Abstract
Human herpesvirus 8 (HHV-8) infection has been implicated in the etiology of Kaposi’s sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman’s disease (MCD), three diseases that frequently develop in immunocompromised, human immunodeficiency virus-positive individuals. One hypothesis that would account for different pathological manifestations of infection by the same virus is that viral genes are differentially expressed in heterogeneous cell types. To test this hypothesis, we analyzed the localization and levels of expression of two viral genes expressed in latent and lytic infections and the viral homologue of interleukin-6 (vIL-6). We show that PEL parallels KS in the pattern of latent and lytic cycle viral gene expression but that the predominant infected cell type is a B cell. We also show that MCD differs from KS not only in the infected cell type (B-cell and T-cell lineage) but also in the pattern of viral gene expression. Only a few cells in the lesion are infected and all of these cells express lytic-cycle genes. Of possibly greater significance is the fact that in a comparison of KS, PEL, and MCD, we found dramatic differences in the levels of expression of vIL-6. Interleukin-6 is a B-cell growth and differentiation factor whose altered expression has been linked to plasma cell abnormalities, as well as myeloid and lymphoid malignancies. Our findings support the hypothesis that HHV-8 plays an important role in the pathogenesis of PEL and MCD, in which vIL-6 acts as an autocrine or paracrine factor in the lymphoproliferative processes common to both.
Since the discovery of human herpesvirus 8 (HHV-8) (8), a substantial body of serological and molecular evidence has pointed to HHV-8 as the etiological agent of Kaposi’s sarcoma (KS) in the immunocompromised host, with or without human immunodeficiency virus (HIV) infection (6, 13). In addition to KS, HHV-8 has also been identified in two other relatively rare diseases of lymphoproliferative nature and unknown etiology in immunocompromised individuals infected with HIV (1, 7, 10, 28). Primary effusion lymphoma (PEL) and multicentric Castleman’s disease (MCD) are two diseases often seen in individuals who have had or will have KS (19, 24, 31). PEL, a relatively late manifestation of HIV infection, is a malignant effusion in the absence of a contiguous solid tumor mass that develops in the pleural, pericardial, or peritoneal cavity. Although this lymphoma is of an indeterminate phenotype, from analysis of immunoglobulin gene rearrangements, these immunoblastic or anaplastic large-cell lymphomas are of B-cell origin (14, 18, 20, 30). Castleman’s disease (CD) (5, 12) is a nonmalignant, atypical lymphoproliferative disorder characterized clinically by systemic manifestations that include fever, anemia, and hypergammaglobulinemia due in part to elevated levels of interleukin-6 (IL-6) in the serum (4, 32). CD refers to a somewhat diverse group of lesions that are clinically classified as one of two forms: (i) localized CD seen as a single, usually mediastinal, lymph node hyperplasia which resolves upon surgical resection or (ii) multifocal or multicentric CD with multisystem involvement accompanied by generalized lymphadenopathy. Histologically, CD is classified as one variant or a combination of two variants, a more common hyaline vascular type and a plasma cell (PC) type. Unlike KS and PEL, not all CD has been found to contain HHV-8 DNA (3, 25, 28), and thus, there is likely more than one etiology for this somewhat diverse group of lesions. However, it is the multifocal or multicentric form of the PC variant or PC-hyaline vascular combination that often arises in HIV-infected individuals and that has been frequently shown to contain HHV-8 DNA (24, 25, 28).
Among the genes encoded by HHV-8, there is a functional viral homologue of IL-6 (vIL-6) (22, 26). Because of the relationship between IL-6 and CD, we hypothesized that expression of this gene might be involved in the pathogenesis of some cases of MCD and perhaps in PEL as well and, further, that the different pathological manifestations of HHV-8 infection might be the result of different patterns of viral gene expression in different cell types. Under this hypothesis, MCD and PEL would be the result of higher levels of vIL-6 expression in B cells than the lower levels in the spindle tumor cells in KS. In this report, we describe the results of an analysis of vIL-6 gene expression in defined cell types in KS, PEL, and MCD that provide strong evidence in support of this hypothesis.
MATERIALS AND METHODS
Tissue specimens.
The KS dermal biopsy and the PEL were derived from two different HIV-negative individuals. The MCD specimen was from an HIV-infected individual with AIDS who also had a history of KS. This patient presented with a clinical and pathological picture beyond that which is seen in the usual HIV patient and which is consistent with CD. This included anemia, unexplained fevers and chills, severe myalgia, arthralgias, massive lymphadenopathy, and hepatosplenomegaly. Constitutional symptoms and lymphadenopathy dramatically decreased upon treatment with prednisone but recurred upon its discontinuation. This pattern was repeated upon multiple rounds of prednisone treatment. Histopathologically, the specimen has features of both the persistent generalized lymphadenopathy of HIV in the follicular involutional stage and of multicentric angiofollicular hyperplasia, i.e., enlarged lymph nodes involving significant follicular hyperplasia and perifollicular lymphoplasmacytic (predominantly plasmacytic) reaction, interfollicular plasmacytosis, and few immunoblasts (12). Immunoperoxidase staining for the kappa and lambda light chains shows that the plasmacytic infiltrate is polyclonal.
From these specimens, which are especially now rare and difficult to obtain since the introduction of HIV protease inhibitors for AIDS therapy, we derived and here present results that are representative of two cases of PEL, five cases of HHV-8-positive MCD, and numerous cases of KS that we have studied. Surgical specimens were fixed in buffered formalin and subsequently processed and embedded in paraffin following standard histological protocols. Thin sections (6 to 8 μm) were cut and attached to silanized slides, dried, heated for 1 h at 60°C, deparaffinized with two 10-min incubations in xylene, cleared with two 10-min incubations in absolute ethanol, and dried.
Hybridization probes.
Human IL-6 (hIL-6) expression plasmid pT7.7/hIL-6 (2), obtained from the American Type Culture Collection, was digested with the HaeIII and BamHI restriction enzymes, and a 1,200-bp fragment containing 87% of the IL-6 protein-coding region was subcloned into a pBluescript SK− transcription vector (Stratagene). The hIL-6 subclone, HHV-8 nut-1 and T0.7 cDNA clones in pBluescript SK+ (33), and a PCR-generated HHV-8 vIL-6 DNA clone in pCR3.1 (Invitrogen) were linearized on either side of the insert to produce templates suitable for runoff transcription of antisense and sense RNAs. Radiolabeled RNAs with specific activities of ∼109 cpm/μg were synthesized with [35S]UTP and the Promega transcription system, and digoxigenin-labeled nut-1 antisense RNA was synthesized with the Boehringer Mannheim Genius transcription system. All RNA transcripts were subjected to controlled alkaline hydrolysis to yield fragments with an average size of 350 ribonucleotides (9).
In situ hybridization (ISH) and quantitative image analysis.
The pretreatment and hybridization methods used have been described in detail elsewhere (15). Briefly, dried, deparaffinized slides were incubated for 30 min in 0.2 N HCl, neutralized with 0.15 M triethanolamine (pH 7.4), incubated in a 0.005% digitonin-containing solution for 5 min, treated with 5-μg/ml proteinase K in a CaCl2-containing buffer for 15 min at 37°C, acetylated, and dehydrated with graded alcohols. Hybridization mixtures containing 10% dextran sulfate, 50% deionized formamide, 20 mM HEPES (pH 7.4), 1 mM EDTA, 1× Denhardt’s medium, 1-mg/ml poly(A), 0.6 M NaCl, 100 mM dithiothreitol (DTT), 250-μg/ml yeast RNA, and 105-cpm/μl 35S-labeled riboprobe were applied to specimens, which were then covered with siliconized coverslips and sealed with rubber cement. Hybridization was carried out for 16 to 18 h at 45°C, after which time the coverslips were removed under 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature. The slides were washed with 5× SSC–10 mM DTT for 30 min at 42°C, with 2× SSC–50% formamide–10 mM DTT for 20 min at 60°C; twice in HWB (0.1 M Tris-HCl [pH 7.4], 0.4 M NaCl, 0.05 M EDTA) for 10 min each time at 37°C; with 25-μg/ml RNase A and 25-U/ml RNase T1 in HWB for 30 min at 37°C; and with HWB, 2× SSC, and 0.1× SSC for 15 min each at 37°C and dehydrated through graded alcohols containing 0.3 M ammonium acetate. The slides were coated with Kodak NTB-2 photographic emulsion, exposed at 4°C for specified periods of time, developed, and counterstained with hematoxylin and eosin. The transcript content of individual cells is directly proportional to the quantity of silver grains that develop in the layer of photographic emulsion covering cells that have bound a specific radiolabeled probe (15). To quantitate the silver grains, we captured video images of cells visualized by epipolarization microscopy and used Metamorph image analysis software (Universal Imaging, West Chester, Pa.) to enumerate the silver grains as previously described (16). When possible, grain counts were determined for at least 100 hybridization-positive cells from randomly selected fields of view for each specimen. Copy numbers were calculated based on transcript calibrations performed on HIV-infected cells (16).
Double-label ISH and immunohistochemistry (IHC).
Specimens were pretreated for double labeling by following a protocol that incorporates antigen retrieval methodology (27). Slides were deparaffinized as previously described. However, after clearing in absolute ethanol, the slides were rehydrated by brief passage through baths of 90, 80, and 70% ethanol and into RNase-free water. The slides were then placed in 10 mM sodium citrate, pH 6.0, exposed to microwaves for 10 min (medium-high setting, 1,200-W oven), allowed to cool slowly to room temperature, and acetylated. The slides were prehybridized at 37°C for 1 h with a hybridization solution as previously described but lacking DTT and a probe. Prehybridization solution was replaced with hybridization solution containing 0.15-μg/ml digoxigenin-labeled nut-1 antisense RNA. Siliconized coverslips were applied and sealed with rubber cement, and the slides were incubated for 16 to 18 h at 45°C. Coverslips were removed under 2× SSC, and the slides were washed as follows: twice in 2× SSC for 5 min each time at room temperature, in STE (0.5 M NaCl, 1 mM EDTA, 20 mM Tris-HCl [pH 7.5]) for 5 min at room temperature, in 2× SSC–50% formamide for 5 min at 50°C, in 1× SSC–0.1% sodium dodecyl sulfate for 10 min at 50°C, and in 0.5× SSC–0.1% sodium dodecyl sulfate for 15 min at 50°C. The slides were equilibrated for 5 min in 0.1 M Tris-HCl (pH 7.4)–0.15 M NaCl and then carried through the Boehringer Mannheim digoxigenin detection protocol using an alkaline phosphatase-conjugated antibody. Nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate substrate development was monitored under the microscope and terminated by incubating the slides for 3 min in TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). For IHC, the slides were transferred to phosphate-buffered saline (PBS) containing 5% (wt/vol) nonfat dry milk for 20 min and then reacted for 16 to 18 h at 4°C with the primary antibody (diluted with PBS), which was a mouse monoclonal antibody to human lambda light chains (DAKO M0614, 1:400), a rabbit polyclonal antibody to human CD3 (DAKO A452, 1:100), or a mouse monoclonal antibody to human CD34 (QB-END/10, 1:50 Vector Laboratories). The slides were washed with PBS and reacted with a horseradish peroxidase-conjugated secondary antibody (ABC Elite mouse kit; Vector), developed with diaminobenzidine (DAB peroxidase substrate kit; Vector), and counterstained briefly with hematoxylin.
For colocalization of two different viral transcripts, a hybridization mixture containing both 35S-labeled vIL-6 and digoxigenin-labeled nut-1 riboprobes was applied to slides pretreated for hybridization as described above. Following posthybridization washes, the slides were processed to detect the digoxigenin-labeled probe, dehydrated through graded alcohols containing 0.3 M ammonium acetate, and then coated with a photographic emulsion. After development, the slides were counterstained for 1 to 5 s in hematoxylin.
RESULTS
Identification and quantitation of infected cells in KS, PEL, and MCD.
We have previously shown that in KS, the spindle-shaped tumor cell is the predominant HHV-8-infected cell within the lesion (29). Using a combination of ISH with a riboprobe specific for the HHV-8 T0.7 gene (open reading frame K12), which is transcribed during viral latency as well as productive infection (33), and IHC with an antibody to CD34, an antigen that is present on the KS spindle tumor cell and endothelial cells from which the tumor cell is thought to be derived (23), we demonstrated that most, if not all, of the tumor cells are infected, regardless of the stage of lesion development. We also demonstrated by ISH detection of HHV-8 lytic transcripts, nut-1 and MCP, that only a small percentage of the total infected cell population of the KS lesions transcribe lytic genes suggestive of productive infection in these cells.
In many cases of PEL from HIV-infected individuals, the tumor cells lack known antigens for immunophenotyping, but the tumors have been shown by analysis of immunoglobulin gene rearrangements to be of B-cell origin (14, 18, 20, 30). Conversely, liquid-phase lymphomas of the HIV-negative cohort are generally not of the null phenotype and can be shown to belong to B-cell or, in a few cases, T-cell lineages. Despite the HIV-negative background of our PEL specimen, we were unable to phenotype the tumor cell and consequently could not perform double-label ISH-IHC to simultaneously phenotype and quantitate those cells infected with HHV-8. ISH alone, however, with a probe to identify T0.7 transcripts revealed infection in a majority of the cells of the effusion, presumably the tumor cells (Fig. 1A). Hybridization with the nut-1-specific probe revealed that only a minority of these cells were potentially productively infected (Fig. 1B). As we have previously shown for KS (29), there is a variable level of T0.7 expression across the population of infected cells, and those cells with the highest T0.7 content correspond to the nut-1-positive cells. Longer exposure times enhanced the signal over cells at the low end of T0.7 expression but did not reveal additional nut-1-positive cells (data not shown).
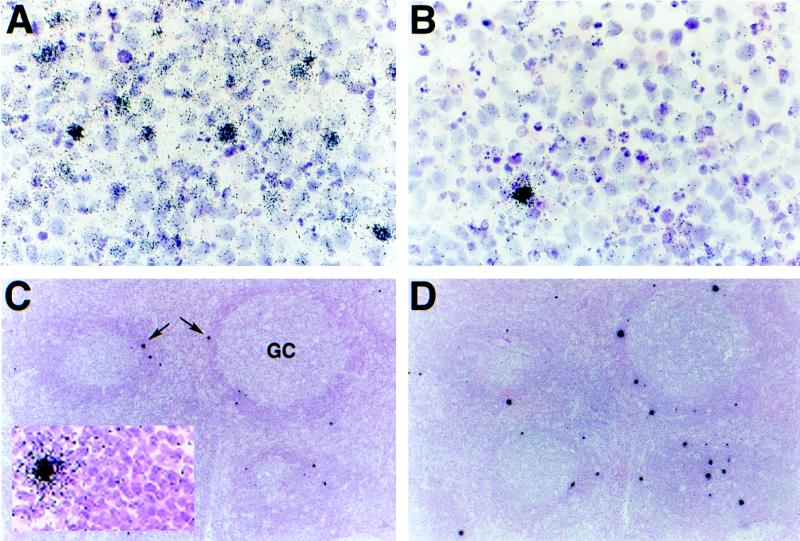
FIG. 1.
In situ localization of HHV-8-infected cells in PEL and MCD. Thin sections of PEL (A and B) and MCD (C and D) specimens were hybridized with 35S-labeled riboprobes specific for HHV-8 genes expressed during latency (T0.7) and productive infection (nut-1). PEL displays a hybridization pattern similar to that which we have previously reported for KS: T0.7 (A, 3-day exposure) is expressed in a majority of cells of the effusion and to various levels across the population of infected cells (visualized as various amounts of silver grains that have developed in the photographic emulsion coating the specimen), whereas nut-1, which potentially indicates lytic infection, is transcribed in only a few of the total infected cells (B, 18-h exposure). Hybridizations to MCD reveal that, relative to PEL, few cells express T0.7; that expression is to relatively high levels (collections of silver grains over individual cells seen as black dots on this overexposed slide at this low magnification; arrows); and that these cells are localized mainly to the “onionskin” perifollicular collections of lymphocytes surrounding the germinal centers (GC) (C, 3-day exposure). Unlike KS and PEL, the pattern of nut-1 hybridization, with respect to location and numbers of positive cells, is similar to that of T0.7 in the same tissue (D, 18-h exposure). Like KS and PEL, these cells express nut-1 to much higher levels than T0.7. Higher magnifications of panels C (inset) and D do not reveal a significant number of additional positive cells that cannot be seen at this magnification. Counterstaining was done with hematoxylin and eosin.
In MCD, by contrast, with the same exposure time, there are a few cells in which T0.7 RNA can be detected and these contain relatively high levels of transcripts (similar to the high end of the spectrum in KS and PEL). Increased exposure times enhanced the signal over these cells but did not reveal a significant number of additional cells with lower transcript content. These cells are located mainly within the follicular mantle of small lymphocytes surrounding the germinal centers with fewer cells in the germinal center and interfollicular plasma cell-containing region (Fig. 1C). The number and distribution of cells containing nut-1 RNA (Fig. 1D) or MCP RNA (data not shown) were similar, and double-label experiments with digoxigenin- and radiolabeled probes for T0.7, nut-1, and MCP (in various pair combinations) resulted in colocalization of these transcripts (data not shown). We interpret this observation as evidence that all of these cells are potentially productively infected.
In this particular specimen, in the subcapsular sinus and extending into the interfollicular spaces, we also detected nests of cells with T0.7 RNA that, with respect to the abundance and cellular distribution of the hybridization signal, are reminiscent of KS. Almost all of the cells within this area were T0.7 positive. There was a gradient of T0.7 transcript content across the population of these infected cells, and we detected nut-1 RNA in very few of these cells. Cells in this region of the MCD lymph node were spindle shaped with elongated nuclei, were colabeled with the nut-1 riboprobe and the antibody to CD34 (Fig. 2A and B), and represent an early stage of KS in the same tissue.
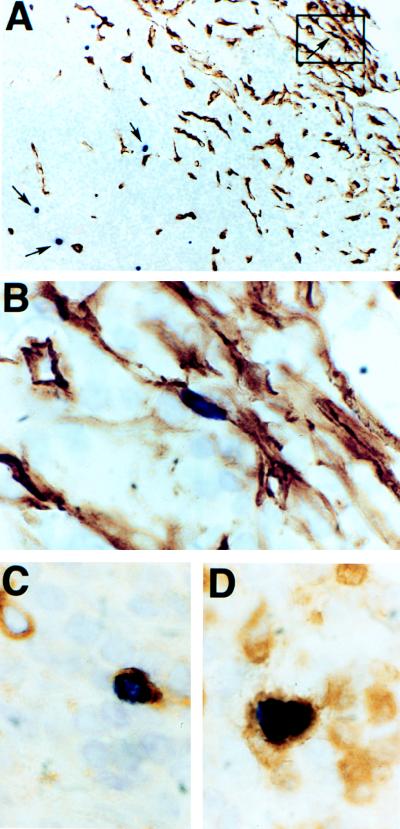
FIG. 2.
Identification of HHV-8-infected cells in MCD. Combined ISH with a digoxigenin-labeled nut-1 riboprobe (dark purple nuclei) and IHC with a monoclonal antibody to human CD34 (brown peroxidase reaction product) reveals the presence of an infected spindle-shaped cell, similar to the KS tumor cell, in a collection of CD34-staining cells in the subcapsular sinus of a lymph node with MCD seen at low (A, boxed area, arrow, upper right) and high (B) magnifications. Other infected cells (A, arrows, center and lower left) in the perifollicular lymphocyte layer do not express CD34. Colocalization of the digoxigenin-labeled nut-1 riboprobe and antibody to the human lambda light chain (C) or CD3 (D) shows that some of these infected CD34-negative cells are plasma cells and T cells, respectively. Counterstaining was done briefly with hematoxylin.
Double-label experiments showed that many CD34-negative HHV-8-infected cells in the perifollicular region reacted with antibody to human lambda light chains (Fig. 2C). In addition to these infected plasma cells, we detected a few infected T cells, which were identified with antibody to CD3 (Fig. 2D). There were a significant number of cells, however, that did not react with any of the antibodies tested (specific for B cells, T cells, plasma cells, κ, and λ light chains, dendritic cells, macrophages, and endothelial cells). We speculate that, like PEL, these may represent B cells in some stage of differentiation or transformation where detectable B-cell-associated antigens have been lost.
Comparison of vIL-6 transcription in KS, PEL, and MCD.
A number of viral homologues of interesting human genes, including those for cytokines, growth factors, and receptors, have been identified in the HHV-8 genome. Among these is a homologue of the gene for IL-6 that has clearly been associated with clinical and pathological abnormalities of MCD (4, 32) and has been speculated to play a role in KS and PEL (11). We tested the hypothesis that differential expression of vIL-6 might be responsible for the different diseases by evaluating vIL-6 gene expression. In comparisons of either the latently or productively infected cells across the three lesion types, we found that the single-cell levels of T0.7 and nut-1 RNAs were similar; however, vIL-6 was expressed at very different and characteristic levels. In MCD (Fig. 3A), vIL-6 was highly expressed in cells corresponding in number and location to those containing both T0.7 (Fig. 1C) and nut-1 (Fig. 1D) RNAs. Double-label ISH with 35S-labeled vIL-6 and digoxigenin-labeled nut-1 probes showed colocalization of these transcripts (Fig. 3A, inset).
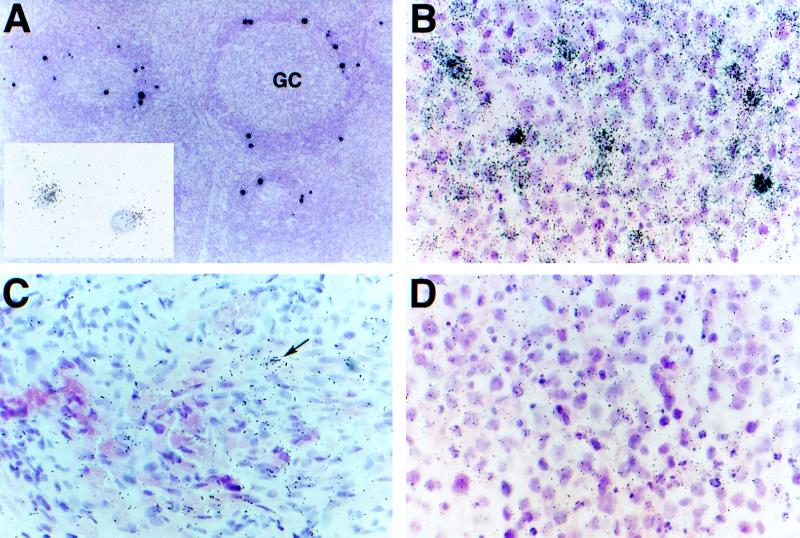
FIG. 3.
vIL-6 expression in KS, PEL, and MCD. Hybridization of MCD with a 35S-labeled vIL-6 riboprobe reveals high levels of transcripts in cells localized mainly to the perifollicular lymphocyte layer of the specimen (A, 3-day exposure). These cells are similar in quantity and location to those transcribing T0.7 and nut-1 RNAs in subjacent sections (Fig. 1C and D) and double ISH with 35S-labeled vIL-6 and digoxigenin-labeled nut-1 riboprobes show colocalization to the same cells (A, inset, 2-h exposure). A higher magnification of this overexposed slide does not reveal additional vIL-6-positive cells. Hybridization of 35S-labeled vIL-6 to PEL shows that, like hybridization for T0.7 in PEL, a majority of cells are transcribing this gene and to various levels within the population (B, 3-day exposure) but to a visibly lesser extent than in MCD. KS expresses the least vIL-6 (C, 7-day exposure) with an infrequent cell containing few transcripts (arrow) among a majority of T0.7-containing cells (hybridization not shown). Control hybridization of a 35S-labeled riboprobe for human IL-6 to PEL shows lack of cross-reactivity (D, 3-day exposure). Counterstaining was done with hematoxylin and eosin.
In PEL, the profiles of vIL-6 (Fig. 3B) and T0.7 (Fig. 1A) were similar. Most cells of the effusion had detectable vIL-6 RNA with a gradient of expression levels. The frequency of those at the high end of expression was similar to the frequency of nut-1-positive cells (Fig. 1B). At the other end of the spectrum, we found in the KS lesion only a few cells with a few copies of vIL-6 RNA (Fig. 3C). In this central part of the lesion, filled with spindle tumor cells that contain detectable levels of T0.7 (not shown), these cells were about as infrequent as the lytic nut-1-containing cells, and we predicted that vIL-6 and nut-1 would colocalize. The collections of HHV-8-infected cells in the subcapsular sinus of the MCD specimen that hybridized to T0.7- and nut-1-specific probes in a manner similar to KS also produced a KS-like hybridization pattern with vIL-6. Specificity of hybridization to the viral homologue of IL-6 was confirmed by lack of hybridization of the hIL-6 probe to a subjacent section of PEL (Fig. 3D).
Quantitative image analysis of appropriately exposed slides confirmed the visual similarities and differences in single-cell levels of viral transcripts among the three diseases. Single-cell levels of T0.7 within the populations of latent or putative productively infected cells were similar in KS, PEL, and MCD. The latent populations in KS and PEL ranged from 30 to 300 copies of T0.7 per cell, and the high-end cells in all three diseases contained an average of 400 copies per cell. There were dramatic differences in the levels of vIL-6 among the three diseases. Cells expressing high levels of vIL-6 in MCD contained an average of 1,800 (range, 270 to 7,942) copies per cell, whereas in PEL and KS, the hybridization-positive cells contained roughly 1/3 (mean, 648; range, 66 to 2,371) and 1/40 (mean, 40; range, 9 to 156) of that number of copies per cell, respectively (Fig. 4).
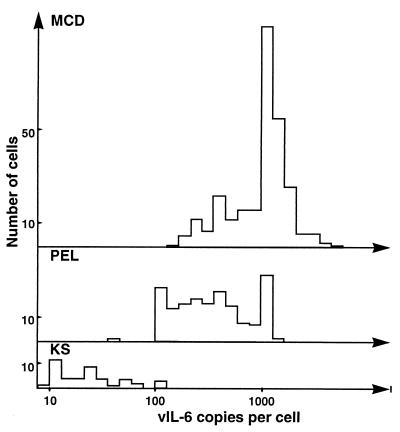
FIG. 4.
Frequency distribution of vIL-6 copy number in MCD, PEL, and KS. The number of copies of vIL-6 RNA in individual infected cells was determined by using computerized image analysis to count grains over randomly selected cells. Grain counts were converted to copy numbers as previously described (6). MCD: range, 270 to 7,942 copies per cell; median, 1,415 copies per cell; mean, 1,790 copies per cell; 281 cells counted. PEL: range, 66 to 2,371 copies per cell; median, 555 copies per cell; mean, 648 copies per cell; 168 cells counted. KS: range, 9 to 156 copies per cell; median, 46 copies per cell; mean, 40 copies per cell; 44 cells counted.
DISCUSSION
We had found previously that virtually all of the KS spindle tumor cells in the KS lesion were infected with HHV-8 but that only a minor population had a transcript profile expected of lytic and productive infections. Similarly, in this study, we found evidence of HHV-8 gene expression in most cells of the PEL. Few of these cells contained detectable levels of the nut-1 gene and other genes, such as that for MCP, that are expressed in the lytic cycle. In dramatic contrast to KS and PEL, MCD appears to contain relatively few infected cells within the lesion and most of these are potentially lytic infections. In this study, the identification and quantitation of infected cells are based on our ability to detect T0.7 RNA, a transcript that has been shown to be expressed in a majority, if not all, of the latently infected cells in both KS lesions and PEL-derived cell lines. It is possible, however, that in MCD, HHV-8 exhibits an alternative program of latency that is characterized by much lower and undetectable levels of T0.7 and that many more cells of the lesion are infected than we describe here. This will be clarified by the refinement and application of a reliable single-cell method for low-copy DNA detection.
Other striking differences that we documented in these pathological states are the types of cells infected and the levels of expression of vIL-6. We were not able to phenotype the PEL, but we determined that in MCD, the majority of infected cells were positive for immunoglobulin light chains and were therefore of B-lymphocyte lineage. We also found HHV-8 infection of a minor population of T cells in MCD. The average levels of vIL-6 RNA in the populations of infected cells in both PEL and MCD was greater by at least an order of magnitude than the low but detectable levels found in a subpopulation of the total KS spindle tumor cells. These data are generally consistent with those of Moore et al. (21) and Parravicini et al. (25), who used a specific antibody to detect relatively high levels of HHV-8 vIL-6 in PEL and MCD, respectively. However, they were unable to consistently detect the vIL-6-containing subpopulation of spindle tumor cells in KS lesions, most likely due to the limited sensitivity of their technology. The morphological characterization of the vIL-6-positive cells of MCD by Parravicini et al. is also consistent with our predominant phenotypic data, but they did not detect vIL-6 in CD3-positive cells as we have shown here. One clear difference between our two studies that warrants further investigation in this regard is the HIV status of the donors. Lastly, of considerable interest is the difference in vIL-6 levels between the latently infected populations of KS and PEL. This finding may signify the existence of at least two cell type-specific programs of latency for HHV-8, a precedent for which exists in Epstein-Barr virus. Variation in the levels of vIL-6 expression is potentially due to the tissue- and cell-specific environments of infection (transcription factors, cytokines, etc.), as well as interactions with other infectious agents. For example, PEL is frequently coinfected with Epstein-Barr virus.
IL-6 is expressed in many types of B-cell lymphomas (4, 11), and we believe that expression of vIL-6, perhaps in conjunction with other potential oncogenes, such as the viral homologue of cyclin D, could play an important role in tumor formation. The elevated levels of IL-6 clinically described for MCD are thought to contribute to the characteristic polyclonal plasmacytosis, hypergammaglobulinemia, and follicular hyperplasia (17, 32). The high levels of vIL-6 in a relatively few B cells documented in this study could act by a paracrine mechanism to drive proliferation and differentiation of B cells in HHV-8-associated MCD. More generally, differential expression of HHV-8 genes in different cell types could be responsible for the heterogeneity of pathological states in these and perhaps as yet undiscovered diseases.
ACKNOWLEDGMENTS
We thank M. Rigsby for specimen acquisition and for critical reading of the manuscript and T. Leonard for preparation of the figures.
This work was supported by Public Health Service grants CA-75172 (K.A.S. and A.T.H.) and CA-70036 (G.M.) from the National Institutes of Health.
REFERENCES
- 1.Ansari M Q, Dawson D B, Nador R, Rutherford C, Schneider N R, Latimer M J, Ricker L, Knowles D M, McKenna R W. Primary body cavity-based AIDS-related lymphomas. Am J Clin Pathol. 1996;105:141–143. doi: 10.1093/ajcp/105.2.221. [DOI] [PubMed] [Google Scholar]
- 2.Arcone R, Pucci P, Zappacosta F, Fontaine V, Malorni A, Marino G, Ciliberto G. Single-step purification and structural characterization of human interleukin-6 produced in Escherichia coli from a T7 RNA polymerase expression vector. Eur J Biochem. 1991;198:541–547. doi: 10.1111/j.1432-1033.1991.tb16048.x. [DOI] [PubMed] [Google Scholar]
- 3.Barozzi P, Luppi M, Masini L, Marasca R, Savarino M, Morselli M, Ferrari M G, Bevini M, Bonacorsi G, Torelli G. Lymphotropic herpes virus (EBV, HHV-6, HHV-8) DNA sequences in HIV negative Castleman’s disease. J Clin Pathol. 1996;49:M232–M235. doi: 10.1136/mp.49.4.m232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burger R, Wendler J, Antoni K, Helm G, Kalden J R, Gramatzki M. Interleukin-6 production in B-cell neoplasias and Castleman’s disease: evidence for an additional paracrine loop. Ann Hematol. 1994;69:25–31. doi: 10.1007/BF01757344. [DOI] [PubMed] [Google Scholar]
- 5.Castleman B, Iverson L, Menendez V P. Localized mediastinal lymph-node hyperplasia resembling thymoma. Cancer. 1956;9:822–830. doi: 10.1002/1097-0142(195607/08)9:4<822::aid-cncr2820090430>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 6.Cesarman E, Knowles D M. Kaposi’s sarcoma-associated herpesvirus: a lymphotropic human herpesvirus associated with Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Semin Diagn Pathol. 1997;14:54–66. [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences are present in AIDS-related body cavity based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 9.Cox K H, DeLeon D V, Angerer L M, Angerer R C. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- 10.Dupin N, Gorin I, Deleuze J, Agut H, Huraux J, Escande J. Herpes-like DNA sequences, AIDS-related tumors, and Castleman’s disease. N Engl J Med. 1995;333:797–799. . (Letter.) [PubMed] [Google Scholar]
- 11.Emilie D, Zou W, Fior R, Llorente L, Durandy A, Crevon M, Maillot M, Durand-Gasselin I, Raphael M, Peuchmaur M, Galamaud P. Production and roles of IL-6, IL-10, and IL-13 in B-lymphocyte malignancies and in B-lymphocyte hyperactivity of HIV infection and autoimmunity. Methods. 1997;11:133–142. doi: 10.1006/meth.1996.0397. [DOI] [PubMed] [Google Scholar]
- 12.Frizzera G. Castleman’s disease and related disorders. Semin Diagn Pathol. 1988;5:346–364. [PubMed] [Google Scholar]
- 13.Ganem D. Human herpesvirus 8 and the biology of Kaposi’s sarcoma. Semin Virol. 1996;7:325–332. [Google Scholar]
- 14.Green I, Espiritu E, Ladanyi M, Chaponda R, Wieczorek R, Gallo L, Feiner H. Primary lymphomatous effusions in AIDS: a morphological, immunophenotypic, and molecular study. Mod Pathol. 1995;8:39–45. [PubMed] [Google Scholar]
- 15.Haase A T. Analysis of viral infections by in situ hybridization. In: Valentino K L, Eberwine J H, Barchas J D, editors. Symposium on in situ hybridization: applications to neurobiology. New York, N.Y: Oxford University Press; 1987. pp. 197–219. [Google Scholar]
- 16.Haase A T, Henry K, Zupancic M, Sedgewick G, Faust R A, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang Z, Dailey P J, Balfour H H, Erice A, Perelson A S. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science. 1996;274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 17.Hsu S, Waldron J A, Xie S, Barlogie B. Expression of interleukin-6 in Castleman’s disease. Hum Pathol. 1993;24:833–839. doi: 10.1016/0046-8177(93)90132-z. [DOI] [PubMed] [Google Scholar]
- 18.Karcher D, Dawkins F, Garret C T, Schulof R F. Body cavity-based non-Hodgkin’s lymphoma (NHL) in HIV-infected patients: B-cell lymphoma with unusual clinical, immunophenotypic, and genotypic features. Lab Investig. 1992;66:80. . (Abstract.) [Google Scholar]
- 19.Kessler E, Beer R. Multicentric giant lymph node hyperplasia clinically simulating angioimmunoblastic lymphadenopathy. Associated Kaposi’s sarcoma in two of three cases. Isr J Med Sci. 1983;19:230–234. [PubMed] [Google Scholar]
- 20.Knowles D M, Inghirami G, Ubriaco A, Dalla-Favera R. Molecular genetic analysis of three AIDS-associated neoplasms of uncertain lineage demonstrates their B-cell derivation and the possible pathogenic role of the Epstein-Barr virus. Blood. 1989;73:792–799. [PubMed] [Google Scholar]
- 21.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 22.Neipel F, Albrecht J, Ensser A, Huang Y, Li J J, Friedman-Kien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nickoloff B J. The human progenitor cell antigen (CD34) is localized on endothelial cells, dermal dendritic cells, and perifollicular cells in formalin-fixed normal skin, and on proliferating endothelial cells and stromal spindle-shaped cells in Kaposi’s sarcoma. Arch Dermatol. 1991;127:523–529. [PubMed] [Google Scholar]
- 24.Oksenhendler E, Duarte M, Soulier J, Cacoub P, Welker Y, Cadranel J, Cazals-Hatem D, Autran B, Clauvel J, Raphael M. Multicentric Castleman’s disease in HIV infection: a clinical and pathological study of 20 patients. AIDS. 1996;10:61–67. [PubMed] [Google Scholar]
- 25.Parravicini C, Corbellino M, Paulli M, Magrini U, Lazzarino M, Moore P S, Chang Y. Expression of a virus-derived cytokine, KSHV vIL-6, in HIV-seronegative Castleman’s disease. Am J Pathol. 1997;151:1517–1522. [PMC free article] [PubMed] [Google Scholar]
- 26.Russo J J, Bohenzky R A, Chien M, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi’s sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi S R, Chaiwun B, Young L, Cote R J, Taylor C R. Antigen retrieval technique utilizing citrate buffer or urea solution for immunohistochemical demonstration of androgen receptor in formalin-fixed paraffin sections. J Histochem Cytochem. 1993;41:1599–1604. doi: 10.1177/41.11.7691930. [DOI] [PubMed] [Google Scholar]
- 28.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M, Clauvel J, Raphael M, Degos L, Sigaux F. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 29.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watts A E, Shintaku I P, Said J W. Diagnosis of malignant lymphoma in effusions from patients with AIDS by gene rearrangement. Am J Clin Pathol. 1990;94:170–175. doi: 10.1093/ajcp/94.2.170. [DOI] [PubMed] [Google Scholar]
- 31.Weisenburger D D, Nathwani B N, Winberg C D, Rappaport H. Multicentric angiofollicular lymph node hyperplasia: a clinicopathologic study of 16 cases. Hum Pathol. 1985;16:162–172. doi: 10.1016/s0046-8177(85)80065-4. [DOI] [PubMed] [Google Scholar]
- 32.Yoshizaki K, Matsuda T, Nishimoto N, Kuritani T, Taeho L, Aozasa K, Nakahata T, Kawai H, Tagoh H, Komori T, Kishimoto S, Hirano T, Kishimoto T. Pathogenic significance of interleukin-6 (IL-6/BSF-2) in Castleman’s disease. Blood. 1989;74:1360–1367. [PubMed] [Google Scholar]
- 33.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi’s sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]