Abstract
An in vitro system is described for the assembly of herpes simplex virus type 1 (HSV-1) procapsids beginning with three purified components, the major capsid protein (VP5), the triplexes (VP19C plus VP23), and a hybrid scaffolding protein. Each component was purified from insect cells expressing the relevant protein(s) from an appropriate recombinant baculovirus vector. Procapsids formed when the three purified components were mixed and incubated for 1 h at 37°C. Procapsids assembled in this way were found to be similar in morphology and in protein composition to procapsids formed in vitro from cell extracts containing HSV-1 proteins. When scaffolding and triplex proteins were present in excess in the purified system, greater than 80% of the major capsid protein was incorporated into procapsids. Sucrose density gradient ultracentrifugation studies were carried out to examine the oligomeric state of the purified assembly components. These analyses showed that (i) VP5 migrated as a monomer at all of the protein concentrations tested (0.1 to 1 mg/ml), (ii) VP19C and VP23 migrated together as a complex with the same heterotrimeric composition (VP19C1-VP232) as virus triplexes, and (iii) the scaffolding protein migrated as a heterogeneous mixture of oligomers (in the range of monomers to ∼30-mers) whose composition was strongly influenced by protein concentration. Similar sucrose gradient analyses performed with mixtures of VP5 and the scaffolding protein demonstrated the presence of complexes of the two having molecular weights in the range of 200,000 to 600,000. The complexes were interpreted to contain one or two VP5 molecules and up to six scaffolding protein molecules. The results suggest that procapsid assembly may proceed by addition of the latter complexes to regions of growing procapsid shell. They indicate further that procapsids can be formed in vitro from virus-encoded proteins only without any requirement for cell proteins.
Herpes simplex virus type 1 (HSV-1) is well known as the etiological agent of cold sores, venereal lesions, and neonatal encephalitis (38). Like all herpesviruses, the HSV-1 virion consists of an icosahedral capsid surrounded by a membrane envelope. The viral double-stranded DNA is contained inside the capsid, while a layer of protein called the tegument is found between the capsid and the membrane. The mature capsid is an icosahedral protein shell approximately 15 nm thick and 126 nm in diameter. Its principal structural features are 162 capsomers (150 hexons and 12 pentons) that lie on a T=16 icosahedral lattice. Each capsomer consists of a roughly cylindrical protruding domain that is extended laterally at its proximal end to create the capsid floor layer (3 to 4 nm thick). The floor is the only place where capsomers make direct contact with each other. Capsomers, however, are also connected indirectly by way of the triplexes, trigonal structures (320 in all) that lie above the floor layer with one triplex found at the local threefold position created by each group of three capsomers (7, 25, 27, 36, 41, 42).
VP5, the HSV-1 major capsid protein (molecular weight [MW], 149,075), is the predominant polypeptide component of the capsid; it is the structural subunit of both the hexons and pentons (16). Hexons are hexamers of VP5, while pentons are pentamers. The triplexes are composed of two minor capsid proteins, VP19C and VP23. Most, if not all, triplexes contain one molecule of VP19C and two of VP23 (16).
HSV-1 capsids are formed in the infected-cell nucleus, where they are also packaged with DNA prior to further virus maturation (7, 27). Assembly requires the three capsid structural proteins mentioned above plus a scaffolding protein. In cells infected with wild-type HSV-1, the primary scaffolding protein is pre-VP22a (also called ICP35; product of the UL26.5 gene), although VP21, a cleavage product of the polypeptide encoded by UL26, can also serve effectively as a scaffolding protein (34). During capsid assembly, the scaffolding protein binds to VP5 and forms a core internal to the shell proteins. Pre-VP22a is cleaved to VP22a and exits the capsid at or near the time DNA enters and is not found in the mature virion.
Intermediates in the capsid assembly process have been identified in studies involving use of a cell-free assembly system (14, 15, 35). The system is based on the use of a panel of recombinant baculoviruses (rBV) encoding HSV-1 capsid proteins. It is constituted by mixing extracts of rBV-infected insect cells containing HSV-1 proteins and incubating the mixture in vitro. Studies with the system have demonstrated that capsids are formed by way of partial and complete procapsid intermediates. Partial procapsids are arc- or dome-shaped structures in which a region of capsid shell partially surrounds a region of core. They grow into complete procapsids as the shell is enlarged and closed. The procapsid has T=16 icosahedral symmetry and the same diameter as the mature capsid, but the two differ in several important ways. First, procapsids are spherical, while the mature capsid is icosahedral. Second, the floor layer in the procapsid is not continuous, resulting in a structure that has large gaps between the capsomers. No comparable gaps are found in the mature capsid floor layer. Third, hexons are oval shaped in the procapsid rather than hexagonal as they are in the mature form. Fourth, the procapsid is a more fragile structure. For example, it is disassembled during incubation at 2°C, a treatment that does not affect the integrity of mature capsids.
Although the cell-free system described above has been employed productively to clarify features of the assembly pathway, the presence of insect cell proteins prevents it from being used to address the issue of whether cell-encoded proteins are required for capsid formation. Studies of pairwise interactions between HSV-1 capsid proteins are also complicated by the presence of cell proteins. In order to evaluate the potential involvement of cell-encoded proteins in capsid formation, we have undertaken an effort to purify HSV-1 capsid proteins biochemically from rBV-infected insect cells expressing them and to test the purified proteins for the ability to assemble into procapsids in vitro. The results described here demonstrate that procapsids can be formed readily from purified capsid proteins. They also suggest the nature of VP5-scaffolding protein complexes that may be involved in capsid assembly.
MATERIALS AND METHODS
Protein purification.
All protein purification was initiated with Sf9 cells expressing the relevant herpesvirus protein(s) after infection with an appropriate rBV vector(s). VP5 and pUL80.5-H were purified from cells infected with BAC-UL19 and BAC-UL80.5-H, respectively, while triplexes were purified from cells coinfected with BAC-UL18 and BAC-UL38. Construction of the four rBV vectors and the methods employed for their propagation and growth on Sf9 cells have been described previously (19, 34). Protein purification was carried out beginning with cell lysates that were produced from pellets of rBV-infected cells. Cell pellets were diluted twofold with phosphate-buffered saline (PBS) containing 20 mM EDTA and protease inhibitors (1 tablet/5 ml; complete, mini; Boehringer Mannheim) and then subjected to three cycles of freezing and thawing to lyse the cells. Lysates were stored in 1-ml aliquots at −80°C until used. The lysate protein concentration was in the range of 15 to 20 mg/ml. Further purification steps were carried out at 4°C beginning with 2 ml of lysates that were thawed and clarified by centrifugation for 5 min at 16,000 × g.
(i) VP5.
The clarified lysate was first subjected to centrifugation at 35,000 rpm (43,000 × g) for 30 min in a Beckman TL100 tabletop ultracentrifuge (TLA100.3 rotor). Sufficient saturated ammonium sulfate was then added to the resulting supernatant to yield an ammonium sulfate concentration of 29% saturation. The mixture was incubated at 4°C for 30 min and then centrifuged at 16,000 × g for 10 min to collect the precipitate which contained VP5. The precipitate was dissolved in 3 ml of PBS–10 mM EDTA and desalted on a Bio-Rad EconoPac 10G desalting column (10 ml volume) eluted with 20 mM Tris-HCl (pH 8). The sample was then filtered (0.22 μm pore size; Millipore) and applied to a 1-ml Pharmacia Resource Q anion-exchange column. The column was eluted with a 20-ml gradient of 0 to 1.0 M NaCl prepared in 20 mM Tris-HCl (pH 8), and fractions (1.0 ml each) containing VP5, which eluted at approximately 0.3 M NaCl, were pooled.
VP5 from the column eluate was further purified by sucrose density gradient ultracentrifugation. A 0.5-ml aliquot of the Resource Q column eluate was applied to a 10 to 30% linear sucrose gradient containing PBS plus 10 mM EDTA prepared in a 5-ml centrifuge tube. Gradients were centrifuged for 20 h at 35,000 rpm in a Beckman LE 80K preparative ultracentrifuge (SW 50.1 rotor; 115,000 × g) operated at 4°C. The gradient was fractionated, and VP5-containing fractions were identified by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Relevant fractions were pooled, adjusted to 50% saturated ammonium sulfate to precipitate VP5, and centrifuged (35,000 rpm, SW50.1 rotor, 30 min) to recover the precipitate. The precipitate was dissolved in sufficient PBS–10 mM EDTA (plus protease inhibitors) to yield a protein concentration of 1 to 2 mg/ml, and this solution was employed for procapsid assembly and other operations as described below.
(ii) Triplexes.
The clarified lysate of BAC-UL18/BAC-UL38-infected Sf9 cells was subjected to high-speed clarification in the TL100 ultracentrifuge, precipitated with ammonium sulfate, desalted, and filtered as described above for purification of VP5, except that the ammonium sulfate precipitate was dissolved in 20 mM Tris-HCl (pH 7.). Triplexes were further purified by chromatography on a 1-ml Pharmacia Resource S cation-exchange column. The sample was applied in 20 mM Tris-HCl (pH 7) and eluted with a gradient of 0 to 1.0 M NaCl in the same buffer. Triplex-containing fractions, as identified by SDS-polyacrylamide gel electrophoresis, were pooled, precipitated with ammonium sulfate, and dissolved in PBS–10 mM EDTA as described above for VP5.
(iii) pUL80.5-H.
Sf9 cell lysates containing pUL80.5-H (MW, 39,855) were clarified by centrifugation at 16,000 × g for 15 min. Lysates were then adjusted to 29% saturated ammonium sulfate and incubated at 4°C, and the precipitate was collected as described above for VP5. The precipitate, which contained pUL80.5-H, was dissolved in 0.5 ml of PBS and fractionated further by sucrose density gradient ultracentrifugation. The sample was applied to the top of a 5.0-ml linear 10 to 30% sucrose gradient containing PBS prepared in a 5-ml Beckman Ultra-Clear SW 50.1 ultracentrifuge tube. Gradients were centrifuged at 35,000 rpm (115,000 × g) in an SW 50.1 rotor for 4 h at 4°C. After centrifugation, gradients were separated into 0.4-ml fractions which were examined by SDS-polyacrylamide gel electrophoresis for the presence of pUL80.5-H. The desired fractions were pooled, and pUL80.5-H was precipitated by adjusting the solution to 50% saturated ammonium sulfate. The precipitate was dissolved in sufficient PBS–10 mM EDTA to yield a protein concentration of 2 to 3 mg/ml and used in procapsid assembly and other studies as described below.
Procapsid assembly.
Procapsids were assembled in reaction mixtures containing 10 μl of VP5 (1 to 2 mg/ml), 10 μl of triplexes (∼1 mg/ml), and 10 μl of pUL80.5-H (2 to 3 mg/ml). Typical reaction mixtures therefore contained approximately 70 pmol of VP5, 84 pmol of triplexes and 500 pmol of pUL80.5-H. Reaction mixtures were adjusted to 25 mM EDTA–10 mM dithiothreitol, and protease inhibitors were added before incubation for 1 h at 37°C. After incubation, reaction mixtures were centrifuged for 2 min at low speed (16,000 × g), and product procapsids were then precipitated by addition of 1 μl of purified monoclonal antibody (MAb) 6F10 (4 mg/ml) (14, 29). Precipitates were allowed to form during 5 min of incubation at room temperature and then harvested by centrifugation at 16,000 × g for 30 s. The precipitate was resuspended in 50 μl of PBS, and the procapsids were dispersed by sonication before further operations were performed.
Sucrose gradient analysis of purified proteins.
The oligomeric state of purified capsid proteins and protein complexes was analyzed by centrifugation on 0.7-ml sucrose density gradients. Linear 10 to 30% sucrose gradients containing PBS were prepared in Beckman Ultra-Clear ultracentrifuge tubes (5 by 41 mm (0.7-ml capacity). A 20-μl sample (30 μl in the case of pUL80.5-H) of the specimen to be analyzed was adjusted to 25 mM EDTA–10 mM dithiothreitol, appropriate protein standards were added (see below), and the mixture was layered on top of the gradient, which was centrifuged in a Beckman SW50.1 rotor operated at 35,000 rpm. Centrifugation was for 14 to 18 h in the case of VP5 and triplex specimens and 2.5 to 4 h in the case of pUL80.5-H and VP5–pUL80.5-H complexes. Most gradients were centrifuged at 4°C, although all specimens were also examined at 26 and 34°C. After centrifugation, gradients were separated into 14 equal fractions and an aliquot (20 μl) of each was analyzed by SDS-polyacrylamide gel electrophoresis followed by Coomassie blue staining. Stained gels were scanned while wet on a flatbed scanner (Hewlett-Packard ScanJet IIc; reflected light), and bands were determined quantitatively by use of ImageQuant (Molecular Dynamics) software. The integrated density of individual bands was plotted as a function of the gradient fraction number. The approximate MWs of proteins and protein complexes were determined with reference to protein standards by the use of the treatment of Martin and Ames (10): distance sedimented1/distance sedimented2 = (MW1/MW2)2/3. The protein standards employed were bovine serum albumin (BSA; MW, 68,000); β-amylase (MW, 200,000), and apoferritin (MW, 443,000).
Cryoelectron microscopy.
Freshly assembled procapsids in PBS were concentrated by adding MAb 6F10 (29), and the resulting precipitate was prepared for cryoelectron microscopy by adsorption to a continuous thin carbon film supported on a thick holey carbon film. The drop was blotted onto a thin film, quenched in liquid ethane cooled by liquid nitrogen in a Reichert FC4 cryostation, transferred into a Gatan 626 cryoholder, and observed on a Philips CM200-FEG electron microscope as described by Zlotnick et al. (43). Micrographs were recorded at a nominal magnification of ×38,000 by using minimal electron dose techniques, producing radiation levels of ∼8 electrons/Å2.
Image processing.
A three-dimensional reconstruction of the procapsid was computed beginning with micrographs that were selected for analysis by visual appraisal (e.g., to assess density of particles and contrast) and by optical diffraction to assess the state of defocus and resolution. Four micrographs whose first contrast transfer function (CTF) zeroes were in the range of 1/22 to 1/24 Å were scanned at 26 μm/pixel on a Perkin-Elmer 1010MG microdensitometer yielding an effective pixel size of ∼7 Å. A total of 94 capsid images were processed as previously described (35). The structure was solved by using the polar Fourier transform (PFT) method (1), with our earlier result for extract-assembled procapsids (35) as the starting model. As a control against imprinting the features of the starting model on the reconstruction, the calculation was repeated by using a density map of the mature B capsid (2) as a starting model. Identical results were obtained. After iterative cycles of refinement of orientation angles and origins, the 72 particles with the highest correlation coefficients were selected and a density map was calculated (5) to 25-Å resolution as assessed by the FRC3D criterion (2).
Other methods.
Procapsid-containing precipitates were prepared for thin-section electron microscopy by fixation, embedding in Epon 812, and sectioning as previously described (11, 14). For negative-stain electron microscopy, specimens were stained with 1% (wt/vol) uranyl acetate as described by Thomas et al. (32). All thin-section and negative-stain electron micrographs were recorded on a JEOL 100CX transmission electron microscope operated at 80 keV. SDS-polyacrylamide gel electrophoresis was carried out on slabs of 12% polyacrylamide (6 by 9 cm) as described previously (13). Protein bands were identified by Coomassie blue staining and determined quantitatively by scanning as described above. Scans were taken in the dynamic range of the scanner, and the error of the measurement was estimated to be ±5%. Western immunoblotting was performed with similar gels in which proteins were transferred electrophoretically to Immobilon membranes and stained as described by Spencer et al. (29). Specific staining was carried out with (i) rabbit polyclonal antisera specific for VP5, VP19C, or VP23 (generously donated by G. Cohen and R. Eisenberg) or (ii) a rabbit polyclonal antiserum specific for the human cytomegalovirus (HCMV) scaffolding protein (UL80.5 gene product) (19). All specific antisera were used at a 1:5,000 dilution. HSV-1 B capsids, used as reference standards in SDS-polyacrylamide gel electrophoresis and immunoblotting studies, were prepared from BHK-21 cells infected with the 17MP strain of HSV-1 as previously described (16).
RESULTS
Proteins employed for procapsid assembly.
Procapsid assembly was carried out with VP5, triplexes (composed of VP19C and VP23), and a hybrid scaffolding protein called pUL80.5-H (MW, 39,855) (19). The latter consists of the N-terminal 364 amino acids of the HCMV scaffolding protein (UL80.5 gene product) linked to the C-terminal 25 amino acids of the corresponding HSV-1 protein (pre-VP22a; UL26.5 gene product). Our strategy of using the hybrid HCMV–HSV-1 scaffolding protein was based on our experience that although assembly of HSV-1 procapsids was accomplished with the endogenous HSV-1 scaffolding protein in unpurified form in cell extracts (14), our yields of this protein in subsequent attempts at expression and purification were insufficient for the current project. Since procapsids had also been assembled with the hybrid scaffolding protein in vivo and in vitro from cell extracts (19) and this protein proved more tractable in purification trials, it was used throughout the project. All proteins were purified from Sf9 cells expressing the relevant herpesvirus protein(s) as a result of infection with an appropriate rBV vector(s). In each case, protein purification began with a clarified cell lysate produced as described in Materials and Methods.
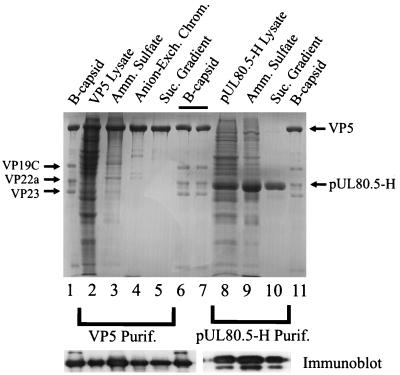
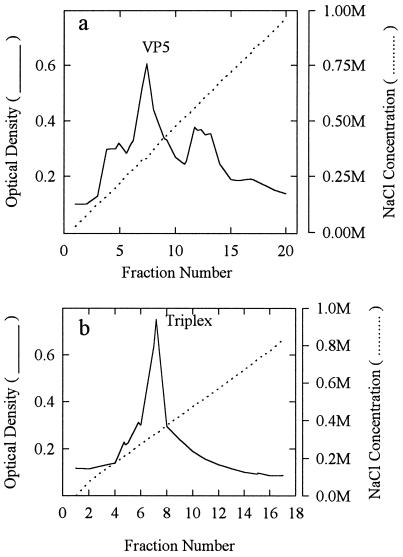
VP5 was purified by ammonium sulfate precipitation followed by anion-exchange chromatography and sucrose density gradient centrifugation. After each stage of purification, VP5-containing fractions were analyzed by SDS-polyacrylamide gel electrophoresis (Fig. 1). Densitometric scanning of the stained gels was employed to estimate the amount of VP5 as a proportion of the total protein present, and the results are shown in Table 1. As both gel and densitometric analyses show, VP5 represented quite a large percentage (17%) of the original clarified lysate, indicating that it was expressed at a high level from the BAC-UL19 vector and that it accumulated in a soluble form. Ammonium sulfate precipitation and anion-exchange chromatography resulted in enrichments of approximately 2.5- and 2-fold, respectively. The anion-exchange column eluate had a prominent VP5-containing peak which was resolved from bands of cell proteins eluting at lower and higher NaCl concentrations, respectively (Fig. 2a). The sucrose density gradient step separated VP5 from several proteins migrating between VP5 and VP23 (Fig. 1, lanes 4 and 5), resulting in a product in which VP5 accounted for 96% of the protein present, as shown in Table 1. Western immunoblot analyses of VP5-containing fractions demonstrated that the gel band identified as VP5 was able to react with an antiserum specific for VP5 after each of the three purification steps (Fig. 1, bottom left bands). The faint Coomassie-staining bands between VP5 and VP19C (Fig. 1, lane 5) did not react with VP5-specific antibody.
FIG. 1.
SDS-polyacrylamide gel electrophoresis of fractions obtained during purification of VP5 (lanes 2 to 5) and pUL80.5-H (lanes 8 to 10). For reference, HSV-1 B-capsid proteins are shown in lanes 1, 6, 7, and 11. The upper panel shows a gel after staining with Coomassie blue, while the lower two panels show Western immunoblots of the VP5 (left) and pUL80.5-H (right) regions of a comparable gel after staining with a rabbit polyclonal antibody specific for VP5 (lanes 1 to 6) or pUL80.5 (lanes 8 to 10), respectively. Amm., ammonium; Exch. Chrom., exchange chromatography; Suc., sucrose; Purif., purification.
TABLE 1.
Purification summary for VP5, triplexes, and pUL80.5-H
| Purification step | % of total protein presenta
|
||
|---|---|---|---|
| VP5 | Triplex | pUL80.5-H | |
| Clarified lysate | 17 | 11 | 24 |
| Ammonium sulfate precipitation | 41 | 48 | 46 |
| Anion-exchange chromatography | 79 | NA | NA |
| Cation-exchange chromatography | NA | 95 | NA |
| Sucrose density gradient | 96 | NA | 93 |
The percentage of each capsid protein present was determined by densitometric scanning of a Coomassie-blue-stained SDS-polyacrylamide gel. NA, not applicable.
FIG. 2.
Column eluate obtained during the ion-exchange chromatography step in the purification of VP5 (a) and triplexes (b). VP5 anion-exchange chromatography was performed on a Pharmacia Resource Q column eluted with a gradient of NaCl (0 to 1.0 M). Triplex chromatography was carried out on a Pharmacia Resource S cation-exchange column eluted with a gradient of NaCl (0 to 1.0 M) as described in Materials and Methods. Optical density at 280 nm was measured. The positions of VP5- and triplex-containing fractions, as judged by SDS-polyacrylamide gel analysis are indicated.
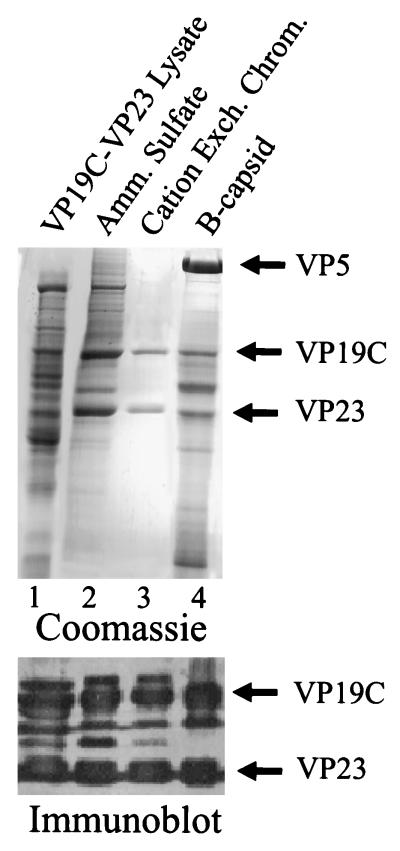
Triplexes were purified from Sf9 cells coinfected with rBV encoding VP19C and VP23. The goal was to isolate triplexes as a unit, since previous studies had shown that triplexes are stable in Sf9 cell extracts (28). Purification was accomplished by ammonium sulfate precipitation followed by cation-exchange chromatography, the two steps resulting in enrichments of approximately four- and twofold, respectively, as shown in Fig. 3 and Table 1. The success of the cation-exchange chromatography step appeared to be due, in part, to the fact that many nontriplex proteins in the ammonium sulfate precipitate did not bind to the column. Only small amounts of nontriplex proteins were recovered in the column eluate (weak bands eluting prior to triplexes in Fig. 2b). The molar ratio of VP23 to VP19C in the purified triplexes was determined to be 1.7 (average of three preparations) from quantitative analysis of stained bands on SDS-polyacrylamide gels. We interpret this as agreeing satisfactorily with 2.0, the expected ratio for heterotrimeric virion triplexes.
FIG. 3.
SDS-polyacrylamide gel electrophoresis of fractions obtained during triplex purification (lanes 1 to 3). For comparison, HSV-1 B-capsid proteins are shown in lane 4. The upper panel shows the gel after staining with Coomassie blue, while the bottom panel shows the results of Western immunoblotting of the VP19C-VP23 region in a comparable gel after staining with rabbit polyclonal antibodies specific for VP19C and VP23. Amm., ammonium; Exch. Chrom., exchange chromatography.
The bands identified as VP19C and VP23 in purified triplex preparations were shown to react with polyclonal antisera specific for the two proteins, as shown in Fig. 3 (bottom panel). Bands found between VP19C and VP23 in the immunoblot are suggested to correspond to small amounts of VP19C degradation products detectable by immunostaining but not by the more quantitative (but less sensitive) staining with Coomassie blue.
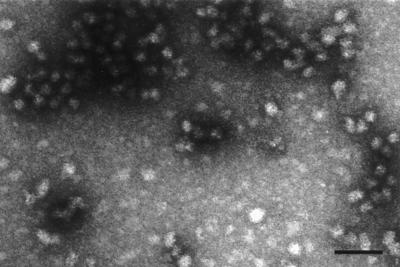
The scaffolding protein pUL80.5-H was purified from the clarified lysate by ammonium sulfate precipitation followed by sucrose density gradient ultracentrifugation as described in Materials and Methods. SDS-polyacrylamide gel analysis of the relevant enriched fractions is shown in Fig. 1 (lanes 8 to 10). During sucrose density gradient centrifugation, pUL80.5-H migrated as a very broad band from the top of the gradient nearly to the bottom. The most rapidly migrating material was found in a peak whose sedimentation rate (compared to that of protein standards) suggested a structure with an MW of approximately 106. This most rapidly migrating material was collected from the gradient and employed for capsid assembly studies. Electron microscopic analysis of negatively stained specimens showed that this material consists of roughly spherical, variably sized particles with a diameter of approximately 28 nm (Fig. 4). For example, measurement of 39 particles yielded an average diameter (longest dimension) of 28 ± 4 nm. In a Western immunoblot, purified pUL80.5-H was found to react with a polyclonal antibody specific for pUL80.5-H (Fig. 1, bottom panel, lane 10). Immunoblotting also revealed the presence of a pUL80.5-H degradation product that accompanied the full molecule through the purification procedure (Fig. 1, bottom panel, lanes 8 to 10). This species must be present at a low level, as it was not observed in gels stained with Coomassie blue (Fig. 1, lane 10).
FIG. 4.
Electron microscopic analysis of purified pUL80.5-H. Material from the most rapidly migrating band of pUL80.5-H observed during the sucrose gradient purification step (see text) was examined by electron microscopy after negative staining as described in Materials and Methods. Note that pUL80.5-H particles are heterogeneous in size and shape. Bar, 100 nm.
Procapsid assembly.
Procapsids were formed by mixing VP5, triplexes, and pUL80.5-H together and incubating them as described in Materials and Methods. The order of component addition did not appear to affect the quality or quantity of the procapsids formed. During the incubation period, the reaction mixture became visibly turbid as procapsids were assembled (data not shown). Turbidity, presumed to be due to procapsids, could not be removed by low-speed centrifugation (16,000 × g) at this stage. Procapsid formation required incubation at temperatures of approximately 26°C or higher. No procapsids formed if reaction components were maintained at 4°C.
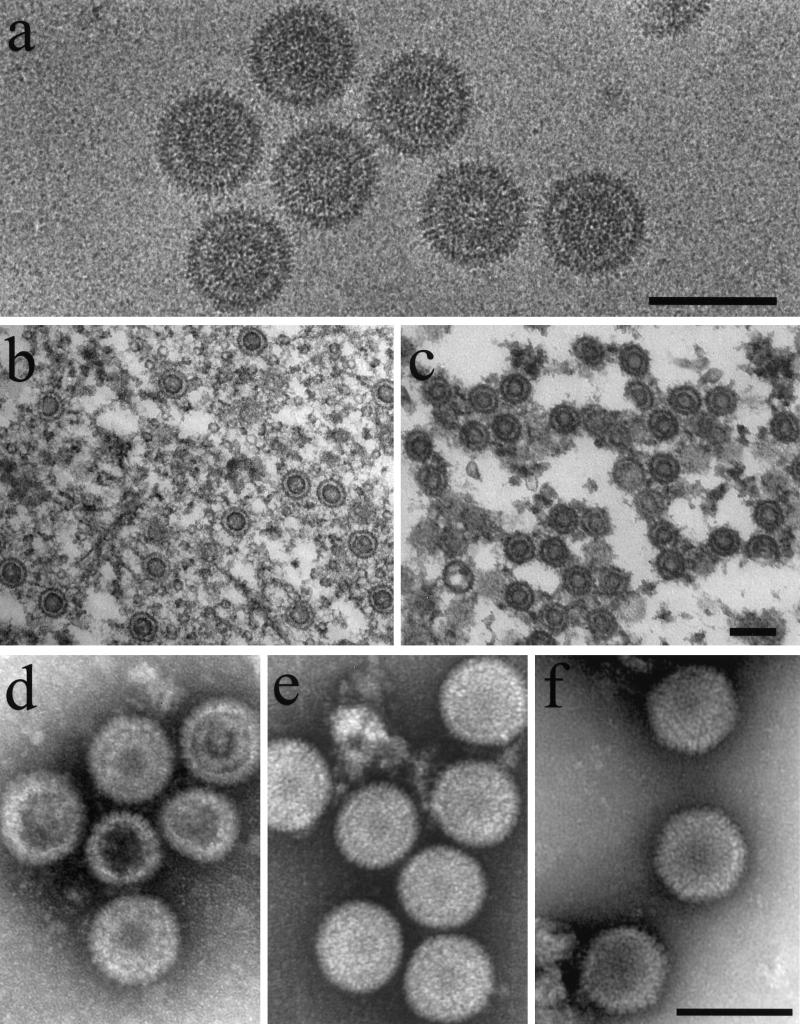
After harvesting by antibody precipitation, procapsids were examined by electron microscopy of frozen-hydrated, thin-sectioned, and negatively stained preparations. Frozen-hydrated specimens appeared uniform in morphology and round in profile, suggesting that the procapsids are spherical (Fig. 5a). The measured diameter was 126 ± 1 nm (n = 12). Distinct shell and core layers were visible, and there was a region of lower density between the two. A low-density region was seen at the center of nearly every image. Capsomers could often be resolved at the periphery. In the leftmost image of Fig. 5a, for example, capsomers are visible near 12 o’clock.
FIG. 5.
Electron microscopy of procapsids assembled from purified proteins (a, b, and d), procapsids assembled from Sf9 cell extracts containing HSV-1 proteins as described previously (c and e) (14), and control HSV-1 B-capsids (f). Specimens were observed after cryopreservation (a), thin sectioning (b and c), or negative staining (d, e, and f) as described in Materials and Methods. Note that procapsids appear round in all of the preparations, indicating they are spherical. Note also that distinct shell and core layers can be seen in frozen hydrated (a) and thin-sectioned (b and c) preparations. Small procapsids with reduced diameters compared to the majority population (e.g., d; middle procapsids) were observed as a small and variable proportion of procapsids formed both from extracts and from purified components. Bars, 150 nm.
Distinct shell and core layers were also observed in thin-sectioned preparations (Fig. 5b). In most images, the core stained somewhat more darkly than the shell, and weakly staining regions were found at the center of the image and between the shell and core layers. Thin-sectioned preparations showed that procapsids formed from purified proteins were similar in structure to procapsids formed in vitro from Sf9 cell extracts containing HSV-1 proteins (Fig. 5, compare b with c) (14).
Surface features of the procapsid were emphasized in images of negatively stained specimens (Fig. 5d). Capsomers were visible both en face at the center of the image and in profile at the procapsid periphery. As in the case of thin-sectioned specimens, negatively stained procapsids assembled from purified proteins resembled their counterparts prepared from cell extracts (Fig. 5, compare d and e). The round morphology observed for procapsids contrasts with the angular, icosahedral shape seen in mature HSV-1 B capsids (Fig. 5f).
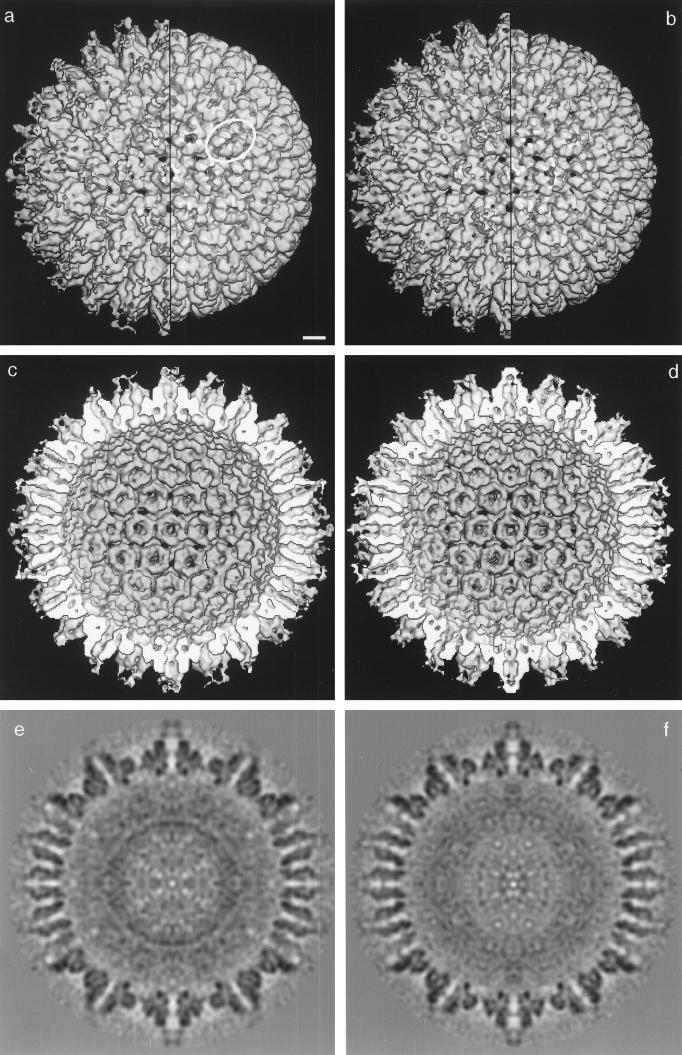
A three-dimensional reconstruction of the procapsid was computed beginning with cryoelectron micrographs such as those shown in Fig. 5a. A total of 72 procapsid images were employed, and the reconstruction was calculated to a resolution of 25 Å by using the PFT method (1). The reconstruction, as viewed along the twofold axis of symmetry, is shown as an outside view (Fig. 6b), an inside view (Fig. 6d), and a central thin section (Fig. 6f). For comparison, the reconstruction computed previously of procapsids formed from HSV-1 proteins in Sf9 cell extracts is shown in Fig. 6a, c, and e (35). Comparison of the two reconstructions shows that procapsids prepared in the two ways are closely similar in nearly all respects. For example, both kinds of procapsids are spherical and have holes through the capsid wall (black spots in Fig. 6b) that do not exist in the mature capsid. In both cases, P and E hexons (30) are oval (Fig. 6a and b; an oval hexon is circled in Fig. 6a). This asymmetry is not expressed on the inner surface of the procapsid shell, where each capsomer has a continuous rim of density that is hexagonal for hexons and pentagonal for pentons (Fig. 6d).
FIG. 6.
Three-dimensional reconstruction of procapsids assembled from purified components (b, d, and f). Also shown is the reconstruction computed previously of HSV-1 procapsids assembled in vitro from insect cell extracts containing VP5, VP19C, VP23, and pre-VP22a (a, c, and e; see reference 14). Both reconstructions are computed to approximately 2.5-nm resolution. In all cases, the reconstructions are shown as viewed along the icosahedral twofold axis of symmetry. The reconstructions are shown as outside views (a and b), inside views (c and d) and central thin sections (e and f). Outside views (a and b) are shown before (left half of each capsid) and after (right half) computational removal of MAb 6F10 from the distal tips of the capsomers (29). In the procapsid hemispheres (c and d), the scaffolding protein has been computationally removed from the cavity to expose the inner surface of the shell. Note the overall similarity between procapsids assembled in the two different ways. For example, in both cases, the procapsid is spherical rather than icosahedral, the hexons are oval rather than hexagonal (one is circled in panel a), and the floor layer is rudimentary (the inner shell surfaces in panels c and d). In both reconstructions, the porous nature of the procapsid shell is apparent from the holes (black patches) through it. Note that in both reconstructions, adjacent capsomers do not make direct contact with each other, but rather interact by way of the triplexes (a and b). Note also that capsomer channels appear closed in procapsids assembled from purified components while they are more open in procapsids formed from cell extracts. Bar, 10 nm.
A minor difference between the two procapsids was observed in the morphology of the core. As shown in central thin sections, the cores of the two kinds of procapsids differ in their radial density distributions (Fig. 6, compare e and 6f). In particular, the core of the extract-assembled procapsid, which is composed primarily of pre-VP22a, has a sharply defined dense ring at a radius of ∼270 Å (Fig. 6e). This ring is absent from the pUL80.5-H-containing core of the procapsids assembled from purified proteins (Fig. 6f). The cores appear to be comparably well preserved in the two kinds of procapsids, so the above differences relate to the respective substructures of the two scaffolding proteins.
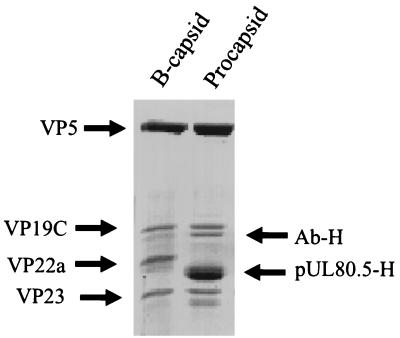
Analysis of the procapsid protein composition by SDS-polyacrylamide gel electrophoresis showed that all four input proteins were present in the product procapsids (Fig. 7). Except for the scaffolding protein, their relative proportions were similar to the proportions found in B capsids. In a representative experiment, for example, the proportions of VP5 to VP19C to scaffolding protein to VP23, as measured by densitometric analysis of a stained gel, were 1.00:0.24:1.23:0.37 for the procapsid and 1.00:0.31:0.45:0.35 for B capsids. The larger amount of scaffolding protein present in procapsids than in B capsids was also observed earlier with procapsids formed in cell extracts (14).
FIG. 7.
SDS-polyacrylamide gel electrophoresis of proteins present HSV-1 B capsids (left) and in procapsids formed from purified proteins (right). Proteins were stained with Coomassie blue. Note that the amount of scaffolding protein is greater in procapsids (pUL80.5-H) than in B capsids (VP22a). Ab-H indicates the position of antibody heavy chain.
As an overall measure of the efficiency of procapsid assembly, we determined the proportion of VP5 that was incorporated into the antibody-precipitable procapsid fraction. VP5 was employed for this determination because it is the reaction component present in limiting quantity. Reaction mixtures were constituted, incubated, and precipitated with MAb 6F10 as described in Materials and Methods. The precipitate was harvested by low-speed centrifugation, and samples of both the precipitate and the supernatant were analyzed by SDS-polyacrylamide gel electrophoresis. Densitometric scanning of the stained gel found 88% of the input VP5 in the precipitate and 12% in the supernatant. In contrast, less than 5% of the input VP5 was found in the precipitate if pUL80.5-H was omitted from the reaction mixture.
Oligomeric state of purified components.
The oligomeric state of purified procapsid components was examined by sucrose density gradient ultracentrifugation. Samples of purified components were mixed with appropriate protein standards and centrifuged on sucrose gradients, and the gradients were separated into equal fractions as described in Materials and Methods. The positions of procapsid proteins in the gradient were determined by SDS-polyacrylamide gel electrophoresis of gradient fractions followed by densitometric scanning of the stained gel. The integrated density corresponding to each protein was then plotted as a function of the fraction number.
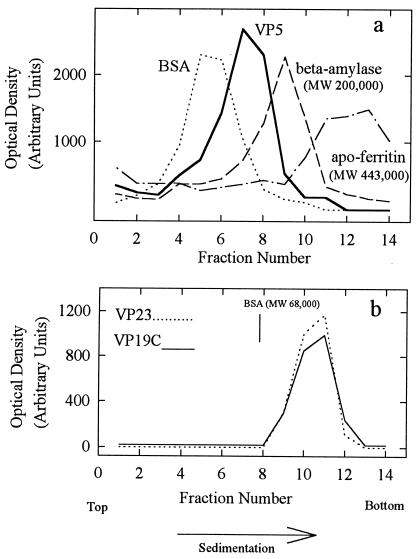
Gradient analysis of VP5 (MW, 149,075) is shown in Fig. 8a. VP5 was found to sediment between the BSA (MW, 68,000) and β-amylase (MW, 200,000) markers, suggesting that it is a monomer under the conditions employed. Only a trace of VP5 was found in material sedimenting more rapidly than β-amylase, where any dimer or higher oligomer is expected. The sedimentation behavior of VP5 was not strongly affected by protein concentration over the range of 0.1 to 1.5 mg/ml or by temperature in the range of 4 to 26°C (data not shown).
FIG. 8.
Sucrose density gradient analysis of purified VP5 (a) and triplexes (b). Gradients were prepared, and the fractions were analyzed by SDS-polyacrylamide gel electrophoresis as described in Materials and Methods. Centrifugation was done at 4°C for 14 (a) and 18 (b) h, respectively. Note that VP5 migrated between the BSA (MW, 68,000) and β-amylase (MW, 200,000) markers.
Figure 8b shows the results obtained with triplexes. VP19C and VP23 sedimented together in a single band between the BSA and β-amylase (not shown) markers. The MW of the triplexes, as estimated with reference to the protein standards, was 107,000, a value in satisfactory agreement with the MW of 118,796 expected of a structure containing one VP19C plus a dimer of VP23. Gradients showed no evidence of structures migrating more slowly than the triplex band. As in the case of VP5, sedimentation of the triplexes showed little dependence on protein concentration in the range of 0.5 to 1.5 mg/ml or temperature (4 to 34°C; data not shown).
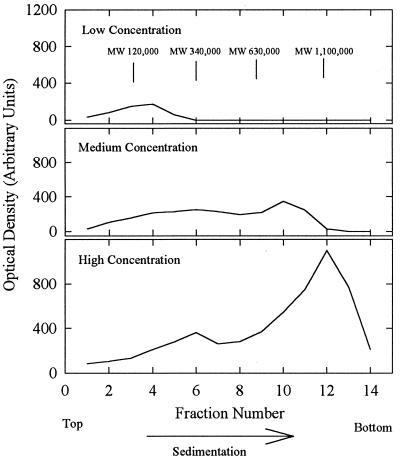
Gradient analyses indicated that the oligomeric state of pUL80.5-H was strongly dependent on protein concentration. For example, the results shown in Fig. 9 were obtained when analyses were performed as described above with 30-μl samples containing 5, 25, and 75 μg of pUL80.5-H (low, medium, and high concentrations, respectively). At a low concentration, pUL80.5-H migrated as a broadly distributed band extending from the top of the gradient to approximately fraction 6 with a peak at fraction 4. The distribution was even broader at medium and high concentrations, extending from the top of the gradient to fractions 11 to 13 at the highest concentration tested. The MWs of the pUL80.5-H complexes, when calculated with respect to those of the BSA, β-amylase, and apoferritin standards, were found to extend from the monomer up to an MW of approximately 106 or more, corresponding to oligomers containing 20 to 30 or more pUL80.5-H molecules. The complexes found at fractions 12 and 13 correspond in their sedimentation rate to the material harvested in the final step of pUL80.5-H purification (Fig. 4). pUL80.5-H did not sediment more rapidly than the fraction 12 and 13 band at any concentration tested. When material from the fraction 12 and 13 band was diluted and recentrifuged, its migration was characteristic of the new, lower concentration (data not shown), suggesting that the distribution of pUL80.5-H oligomers re-equilibrates readily. The sedimentation behavior of pUL80.5-H was not strongly influenced by temperature in the range of 4 to 34°C (data not shown).
FIG. 9.
Sucrose density gradient analysis of purified pUL80.5-H. Gradients contained 5 (top panel), 25 (middle), and 75 (bottom) μg of purified protein, respectively. The gradients were prepared, and the fractions were analyzed by SDS-polyacrylamide gel electrophoresis as described in Materials and Methods. Centrifugation was done for 2.5 h at 4°C. The MW positions shown in the top panel were calculated with reference to the sedimentation of protein standards using the equation of Martin and Ames (10). Note that the sedimentation behavior of pUL80.5-H was strongly influenced by protein concentration.
VP5–pUL80.5-H complexes.
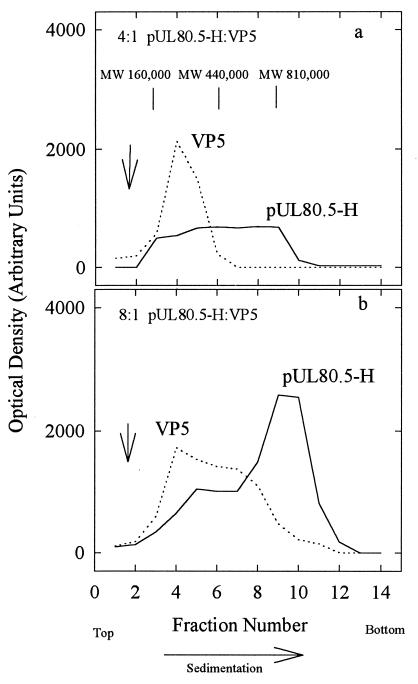
Sucrose gradient ultracentrifugation was also employed to examine complexes formed between VP5 and pUL80.5-H. Analysis was carried out at 4°C as described above for purified proteins, except that VP5 and pUL80.5-H were mixed prior to centrifugation. All experiments were carried out at a molar excess of pUL80.5-H over VP5, with pUL80.5-H-to-VP5 ratios in the range of 10:1 to 2:1. Figure 10 shows the results obtained with two specimens containing pUL80.5-H and VP5 at molar ratios of 4:1 and 8:1, respectively. In both cases, pUL80.5-H migrated as a broad band extending across most of the gradient, as it did in the absence of VP5. Migration of VP5, however, was found to be affected by the presence of pUL80.5-H, indicating an interaction between the two proteins. At a 4:1 ratio of pUL80.5-H to VP5, VP5 migrated as a band corresponding to a complex with an estimated MW of 200,000 to 270,000 (Fig. 10a), which was distinct from the band of VP5 sedimented in the absence of pUL80.5-H (arrow in Fig. 10a). The 200,000 to 270,000-MW band was also observed at an 8:1 pUL80.5-H–VP5 ratio, but in this case, the VP5 distribution extended further down the gradient, suggesting the existence of larger complexes, the largest corresponding to an estimated MW of ∼500,000 to 600,000 (Fig. 10b). Only trace amounts of VP5 were found to migrate coincidentally with the major 106-MW pUL80.5-H band (Fig. 10b, fractions 10 and 11). No VP5-containing complexes were found outside the region between approximately fractions 3 and 9 under any of the experimental conditions tested.
FIG. 10.
Sucrose density gradient analysis of VP5–pUL80.5-H mixtures. Mixtures contained 10 μg of pUL80.5-H plus 10 μg of VP5 (a; ∼4:1 molar ratio of pUL80.5-H to VP5) and 30 μg of pUL80.5-H plus 18 μg of VP5 (b; ∼8:1 molar ratio). Gradients were prepared, and the fractions were analyzed by SDS-polyacrylamide gel electrophoresis as described in Materials and Methods. Centrifugation was done for 2.5 h at 26°C. The MW positions indicated in the top panel were calculated with reference to the sedimentation of protein standards by use of the equation of Martin and Ames (10). Note that VP5 migrated in the form of complexes with estimated MWs in the range of 200,000 to 600,000, and the most rapidly migrating peak of pUL80.5-H bound VP5 poorly. Vertical arrows indicate the position of VP5 migration in companion gradients in which no pUL80.5-H was present.
DISCUSSION
Protein purification.
It was attractive to initiate protein purification beginning with extracts of rBV-infected Sf9 cells because capsid proteins were known to be competent for procapsid assembly in such preparations. Procapsids are formed after mixing Sf9 cell extracts containing VP5, VP19C, VP23, and pre-VP22a (14, 15). The decision to purify VP19C and VP23 as a complex from cells coinfected with rBV encoding both proteins rather than as separate species was made for three reasons. First, it was observed that VP19C and VP23 participate in assembly only after they associate with each other to form triplexes (28). Neither protein can bind to nascent capsids without the other. Second, preformed triplexes were found to remain intact and to retain their assembly competence during purification. Third, we were unsuccessful in attempts to purify assembly-competent VP19C from Sf9 cells containing it. In extracts, VP19C proved sensitive to proteolytic digestion and the purified protein did not support procapsid formation (17). We suggest, therefore, that association with VP23 serves to stabilize VP19C against proteolysis or other processes that denature or inactivate it.
Purification of pUL80.5-H by sucrose density gradient ultracentrifugation as described here depended critically on its presence in the form of large (28-nm diameter) oligomers (Fig. 4). When the ammonium sulfate-fractionated lysate containing pUL80.5-H was centrifuged on sucrose gradients, the large scaffolding protein oligomers migrated more rapidly than contaminating cellular proteins, affording a substantial enrichment in pUL80.5-H (Fig. 1, lanes 9 and 10). Similar sucrose gradient analyses of the HSV-1 scaffolding protein, pre-VP22a, have shown that it also forms irregularly shaped oligomers with approximately the same size as those formed by pUL80.5-H (17, 21).
Protein oligomeric state.
Sucrose density gradient analysis of purified VP5 demonstrated that it migrated between the BSA and β-amylase markers, suggesting that it is a monomer in solution. No larger structures were observed at any concentration tested. Since MAbs cannot ordinarily precipitate protein monomers (9), the presence of VP5 as a monomer in solution accounts for the observation that it is not precipitated by specific MAbs such as 6F10 unless it is first complexed with scaffolding or scaffolding-plus-triplex proteins (15).
Analyses of purified triplexes on sucrose gradients were in agreement with earlier studies of triplexes in Sf9 cell extracts (28). The two triplex proteins were found to migrate together in a structure with a composition and estimated MW compatible with a heterotrimer consisting of one VP19C and two VP23 molecules. Gradients containing triplexes were remarkable for the fact that they showed little or no evidence of dissociation of triplexes into individual proteins (Fig. 8b), suggesting a very strong association between VP19C and VP23. Studies are under way to measure the affinity of the triplex proteins for each other.
The oligomeric state of pUL80.5-H, as determined by sucrose gradient analysis, was found to be strongly dependent on protein concentration. Larger structures were favored at higher protein concentrations. The largest (Fig. 4) were estimated, on the basis of their sedimentation rates, to correspond to oligomers of 25 to 30 pUL80.5-H molecules (i.e., the 28-nm-diameter particles), while the smallest, found near the tops of the gradients, corresponded to monomers or dimers. We interpret the broad range of pUL80.5-H oligomers observed by sucrose gradient analysis to be due to the ability of pUL80.5-H to self-associate in many different proportions in an equilibrium in which larger oligomers are favored by higher protein concentrations.
It was expected that pUL80.5-H would be found to self-associate, as studies involving the yeast two-hybrid system have demonstrated self-interactions in both HCMV and HSV-1 scaffolding proteins (20, 39). Specific regions involved in self-interaction have been identified in both cases, and an α-helical, coiled-coil motif has been proposed for the HSV-1 pre-VP22a self-association. In both HCMV and HSV-1, the ability of the scaffolding protein to self-associate has been found to be required for its interaction with the major capsid protein (20, 39).
The importance of scaffold self-association for procapsid formation is emphasized by the observation that VP5 molecules do not interact in the absence of a scaffolding protein (Fig. 8a). It suggests that either (i) binding of VP5 to a scaffolding protein causes VP5 to acquire the ability to self-associate or (ii) procapsid formation involving VP5-scaffolding protein complexes depends critically on scaffold-scaffold interactions to concentrate VP5 molecules in the proper physical relationship to each other, forming loosely associated precursor capsids that become more tightly bound when angularization takes place.
Procapsid assembly.
Procapsids formed readily when purified VP5, triplexes, and pUL80.5-H were mixed and incubated. The ability of procapsids to assemble from purified proteins indicates that cell-encoded proteins are not required for assembly. The ability of procapsids to form without involvement of cell proteins is further suggested by the fact that only input virus proteins (and antibody) can be identified by SDS-polyacrylamide gel electrophoresis of the product procapsids (Fig. 7). As with cell proteins, cellular small molecules are also unlikely to play a major role in procapsid assembly from purified components. Purification of each of the three reaction components involves a desalting step that is expected to remove small molecules present in the original cell lysate.
Electron microscopic analysis of capsids assembled from purified proteins demonstrated that nearly all were round, not angular in profile, suggesting they correspond to the spherical procapsid rather than to the mature, icosahedral capsid (Fig. 5). The proportion of angular capsids was less than 15%, for example, even when incubations were carried out for 4 h at 37°C. Reaction mixtures containing purified proteins differ significantly in this respect from assembly in Sf9 cell extracts, where nearly all capsids are angular after 2 h or more of incubation at 37°C (14, 15, 17). Possible explanations for the lower number of angular capsids in the purified system include (i) use of the hybrid HCMV–HSV-1 scaffold rather than the homologous HSV-1 form, (ii) the potential role for a cell-encoded protein (e.g., a protease) in angularization, and (iii) the possibility that ions or other small molecules required for angularization were not present in reaction mixtures.
Structural analyses by electron microscopy demonstrated a close resemblance between procapsids assembled from purified components and those formed in Sf9 cell extracts (Fig. 5 and 6). Like extract procapsids, those assembled from purified proteins were found to be spherical in overall shape with distinct shell and core layers. Structural features of the shell, as revealed at 2.5-nm resolution in the three-dimensional reconstruction (Fig. 6), were indistinguishable from those of extract procapsids. There can be little doubt, therefore, that the structures formed from purified components are authentic procapsids. The close structural similarity between the two procapsids is additionally noteworthy because different scaffolding proteins were employed in the two cases, pre-VP22a in extract procapsids and the hybrid scaffold (pUL80.5-H) in purified protein procapsids. The structural similarity of the shells suggests that the identity of the scaffolding protein does not have a pronounced effect on shell morphology. Like procapsids formed in cell extracts, those assembled from purified components were sensitive to disruption at 2°C (17).
The protein composition of procapsids, as determined by SDS-polyacrylamide gel analysis (Fig. 7), demonstrated that procapsids contain all four of the herpesvirus proteins added. Their relative proportions were found to be similar to those of B capsids, except for that of the scaffolding protein, which was regularly observed to be higher in procapsids (compare the left and right lanes in Fig. 7). We considered the possibility that the greater amount of scaffolding protein in the procapsid results from its presence in immune precipitates in a form other than procapsids. This possibility was addressed by analyzing the procapsid reconstruction (Fig. 6). Densities corresponding to the shell and core layers were integrated separately and found at a ratio of 2.62 parts shell to 1 part core (37). Assuming that the core is entirely pUL80.5-H, this ratio indicates a pUL80.5-H copy number of 1,736, a value significantly higher than the 1,153 ± 169 reported for B capsids (16).
The efficiency of procapsid formation, as estimated from the proportion of input VP5 incorporated into material precipitated by MAb 6F10 (88%), represents an upper limit to the procapsid assembly yield. In addition to completed procapsids, MAb 6F10 also precipitates VP5-scaffolding complexes (17) that may be formed in addition to procapsids in reaction mixtures. Analysis of procapsid precipitates by thin-section electron microscopy (Fig. 5b) demonstrated the presence of non-procapsid material that could correspond to VP5–pUL80.5-H complexes. Less non-procapsid material was observed, however, in negatively stained and frozen-hydrated preparations (Fig. 5d and a), suggesting that procapsids account for a large proportion of the overall antibody precipitate.
VP5-scaffolding protein complexes.
Sucrose gradient analyses of VP5–pUL80.5-H mixtures demonstrated that the two proteins interact readily to form complexes with estimated MWs in the range of 200,000 to 600,000 (Fig. 10). Taking into account the protein composition of the complexes as shown in Fig. 10, we interpreted them to be a population of oligomers in which individual species contain one or two VP5 and one to five or six pUL80.5-H molecules. Little or no free VP5 was observed in the gradients, suggesting that interaction between the two proteins is strong enough that complexes do not dissociate during sucrose gradient centrifugation. Interaction between VP5 and pUL80.5-H has been demonstrated in vitro (19), and interaction of VP5 with the HSV-1 scaffolding protein has been documented both in vivo and in vitro (8, 12, 20, 33). The present studies extend the earlier work by indicating the molecular size and composition of VP5-scaffolding protein complexes.
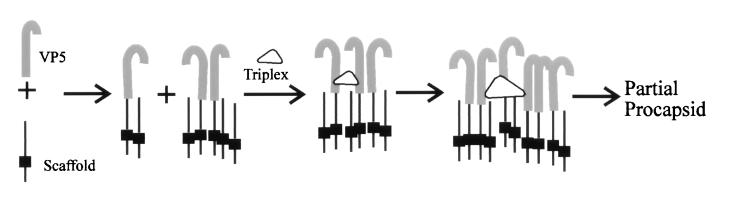
The VP5–pUL80.5-H complexes observed by gradient analysis suggest themselves as functional subunits involved in procapsid assembly. Complexes could be cross-linked to each other by way of the triplexes, as shown diagramatically in Fig. 11, to initiate or extend the growth of the nascent procapsid. Involvement of VP5–pUL80.5-H complexes in procapsid formation is consistent with the observation that procapsids appear to form by incremental addition of both VP5 and scaffolding protein to partial capsid intermediates (14).
FIG. 11.
Schematic representation of the pathway proposed for HSV-1 procapsid formation. VP5 (gray) and the scaffolding protein (black) are considered to interact to form complexes involving one or two VP5 and one to six scaffolding molecules, as suggested by the results of the sucrose gradient analyses shown in Fig. 10. VP5-scaffolding protein complexes are then cross-linked to each other by triplexes to initiate procapsid formation or to extend regions of growing procapsid shell as found in partial procapsids. The thickening in each scaffolding protein molecule represents the domain by which scaffolding molecules interact with each other (20, 39).
The VP5-scaffolding protein complexes observed here could suggest the form in which VP5 and pre-VP22a are transported to the nucleus in HSV-1-infected cells. Although VP5 can enter the nucleus in other ways (26), VP5 and pre-VP22a can enter the nucleus as a complex (18), and those described here are small enough that they should be able to be transported through nuclear pores (6).
A consistent feature of the sucrose gradient analyses was the observation that VP5 interacted weakly or not at all with the large (MW, 1.1 × 106; 28-nm diameter) pUL80.5-H particles (Fig. 10). If they occur in infected cells, therefore, the large particles are likely to serve as a reservoir of scaffolding protein rather than as direct participants in the assembly process.
Assembly of HSV-1 procapsids from purified components as described here has important features in common with assembly of Salmonella typhimurium phage P22 procapsids from purified major capsid (gp5) and scaffolding (gp8) proteins as described by King, Prevelige, and their colleagues (3, 4, 22, 23, 24). In both cases, the major capsid protein is a monomer in solution, and it does not form larger structures in the absence of the scaffolding protein. In both cases, procapsid assembly is considered to take place by incremental addition of major capsid and scaffolding proteins to partial capsid intermediates, and in both cases, experimental studies have defined the nature of small major capsid-scaffolding protein complexes thought to serve as assembly subunits (24; this study).
Major differences between P22 and HSV-1 in procapsid formation have to do with the triplexes and the scaffolding protein. Triplexes are absolutely required for HSV-1 procapsid assembly (15, 31, 34). They are suggested to mediate association between small major capsid-scaffolding protein complexes and between complexes and regions of growing procapsid wall (14). Triplexes may also be involved in organizing major capsid protein molecules into capsomers (35). In contrast, P22 procapsids assemble from purified gp5 and gp8 only. The two systems also differ in the amount of scaffolding protein involved in procapsid assembly. Whereas the molar ratio of scaffolding protein to major capsid protein in HSV-1 procapsids is greater than one (and approaches two), in P22, the comparable ratio is 0.71 (300 scaffolding protein molecules to 420 major capsid protein molecules; 4, 22).
Purified pUL80.5-H and the P22 scaffolding protein differ in the ability to oligomerize in solution. Whereas purified gp8 exists as a monomer or a dimer in solution and does not further oligomerize in the absence of gp5 (23), pUL80.5-H is found to self-associate to form a wide variety of structures containing up to 25 to 30 pUL80.5-H molecules (Fig. 9). It would be of interest to test whether pre-VP22a, the fully homologous HSV-1 scaffolding protein, oligomerizes as described here for pUL80.5-H.
The ability of procapsids to assemble in vitro from purified VP5, triplexes, and pUL80.5-H as described here suggests that similar structures will prove to be involved as capsid assembly intermediates in HSV-1-infected cells. Efforts to identify and isolate in vivo procapsids can, with some confidence, now be guided by the observed properties (e.g., spherical morphology and cold sensitivity) of in vitro procapsids. Although in vivo procapsids are expected to resemble their in vitro counterparts, it is reasonable to expect that there may be differences as well. Proteins involved in DNA processing and packaging, for instance, may be present in in vivo procapsids in addition to shell and scaffolding proteins (7, 40). The identity and abundance of such additional polypeptides may provide clues to the events of DNA packaging and subsequent capsid maturation.
ACKNOWLEDGMENTS
We thank Arnita Barber and David Burkwall for help with the sucrose density gradient analyses.
This work was supported in part by research grants (AI41644 and AI37549) from the National Institutes of Health.
REFERENCES
- 1.Baker T S, Cheng R H. A model-based approach for determining orientations of biological molecules imaged by cryoelectron microscopy. J Struct Biol. 1998;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 2.Conway J F, Trus B L, Booy F P, Newcomb W W, Brown J C, Steven A C. Visualization of three-dimensional density maps reconstructed from cryo-electron micrographs of viral capsids. J Struct Biol. 1996;116:200–208. doi: 10.1006/jsbi.1996.0031. [DOI] [PubMed] [Google Scholar]
- 3.Fuller M T, King J. Purification of the coat and scaffolding proteins from procapsids of bacteriophage P22. Virology. 1981;112:529–547. doi: 10.1016/0042-6822(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 4.Fuller M T, King J. Assembly in vitro of bacteriophage P22 procapsids from purified coat and scaffolding subunits. J Mol Biol. 1982;156:633–665. doi: 10.1016/0022-2836(82)90270-4. [DOI] [PubMed] [Google Scholar]
- 5.Fuller S D, Butcher S J, Cheng R H, Baker T S. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 6.Gorlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Homa F L, Brown J C. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 8.Hong Z, Beaudet-Miller M, Durkin J, Zhang R, Kwong A D. Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein. J Virol. 1996;70:533–540. doi: 10.1128/jvi.70.1.533-540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janeway C A, Jr, Travers P. Immunobiology: the immune system in health and disease. New York, N.Y: Garland; 1997. pp. 2–13. [Google Scholar]
- 10.Martin R G, Ames B N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961;236:1372–1379. [PubMed] [Google Scholar]
- 11.Matusick-Kumar L, Hurlburt W, Weinheimer S P, Newcomb W W, Brown J C, Gao M. Phenotype of the herpes simplex virus type 1 protease substrate ICP35 mutant virus. J Virol. 1994;68:5384–5394. doi: 10.1128/jvi.68.9.5384-5394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matusick-Kumar L, Newcomb W W, Brown J C, McCann III P J, Hurlburt W, Weinheimer S P, Gao M. The C-terminal 25 amino acids of the protease and its substrate ICP35 of herpes simplex virus type 1 are involved in the formation of sealed capsids. J Virol. 1995;69:4347–4356. doi: 10.1128/jvi.69.7.4347-4356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newcomb W W, Brown J C. Use of Ar+ plasma etching to localize structural proteins in the capsid of herpes simplex virus type 1. J Virol. 1989;63:4697–4702. doi: 10.1128/jvi.63.11.4697-4702.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid assembly. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 15.Newcomb W W, Homa F L, Thomsen D R, Ye Z, Brown J C. Cell-free assembly of the herpes simplex virus capsid. J Virol. 1994;68:6059–6063. doi: 10.1128/jvi.68.9.6059-6063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 17.Newcomb, W. W., F. L. Homa, and J. C. Brown. Unpublished observations.
- 18.Nicholson P, Addison C, Cross A M, Kennard J, Preston V G, Rixon F J. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J Gen Virol. 1994;75:1091–1099. doi: 10.1099/0022-1317-75-5-1091. [DOI] [PubMed] [Google Scholar]
- 19.Oien N L, Thomsen D R, Wathen M W, Newcomb W W, Brown J C, Homa F L. Assembly of herpes simplex virus capsids using the human cytomegalovirus scaffold protein: critical role of the C terminus. J Virol. 1997;71:1281–1291. doi: 10.1128/jvi.71.2.1281-1291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelletier A, Do F, Brisebois J J, Lagace L, Cordingly M G. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J Virol. 1997;71:5197–5208. doi: 10.1128/jvi.71.7.5197-5208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preston V G, Al-Kobaisi M F, McDougall I M, Rixon F J. The herpes simplex virus gene UL26 proteinase in the presence of the UL26.5 gene product promotes the formation of scaffold-like structures. J Gen Virol. 1994;75:2355–2366. doi: 10.1099/0022-1317-75-9-2355. [DOI] [PubMed] [Google Scholar]
- 22.Prevelige P E, King J. Assembly of bacteriophage P22: a model for ds-DNA virus assembly. Prog Med Virol. 1993;40:206–221. [PubMed] [Google Scholar]
- 23.Prevelige P E, Thomas D, King J. Scaffolding protein regulates the polymerization of P22 coat subunits into icosahedral shells in vitro. J Mol Biol. 1988;202:743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- 24.Prevelige P E, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 26.Rixon F J, Addison C, McGregor A, Macnab S J, Nicholson P, Preston V G, Tatman J D. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J Gen Virol. 1996;77:2251–2260. doi: 10.1099/0022-1317-77-9-2251. [DOI] [PubMed] [Google Scholar]
- 27.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Strauss S E, Roizman B, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2221–2230. [Google Scholar]
- 28.Spencer J V, Newcomb W W, Thomsen D R, Homa F L, Brown J C. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J Virol. 1998;72:3944–3951. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spencer J V, Trus B L, Booy F P, Steven A C, Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: peptide A862-H880 of the major capsid protein is displayed on the rim of the capsomer protrusions. Virology. 1997;228:229–235. doi: 10.1006/viro.1996.8392. [DOI] [PubMed] [Google Scholar]
- 30.Steven A C, Roberts C R, Hay J, Bisher M E, Pun T, Trus B L. Hexavalent capsomers of herpes simplex virus type 2: symmetry, shape, dimensions, and oligomeric status. J Virol. 1986;57:578–584. doi: 10.1128/jvi.57.2.578-584.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatman J D, Preston V G, Nicholson P, Elliott R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 32.Thomas D, Newcomb W W, Brown J C, Wall J S, Hainfeld J F, Trus B L, Steven A C. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J Virol. 1985;54:598–607. doi: 10.1128/jvi.54.2.598-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomsen D R, Newcomb W W, Brown J C, Homa F L. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J Virol. 1995;69:3690–3703. doi: 10.1128/jvi.69.6.3690-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19C and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 36.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trus, B. L., and A. C. Steven. Unpublished observations.
- 38.Whitley R J. Herpes simplex viruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Strauss S E, Roizman B, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2297–2342. [Google Scholar]
- 39.Wood L J, Baxter M K, Plafker S M, Gibson W. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J Virol. 1997;71:179–190. doi: 10.1128/jvi.71.1.179-190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu D, Weller S K. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids. J Virol. 1998;72:7428–7439. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z H, Chiu W, Haskell K, Spears H, Jr, Jakana J, Rixon F J, Scott L R. Refinement of herpesvirus B-capsid structure on parallel supercomputers. Biophys J. 1998;74:576–588. doi: 10.1016/S0006-3495(98)77816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Z H, Prasad B V V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in herpes simplex virus A-capsid determined from 400kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]
- 43.Zlotnick A, Cheng N, Conway J F, Booy F P, Steven A C, Stahl S J, Wingfield P T. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]