Abstract
Varicella-zoster virus (VZV) is an alphaherpesvirus that is the causative agent of chickenpox and herpes zoster. VZV open reading frame 5 (ORF5) encodes glycoprotein K (gK), which is conserved among alphaherpesviruses. While VZV gK has not been characterized, and its role in viral replication is unknown, homologs of VZV gK in herpes simplex virus type 1 (HSV-1) and pseudorabies virus (PRV) have been well studied. To identify the VZV ORF5 gene product, we raised a polyclonal antibody against a fusion protein of ORF5 codons 25 to 122 with glutathione S-transferase and used it to study the protein in infected cells. A 40,000-molecular-weight protein was detected in cell-free virus by Western blotting. In immunogold electron microscopic studies, VZV gK was in enveloped virions and was evenly distributed in the cytoplasm in infected cells. To determine the function of VZV gK in virus growth, a series of gK deletion mutants were constructed with VZV cosmid DNA derived from the Oka strain. Full and partial deletions in gK prevented viral replication when the gK mutant cosmids were transfected into melanoma cells. Insertion of the HSV-1 (KOS) gK gene into the endogenous VZV gK site did not compensate for the deletion of VZV gK. The replacement of VZV gK at a nonnative AvrII site in the VZV genome restored the phenotypic characteristics of intact recombinant Oka (rOka) virus. Moreover, gK complementing cells transfected with a full gK deletion mutant exhibited viral plaques indistinguishable from those of rOka. Our results are consistent with the studies of gK proteins of HSV-1 and PRV showing that gK is indispensable for viral replication.
Varicella-zoster virus (VZV) is a human herpesvirus. It is the causative agent of chickenpox and shingles; the latter disease is a recurrent infection after a prolonged latency (2). VZV contains a 125-kb genome which encodes the six glycoproteins gB, gC, gE, gH, gI, and gL as well as two putative glycoproteins, gK and gM (9). VZV open reading frame 5 (ORF5) is the homolog of gK proteins in alphaherpesviruses. Although gK proteins in herpes simplex virus type 1 (HSV-1) and pseudorabies virus (PRV) have been characterized and their functions in viral replication have been studied (17, 20, 22), the VZV ORF5 gene product has not been described previously. Glycoprotein K is considered to be essential for viral replication in HSV-1 and PRV, since mutant viruses with insertions or deletions in the UL53 ORF cannot grow in tissue culture (17, 22). Viruses with mutations in gK genes can be recovered only from gK complementing cells, indicating that gK proteins in HSV-1 and PRV are indispensable for viral replication. Function studies of HSV-1 gK partial deletion mutants indicated that gK is required for viral egress as well as for capsid envelopment (17, 20). Two PRV gK insertion mutants, in which the UL53 ORF was truncated after codon 164, produced very few extracellular virions. Although the plating efficiencies of wild-type and gK mutant PRV were similar, the gK mutant formed only single infected cells or smaller plaques, indicating that PRV gK is essential for virus egress but not for entry. A possible role for gK in blocking viral reinfection was also suggested (22).
The formation of syncytia is the hallmark of VZV infection in tissue culture cells. VZV spreads only by cell-cell fusion in tissue culture. VZV glycoproteins gE, gH, gI, and gL are thought to be involved in cell fusion. Full or partial deletions of gI resulted in the inhibition of syncytium formation, indicating that gI acts as a fusion regulator or is directly involved in membrane fusion (24). VZV gH is defined as a fusogenic protein, since an anti-gH monoclonal antibody blocks cell-cell spread (32). The fusogenic property of gH may be regulated by gL (12). In addition to viral glycoproteins, disruption of the VZV dUTPase and the adjacent ORF9A also results in reduced syncytium formation in vitro (34). In contrast to VZV, wild-type HSV does not form syncytia in cell culture. In some HSV strains, the majority of syncytial (syn) mutations analyzed were mapped to the gK gene, although other syn mutations are located in the gene products gB, UL20, and UL24 (3, 5, 6, 11, 19, 25, 31). HSV-1 gK may play a role in regulating the cell fusion process, but it is still unclear how the gK syn mutations affect other functions of the protein.
The predicted amino acid sequence of the VZV ORF5 protein contains putative glycosylation sites and hydrophobic domains that are similar in structure and position to those of gK homologs (Table 1) in the alphaherpesvirus subfamily (26). VZV gK has 28 and 33% amino acid identity with its HSV-1 and PRV homologs, respectively (4, 10). The purpose of this study was to characterize the homolog of HSV-1 gK by in vitro translation, by using anti-gK antibody to identify the ORF5 gene product in infected cells, and to study the functions of VZV glycoprotein K in viral replication and its effects on the formation of syncytia. Our data suggest that VZV gK is essential for viral replication and is a component of the virion. In cell culture or skin tissue, gK is located in the cytoplasm as well as on the cell surface. Transfection of recombinant Oka (rOka) or gK deletion cosmids into gK-expressing cells produced smaller plaques, suggesting that the constitutive expression of gK may inhibit cell-to-cell spread and cell fusion.
TABLE 1.
VZV ORF5 homologsa
| Virusb | Designation | Length (aa) | % Identity/aa | % Similarity/aa | Scorec |
|---|---|---|---|---|---|
| EHV-1 | ORF6 | 343 | 41/139 | 61/209 | 698 |
| BHV-1 | UL53 | 338 | 30/100 | 48/163 | 457 |
| HSV-1 | UL53 | 338 | 28/93 | 52/172 | 446 |
| MDV | ORF53 | 354 | 30/93 | 50/158 | 423 |
| PRV | UL53 | 312 | 33/80 | 56/134 | 420 |
| HSV-2 | UL53 | 338 | 26/87 | 50/165 | 378 |
| GHV-1 | gK | 337 | 34/66 | 51/165 | 257 |
| HCMV | UL22 | 128 | 32/13 | 50/20 | 59 |
Alignment of gK proteins from alphaherpesviruses with the DeCypher II Smith-Waterman Example program (Center for Molecular and Genetic Medicine, Stanford University) shows substantial similarity to VZV. aa, amino acid(s).
EHV-1, equine herpesvirus 1; BHV-1, bovine herpesvirus 1; MDV, Marek’s disease virus; GHV-1, gallid herpesvirus 1; HCMV, human cytomegalovirus.
Optimized score.
MATERIALS AND METHODS
Cells and gK-expressing cell line.
Human melanoma cells were grown in Dulbecco minimal essential medium supplemented with 12% fetal calf serum, penicillin-streptomycin, and amphotericin B (Fungizone). The complementing cell line containing the ORF5 gene was constructed by inserting a PCR fragment containing the VZV gK-coding region into TA cloning sites of the pCR3.1 plasmid vector (Invitrogen, Carlsbad, Calif.) to make the plasmid pCRgK. Subconfluent melanoma cell monolayers were transfected with pCRgK by the Lipofectin method. G418 was added to the medium, and colonies of G418-resistant cells were isolated. Genomic DNAs from gK-transformed cells were analyzed by PCR by using primers gKf and gKb. The expected 1.1-kb ORF5 DNA fragment was detected. K9 is one the cell lines used for cosmid transfection.
Construction of plasmids.
The entire VZV genome of the Oka strain is contained in the following four overlapping SuperCos 1 cosmid vectors: pvFsp4 (nucleotides [nt] 1 to 33211), pvSpe5 (nt 21875 to 62008), pvPme19 (nt 53877 to 96188), and pvSpe21 nt 94208 to 124884) (24). ORF5 spans VZV nt 4252 to 5274, located within the unique long region in the cosmid pvFsp4. An XhoI-SacI 11.4-kb fragment (nt 23 to 11433) containing the gK ORF was subcloned into the plasmid vector pBS to generate pBS-XhoSac11.4. VZV gK was amplified from pBS-XhoSac11.4 with a T7 promoter sequence added at the upstream of ORF5, using primers gKf and gKb (5′-TAATACGACTCACTATAGGGATGCAGGCTTTAGGAATC-3′ and 5′-TTAATGCTTCTGGGAGTTTTC-3′). The PCR DNA was ligated into the pCR3.1 TA cloning vector, forming plasmid pCRgK.
In vitro transcription.
Plasmid DNA pCRgK was digested with EcoRI, and the fragment containing the T7 promoter and gK ORF was used as the template; mRNA was transcribed by using an in vitro T7 transcription kit (MEGAscript; Ambion, Austin, Tex.). Briefly, each transcription reaction contained 1 μg of DNA template, 1× transcription buffer, 7.5 mM (each) nucleoside triphosphate, 1× T7 RNA polymerase mix, and nuclease-free water in a total volume of 20 μl. The transcription mixture was incubated at 37°C for 2 h. Then mRNAs were precipitated by 10 μl of LiCl at −80°C. RNA pellets were washed with 50 μl of 80% ethanol and redissolved in nuclease-free water. RNA transcripts were analyzed by agarose gel electrophoresis before being used for in vitro translation reactions.
In vitro translation.
RNAs were translated in vitro by using a reticulocyte lysate system (Promega, Madison, Wis.). Typically, a 25-μl translational mixture contained 1 to 2 μg of RNA, 2.5 μCi of [35S]methionine (Amersham, Arlington Heights, Ill.), 20 U of RNasin ribonuclease inhibitor, 0.01 mM amino acid mixture (without methionine), and 12.5 μl of rabbit reticulocyte lysate (30). Nuclease-treated microsome membranes (3.5 μl) were added to certain reaction mixtures when posttranslational processing was needed. After 90 min of incubation at 30°C, samples were analyzed on sodium dodecyl sulfate (SDS)-polyacrylamide gels.
Glycosidase treatment.
Endoglycosidase H (endo H) and O-glycosidase (Boehringer Mannheim, Indianapolis, Ind.) were used for carbohydrate digestion of gK proteins. To test for endo H sensitivity, samples of translation reactions were digested by adding 1 μl of 1-U/μl endo H or O-glycosidase with 1 M β-mercaptoethanol and 1% Triton X-100. These samples were incubated at 37°C for 2 h before SDS-polyacrylamide gel electrophoresis (PAGE) analysis.
Preparation of GST-gK fusion proteins and antisera.
The gK amino-terminal region was amplified from pBS-XhoSac11.4 by PCR with primers containing BamHI sites. The PCR product was digested with BamHI and inserted into the expression vector pGEX-2T (Pharmacia Biotech, Milwaukee, Wis.). The resulting plasmid, pGSTNgK, contained nt 75 to 366 of ORF5 fused to the glutathione S-transferase (GST) gene. Plasmid DNA with the proper insert orientation was used to transform Escherichia coli DH5α for expression of GST fusion proteins. Induced proteins were obtained by the method of Frangioni and Neel (14). The GSTNgK fusion protein was separated on a preparative SDS-polyacrylamide gel. The gel fragment containing the 36-kDa fusion protein was excised, homogenized in phosphate-buffered saline (PBS), and used to raise anti-gK antibody. After an initial intramuscular injection of 200 μg of 36-kDa fusion protein, one rabbit was immunized four times at 2-week intervals with 100 μg of the fusion protein. Sera were collected before and after immunization and stored at −20°C.
Western blot analysis.
Protein samples were separated on a 15% Laemmli gel and electrotransferred to a nitrocellulose membrane (Immobilon P; Millipore, Bedford, Mass.). Polyclonal rabbit antiserum (αgKN) at a dilution of 1:500 was used as the probe and detected with donkey anti-rabbit antibody conjugated with horseradish peroxidase (HRP; Amersham, Buckinghamshire, England) at a dilution of 1:1,000.
Virion purification.
At 48 h postinfection, rOka-infected cells were harvested by trypsinization and divided into two portions. Part of the cell suspension was sonicated to release the highly cell-associated virus and then filtered through a 0.2-μm-pore-size filter. Cell-free virions were recovered from the filtrate. The purity of the cell-free virus preparation was verified by Western blot analysis with antibodies specific for VZV gE, the most abundant VZV glycoprotein on the cell surface (28), or ORF61, a heterogeneous phosphoprotein in the nucleus of infected cells (36).
Fluorescence microscopy.
Melanoma cell monolayers growing on glass coverslips were either mock infected or inoculated with VZV-infected cells at a dilution of 1:8. After 48 h, the cells were fixed with 1% formaldehyde and permeabilized with 0.2% Triton X-100. The cells were incubated with rabbit preimmune serum or polyclonal rabbit anti-gK serum (αgKN) before the second antibody, fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G (IgG) (Molecular Probes, Eugene, Oreg.), was added. Cells were examined with a Nikon fluorescence microscope.
Virus-infected skin sections were obtained as described elsewhere (27). The tissue sections were fixed and permeablized. The antibodies used for fluorescence microscopy were αgKN and goat anti-rabbit FITC-conjugated antibody.
Confocal microscopy.
Melanoma cell monolayers growing on glass coverslips were inoculated with VZV-infected cells at a dilution of 1:8. After 48 h, the cells were fixed with 1% formaldehyde and either permeabilized with 0.2% Triton X-100 or not permeabilized. The cells were incubated with primary anti-gI (6B5) and anti-gK sera containing 10% goat normal serum and subsequently washed with PBS-bovine serum albumin before incubation with Texas red-conjugated goat anti-mouse IgG and FITC-conjugated goat anti-rabbit IgG (Molecular Probes). The coverslips were washed with PBS and mounted on glass slides. Cells were examined with a Molecular Dynamics MultiProbe 2010 laser scanning confocal microscope.
Immunogold electron microscopy of VZV-infected melanoma cells.
Melanoma cell monolayers were inoculated with VZV-infected cells at a dilution of 1:8. After incubation for 72 h, cells with 4+ cytopathic effect (CPE) were harvested and resuspended in PBS buffer. To make cryosections for immunogold labeling, cells were fixed with 4% paraformaldehyde and 0.5% glutaraldehyde. Then, the cell pellet was embedded in 10% gelatin. The sample was cut into small pieces and incubated in 2.6 M sucrose and 10% polyvinylpyrrolidone overnight. The sample on the specimen stub was frozen down and cut in the cryochamber. The section was placed on a coated specimen grid covered with a polyvinyl Formvar support film. The section-mounted grids were incubated in primary antibody αgKN at 1:50 or 1:100 dilution in PBS. After the section was washed four times with PBS, 10-nm-diameter gold-conjugated anti-rabbit goat serum (TED PELLA, Inc., Redding, Calif.) at 1:10 dilution was added.
Construction of VZV cosmids.
Unlike UL53 in HSV-1 or PRV, the gK gene sequence does not overlap with its adjacent genes, ORF4 and ORF6. For full deletion of VZV gK, primers Eag@2893 (GCAAAACGGCTGTAGGTCAA) and gKstop4896 (GAAGCATTGGCCATGTGACT) and primers gK@5919Msc (TGCATGGCCAGTGGAAGA) and Hind3@8850 (CCCACTCGCTGTTGTTGCTT) were designed to amplify two sequence fragments adjacent to the gK coding region. Mutagenized sequences are underlined. Nucleotides were mutated to introduce MscI restriction sites into primers gKstop4896 and gK@5919Msc. The PCR product generated by Eag@2893 and gKstop4896 was digested with EagI and MscI. The PCR product of gK@5919Msc and Hind3@8850 was digested with MscI and HindIII. pBS-XhoSac11.4 was digested with EagI and HindIII, and the 8.3-kb fragment containing the plasmid backbone was isolated. These three fragments were ligated together to generate pBSΔgK, which is deleted for the entire gK coding region (VZV nt 4251 to 5279).
Cosmids with partial deletions of VZV gK were made as follows. Regions on either side of the desired deletion were amplified with primers Eag@2893 and delNgK5380 (AAGCTAGCAAGGTTGTTATG) and primers delNgK5910 (TTCCTAAAGCTAGCATGACC) and Hind3@8850. The PCR products were digested with EagI/NheI and NheI/HindIII, respectively. pBS-XhoSac11.4 was digested with EagI and HindIII, and the 8.3-kb fragment was isolated. The product of this triple ligation, pBSΔNgK, has a deletion in VZV nt 4738 to 5271, which encodes 178 amino acids of the N terminus. One hundred eighty-nine amino acids of the C terminus were deleted by PCR mutagenesis with primers Eag@2893 and delCgK4902 (CTCCCGCTAGCATTAATCAT) and primers delCgK5460 (TGGGGGATGCTAGCTAACAG) and Hind3@8850. The PCR products were digested with EagI/NheI and NheI/HindIII, respectively. Ligation of the two PCR fragments with the 8.3-kb fragment from pBS-XhoSac11.4 yielded pBSΔCgK, containing a deletion of VZV nt 4259 to 4822. Similarly, primers Eag@2893 and delCgK4902 and primers delCgK5251 (ATGAGCGCTAGCCTACAAAA) and Hind3@8850 were used to delete 122 amino acids of the C terminus of gK. Triple ligation of the PCR fragments with the plasmid backbone generated pBSΔCgK5251, containing a deletion of VZV nt 4259 to 4620.
Cosmid Fsp-HSVgK was constructed to replace VZV gK with HSV-1 gK. The endogenous VZV gK coding region within pvFsp4 was deleted in a similar manner, except for the creation of an NheI site at the deleted gK site. PCR amplification of the surrounding regions was performed with primers Eag@2893 and delCgK4902 and primers delNgK5910 and Hind3@8850. Triple ligation with the 8.3-kb EagI-HindIII fragment from pBS-XhoSac11.4 generated a deletion of the VZV gK coding region (nt 4259 to 5271), with an NheI restriction site introduced at the site of deletion. The HSV gK coding region was PCR amplified from the plasmid vector CMVgK PCR3.1, a kind gift from K. G. Kousoulas, Louisiana State University. PCR primers HSVgK@2083 (CCATGCTAGCCGTCCGTTCC) and HSVgK@3104 (TTCCGCTAGCCTGGATGTGA) were designed to amplify the 1.0-kb HSV gK coding region, with NheI sites introduced at both termini. The HSV gK coding region was inserted into the endogenous VZV gK site in the negative orientation to generate pBS-HSVgK. The XhoI-SacI fragments within plasmids pBSΔgK, pBSΔNgK, pBSΔCgK, pBSΔCgK5251, and pBS-HSVgK were then subcloned back into the cosmid vector to generate FspΔgK, FspΔNgK, FspΔCgK, FspΔCgK5251, and Fsp-HSVgK, respectively.
Cosmid gK-Spe21 was constructed to place the VZV gK coding region into a unique AvrII site in pvSpe21 (24). The VZV gK coding region as well as approximately 400 bp of surrounding noncoding sequence (nt 4144 to 5559) containing putative promoter sequences and a polyadenylation site was amplified from the template pBS-XhoSac13.4, with AvrII sites introduced at the termini with primers gK@4779Avr (GAGGCCTAGGCTGCAAAATA) and gK@6200Avr (ATCGCCATCCCTAGGGACTG). The PCR fragment and pvSpe21 were both digested with AvrII, and ligation of two AvrII fragments generated gK-Spe21.
Transfections.
Twenty-four hours prior to transfection, 106 human melanoma cells were seeded into a 25-cm2 flask to generate a 25 to 50% confluency on the day of transfection. Five micrograms of pvFsp4, pvSpe5, and pvPme19 and 2.5 μg of pvSpe21 were digested with AscI to release the inserted VZV fragments from the cosmids. DNA from the four cosmids was pooled to a final volume of 200 μl. Transfection was performed with 80 or 120 μl of cosmid mix in 31.5 μl of 2 M CaCl2 in water and HEPES buffer. Cells were incubated at 37°C and passaged every 3 to 5 days for 4 weeks or until plaques were detected. DNAs from transfected cells were harvested when flasks showed a CPE of 3 to 4+ or at 3 weeks if no plaques were visible. DNA isolation was performed with DNazol reagent (Gibco BRL, Gaithersburg, Md.) as per manufacturer instructions.
Sequencing.
Transfections of the full gK deletion mutant yielded no plaques. No viral DNA was detected by PCR amplification with primers Eag@2893 and Hind3@8850. Consequently, cosmid DNA from FspΔgK was used to amplify the gK region for sequencing. The PCR product was purified on Qiagen columns and quantified by gel electrophoresis. The gK region of the PCR product was sequenced with primer gKseq6200 (CGCCATCAAAAGGGACTG), a lower-strand oligonucleotide primer designed to anneal 280 nt upstream of the gK start codon. The gK regions of FspΔNgK, FspΔCgK, FspΔCgK5251, and FspHSVgK were also amplified from cosmid DNA with primers Eag@2893 and Hind3@8850. FspΔNgK was sequenced with primer gKseq6200. FspΔCgK and FspΔCgK5251 were sequenced with primer gKseq4705 (AAGGTTCGTCTGGTAGCA), an upper-strand primer starting 200 nt downstream of the gK stop codon. FspHSVgK was sequenced with both primers gKseq6200 and gKseq4705. Cotransfection of FspΔgK, pvSpe5, pvPme19, and gK-Spe21 resulted in live virus. The gK region was PCR amplified with primers Eag@2893 and Hind3@8850 to verify the deletion in gK at the endogenous site. This region was sequenced with primer gKseq6200. The AvrII region was PCR amplified with primers VZV116194 and VZV117317 to verify the insertion of the gK cassette. The DNA product was sequenced with primer Seq116936 as previously described (24).
RESULTS
In vitro characterization of VZV ORF5 protein (gK).
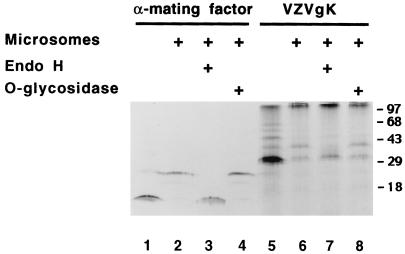
The amino acid sequence predicted from nucleotide sequence analysis of VZV ORF5 consists of 340 amino acids with putative glycosylation sites and several hydrophobic domains (Fig. 1). To study the ORF5 gene product by in vitro translation, the EcoRI DNA fragment from pCRgK containing the T7 promoter and the ORF5 sequence was used as a template for in vitro transcription. The mRNA containing ORF5 was translated in vitro in the presence or absence of microsomal membranes. Protein samples were analyzed by SDS-PAGE 15% (gel) (Fig. 2). A glycosylation control (α-mating factor) was shown in lanes 1 to 4. In the absence of microsomes, gK was synthesized as a 32,000-molecular-weight (32K) protein (lane 5). A few bands larger than 32 kDa were observed in the absence of microsomes. These appear to be ubiquitin-conjugated gK proteins as described in HSV-1 gK studies (26). In the presence of microsomes, the majority of this protein was processed to a 37K form (lane 6).
FIG. 1.
Predicted amino acid sequence of the VZV ORF5 protein. gK is encoded by VZV ORF5 and is homologous to HSV-1 gK (9). The predicted N-terminal signal sequence, two potential sites for N-linked glycosylation, and four hydrophobic domains are underlined (23, 37).
FIG. 2.
Characterization of VZV ORF5 (gK) in vitro. ORF5 transcripts were translated in vitro in the absence or presence (+) of microsomes. A portion of the product translated in the presence of microsomes was treated with endo H or O-glycosidase as indicated (+). α-Mating factor mRNA was used as a control.
To determine whether the increase in molecular weight of the processed form of the ORF5 protein was due to glycosylation, the processed form of protein was treated with endo H or O-glycosidase. The processed form of gK was sensitive to endo H but not to O-glycosidase digestion. Its apparent molecular weight was decreased from 37K to 32K (lane 7). These results indicated that in vitro-translated and processed ORF5 contained N-linked oligosaccharides.
Identification of the VZV ORF5 gene product, gK.
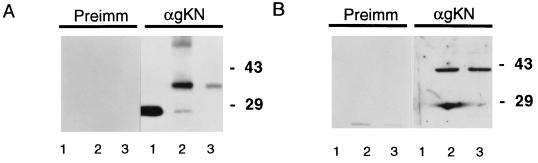
To make antiserum against the ORF5 gene product, the BamHI fragment containing the amino-terminal amino acids 25 to 122 of gK was cloned into the pGEX-2T vector and the GSTgKN fusion protein was expressed in E. coli. The 36K fusion protein was gel purified and used for raising anti-gK antibody. This serum was tested by Western blot analysis. GST protein was detected at 26K in the extract from cells transformed with a GST expression vector (Fig. 3A, lane 1). GSTgKN is detected at 36K in the extract from GSTgKN-expressing cells and gel-purified gK fusion proteins (Fig. 3A, lanes 2 and 3). Only a single protein band at 40K is observed in the extracts from gK-transformed cells (Fig. 3B, lane 2) or rOKa-infected cells (lane 3). These proteins were not recognized by the preimmune serum. The results indicated that the rabbit polyclonal antibody can specifically recognize VZV gK as well as GST fusion protein. To determine whether gK was glycosylated, protein lysates from gK-expressing cells or virus-infected cells were subjected to endo H digestion. Most of the gK protein was resistant to glycosidase digestion as determined by Western blot analysis, indicating that N-linked oligosaccharides on gK are processed (data not shown).
FIG. 3.
The purified GST-gK fusion protein was used to immunize one rabbit. Sera collected before or after immunization were analyzed. Samples were run on a 15% Laemmli gel and transferred to a nitrocellulose membrane. Preimmune sera or polyclonal rabbit antisera (αgKN) against gK were used as probes and detected with anti-rabbit antibody conjugated with HRP. (A) Western blot analysis of GST or GST-gK fusion proteins: GST (lanes 1), GSTgKN (lanes 2), and gel-purified GSTgKN (lanes 3). (B) Expression of VZV gK in gK-transformed cells or rOKa-infected melanoma cells at 48 h: mock-infected cells (lanes 1), gK-transformed cells (lanes 2), and infected cells (lanes 3).
VZV gK is distributed throughout the cytoplasm and surface membrane.
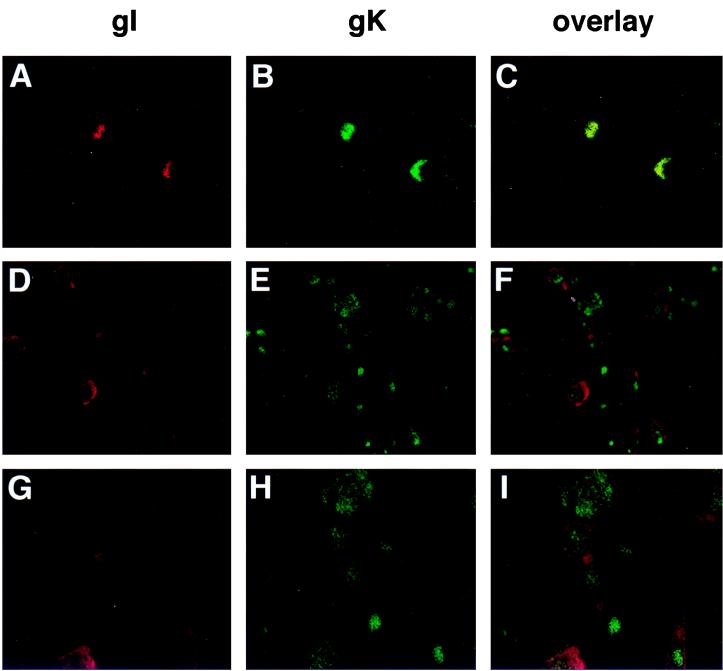
Confocal microscopy was used to compare the localization of VZV gK and gI. VZV gI is found in the plasma membrane, the trans-Golgi network, and the endoplasmic reticulum (ER) (1). rOka-infected melanoma cells were grown on glass coverslips for 2 days, and cells were tested for surface immunofluorescence (Fig. 4A to C) or permeabilized to examine the internal distribution of gK and gI (Fig. 4D to I). When the cells were not permeabilized, there was granular, surface fluorescence (Fig. 4A and B) and gK was found to colocalize with gI (Fig. 4C). In permeabilized cells, gK was evenly distributed throughout the cytoplasm and surface membranes, whereas gI was localized primarily on the cell surface (F and I). VZV gK could be detected in infected skin tissue sections by anti-gK antibody. The staining pattern is the same as that seen in tissue culture cells (data not shown).
FIG. 4.
Immunofluorescence analysis of VZV gI and gK distribution. Melanoma cell monolayers growing on glass coverslips were inoculated with VZV-infected cells. The cells were fixed and either not permeabilized (A to C) or permeabilized (D to I). The cells were incubated with primary mouse anti-gI and rabbit anti-gK sera before incubation with Texas red-conjugated goat anti-mouse IgG and FITC-conjugated goat anti-rabbit IgG. Cells were examined with a Molecular Dynamics MultiProbe 2010 laser scanning confocal microscope. Magnification, ×200 (A to F) and ×400 (G to I).
VZV gK is a component of the virion.
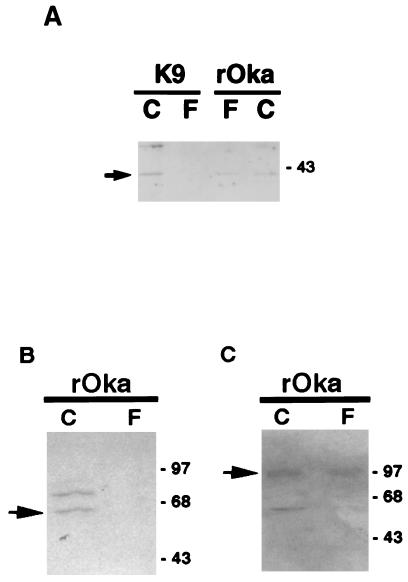
Since VZV is highly associated with cells, cell-free virions were recovered from the filtrate passing through a 0.2-μm-pore-size filter. The purity of cell-free virus was verified by Western blot analysis with antibodies specific for gE, an envelope glycoprotein, or ORF61, a heterogeneous phosphoprotein in VZV-infected cell nuclei. The 98K gE protein was detected in the virion preparation as well as in infected cell lysate (Fig. 5C). On the other hand, a 62K protein was recognized by anti-ORF61 antibody in cell lysate but not in cell-free virus (Fig. 5B). To determine whether gK exists in the virion, Western blot analysis was done with anti-gK serum as the probe (Fig. 5A). The gK protein (arrow) was detected at 40K in the purified virions (rOka, F) as well as in infected cells (rOka, C). A gK-transformed cell line (K9) which expresses cell-associated gK was used as a control. A 40K protein was detected only in the cell lysate of K9 (C) but not in the filtrate of K9 (F). These data indicate that VZV gK may be a component of the virion.
FIG. 5.
gK is a component of the virion. Protein lysates were run on an SDS–12.5% polyacrylamide gel and transferred to a nitrocellulose membrane. (A) Polyclonal rabbit antiserum αgKN was used as the probe and detected with anti-rabbit antibody conjugated with HRP. A gK-transformed cell line (K9) which expresses cell-associated gK was used as a control. (B and C) The purity of cell-free virus was verified with anti-ORF61 and anti-gE sera.
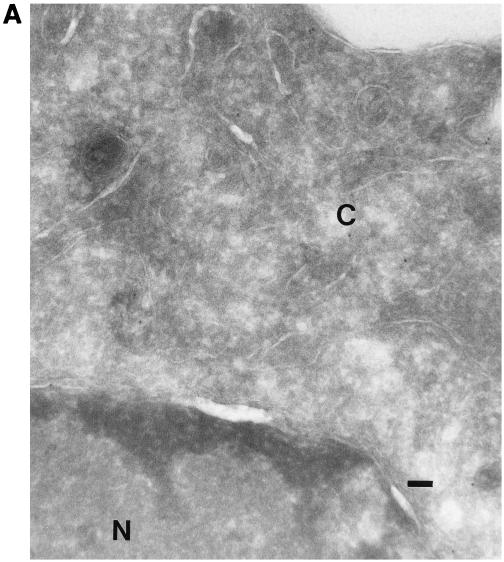
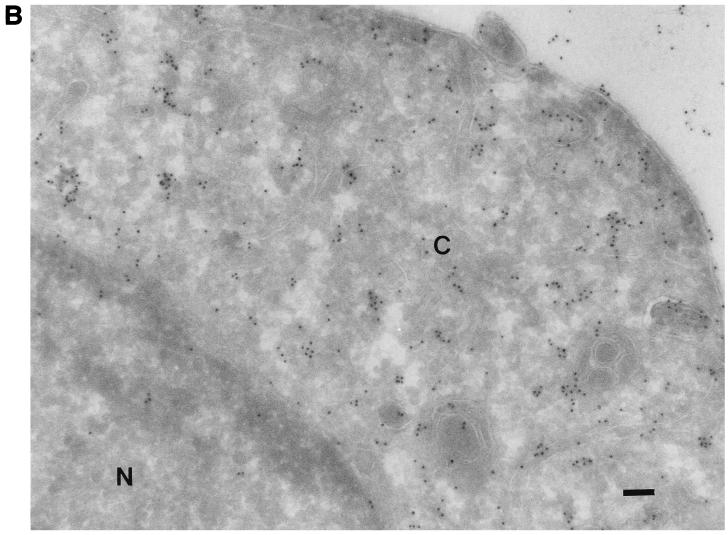
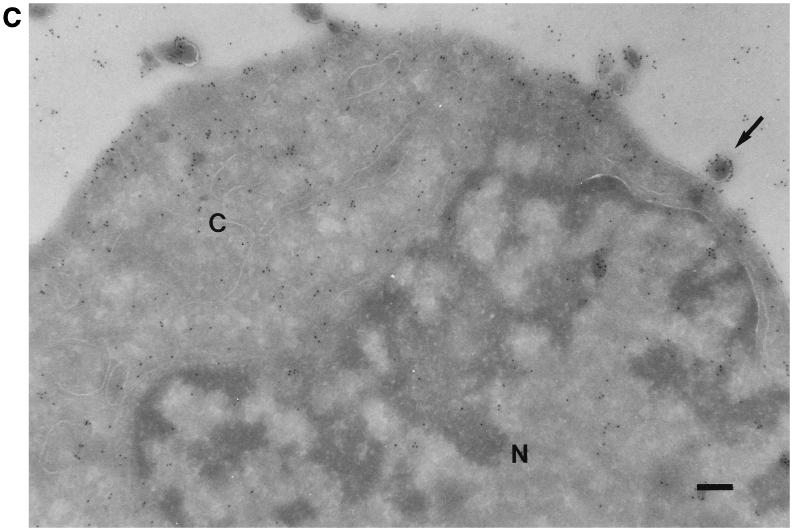
All of the known VZV glycoproteins are inserted into the envelope of the virus (8). To determine whether VZV gK is an envelope glycoprotein, rOka-infected cells were examined by immunogold electron micrography. There was little or no label when preimmune serum was used (Fig. 6A). The gK proteins were seen as small particles with anti-gK antibody. They were mostly found in the cytoplasm and on the cell surface. Little background labeling in the nucleus was observed (Fig. 6B). gK was also detected on the surface of enveloped virions (Fig. 6C), suggesting that gK may be an envelope glycoprotein.
FIG. 6.
Immunogold electron microscopy of VZV-infected melanoma cells. Melanoma cell monolayers were inoculated with VZV-infected cells. The section-mounted grids were incubated in preimmune serum (A) or primary antibody αgKN (B and C) and then in 10-nm-diameter gold-conjugated anti-rabbit goat serum. The cytoplasm (C) and the nucleus (N) are indicated. An enveloped virion released from the cell is indicated by the arrow. Bar, 1 μm (A and B); 2 μm (C).
Failure to generate infectious VZV from cosmids with full or partial deletions of gK.
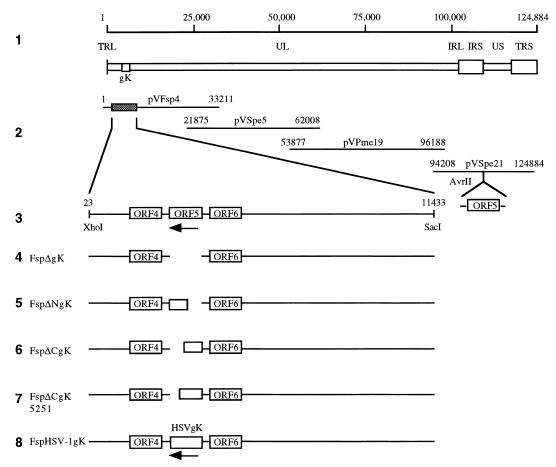
To determine the requirement of VZV gK for virus growth, a series of gK deletion mutants were constructed with VZV cosmid DNA derived from the Oka strain. Removal of VZV nt 4251 to 5279 from pvFsp4 resulted in a complete deletion of the gK coding region (Fig. 7, line 4). Cotransfection of cosmid clones containing full deletions of gK (FspΔgK) with pvSpe5, pvPme19, and pvSpe21 yielded no detectable viral plaques. Transfections of FspΔgK clones were repeated three times, with the same negative result. DNA harvested from transfected cells at 3 weeks posttransfection showed no detectable VZV DNA by PCR analysis. As a positive control, intact cosmids pvFsp4, pvSpe5, pvPme19, and pvSpe21 were cotransfected in parallel with FspΔgK. The wild-type cosmids consistently yielded infectious virus, with plaques visible 6 days posttransfection.
FIG. 7.
Construction of cosmid vectors with deletions in VZV ORF5. To determine the function of VZV gK in virus growth, a series of gK deletion mutants were constructed with the VZV cosmid DNA derived from the Oka strain. Lines: 1, schematic diagram of the VZV genome with the location of the gK gene; 2, overlapping segments of the VZV genome used to construct the VZV cosmids; 3, the subcloned XhoI-SacI fragment from pvFsp4 containing ORF5 (gK); 4 to 7, gK deletion mutants; 8, insertion of HSV-1 gK at the endogenous gK site. Also shown is the unique AvrII site within pvSpe21, where ORF5 as well as 400 bp of noncoding sequence was inserted to generate gK-Spe21.
FspΔNgK has a 530-nt N-terminal deletion, removing 178 of the 340 amino acids of the gK coding region, including the first putative transmembrane domain. The methionine start site is followed by leucine-180. While several nucleotides were mutagenized to create the NheI site, the leucine was preserved due to the degeneracy of the genetic code. The remainder of the triplet coding sequence was retained in frame (Fig. 7, line 5). FspΔCgK has a 560-nt/189-aa deletion of the C terminus, encompassing the second and third putative transmembrane regions. A single base mutation designed in primer delCgK5460 resulted in a stop codon immediately following cysteine-151 (Fig. 7, line 6). FspΔCgK5251 possesses a 360-nt/122-aa C-terminal deletion that also includes the second and third transmembrane regions. A nucleotide substitution in primer delCgK5251 introduced a stop codon immediately after leucine-218 (Fig. 7, line 7). FspΔNgK, FspΔCgK, and FspΔCgK5251 were transfected in parallel along with positive and negative controls. As with the full deletion experiments, only the transfection of overlapping intact rOka cosmids yielded plaques. These results suggested that the intact gK protein was essential for viral replication in melanoma cells.
To examine the possibility of overlapping function(s) between HSV and VZV gK, VZV gK was replaced by the HSV gK coding sequence, generating a transgenic clone that used the promoter and regulatory sequence of VZV gK to drive the transcription of HSV gK. The HSV gK fragment was inserted in a negative orientation because VZV ORF5 is transcribed from the lower strand. Nucleotide substitutions to generate NheI sites for insertional purposes did not alter the amino acid sequence (Fig. 7, line 8). Cotransfection of Fsp-HSVgK was performed twice and yielded no detectable viral plaques up to 4 weeks posttransfection, indicating that HSV gK may not compensate for the deletion of VZV gK. It should be noted that the transcription and translation of HSV gK were not evaluated.
Generation and replication characteristics of rOka gK@AvrII with insertion of the gK gene into a nonnative AvrII site.
gK-Spe21 contains ORF5 with 0.4 kb of surrounding noncoding sequence (282 bp of upstream sequence) inserted into the AvrII site at VZV nt 112853 in the negative orientation (Fig. 7, line 3). Cotransfection of cosmids FspΔgK (containing the full deletion of gK), pvSpe5, pvPme19, and gK-Spe21 resulted in plaques that appeared indistinguishable from those of rOka. PCR detection of DNA harvested from these transfected cells showed the expected 1.0-kb deletion at the endogenous gK site in pvFsp4 (PCR fragment size decreased from 6.0 to 5.0 kb) and the expected 1.4-kb insertion at the AvrII site in pvSpe21 (PCR fragment size increased from 1.1 to 2.5 kb [data not shown]). Our data suggest that gK might be indispensable for viral replication.
Plaque morphology of rOka and rOka gK deletion mutant.
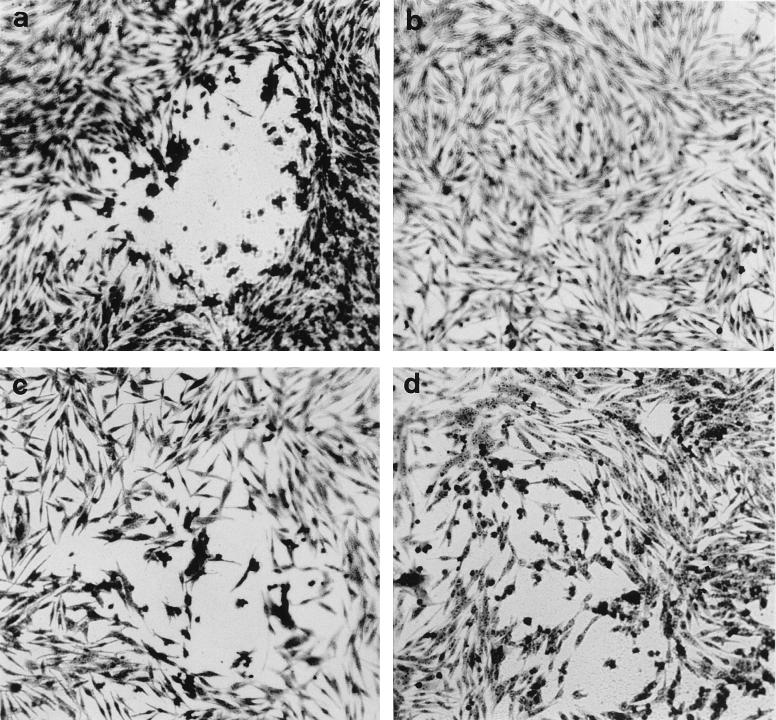
To further prove that VZV gK is essential for viral replication, we made gK-expressing cell lines. Genomic DNAs from gK-transformed cells were analyzed by PCR with primers gKf and gKb. The expected 1.1-kb ORF5 DNA fragment was detected (data not shown). When the protein lysate from gK-transformed cells was analyzed by Western blot analysis, a 40K protein band was observed with anti-gK antibody (Fig. 3B). Cotransfection of gK deletion cosmids into gK-complementing cells yielded viral plaques indistinguishable from those of intact rOka cosmids. Compared with rOka in melanoma cells (Fig. 8a), rOka or ΔgKrOka replication in gK-complementing cells formed smaller plaques and caused less CPE (Fig. 8c and d), suggesting that constitutive expression of gK may inhibit syncytium formation.
FIG. 8.
Plaque morphology of rOka and rOka gK-deletion mutant. Melanoma cells or K9 cells were cotransfected with the cosmid mix by the CaPO4 method. The transfected cells were passaged at a 1:3 ratio every 3 to 4 days until plaques were observed. Then the cells were fixed and stained with crystal violet. (a) rOka in melanoma cells, (b) mock-transfected gK-transformed cells (K9), (c) rOka in K9 cells, (d) ΔgKrOka in K9 cells. Magnification, ×100.
DISCUSSION
In this first characterization of VZV ORF5, we found that it encoded a 40K glycosylated protein in VZV-infected cells and in gK-transformed melanoma cells. The apparent molecular weight of the protein was 32,000 when ORF5 was translated in vitro. In the presence of microsomes, most of the 32K product was processed to a form with a mass of 37K. Processed gK was sensitive to heat denaturation and contained N-linked oligosaccharides, as predicted from the protein sequence. VZV gK was distributed throughout the cytoplasm and was detected at cell surfaces after infection and in constitutively expressing cells. VZV gK was localized in a more punctate pattern, extending from the ER to the cell surface, compared with VZV gI, which was more diffuse on the surface of VZV-infected cells. The detection of VZV gK on the cell surface differed from the perinuclear pattern observed for HSV-1 gK (18). VZV gK was demonstrated to be a component of the virion, indicating that it could play a role in fusion of the virion envelope with the uninfected cell membrane. PRV gK has also been identified as a virion structural component, whereas HSV-1 gK has not been detected in virions (18, 22).
The differences in patterns of VZV and HSV-1 gK localization in cells and virions may imply that these related proteins contribute differently to the viral replication cycle and that some pathways required for viral assembly and egress are not common to these two human alphaherpesviruses (7, 15, 21). However, the characterization of herpesvirus gK gene products is hampered by the technical difficulties of generating potent antibodies that bind to these very hydrophobic membrane proteins. The antiserum against VZV gK, which was elicited with a fusion protein containing 98 amino acids of the gene product, may have enhanced detection of the relatively small quantities of this viral protein present in infected cells and virions by immunofluorescence and Western blot methods. HSV-1 gK has been detected primarily in perinuclear sites, consistent with the accepted model for HSV-1 assembly, in which viral nucleocapsids are enveloped at the inner nuclear membrane and then transported to the cell surface within vacuoles (7, 33). HSV gK appears to remain sensitive to endo H, indicating that it does not reach the Golgi but is retained in the ER and nuclear envelope (18). In our experiments, VZV gK was sensitive to endo H, but mature, resistant forms of the protein were also detected. The analysis of VZV replication indicates that the initial viral envelope, obtained at the inner nuclear membrane, does not contain any VZV glycoproteins (15, 16). Instead, the nascent viral particles appear to acquire VZV glycoproteins during transit through the trans-Golgi network. The generalized distribution of gK was similar to that of other VZV glycoproteins in infected cells and implies that gK is accessible for incorporation into virions at sites beyond the perinuclear area. The detection of VZV gK at multiple sites, including the ER, Golgi, and cytoplasm, indicates that VZV gK could act as a chaperone protein during viral egress. VZV gK was also detected diffusely in cutaneous epithelial cells in biopsies of varicella skin lesions, suggesting that the observations of gK trafficking in tissue culture cells were representative of its distribution during productive infection in vivo and were not associated only with the limited replication of VZV in vitro.
Although fusion has been considered to be a major function of the herpesvirus gK gene products, based on observations about the HSV-1 syn mutations, the biological activities of these proteins are not well understood. Recent HSV-1 gK and PRV gK experiments demonstrate that gK affects the movement of infectious virions from the regions of assembly to the cell surface and their release into extracellular spaces (20, 22). Our analysis of full and partial deletions of ORF5 demonstrated that VZV gK, like its homologs in HSV-1 and PRV, was critical for viral replication (17, 20, 22). HSV-1 gK mutants replicated to a limited degree in rapidly dividing cells, but these mutations were associated with accumulation of unenveloped virions in the cytoplasm (20). The requirement for VZV gK expression was documented by the restoration of infectivity by the insertion of the ORF5 sequence into a nonnative site in the Us region of the genome or by transfection into the gK-complementing cell line. The failure to generate infectious VZV from cosmid transfections when gK was deleted indicates that this glycoprotein must have an essential role in the VZV replication cycle in addition to any functions it may have related to viral entry. A connection between the occurrence of the HSV-1 syn mutation and the failure to release virus has been suggested (17). In the case of VZV, syncytium formation and cytoplasmic retention of virions, with extensive intracellular degradation, are characteristic of low-passage as well as tissue culture-adapted virus (2). Whether this phenotype reflects some specific interference with normal VZV gK function in vitro is of interest, since VZV replicates efficiently and infectious virus is released from differentiated T cells and skin in vivo (27). Of note, the residues to which HSV-1 gK syn mutations have been mapped are the same in VZV and wild-type HSV-1.
VZV gK and gB and the products of ORF35 and ORF39 are the homologs of HSV-1 gB and gK and the UL20 and UL24 proteins, to which fusion effects have been mapped (8, 35). In the case of HSV-1 gK, the fusion regulatory effect is considered to be inhibitory in nature, protecting against fusion with cellular membranes during intracellular transport. The syn mutation is thought to block an essential function of gK, allowing the enhanced expression of other HSV-1 proteins that make up the fusion complex on the cell surface. Our experiments indicate that VZV gK has some of the fusion inhibitory effects of HSV-1 gK. Plaques were smaller and cytopathic changes were more limited when infectious virus was generated by transfecting gK-expressing cells with intact rOka or with gK deletion cosmids. The overexpression of VZV gK in this cell line was associated with an inhibition of the characteristic syncytium formation expected with VZV replication in tissue culture cells.
The products of several VZV immediate-early genes, such as ORF61, ORF62, and ORF51, can complement the functions of their HSV-1 homologs (13, 29, 38). In contrast, the insertion of the HSV-1 gK sequence did not compensate for the deletion of VZV gK by restoring viral replication. This observation provides further evidence that while the glycoproteins of VZV and their homologs in HSV-1 have many structural similarities, their biological functions have important differences (8).
Our studies suggest an essential role for VZV gK, consistent with the observation that the gK homologs are almost as highly conserved as the gB homologs among alphaherpesvirus glycoproteins. VZV gK has four hydrophobic domains, indicating that it has a complex transmembrane structure. In HSV-1 gK, this complex tertiary structure was shown to be critical for its biological function (26). We found that relatively large partial deletions of the N-terminal or C-terminal residues of VZV gK which interfered with predicted extracellular domains were incompatible with viral replication. Important questions about gK function in VZV as well as in the other alphaherpesviruses remain to be addressed, including the role of gK in viral entry, membrane fusion between infected and adjacent cells, and virion transport out of the infected cell and the relationship of these putative functions to each other. The functional domains of gK that are required for VZV replication are being defined by site-directed mutagenesis in ORF5 constructs.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI20459 to A.M.A. C.M. was supported by a Medical School Dean’s fellowship.
We thank Nafisa Ghori, Stanford University Department of Microbiology, for help with electron microscopy. We thank Chris Canfield and Susan Palmieri, Stanford University Cell Sciences Imaging Facility, for assistance with the confocal microscopy. We thank Charles Grose, University of Iowa, for providing anti-gI antibody and George Kemble, Aviron, Inc., for providing the VZV cosmids.
REFERENCES
- 1.Alconada A, Bauer U, Baudoux L, Piette J, Hoflack B. Intracellular transport of the glycoproteins gE and gI of the varicella-zoster virus. gE accelerates the maturation of gI and determines its accumulation in the trans-Golgi network. J Biol Chem. 1998;273:13430–13436. doi: 10.1074/jbc.273.22.13430. [DOI] [PubMed] [Google Scholar]
- 2.Arvin A M. Varicella-zoster virus. In: Fields B N, Knipe D N, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2547–2585. [Google Scholar]
- 3.Baines J D, Ward P L, Campadelli-Fiume G, Roizman B. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol. 1991;65:6414–6424. doi: 10.1128/jvi.65.12.6414-6424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumeister J, Klupp B G, Mettenleiter T C. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J Virol. 1995;69:5560–5567. doi: 10.1128/jvi.69.9.5560-5567.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond V C, Person S. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology. 1984;132:368–376. doi: 10.1016/0042-6822(84)90042-4. [DOI] [PubMed] [Google Scholar]
- 6.Bzik D J, Fox B A, DeLuca N A, Person S. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology. 1984;137:185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- 7.Campadelli-Fiume G, Farabegoli F, Di Gaeta S, Roizman B. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J Virol. 1991;65:1589–1595. doi: 10.1128/jvi.65.3.1589-1595.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J I, Straus S E. Varicella-zoster virus and its replication. In: Fields B N, Knipe D N, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2525–2545. [Google Scholar]
- 9.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 10.Debroy C, Pederson N, Person S. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology. 1985;145:36–48. doi: 10.1016/0042-6822(85)90199-0. [DOI] [PubMed] [Google Scholar]
- 11.DeLuca N, Bzik D J, Bond V C, Person S, Snipes W. Nucleotide sequences of herpes simplex virus type 1 (HSV-1) affecting virus entry, cell fusion, and production of glycoprotein gB (VP7) Virology. 1982;122:411–423. doi: 10.1016/0042-6822(82)90240-9. [DOI] [PubMed] [Google Scholar]
- 12.Duus K M, Grose C. Multiple regulatory effects of varicella-zoster virus (VZV) gL on trafficking patterns and fusogenic properties of VZV gH. J Virol. 1996;70:8961–8971. doi: 10.1128/jvi.70.12.8961-8971.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felser J M, Straus S E, Ostrove J M. Varicella-zoster virus complements herpes simplex virus type 1 temperature-sensitive mutants. J Virol. 1987;61:225–228. doi: 10.1128/jvi.61.1.225-228.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frangioni J V, Neel B G. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 15.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harson R, Grose C. Egress of varicella-zoster virus from the melanoma cell: a tropism for the melanocyte. J Virol. 1995;69:4994–5010. doi: 10.1128/jvi.69.8.4994-5010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchinson L, Johnson D C. Herpes simplex virus glycoprotein K promotes egress of virus particles. J Virol. 1995;69:5401–5413. doi: 10.1128/jvi.69.9.5401-5413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchinson L, Roop-Beauchamp C, Johnson D C. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J Virol. 1995;69:4556–4563. doi: 10.1128/jvi.69.7.4556-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobson J G, Martin S L, Coen D M. A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J Virol. 1989;63:1839–1843. doi: 10.1128/jvi.63.4.1839-1843.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayachandra S, Baghian A, Kousoulas K G. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J Virol. 1997;71:5012–5024. doi: 10.1128/jvi.71.7.5012-5024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp B G, Baumeister J, Dietz P, Granzow H, Mettenleiter T C. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J Virol. 1998;72:1949–1958. doi: 10.1128/jvi.72.3.1949-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Mallory S, Sommer M, Arvin A M. Mutational analysis of the role of glycoprotein I in varicella-zoster virus replication and its effects on glycoprotein E conformation and trafficking. J Virol. 1997;71:8279–8288. doi: 10.1128/jvi.71.11.8279-8288.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGeoch D J, Dalrymple M A, Davison A J, Donlan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 26.Mo C, Holland T C. Determination of the transmembrane topology of herpes simplex virus type 1 glycoprotein K. J Biol Chem. 1997;272:33305–33311. doi: 10.1074/jbc.272.52.33305. [DOI] [PubMed] [Google Scholar]
- 27.Moffat J F, Zerboni L, Kinchington P R, Grose C, Kaneshima H, Arvin A M. Attenuation of the vaccine Oka strain of varicella-zoster virus and role of glycoprotein C in alphaherpesvirus virulence demonstrated in the SCID-hu mouse. J Virol. 1998;72:965–974. doi: 10.1128/jvi.72.2.965-974.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montalvo E A, Parmley R T, Grose C. Structural analysis of the varicella-zoster virus gp98-gp62 complex: posttranslational addition of N-linked and O-linked oligosaccharide moieties. J Virol. 1985;53:761–770. doi: 10.1128/jvi.53.3.761-770.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriuchi H, Moriuchi M, Smith H A, Straus S E, Cohen J I. Varicella-zoster virus open reading frame 61 protein is functionally homologous to herpes simplex virus type 1 ICP0. J Virol. 1992;66:7303–7308. doi: 10.1128/jvi.66.12.7303-7308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelham H R, Jackson R J. An efficient mRNA-dependent translational system from reticulocyte lysates. Eur J Biochem. 1976;67:247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- 31.Pogue-Guile K L, Spear P G. The single base pair substitution responsible for the syn phenotype of herpes simplex virus type 1, strain MP. Virology. 1987;157:67–74. doi: 10.1016/0042-6822(87)90314-x. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez J E, Moninger T, Grose C. Entry and egress of varicella virus blocked by same anti-gH monoclonal antibody. Virology. 1993;196:840–844. doi: 10.1006/viro.1993.1543. [DOI] [PubMed] [Google Scholar]
- 33.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1043–1107. [Google Scholar]
- 34.Ross J, Williams M, Cohen J I. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology. 1997;234:186–195. doi: 10.1006/viro.1997.8652. [DOI] [PubMed] [Google Scholar]
- 35.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press; 1993. pp. 201–232. [Google Scholar]
- 36.Stevenson D, Colman K L, Davison A J. Characterization of the varicella-zoster virus gene 61 protein. J Gen Virol. 1992;73:521–530. doi: 10.1099/0022-1317-73-3-521. [DOI] [PubMed] [Google Scholar]
- 37.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster C B, Chen D, Horgan M, Olivo P D. The varicella-zoster virus origin-binding protein can substitute for the herpes simplex virus origin-binding protein in a transient origin-dependent DNA replication assay in insect cells. Virology. 1995;206:655–660. doi: 10.1016/s0042-6822(95)80084-0. [DOI] [PubMed] [Google Scholar]