Abstract
A sustainable enzymatic strategy for the preparation of amides by using Candida antarctica lipase B as the biocatalyst and cyclopentyl methyl ether as a green and safe solvent was devised. The method is simple and efficient and it produces amides with excellent conversions and yields without the need for intensive purification steps. The scope of the reaction was extended to the preparation of 28 diverse amides using four different free carboxylic acids and seven primary and secondary amines, including cyclic amines. This enzymatic methodology has the potential to become a green and industrially reliable process for direct amide synthesis.
Keywords: sustainable enzymatic strategy, direct amide synthesis, green solvent, CALB, carboxylic acid, amine
1. Introduction
The amide bond is a fundamental linkage in nature. It is the main chemical bond that links amino acid building blocks together to give peptides and proteins, which occur worldwide [1,2,3]. Furthermore, as an important moiety of pharmaceutically active compounds, it can be found in a significant array of commercial drugs worldwide. For example, Acetaminophen, a common pain reliever and an antipyretic agent, is used to treat various conditions such as headache, muscle aches, and arthritis [4]. Amide-based local anesthetics are applied to numb a specific area of the body, before a medical procedure or surgery [5,6]. β-Lactam antibiotics are a group of antibiotics that are used to treat bacterial infections, including pneumonia, bronchitis, and urinary tract infections [7,8,9]. Celecoxib is a nonsteroidal anti-inflammatory drug (NSAID) utilized to treat pain and inflammation associated with conditions, such as arthritis, menstrual cramps, and sport injuries [10,11]. These are just a few examples of amide drugs, and there are many others used in the treatment of various medical conditions.
A large number of synthetic methods resulting in the formation of amide bonds have been devised in the last decade [12,13,14,15,16,17,18,19]. However, there are only a limited number of strategies that are both efficient and environmentally benign [20,21,22]. The most common processes utilize coupling reagents or activating agents with larger stoichiometric ratio to couple a free carboxylic acid with an amine. However, these are generally hazardous/poisonous reagents and, consequently, they put a heavy burden on the environment. Furthermore, the purification of the crude products is problematic, since it requires a large quantity of organic solvents, due to the formation of large quantities of by-products [23,24]. Therefore, there is a great demand to develop simple amide-bond-forming reactions to access amides from free carboxylic acids and amines in a green and efficient way.
The green chemistry concept has 12 principles aimed at the design of chemical products and processes, that reduce or eliminate the application and generation of hazardous substances, which are harmful for human health or the environment [25,26]. Using enzymes in synthetic chemistry has always been a hot topic due to the ability of enzymes to catalyze chemical transformations with high catalytic efficiency and specificity [27,28,29,30,31]. For instance, the reactions mentioned above carried out under harsh conditions can be induced to proceed faster under mild conditions (lower temperatures and pressures, neutral pH), with fewer work-up steps and higher yields. All of these result in an improvement in efficiency and save energy. Enzymes have emerged as preferred tools in green chemistry by replacing hazardous/poisonous reagents, generating the formation of fewer by-products [32,33,34,35].
In the case of amide bond formation, enzymes, particularly the members of the lipase family, are powerful and effective biocatalysts for esterification reactions [36,37,38]. Several lipases have been reported to exhibit high catalytic activity and stability in organic solvents [39,40]. In particular, Candida antarctica lipase B (CALB; Novozyme 435) prefers anhydrous conditions and it has been widely applied in esterification and hydrolysis studies [41,42,43,44,45,46]. CALB could also catalyze amidation reactions [47,48,49,50,51,52] when the amine is used as a nucleophile in anhydrous organic media. In these reactions, amine amidation with a free carboxylic acid takes place, resulting in the amide product. A previous study reported by the Manova group [53] showed that CALB could be a simple and convenient biocatalyst for the efficient, direct amidation of free carboxylic acids with amines applied for a wide range of substrates, including lipoic acid.
Thus, the CALB enzymatic approach could offer the possibility of accomplishing direct amide coupling in an efficient and sustainable way without any additives in green organic solvents, providing amides with high yields and excellent purity.
Herein, we report a sustainable amidation strategy through CALB-catalyzed coupling of free acyclic carboxylic acids with different primary and secondary amines in a prominent green solvent (Scheme 1). In order to follow the progress of the enzymatic reactions, we also develop an adequate gas chromatography–mass spectrometry (GC-MS) analytic method.
Scheme 1.
CALB-catalyzed synthesis of amides.
2. Results and Discussion
In view of the results on the enzymatic amidation of free carboxylic acids with amines [49], CALB-catalyzed amidation of octanoic acid (1) with benzylamine (5) in the presence of a molecular sieve in toluene at 60 °C was performed (Figure 1a curve I). An excellent conversion of >99% after 30 min was observed.
Figure 1.
The effect of temperature (a) and solvents (b) on the reaction conversion catalyzed by CALB.
In order to increase the reaction rate, further preliminary experiments were performed at different temperatures (Figure 1a curves II–IV). When the amidation was performed at 25 °C, the desired product with 78% conversion was obtained after 30 min (curve IV). By applying a higher temperature of 50 °C, the conversion of the reaction improved remarkably, reaching a >99% conversion in 60 min (curve III). However, at temperatures higher than 60 °C, the conversion decreased slightly, because of the thermal denaturation process of CALB protein chains (curve II). The optimal temperature of 60 °C was chosen for further reactions.
Next, we screened organic solvents with different types of polarity such as acetonitrile and N,N-dimethylformamide (DMF). In addition, we focused on the application of greener alternative solvents, such as propylene carbonate (PC), 2-methyltetrahydrofuran (2-MeTHF), diisopropyl ether (DIPE), and cyclopentyl methyl ether (CPME). While the reaction rate in acetonitrile was relatively low, CPME and PC were the most promising green solvents with conversions reaching >99% in 30 min (Figure 1b).
In an attempt to increase the efficiency of the present method, the initial concentration of the substrate of 46 mM was increased to 92, 460, and 920 mM (Figure 2a,b). Reactions were performed in CPME and PC solvents with a constant enzyme concentration of 50 mg mL–1. Slightly lower reaction rates with 92 mM concentration (curves II(a) and (b)) were observed. Despite the much lower reaction rates found at concentrations of 460 and 920 mM, the amide formation was still significant after 60 min (curves III(a) and (b) and IV(a) and (b)). Such robust behavior of commercially available CALB might be an important parameter not only for laboratory-scale but also industrial-scale reactions [54,55].
Figure 2.
The efficiency of amide formation catalyzed by CALB in CPME (a) and PC (b).
Having in hand the optimal conditions (CALB, molecular sieve 3 Å, CPME solvent, 60 °C, substrate concentration 920 mM), we performed further amidation reactions and obtained 28 different amides (12–39) with excellent conversions (>92%) and yields (>90%) in 90 min (Table 1). The study involved the use of four different free carboxylic acids and seven amines including primary, secondary, and secondary cyclic amines. According to GC-MS analysis, all reactions were completed in 90 min. The product molecules might be used as potential intermediates or building blocks in the synthesis of biologically active compounds [56,57].
Table 1.
Substrate scope of amide formation in CPME solvent with conversion data.
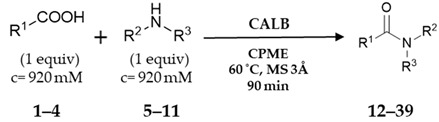
| ||||
|---|---|---|---|---|
| Substrates | Octanoic Acid (1) | Hexanoic Acid (2) | Butyric Acid (3) | 4-Phenylbutyric Acid (4) |
| benzylamine (5) |
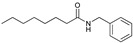 12, 99% |
 13, 98%, |
 14, 98% |
 15, 99% |
| allylamine (6) |
 16, 96% |
 17, 96% |
 18, 94% |
 19, 98% |
| propargylamine (7) |
 20, 97% |
 21, 98% |
 22, 92% |
 23, 97% |
| piperidine (8) |
 24, 95% |
 25, 93% |
 26, 92% |
 27, 97% |
| morpholine (9) |
 28, 99% |
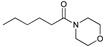 29, 98% |
 30, 94% |
 31, 96% |
| N1,N1-dimethylethane-1,2-diamine (10) |
 32, 96% |
 33, 98% |
 34, 93% |
 35, 97% |
| N1,N1-dimethylpropane-1,3-diamine (11) |
 36, 99% |
 37, 98% |
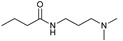 38, 92% |
 39, 99% |
3. Materials and Methods
3.1. General
All solvents and reagents were of analytical grade and used directly without further purification. The lipase acrylic resin (CALB, ≥5000 U/g, recombinant, expressed in Aspergillus niger, quality level 200, Catalogue no. L4777), cyclopentyl methyl ether (inhibitor-free, anhydrous, ≥99.9%), propylene carbonate (ReagentPlus®, 99%), toluene (anhydrous, 99.8%), 2-methyltetrahydrofuran (BioRenewable, anhydrous, ≥99%, inhibitor-free), acetonitrile (suitable for HPLC, gradient grade, ≥99.9%), N,N-dimethylformamide (suitable for HPLC, ≥99.9%), and 3 Å molecular sieve (beads, 8–12 mesh) used in this study were purchased from Merck Life Science Kft., an affiliate of Merck KGaA, Darmstadt, Germany (Budapest, Hungary). All reactions were carried out in an Eppendorf™ Innova™ 40R Incubator Shaker. Melting points were determined on a Kofler apparatus. 1H NMR and 13C NMR spectra were recorded on a Bruker Avance NEO 500.1 spectrometer, in CDCl3 as solvent, with tetramethylsilane as an internal standard at 500.1 and 125 MHz, respectively. GC-MS analyses were performed on a Thermo Scientific Trace 1310 Gas Chromatograph coupled with a Thermo Scientific ISQ QD Single-Quadrupole Mass Spectrometer using a Thermo Scientific TG-SQC column (15 m × 0.25 mm ID × 0.25 μm film). Measurement parameters were as follows: column oven temperature: from 50 to 300 °C at 15 °C min−1; injection temperature: 240 °C; ion source temperature: 200 °C; electrospray ionization: 70 eV; carrier gas: He at 1.5 mL min−1; injection volume: 5 μL; split ratio: 1:50; and mass range: 25–500 m/z.
3.2. General Procedure for CALB-Catalyzed Amidation
In preliminary amidation experiments, the carboxylic acid and amine substrates (1:1 equiv) were dissolved in an organic solvent (1 mL) to provide a solution with a given concentration (46, 92, 460, or 920 mM). CALB (50 mg), molecular sieve (50 mg 3 Å size) to avoid the reversible hydrolysis reaction, and n-heptadecane (2 µL) as an internal standard were added to the above solution. The mixture was shaken at a selected temperature (25, 50, 60 or 70 °C) in an incubator shaker. The progress of the reaction was followed by taking samples from the reaction mixture at intervals and analyzing them by GC-MS measurements. Conversion of the starting materials (moles of the converted molecules/moles of the initial starting materials × 100) was calculated by n-heptadecane as an internal standard, in percent yield of the desired amides (actual yield/theoretical yield × 100) (see Supplementary Information). In order to obtain reliable yields, we filtered the CPME samples through a silica gel plug followed by vacuum evaporation of the solvent. All samples were analyzed by 1H and 13C NMR spectroscopy without any prior purification. Amidations, to form amides 12–39, were performed with the above protocol under the optimized conditions (50 mg CALB, 920 mM substrate concentration, 1 mL solvent CPME, 50 mg molecular sieve, 2 µL n-heptadecane, 60 °C).
3.2.1. N-Benzyloctanamide (12)
CAS number: 70659-87-9, white solid, mp = 65.1–66.3 °C [58]. 1H-NMR (CDCl3, 500.1 MHz): δ = 7.25–7.34 (m, 5H, Ar), 5.69 (s, 1H, NH), 4.44 (d, J = 5.69 Hz, 2H, CH2), 2.20 (t, J = 7.86 Hz, 2H, CH2CO), 1.62–1.68 (m, 2H, CH2), 1.27–1.33 (m, 8H, CH2CH2CH2CH2), 0.87 (t, J = 7.16 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 14.05, 22.59, 25.77, 28.99, 29.27, 29.69, 31.68, 36.84, 43.60, 127.50, 127.84, 128.71, 138.45, 172.93.
3.2.2. N-Benzylhexanamide (13)
CAS number: 6283-98-3, yellowish white solid, mp = 55.1–55.5 °C [59]. 1H-NMR (CDCl3, 500.1 MHz): δ = 7.25–7.33 (m, 5H, Ar), 5.84 (s, 1H, NH), 4.42 (d, J = 5.73 Hz, 2H, CH2), 2.19 (t, J = 7.87 Hz, 2H, CH2CO), 1.62–1.68 (m, 2H, CH2), 1.27–1.34 (m, 4H, CH2CH2), 0.88 (t, J = 7.04 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.92, 22.39, 25.45, 29.69, 31.48, 36.75, 43.56, 53.42, 127.46, 127,80, 128.68, 138.47, 173.03.
3.2.3. N-Enzylbutyramide (14)
CAS number: 10264-14-9, yellowish solid, mp = 43.0–43.6 °C [60]. 1H-NMR (CDCl3, 500.1 MHz): δ = 7.23–7.31 (m, 5H, Ar), 6.14 (s, 1H, NH), 4.39 (d, J = 5.75 Hz, 2H, CH2), 2.16 (t, J = 7.64 Hz, 2H, CH2CO), 1.62–1.69 (m, 2H, CH2), 0.93 (t, J = 7.42 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.78, 19.18, 29.69, 38.58, 43.46, 53.45, 127.38, 127.73, 128.63, 138.54, 173.00.
3.2.4. N-Benzyl-4-phenylbutanamide (15)
CAS number: 179923-27-4, yellowish solid, mp = 79.2–80.2 °C [61]. 1H-NMR (CDCl3, 500.1 MHz): δ = 7.13–7.32 (m, 10H, Ar), 5.84 (s, 1H, NH), 4.39 (d, J = 5.73 Hz, 2H, CH2), 2.63 (t, J = 7.54 Hz, 2H, CH2), 2.18 (t, J = 7.78 Hz, 2H, CH2CO), 1.94–2.00 (m, 2H, CH2), 13C-NMR (CDCl3, 125 MHz): δ = 27.13, 29.72, 35.22, 35.85, 43.59, 46.42, 70.52, 72.51, 125.99, 127.51, 127.83, 128.41, 128.51, 128.71, 138.41, 141.48, 172, 57.
3.2.5. N-Allyloctanamide (16)
CAS number: 70659-85-7, yellow solid, mp = 27.6–28.3 °C [62]. 1H-NMR (CDCl3, 500.1 MHz): δ = 5.84 (ddt, J = 15.97 Hz, 10.29 Hz, 5.09 Hz, 1H, CH), 5.53 (s, 1H, NH), 5.11–5.19 (m, 2H, CH2), 3.88 (t, J = 5.73 Hz, 2H, CH2), 2.19 (t, J = 7.79 Hz, 2H, CH2CO), 1.61–1.66 (m, 2H, CH2), 1,25–1.31 (m, 8H, CH2CH2CH2CH2), 0.87 (t, J = 7.03 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 14.04, 22.59, 25.77, 29.00, 29.27, 31.67, 36.81, 41.86, 116.27, 134.42, 172.94.
3.2.6. N-Allylhexanamide (17)
CAS number: 128007-44-3, yellowish oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 5.84 (ddt, J = 15.96 Hz, 10.27 Hz, 5.04 Hz, 1H, CH), 5.55 (s, 1H, NH), 5.11–5.20 (m, 2H, CH2), 3.88 (t, J = 5.74 Hz, 2H, CH2), 2.19 (t, J = 7.81 Hz, 2H, CH2CO), 1.61–1.67 (m, 2H, CH2), 1.29–1.35 (m, 4H, CH2CH2), 0.89 (t, J = 7.08 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.91, 22.39, 25.45, 31.47, 36.77, 41.86, 116.29, 134.41, 172.97.
3.2.7. N-Allylbutyramide (18)
CAS number: 2978-29-2, yellowish oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 5.84 (ddt, J = 16.00 Hz, 10.32 Hz, 5.06 Hz, 1H, CH), 5.46 (s, 1H, NH), 5.12–5.20 (m, 2H, CH2), 3.89 (t, J = 5.74 Hz, 2H, CH2), 2.17 (t, J = 7.68 Hz, 2H, CH2CO), 1.64–1.71 (m, 2H, CH2), 0.96 (t, J = 7.36 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.77, 19.16, 29.69, 38.72, 41.86, 116.33, 134.40.
3.2.8. N-Allyl-4-phenylbutanamide (19)
CAS number: 430450-20-7, yellow oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 7.17–7.29 (m, 5H, Ar), 5.83 (ddt, J = 16.00 Hz, 10.28 Hz, 5.06 Hz, 1H, CH), 5.40 (s, 1H, NH), 5.11–5.19 (m, 2H, CH2), 3.87 (t, J = 5.77 Hz, 2H, CH2), 2.66 (t, J = 7.53 Hz, 2H, CH2), 2.19 (t, J = 7.84 Hz, 2H, CH2CO), 1.94–2.02 (m, 2H, CH2), 13C-NMR (CDCl3, 125 MHz): δ = 27.08, 29.70, 35.19, 35.86, 41.91, 116.44, 125.98, 128.40, 128.49, 134.30, 141.46, 172.42.
3.2.9. N-(Prop-2-yn-1-yl)octanamide (20)
CAS number: 422284-34-2, white solid, mp = 72.4–73.4 °C [63]. 1H-NMR (CDCl3, 500.1 MHz): δ = 5.60 (s, 1H, NH), 4.06 (dd, J = 5.18 Hz, J = 2.53 Hz 1H, CH2), 4.04 (dd, J = 5.18 Hz, J = 2.53 Hz, 1H, CH2), 2.22 (t, J = 2.55 Hz, 1H, CH), 2.19 (t, J = 7.77 Hz, 2H, CH2CO), 1.62–1.66 (m, 2H, CH2), 1.25–1.30 (m, 8H, CH2CH2CH2CH2), 0.87 (t, J = 7.05 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 14.04, 22.58, 25.54, 28.97, 29.14, 29.20, 31.65, 36.49, 71.53, 79.67, 172.70.
3.2.10. N-(Prop-2-yn-1-yl)hexanamide (21)
CAS number: 62899-12-1, white solid, mp = 47.3–48.0 °C [64]. 1H-NMR (CDCl3, 500.1 MHz): δ = 5.74 (s, 1H, NH), 4.05 (dd, J = 5.21 Hz, J = 2.54 Hz 1H, CH2), 4.04 (dd, J = 5.21 Hz, J = 2.54 Hz, 1H, CH2), 2.22 (t, J = 2.53 Hz, 1H, CH), 2.19 (t, J = 7.84 Hz, 2H, CH2CO), 1.61–1.67 (m, 2H, CH2), 1.29–1.35 (m, 4H, CH2CH2), 0.89 (t, J = 6.98 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.89, 22.36, 25.23, 29.13, 31.40, 36.42, 71.49, 79.68, 172.80.
3.2.11. N-(Prop-2-yn-1-yl)butyramide (22)
CAS number: 2978-28-1, yellowish solid, mp = 26.1–26.6 °C [65]. 1H-NMR (CDCl3, 500.1 MHz): δ = 5.56 (s, 1H, NH), 4.06 (dd, J = 5.21 Hz, J = 2.54 Hz 1H, CH2), 4.05 (dd, J = 5.19 Hz, J = 2.52 Hz, 1H, CH2), 2.22 (t, J = 2.55 Hz, 1H, CH), 2.17 (t, J = 7.66 Hz, 2H, CH2CO), 1.64–1.71 (m, 2H, CH2), 0.95 (t, J = 7.42 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.71, 18.97, 29.14, 29.69, 38.37, 71.55.
3.2.12. 4-Phenyl-N-(prop-2-yn-1-yl)butanamide (23)
CAS number: 1250568-47-8, colorless oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 7.16–7.29 (m, 5H, Ar), 5.54 (s, 1H, NH), 4.04 (dd, J = 5.24 Hz, J = 2.56 Hz 1H, CH2), 4.03 (dd, J = 5.19 Hz, J = 2.51 Hz, 1H, CH2), 2.66 (t, J = 7.53 Hz, 2H, CH2), 2.22 (t, J = 2.58 Hz, 1H, CH), 2.19 (t, J = 7.83 Hz, 2H, CH2CO), 1.95–2.01 (m, 2H, CH2), 13C-NMR (CDCl3, 125 MHz): δ = 26.84, 29.15, 29.17, 35.08, 35.49, 71.59, 79.58, 126.02, 128.42, 128.50, 141.33, 172.22.
3.2.13. 1-(Piperidin-1-yl)octan-1-one (24)
CAS number: 20299-83-6, colorless oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 3.54 (t, J = 5.47 Hz, 2H, CH2N), 3.39 (t, J = 5.36 Hz, 2H, CH2N), 2.30 (t, J = 7.91 Hz, 2H, CH2CO), 1.50–1.66 (m, 8H, CH2CH2CH2CH2), 1.25–1.32 (m, 8H, CH2CH2CH2CH2), 0.87 (t, J = 7.01 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 14.05, 22.60, 24.60, 25.50, 25.59, 26.58, 29.10, 29.50, 31.72, 33.49, 42.58, 46.72, 171.52.
3.2.14. 1-(Piperidin-1-yl)hexan-1-one (25)
CAS number: 15770-38-4, yellowish oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 3.54 (t, J = 5.47 Hz, 2H, CH2N), 3.39 (t, J = 5.47 Hz, 2H, CH2N), 2.30 (t, J = 7.77 Hz, 2H, CH2CO), 1.51–1.65 (m, 8H, CH2CH2CH2CH2), 1.30–1.35 (m, 4H, CH2CH2), 0.90 (t, J = 6.91 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.96, 22.49, 24.61, 25.18, 25.60, 26.59, 31.72, 33.45, 42.59, 46.72, 171.53.
3.2.15. 1-(Piperidin-1-yl)butan-1-one (26)
CAS number: 4637-70-1, yellow oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 3.54 (t, J = 5.48 Hz, 2H, CH2N), 3.39 (t, J = 5.34 Hz, 2H, CH2N), 2.29 (t, J = 7.78 Hz, 2H, CH2CO), 1.50–1.69 (m, 8H, CH2CH2CH2CH2), 0.96 (t, J = 7.41 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 14.04, 18.88, 24.61, 25.61, 26.58, 35.40, 42.58, 46.71, 171.34.
3.2.16. 4-Phenyl-1-(piperidin-1-yl)butan-1-one (27)
CAS number: 41208-51-9, white solid, mp = 153.9–154.4 °C [66]. 1H-NMR (CDCl3, 500.1 MHz): δ = 7.16–7.29 (m, 5H, Ar), 3.54 (t, J = 5.53 Hz, 2H, CH2N), 3.31 (t, J = 5.47 Hz, 2H, CH2N), 2.67 (t, J = 7.66 Hz, 2H, CH2), 2.32 (t, J = 7.84 Hz, 2H, CH2CO), 1.93–1.99 (m, 2H, CH2), 1.49–1.64 (m, 6H, CH2CH2CH2), 13C-NMR (CDCl3, 125 MHz): δ = 24.58, 25.60, 26.54, 26.84, 29.69, 33.54, 35.42, 42.63, 46.61, 125.86, 128.33, 128.49, 141.85, 171.01.
3.2.17. 1-Morpholinooctan-1-one (28)
CAS number: 5338-65-8, colorless oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 3.66 (t, J = 5.14 Hz, 4H, CH2OCH2), 3.61 (t, J = 3.75 Hz, 2H, CH2N), 3.46 (t, J = 4.53 Hz, 2H, CH2N), 2.30 (t, J = 7.83 Hz, 2H, CH2CO), 1.59–1.65 (m, 2H, CH2), 1.25–1.34 (m, 8H, CH2CH2CH2CH2), 0.88 (t, J = 7.08 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 14.03, 22.58, 25.25, 29.05, 29.40, 31.68, 33.11, 41.85, 46.06, 66.68, 66.95, 171.89.
3.2.18. 1-Morpholinohexan-1-one (29)
CAS number: 17598-10-6, yellowish oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 3.66 (t, J = 5.18 Hz, 4H, CH2OCH2), 3.61 (t, J = 3.75 Hz, 2H, CH2N), 3.46 (t, J = 4.56 Hz, 2H, CH2N), 2.30 (t, J = 7.85 Hz, 2H, CH2CO), 1.60–1.66 (m, 2H, CH2), 1.30–1.36 (m, 4H, CH2CH2), 0.90 (t, J = 6.95 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.92, 22.45, 24.93, 31.62, 33.07, 41.86, 46.06, 66.68, 66.96, 171.90.
3.2.19. 1-Morpholinobutan-1-one (30)
CAS number: 5327-51-5, colorless oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 3.66 (t, J = 5.20 Hz, 4H, CH2OCH2), 3.61 (t, J = 4.56 Hz, 2H, CH2N), 3.46 (t, J = 4.56 Hz, 2H, CH2N), 2.29 (t, J = 7.70 Hz, 2H, CH2CO), 1.63–1.70 (m, 2H, CH2), 0.97 (t, J = 7.46 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.96, 18.66, 35.02, 41.86, 46.05, 66.69, 66.97, 171.75.
3.2.20. 1-Morpholino-4-phenylbutan-1-one (31)
CAS number: 61123-44-2, white solid, mp = 40.7–42.5 °C [67]. 1H-NMR (CDCl3, 500.1 MHz): δ = 7.17–7.29 (m, 5H, Ar), 3.59–3.66 (m, 6H, NCH2CH2OCH2), 3.37 (t, J = 4.68 Hz, 2H, NCH2), 2.68 (t, J = 7.59 Hz, 2H, CH2), 2.30 (t, J = 7.71 Hz, 2H, CH2CO), 1.95–2.01 (m, 2H, CH2), 13C-NMR (CDCl3, 125 MHz): δ = 26.54, 29.70, 32.10, 35.26, 45.89, 45.92, 66.63, 66.95, 125.98, 128.40, 128.48, 141.57, 171.42.
3.2.21. N-(2-(Dimethylamino)ethyl)octanamide (32)
CAS number: 114011-26-6, colorless oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 6.14 (s, 1H, NH), 3.30–3.34 (m, 2H, CONHCH2), 2.41 (t, J = 5.90 Hz, 2H, CH2), 2.23 (s, 6H, CH3CH3), 2.17 (t, J = 7.82 Hz, 2H, CH2CO), 1.59–1.65 (m, 2H, CH2), 1.27–1.30 (m, 8H, CH2CH2CH2CH2), 0.87 (t, J = 7.09 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 14.04, 22.58, 25.78, 29.00, 29.26, 29.68, 31.69, 36.64, 36.73, 45.07, 57.94, 173.37.
3.2.22. N-(2-(Dimethylamino)ethyl)hexanamide (33)
CAS number: 114011-25-5, colorless oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 6.07 (s, 1H, NH), 3.31–3.34 (m, 2H, CONHCH2), 2.41 (t, J = 5.90 Hz, 2H, CH2), 2.23 (s, 6H, CH3CH3), 2.17 (t, J = 7.81 Hz, 2H, CH2CO), 1.60–1.66 (m, 2H, CH2), 1.28–1.36 (m, 4H, CH2CH2), 0.89 (t, J = 7.04 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.94, 22.41, 25.46, 29.69, 31.49, 36.62, 36.72, 45.08, 57.91, 173.30.
3.2.23. N-(2-(Dimethylamino)ethyl)butyramide (34)
CAS number: 63224-16-8, yellowish oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 6.12 (s, 1H, NH), 3.31–3.35 (m, 2H, CONHCH2), 2.42 (t, J = 5.88 Hz, 2H, CH2), 2.24 (s, 6H, CH3CH3), 2.15 (t, J = 7.69 Hz, 2H, CH2CO), 1.62–1.70 (m, 2H, CH2), 0.94 (t, J = 7.36 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.77, 19.19, 29.69, 36.57, 38.65, 45.03, 57.19, 173.14.
3.2.24. N-(2-(Dimethylamino)ethyl)-4-phenylbutanamide (35)
CAS number: 63224-25-9, yellowish oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 7.17–7.28 (m, 5H, Ar), 3.30–3.33 (m, 2H, CONHCH2), 2.65 (t, J = 7.60 Hz, 2H, CH2), 2.40 (t, J = 5.88 Hz, 2H, CH2), 2.22 (s, 6H, CH3CH3), 2.18 (t, J = 7.79 Hz, 2H, CH2CO), 1.94–2.00 (m, 2H, CH2), 13C-NMR (CDCl3, 125 MHz): δ = 22.68, 27.13, 29.35, 29.69, 35.23, 25.84, 36.66, 45.07, 57.90, 125.91, 128.35, 128.51, 141.61, 172.85.
3.2.25. N-(3-(Dimethylamino)propyl)ctanamide (36)
CAS number: 22890-10-4, colorless oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 6.89 (s, 1H, NH), 3.30–3.34 (m, 2H, CONHCH2), 2.37 (t, J = 6.41 Hz, 2H, CH2), 2.23 (s, 6H, CH3CH3), 2.14 (t, J = 7.80 Hz, 2H, CH2CO), 1.57–1.68 (m, 4H, CH2CH2), 1.25–1.30 (m, 8H, CH2CH2CH2CH2), 0.87 (t, J = 7.04 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 14.04, 22.59, 25.75, 26.14, 29.02, 29.26, 29.68, 31.68, 36.96, 39.15, 45.34, 58.50, 173.16.
3.2.26. N-(3-(Dimethylamino)propyl)hexanamide (37)
CAS number: 73603-23-3, yellowish oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 6.92 (s, 1H, NH), 3.30–3.34 (m, 2H, CONHCH2), 2.37 (t, J = 6.45 Hz, 2H, CH2), 2.22 (s, 6H, CH3CH3), 2.14 (t, J = 7.77 Hz, 2H, CH2CO), 1.57–1.67 (m, 4H, CH2CH2), 1.28–1.34 (m, 4H, CH2CH2), 0.89 (t, J = 7.16 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.90, 22.40, 25.42, 26.17, 29.67, 31.45, 36.89, 39.15, 45.35, 58.49, 173.18.
3.2.27. N-(3-(Dimethylamino)propyl)butyramide (38)
CAS number: 53201-67-5, colorless oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 6.89 (s, 1H, NH), 3.32–3.35 (m, 2H, CONHCH2), 2.43 (t, J = 6.43 Hz, 2H, CH2), 2.27 (s, 6H, CH3CH3), 2.13 (t, J = 7.63 Hz, 2H, CH2CO), 1.60–1.70 (m, 4H, CH2CH2), 0.94 (t, J = 7.39 Hz, 3H, CH3), 13C-NMR (CDCl3, 125 MHz): δ = 13.80, 19.12, 26.01, 29.69, 38.87, 38.98, 45.16, 58.36, 172.97.
3.2.28. N-(3-(Dimethylamino)propyl)-4-phenylbutanamide (39)
CAS number: 885912-19-6, yellowish oil. 1H-NMR (CDCl3, 500.1 MHz): δ = 7.16–7.28 (m, 5H, Ar), 6.88 (s, 1H, NH), 3.30–3.34 (m, 2H, CONHCH2), 2.64 (t, J = 7.64 Hz, 2H, CH2), 2.38 (t, J = 6.42 Hz, 2H, CH2), 2.22 (s, 6H, CH3CH3), 2.16 (t, J = 7.78 Hz, 2H, CH2CO), 1.92–1.98 (m, 2H, CH2), 1.62–1.67 (m, 2H, CH2), 13C-NMR (CDCl3, 125 MHz): δ = 14.11, 26.11, 27.19, 29.69, 35.25, 36.09, 39.17, 45.30, 53.42, 58.52, 125.89, 128.34, 128.46, 141.64, 172.61.
4. Conclusions
We successfully developed a sustainable and green enzymatic strategy for the synthesis of amides from free carboxylic acids and amines by using Candida antarctica lipase B as a biocatalyst, using GC-MS analysis to monitor the reaction progress. The green enzymatic amidation is simple and efficient without any additives, with the application of cyclopentyl methyl ether as the solvent, which is a greener and safer solvent alternative in comparison with the usual organic solvents. The scope of the reaction was extended to the preparation of 28 diverse amides, by using four different free carboxylic acids and seven amines, including primary and secondary amines as well as cyclic amines. In every case, excellent conversions and yields were achieved without the need of any intensive purification step. This enzymatic methodology offers a way to synthetize pure amides. That is, this synthetic approach is both eco-friendly and practical for large-scale production.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/molecules28155706/s1, Figures S1–S62: 1H-NMR and 13C-NMR spectra of 12–39 amide products and GC-MS measurement.
Author Contributions
E.F. and G.O. planned and designed the project; G.O. and S.S. performed the syntheses and characterized the synthesized compounds; E.F. and G.O. prepared the manuscript for publication. All authors discussed the results and commented on the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of compounds 12–39 are available from the authors.
Funding Statement
The authors’ thanks are due to the Hungarian Research Foundation (OTKA No. K-138871), the Ministry of Human Capacities, Hungary (grant TKP-2021-EGA-32).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Mahesh S., Tang K.-C., Raj M. Amide Bond Activation of Biological Molecules. Molecules. 2018;23:2615. doi: 10.3390/molecules23102615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pattabiraman V.R., Bode J.W. Rethinking amide bond synthesis. Nature. 2011;480:471–479. doi: 10.1038/nature10702. [DOI] [PubMed] [Google Scholar]
- 3.Wieland T., Bodanszky M. The World of Peptides: A Brief History of Peptide Chemistry. Springer; Berlin/Heidelberg, Germany: 2012. pp. 1–22. [Google Scholar]
- 4.Ohashi N., Kohno T. Analgesic Effect of Acetaminophen: A Review of Known and Novel Mechanisms of Action. Front. Pharmacol. 2020;11:580289. doi: 10.3389/fphar.2020.580289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Day T.K., Skarda R.T. The pharmacology of local anesthetics. Vet. Clin. N. Am. Large Anim. Pract. 1991;7:489–500. doi: 10.1016/S0749-0739(17)30482-0. [DOI] [PubMed] [Google Scholar]
- 6.Becker D.E., Reed K.L. Local anesthetics: Review of pharmacological considerations. Anesth. Prog. 2012;59:90–102. doi: 10.2344/0003-3006-59.2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lima L.M., Silva B., Barbosa G., Barreiro E.J. β-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 2020;208:112829. doi: 10.1016/j.ejmech.2020.112829. [DOI] [PubMed] [Google Scholar]
- 8.Turner J., Muraoka A., Bedenbaugh M., Childress B., Pernot L., Wiencek M., Peterson Y.K. The Chemical Relationship Among Beta-Lactam Antibiotics and Potential Impacts on Reactivity and Decomposition. Front. Microbiol. 2022;13:807955. doi: 10.3389/fmicb.2022.807955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King D.T., Sobhanifar S., Strynadka N.C.J. One ring to rule them all: Current trends in combating bacterial resistance to the β-lactams. Protein Sci. 2016;25:787–803. doi: 10.1002/pro.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxena P., Sharma P.K., Purohit P. A journey of celecoxib from pain to cancer. Prostaglandins Other Lipid Mediat. 2020;147:106379. doi: 10.1016/j.prostaglandins.2019.106379. [DOI] [PubMed] [Google Scholar]
- 11.Sadée W., Bohn L. How specific are “target-specific” drugs? Celecoxib as a case in point. Mol. Interv. 2006;6:196–198. doi: 10.1124/mi.6.4.5. [DOI] [PubMed] [Google Scholar]
- 12.Bering L., Craven E.J., Sowerby Thomas S.A., Shepherd S.A., Micklefield J. Merging enzymes with chemocatalysis for amide bond synthesis. Nat. Commun. 2022;13:380. doi: 10.1038/s41467-022-28005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen C.L., Williams J.M.J. Metal-catalysed approaches to amide bond formation. Chem. Soc. Rev. 2011;40:3405–3415. doi: 10.1039/c0cs00196a. [DOI] [PubMed] [Google Scholar]
- 14.Massolo E., Pirola M., Benaglia M. Amide Bond Formation Strategies: Latest Advances on a Dateless Transformation. Eur. J. Org. Chem. 2020;2020:4641–4651. doi: 10.1002/ejoc.202000080. [DOI] [Google Scholar]
- 15.Todorovic M., Perrin D.M. Recent developments in catalytic amide bond formation. Pept. Sci. 2020;112:e24210. doi: 10.1002/pep2.24210. [DOI] [Google Scholar]
- 16.El-Faham A., Albericio F. Peptide Coupling Reagents, More than a Letter Soup. Chem. Rev. 2011;111:6557–6602. doi: 10.1021/cr100048w. [DOI] [PubMed] [Google Scholar]
- 17.Valeur E., Bradley M. Amide bond formation: Beyond the myth of coupling reagents. Chem. Soc. Rev. 2009;38:606–631. doi: 10.1039/B701677H. [DOI] [PubMed] [Google Scholar]
- 18.Orsy G., Fülöp F., Mándity I.M. Direct amide formation in a continuous-flow system mediated by carbon disulfide. Catal. Sci. Technol. 2020;10:7814–7818. doi: 10.1039/D0CY01603A. [DOI] [Google Scholar]
- 19.Lundberg H., Tinnis F., Selander N., Adolfsson H. Catalytic amide formation from non-activated carboxylic acids and amines. Chem. Soc. Rev. 2014;43:2714–2742. doi: 10.1039/C3CS60345H. [DOI] [PubMed] [Google Scholar]
- 20.Hazra S., Gallou F., Handa S. Water: An Underestimated Solvent for Amide Bond-Forming Reactions. ACS Sustain. Chem. Eng. 2022;10:5299–5306. doi: 10.1021/acssuschemeng.2c00520. [DOI] [Google Scholar]
- 21.Sabatini M.T., Boulton L.T., Sneddon H.F., Sheppard T.D. A green chemistry perspective on catalytic amide bond formation. Nat. Catal. 2019;2:10–17. doi: 10.1038/s41929-018-0211-5. [DOI] [Google Scholar]
- 22.Zhang R., Yao W.-Z., Qian L., Sang W., Yuan Y., Du M.-C., Cheng H., Chen C., Qin X. A practical and sustainable protocol for direct amidation of unactivated esters under transition-metal-free and solvent-free conditions. Green Chem. 2021;23:3972–3982. doi: 10.1039/D1GC00720C. [DOI] [Google Scholar]
- 23.Dunetz J.R., Magano J., Weisenburger G.A. Large-Scale Applications of Amide Coupling Reagents for the Synthesis of Pharmaceuticals. Org. Process Res. Dev. 2016;20:140–177. doi: 10.1021/op500305s. [DOI] [Google Scholar]
- 24.Magano J. Large-Scale Amidations in Process Chemistry: Practical Considerations for Reagent Selection and Reaction Execution Organic. Org. Process Res. Dev. 2022;26:1562–1689. doi: 10.1021/acs.oprd.2c00005. [DOI] [Google Scholar]
- 25.Ganesh K.N., Zhang D., Miller S.J., Rossen K., Chirik P.J., Kozlowski M.C., Zimmerman J.B., Brooks B.W., Savage P.E., Allen D.T., et al. Green Chemistry: A Framework for a Sustainable Future. Org. Process Res. Dev. 2021;25:1455–1459. doi: 10.1021/acs.oprd.1c00216. [DOI] [PubMed] [Google Scholar]
- 26.Anastas P., Eghbali N. Green Chemistry: Principles and Practice. Chem. Soc. Rev. 2010;39:301–312. doi: 10.1039/B918763B. [DOI] [PubMed] [Google Scholar]
- 27.Meyer H.-P., Eichhorn E., Hanlon S., Lütz S., Schürmann M., Wohlgemuth R., Coppolecchia R. The use of enzymes in organic synthesis and the life sciences: Perspectives from the Swiss Industrial Biocatalysis Consortium (SIBC) Catal. Sci. Technol. 2013;3:29–40. doi: 10.1039/C2CY20350B. [DOI] [Google Scholar]
- 28.Silverman R.B. Organic Chemistry of Enzyme-Catalyzed Reactions. 2nd ed. Academic Press; San Diego, SD, USA: 2002. pp. 1–38. [Google Scholar]
- 29.Strohmeier G.A., Pichler H., May O., Gruber-Khadjawi M. Application of Designed Enzymes in Organic Synthesis. Chem. Rev. 2011;111:4141–4164. doi: 10.1021/cr100386u. [DOI] [PubMed] [Google Scholar]
- 30.Winkler C.K., Schrittwieser J.H., Kroutil W. Power of Biocatalysis for Organic Synthesis. ACS Cent. Sci. 2021;7:55–71. doi: 10.1021/acscentsci.0c01496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forró E., Fülöp F. Advanced procedure for the enzymatic ring opening of unsaturated alicyclic β-lactams. Tetrahedron Asymmetry. 2004;15:2875–2880. doi: 10.1016/j.tetasy.2004.05.029. [DOI] [Google Scholar]
- 32.Shoda S.-i., Uyama H., Kadokawa J.-i., Kimura S., Kobayashi S. Enzymes as Green Catalysts for Precision Macromolecular Synthesis. Chem. Rev. 2016;116:2307–2413. doi: 10.1021/acs.chemrev.5b00472. [DOI] [PubMed] [Google Scholar]
- 33.Sheldon R.A. Engineering a more sustainable world through catalysis and green chemistry. J. R. Soc. Interface. 2016;13:20160087. doi: 10.1098/rsif.2016.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Augustin M.M., Augustin J.M., Brock J.R., Kutchan T.M. Enzyme morphinan N-demethylase for more sustainable opiate processing. Nat. Sustain. 2019;2:465–474. doi: 10.1038/s41893-019-0302-6. [DOI] [Google Scholar]
- 35.Sheldon R.A., Brady D. Green Chemistry, Biocatalysis, and the Chemical Industry of the Future. ChemSusChem. 2022;15:e202102628. doi: 10.1002/cssc.202102628. [DOI] [PubMed] [Google Scholar]
- 36.Stergiou P.Y., Foukis A., Filippou M., Koukouritaki M., Parapouli M., Theodorou L.G., Hatziloukas E., Afendra A., Pandey A., Papamichael E.M. Advances in lipase-catalyzed esterification reactions. Biotechnol. Adv. 2013;31:1846–1859. doi: 10.1016/j.biotechadv.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Gamayurova V.S., Zinov’eva M.E., Shnaider K.L., Davletshina G.A. Lipases in Esterification Reactions: A Review. Catal. Ind. 2021;13:58–72. doi: 10.1134/S2070050421010025. [DOI] [Google Scholar]
- 38.Singhania V., Cortes-Clerget M., Dussart-Gautheret J., Akkachairin B., Yu J., Akporji N., Gallou F., Lipshutz B.H. Lipase-catalyzed esterification in water enabled by nanomicelles. Applications to 1-pot multi-step sequences. Chem. Sci. 2022;13:1440–1445. doi: 10.1039/D1SC05660C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar A., Dhar K., Kanwar S.S., Arora P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online. 2016;18:2. doi: 10.1186/s12575-016-0033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ingenbosch K.N., Vieyto-Nuñez J.C., Ruiz-Blanco Y.B., Mayer C., Hoffmann-Jacobsen K., Sanchez-Garcia E. Effect of Organic Solvents on the Structure and Activity of a Minimal Lipase. J. Org. Chem. 2022;87:1669–1678. doi: 10.1021/acs.joc.1c01136. [DOI] [PubMed] [Google Scholar]
- 41.Ortiz C., Ferreira M.L., Barbosa O., dos Santos J.C.S., Rodrigues R.C., Berenguer-Murcia Á., Briand L.E., Fernandez-Lafuente R. Novozym 435: The “perfect” lipase immobilized biocatalyst? Catal. Sci. Technol. 2019;9:2380–2420. doi: 10.1039/C9CY00415G. [DOI] [Google Scholar]
- 42.Mangiagalli M., Carvalho H., Natalello A., Ferrario V., Pennati M.L., Barbiroli A., Lotti M., Pleiss J., Brocca S. Diverse effects of aqueous polar co-solvents on Candida antarctica lipase B. Int. J. Biol. Macromol. 2020;150:930–940. doi: 10.1016/j.ijbiomac.2020.02.145. [DOI] [PubMed] [Google Scholar]
- 43.Zieniuk B., Fabiszewska A., Białecka-Florjańczyk E. Screening of solvents for favoring hydrolytic activity of Candida antarctica Lipase B. Bioprocess Biosyst. Eng. 2020;43:605–613. doi: 10.1007/s00449-019-02252-0. [DOI] [PubMed] [Google Scholar]
- 44.Forró E., Fülöp F. Enzymatic Strategies for the Preparation of Pharmaceutically Important Amino Acids through Hydrolysis of Amino Carboxylic Esters and Lactams. Curr. Med. Chem. 2022;29:6218–6227. doi: 10.2174/0929867329666220718123153. [DOI] [PubMed] [Google Scholar]
- 45.Megyesi R., Forró E., Fülöp F. Substrate engineering: Effects of different N-protecting groups in the CAL-B-catalysed asymmetric O-acylation of 1-hydroxymethyl-tetrahydro-β-carbolines. Tetrahedron. 2018;74:2634–2640. doi: 10.1016/j.tet.2018.04.012. [DOI] [Google Scholar]
- 46.Shahmohammadi S., Faragó T., Palkó M., Forró E. Green Strategies for the Preparation of Enantiomeric 5–8-Membered Carbocyclic β-Amino Acid Derivatives through CALB-Catalyzed Hydrolysis. Molecules. 2022;27:2600. doi: 10.3390/molecules27082600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun M., Nie K., Wang F., Deng L. Optimization of the Lipase-Catalyzed Selective Amidation of Phenylglycinol. Front. Bioeng. Biotechnol. 2020;7:486. doi: 10.3389/fbioe.2019.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pitzer J., Steiner K., Schmid C., Schein V.K., Prause C., Kniely C., Reif M., Geier M., Pietrich E., Reiter T., et al. Racemization-free and scalable amidation of l-proline in organic media using ammonia and a biocatalyst only. Green Chem. 2022;24:5171–5180. doi: 10.1039/D2GC00783E. [DOI] [Google Scholar]
- 49.Hassan S., Tschersich R., Müller T.J.J. Three-component chemoenzymatic synthesis of amide ligated 1,2,3-triazoles. Tetrahedron Lett. 2013;54:4641–4644. doi: 10.1016/j.tetlet.2013.06.051. [DOI] [Google Scholar]
- 50.Irimescu R., Kato K. Lipase-catalyzed enantioselective reaction of amines with carboxylic acids under reduced pressure in non-solvent system and in ionic liquids. Tetrahedron Lett. 2004;45:523–525. doi: 10.1016/j.tetlet.2003.10.210. [DOI] [Google Scholar]
- 51.Prasad A.K., Husain M., Singh B.K., Gupta R.K., Manchanda V.K., Olsen C.E., Parmar V.S. Solvent-free biocatalytic amidation of carboxylic acids. Tetrahedron Lett. 2005;46:4511–4514. doi: 10.1016/j.tetlet.2005.04.121. [DOI] [Google Scholar]
- 52.Gotor V. Non-conventional hydrolase chemistry: Amide and carbamate bond formation catalyzed by lipases. Bioorg. Med. Chem. 1999;7:2189–2197. doi: 10.1016/S0968-0896(99)00150-9. [DOI] [PubMed] [Google Scholar]
- 53.Manova D., Gallier F., Tak-Tak L., Yotava L., Lubin-Germain N. Lipase-catalyzed amidation of carboxylic acid and amines. Tetrahedron Lett. 2018;59:2086–2090. doi: 10.1016/j.tetlet.2018.04.049. [DOI] [Google Scholar]
- 54.Dorr B.M., Fuerst D.E. Enzymatic amidation for industrial applications. Curr. Opin. Chem. Biol. 2018;43:127–133. doi: 10.1016/j.cbpa.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 55.Lima R.N., dos Anjos C.S., Orozco E.V.M., Porto A.L.M. Versatility of Candida antarctica lipase in the amide bond formation applied in organic synthesis and biotechnological processes. Mol. Catal. 2019;466:75–105. doi: 10.1016/j.mcat.2019.01.007. [DOI] [Google Scholar]
- 56.Lo Y.C., Lin C.L., Fang W.Y., Lőrinczi B., Szatmári I., Chang W.H., Fülöp F., Wu S.N. Effective Activation by Kynurenic Acid and Its Aminoalkylated Derivatives on M-Type K+ Current. Int. J. Mol. Sci. 2021;22:1300. doi: 10.3390/ijms22031300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hollmann F., Kara S., Opperman D.J., Wang Y. Biocatalytic synthesis of lactones and lactams. Chem.-Asian J. 2018;13:3601–3610. doi: 10.1002/asia.201801180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kunishima M., Watanabe Y., Terao K., Tani S. Substrate-Specific Amidation of Carboxylic Acids in a Liquid-Liquid Two-Phase System Using Cyclodextrins as Inverse Phase-Transfer Catalysts. Eur. J. Org. Chem. 2004;2004:4535–4540. doi: 10.1002/ejoc.200400470. [DOI] [Google Scholar]
- 59.Kim S.E., Hahm H., Kim S., Jang W., Jeon B., Kim Y., Kim M. A Versatile Cobalt Catalyst for Secondary and Tertiary Amide Synthesis from Various Carboxylic Acid Derivatives. Asian J. Org. Chem. 2016;5:222–231. doi: 10.1002/ajoc.201500444. [DOI] [Google Scholar]
- 60.Ojeda-Porras A., Hernandez-Santana A., Gamba-Sanchez D. Direct amidation of carboxylic acids with amines under microwave irradiation using silica gel as a solid support. Green Chem. 2015;17:3157–3163. doi: 10.1039/C5GC00189G. [DOI] [Google Scholar]
- 61.Métro T.-X., Bonnamour J., Reidon T., Duprez A., Sarpoulet J., Martinez J., Lamaty F. Comprehensive Study of the Organic Solvent-Free CDI-Mediated Acylation of Various Nucleophiles by Mechanochemistry. Chem. Eur. J. 2015;21:12787–12796. doi: 10.1002/chem.201501325. [DOI] [PubMed] [Google Scholar]
- 62.Roe E.T., Stutzman J.M., Swern D. Fatty Acid Amides. III.2a N-Alkenyl and N,N-Dialkenyl Amides2b. J. Am. Chem. Soc. 1951;73:3642–3643. doi: 10.1021/ja01152a024. [DOI] [Google Scholar]
- 63.Willand N., Desroses M., Toto P., Dirié B., Lens Z., Villeret V., Rucktooa P., Locht C., Baulard A., Deprez B. Exploring Drug Target Flexibility Using in Situ Click Chemistry: Application to a Mycobacterial Transcriptional Regulator. ACS Chem. Biol. 2010;5:1007–1013. doi: 10.1021/cb100177g. [DOI] [PubMed] [Google Scholar]
- 64.Tian J., Gao W.-C., Zhou D.-M., Zhang C. Recyclable Hypervalent Iodine(III) Reagent Iodosodilactone as an Efficient Coupling Reagent for Direct Esterification, Amidation, and Peptide Coupling. Org. Lett. 2012;14:3020–3023. doi: 10.1021/ol301085v. [DOI] [PubMed] [Google Scholar]
- 65.Ouerghui A., Elamari H., Dardouri M., Ncib S., Meganem F., Girard C. Chemical modifications of poly(vinyl chloride) to poly(vinyl azide) and “clicked” triazole bearing groups for application in metal cation extraction. Adv. Mater. Res. 2016;100:191–197. doi: 10.1016/j.reactfunctpolym.2016.01.016. [DOI] [Google Scholar]
- 66.Wei W., Hu X.-Y., Yan X.-W., Zhang Q., Cheng M., Ji J.-X. Direct use of dioxygen as an oxygen source: Catalytic oxidative synthesis of amides. Chem. Commun. 2012;48:305–307. doi: 10.1039/C1CC14640H. [DOI] [PubMed] [Google Scholar]
- 67.Cromwell N.H., Creger P.L., Cook K.E. Studies with the Amine Adducts of β-Benzoylacrylic Acid and its Methyl Ester. J. Am. Chem. Soc. 1956;78:4412–4416. doi: 10.1021/ja01598a058. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the article and Supplementary Materials.





