Abstract
The formation of an infectious retrovirus particle requires several RNA-RNA interaction events. In particular, the genomic RNA molecules form a dimeric structure, and a cellular tRNA molecule is annealed to an 18-base complementary region (the primer binding site, or PBS) on the genomic RNA, where it will serve as primer for reverse transcription. tRNAs normally possess a highly stable secondary and tertiary structure; it seems unlikely that annealing of a tRNA molecule to the PBS, which involves unwinding of this structure, could occur efficiently at physiological temperatures without the assistance of a cofactor. Many prior studies have shown that the viral nucleocapsid (NC) protein can act as a nucleic acid chaperone (i.e., facilitate annealing events between nucleic acids), and the assays used to demonstrate this activity include its ability to catalyze dimerization of transcripts representing retroviral genomes and the annealing of tRNA to the PBS in vitro. However, mature NC is not required for these events in vivo, since protease-deficient viral mutants, in which NC is not cleaved from the parental Gag polyprotein, are known to contain dimeric RNAs with tRNA annealed to the PBS. In the present experiments, we have tested recombinant human immunodeficiency virus type 1 Gag polyprotein for nucleic acid chaperone activity. The protein was positive by all of our assays, including the ability to stimulate dimerization and to anneal tRNA to the PBS in vitro. In quantitative experiments, its activity was approximately equivalent on a molar basis to that of NC. Based on these results, we suggest that the Gag polyprotein (presumably by its NC domain) catalyzes the annealing of tRNA to the PBS during (or before) retrovirus assembly in vivo.
A single viral protein, the Gag polyprotein, is sufficient for assembly of retrovirus particles in cells of higher eukaryotes. Normally, after the particle is released from the cell, the polyprotein is cleaved by the virus-coded protease (PR), liberating a series of cleavage products. This series of cleavage events causes a major structural reorganization of the particle called maturation; PR-deficient particles, in which Gag is not cleaved, are termed immature particles (reviewed in reference 10).
Among the molecular events which occur during virus assembly and maturation are several instances of RNA-RNA interaction, including the formation of a dimer from two identical, plus-strand molecules of genomic RNA; maturation or condensation of the dimer to a more compact, more thermostable dimer; and placement of a cellular tRNA molecule at a specific site, the primer binding site (PBS), on the genomic RNA, where it will serve as the primer for viral DNA synthesis by reverse transcriptase when the particle infects a new host cell (reviewed in reference 10). It seems likely that some or all of these RNA-RNA interactions are facilitated by viral or cellular cofactors. In particular, it appears extremely improbable that the primer tRNA could spontaneously bind to the PBS with high efficiency under physiological conditions. This binding involves the annealing of the 18 3′ nucleotides of the tRNA to the PBS (to which they are complementary) and additional interactions between other regions of the tRNA and other sites in the genomic RNA (2, 24, 27); thus, formation of this complex requires the disruption of the normal secondary and tertiary structure of the tRNA.
What is the identity of this putative cofactor? In fact, a virus-coded protein, termed nucleocapsid (NC), is an obvious candidate, since it is capable of facilitating all of these RNA-RNA interactions in vitro (12). NC is one of the products formed as a result of cleavage of the Gag polyprotein during virus maturation. NC proteins are small, basic proteins which bind single-stranded nucleic acids in vitro and are associated with the genomic RNA in the interior of mature retrovirus particles. They have been shown to possess nucleic acid chaperone activity in a wide variety of assays; that is, they catalyze the conformational rearrangement of nucleic acids into optimally base-paired structures. The molecular mechanism of this effect is not well understood, but NC presumably acts by lowering the energy barrier for breakage and reformation of base pairs. Recent evidence strongly suggests that this activity gives NC a crucial accessory role during the reverse transcription of retroviral RNA into DNA (reviewed in references 12 and 31).
Further evidence supporting the idea that NC assists in the placement of tRNA on the PBS in vivo is the fact that mutations in the NC coding region prevent the placement of the primer tRNA on the PBS in packaged viral RNA (22, 30).
On the other hand, well-documented biological observations are difficult to reconcile with the hypothesis that the mature NC protein performs these functions in vivo. In particular, immature retrovirus particles, in which NC is not formed from the Gag polyprotein because of a defect in PR, contain dimeric RNA (16, 18, 32). Further, primer tRNA is annealed to the PBS on these genomic RNA molecules (11, 17, 23, 32).
One hypothesis which is consistent with all of these observations is that the retroviral Gag polyprotein, like the NC protein derived from it by proteolytic cleavage, possesses nucleic acid chaperone activity. The present experiments have tested this possibility, and we now report that recombinant human immunodeficiency virus type 1 (HIV-1) Gag polyprotein indeed exhibits nucleic acid chaperone activity. The data presented here show that this protein is able to catalyze both the dimerization of transcripts containing HIV-1 genomic sequences and the annealing of primer tRNA to the PBS in vitro. In view of these results, we suggest that Gag catalyzes these two events in vivo, probably during assembly of the retrovirus particle.
MATERIALS AND METHODS
Reagents.
The production of recombinant HIV-1 Gag polyprotein and of Gag polyprotein lacking the p6 domain (GagΔp6) have been described previously (7). Briefly, the coding regions from the BH10 isolate of HIV-1 were expressed by using the pET 3xc vector. Some experiments used proteins with two point mutations, viz., A37V and Q63D, but control experiments indicated that these differences from the wild-type sequence did not affect the results of our studies. The two polyproteins were purified as previously described for derivatives of the Gag protein of Rous sarcoma virus (8); in addition, the full-length protein was purified with an affinity column containing rabbit antibodies against the 21 C-terminal amino acids of Gag. We estimate, based on inspection of Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels, that the proteins were ≥90% pure. One microgram of GagΔp6 corresponds to 20 pmol of the protein.
Recombinant HIV-1 NC protein (6, 35), whose sequence was from the NL4-3 isolate of HIV-1 (1), was a kind gift from Robert Gorelick and Louis Henderson, National Cancer Institute-Frederick Cancer Research and Development Center. One microgram of NC represents 170 pmol.
tRNA3Lys was purified from beef liver by an adaptation of the procedure described by Isel et al. (25) and was then further purified by two-dimensional electrophoresis (19). Both the primary sequence and the posttranscriptional modifications of bovine tRNA3Lys are identical to those of human tRNA3Lys (25a).
The template for synthesis of RNA representing nucleotides (nt) 1 to 331 of NL4-3 HIV-1 genomic RNA was constructed as follows. pJD DNA (20), which contains the 5′ 131 nt of HIV-1 genomic RNA sequence after a phage T7 promoter, was altered by addition, at the 3′ end of the viral sequence, of a PCR product containing the next 200 bases of the NL4-3 genome, followed by an NcoI site. The plasmid was linearized by digestion with NcoI before transcription with T7 RNA polymerase (Promega) in accordance with the manufacturer’s instructions.
HaSV 34-378 RNA was synthesized with SP6 RNA polymerase (Promega) exactly as described previously (14).
Selective annealing of oligonucleotides.
Selective annealing experiments were performed exactly as described by Tsuchihashi and Brown (34), except that the sequences of the oligonucleotides were slightly altered: the labeled oligonucleotide was 5′GACTAAAAAAAAATCTCTAGCAGTGCAT3′, and the two competing oligonucleotides were 5′ATGCACTGCTAGAGATTTTTTTTTAGTC3′ (28-base perfect match) and 5′ACTGCTAGAGATTTTTTTTTT3′ (21-base oligonucleotide, which can form 20 base pairs with the labeled oligonucleotide). This change in sequence had no detectable effect on the outcome of selective annealing experiments with NC protein (data not shown). Briefly, 20 fmol of the labeled oligonucleotide was mixed with 20 fmol of the complementary 28-base oligonucleotide and 1 pmol of the complementary 21-base oligonucleotide, along with the protein being tested, in 10 μl of 50 mM Tris (pH 7.9)–1 mM EDTA–3 mM MgCl2–1 mM dithiothreitol–0.1% Triton X-100–0.2 μg of BSA/ml. After incubation as indicated, the reactions were terminated by the addition of SDS (final concentration, 2%), extracted with phenol, and analyzed by electrophoresis in 15% polyacrylamide and by autoradiography.
RNA-RNA annealing.
The ability of proteins to stimulate the annealing of complementary RNA molecules was assayed as follows. A 205-base RNA molecule, consisting of nucleotides 34 to 180 of the sense strand of the Neor-encoding open reading frame plus 58 vector nucleotides, was synthesized by using T7 RNA polymerase in the presence of [α-32P]CTP. The second RNA, also synthesized with T7 RNA polymerase, was 833 nt long, consisting of the antisense strand of the Neor-encoding open reading frame from residues 790 to 34, plus 76 residues from the 3′ untranslated region of the gene. Twenty fmol of the first RNA were mixed with 200 femtomol of the second in 10 μl of 5 mM NaCl–0.1 mM EDTA–10 mM Tris [pH 8.0]–0.1 M β-mercaptoethanol–0.2 μg of bovine serum albumin (BSA) (New England Biolabs)–8 U of RNasin (Promega)/μl. Each tube thus contained a total of ∼57 ng of RNA. The RNAs were first denatured by heating the mixture to 90°C for 5 min and chilling it on ice. Proteins were then added to the mixture as indicated, and the reactions were incubated at 0°C. (Incubation of these reactions at 37°C frequently resulted in aggregation of the RNA samples in the presence of NC). After incubation times as indicated, the reactions were terminated by the addition of 2 μl 10% SDS and 10 μl H2O, followed by extraction with phenol-CHCl3 (1:1) and analysis by electrophoresis in agarose under nondenaturing conditions and by autoradiography.
Dimerization of HIV-1 RNA.
An RNA transcript representing nt 1 to 331 of the NL4-3 clone of HIV-1 (1) was synthesized by T7 RNA polymerase in the presence of [α-32P]CTP. The transcript was purified on a G-50 spun column (Stratagene) and by electrophoresis in polyacrylamide. For dimerization, 1.5 μg of RNA was heated in water to 85°C for 2 min and chilled on ice. It was then incubated in 20 μl of 250 mM NaCl–5 mM MgCl2–20 mM Tris (pH 7.0) at 37°C for 3 h, in the presence of NC or Gag. The RNAs were then treated with 2% SDS, extracted with phenol, and analyzed by electrophoresis through 2.5% Metaphor (FME) under nondenaturing conditions and by autoradiography.
Stabilization of dimers of HaSV RNA.
The thermostability of dimers of HaSV 34-378 RNA was assessed as described previously (13), except that in some experiments the dimers were formed in the presence of Gag, NC, or BSA rather than being formed first and then treated with the proteins. Briefly, labeled RNA was allowed to dimerize by incubation for 30 min at 37°C in 0.25 M NaCl–1 mM MgCl2–0.01 M Tris (pH 7.0). After deproteinization by phenol extraction, it was then microdialyzed into 0.05 M NaCl–1 mM EDTA–0.01 M Tris (pH 7.0). The thermostabilities of the dimers were then determined by incubating them for 10 min at the indicated temperatures and analyzing them by electrophoresis in agarose, followed by autoradiography.
Placement of tRNA3Lys on PBS of HIV-1 RNA.
Purified tRNA3Lys was labeled at its 5′ end by incubation with T4 polynucleotide kinase (Boehringer Mannheim) and [γ-32P]ATP. The labeled tRNA was repurified by electrophoresis in a polyacrylamide gel. After elution from the gel, it was precipitated with ethanol and redissolved in water. Unlabeled RNA transcripts representing nt 1 to 331 of the NL4-3 clone of HIV-1 or nt 34 to 378 of HaSV were synthesized using T7 or SP6 RNA polymerase, respectively. The transcripts were heated to 85°C in water and placed on ice. Fifty nanograms of radioactive tRNA was mixed with 250 ng of transcript in 20 μl of 50 mM NaCl–5 mM MgCl2–20 mM Tris (pH 7.0) in the presence or absence of GagΔp6 protein. After incubation for 30 min at 37°C, the samples were deproteinized and analyzed as in the dimerization experiments (see above).
Extension of tRNA3Lys with reverse transcriptase after annealing to PBS of HIV-1 RNA.
Two hundred fifty nanograms of an RNA transcript representing nucleotides 1 to 331 of pNL4-3 HIV-1 was heated to 100°C and placed on ice. It was then mixed with 50 ng of unlabeled tRNA3Lys in 20 μl of 250 mM NaCl–1 mM MgCl2–10 mM Tris (pH 7.0). After incubation for 60 min at 37°C in the presence or absence of GagΔp6 protein, the mixture was deproteinized by treatment with 2% SDS and phenol extraction. After ethanol precipitation, the RNAs were redissolved in 20 μl of 75 mM KCl–5 mM MgCl2–0.1 mM dithiothreitol–50 mM Tris (pH 7.5) containing 57 U of HIV-1 reverse transcriptase (Worthington) and 0.1 mM (each) deoxynucleoside triphosphate (dNTP), including 1 μCi of [α-32P]dCTP (Amersham). After 30 min at 37°C, 27 additional U of reverse transcriptase was added and the incubation was continued for 30 min more. The reaction mixture was then heated to 85°C for 5 min and digested for 60 min at 37°C with 20 μg of pancreatic RNase (Boehringer Mannheim) and 8 U of RNase H (Stratagene). The digest was then extracted with phenol, and the DNA was purified with a G-50 spun column (Boehringer Mannheim). Finally, the product was analyzed by electrophoresis on a 15% polyacrylamide gel in the presence of 7 M urea, followed by autoradiography.
RESULTS
Selective annealing of oligodeoxynucleotides.
The nucleic acid chaperone activity of NC protein has previously been demonstrated by the selective annealing of oligonucleotides (34). In this assay, a radioactively labeled 28-base oligodeoxynucleotide is mixed with two other oligodeoxynucleotides. Both of these oligonucleotides are complementary to the labeled oligonucleotide, but one is a 28-base, completely complementary molecule, while the other is shorter and can form only 20 base pairs with the labeled oligonucleotide. The shorter of these two molecules is in 50-fold excess over the longer one; therefore, the less stable hybrid which can form in this mixture is kinetically favored over the more stable one. As previously shown by Tsuchihashi and Brown (34), labeled oligonucleotide is found annealed to the shorter complementary oligonucleotide if the mixture is incubated in the absence of NC but is annealed to the longer oligonucleotide if NC is present. This shift presumably reflects the ability of NC to destabilize the 20-bp hybrid, enabling the labeled DNA strand to enter into the more stable, 28-bp hybrid.
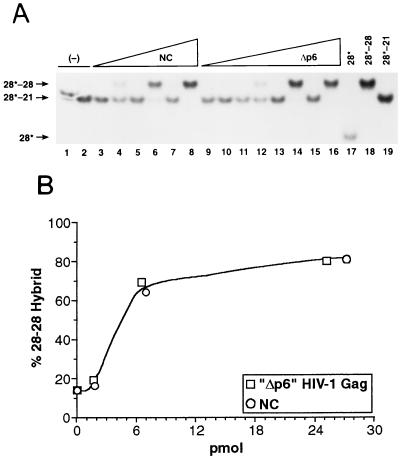
We tested the ability of HIV-1 Gag Δp6 polyprotein to promote the selective formation of the more stable hybrid in this assay. As shown in Fig. 1A, incubation with Gag Δp6 at 37°C did cause the labeled oligonucleotide to shift from the kinetically favored, 20-bp hybrid to the thermodynamically favored 28-bp hybrid; in fact, analysis of the data (Fig. 1B) indicated that the activities of the polyprotein and of NC in this assay were, on a molar basis, indistinguishable.
FIG. 1.
Demonstration of nucleic acid chaperone activity by selective annealing of oligonucleotides. (A) Oligonucleotides were incubated alone (lanes 1 and 2) or with 0.01, 0.04, or 0.16 μg of NC (lanes 3 and 4, 5 and 6, 7 and 8, respectively) or 0.02, 0.08, 0.32, or 1.3 μg of GagΔp6 (lanes 9 and 10, 11 and 12, 13 and 14, and 15 and 16, respectively) at 0° (odd-numbered lanes) or 37°C (even-numbered lanes) for 60 min. Lanes 17 to 19 represent markers, in which the labeled oligonucleotide was mixed with nothing (lane 17), with the 28-base complementary oligonucleotide alone (lane 18), or with the 21-base complementary oligonucleotide alone (lane 19), heated to 90°C for 5 min, and allowed to cool slowly to room temperature. (B) Data from incubations at 37°C in panel A as analyzed by phosphorimaging.
Annealing of complementary RNA molecules.
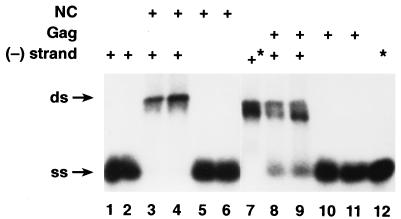
We also tested the ability of the Gag polyprotein to stimulate the annealing of complementary RNAs. These RNA molecules were transcripts of portions of the Neor gene. Figure 2 (lanes 8 and 9) shows that in the presence of Gag, a large fraction of the labeled RNA (although less than in the sample incubated with NC) was hybridized to the complementary strand, while under these conditions no detectable hybrid was formed in the absence of Gag.
FIG. 2.
Annealing of complementary RNAs in the presence of Gag or NC. RNAs were incubated with nothing (lane 12) or with 200 ng of BSA/μl (lanes 1 and 2), 330 ng of NC/μl (lanes 3 to 6), or 2.7 μg of Gag/μl (lanes 8 to 11) for 1 h (lanes 1, 3, 5, 8, and 10) or 4 h (lanes 2, 4, 6, 9, 11, and 12) and analyzed as described in Materials and Methods. RNA which was complementary to the labeled RNA was present in lanes 1 to 4 and 7 to 9. The samples were heated to 90°C for 5 min; those shown in lanes 7 and 12 (indicated by an asterisk) were allowed to cool slowly to room temperature, while the others were quickly chilled on ice before incubation as indicated. ss, single stranded; ds, double stranded.
Dimerization of retroviral RNAs in the presence of Gag.
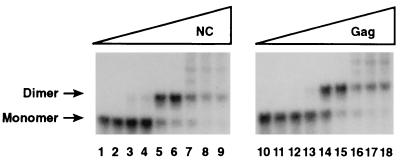
As originally demonstrated by Darlix and coworkers (12, 29), RNA transcripts containing sequences from the 5′ portion of a retroviral genome can form dimers in vitro, even in the absence of any added proteins or other macromolecular cofactors. However, as noted above, NC has been shown to accelerate this dimerization process in vitro (12). We tested the ability of the Gag polyprotein to promote the dimerization of RNA molecules consisting of nt 1 to 331 of the HIV-1 genome, using a low RNA concentration at which there was virtually no spontaneous dimerization during the incubation period. As shown in Fig. 3, this RNA was converted to a dimeric structure in the presence of Gag; the amount of Gag required for this effect, ∼0.8 μg (lane 14), was equivalent in molar terms to the level of NC (∼0.1 μg) (lane 5) which exerted a similar effect.
FIG. 3.
Dimerization of HIV 1-331 transcripts in the presence of Gag or NC. 32P-labeled HIV-1 1-331 RNA, prepared as described in Materials and Methods, was incubated with 0, 0.01, 0.02, 0.05, 0.10, 0.50, 1.0, 1.25, or 1.5 μg of NC (lanes 1 to 9) or 0, 0.08, 0.16, 0.4, 0.8, 4.0, 8.0, 10.0, or 12.0 μg of Gag (lanes 10 to 18). The results were analyzed by electrophoresis and autoradiography as described in Materials and Methods.
Stabilization of a dimer of retroviral RNA by Gag.
The genomic RNA in an immature retrovirus particle is dimeric. However, during maturation of the particle, the dimer is converted to a more thermostable dimeric structure (16, 18, 33). We previously described conditions in which a 345-base transcript of HaSV RNA formed dimers in vitro, but some of these dimers were relatively thermolabile. Incubation of these preparations of dimeric RNA with recombinant HIV-1 NC was shown to convert the thermolabile dimers into more stable dimers; this conversion thus appeared to replicate, in a defined system in vitro, the stabilization of dimeric RNAs which occurs during the maturation of a retrovirus particle (13).
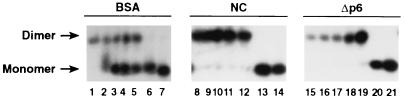
We also tested the ability of the HIV-1 Gag polyprotein to induce this stabilization in dimers of HaSV transcripts. Figure 4 shows the results of this experiment. As can be seen, the dissociation profile of the dimers after exposure to the GagΔp6 polyprotein (lanes 15 to 21), like that seen after exposure to NC (lanes 8 to 14), showed that the dimers were uniformly dissociated between 55 and 65°C; in contrast, controls incubated with BSA (lanes 1 to 7) were a mixture in which some molecules dissociated between 25 and 37°C, and others were stable to a temperature between 55 and 65°C. Thus, the Gag polyprotein is evidently able to induce the stabilization of dimers of retroviral RNAs in vitro. Moreover, its activity per mole appears to be at least as high as that of NC in this assay, since NC and Gag were equimolar in these experiments and since the dose of NC is only slightly more than sufficient for full stabilization of the dimers (13).
FIG. 4.
Stabilization of dimers of HaSV 34-378 RNA by incubation with Gag or NC. A total of 0.5 μg of [32P]-labeled HaSV 34-378 RNA was incubated at 37°C with 2 μg of BSA (lanes 1 to 7), 1.5 μg of NC (lanes 8 to 14), or 12 μg of GagΔp6 (lanes 15 to 21). Thermostabilities of the dimers were then determined by incubation for 10 min at 0° (lanes 1, 8, and 15), 25° (lanes 2, 9, and 16), 37° (lanes 3, 10, and 17), 45° (lanes 4, 11, and 18), 55° (lanes 5, 12, and 19), 65° (lanes 6, 13, and 20), or 70° (lanes 7, 14, and 21), followed by electrophoresis and autoradiography as described in Materials and Methods. Some RNA remained at the top of the gel in lanes 15 to 17, but this aggregation was not a consistent feature of these experiments.
Placement of tRNA3Lys on the PBS by Gag.
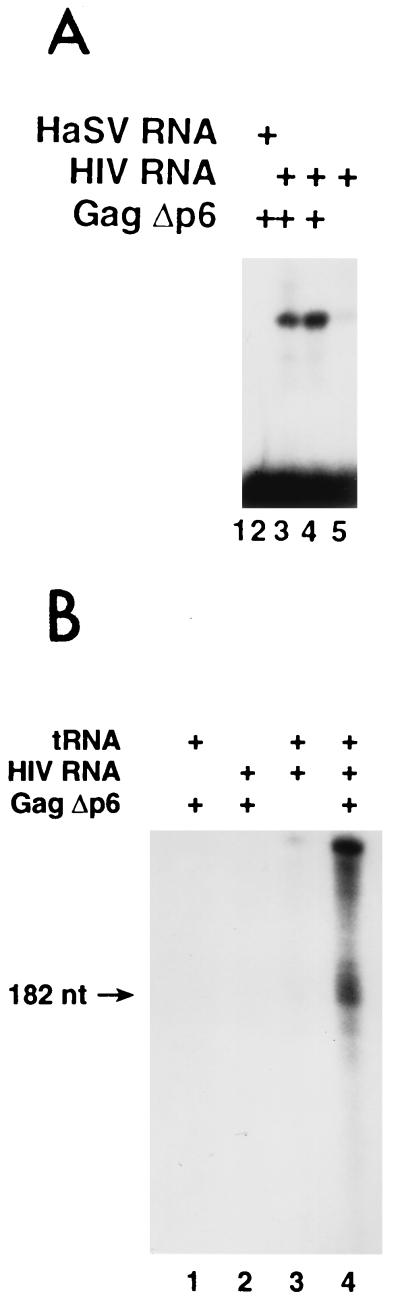
Finally, we also tested the ability of the GagΔp6 polyprotein to anneal tRNA3Lys to the PBS on transcripts of the HIV-1 genome. We used natural tRNA3Lys, rather than tRNA produced by transcription in vitro, since the posttranscriptional modifications in natural tRNA play a significant role in the interaction of the primer with the PBS (25). As shown in Fig. 5A, incubation of labeled tRNA3Lys with RNA molecules representing nt 1 to 331 of HIV RNA, together with GagΔp6, caused a shift of radioactive tRNA to a high-molecular-weight complex (lanes 3 and 4). This shift was not observed in the absence of GagΔp6 (lane 5) and reflected a true annealing reaction, rather than aggregation of the RNAs, since it was also not seen in a control reaction in which the tRNA was incubated with a 345-base HaSV transcript, which contains a PBS complementary to tRNAPro rather than tRNA3Lys, in place of the HIV transcript (lane 2). In other experiments (data not shown), there was no shift of the labeled tRNA into a complex with the HaSV RNA, even when the latter was present at a fourfold-higher level than that of HIV-1 RNA used in these experiments.
FIG. 5.
Placement of tRNA3Lys on HIV-1 1-331 RNA by GagΔp6. (A) 32P-labeled tRNA3Lys was incubated with retroviral transcript and GagΔp6, and the reactions were analyzed as described in Materials and Methods. Lanes: 1, tRNA alone; 2, tRNA plus HaSV 34-378 plus 6 μg of GagΔp6; 3, tRNA plus HIV-1 1-331 plus 3 μg of GagΔp6; 4, tRNA plus HIV-1 1-331 plus 6 μg of GagΔp6; 5, tRNA plus HIV-1 1-331. (B) Unlabeled tRNA3Lys was annealed to the PBS on HIV-1 1-331 RNA by incubation as indicated. The mixtures were deproteinized as described in Materials and Methods and then tested for the presence of primer on the HIV-1 template RNA by addition of reverse transcriptase and dNTPs, including [α-32P]dCTP. The reactions were analyzed as described in Materials and Methods. Size of the labeled DNA product was determined ± 10 nt compared with labeled DNAs of known sizes (data not shown). Lanes: 1, tRNA plus 6 μg of GagΔp6; 2, 6 μg of GagΔp6 plus HIV-1 1-331; 3, tRNA plus HIV-1 1-331; 4, tRNA plus 6 μg of GagΔp6 plus HIV-1 1-331.
As an additional test of the proper placement of the tRNA on the PBS in these experiments, we annealed unlabeled tRNA to HIV-1 transcripts as in Fig. 5A and then extended it by adding reverse transcriptase and dNTPs, including [α-32P]dCTP, to the reaction mixtures. The products were then digested with RNase and analyzed by polyacrylamide gel electrophoresis and autoradiography. As shown in Fig. 5B (lane 4), a major DNA product was seen at ∼182 nucleotides in the reaction mixture containing HIV-1 RNA, tRNA3Lys, and GagΔp6. This product corresponds to minus-strand strong stop DNA. (Higher-molecular-weight products were also observed in this lane; these products presumably arise by self-priming of DNA synthesis from the minus-strand strong stop DNA [20, 26]). These three reactants are all required for the synthesis of significant amounts of DNA under these conditions, since very little product was observed if any of them was omitted (lanes 1 to 3). The formation of a DNA product of the correct size is strong evidence that GagΔp6 faithfully anneals the natural primer molecule to the PBS under our in vitro conditions.
DISCUSSION
The basic conclusion which can be drawn from the results presented here is that the HIV-1 Gag polyprotein, like its cleavage product NC, exhibits nucleic acid chaperone activity. That is, it is capable of catalyzing the rearrangement of nucleic acid molecules into the conformation with the maximum number of base pairs. As discussed below, these results support the suggestion that in vivo, the Gag polyprotein catalyzes the dimerization of the genomic RNA of the virus and the placement of the primer tRNA on the PBS, probably during assembly of the virion.
It is striking to note that the Gag polyprotein is evidently capable of exquisitely specific interaction with RNA, i.e., selection of the genomic RNA for packaging during virus assembly in vivo (reviewed in reference 5), and also of nonspecific interactions, as demonstrated here by the nucleic acid chaperone activity observed with nonviral RNA substrates (Fig. 1 and 2). Gag resembles NC in this respect, since NC also exhibits profound sequence preferences in binding to DNA or RNA (3–5, 15) but displays nucleic acid chaperone activity in a wide variety of assays with nonviral substrates (12, 21, 31). Remarkably, still another outcome of the interaction between Gag and nucleic acid molecules is the assembly of minute virus-like particles in vitro, which occurs with short oligodeoxynucleotides as well as larger RNA molecules and shows almost no sequence dependence (7). It is conceivable that these structures formed during some of the experiments described above.
In some of the nucleic acid chaperone assays presented here, it was possible to make a direct comparison between the dose-response curve of the Gag polyprotein and that of NC. It was found that the two proteins have very similar activities per mole in these assays (Fig. 1 and 3). Thus, the NC domain of the Gag polyprotein, which is near the C terminus of the polyprotein (and only 16 residues from the C terminus of GagΔp6, which was used in many of these experiments), probably functions as a nucleic acid chaperone independently of the remainder of the polyprotein.
What is the biological significance of the nucleic acid chaperone activity of Gag? Analysis of protease-deficient, immature virions, in which the Gag polyprotein is not cleaved, shows that they contain dimers of genomic RNA (16, 18, 32) and that the primer tRNA is annealed to the PBS on this RNA (11, 17, 23, 32). Thus, cleavage of the polyprotein, which releases NC protein, is not required for these RNA-RNA interaction events in vivo. We have also demonstrated (17) that in murine leukemia virus, expression of the pol gene product is unnecessary for these interactions. Thus the Gag polyprotein is the only internal viral protein which might be required to facilitate these events. In light of the present results (Fig. 3 and 5), showing that Gag can catalyze them in vitro, we consider it likely that it performs this function in vivo. (It is also possible that RNA dimerization occurs without the aid of a cofactor in vivo, as has been shown with transcripts of retroviral RNAs in vitro [12, 29]).
There is no direct information as to whether RNA dimerization and tRNA annealing precede or occur simultaneously with virus assembly, and one could imagine that the cytoplasm contains genomic RNA molecules which are dimeric and/or contain primer tRNA annealed to the PBS as a result of interactions with Gag. However, several observations seem to argue against this possibility. First, an early study suggested that tRNA annealing was incomplete in newly released particles of avian leukosis virus (9), and secondly, a recent report indicated that the placement was also incomplete in immature (PR-deficient) HIV-1 particles (28). The fact that the annealing is not complete at the moment of virus release suggests that it occurs simultaneously with virus assembly; perhaps under normal circumstances it is catalyzed in part by the Gag polyprotein and in part by NC. It is also striking that that the HIV-1 Gag polyprotein can induce the stabilization of a dimer of retroviral RNA in vitro (Fig. 4). This finding was somewhat unexpected, since stabilization of dimeric RNAs within the retroviral particle in vivo depends upon the release of NC from the polyprotein by the viral PR (16, 18). Thus, the polyprotein is evidently capable of inducing stabilization in solution but not within the virion. One possible explanation for this discrepancy is that the polyprotein does not interact with viral RNAs within the cytoplasm, but only during the formation of the virion, and that the structure of the nascent virus particle somehow restricts the ability of the protein to interact with the RNA. Indeed, contact between Gag and RNA in the immature particle is probably very limited: a careful analysis of avian leukosis virus particles (in which the maturation of dimeric RNA was first observed [33]) showed that UV-induced cross-linking of Gag to RNA in immature particles is extremely inefficient compared to the cross-linking of NC to RNA in mature particles (32).
In summary, the initial assembly of a retrovirus particle involves at least two RNA-RNA interaction events: dimerization of genomic RNA and annealing of a cellular tRNA to a specific site on the genomic RNA. The present results strongly suggest that the Gag polyprotein (the major structural protein of the immature retrovirus particle) is responsible not only for formation of the particle but also for catalyzing these associations between RNA molecules. This catalysis is presumably carried out by the NC domain of the polyprotein, but the detailed molecular mechanism of the catalytic reaction is not yet understood.
ACKNOWLEDGMENTS
We thank Guillaume Bec and Gerard Keith for the gift of purified bovine tRNA3Lys, Robert Gorelick and Louis Henderson for the gift of HIV-1 NC protein, Jianhui Guo and Judith Levin for the pJD plasmid, and Judith Levin for help with the experiments and for a thoughtful reading of the manuscript.
Research was supported in part by the National Cancer Institute, DHHS, under contract with ABL, and also by grants from the French Agence Nationale de Recherches sur le SIDA and the CNRS.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar A, Cobrinik D, Ge Z, Kung H J, Leis J. Interaction between retroviral U5 RNA and the T psi C loop of the tRNA(Trp) primer is required for efficient initiation of reverse transcription. J Virol. 1992;66:2464–2472. doi: 10.1128/jvi.66.4.2464-2472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen P, Collins B, Brown D, Hostomsky Z, Gold L. A specific RNA structural motif mediates high affinity binding by the HIV-1 nucleocapsid protein (NCp7) Virology. 1996;225:306–315. doi: 10.1006/viro.1996.0605. [DOI] [PubMed] [Google Scholar]
- 4.Berglund J A, Charpentier B, Rosbash M. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 1997;25:1042–1049. doi: 10.1093/nar/25.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 6.Busch L K. Production of HIV-1 (MN) nucleocapsid protein (p7) by recombinant DNA technology. M.S. thesis. Frederick, Md: Hood College; 1994. [Google Scholar]
- 7.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canaani E, Helm K V, Duesberg P. Evidence for 30-40S RNA as precursor of the 60-70S RNA of Rous sarcoma virus. Proc Natl Acad Sci USA. 1973;70:401–405. doi: 10.1073/pnas.70.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 11.Crawford S, Goff S P. A deletion mutation in the 5′ part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J Virol. 1985;53:899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y X, Fu W, Winter A J, Levin J G, Rein A. Multiple regions of Harvey sarcoma virus RNA can dimerize in vitro. J Virol. 1995;69:2486–2490. doi: 10.1128/jvi.69.4.2486-2490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher R J, Rein A, Fivash M, Urbaneja M A, Casas-Finet J R, Medaglia M, Henderson L E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu W, Ortiz-Conde B A, Gorelick R J, Hughes S H, Rein A. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J Virol. 1997;71:6940–6946. doi: 10.1128/jvi.71.9.6940-6946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross H J, Krupp G, Domdey H, Raba M, Jank P, Lossow C, Alberty H, Ramm K, Sanger H L. Nucleotide sequence and secondary structure of citrus exocortis and chrysanthemum stunt viroid. Eur J Biochem. 1982;121:249–257. doi: 10.1111/j.1432-1033.1982.tb05779.x. [DOI] [PubMed] [Google Scholar]
- 20.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Khorchid A, Gabor J, Wang J, Li X, Darlix J L, Wainberg M A, Kleiman L. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA(3Lys) genomic placement and initiation of reverse transcription in human immunodeficiency virus type 1. J Virol. 1998;72:3907–3915. doi: 10.1128/jvi.72.5.3907-3915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Wang J, Shalom A, Li Z, Khorchid A, Wainberg M A, Kleiman L. Primer tRNA3Lys on the viral genome exists in unextended and two-base extended forms within mature human immunodeficiency virus type 1. J Virol. 1997;71:726–728. doi: 10.1128/jvi.71.1.726-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer) J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 25.Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J Biol Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- 25a.Keith, G. Personal communication.
- 26.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J L. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 27.Liang C, Li X, Rong L, Inouye P, Quan Y, Kleiman L, Wainberg M A. The importance of the A-rich loop in human immunodeficiency virus type 1 reverse transcription and infectivity. J Virol. 1997;71:5750–5757. doi: 10.1128/jvi.71.8.5750-5757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang C, Rong L, Morin N, Cherry E, Huang Y, Kleiman L, Wainberg M A. The roles of the human immunodeficiency virus type 1 Pol protein and the primer binding site in the placement of primer tRNA(3Lys) onto viral genomic RNA. J Virol. 1997;71:9075–9086. doi: 10.1128/jvi.71.12.9075-9086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prats A-C, Roy C, Wang P, Erard M, Housset V, Gabus C, Paoletti C, Darlix J-L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prats A C, Sarih L, Gabus C, Litvak S, Keith G, Darlix J L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rein A, Henderson L E, Levin J G. Nucleic acid chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 32.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoltzfus C M, Snyder P N. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J Virol. 1975;16:1161–1170. doi: 10.1128/jvi.16.5.1161-1170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]