DOI] [PubMed] [Google Scholar]
- 58.Rickinson A B, Young L S, Rowe M. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J Virol. 1987;61:1310–1317. doi: 10.1128/jvi.61.5.1310-1317.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sixbey J W, Shirley P, Chesney P J, Buntin D M, Resnick L. Detection of a second widespread strain of Epstein-Barr virus. Lancet. 1989;ii:761–765. doi: 10.1016/s0140-6736(89)90829-5. [DOI] [PubMed] [Google Scholar]
- 61.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, Neel B G, Birge R B, Fajardo J E, Chou M M, Hanafusa H, Schaffhausen B, Cantley L C. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 62.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 63.Starcich B R, Hahn B H, Shaw G M, McNeely P D, Modrow S, Wolf H, Parks E S, Parks W P, Josephs S F, Gallo R C, Wong-Staal F. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 64.Su I-J, Hsu Y-S, Chang Y-C, Wang I-W. Herpesvirus-like DNA sequence in Kaposi’s sarcoma from AIDS and non-AIDS patients in Taiwan. Lancet. 1995;345:722–723. [PubMed] [Google Scholar]
- 65.Wegener A M, Letourneur F, Hoeveler A, Brocker T, Luton F, Malissen B. The T cell receptor/CD23 complex is composed of at least two autonomous transduction modules. Cell. 1992;68:83–95. doi: 10.1016/0092-8674(92)90208-t. [DOI] [PubMed] [Google Scholar]
- 66.Weiss A, Littman D R. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 67.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatzioannou T, Suggett R E, Aldam D M, Denton A S, et al. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi’s sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 68.Whitby D, Luppi M, Barozzi P, Boshoff C, Weiss R A, Torelli G. Human herpesvirus 8 seroprevalence in blood donors and lymphoma patients from different regions of Italy. J Natl Cancer Inst. 1998;90:395–397. doi: 10.1093/jnci/90.5.395. [DOI] [PubMed] [Google Scholar]
- 68a.Whitby, D., and T. Schulz. Personal communication.
- 69.Wrightham M N, Stewart J P, Janjua N J, Pepper S D, Sample C, Rooney C M, Arrand J R. Antigenic and sequence variation in the C-terminal unique domain of the EBV EBNA1. Virology. 1995;208:521–530. doi: 10.1006/viro.1995.1183. [DOI] [PubMed] [Google Scholar]
- 70.Zong J-C, Metroka C, Reitz M S, Nicholas J, Hayward G S. Strain variability among Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genomes: evidence that a large cohort of United States AIDS patients may have been infected by a single common isolate. J Virol. 1997;71:2505–2511. doi: 10.1128/jvi.71.3.2505-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]