Abstract
A two-cell system for the stimulation of herpes simplex virus type 1 (HSV-1) from an in vitro model of long-term (quiescent) infection is described. Rat pheochromocytoma (PC12) cells differentiated with nerve growth factor were infected with HSV-1 strain 17. Little, if any, cytotoxicity was observed, and a quiescent infection was established. The long-term infection was characterized by the absence of all detectable virus in the culture medium and little, if any, detectable early or late viral-gene expression as determined by reverse transcriptase PCR analysis. The presence of HSV-1 DNA was determined by PCR analysis. This showed that approximately 180 viral genomes were present in limiting dilutions where as few as 16 cells were examined. The viral DNA was infectious, since cocultivation with human corneal fibroblasts (HCF) or human corneal epithelial cells (HCE) resulted in recovery of virus from most, if not all, clusters of PC12 cells. Following cocultivation, viral antigens appeared first on PC12 cells and then on neighboring inducing cells, as determined by immunofluorescent staining, demonstrating that de novo viral protein synthesis first occurred in the long-term-infected PC12 cells. Interestingly, the ability to induce HSV varied among the cell lines tested. For example, monkey kidney CV-1 cells and human hepatoblastoma HepG2 cells, but not mouse neuroblastoma cells or undifferentiated PC12 cells, mediated stimulation. This work thus shows that (i) quiescent HSV infections can be maintained in PC12 cells in vitro, (ii) HSV can be induced from cells which do not accumulate significant levels of latency-associated transcripts, and (iii) the activation of HSV gene expression can be induced via neighboring cells. The ability of adjacent cells to stimulate HSV gene expression in neuron-like cells represents a novel area of study. The mechanism(s) whereby HCF, HCE, and HepG2 and CV-1 cells communicate with PC12 cells and stimulate viral replication, as well as how this system compares with other in vitro models of long-term infection, is discussed.
All herpesviruses establish latent infections in their natural hosts. Following productive infection of permissive cells at the periphery, herpes simplex virus type 1 (HSV-1), for example, usually colonizes neurons of the peripheral nervous system (reviewed in reference 40). The virus may, from time to time and by unclear mechanisms, reactivate from latency and cause productive infection at or near the site of initial entry into the host.
The molecular and cellular mechanisms involved in establishing, maintaining, and mediating reactivation from latency are unclear. In recent years, studies have centered on the role of latency-associated transcripts (LAT) in latency (1, 3, 10, 15, 22, 23, 29, 31, 32, 35, 42, 43, 45, 53). Since LAT are the only family of transcripts detected by Northern blotting or conventional in situ hybridization in the latently infected neurons, expression of the more than 70 genes of HSV-1 is repressed (6, 9, 11, 14, 15, 46, 47). As many as 40,000 LAT copies may be present in each latently infected cell (51). However, it was shown that there is a group of neurons latently infected with HSV that have such low LAT levels that reverse transcription-PCR (RT-PCR) amplification is necessary for detection (38). Despite the convenience of LAT as a marker of latently infected cells in mice and rabbits, the percent HSV DNA-containing neurons in which abundant LAT have accumulated is 30% or less (12, 13, 19, 21, 33, 34a). Thus, there appear to be at least two populations of latently infected neurons with respect to LAT levels: those with abundant LAT and those with few or no detectable LAT. Although HSV mutants unable to produce LAT can reactivate, the reason that some, but not all, cells accumulate vast amounts of LAT and whether subpopulations of cells unable to accumulate LAT are competent to support reactivation, are not known. Most studies of latency and reactivation are conducted in animal models. However, the small number of neurons which harbor the viral genome, the complexity of the in vivo setting, and ethical constraints place limits on animal studies. Thus, an in vitro model which resembles natural latency is appealing. There have been a number of reports of tissue culture models of latency (20, 24, 41, 44, 50), some of which involve B lymphocytes or fibroblasts as the host cells (24, 50). However, studies with fibroblasts or lymphocytes may not be representative of neuronal-cell viral latency seen following natural infection. Perhaps the best-studied in vitro system is primary explant rat neonatal dorsal root ganglia, which has been shown to be dependent on nerve growth factor (NGF) for the maintenance of the latent viral state (44, 54, 55). However, preparation of dissected dorsal root ganglia is inconvenient, and they contain heterogeneous groups of cells, including support tissue as well as neurons. Furthermore, in all of the in vitro models mentioned above, either antiviral agents, such as acyclovir, nonpermissive temperatures, or low infectious doses were needed to facilitate the establishment of latency.
We, therefore, further explored the possibility of using the rat pheochromocytoma (PC12) tissue culture line as a model of long-term in vitro HSV infections which might resemble in vivo latency or other aspects of pathogenesis. In the presence of NGF, PC12 cells cease division, extend long processes which have been shown to support action potentials, and acquire many biochemical properties characteristic of the peripheral nervous system (17, 18). We have previously shown that HSV can establish a long-term infection of NGF-differentiated PC12 cells which in some respects resembles in vivo latency (2). In that system, while the level of virus in the medium was low, it was not zero. This suggested an occurrence of either a persistent infection or periodic reactivation. Moreover, reactivation from the majority of cells in the population involved NGF deprivation following the return of PC12 cells to a morphologically undifferentiated state. The NGF-deprived PC12 cells were then destroyed by HSV replication. Since reactivation of HSV from latently infected neurons probably occurs in an environment where neurons do not lose their differentiated phenotype, it was of interest to use the long-term-infected PC12 system to define conditions resulting in a complete absence of virus production. These conditions would then be used to pursue alternative mechanisms of reactivation of the viral genome. This article describes modifications of the PC12 cell system in which HSV-1 infection resolves into completely quiescent infection with respect to virus production.
Herpes simplex viruses are the leading cause of blindness among the infectious agents in the United States (34a). Herpes eye diseases may be the result of primary or recurrent infection (8, 52). In both cases, permissive corneal cells such as stromal fibroblasts and epithelial cells play a role either in supporting viral replication or in communicating with surrounding cells, perhaps neurons. The role of interaction between permissive corneal cells and latently infected neurons is uncertain.
In this report, a two-cell system of long-term infection from which HSV can apparently be stimulated is described. “Long-term infection” and “stimulation of virus” are used to describe this system instead of “latency” and “reactivation” in order to avoid confusion or overstatements. Long-term infection, as discussed here, can be achieved in NGF-treated PC12 cells with a high infectious dose (a multiplicity of infection [MOI] of 20), in the absence of antiviral agents, and at an incubation temperature of 37°C. Two weeks after infection, there was no detectable virus in the culture medium, and the cultures are therefore called long-term infections. However, stimulation of HSV, as defined by the appearance of infectious virus, could be induced by cocultivation with primary human corneal fibroblasts (HCFs), or human corneal epithelial cells (HCEs), and human hepatoblastoma cells (HepG2). Monkey CV-1 cells, under appropriate conditions, could also stimulate viral replication. However, mouse neuroblastoma (NA) or undifferentiated PC12 cells were ineffective. To our knowledge, this is the first report directly demonstrating a role of adjacent or supporting cells in influencing HSV gene expression in neuron-like cells. The possibility that information from some, but not all, types of permissive cells stimulates HSV gene activity in the long-term-infected PC12 cells is discussed, and the way in which this system compares to in vivo models and other in vitro models is considered.
MATERIALS AND METHODS
Virus and cells.
CV-1 cells (from the American Type Culture Collection [ATCC]) were maintained in Eagle’s minimal medium plus 5% calf serum. HepG2 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. HSV-1 strain 17 was prepared in CV-1 cells. Virus titers were determined by a standard plaque assay on a CV-1 monolayer under methylcellulose. PC12 cells from ATCC were grown in RPMI 1640 supplemented with 10% heat-inactivated horse serum and 5% heat-inactivated fetal bovine serum (PC12 medium). HCF and HCE cultures were prepared as previously described (7). NA cells (a gift from Bernard Dietzshold, Thomas Jefferson University) were maintained as described elsewhere (50). The medium used in cocultivations was the medium used for inducer cells.
Differentiation of PC12 cells.
To differentiate PC12 cells, 105 cells were seeded on poly-l-orinithine (Sigma, St. Louis, Mo.)-coated 25-cm2 culture flasks. The following day, cells were incubated in PC12 medium containing 100 ng of 2.5S NGF (Collaborative Biomedical Products, Bedford, Mass.)/ml for 1 week. Medium was replaced every 3 days. On day 7, 20 μM fluorodeoxyuridine (FdUrd) (Sigma) was added for 2 to 3 days to eliminate undifferentiated PC12 cells. Fresh NGF-supplemented medium was supplied thereafter.
Establishment of long-term HSV-1 infection.
Differentiated PC12 cultures were infected with HSV-1 strain 17 at an MOI of 20 (2 × 106 PFU/flask). Following a 1-h incubation at 37°C, cultures were treated with 3 ml of sodium citrate buffer (pH 3), modified as described elsewhere (4), for 30 s to 1 min to inactivate residual virus. Buffer was removed and flasks were rinsed with PC12 medium once. After low-pH treatment, cultures were incubated at 37°C with fresh medium containing NGF. To monitor for the release of HSV-1 progeny, the culture medium was collected and titered on CV-1 cells by a standard plaque assay.
RNA isolation and RT-PCR.
Total cellular RNA was isolated from cell culture by using the Trizol reagent (Gibco-BRL, Rockville, Md.). RNA was treated with DNase I (Boehringer Mannheim) to eliminate DNA contamination. One microgram of RNA was denatured with glyoxal and subjected to a 1% agarose gel for electrophoresis to determine the quality of RNA. cDNA was synthesized from 0.5 μg of total RNA isolated from each T25 flask in a total 20-μl volume with the SuperScript Pre-amplification System (Gibco-BRL) according to the manufacturer’s instructions. PCR amplifications were carried out with 2.5 U of Taq polymerase (Fisher Scientific, Pittsburgh, Pa.), primers, and 1 μl of cDNA in a 50-μl reaction volume. Primers and internal oligonucleotide probes used in this study are listed in Table 1. Amplifications consisted of 40 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 45 s, and extension at 72°C for 1 min. PCR products were resolved by electrophoresis through 1% agarose gels, visualized by ethidium bromide staining, and then transferred onto a Genescreen membrane (NEN Life Science Products, Boston, Mass.). The membranes were hybridized with the specific probe of interest by using the Renaissance CDP-Star Chemiluminescence detection system (NEN Life Science Products).
TABLE 1.
Primer and internal oligonucleotide probe sequences used in this study
| Primer | Sequences | Product size (bp) | Reference |
|---|---|---|---|
| Rat GAPDH | Sense: 5′ AAC CCT TCA TTG ACC TCA ACT A 3′ | 622 | 37 |
| Antisense: 5′ CTT CTC CAT GGT GGT GAA GAC 3′ | |||
| HSV ICP27 | Sense: 5′ TTT CTC CAG TGC TAC CTG AAG G 3′ | 283 | 11 |
| Antisense: 5′ TCA ACT CGC AGA CAC GAC TCG 3′ | |||
| Probe: 5′ TCC TTA ATG TCC GCC AGA CGC 3′ | |||
| HSV TK | Sense: 5′ ATG GCT TCG TAC CCC TGC CAT 3′ | 531 | 48 |
| Antisense: 5′ GGT ATC GCG CGG CCG GGT A 3′ | |||
| Probe: 5′ AAC CG CGT CTG CGT TCG ACC A 3′ | |||
| HSV gC | Sense: 5′ GAA ACT GCC TCC ACC GGG C 3′ | 628 | 48 |
| Antisense: 5′ GGC GTC ACC TCG CCG ATA ATC 3′ | |||
| Probe: 5′ CTC CGT TGT ATT CTG TCA 3′ | |||
| HSV LAT | Sense: 5′ CGG CGA CAT CCT CCC CCT AAG C 3′ | 149 | 11 |
| Antisense: 5′ GAC AGA CGA ACG AAA CGT TCC G 3′ | |||
| Probe: 5′ GTT TCT TTA ACC CGT CTG GGG 3′ |
Determination of the RT-PCR assay sensitivity.
To determine the RT-PCR assay sensitivity for each primer set, PCR fragments of each primer set were cloned into the pCR-Script cloning vector (Stratagene, La Jolla, Calif.) according to the manufacturer’s specifications. Each clone was then transcribed by in vitro transcription (Promega, Madison, Wis.) according to the manufacturer’s specifications. RNA was quantitated by spectrophotometry. Serial fivefold dilutions of a known amount of RNA mixed with 0.5 μg of PC12 total RNA were reverse transcribed by the first-strand cDNA synthesis system (Gibco-BRL), followed by PCR amplification with specific primers. After 40 cycles of amplification, the PCR product was resolved by agarose gel electrophoresis, visualized by ethidium bromide staining, and then transferred onto a Genescreen membrane (NEN Life Science Products). The membranes were hybridized with a specific internal oligonucleotide probe by using the Renaissance CDP-Star Chemiluminescence detection system (NEN Life Science Products). The assay sensitivity was determined as the highest dilution which yields a visible band after hybridization.
Quantification of HSV-1 genomes in long-term-infected PC12 culture.
One 25-cm2 flask of a PC12 culture with a long-term HSV-1 infection was trypsinized, and cells were collected by centrifugation, dispersed by passing through a 22-gauge needle three times, and then counted by trypan blue exclusion. A series of 10-fold dilutions of cells were prepared, treated with low pH (pH 2.4) as described previously (30), and then subjected to 40 cycles of PCR with a series of dilutions of known amounts of HSV-1 strain 17 DNA by using the primers specific for HSV-1 thymidine kinase (TK) gene. The PCR products were then resolved by agarose gel electrophoresis, visualized by ethidium bromide staining, and photographed. The photograph was then scanned with a Hewlett-Packard Scanjet 3C, and the band intensity was analyzed by using BioMax software (Kodak Scientific Imaging System). The quantity of HSV-1 genome in long-term-infected cells was determined by comparing the intensity readings of PCR-amplified bands.
Immunofluorescence and antibodies.
A total of 104 PC12 cells were seeded and differentiated on poly-l-orithine-coated one-well chamber slides. A long-term HSV-1 infection was established as described above. HepG2 cells were used to induce the quiescent viruses. To prevent virus spread from cell to cell, methylcellulose medium was added 4 h after overlay. At 48 h after stimulation, the culture was rinsed twice with phosphate-buffered saline (PBS) and then fixed with acetone-methanol (1:1) at −20°C for 10 min. After two washes with PBS and one wash with wash buffer (0.8% bovine serum albumin [BSA]–0.1% gelatin in PBS), the slides were incubated with the blocking reagent (0.8% BSA–0.1% gelatin–5% goat serum in PBS) at room temperature for 20 min. This was followed by a 1:100 dilution of rabbit polyclonal antibodies to the extreme C terminus of neurofilament H (Chemicon International Inc., Temecula, Calif.) and Cy3-conjugated affinity-purified anti-rabbit antibodies (Chemicon) with intervening washing steps with wash buffer and PBS. Finally, slides were incubated with a 1:50 dilution of fluorescein isothiocyanate (FITC)-conjugated polyclonal rabbit anti-HSV-1 and -2 antibody (Chemicon) and examined with a fluorescence microscope (Olympus BX-FLA system). The anti-HSV monoclonal antibody (MAb) 8D2 was prepared and purified as described previously (28). A 1:500 dilution of ascites fluid of MAb 8D2 was used; this dilution was shown by standard virus neutralization assays (28) to be sufficient to neutralize all 1,000 PFU of HSV in a reconstruction experiment in which 1,000 PFU of HSV were incubated with MAb.
RESULTS
Virus production following infection of NGF-treated PC12 cells as a function of time.
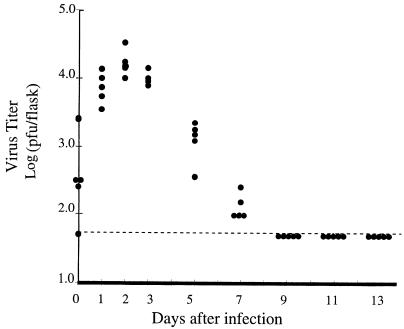
PC12 cells were seeded onto poly-l-ornithine coated flasks and differentiated with 100 ng of NGF/ml as described in Materials and Methods. One week later, almost all (more than 99%) of the PC12 cells in the culture were differentiated, on the basis of morphological evaluation (e.g., they contained processes at least 2 cell diameters in length). Since, in previous studies of NGF-differentiated cells (2), the population of undifferentiated cells appeared to persist, undifferentiated cells were eliminated from the cultures following 2 to 3 days of incubation in medium containing 20 μM 5-fluorodeoxyuridine. Differentiated cultures were then infected at an MOI of 20 with HSV-1 strain 17. One hour after inoculation, cultures were rinsed with sodium citrate, pH 3.0, to inactivate residual viruses. The efficiency of inactivation by sodium citrate buffer was greater than 99%, as shown in Table 2. After sodium citrate buffer treatment, the infected NGF-differentiated PC12 cultures were maintained continuously in NGF-supplemented medium. To monitor for virus growth, medium was collected and assayed for infectious particles on a CV-1 monolayer by a standard plaque assay. As shown in Fig. 1, in each of the five flasks examined, there was production of progeny virus, peaking and dropping at 2 to 3 days after infection, although the titer never exceeded 105 PFU per flask of 105 cells. Since each flask contains 105 cells, a low level of virus production could represent very inefficient viral production from a minority of the population, which tolerate synthesis and release of virus and then recover. Another possibility is that an antimitotic agent did not completely eliminate undifferentiated cells, and it is these cells which synthesize progeny after viral infection and are eventually eliminated from the culture by virus-induced cytopathic effect. In any event, following the initial period of virus production, there is a complete lack of detectable virus in the culture medium. This was true for each of the 25 flasks examined over more than three experiments. Cultures surviving infection with HSV-1 are, at this point, designated as long-term or quiescently infected cultures.
TABLE 2.
Inactivation of infectivity following sodium citrate (pH 3) incubationa
| Treatment | Virus titer (PFU/ml)b |
|---|---|
| Medium onlyc | 2.3 × 105 |
| 2.0 × 105 | |
| 2.7 × 105 | |
| 2.7 × 105 | |
| 3.6 × 105 | |
| Sodium citrate (pH 3)d | 8 × 102 |
| <10 | |
| 5 × 102 | |
| 5 × 101 | |
| 7 × 101 |
An NGF-differentiated PC12 culture was infected with 2 × 106 PFU of HSV-1 strain 17. After 1 h of incubation at 37°C, five infected flasks were treated with sodium citrate (pH 3), 3 ml each, for 1 min, which was then replaced with normal NGF medium. Five control flasks received only normal NGF medium. Two hours after treatment, 0.5 ml was collected from each flask and the infectious virus titer was determined on a CV-1 monolayer.
Each value represents the infectious virus titer from 1 of the 10 flasks described in footnote a.
Average infectious virus titer, 2.66 × 105 PFU/ml.
Inactivation efficiency, 99.7 to 99.99%.
FIG. 1.
HSV-1 production in NGF-treated PC12 cells as a function of time following infection. PC12 cells maintained in five separate flasks were differentiated with NGF and infected with HSV-1 as described in Materials and Methods. At the indicated times after infection, aliquots of medium were removed and the number of PFU was determined by a standard plaque assay on monolayers of CV-1 cells. Each value is presented as total PFU in each flask. Each dot represents the results from a different flask. The dashed line represents the minimum level of detection by the standard plaque assay, which is 40 PFU/flask.
HSV-1 DNA is detected in long-term-infected PC12 cultures.
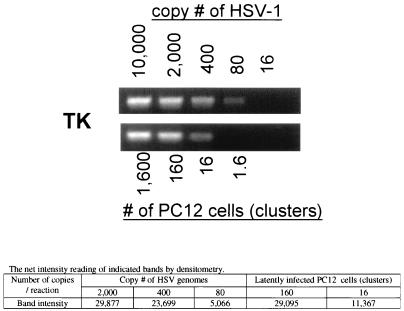
To investigate if the lack of virus production in long-term-infected cultures was due to the disappearance of viral genomes from the cultures, the presence of viral DNA was examined. Long-term HSV-infected PC12 cells were trypsinized, collected, passed through a 22-gauge needle three times, and counted. It was noticed that there were still cell clusters in cell suspensions, even after passing through the needle, which could be due to the aggregation of PC12 cells after NGF treatment. By counting cells from three independent flasks, it was determined that there was a range of 2 × 104 to 6 × 104 cells per flask, which were mostly within clusters. PCR analysis with primers specific for the TK gene was performed to quantify HSV genomes in limiting numbers of long-term-infected cells. Serial fivefold dilutions of known amounts of HSV-1 strain 17 DNA were used as a reference in the PCR analysis. As shown in Fig. 2, in this experiment, HSV DNA was detected in the dilutions containing as few as 16 cells per reaction. As shown by densitometry, the amplification of 80 to 400 copies of HSV-1 genome was within the linear range. The intensity reading of the ethidium bromide-stained band from 16 cells suggests that the quantity of HSV-1 genomes is approximately 180 copies. Therefore, 16 PC12 cells in long-term-infected cultures harbor a minimum of 11 genomes per cell on average, and approximately 22,000 to 67,000 copies of the HSV-1 genome are present per culture flask.
FIG. 2.
Detection of HSV-1 DNA in long-term-infected PC12 cells. A long-term-infected PC12 culture lacking detectable virus in the medium was trypsinized. Cells were collected, passed through a 22-gauge needle three times, counted, and diluted as indicated. Each cell dilution was treated with sodium acetate (pH 2.2) and subjected to PCR with the primer specific for the TK gene of HSV-1. A fivefold dilution of a known amount of HSV-1 DNA was used under the same PCR condition to estimate the number of HSV genomes in each reaction mixture as described in Materials and Methods. A densitometric analysis of the gel is given at the bottom.
HSV gene expression in long-term-infected cells.
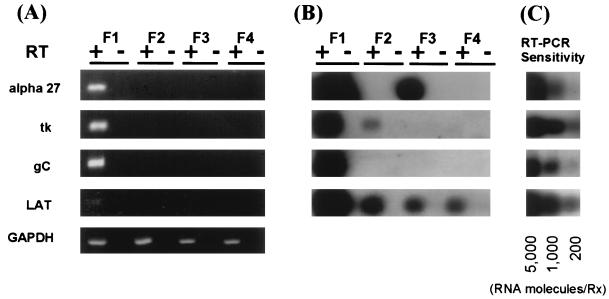
The hallmark of HSV latency is limited, if any, viral gene expression (47). The LAT appear to be the only viral-gene products which accumulate abundantly in neurons derived from latently infected humans and animals (15), although low levels of non-LAT viral gene expression have been observed by using extremely sensitive methods (26, 27). It was therefore of interest to determine which, if any, viral-gene products were present in long-term-infected PC12 cell cultures. RNA was isolated from long-term-infected PC12 cultures 2 weeks after infection, a time at which no virus was detected in the culture medium (as determined by sampling at three separate times). This was compared with RNA isolated from productively infected NGF-differentiated PC12 cells 24 h after infection with HSV-1. The expression of four key viral transcripts (alpha 27, TK, gC, and LAT) representing the different kinetic classes was examined. Primers and amplification schemes are described in Materials and Methods. Figure 3A shows the results of RT-PCR analysis of RNA isolated from the long-term-infected cultures. Clearly, although viral RNA is detected in samples from productively infected PC12 cells (Fig. 3A, lane F1), no HSV-1 transcripts were detected in samples from long-term-infected cultures (lanes F2 through F4). That is, neither alpha 27 transcripts, TK transcripts, gC transcripts, nor LAT had accumulated in the long-term-infected PC12 cells even to levels detectable by RT-PCR. Since glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA is a constitutively produced cellular (host) transcript, it was used to indicate the relative amount of RNA in each sample. The relatively equal amounts of GAPDH RNA detected in each sample suggest that similar amounts of RNA were present in each experimental set. Also, the failure to detect a PCR product from any samples when RT was omitted confirms that RNA and not DNA was being detected in the samples. Parenthetically, viral transcripts could not be detected in RNA derived from long-term-infected cultures, as determined by Northern hybridization (data not shown).
FIG. 3.
HSV-1 gene expression in long-term-infected PC12 cells. Total RNA was extracted from HSV-1-infected PC12 culture as described in Materials and Methods. A total of 0.5 μg of total RNA was reverse transcribed (RT), and the cDNA was then subjected to a PCR with the primers specific for HSV-1 alpha 27, TK, gC, and LAT genes as listed in Table 1. After 40 cycles of amplification, PCR products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining (A). The gel was then transferred onto a membrane and hybridized with specific probes as described in Materials and Methods. (B) Chemiluminescence exposure of the Southern hybridization of PCR products. Lanes F1 show the results obtained with cDNA from an NGF-differentiated PC12 culture 24 h after infection with HSV-1 strain 17, with (+) and without (−) RT. Lanes F2, F3, and F4, cDNAs from three individual long-term HSV-1-infected flasks. (C) Assay sensitivity of each primer after Southern hybridization of RT-PCR products.
Since extremely low levels of viral-gene expression have been reported to occur from various regions of the viral genome in long-term-infected cells (26, 27), we explored the possibility that HSV gene expression occurred in the long-term-infected PC12 cells at levels beneath the level of even direct RT-PCR detection. The sensitivity of transcript detection was thus enhanced approximately 100-fold by Southern blot hybridization of the products of the RT-PCR, immobilized in membranes, with labeled probes. The primers and specific internal oligonucleotide probes used are listed in Table 1 and described in Materials and Methods, and the assay sensitivity is shown in Fig. 3C. Figure 3B shows that when this sensitive assay was used to detect viral transcripts, gC RNA, a marker of late gene expression, could still not be detected in any samples of RNA from long-term-infected cultures. However, a minority of the long-term-infected cultures did appear to contain approximately 30,000 copies of alpha 27 (Fig. 3B, lane F3) and 1,200 copies of TK (Fig. 3B, lane F2) transcripts per flask. LAT could be detected in all three flasks containing long-term-infected cells by using this sensitive method of detection. These data suggest that viral-gene expression does occur in a limited way in a minority of the long-term-infected PC12 cell cultures, possibly in subpopulations of cells. The meaning of this is considered in the Discussion.
HCFs induce HSV from long-term-infected cultures.
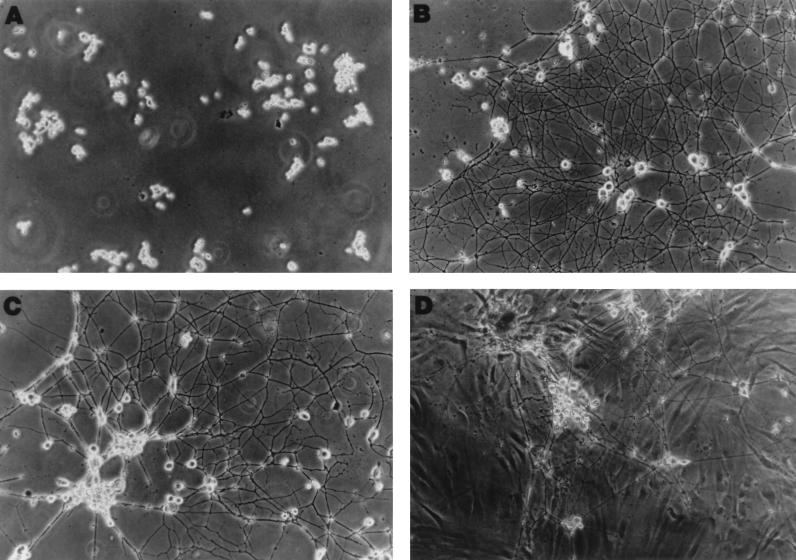
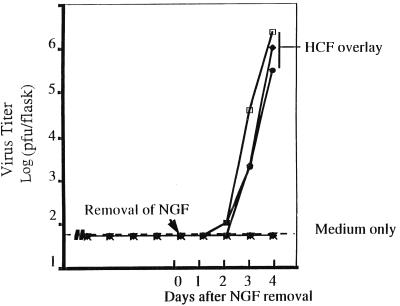
Under physiological settings of in vivo conditions, latently infected cells are in intimate contact with other, noninfected cells. To test the possibility that the HSV genome in long-term-infected PC12 cells could be influenced and even stimulated (“reactivated”) by neighboring noninfected cells, a two-cell system was used. HCFs were chosen as the inducer cells because they are sites of recurrent infection in the eyes and are highly permissive both in vivo and in vitro. In addition, these cells are in proximity to the axon processes of the peripheral ganglia, which may harbor latent HSV genomes in vivo (34). Figure 4A and B show PC12 cells prior to and 10 days after NGF treatment, respectively. No cytopathic effect was detectable in NGF-differentiated PC12 culture 13 days after HSV-1 strain 17 infection (Fig. 4C). In these experiments, NGF removal alone did not induce the appearance of virus in the culture medium (see Discussion). However, long-term-infected cultures that had been shown to be devoid of infectious virus were overlaid with confluent monolayers of primary HCFs (Fig. 4D). Curiously, HCFs appeared to move under PC12 cells to form a monolayer, while the differentiated morphology of PC12 remained. Figure 5 shows the dramatic virus growth following HCF overlay: although most flasks remained devoid of virus by day 2, infectious virus was recovered in the culture medium, with titers reaching 105 to 106 per flask by day 4. Therefore, long-term-infected PC12 cells contain infection-competent HSV genomes which were reactivated by an overlay of corneal fibroblasts.
FIG. 4.
Morphology of NGF-differentiated PC12 cells. A total of 105 undifferentiated PC12 cells were seeded onto a poly-l-ornithine-coated flask (A). The next day, cells were incubated in medium containing NGF for a week. On day 7, the antimitotic agent FdUrd was added to the growth medium for an additional 2 days to eliminate undifferentiated cells. (B) NGF-differentiated PC12 cells 10 days after treatment and prior to HSV-1 strain 17 infection. Thirteen days after infection, the culture was overlaid with HCFs. (C and D) Long-term-infected PC12 culture before and 1 day after HCF overlay, respectively.
FIG. 5.
Reactivation of HSV-1 in long-term-infected cultures. NGF was removed from long-term HSV-1-infected PC12 cultures at the time indicated by the arrow and then replaced with 5 ml of medium/flask or overlaid with HCFs. Culture medium was collected daily to monitor for virus content. There were three flasks for each group. The dashed line represents the minimum level of detection by the standard plaque assay, which is 40 PFU/flask.
The possibility that the ability of overlaying cells to mediate stimulation was restricted in cell type was tested. Long-term-infected cultures were overlaid on day 15 after HSV-1 infection with cells of the following lines: undifferentiated PC12, NA, HCF, HCE, CV-1, or HepG2. As expected, HCFs were able to stimulate virus 2 days after overlay, as shown in Table 3. Interestingly, only CV-1 cells, HCEs, and HepG2 cells could mediate the stimulation of virus from long-term-infected cultures. No virus was detectable in culture media of PC12 cells overlaid with cells of the neuronal lineage (PC12 or NA cells) for 4 days after overlay. Moreover, it is noted that, unlike the situation with HepG2 cells, the ability of CV-1 cells to mediate the stimulation of HSV from long-term-infected cultures declined with the age of the long-term cultures. This is considered further in the Discussion.
TABLE 3.
Long-term-infected cultures produce HSV-1 following overlay with inducer cellsa
| Cell type | Resultb on:
|
|||
|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | |
| No overlay | − | − | − | − |
| PC12 | − | − | − | − |
| NA | − | − | − | − |
| HCF | − | ++ | ++++ | ++++++ |
| HCE | − | ++ | ++++ | ++++++ |
| HepG2 | − | ++++ | ++++++ | ++++++ |
| CV-1 | − | − | ++ | +++++ |
Long-term-infected PC12 cultures were overlaid with cells of the indicated lines on day 15 after infection. Culture medium was collected daily and assayed for infectious virus content on CV-1 cells.
Each plus sign represents 1 log unit of virus detected in culture medium per flask. A minus sign indicates that no virus was detected. The limit of detection was 40 PFU/flask. There were three flasks in each group.
The possibility that the virus stimulated from long-term-infected cells was due to residual virus from the initial inoculum or a persistent infection was tested by determining if the stimulation was reduced by incubation with anti-HSV serum in order to neutralize the virus remaining in the medium of long-term cultures. Briefly, 3 days prior to stimulation with HepG2 cells, long-term-infected PC12 cells were incubated with an amount of MAb 8D2 that could neutralize 1,000 PFU of HSV (see Materials and Methods) (28). HepG2 cells were then added to the cultures, as usual, and the amount of virus stimulated from the long-term cultures was measured. Stimulation of the long-term-infected PC12 cultures by HepG2 cells was not influenced by the presence of the MAb to HSV in the culture medium for a time prior and up to the time of induction. This suggests that the source of virus infecting the HepG2 cells used for induction was within PC12 cells (was intracellular) and inaccessible to the MAb.
To test whether the failure of PC12 and NA cells to mediate reactivation is due to a nonpermissiveness to viral replication, the single-step growth kinetics of HSV-1 strain 17 in PC12, NA, CV-1, and HepG2 cells were compared. Although HSV-1 strain 17 replication in NA, CV-1, or HepG2 cells was comparable, PC12 cells yielded 1 to 2 log units less infectious-virus growth than the others (data not shown). Nevertheless, all these four cell lines were permissive to HSV replication. Thus, the failure of NA and PC12 cells to induce reactivation was not due to nonpermissiveness to viral replication.
In addition, the efficiency of plating (EOP) of HSV-1 strain 17 on each of the relevant cell lines was tested. Briefly, 1,000-PFU aliquots (as defined by plaquing on CV-1 cells) were used to inoculate flasks containing 100,000 PC12, HepG2, NA, or CV-1 cells, and the numbers of infectious centers were determined by plating on CV-1 monolayers. The EOPs of HSV-1 were comparable for CV-1 and HepG2 cells. The EOPs of HSV-1 on NA and PC12 cells were 60 and 10%, respectively, relative to CV-1 cells. These experiments were performed in triplicate, at two different times. Since each long-term-infected PC12 cell flask contained fewer than 40 PFU (Fig. 5), as assessed on CV-1 cells, the EOP data suggest that for residual HSV to have infected and been amplified in the PC12 cells used as stimulator (Table 3), at least 400 PFU would have had to be present.
The stimulation of HSV from long-term-infected cultures occurs ubiquitously throughout the culture.
As previously stated, following overlay with CV-1 cells or HCFs, long-term-infected cultures produced infectious HSV (Table 3). Although medium from long-term-infected cultures overlaid with inducer cells contained substantial amounts of HSV, the virus in the culture medium was likely to have included progeny from PC12 cells which were amplified in the permissive indicator cells. Thus, it was not clear if the source of virus infecting the indicator cells was many or few PC12 cells. To distinguish between these possibilities, long-term-infected PC12 cells were overlaid with CV-1 monolayers under plaquing conditions, in methylcellulose medium, where diffusion of virus was very limited. Five days after overlay, typical HSV-induced plaques were seen widely distributed throughout the flasks. More than 500 plaques were counted, with many merging zones of cytopathic effect. NGF-treated PC12 cells grew in clusters of 5 to 50 cells, and nearly every cluster was associated with a virus plaque.
Detection of induced HSV antigens on long-term-infected PC12 cells following stimulation.
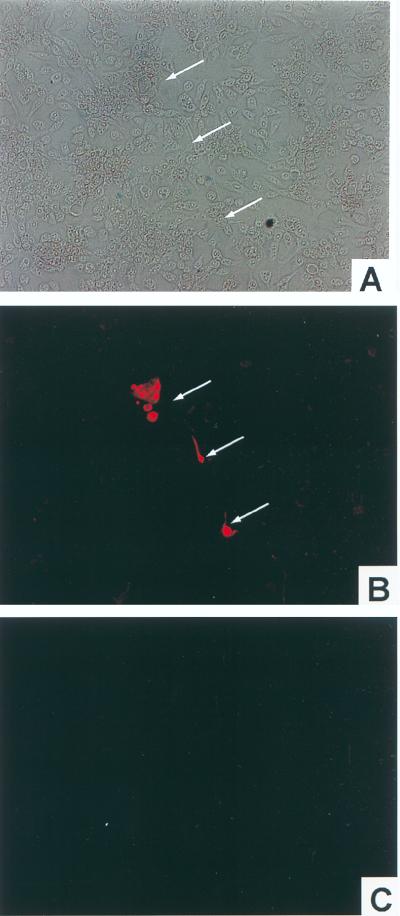
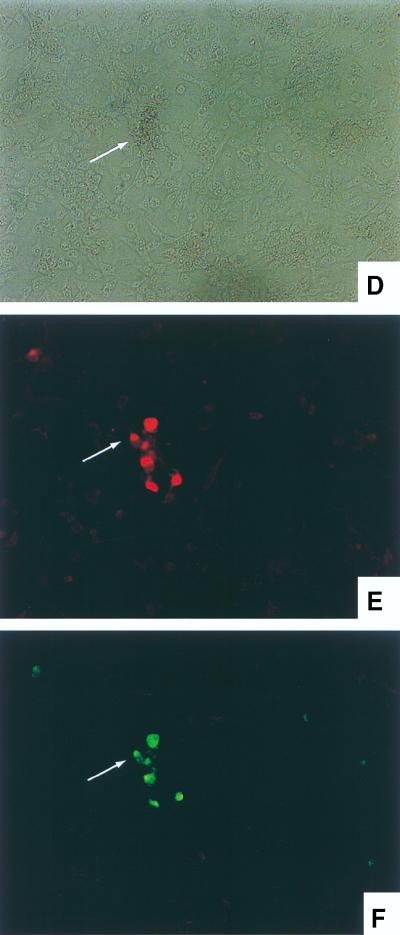
To more directly observe and locate HSV antigens appearing after stimulation of long-term PC12 cells, dual immunofluorescence staining was performed. PC12 cells were seeded and differentiated onto chamber slides as described in Materials and Methods. After long-term infection was established, HepG2 cells were overlaid to stimulate quiescent viruses. Methylcellulose medium was used to impede virus diffusion. Mock-infected NGF-differentiated PC12 cultures were also overlaid with HepG2 cells as a negative control for HSV antigen. Two days after HepG2 overlay, slides were processed for immunofluorescence staining by using antibodies to neurofilaments and HSV antigens as described in Materials and Methods. The specificity of anti-neurofilament antibodies was confirmed by showing binding to NGF-differentiated PC12 cultures and not to HepG2 cells (data not shown). Figure 6 shows immunofluorescence staining of mock-infected (Fig. 6A through C) and long-term-infected (Fig. 6D through F) PC12 cultures overlaid with HepG2 cells. Figure 6A, B, and C all show the same field of mock-infected PC12 cells, and Fig. 6D, E, and F all show the same field of long-term-infected PC12 cells. The photographs in panels A and D were taken under the light microscope, those in panels B and E were taken under the filter which permits the visualization of red fluorescence (anti-neurofilament staining) only, and those in panels C and F were taken under the filter for visualization of green fluorescence (anti-HSV antigen staining). As shown in Fig. 6B, PC12 cells were positively stained by anti-neurofilament antibodies, while HepG2 cells were negative. There was no detectable HSV antigen on either PC12 or HepG2 cells or mock-infected cultures, as shown in Fig. 6C. In contrast, after 2 days of overlay with HepG2 cells, long-term-infected PC12 cells expressed HSV antigens detected by FITC-conjugated polyclonal anti-HSV antibodies (Fig. 6F). By counting the number of neurofilament-positive cells (to restrict the view to PC12-derived cells) which were also HSV antigen positive, it is estimated that at least 70% of the long-term PC12 cell clusters harbored quiescent HSV which had been stimulated. These data demonstrate that (i) infectious HSV detected in the stimulated culture medium (Fig. 5) originates from the long-term-infected PC12 cells and (ii) as implied elsewhere, almost all PC12 cells in the long-term-infected cultures harbor HSV.
FIG. 6.
Immunofluorescence staining of HSV-1 antigens on long-term-infected PC12 cells following reactivation. NGF-differentiated PC12 cultures were prepared in chamber slides. The cultures were infected with HSV-1 strain 17 at an MOI of 20 (D through F) or mock infected (A through C). Twenty-two days after infection, the cultures were overlaid with HepG2 cells in methylcellulose medium to stimulate quiescent viruses. Two days later, the cultures were fixed and stained with anti-neurofilament (red) and anti-HSV (green) antibodies. Panels A, B, and C all show the same field of the mock-infected culture. Panels D, E, and F all show the same field of the long-term-infected culture. The photographs in panels A and D were taken under the light microscope, those in panels B and E were taken under the filter visualizing red fluorescence, and those in panels C and F were taken under the filter in which green fluorescence only was visualized. Arrows point to the stained PC12 cells.
DISCUSSION
HSV reactivation from latent infection normally occurs from neurons in a complex in vivo physiological setting in which multiple cell types interact. In this report, we describe a novel two-cell system for the stimulation of HSV from an in vitro infection in which the virus is quiescent with regard to production of infectious virus. The quiescently infected PC12 cells are called long-term-infected rather than latent cultures in order to avoid confusion with in vivo latency. However, in the long-term-infected cultures described here, there is extremely limited, if any, viral-gene expression. Moreover, the viral genome can be induced by cocultivation with specific cells.
PC12 cells were differentiated with NGF and infected with HSV-1 strain 17. After an initial period in which low-level viral replication occurred (Fig. 1), possibly in a minority of cells, a quiescent infection was established. Northern hybridization and RT-PCR failed to detect any HSV-1 transcripts (including LAT) from RNA derived from long-term-infected PC12 cells. This was surprising because LAT have been shown to accumulate in the nuclei of neurons derived from latently infected animals in great abundance, up to 40,000 copies per neuron (51). However, PC12 cells in the long-term-infected cultures did accumulate low levels of LAT, as determined by Southern blot hybridization of the RT-PCR products (Fig. 3). Thus, by using methods that could detect as few as 200 RNA molecules per reaction, it appears that all long-term-infected cultures tested contained a minority of PC12 cells that were harboring LAT. This level of LAT was too low to be detected by Northern blot hybridization or direct RT-PCR. Although LAT were synthesized following productive infection of NGF-treated PC12 cells, the amount of LAT detected was at least 10 times less than that seen in samples derived from productive infection of undifferentiated PC12 cells (data not shown). Thus, it may also be that LAT stability, accumulation, or synthesis, is different in undifferentiated, NGF-differentiated, and latently infected cells. In this regard, we note that the accumulation of LAT is dependent upon alternative splicing and the failure of the host cell to efficiently debranch the LAT lariat (39, 56). Perhaps long-term-infected PC12 cells have sufficient levels of debranching enzyme to eliminate LAT.
The immediate-early gene alpha 27 and the early tk gene were also detected by Southern blot hybridization to RT-PCR products in a minority of the flasks containing long-term-infected cultures. Transcripts of the late gC gene were not detected in any long-term-infected cultures (Fig. 3). Since there were no detectable gC transcripts and spontaneous release of infectious particles, as measured by plaque assay, did not occur, it is unlikely that the alpha 27 or TK transcripts were from “smoldering” infections. It is possible that, occasionally, abortive viral replication occurs in subpopulations. Perhaps abortive reactivations occur during in vivo latent infection (27). This is under investigation.
The presence of LAT is associated with latency in vivo (9, 15), but abundant LAT are actually not present in most latently infected cells (51). There appear to be at least two populations of latently infected neurons in vivo: (i) those with abundant LAT and (ii) those with few or no detectable LAT. Therefore, it appears that the PC12 long-term infections described in this report are more representative of the subpopulation of latently infected cells which have no detectable LAT or very low levels of LAT.
In spite of the fact that mutant viruses unable to synthesize LAT can reactivate, whether the subpopulation of cells in vivo which are LAT negative, or have very low levels of LAT, can support reactivation remains an open question. We demonstrate here that long-term-infected PC12 cells which harbor few or no LAT can support the stimulation and release of HSV.
NGF deprivation alone did not induce virus appearance in long-term-infected cultures described in this report (data not shown). In a previous report, for example, NGF deprivation alone was sufficient to cause the dedifferentiation of PC12 cells and the appearance of virus in the culture medium (2). In that report, there was evidence that the viral genome was not completely quiescent, since low levels of infectious virus could be detected in the culture medium of the long-term-infected cells. It is possible that the reactivation observed before was a combination of stimulation from latently infected cells and reinfection of dedifferentiated PC12 cells. However, in the system described here, there is no detectable virus in the long-term-infected culture medium. Perhaps all undifferentiated PC12 cells were eliminated by the antimitotic agent FdUrd.
It is possible that the source of virus infecting the CV-1 cells, HepG2 cells, or HCFs (see Fig. 5) was residual virus persisting in the culture medium or cell membranes of long-term-infected cells and not a stimulation event induced by the overlaying cells. This possibility is considered unlikely because (i) HSV antigens were detected only on anti-neurofilament-stained PC12 cells after 48 h after induction by a HepG2 cell overlay in the methylcellulose medium (Fig. 6); (ii) infectious virus was not recovered for more than 2 days following cocultivation with inducer cells (had there been residual virus in the long-term-infected cultures, progeny would have been detected within 1 day following cocultivation with permissive CV-1 or HepG2 cells); (iii) when amounts of an HSV-specific MAb sufficient to neutralize 1,000 PFU of HSV per flask were included in the culture medium prior to stimulation with HepG2 cells, they did not prevent HepG2 cell induction; and (iv) finally, although HepG2 cells could induce stimulation of virus from long-term cultures regardless of the length of time in culture, the ability of CV-1 cells to induce virus declined to zero 20 days after primary infection. Thus, since the EOP of HSV-1 on CV-1 cells was similar to that on HepG2 cells, if residual virus was responsible for the stimulation achieved by HepG2 cells after 20 days of long-term culture, CV-1 cells would also have been effective stimulators, since they would also have been targets of residual virus infection.
The mechanism whereby human corneal cells, HepG2 cells, or CV-1 cells induce the stimulation of HSV from the quiescently infected cultures is unclear. The inducer cells could produce a soluble factor (a hormone or neurotransmitter) which stimulates the viral genome in the long-term-infected PC12 cells. Experiments to test this hypothesis are under way. Another possibility is that cell adhesion molecules or other plasma membrane molecules on the inducer cells interact directly with NGF-treated PC12 cell membranes or cell matrix structures to communicate a signal which stimulates viral-gene activation. In any case, HSV stimulation did not require that PC12 cells dedifferentiate and lose their neuron-like morphology. This is an important distinction from our previous work, in which reactivation occurred predominantly from dedifferentiated PC12 cells, following NGF removal. Stimulation was also independent of the presence or absence of NGF, although NGF could apparently depress stimulation kinetics (data not shown). In addition, cell manipulation alone, such as trypsinization of long-term-infected PC12 cells and overlaying long-term-infected PC12 cells on the monolayer of CV-1 cells, did not induce the appearance of virus in the culture medium.
Thus, although more work clearly needs to be done to determine the extent to which the PC12 cell system can predict features of in vivo HSV latency, the availability of a tissue culture model in which viral-gene expression can be induced by activator cells provides an attractive means to study, at a minimum, how one cell can elicit viral gene expression in another.
ACKNOWLEDGMENTS
This research was supported by NIH grant NS33768-11.
We thank Robert Jordan, Richard Gesser, Bruce Randazzo, and Ruth Tal-Singer for discussion of this work; Mike Moxley for excellent technical assistance; and Mitra Dadmarz for reading the manuscript.
REFERENCES
- 1.Block T, Masonis J, Maggioncalda J, Deshmane S J, Fraser N W. A herpes simplex virus LAT minus mutant that makes small plaques on confluent CV-1 cells. Virology. 1993;192:618–630. doi: 10.1006/viro.1993.1078. [DOI] [PubMed] [Google Scholar]
- 2.Block T, Barney S, Masonis J, Maggioncalda J, Rattan S, Valyi-Nagy T, Fraser N W. Long-term herpes simplex virus infections of nerve growth factor-differentiated PC12 cells. J Gen Virol. 1994;75:2481–2487. doi: 10.1099/0022-1317-75-9-2481. [DOI] [PubMed] [Google Scholar]
- 3.Block T M, Spivack J G, Steiner I, Deshmane S, McIntosh M T, Lirette R P, Fraser N W. A herpes simplex virus type 1 latency-associated transcript mutant reactivates with normal kinetics from latent infection. J Virol. 1990;64:3417–3426. doi: 10.1128/jvi.64.7.3417-3426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai W, Person S, Debroy C, Gu B. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. J Mol Biol. 1988;201:575–588. doi: 10.1016/0022-2836(88)90639-0. [DOI] [PubMed] [Google Scholar]
- 5.Cook M L, Bastone V B, Stevens J G. Evidence that neurons harbor latent herpes simplex virus. Infect Immun. 1974;9:946–951. doi: 10.1128/iai.9.5.946-951.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croen K D, Ostrove J M, Dragovic L J, Smialek J E, Straus S E. Latent herpes simplex virus in human trigeminal ganglia. Detection of an immediate-early gene “anti-sense” transcript by in situ hybridization. N Engl J Med. 1988;317:1427–1432. doi: 10.1056/NEJM198712033172302. [DOI] [PubMed] [Google Scholar]
- 7.Cubbit C L, Lausch R N, Oakes J E. Differential regulation of granulocyte-macrophage colony-stimulating factor gene expression in human corneal cells by pro-inflammatory cytokines. J Immunol. 1994;153:232–240. [PubMed] [Google Scholar]
- 8.Dawson C R, Togni B. Herpes simplex eye infections: clinical manifestations, pathogenesis and management. Surv Ophthalmol. 1976;21:121. doi: 10.1016/0039-6257(76)90090-4. [DOI] [PubMed] [Google Scholar]
- 9.Deatly A M, Spivack J G, Lavi E, Fraser N W. RNA from an immediate early region of the HSV-1 genome is present in the trigeminal ganglia of latently infected mice. Proc Natl Acad Sci USA. 1987;84:3204–3208. doi: 10.1073/pnas.84.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshmane S L, Nicosia M, Valyi-Nagy T, Feldman L T, Dillner A, Fraser N W. An HSV-1 mutant lacking the LAT TATA element reactivated normally in explant cocultivation. Virology. 1993;196:868–872. doi: 10.1006/viro.1993.1548. [DOI] [PubMed] [Google Scholar]
- 11.Devi-Rao G B, Bloom D C, Stevens J G, Wagner E K. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J Virol. 1994;68:1271–1282. doi: 10.1128/jvi.68.3.1271-1282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ecob-Prince M S, Hassan K, Denheen M T, Preston C M. Expression of beta-galactosidase in neurons of dorsal root ganglia which are latently infected with herpes simplex virus type 1. J Gen Virol. 1995;76:1527–1532. doi: 10.1099/0022-1317-76-6-1527. [DOI] [PubMed] [Google Scholar]
- 13.Ecob-Prince M S, Rixon F J, Preston C M, Hassan K, Kennedy P G E. Reactivation in vivo and in vitro of herpes simplex virus from mouse dorsal root ganglia which contain different levels of latency-associated transcripts. J Gen Virol. 1993;74:446–455. doi: 10.1099/0022-1317-74-6-995. [DOI] [PubMed] [Google Scholar]
- 14.Feldman L T. Transcription of the HSV-1 genome in neurons in vivo. Semin Virol. 1994;5:207–212. [Google Scholar]
- 15.Fraser N W, Block T M, Spivack J G. The latency-associated transcripts of herpes simplex virus: RNA in search of function. Virology. 1992;191:1–8. doi: 10.1016/0042-6822(92)90160-q. [DOI] [PubMed] [Google Scholar]
- 16.Frazier D P, Cox D, Godshalk E M, Schaffer P A. The herpes simplex virus type 1 latency-associated transcript promoter is activated through Ras and Raf by nerve growth factor and sodium butyrate in PC12 cells. J Virol. 1996;70:7424–7432. doi: 10.1128/jvi.70.11.7424-7432.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene L A, Rukenstein A. The quantitative bioassay of nerve growth factor with PC12 cells. In: Rush R A, editor. Nerve growth factors. New York, N.Y: John Wiley; 1989. pp. 139–147. [Google Scholar]
- 18.Greene L A, Tischler A S. Establishment of a nonadrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gressens P, Martin J R. In situ polymerase chain reaction: localization of HSV-2 DNA sequences in infections of the nervous system. J Virol Methods. 1994;46:61–83. doi: 10.1016/0166-0934(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 20.Harris R A, Preston C M. 1814. J. Gen. Virol. 72:907–913. 1991. Establishment of latency in vitro by the herpes simplex virus type 1 mutant. [DOI] [PubMed] [Google Scholar]
- 21.Hill J M, Gebhardt B M, Wen R J, Bouterie A M, Thompson H W, O’Callaghan R J, Halford W P, Kaufman H E. Quantitation of herpes simplex virus type 1 DNA and latency-associated transcripts in rabbit trigeminal ganglia demonstrates a stable reservoir of viral nucleic acids during latency. J Virol. 1996;70:3137–3141. doi: 10.1128/jvi.70.5.3137-3141.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill J M, Maggioncalda J B, Garza H H, Su Y-H, Fraser N W, Block T M. In vivo epinephrine reactivation of ocular herpes simplex virus type 1 in the rabbit is correlated to a 370-base pair region located between the promoter and the 5′ end of the 2.0-kilobase latency-associated transcript. J Virol. 1996;70:7270–7274. doi: 10.1128/jvi.70.10.7270-7274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho D Y, Mocarski E S. Herpes simplex virus latent RNA (LAT) is not required for latent infection in the mouse. Proc Natl Acad Sci USA. 1989;86:7596–7600. doi: 10.1073/pnas.86.19.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamieson D R S, Robinson L H, Daksis J I, Nicholl M J, Preston C M. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus type 1 Vmw65 mutants. J Gen Virol. 1995;76:1417–1431. doi: 10.1099/0022-1317-76-6-1417. [DOI] [PubMed] [Google Scholar]
- 25.Kosz-Vnenchak M, Jacobson J, Coen D M, Knipe D M. Evidence for a novel regulatory pathway for herpes simplex virus gene expression in trigeminal ganglion neurons. J Virol. 1993;67:5383–5393. doi: 10.1128/jvi.67.9.5383-5393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer M F, Coen D M. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1995;69:1389–1399. doi: 10.1128/jvi.69.3.1389-1399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer M F, Chen S-H, Knipe D M, Coen D M. Accumulation of viral transcripts and DNA during establishment of latency by herpes simplex virus. J Virol. 1998;72:1177–1185. doi: 10.1128/jvi.72.2.1177-1185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lausch R N, Oakes J E, Metcalf J F, Scimeca J M, Smith L A, Robertson S M. Quantitation of purified monoclonal antibody needed to prevent HSV-1-induced stromal keratitis in mice. Curr Eye Res. 1989;8:499–506. doi: 10.3109/02713688909000030. [DOI] [PubMed] [Google Scholar]
- 29.Leib D A, Bogard C L, Kosz-Vnenchak M, Hicks K A, Coen D M, Knipe D M, Schaffer P A. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol. 1989;63:2893–2900. doi: 10.1128/jvi.63.7.2893-2900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X, Block T M, Gerlich W H. Protease-induced infectivity of hepatitis B virus for a human hepatoblastoma cell line. J Virol. 1996;70:2277–2285. doi: 10.1128/jvi.70.4.2277-2285.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maggioncalda J, Mehta A, Fraser N W, Block T M. Analysis of a herpes simplex virus type 1 LAT mutant with a deletion between the putative promoter and the 5′ end of the 2.0-kilobase transcript. J Virol. 1994;68:7816–7824. doi: 10.1128/jvi.68.12.7816-7824.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta A, Maggioncalda J, Bagasra O, Block T M. A PCR DNA hybridization method to detect HSV-1 DNA neuronal cells, in situ, derived from latently infected people. J Anal Morphol. 1994;1:110–115. [Google Scholar]
- 33.Mehta A, Maggioncalda J, Bagasra O, Thikkavarapu S, Saikumari P, Valyi-Nagy T, Fraser N W, Block T M. In situ DNA PCR and RNA hybridization detection of herpes simplex virus sequences in trigeminal ganglia of latently infected mice. Virology. 1995;206:633–640. doi: 10.1016/s0042-6822(95)80080-8. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell W J, Gressens P, Martin J R, DeSanto R. Herpes simplex virus type 1 DNA persistence, progressive disease and transgenic immediate early gene promoter activity in chronic corneal infections in mice. J Gen Virol. 1994;75:1201–1210. doi: 10.1099/0022-1317-75-6-1201. [DOI] [PubMed] [Google Scholar]
- 34a.National Eye Advisory Council. Vision research—a national plan, 1994–1998. NIH publication 93–3186. Bethesda, Md: National Institutes of Health; 1994. [Google Scholar]
- 35.Perng G-C, Ghiasi H, Slanina S, Nesburn A B, Wechsler S L. The spontaneous reactivation function of the herpes simplex virus type 1 LAT genes resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J Virol. 1996;70:976–984. doi: 10.1128/jvi.70.2.976-984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramakrishnan R, Fink D J, Jiang G, Desai G P, Glorioso J, Levine M. Competitive quantitative PCR analysis of herpes simplex virus type 1 DNA and latency-associated transcript RNA in latently infected cells of the rat brain. J Virol. 1994;68:1864–1873. doi: 10.1128/jvi.68.3.1864-1873.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramakrishnan R, Levine M, Fink D J. PCR-based analysis of herpes simplex virus type 1 latency in the rat trigeminal ganglion established with a ribonucleotide reductase-deficient mutant. J Virol. 1994;68:7083–7091. doi: 10.1128/jvi.68.11.7083-7091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramakrishnan R, Poliani P L, Levine M, Glorioso J C, Fink D J. Detection of herpes simplex virus type 1 latency-associated transcript expression in trigeminal ganglia by in situ reverse transcriptase PCR. J Virol. 1996;70:6519–6523. doi: 10.1128/jvi.70.9.6519-6523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rødahl E, Haarr L. Analysis of the 2-kilobase latency-associated transcript expressed in PC12 cells productively infected with herpes simplex virus type 1: evidence for a stable, nonlinear structure. J Virol. 1997;71:1703–1707. doi: 10.1128/jvi.71.2.1703-1707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, editors. Fundamental virology. New York, N.Y: Raven Press, Ltd.; 1996. pp. 2231–2296. [Google Scholar]
- 41.Russell J, Preston C M. An in vitro latency system for herpes simplex virus type 2. J Gen Virol. 1986;67:397–403. doi: 10.1099/0022-1317-67-2-397. [DOI] [PubMed] [Google Scholar]
- 42.Sawtell N, Thompson R L. Herpes simplex virus latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol. 1992;66:2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sedarati F, Izumi K M, Wagner E K, Stevens J G. Herpes simplex virus type 1 latency-associated transcription plays no role in establishment of maintenance of a latency infection in murine sensory neurons. J Virol. 1989;63:4455–4458. doi: 10.1128/jvi.63.10.4455-4458.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith R L, Escudero J M, Wilcox C L. Regulation of the herpes simplex virus latency-associated transcripts during establishment of latency in sensory neurons in vitro. Virology. 1994;202:49–60. doi: 10.1006/viro.1994.1321. [DOI] [PubMed] [Google Scholar]
- 45.Steiner I, Spivack J G, Lirette R P, Brown S M, Maclean A R, Subak-Sharpe J, Fraser N W. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 1989;8:505–511. doi: 10.1002/j.1460-2075.1989.tb03404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevens J G. Human herpesviruses: a consideration of the latent state. Microbiol Rev. 1989;53:318–332. doi: 10.1128/mr.53.3.318-332.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpes virus gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 48.Tal-Singer R, Lasner T M, Podrzucki W, Skokotas A, Leary J J, Berger S L, Fraser N W. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J Virol. 1997;71:5268–5276. doi: 10.1128/jvi.71.7.5268-5276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiele K, Mittnacht S, Kirchner H. Persistent replication of herpes simplex virus type 1 in JOK-1 cells. J Gen Virol. 1989;70:1907–1911. doi: 10.1099/0022-1317-70-7-1907. [DOI] [PubMed] [Google Scholar]
- 50.Vahlne A, Lycke E. Herpes simplex virus infection on in vitro cultured neuronal (mouse neuroblastoma C1300) cells. J Gen Virol. 1978;39:321–332. doi: 10.1099/0022-1317-39-2-321. [DOI] [PubMed] [Google Scholar]
- 51.Wagner E K, Bloom D C. Experimental investigation of herpes simplex virus latency. Clin Microbiol Rev. 1997;10:419–443. doi: 10.1128/cmr.10.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wander A H, Centifanto Y M, Kaufman H E. Strain specificity of clinical isolates of herpes simplex virus. Arch Ophthalmol. 1980;98:1458–1467. doi: 10.1001/archopht.1980.01020040310020. [DOI] [PubMed] [Google Scholar]
- 53.Wigdahl B L, Ziegler R J, Sneve M, Rapp F. Herpes simplex virus latency and reactivation in isolated rat sensory neurons. Virology. 1983;127:159–167. doi: 10.1016/0042-6822(83)90380-x. [DOI] [PubMed] [Google Scholar]
- 54.Wilcox C L, Johnson E M. Characterization of nerve growth factor-dependent latency in neurons in vitro. J Virol. 1988;62:393–399. doi: 10.1128/jvi.62.2.393-399.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilcox C L, Smith R L, Freed C R, Johnson E M., Jr Nerve growth factor-dependence of herpes simplex virus latency in peripheral sympathetic and sensory neurons in vitro. J Neurosci. 1990;10:1268–1275. doi: 10.1523/JNEUROSCI.10-04-01268.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu T-T, Su Y-H, Block T M, Taylor J M. Evidence that two latency-associated transcripts of herpes simplex virus type 1 are nonlinear. J Virol. 1996;70:5962–5967. doi: 10.1128/jvi.70.9.5962-5967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]