Abstract
The WRKY transcription factors play important roles in plant growth and resistance, but only a few members have been identified in strawberry. Here we identified a WRKY transcription factor, FvWRKY50, in diploid strawberry which played essential roles in strawberry vegetative growth, and reproductive growth. Knocking out FvWRKY50 by genome editing accelerated flowering time and leaf senescence but delayed anthocyanin accumulation in fruit. Further analysis showed that FvWRKY50 acted as a transcriptional repressor to negatively regulate the expression of flowering- and leaf senescence-related genes, including FvFT2, FvCO, FvFT3, and FvSAUR36. Notably, FvWRKY50 directly upregulated the expression of FvCHI and FvDFR by binding their promoter under normal conditions, but at low temperature FvWRKY50 was phosphorylated by FvMAPK3 and then induced protein degradation by ubiquitination, delaying anthocyanin accumulation. In addition, the homozygous mutant of FvWRKY50 was smaller while the biallelic mutant showed normal size. These new findings provide important clues for us to further reveal the regulatory mechanisms of strawberry growth and fruit ripening
Introduction
Strawberry (Fragaria × ananassa Duch.) is one of the most important fruit crops worldwide because of its unique flavor and high nutritional value. Strawberry fruit is non-climacteric because its respiration rate and ethylene production have not dramatically changed during fruit development and ripening [5, 38]. Fruit ripening is a very complex process during which various biochemical and physiological changes happen in various traits, including fruit color, texture, aroma, and other quality aspects [17, 21, 36]. During the complex fruit ripening progress transcriptional regulation is indispensable because expressions of the ripening-related genes are modulated by various transcription factors [23, 35,56].
Previous studies have reported some transcription factors that could regulate strawberry fruit development and ripening, including RIF (an NAC transcription factor), TCP9 (a TCP transcription factor), WRKY48 and WRKY71 (WRKY transcription factors), MYB10, MYB44.2 and MYB79 (MYB transcription factors), RAV (an AP2/ERF transcription factor), and MADS9 [1, 3, 4, 22, 27, 39, 43, 48, 51, 52]. Among the different kinds of transcription factors, plant-specific WRKY transcription factors play an important role in the process of strawberry fruit ripening. However, to date only a few members of the WRKY transcription factors have been reported, and most of them are not well known in strawberry.
There are always one or two conserved WRKY domains in WRKY proteins that can bind the W-box of target gene promoter sequences to regulate their expression in various physiological processes [2, 32, 34]. It is well known that WRKY transcription factors can modulate defensive responses to biotic or abiotic stresses [13, 42]. WRKY transcription factor family genes have been analyzed in the diploid woodland strawberry and their expression patterns in various stress responses or the fruit development process have been determined [46, 53]. Among them, FvWRKY42 was reported to regulate various stress responses, and overexpression of FvWRKY42 in Arabidopsis thaliana could affect powdery mildew infection resistance and salt stress response in plants [45]. FaWRKY25 and FvWRKY50 (the homolog gene of AtWRKY50) were reported to regulate resistance to Botrytis cinerea in strawberry fruits [14, 25]. WRKYs were also found to function in the plant development and fruit ripening processes by regulating related target genes. For example, overexpression FvWRKY71 in woodland strawberry could promote flowering by directly activating FvFUL, FvAGL42, FvSEP1, FvLFY, and FvFPF1 [22], while transient overexpression of FaWRKY71 in octoploid strawberry could affect strawberry fruit ripening [49]. FvWRKY48 could regulate strawberry fruit softening through directly binding to the pectate lyase FvPLA promoter, resulting in pectin degradation [51]. However, besides FvWRKY48 and FaWRKY71, knowledge of the roles of other WRKY transcription factors in strawberry fruit ripening is still limited.
Numerous external environmental factors and internal regulators were regarded as signals to induce transcription factor function in the transcriptional regulation network [19]. MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) cascades were reported to govern diverse biological functions by activating downstream response factors like kinases and transcription factors [11, 18, 41] . A typical MAPK cascade is composed of three consecutive kinases, MAPK kinase kinases (MKKKs), MKKs, and MAPKs. In diploid woodland strawberry 12 MAPK genes have been identified [44, 54]. Strawberry fruit is highly sensitive to temperature and low temperature results in poor color of strawberry fruits. Our previous work indicated that FvMAPK3 could be activated by low temperature. On the one hand, FvMAPK3 phosphorylated and reduced the activity of FvMYB10. On the other hand, FvMAPK3 phosphorylated and degraded the FvCHS1 protein, both of which repressed the anthocyanin accumulation of strawberry fruits during the ripening process [26]. Besides FvMAPK3, a previous study also demonstrated that FaSnRK2.6 acted as a negative regulator to regulate strawberry fruit ripening [10]. However, whether FvMAPKs participate in the WRKY transcriptional regulation pathway in the strawberry fruit ripening process has not been explored.
Previously, FvWRKY50 (the homolog gene of AtWRKY75) was reported to be induced by various stresses and hormone signals; however, its biological function remained unknown [46]. In this study we analyzed the function of FvWRKY50 by generating gene-edited mutants in the diploid woodland strawberry ‘di Bosco’. The results demonstrated that FvWRKY50 played essential roles in plant growth, leaf senescence, flowering and fruit ripening in strawberry. In addition, FvWRKY50 could be phosphorylated and degraded by FvMAPK3 to regulate anthocyanin accumulation in strawberry fruit at low temperature.
Results
Identification of FvWRKY50 characteristics in diploid strawberry plants
Previous study has showed that FvWRKY50 belongs to subgroup IIc of the FvWRKY family and responds dramatically to various biotic and abiotic stresses in the diploid strawberry ‘Heilongjiang-3’ [46]. To fully explore the function of FvWRKY50 in strawberry, we further analyzed the bioinformatics and expression characteristic of FvWRKY50 using the diploid strawberry ‘di Bosco’.
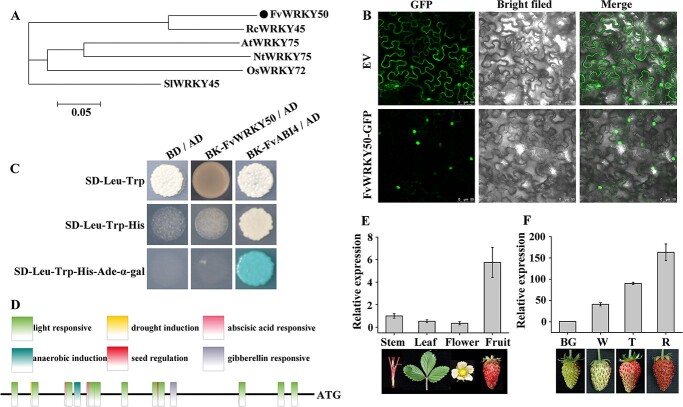
To examine the evolutionary relationship between FvWRKY50 and its orthologs from other species, a phylogenetic tree was constructed by neighbor-joining analysis, which indicated that FvWRKY50 had a close genetic relationship to the ortholog RcWRKY45 from Rosa chinensis and AtWRKY75 from A. thaliana (Fig. 1A).
Figure 1.
Bioinformatics and expression characteristic of FvWRKY50. (A) Phylogenetic analysis of WRKY50 in strawberry and other species. Fv, F. vesca; Rc, R. chinensis; At, A. thaliana; Nt, N. tabacum; Os, Oryza sativa; Sl, Solanum lycopersicum. (B) Subcellular localization of FvWRKY50. pSuper:GFP (top) or pSuper:FvWRKY50-GFP (bottom) was transformed into tobacco and the fluorescence signal was observed. (C) Transcriptional activation activity analysis of FvWRKY50 in yeast. (D) Motif analysis of the FvWRKY50 promoter. (E) FvWRKY50 gene expression in stem, leaf, flower, and fruit of diploid strawberry. qRT–PCR was used to determine the expression levels of FvWRKY50. (F) FvWRKY50 gene expression pattern in the strawberry fruit developmental process. Diploid strawberry fruits were harvested at big green (BG), white (W), turning (T), and red ripening (R) stages and qRT–PCR was used to analyze the expression levels of FvWRKY50.
To identify the subcellular localization of FvWRKY50 protein, FvWRKY50-GFP recombinant protein and GFP control protein were expressed in leaves of Nicotiana benthamiana. The fluorescence signal indicated that FvWRKY50 protein localized only in the nucleus and it was consistent with FvWRKY50 as a transcription factor (Fig. 1B). We further detected the activation activity of FvWRKY50 by transforming pGBKT7-FvWRKY50 into yeast cells and culturing on SD/−Leu−Trp, SD/−Leu−Trp−His, and SD/−Leu−Trp−His−Ade medium. pGBKT7-FvABI4 was used as the positive control. As shown in Fig. 1C, pGBKT7-FvWRKY50 could not grow while pGBKT7-FvABI4 grew well on different mediums, indicating that FvWRKY50 has no independent transcriptional activation activity. We then analyzed the sequence of the FvWRKY50 promoter; several cis-acting regulatory elements involved in light responsiveness, abscisic acid responsiveness, anaerobic induction, gibberellin response, and seed regulation were identified (Fig. 1D). All these results suggested that FvWRKY50 may be involved in the regulation of various physiological processes and may need to recruit other transcription factors to perform transcriptional activation functions.
We further identified the expression pattern of FvWRKY50 in different tissues and during different developmental stages of fruit in diploid strawberry. Real-time quantitative PCR (qRT–PCR) was used to determine the expression level of FvWRKY50 in stem, leaf, flower, and fruit (Fig. 1E). The expression level of FvWRKY50 increased gradually from big green fruit and showed the highest level in red fruit, indicating that FvWRKY50 may be related to the regulation of strawberry fruit ripening (Fig. 1F).
FvWRKY50 regulated strawberry plant growth and auxin biosynthesis
To fully analyze the biological functions of FvWRKY50 in strawberry, we used CRISPR/Cas9 to edit FvWRKY50 in ‘di Bosco’ strawberry (Supplementary Data Fig. S1A). As shown in Supplementary Data Fig. S1B, two genome-edited lines were obtained. The biallelic mutant FvWRKY50 CR 2-9 has a deletion of 5-bp/131-bp in one allele and a deletion of 114-bp in the other allele, respectively. The homozygous mutant FvWRKY50 CR 3-16 has a deletion of 114-bp in both alleles (Supplementary Data Fig. S1B). Surprisingly, we noticed that FvWRKY50 CR 3-16 grew slowly and showed a distinct dwarf phenotype while FvWRKY50 CR 2-9 showed no significant difference compared with wild-type (WT) strawberry (Fig. 2A). Consistently, the crown diameter and plant height of FvWRKY50 CR 3-16 were significantly lower than WT while FvWRKY50 CR 2-9 showed no significant difference (Fig. 2B). These results indicated that FvWRKY50 was involved in the regulation of strawberry plant growth.
Figure 2.
FvWRKY50 regulated auxin biosynthesis and signaling transduction. (A) Phenotypes of WT, FvWRKY50 CR 2-9, and FvWRKY50 CR 3-16 in growth chambers. Seedlings of 30, 60, and 105 days were observed and photographed. (B) Crown diameter and plant height of WT, FvWRKY50 CR 2-9, and FvWRKY50 CR 3-16 plants at the same stage. (C) Auxin content of WT, FvWRKY50 CR 2-9, and FvWRKY50 CR 3-16 leaves. (D) Expression profiles of genes in auxin biosynthesis and signaling transduction pathways regulated by FvWRKY50. The heat map is based on transcriptome sequencing data of WT, FvWRKY50 CR 2-9, and FvWRKY50 CR 3-16 leaves. (E) Expression levels of partial DEGs confirmed by qRT–PCR.
In order to explore the regulation function of FvWRKY50, RNA sequencing (RNA-seq) experiments were conducted to analysis the transcriptome of WT, FvWRKY50 CR 2-9, and 3-16 leaves. Analysis results showed that there were 747 differentially expressed genes (DEGs) between WT and FvWRKY50 CR 2-9, among which 313 were upregulated and 434 were downregulated (Supplementary Data Fig. S2A), while between WT and FvWRKY50 CR 3-16 there were 2787 DEGs, among which 1483 were upregulated and 1304 were downregulated (Supplementary Data Fig. S2B). Gene ontology (GO) enrichment analysis of the DEGs between WT and FvWRKY50 CR 3-16 were conducted, which revealed that many DEGs were significantly enriched in biological process related to response to auxin (Supplementary Data Fig. S2C). As the phytohormone auxin plays an important role in plant growth by regulating cell expansion, division, elongation, and differentiation [40], we determined the auxin content and found that auxin content was decreased significantly in both FvWRKY50 CR 2-9 and 3-16 leaves compared with WT leaves (Fig. 2C). Consistent with these results, transcript levels of FvYUCCA6 (FvYUC6) and FvYUCCA3 (FvYUC3), which catalyze a rate-limiting step in auxin biosynthesis [24], were decreased in FvWRKY50 CR 2-9 and FvWRKY50 CR 3-16 plants respectively (Supplementary Data Fig. S3). However, the transcript levels of genes in the auxin signaling pathway were differently regulated between FvWRKY50 CR 2-9 and FvWRKY50 CR 3-16. For example, genes for many auxin-responsive proteins, like SAMLL AUXIN UP RNA (SAURs), ARGs, IAAs, auxin transporters (ATLs) and auxin binding proteins (ABPs), were downregulated significantly in FvWRKY50 CR 3-16 plants relative to WT plants, while the expression levels of most of these genes in FvWRKY50 CR 2-9 did not decline significantly compared with WT (Fig. 2D). These results suggested that the dramatic changes in genes in the auxin signaling pathway other than the auxin synthesis pathway might cause the dwarf phenotype of FvWRKY50 CR 3-16 plants.
In addition to auxin-responsive genes, the transcript levels of many other phytohormone genes, like abscisic acid-, jasmonate- and gibberellin-related genes, transcription factor-encoding genes belonging to the AP2/ERF, bZIP, bHLH, MYB, NAC, and WRKY families, and light-responsive genes, were also changed significantly in FvWRKY50 CR 3-16 plants (Table 1). qRT–PCR results confirmed the changes of some DEGs between WT and FvWRKY50 CR plants (Fig. 2E). All these results indicated that FvWRKY50 might play essential roles in diverse biological processes in strawberry.
Table 1.
Selected DEGs in FvWRKY50 CR 3-16 leaves compared with WT leaves, and comparison of these genes between WT and FvWRKY50 CR 2-9
| Functional category | Gene ID | Annotation | CR 3-16 vs WT log 2 fold change | CR 2-9 vs WT log 2 fold change |
|---|---|---|---|---|
| Plant hormone | ||||
| Auxin | FvH4_2g20150 | Indole-3-pyruvate monooxygenase YUCCA3 | −2.63 | −1.73 |
| FvH4_7g11280 | Auxin responsive protein SAUR36 | 1.05 | 0.66 | |
| FvH4_2g10850 | Auxin responsive protein ARG7 | −2.89 | −0.09 | |
| FvH4_5g22810 | Auxin responsive protein ARG7 | −2.36 | −0.53 | |
| FvH4_5g22620 | Auxin responsive protein ARG7 | −1.66 | −0.32 | |
| FvH4_5g22780 | Auxin responsive protein ARG7 | −1.73 | −1.65 | |
| FvH4_6g02870 | Auxin-responsive protein IAA4 | −1.45 | −0.38 | |
| FvH4_4g04700 | Auxin-responsive protein IAA13 | −1.15 | −0.45 | |
| FvH4_6g30850 | Auxin-responsive protein IAA17 | −1.41 | −0.14 | |
| FvH4_2g28970 | Auxin-responsive protein IAA29 | −2.09 | −0.38 | |
| FvH4_4g26700 | Auxin-responsive protein IAA32 | −3.14 | −1.25 | |
| FvH4_2g38760 | Auxin response factor 5 | 1.13 | −0.23 | |
| FvH4_3g32000 | Auxin response factor 18 | −1.79 | −0.73 | |
| FvH4_6g30860 | Auxin-induced protein AUX22 | −2.04 | −0.89 | |
| Abscisic acid | FvH4_3g16730 | 9-cis-epoxycarotenoid dioxygenase NCED | 1.98 | 1.54 |
| FvH4_3g36810 | Abscisic acid 8′-hydroxylase 2 | 2.49 | 1.50 | |
| FvH4_2g14100 | Abscisic acid 8′-hydroxylase 3 | −2.30 | −0.57 | |
| FvH4_6g53030 | β-Glucosidase 13 | 1.36 | −0.11 | |
| FvH4_6g39430 | UDP-glucuronosyl and UDP-glucosyl transferase | 3.04 | 0.95 | |
| FvH4_3g16470 | Abscisic acid receptor PYL3 | −3.29 | −2.15 | |
| FvH4_3g07720 | Abscisic acid receptor PYL13 | −3.12 | −3.87 | |
| Jasmonate | FvH4_2g40510 | Allene oxide synthase 1, AOS1 | 2.39 | 0.11 |
| FvH4_2g07410 | Allene oxide synthase 3, AOS3 | 3.04 | 0.84 | |
| FvH4_5g32640 | Oxophytodienoate reductase 1, OPR1 | 1.72 | 0.38 | |
| FvH4_5g32690 | NADH oxidase family, OPR2 | 4.16 | 1.73 | |
| FvH4_1g08454 | NADH oxidase family, OPR3 | 3.32 | 2.14 | |
| FvH4_7g00440 | Lipoxygenase, LOX | 2.85 | 2.18 | |
| FvH4_2g39441 | Jasmonic acid carboxyl methyltransferase 2 | 1.97 | 0.77 | |
| Gibberellin | FvH4_3g02670 | Gibberellin 2-β-dioxygenase 8 | 1.33 | 0.91 |
| FvH4_2g38480 | Gibberellin-regulated protein 1 | 1.02 | −0.31 | |
| FvH4_6g48590 | Gibberellin-regulated protein 4 | −2.63 | −0.49 | |
| FvH4_5g14950 | Gibberellin-regulated protein 6 | −3.01 | −1.97 | |
| Transcription factors | ||||
| AP2/ERF family | FvH4_7g30930 | Ethylene-responsive transcription factor CRF3 | 1.89 | 1.72 |
| FvH4_1g03190 | Ethylene-responsive transcription factor WIN1 | −1.11 | −0.49 | |
| FvH4_6g26090 | Ethylene-responsive transcription factor 53 | 1.36 | 0.56 | |
| FvH4_4g03460 | Ethylene-responsive transcription factor 98 | −1.11 | −0.06 | |
| FvH4_2g13240 | Ethylene-responsive transcription factor 110 | 3.52 | −0.73 | |
| bZIP family | FvH4_2g36400 | bZIP transcription factor 9 | −1.47 | −0.88 |
| FvH4_7g25870 | bZIP transcription factor 18 | −1.05 | −1.01 | |
| FvH4_6g26970 | bZIP transcription factor 34 | −2.3 | −1.68 | |
| FvH4_3g24830 | bZIP transcription factor TGA | 1.25 | 0.34 | |
| bHLH family | FvH4_4g07090 | Transcription factor bHLH35 | 1.61 | 0.18 |
| FvH4_7g24720 | Transcription factor bHLH51 | −1.27 | 0.10 | |
| FvH4_2g09030 | Transcription factor bHLH63 | −1.29 | −0.61 | |
| FvH4_7g17790 | Transcription factor bHLH67 | −1.03 | −0.65 | |
| FvH4_1g18930 | Transcription factor bHLH93 | −1.63 | 0.09 | |
| FvH4_2g39400 | Transcription factor bHLH110 | 1.75 | 0.52 | |
| FvH4_4g20270 | Transcription factor bHLH148 | −1.14 | −0.95 | |
| FvH4_3g09050 | Transcription factor HLH162 | −3.85 | −0.16 | |
| MYB family | FvH4_5g19400 | Transcription factor MYB3 | −1.62 | −2.35 |
| FvH4_7g16990 | MYB-like transcription factor 4 | 1.74 | 1.76 | |
| FvH4_4g03610 | Transcription factor MYB14 | 1.99 | 0.43 | |
| FvH4_6g08620 | Transcription factor MYB61 | −1.84 | −0.75 | |
| FvH4_5g11930 | Transcription factor MYB62 | 3.12 | 1.65 | |
| FvH4_1g02690 | Transcription factor MYB82 | −3.62 | −0.39 | |
| FvH4_2g31100 | Transcription factor MYB123-like | 2.43 | 2.34 | |
| NAC family | FvH4_3g20700 | NAC transcription factor RIF | 1.68 | −0.74 |
| FvH4_2g16180 | NAC transcription factor 6 | 1.55 | 0.53 | |
| FvH4_6g51660 | NAC transcription factor 8 | 1.72 | 0.70 | |
| FvH4_6g33050 | NAC transcription factor 35 | −1.21 | −0.12 | |
| FvH4_5g21670 | NAC transcription factor 71 | 1.03 | 1.11 | |
| FvH4_6g19430 | NAC transcription factor 90 | 1.73 | 0.46 | |
| WRKY family | FvH4_1g16480 | WRKY transcription factor 15 | 1.24 | −0.09 |
| FvH4_3g39850 | WRKY transcription factor 24 | 1.15 | 0.02 | |
| FvH4_2g41070 | WRKY transcription factor 40 | 3.35 | 0.52 | |
| FvH4_1g26200 | WRKY transcription factor 42 | 1.87 | −0.54 | |
| FvH4_2g31400 | WRKY transcription factor 50 | 2.70 | 0.92 | |
| FvH4_5g15340 | WRKY transcription factor 72 | 2.42 | 0.97 | |
| Light responsive | FvH4_4g22790 | Light-regulated protein 1 | −2.06 | −0.39 |
| FvH4_2g20400 | Early light-induced protein 1 | 1.51 | 2.71 | |
| FvH4_7g15980 | Light-sensor protein kinase PHY1 | 1.07 | 0.17 | |
| FvH4_6g02940 | Light-inducible protein CPRF2 | −1.73 | −1.03 | |
| FvH4_7g26541 | Light-harvesting complex-like protein 3 | 1.36 | 2.21 |
FvWRKY50 regulated strawberry leaf senescence by directly targeting SAUR36
SAUR (small auxin up RNA) genes have been reported to be the key regulators of leaf senescence [12, 37]. RNA-seq data and qRT–PCR results showed that SAUR36 and SAUR72 were significantly upregulated in FvWRKY50 CR 2-9 and 3-16 plants (Fig. 2D and E). Further correlation analysis showed that FvWRKY50 was co-expressed with SAUR36 and other senescence-related genes, including senescence-associated carboxylesterase 101 (SG101), senescence-specific cysteine protease 39 (SAG39), senescence-associated transcription factor JUB1, WRKY70 [15, 16] and so on (Supplementary Data Fig. S4A; Supplementary Data Table S1). These results suggested that FvWRKY50 might be involved in leaf senescence in strawberry.
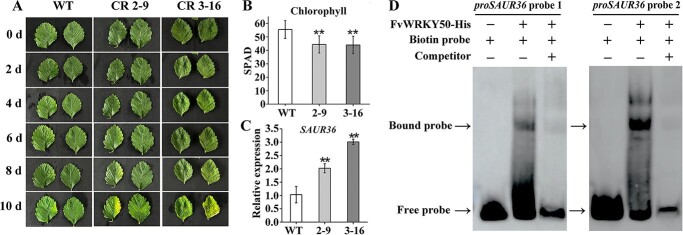
We then used detached leaves from WT, FvWRKY50 CR 2-9, and 3-16 lines to observe the process of leaf senescence. At 8 days after detachment FvWRKY50 CR 2-9 and 3-16 leaves began to turn yellow while WT leaves were still green, indicating that FvWRKY50 CR leaves initiated senescence more quickly than WT leaves. At 10 days after detachment the WT leaves began to turn yellow (Fig. 3A). We determined the chlorophyll contents at 10 days, and the results showed that chlorophyll content decreased more rapidly in FvWRKY50 CR leaves than WT leaves (Fig. 3B). The expression level of SAUR36 was significantly increased in FvWRKY50 CR plants compared with WT (Fig. 3C). These results indicated that FvWRKY50 regulated leaf senescence in strawberry, possibly by modulating the expression of senescence-related genes.
Figure 3.
. FvWRKY50 regulated strawberry leaf senescence by directly targeting the SAUR36 promoter. (A) Leaf senescence progress of WT, FvWRKY50 CR 2-9, and FvWRKY50 CR 3-16 leaves. Detached leaves of WT and FvWRKY50 CR plants were placed in Petri dishes with wet filter paper to observe leaf senescence phenotypes and were photographed every 2 days. (B) Chlorophyll content of detached leaves of WT, FvWRKY50 CR 2-9, and FvWRKY50 CR 3-16. A chlorophyll meter was used to determine chlorophyll content 10 days after detachment. (C) Expression level of SAUR36 in WT, FvWRKY50 CR 2-9, and FvWRKY50 CR 3-16 leaves at 10 days after detachment. qRT–PCR assays were used in this experiment. (D) EMSA assays showing the direct binding of FvWRKY50 to the SAUR36 promoter. The proSAUR36 probes 1 and 2 containing W-box were used as biotin probes. Unlabeled probes (200-fold excess) were used as competitors.
WRKY transcription factors regulate target genes by binding to the W-box elements of target gene promoter sequences. To determine whether FvWRKY50 could regulate senescence-related genes directly, we analyzed the promoter sequences of these genes and found there were two W-box elements at 1483 and 1636 bp upstream of ATG in the SAUR36 promoter. Electrophoretic mobility shift assay (EMSA) results showed that the recombinant FvWRKY50-His fusion protein could bind to the DNA probes containing the W-box motif 1 and 2 of the SAUR36 promoter, and the unlabeled probe could compete this binding effectively (Fig. 3D). These results suggested that FvWRKY50 negatively regulated strawberry leaf senescence by directly targeting senescence-related genes like SAUR36 to modulate their expression level.
FvWRKY50 mutation affected flowering time of strawberry by regulating expression of FvFT2, FvFT3, and FvCO
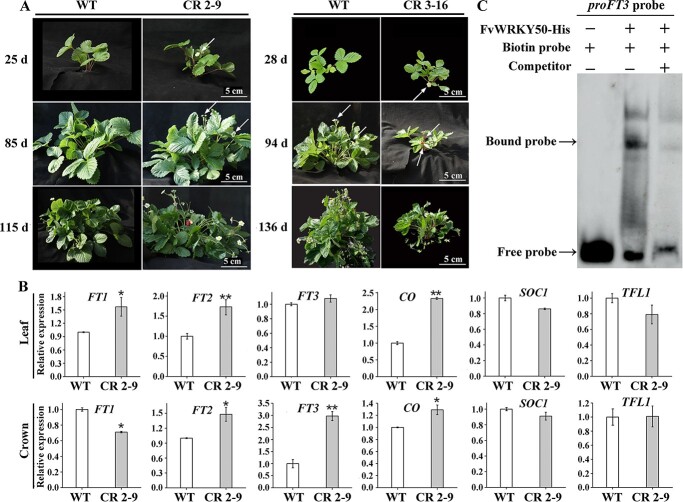
By continuous phenotype observation we found that the FvWRKY50 mutation induced an early flowering phenotype compared with WT plants. It took ~30 days for FvWRKY50 2-9 and 3-16 plants to come into the anthesis stage while for WT it took ~100 days (Fig. 4A). Previous studies have identified flowering-related genes in strawberry, including florigen FTs, TFL1, CO, and SOC1 [8, 28, 47]. We examined the expression of FvFT1/2/3, FvCO, FvSOC1, and FvTFL1 in leaf and crown. qRT–PCR results showed that the transcript levels of positive flowering regulators FvFT1, FvFT2, and FvCO were higher in FvWRKY50 CR 2-9 leaf than in WT leaf, while in the crown FvFT3 gene expression was higher in FvWRKY50 CR 2-9 than in WT (Fig. 4B). Promoter motif analysis showed that there were W-box motifs in the promoter sequence of FT3, and EMSA results showed that the recombinant FvWRKY50-His fusion protein could bind to a DNA probe containing the W-box motif of the FT3 promoter (Fig. 4C). These results suggested that FvWRKY50 played a negatively role in strawberry flowering by directly or indirectly regulating FT and CO gene expressions.
Figure 4.
FvWRKY50 mutation influenced flowering time of strawberry plant. (A) Phenotypes of WT and FvWRKY50 CR plants at different developmental stages. WT and FvWRKY50 CR 2-9 plant phenotypes were observed and photographed 25, 85, and 115 days after planting. WT and FvWRKY50 CR 3-16 plant phenotypes were observed and photographed 28, 94 and 136 days after planting. Flowers and fruits at 25, 85, 28 and 94 days are indicated by white arrows. (B) Relative expression levels of flowering-related genes in WT and FvWRKY50 CR 2-9. qRT–PCR assay was used to analyze the expression levels of FT1, FT2, FT3, CO, SOC1, and TFL1 in leaves and crown of WT and FvWRKY50 CR 2-9. (C) EMSA assays showing that FvWRKY50 could bind to the FT3 promoter directly. proFT3 probes containing W-box were used as biotin probes. Unlabeled probes (200-fold excess) were used as competitors.
FvWRKY50 was involved in the regulation of fruit ripening and anthocyanin accumulation in strawberry fruit
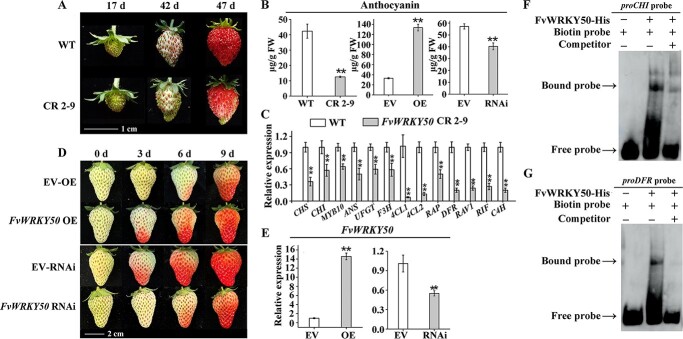
To evaluate the effects of FvWRKY50 mutation on strawberry fruit ripening, we observed the fruit phenotype of FvWRKY50 CR and WT plants. As homozygous mutation of FvWRKY50 made the fruits of FvWRKY50 CR 3-16 too small and severely deformed, which might affect observations (Supplementary Data Fig. S5), FvWRKY50 CR 2-9 and WT fruit phenotypes at different developmental stages were observed. We found that anthocyanin accumulation of FvWRKY50 CR 2-9 fruits was delayed compared with WT fruits (Fig. 5A), consistent with the lower anthocyanin content in FvWRKY50 CR 2-9 fruits (Fig. 5B). The expression levels of anthocyanin accumulation-related genes were determined by qRT–PCR and results showed that CHS, CHI, MYB10, ANS, UFGT, F3H, 4CL1, 4CL2, RAP, DFR, RAV1, RIF, and C4H were all downregulated significantly in FvWRKY50 CR 2-9 fruit compared with WT (Fig. 5C). Since RIF is also a key transcription factor affecting the formation of sugar, acid, and other qualities of strawberry fruit, FvWRKY50 may play a more important role rather than specifically regulating anthocyanin accumulation. We transiently overexpressed FvWRKY50 (FvWRKY50-OE) and knocked down FvWRKY50 (FvWRKY50-RNAi) in cultivated strawberry fruits. We observed an acceleration in anthocyanin accumulation in FvWRKY50-OE fruits and delayed anthocyanin accumulation of FvWRKY50-RNAi fruits during the ripening process compared with WT fruits (Fig. 5B and D). qRT–PCR results validated the overexpression and downregulation expression level of FvWRKY50 in FvWRKY50-OE and FvWRKY50-RNAi fruits respectively (Fig. 5E). Furthermore, EMSA results showed that FvWRKY50 could regulate anthocyanin accumulation in strawberry fruits by directly regulating FvCHI and FvDFR gene expression (Fig. 5F and G). These results showed that FvWRKY50 was an important regulator of fruit ripening, especially in anthocyanin accumulation.
Figure 5.
. FvWRKY50 regulated anthocyanin accumulation in strawberry fruit. (A) Anthocyanin accumulation phenotypes of WT and FvWRKY50 CR 2-9 fruits at different developmental stages. Fruits of WT and FvWRKY50 CR 2-9 were harvested and photographed 17, 42, and 47 days after anthesis. (B) Anthocyanin content of WT and FvWRKY50 CR 2-9 (left), EV-OE, and transient OE (middle), and EV-RNAi and transient RNAi (right) fruits. (C) Anthocyanin accumulation-related gene expression levels of WT and FvWRKY50 CR 2-9 fruits 47 days after anthesis. (D) Phenotypes of FvWRKY50-OE and FvWRKY50-RNAi fruits. Octoploid strawberry fruits at big green stage were selected for transient transformation. Control (EV-OE, EV-RNAi) and transient (FvWRKY50-OE, FvWRKY50-RNAi) fruits were photographed 0, 3, 6 and 9 days after injection. (E) FvWRKY50 gene expression levels in EV-OE and FvWRKY50-OE (transient OE) and EV-RNAi, FvWRKY50-RNAi (transient RNAi). qRT–PCR assay was used. (F, G) EMSAs showing that FvWRKY50-His protein could bind to CHI (F) and DFR (G) promoters directly. Negative control is shown in the first lane. Unlabeled probe (200-fold excess) was used as competitor.
FvWRKY50 was phosphorylated and degraded by FvMAPK3 under low temperature
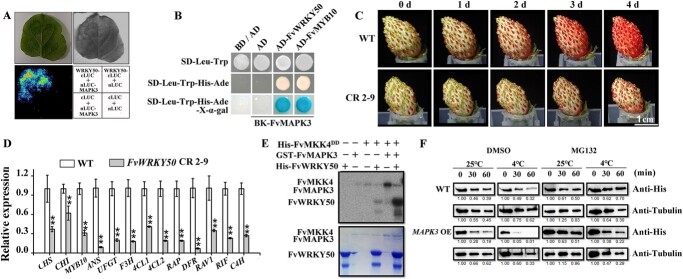
To reveal the molecular mechanism of FvWRKY50 in regulating strawberry fruit ripening, we screened the interacting proteins of FvWRKY50. Bimolecular fluorescence complementation (BiFC) and yeast two-hybrid (Y2H) results both indicated that FvWRKY50 interacted with strawberry MITOGEN-ACTIVATED PROTEIN KINASE3 (FvMAPK3; Fig. 6A and B). FvMAPK3 was reported as an important negative regulator of anthocyanin accumulation in strawberry fruits and could be activated by low temperature to repress anthocyanin accumulation [26].
Figure 6.
FvMAPK3 phosphorylated and degraded FvWRKY50 protein under low temperature to affect anthocyanin accumulation in strawberry fruit. (A) Luciferase complementary imaging assays showing the interaction between FvWRKY50 and FvMAPK3 in vivo. Various constructs were co-infiltrated into N. benthamiana leaves, and LUC activity was captured. (B) Y2H assays showing the interaction between FvWRKY50 and FvMAPK3. The interaction between FvMYB10 and FvMAPK3 was the positive control. (C) Anthocyanin accumulation phenotypes of WT and FvWRKY50 CR 2-9 fruits at 10°C for low temperature treatment. White stage fruits of WT and FvWRKY50 CR 2-9 were harvested and cultured in 88 mM sucrose solution at 10°C. Fruits were photographed at different days after treatment. (D) Anthocyanin accumulation-related gene expression of WT and FvWRKY50 CR 2-9 fruits at 4 days after low temperature treatment. (E) FvMAPK3 phosphorylated FvWRKY50 in vitro. His-FvMKK4DD, GST-FvMAPK3, and His-FvWRKY50 recombinant proteins were incubated in kinase reaction buffer and then detected by SDS–PAGE. The autoradiograph (top) and CBB staining (bottom) of the proteins are indicated. (F) Cell-free degradation assay of recombinant His-FvWRKY50. Total proteins of WT and FvMAPK3-OE fruits were extracted and incubated with recombinant His-FvWRKY50 protein at 25 and 4°C with 50 mM MG132 or DMSO. His-FvWRKY50 protein was detected with anti-His antibody and tubulin was detected with anti-tubulin.
To evaluate whether FvWRKY50 was involved in low temperature-repressed anthocyanin accumulation, we examined the effects of 10°C treatment on anthocyanin accumulation in WT and FvWRKY50 CR 2-9 fruits. As shown in Fig. 6C, the accumulation of anthocyanin in FvWRKY50 CR 2-9 fruits was significantly delayed compared with that in WT fruits, and accordingly the anthocyanin accumulation-related genes were all downregulated significantly (Fig. 6D). These results indicated that FvWRKY50 CR 2-9 fruit was more sensitive to low temperature and FvWRKY50 acted as a downstream component of low temperature signal during anthocyanin accumulation in strawberry fruit.
To test whether FvWRKY50 was involved in FvMAPK3-mediated anthocyanin accumulation as its phosphorylation substrate, we conducted an in vitro kinase assay using GST-FvMAPK3 protein and His-FvWRKY50 protein. The results showed that FvMAPK3 was activated by His-FvMKK4DD and then phosphorylated His-FvWRKY50, indicating that FvWRKY50 was the phosphorylation substrate of FvMAPK3 (Fig. 6E). In addition, the proteasome inhibitor MG132 decelerated the FvMAPK3-mediated degradation of FvWRKY50, which suggested that the degradation of FvWRKY50 was regulated by 26S proteasome-mediated ubiquitination (Fig. 6F). These results indicated that FvWRKY50 was an important downstream regulator of FvMAPK3 in low temperature-mediated anthocyanin accumulation in strawberry fruits.
In conclusion, FvWRKY50 is an important gene regulating vegetative and reproductive growth of strawberry, and plays a key role in strawberry fruit response to low temperature (Fig. 7). The identification of this gene provides valuable clues to reveal the molecular regulation mechanism of strawberry growth and fruit ripening, especially the regulation of plant growth and fruit ripening by low temperature. The creation of FvWRKY50 mutants, especially the biallelic mutant, could lay an important foundation for improving strawberry flowering time by genome editing technology.
Figure 7.
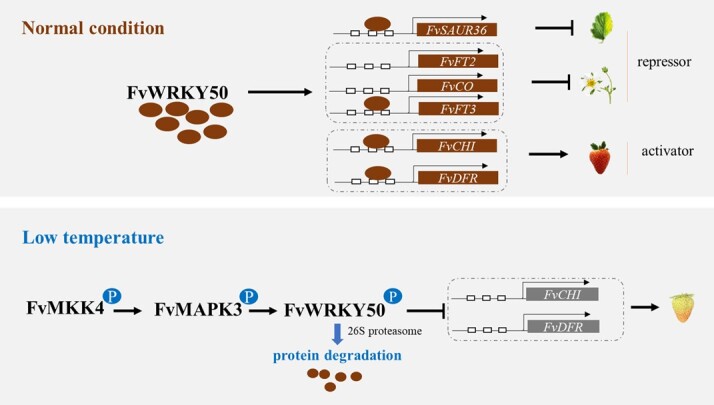
FvWRKY50 is an important gene that regulates both vegetative growth and reproductive growth in strawberry. During vegetative growth, FvWRKY50 negatively regulates leaf senescence by directly targeting FvSAUR36. Then FvWRKY50 affects flowering time mainly by repression the expression of FvFT2, FvFT3, and FvCO. During the fruiting stages, complete mutation of FvWRKY50 leads to abnormal fertility of strawberry. High expression of FvWRKY50 can promote ripening and coloring of strawberry fruit. However, at low temperature FvWRKY50 is phosphorylated by FvMAPK3, and ubiquitination-mediated degradation of FvWRKY50 protein occurs, resulting in delayed fruit coloring. In conclusion, FvWRKY50 is a key gene in the regulation of strawberry growth and fruit ripening. It is of great value to continue to explore the regulatory mechanisms of FvWRKY50 in different biological processes of strawberry.
Discussion
WRKY transcription factors belong to a large gene family and have diverse functions [32,55]. However, relative to other species the regulatory function of specific WRKY transcription factors in strawberry is little known. In this study we identified a WRKY transcription factor, FvWRKY50, in diploid strawberry. Phylogenetic analysis showed that FvWRKY50 was close to RcWRKY45 and AtWRKY75 in genetic relationship (Fig. 1A). The function of RcWRKY45 has not been clear, while several studies have indicated that AtWRKY75 functions in multiple physiological processes, including nutrient starvation response, root development, leaf senescence, and flowering [6, 9, 33, 50]. Our findings suggest that FvWRKY50 regulated strawberry leaf senescence and flowering negatively while regulating strawberry fruit ripening and anthocyanin accumulation positively (Figs 2–5). Interestingly, FvWRKY50 had no transcriptional activation activity (Fig. 1C). It is speculated that FvWRKY50 can recruit different transcription factors to form a transcription complex and act as a transcriptional activator or repressor, which may be an important reason for the diverse functions of FvWRKY50.
Although only FvWRKY50-homozygous plants dwarfed and had smaller plant size, there was no significant difference in the decrease in auxin content between biallelic and homozygous mutants, indicating that the difference did not result from the change in auxin content (Fig. 2A and C). Transcriptome data analysis and qRT–PCR results both revealed that many auxin-related gene transcript levels were more significantly downregulated in FvWRKY50 CR 3-16 plants, including auxin transporters ATL2/3, auxin binding proteins (ABPs), and auxin-responsive protein genes like IAAs and ARGs (Fig. 2D). Based on the above results, we speculated that the differences in plant type between the two mutants might be associated with their sensitivity to auxin or not be related to auxin.
SAUR is an important gene family that has attracted much attention in recent years. In addition to participating in auxin-mediated plant development, SAUR can also play its biological functions independently of auxin [37]. However, little is known about SAURs in strawberry. In this study, we found that the expression of three SAUR genes, SAUR36, SAUR72, and SAUR50, was significantly regulated in FvWRKY50 mutants and that FvWRKY50 bound the promoter of SAUR36 (Fig. 3). The homolog gene of SAUR36 has been demonstrated as an important regulator of leaf senescence, and we found that SARU36 was much more significantly upregulated in senescent leaves of FvWRKY50 mutants [12] (Fig. 3A). Our finding validates that SAUR36 can conservatively regulate leaf senescence in A. thaliana and strawberry, and can also provide valuable clues for further revealing the function of SAUR genes such as SARU72 and SAUR50 in strawberry.
Besides vegetative growth, FvWRKY50 was also involved in the regulation of reproductive growth, including flowering and fruit ripening (Figs 4 and 5). Flowering time is an important economic character of strawberry which determines the ripening period. Previous studies have identified a few important structural genes involved in strawberry flowering using transgenic materials, including FvFT1, FvFT2, FvFT3, FvCO, FvSOC1, and FvTFL1 [8, 20, 28, 31]. However, the transcriptional regulatory mechanism of these genes is not clear. In this study, we demonstrated that FvWRKY50 bound the promoter of FvFT3 and significantly regulated the expression of FvFTs and FvCO in strawberry crown (Fig. 4). Considering that overexpressed FvFT3 regulated strawberry plant branching and FvWRKY50 homozygous mutant showed smaller plant size, FvWRKY50 may also function in the regulation of strawberry plant morphology [8]. In addition to FvWRKY50, FvWRKY71 has also been reported to be involved in flowering regulation by affecting the expression of FvFUL and FvLFY [22], suggesting that different WRKY genes may cooperatively regulate strawberry flowering by regulating different flowering structure genes. Our results also showed that other WRKY genes, such as FvWRKY53 and FvWRKY70, were significantly regulated in FvWRKY50 mutants (Fig. 2). The function of other FvWRKYs in flowering regulation needs to be examined in the future. Notably, overexpressed FvYUC6 or FvARF4 delayed or promoted flowering, suggesting that auxin may be the important hormone involved in flowering regulation in strawberry in a concentration-dependent or sensitive-dependent manner [7, 24]. Given that FvWRKY50 was highly associated with auxin biosynthesis and signaling transduction, whether and how FvWRKY50 participates in auxin-mediated flowering is worth further exploration.
While FvWRKY50 negatively regulated plant growth, flowering time, and leaf senescence, it regulated anthocyanin accumulation positively in strawberry fruit (Fig. 5). Besides structural genes, the transcription factor genes related to anthocyanin accumulation and fruit ripening, such as MYB10, RAP, RAV, and RIF, were also significantly regulated by FvWRKY50, suggesting that FvWRKY50 may be involved in the regulation of fruit ripening upstream of these important transcription factors. The specific roles of FvWRKY50 in fruit ripening and the formation of other quality indexes is worth further study. Additionally, the FvWRKY50 homozygous mutant hardly bore normal fruits, indicating that FvWRKY50 was also involved in the regulation of strawberry fertility, which may affect the pollination and fertilization process (Supplementary Data Fig. S5).
Low temperature is one of the important environmental factors affecting fruit ripening, especially anthocyanin accumulation, in strawberry fruit [26]. However, little is known about how low temperature affects anthocyanin accumulation in strawberry fruits. Recently, we revealed that protein kinase FvMAPK3 was the critical regulator mediating low-temperature repression of anthocyanin accumulation by phosphorylating FvMYB10 and FvCHS1 [26]. In this study, we found that FvWRKY50 acted as the substrate of FvMAPK3 in low-temperature repression of anthocyanin accumulation in strawberry fruit (Figs 5 and 6). FvMAPK3 could induce the degradation by phosphorylating FvWRKY50 (Fig. 6). Previous studies have demonstrated that different members of MAPK and WRKY can form various phosphorylation modules to fine-regulate disease resistance signals in A. thaliana and rice [30, 40]. Considering the multiple regulatory functions of FvWRKY50, further exploration of whether WRKY50 can form phosphorylation modules with other MAPKs will provide clues for the identification of new genes regulating strawberry development and fruit ripening, and lay a foundation for further revealing the post-transcriptional regulatory mechanism of strawberry development and fruit ripening. Additionally, given the important roles of low temperature in the regulation of flowering and fruit ripening, it is necessary to further clarify whether other biological functions of FvWRKY50 are also related to its involvement in low temperature response.
Materials and methods
Plant materials and growing conditions
In this study diploid strawberry Fragaria vesca cultivar ‘Fragola di Bosco’ and octoploid strawberry F. × ananassa Duch. cultivar ‘Benihoppe’ were used. Strawberry plants were grown under controlled photoperiod (12 hours light/12 hours dark) with a light intensity of 200–300 μmol m−2 s−1 (white fluorescent tube, T5, 14 W). The temperature was 25°C in the daytime and 15°C at night, and the relative humidity was controlled at 70%.
After rooting in tissue culture bottles, the transgenic plants were transplanted into 10-cm seedling pots. After 1 month of culture, the transgenic plants were transplanted together with WT into large white pots and filed for phenotype observation.
Low temperature treatment
To observe the effects of low temperature on anthocyanin accumulation, detached white fruits of WT and FvWRKY50 CR 2-9 were cultured in 88 mM sucrose solution at 10°C under a light intensity of 50 μmol m−2 s−1 in a growth chamber. Samples were frozen in liquid nitrogen after 4 days of treatment, and then stored at −80°C for anthocyanin determination and qRT–PCR analysis.
To analyze the effects of low temperature-induced phosphorylation of FvMAPK3 on FvWRKY50 protein stability, white fruits of WT and FvMAPK3 OE were treated at 4°C and 25°C for 1 hour. Samples were frozen in liquid nitrogen after treatment, and then stored at −80°C for protein extraction.
Leaf senescence phenotype observation
Mature leaves of WT, FvWRKY50 CR 2-9, and FvWRKY50 CR 3-16 were detached and placed in Petri dishes with wet filter paper to observe the progress of leaf senescence. The Petri dishes were wrapped in plastic film to keep the high humidity. The leaves were photographed every 2 days and samples were frozen in liquid nitrogen 10 days after detachment.
Transient transformation for strawberry fruits
To transiently overexpress FvWRKY50 in strawberry fruits, the coding sequence of FvWRKY50 was cloned from strawberry cDNA and reconstructed into pH7WG2D vector using the Gateway method. To construct the FvWRKY50 RNAi plasmid, two parts of the FvWRKY50 coding sequence were cloned into pFGC5941 vector in opposite orientation. Agrobacterium EHA105 was used to transfect the fruit cells. When the Agrobacterium culture had reached OD600 0.6–0.8, the bacterial solution was centrifuged at 25°C for 10 minutes at 5000 g and then suspended in a buffer solution containing 10 mM MES pH 5.6, 10 mM MgCl2, and 200 μM acetosyringone, and shaken at 25°C for 1 hour.
In the transient transformation experiments, bacterial suspensions of EV-OE, FvWRKY50-OE, EV-RNAi or FvWRKY50-RNAi were injected into the octoploid strawberry fruits at the big green stage with 1-ml syringes, and 20 fruits were treated each time for biological repeats. The strawberry fruit was continuously photographed. The fruits were harvested 9 days after injection, frozen in liquid nitrogen, and stored at −80°C for subsequent gene expression analysis and anthocyanin detection.
Stable transformation of diploid strawberry
Genome editing of FvWRKY50 was performed using the CRISPR/Cas9 system, and two sgRNAs were constructed in the coding sequence region of FvWRKY50 and cloned into pYLCRISPR/Cas9 vector. Subsequently, the constructed FvWRKY50 CR vector was transformed into diploid strawberry as described previously [29]. After screening in medium containing 2 mg/l hygromycin, DNA was extracted from strawberry seedlings with hygromycin resistance and sequenced to validate the editing method. The specific primers listed in Supplementary Data Table S2 were used for sequencing analysis of target genes by PCR. WT plants and FvWRKY50 mutants were subcultured at the same time, and then transplanted into the growth chamber or greenhouse according to the same procedure. At least three plants of each edited type were used for phenotype analysis.
Yeast two-hybrid assay
A GAL4-based Two-Hybrid System 3 (Clontech, Mountain View, CA, USA) was selected to conduct the Y2H assay. The FvWRKY50 coding sequence was cloned into pGADT7 vector and the FvMAPK3 coding sequence was cloned into pGBKT7 vector. The plasmids were co-transformed into yeast strain AH109 and cultured in −Leu−Trp and −Leu−Trp−His−Ade medium. After 3 days of culture, the colonies were stained with X-α-gal. Primers used for cloning are listed in Supplementary Data Table S2.
Luciferase complementary imaging assay
The full-length coding sequences of FvWRKY50 and FvMAPK3 were cloned into pCAMBIA1300-cLUC vector and pCAMBIA1300-nLUC vector, respectively. Then the nLUC or cLUC construct was transformed into Agrobacterium strain EHA105 and the bacterial solutions were co-transformed at a 1:1 ratio into N. benthamiana leaves to observe the luciferase activity. Primers used for cloning are listed in Supplementary Data Table S2.
Protein subcellular localization
The coding sequence region of FvWRKY50 was cloned into the pSuper-GFP vector, and the constructed vector was transformed into Agrobacterium strain GV3101. Buffer solution with Agrobacterium tumefaciens was infiltrated into N. benthamiana leaves at the seedling age of 50 days. After 2–3 days, tobacco epidermal cells were observed using a confocal laser scanning microscope (Leica SP8). Primers used for constructs are listed in Supplementary Data Table S2.
Recombinant protein production and purification
Full-length cDNAs of FvWRKY50, FvMAPK3, and FvMAPKK4 were amplified and connected to the vectors pET30a, pGEX4T1, and pET30a respectively to generate the vectors His-FvWRKY50, GST-FvMAPK3, and His-FvMAPKK4 (FvMKK4DD, activated state of FvMKK4). The plasmids were then transformed into Escherichia coli BL21 (DE3) cells. Recombinant proteins of His-FvWRKY50, GST-FvMAPK3, and His-FvMAPKK4 were purified using glutathione agarose beads (GE Healthcare) and Ni-NTA agarose (Novagen).
Plant protein extraction and western blotting
Strawberry fruits were ground into powder in liquid nitrogen and then total proteins were extracted with extraction buffer [phosphate buffers pH 7.8, 1 mM EDTA, 10% (v/v) glycerol, 0.5% (v/v) Triton X-100, 1 mM DTT, 1 mM benzoyl sulfonyl fluoride, 1 × protease inhibitor mixture, and 1 × phosphatase inhibitor mixture]. Then, the homogenate was centrifuged at 13 000 g for 10 minutes, and the supernatant was analyzed using 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The antibodies anti-His (1:1000, CWBIO) and anti-tubulin (1:2000; Abmart, Shanghai, China) were used for western blotting.
In vitro phosphorylation analysis
Recombinant GST-FvMAPK3 was activated by His-MAPKK4 (FvMKK4DD) proteins at 30°C for 30 minutes in buffer (20 mM Tris–HCl at pH 7.5, 10 mM MgCl2, and 50 μM ATP). His-FvWRKY50 recombinant protein was then combined with activated GST-FvMAPK3 in kinase reaction buffer [20 mM Tris–HCl at pH 7.5, 10 mM MgCl2 25 μM ATP, and 1 μCi (γ-32p) ATP]. Then the reaction was conducted at 30°C for 30 minutes. The sample was then subjected to 10% SDS–PAGE to isolate the protein. The gel was exposed to a phosphor image screen for 12–24 hours and observed with a Typhoon 9410 imager.
Anthocyanin content determination
Anthocyanin was extracted using a plant anthocyanin content kit (Kemingshengwu, Suzhou, China). To 0.1 g of sample was added 1 ml of extract liquid, and ultrasonic extraction was performed for 2 hours followed by centrifugation at 8000 g for 10 min, and the supernatant was then analyzed. The content of anthocyanins was determined by the pH differential method.
RNA-seq analysis
Total RNA was extracted from WT, FvWRKY50 CR 2-9, and FvWRKY50 CR 3-16 mature leaves of the same seedling age using an E.Z.N.A. Total RNA Kit (Omega, Bienne, Switzerland). Sequencing was performed on an Illumina Novaseq platform and mapped by using the F. vesca subsp. vesca reference genome (https://www.rosaceae.org/rosaceae_downloads/Fragaria_vesca/Fvesca-genome.v4.0.a1/assembly/Fragaria_vesca_v4.0.a1.fasta.gz). Based on DESeq2 analysis, |log2 fold change| >1 and padj < 0.05 was considered as differential expression. Three independent biological replicates were analyzed for this experiment.
Real-time quantitative PCR
Following the instrument instructions, qRT–PCR was performed using Taq Pro Universal SYBR qPCR Master Mix (q712 Vazyme, Nanjing China) on a fluorescence quantitative PCR instrument (QuantStudio 6 Flex, Applied Biosystems, Thermo Fisher Scientific, USA). The internal reference was FvActin and the relative expression was analyzed by the 2-ΔΔCT method. Three biological replicates were analyzed for the qRT–PCR experiments. Primer sequences used for RT–qPCR are shown in Supplementary Data Table S2.
Cell-free degradation assays
Total proteins of WT and FvMAPK3-OE fruits after low temperature treatment were extracted and incubated with recombinant His-FvWRKY50 protein in buffer (50 mM Tris-MES pH 8.0, 500 mM sucrose, 1 mM MgCl2, 10 mM EDTA, and 5 mM DTT). For one treatment, 50 μM MG132 was added to the mixture to inhibit protein degradation while for the other treatment an equal amount of DMSO as control was added into the sample. Both samples were incubated at 37°C and a 30-μl aliquot was taken at 0, 30, and 60 minutes. His-FvWRKY50 protein was detected by western blotting using anti-His antibody.
Sequence analysis and phylogenetic analysis
ClustalX version 2.1 was used for protein sequence alignments. The phylogenetic tree was constructed by using the neighbor-joining method in MEGA version 5.0 software. The protein sequences of FvWRKY50, RcWRKY45 (XP_024181838.1), AtWRKY75 (NP_196812.1), NtWRKY75 (XP_016446514.1), OsWRKY72 (ALB35168.1), and SlWRKY45 (XP_004233585.1) were used to construct the phylogenetic tree.
Supplementary Material
Contributor Information
Yating Chen, Department of Pomology, College of Horticulture, China Agricultural University, Beijing, 10093, China.
Liping Liu, Department of Pomology, College of Horticulture, China Agricultural University, Beijing, 10093, China.
Qianqian Feng, Department of Pomology, College of Horticulture, China Agricultural University, Beijing, 10093, China.
Chuang Liu, Department of Pomology, College of Horticulture, China Agricultural University, Beijing, 10093, China.
Yujuan Bao, Department of Pomology, College of Horticulture, China Agricultural University, Beijing, 10093, China.
Nan Zhang, Department of Pomology, College of Horticulture, China Agricultural University, Beijing, 10093, China.
Ronghui Sun, Department of Pomology, College of Horticulture, China Agricultural University, Beijing, 10093, China.
Zhaonan Yin, Department of Pomology, College of Horticulture, China Agricultural University, Beijing, 10093, China.
Chuanfei Zhong, Beijing Engineering Research Center for Strawberry, Institute of Forestry and Pomology, Beijing Academy of Agriculture and Forestry Sciences, Beijing, 100093, China.
Yuanhua Wang, Department of Agronomy and Horticulture, Jiangsu Vocational College of Agriculture and Forestry, Jiangsu, 212400, China; Engineering and Technical Center for Modern Horticulture, Jiangsu, 212400, China.
Qian Li, Department of Pomology, College of Horticulture, China Agricultural University, Beijing, 10093, China.
Bingbing Li, Department of Pomology, College of Horticulture, China Agricultural University, Beijing, 10093, China.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32222074, 32072551), the National Key Research and Development Program of China (2022YFD2100102-3), 111 Project (B17043), Beijing Innovation Consortium of Agriculture Research System (BAIC04-2022), and the 2115 Talent Development Program of China Agricultural University.
Author contributions
B. L. designed the study. Y. C. carried out the majority of the experiments and analyzed the data. L. L., Q. F., C. L., Y. B., N. Z., R. S., Z. Y. assisted to do some experiments. C. Z. and Y. W. provided advice and support for the study. B. L and Q. L. wrote the manuscript. All authors read and approved the manuscript.
Data availability
Data supporting the results reported in the paper are available in the main text and supplementary data.
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at Horticulture Research online.
References
- 1. Aharoni A, Ric De Vos CH, Wein Met al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001;28:319–32. [DOI] [PubMed] [Google Scholar]
- 2. Bakshi M, Oelmüller R. WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav. 2014;9:e27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cai J, Mo X, Wen Cet al. FvMYB79 positively regulates strawberry fruit softening via transcriptional activation of FvPME38. Int J Mol Sci. 2022;23:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castillejo C, Waurich V, Wanger Het al. Allelic variation of MYB10 is the major force controlling natural variation in skin and flesh color in strawberry (Fragaria spp.) fruit. Plant Cell. 2020;32:3723–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cherian S, Figueroa CR, Nair H. ‘Movers and shakers’ in the regulation of fruit ripening: a cross-dissection of climacteric versus non-climacteric fruit. J Exp Bot. 2014;65:4705–22. [DOI] [PubMed] [Google Scholar]
- 6. Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007;143:1789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dong X, Li Y, Guan Yet al. AUXIN-induced AUXIN RESPONSE FACTOR4 activates APETALA1 and FRUITFULL to promote flowering in woodland strawberry. Hortic Res. 2021;8:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaston A, Potier A, Alonso Met al. The FveFT2 florigen/FveTFL1 antiflorigen balance is critical for the control of seasonal flowering in strawberry while FveFT3 modulates axillary meristem fate and yield. New Phytol. 2021;232:372–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo P, Li Z, Huang Pet al. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell. 2017;29:2854–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han Y, Dang R, Li Jet al. SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2.6, an ortholog of OPEN STOMATA1, is a negative regulator of strawberry fruit development and ripening. Plant Physiol. 2015;167:915–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He X, Wang C, Wang Het al. The function of MAPK cascades in response to various stresses in horticultural plants. Front Plant Sci. 2020;11:952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hou K, Wu W, Gan SS. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013;161:1002–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang JJ, Ma S, Ye Net al. WRKY transcription factors in plant responses to stress. J Integr Plant Biol. 2016;59:86–101. [DOI] [PubMed] [Google Scholar]
- 14. Jia S, Wang Y, Zhang Get al. Strawberry FaWRKY25 transcription factor negatively regulated the resistance of strawberry fruits to Botrytis cinerea. Genes. 2021;12:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim HJ, Nam HG, Lim PO. Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol. 2016;33:48–56. [DOI] [PubMed] [Google Scholar]
- 16. Kim J, Kim JH, Lyu JIet al. New insights into the regulation of leaf senescence in Arabidopsis. J Exp Bot. 2018;69:787–99. [DOI] [PubMed] [Google Scholar]
- 17. Klee HJ, Giovannoni JJ. Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet. 2011;45:41–59. [DOI] [PubMed] [Google Scholar]
- 18. Komis G, Šamajová O, Ovečka Met al. Cell and developmental biology of plant mitogen-activated protein kinases. Annu Rev Plant Biol. 2018;69:237–65. [DOI] [PubMed] [Google Scholar]
- 19. Kou X, Feng Y, Yuan Set al. Different regulatory mechanisms of plant hormones in the ripening of climacteric and non-climacteric fruits: a review. Plant Mol Biol. 2021;107:477–97. [DOI] [PubMed] [Google Scholar]
- 20. Koskela EA, Mouhu K, Albani MCet al. Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiol. 2012;159:1043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar R, Khurana A, Sharma AK. Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot. 2014;65:4561–75. [DOI] [PubMed] [Google Scholar]
- 22. Lei Y, Sun Y, Wang Bet al. Woodland strawberry WRKY71 acts as a promoter of flowering via a transcriptional regulatory cascade. Hortic Res. 2020;7:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li X, Wang X, Zhang Yet al. Regulation of fleshy fruit ripening: from transcription factors to epigenetic modifications. Hortic Res. 2022;9:uhac013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu H, Xie WF, Zhang Let al. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J Integr Plant Biol. 2013;56:350–63. [DOI] [PubMed] [Google Scholar]
- 25. Ma C, Xiong J, Liang Met al. Strawberry WRKY transcription factor WRKY50 is required for resistance to necrotrophic fungal pathogen Botrytis cinerea. Agronomy. 2021;11:2377. [Google Scholar]
- 26. Mao W, Han Y, Chen Yet al. Low temperature inhibits anthocyanin accumulation in strawberry fruit by activating FvMAPK3-induced phosphorylation of FvMYB10 and degradation of chalcone synthase 1. Plant Cell. 2022;34:1226–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martín-Pizarro C, Vallarino JG, Osorio Set al. The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell. 2021;33:1574–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muñoz-Avila JC, Prieto C, Sánchez-Sevilla JFet al. Role of FaSOC1 and FaCO in the seasonal control of reproductive and vegetative development in the perennial crop Fragaria × ananassa. Front Plant Sci. 2022;13:971846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oosumi T, Gruszewski HA, Blischak LAet al. High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta. 2006;223:1219–30. [DOI] [PubMed] [Google Scholar]
- 30. Qiu JL, Fiil BK, Petersen Ket al. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 2008;27:2214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rantanen M, Kurokura T, Jiang Pet al. Strawberry homologue of TERMINAL FLOWER1 integrates photoperiod and temperature signals to inhibit flowering. Plant J. 2015;82:163–73. [DOI] [PubMed] [Google Scholar]
- 32. Rinerson CI, Rabara RC, Tripathi Pet al. The evolution of WRKY transcription factors. BMC Plant Biol. 2015;15:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rishmawi L, Pesch M, Juengst Cet al. Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiol. 2014;165:186–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rushton PJ, Somssich IE, Patricia Ret al. WRKY transcription factors. Plant Signal and Behavior. 2010;15:247–58. [DOI] [PubMed] [Google Scholar]
- 35. Sánchez-Gómez C, Posé D, Martín-Pizarro C. Insights into transcription factors controlling strawberry fruit development and ripening. Front Plant Sci. 2022;13:1022369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seymour GB, Østergaard L, Chapman NHet al. Fruit development and ripening. Annu Rev Plant Biol. 2013;64:219–41. [DOI] [PubMed] [Google Scholar]
- 37. Stortenbeker N, Bemer M. The SAUR gene family: the plant’s toolbox for adaptation of growth and development. J Exp Bot. 2019;70:17–27. [DOI] [PubMed] [Google Scholar]
- 38. Symons GM, Chua YJ, Ross JJet al. Hormonal changes during non-climacteric ripening in strawberry. J Exp Bot. 2012;63:4741–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vallarino JG, Merchante C, Sánchez-Sevilla JFet al. Characterizing the involvement of FaMADS9 in the regulation of strawberry fruit receptacle development. Plant Biotechnol J. 2020;18:929–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang L, Ruan YL. Regulation of cell division and expansion by sugar and auxin signaling. Front Plant Sci. 2013;4:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang S, Han S, Zhou Xet al. Phosphorylation and ubiquitination of OsWRKY31 are integral to OsMKK10-2-mediated defense response in rice. Plant Cell. 2023;00:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wani SH, Anand S, Singh Bet al. WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 2021;40:1071–85. [DOI] [PubMed] [Google Scholar]
- 43. Wei L, Mao W, Jia Met al. FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J Exp Bot. 2018;69:4805–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wei W, Chai Z, Xie Yet al. Bioinformatics identification and transcript profile analysis of the mitogen-activated protein kinase gene family in the diploid woodland strawberry Fragaria vesca. PLoS One. 2017;12:e0178596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei W, Cui MY, Hu Yet al. Ectopic expression of FvWRKY42, a WRKY transcription factor from the diploid woodland strawberry (Fragaria vesca), enhances resistance to powdery mildew, improves osmotic stress resistance, and increases abscisic acid sensitivity in Arabidopsis. Plant Sci. 2018;275:60–74. [DOI] [PubMed] [Google Scholar]
- 46. Wei W, Hu Y, Han YTet al. The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: identification and expression analysis under biotic and abiotic stresses. Plant Physiol Biochem. 2016;105:129–44. [DOI] [PubMed] [Google Scholar]
- 47. Wickland DP, Hanzawa Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol Plant. 2015;8:983–97. [DOI] [PubMed] [Google Scholar]
- 48. Xie YG, Ma YY, Bi PPet al. Transcription factor FvTCP9 promotes strawberry fruit ripening by regulating the biosynthesis of abscisic acid and anthocyanins. Plant Physiol Biochem. 2020;146:374–83. [DOI] [PubMed] [Google Scholar]
- 49. Yue M, Jiang L, Zhang Net al. Importance of FaWRKY71 in strawberry (Fragaria × ananassa) fruit ripening. Int J Mol Sci. 2022;23:12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang L, Chen L, Yu D. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 2018;176:790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang WW, Zhao SQ, Gu Set al. FvWRKY48 binds to the pectate lyase FvPLA promoter to control fruit softening in Fragaria vesca. Plant Physiol. 2022;189:1037–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Z, Shi Y, Ma Yet al. The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol J. 2020;18:2267–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou H, Li Y, Zhang Qet al. Genome-wide analysis of the expression of WRKY family genes in different developmental stages of wild strawberry (Fragaria vesca) fruit. PLoS One. 2016;11:e0154312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou H, Ren S, Han Yet al. Identification and analysis of mitogen-activated protein kinase (MAPK) cascades in Fragaria vesca. Int J Mol Sci. 2017;18:1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu W, Fu P and Lu J.. Grapevine WRKY transcription factors. Fruit Res. 2022; 2: 10. [Google Scholar]
- 56. Zheng Z, Zhang Y, Gao Yet al. GRAS family transcription factor FaSCL8 regulates FaVPT1 expression mediating phosphate accumulation and strawberry fruit ripening. Fruit Res. 2023; 3: 15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the results reported in the paper are available in the main text and supplementary data.