Abstract
Background
We evaluated the associations between baseline influenza virus–specific hemagglutination inhibition (HAI) and microneutralization (MN) titers and subsequent symptomatic influenza virus infection in a controlled human infection study.
Methods
We inoculated unvaccinated healthy adults aged 18–49 years with an influenza A/California/04/2009/H1N1pdm-like virus (NCT04044352). We collected serial safety labs, serum for HAI and MN, and nasopharyngeal swabs for reverse-transcription polymerase chain reaction (RT-PCR) testing. Analyses used the putative seroprotective titer of ≥40 for HAI and MN. The primary clinical outcome was mild-to-moderate influenza disease (MMID), defined as ≥1 postchallenge positive qualitative RT-PCR test with a qualifying symptom/clinical finding.
Results
Of 76 participants given influenza virus challenge, 54 (71.1%) experienced MMID. Clinical illness was generally very mild. MMID attack rates among participants with baseline titers ≥40 by HAI and MN were 64.9% and 67.9%, respectively, while MMID attack rates among participants with baseline titers <40 by HAI and MN were 76.9% and 78.3%, respectively. The estimated odds of developing MMID decreased by 19% (odds ratio, 0.81 [95% confidence interval, .62–1.06]; P = .126) for every 2-fold increase in baseline HAI. There were no significant adverse events.
Conclusions
We achieved a 71.1% attack rate of MMID. High baseline HAI and MN were associated with protection from illness.
Clinical Trials Registration. NCT04044352.
Keywords: H1N1 subtype, clinical trial, human, influenza, influenza A virus
In a multicenter controlled human infection model study, we inoculated 76 participants with influenza A/California/04/2009/H1N1pdm-like virus and safely achieved a 71.1% attack rate of mild-to-moderate influenza disease. High virus-specific hemagglutination inhibition and microneutralization titers were associated with protection from illness.
Controlled human infection models (CHIMs) of influenza virus have been used since the 1930s to advance understanding of infection natural history, clinical characteristics, and immune responses [1–16]. Such studies were critical for advancing the development of influenza antivirals and have helped establish a relative correlate of protection, hemagglutination inhibition (HAI) antibody titer ≥40, used as the regulatory standard for influenza vaccines [17–19]. CHIMs can efficiently evaluate vaccine efficacy, avoiding the high costs of field trials and the uncertainty of community strain circulation and attack rates [20]. While there is a precedent for vaccines to achieve licensure based on a CHIM study [21], it is more likely that they would be used in midstage clinical development of influenza vaccines by informing the best vaccine candidates to advance into pivotal trials conducted to obtain regulatory approval [22].
In 2018, the National Institute of Allergy and Infectious Diseases (NIAID) launched its Strategic Plan for a Universal Influenza Vaccine [22]. This unprecedented public investment in influenza vaccine research and development created a consortium of scientists working in a coordinated effort to develop a universal influenza vaccine. Acknowledging that vaccine field trials are a major bottleneck in the clinical development pathway for new vaccines, NIAID made CHIMs central to its strategic plan for the development of universal influenza vaccines and committed to expanding capacity for conducting them in the United States. In 2019, NIAID selected 6 vaccine and treatment evaluation unit (VTEU) sites to design and execute an influenza virus CHIM trial. The goal of this report is to present all trial primary and secondary outcomes as defined by the study protocol.
METHODS
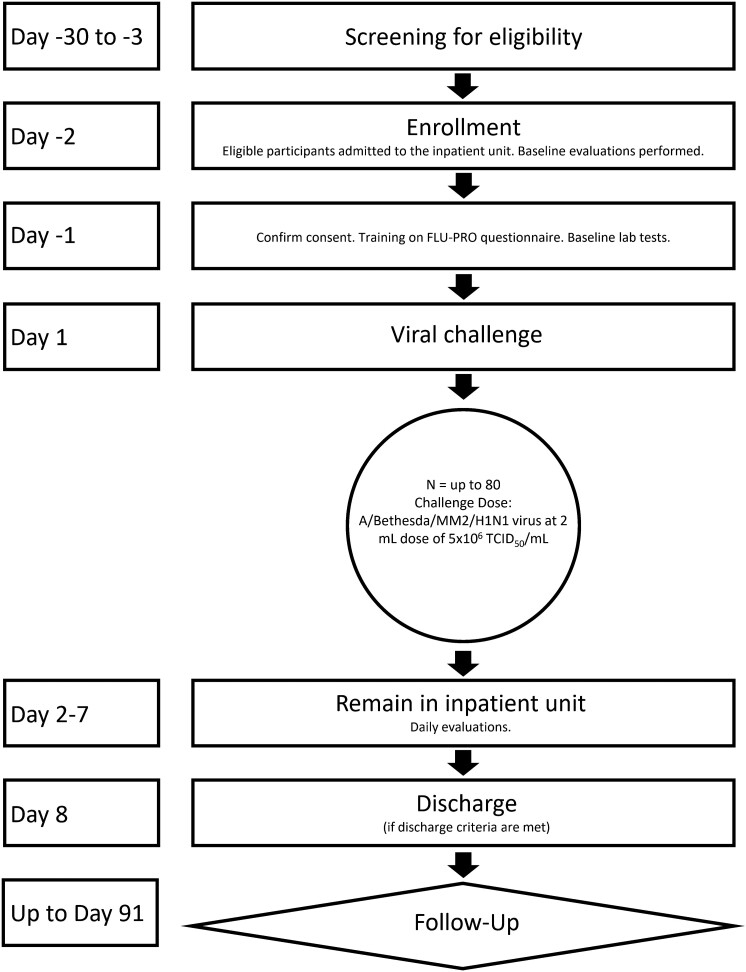
We conducted a CHIM study with influenza A/Bethesda/MM2/H1N1, an A/California/04/2009/H1N1pdm-like virus, at 4 sites among healthy adults aged 18–49 years. The study was designed to assess clinical response, immunological response, and safety of the (H1N1)pdm09 viral challenge. A schematic of the study design is in Figure 1. Expanded study procedures are provided in the Supplementary Material. Participants were screened for good health beginning 1 month prior to viral challenge. Influenza vaccination within the 6 months prior to screening was exclusionary. Two days prior to viral challenge, we admitted participants into an inpatient research unit and collected baseline specimens. We assessed influenza and other respiratory viruses from nasopharyngeal (NP) swabs collected upon admission to the inpatient facility and then daily postchallenge.
Figure 1.
Schematic of study design. Abbreviation: TCID50, median tissue culture infective dose.
The challenge virus was antigenically similar to the 2009 influenza A pandemic virus that emerged in 2009 [23], circulated at the time of the study [24], and continues to circulate globally [24]. The challenge virus was produced under Good Manufacturing Practices (GMP) conditions from a virus seed stock derived by reverse genetics [25]. The 2-mL inoculum of virus contained a 1 × 107 median tissue culture infective dose (TCID50). On study day 1, we delivered 1 mL of virus product in each nostril to semi-recumbent participants using an intranasal mucosal atomization device. Participants were subsequently monitored in the inpatient facility for at least 7 days with regular self-reported symptom assessments, clinical evaluations, safety assessments, and specimen collections. Beginning on study day 8, participants could leave the inpatient unit if they met standardized discharge criteria, which included negative influenza tests. All participants with positive influenza tests on study day 8 were offered 1 dose of baloxavir marboxil. Additional follow-up for clinical evaluation, safety assessment, and clinical specimen collections continued for 3 months postchallenge.
Participants had standardized clinical evaluations daily while in the inpatient unit and during follow-up appointments. Safety laboratory tests were conducted on screening and on study days 2, 4, and 8. Participants reported their symptoms twice daily during their inpatient stay using FLU-PRO, a standardized and validated symptom diary [26–28]. The FLU-PRO total score is computed as a mean score across 32 items and ranges from 0 (symptom free) to 4 (very severe symptoms).
The primary objective was to evaluate the association of baseline HAI antibody titers with the development of mild-to-moderate influenza disease (MMID) postchallenge (Supplementary Table 1). Secondary and exploratory objectives included evaluating the association of baseline microneutralization (MN) or neuraminidase inhibition (NAI) antibody titers with the development of MMID postchallenge, determining the frequency of serious adverse events (SAEs), assessing potential alternative clinical case definitions, and evaluating associations between asymptomatic influenza virus infection or symptomatic influenza-negative status and baseline HAI, MN, and NAI antibody titers (Supplementary Table 1).
MMID required 2 components: (1) evidence of influenza virus by qualitative reverse-transcription polymerase chain reaction (RT-PCR) assay from an NP swab; and (2) any 1 or more of arthralgia, chest tightness, chills, conjunctivitis, coryza, decreased appetite, diarrhea, dry cough, dyspnea/shortness of breath, fatigue/tiredness, fever (>38.0°C), headache, lymphopenia (<1000 cells/mL), myalgia, nasal congestion, nausea, oxygen saturation decrease by ≥3% from baseline, productive cough, rhinorrhea, sinus congestion, sore throat, and sweats [25]. For a secondary analysis, a more stringent outcome measure, MMID-2, required 2 or more positive qualitative RT-PCR tests of NP swabs.
Participant sera were tested by HAI and MN assays against the challenge virus strain and by NAI assay against a reassortant H6N1 virus [29, 30]. Seroprotection for HAI and MN were defined as a titer ≥40, reflecting a putative cut point commonly used to define susceptible versus seroprotective titers [31]. Seroconversion for each assay was defined by a minimum 4-fold rise in titer postchallenge. NP swabs were tested by quantitative RT-PCR to measure peak and total viral load.
Study procedures were in accordance with the ethical standards of the Helsinki Declaration. The protocol and informed consent forms were approved by the institutional review boards at the participating VTEUs. Study participants provided informed consent. The study is registered at ClinicalTrials.gov (NCT04044352).
RESULTS
Participant Demographics and Baseline Immunologic Results
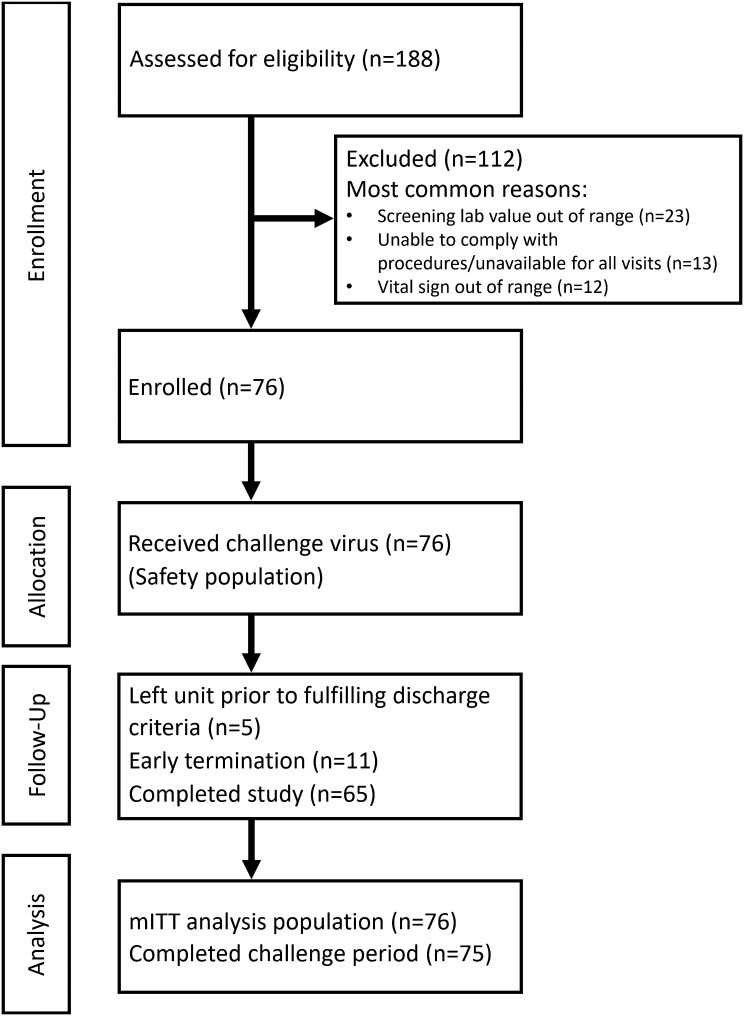
From 8 October 2019 through 8 December 2019, we screened 188 people; 76 received influenza virus challenge (Figure 2). Demographics and baseline influenza antibody titers varied across the 4 study sites (Table 1 and Supplementary Table 2). The mean age was 33.4 years (range, 18–49 years), 46 (61%) were male, 5 (7%) were Hispanic, and 35 (46%) were Black or African American. Sixty-seven (88%) participants reported not receiving the seasonal influenza vaccine during the previous year (2018–2019). Among participants receiving the influenza virus challenge, 75 (99%) completed the inpatient phase and 65 (86%) completed all study visits. The geometric mean titers (GMTs) prior to challenge for HAI, MN, and NAI were 42.8 (95% confidence interval [CI], 5.0–718.4), 79.7 (95% CI, 5.0–1280.0), and 651.8 (95% CI, 51.0–3412.0), respectively (Table 2). Thirty-nine (51.3%) participants had baseline HAI titers <40 and 23 (30.3%) had baseline MN titers <40.
Figure 2.
Consort flow diagram. Abbreviation: mITT, modified intent-to-treat.
Table 1.
Participant Demographics, Baseline Antibody Status, and Postchallenge Outcomes
| Characteristic | Baltimore, Maryland (n = 20) | Cincinnati, Ohio (n = 24) | Durham, North Carolina (n = 15) | St Louis, Missouri (n = 17) | All Participants (N = 76) |
|---|---|---|---|---|---|
| Age, y, mean (SD) | 35.0 (7.9) | 32.3 (8.9) | 34.9 (11.8) | 31.7 (17) | 33.4 (9.2) |
| Sex | |||||
| Male | 14 (70.0) | 17 (70.8) | 6 (40.0) | 9 (52.9) | 46 (60.5) |
| Female | 6 (30.0) | 7 (29.2) | 9 (60.0) | 8 (47.1) | 30 (39.5) |
| Ethnicity | |||||
| Not Hispanic/Latino | 17 (85.0) | 24 (100) | 14 (93.3) | 16 (94.1) | 71 (93.4) |
| Hispanic/Latino | 3 (15.0) | 0 (0.0%) | 1 (6.7) | 1 (5.9) | 5 (6.6) |
| Race | |||||
| Black/African American | 14 (70.0) | 11 (45.8) | 8 (53.3) | 2 (11.8) | 35 (46.1) |
| White | 3 (15.0) | 13 (54.2) | 5 (33.3) | 13 (76.5) | 34 (44.7) |
| Other | 3 (15.0) | 0 (0.0) | 2 (13.3) | 2 (11.8) | 7 (9.2) |
| BMI, mean (SD) | 29.3 (4.1) | 27.2 (4.1) | 28.8 (3.8) | 25.3 (4.1) | 27.6 (4.3) |
| Prior seasonal influenza vaccination (2018–2019) | |||||
| No | 19 (95.0) | 23 (95.8) | 13 (86.7) | 12 (70.6) | 67 (88.2) |
| Yes | 1 (5.0) | 1 (4.2) | 2 (13.3) | 5 (29.4) | 9 (11.8) |
| Baseline hemagglutination inhibition titer ≥40a | |||||
| No | 10 (50.0) | 13 (54.2) | 11 (73.3) | 5 (29.4) | 39 (51.3) |
| Yes | 10 (50.0) | 11 (44.8) | 4 (26.7) | 12 (70.6) | 37 (48.7) |
| Baseline microneutralization titer ≥40a | |||||
| No | 8 (40.0) | 9 (37.5) | 2 (13.3) | 4 (23.5) | 23 (30.3) |
| Yes | 12 (60.0) | 15 (62.5) | 13 (86.7) | 13 (76.5) | 53 (69.7) |
| Clinical outcomes | |||||
| MMIDb | 14 (70.0) | 20 (83.3) | 10 (66.7) | 10 (58.8) | 54 (71.1) |
| MMID-2c | 9 (45.0) | 13 (54.2) | 8 (53.3) | 8 (47.1) | 38 (50.0) |
| Asymptomatic with at least 1 influenza detection | 3 (15.0) | 1 (4.2) | 4 (26.7) | 0 (0.0) | 8 (10.5) |
| No influenza detections | 3 (15.0) | 3 (12.5) | 1 (6.7) | 7 (41.2) | 14 (18.4) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: BMI, body mass index; MMID, mild-to-moderate influenza disease; SD, standard deviation.
We defined seroprotection for hemagglutination inhibition and microneutralization titers by a titer ≥40, reflecting a putative cut point commonly used to define susceptible versus seroprotective titers. We did not define a seroprotective titer for neuraminidase inhibition.
Including at least 1 influenza A detection (primary clinical endpoint).
Including at least 2 influenza A detections (secondary clinical endpoint).
Table 2.
Baseline Hemagglutination Inhibition, Microneutralization, and Neuraminidase Inhibition Antibodies and Postchallenge Outcomes
| Antibody | RT-PCR Negative (None Positive) | RT-PCR Positive (1 or More) |
RT-PCR Positive (2 or More) |
MMIDa | MMID-2b | RT-PCR Positive Asymptomatic |
|---|---|---|---|---|---|---|
| (n = 14) | (n = 62) | (n = 43) | (n = 54) | (n = 38) | (n = 8) | |
| Hemagglutination inhibition | ||||||
| GMT (95% CI) | 117.9 (61.3–227.0) | 34.0 (24.7–47.0) | 26.3 (18.0–38.6) | 36.8 (26.1–52.0) | 27.6 (18.4–41.4) | 20.0 (6.9–58.0) |
| % with titer ≥40 (95% CI)c | 78.6 (49.2–95.3) | 41.9 (29.5–55.2) | 32.6 (19.1–48.5) | 44.4 (30.9–58.6) | 34.2 (19.6–51.4) | 25 (3.2–65.1) |
| Microneutralization | ||||||
| GMT (95% CI) | 276.7 (124.8–613.1) | 60.1 (42.3–85.4) | 45.2 (30.0–68.2) | 62.9 (43.1–91.9) | 45.1 (29.3–69.5) | 44.3 (13.8–142.6) |
| % with titer ≥40 (95% CI)c | 92.9 (66.1–99.8) | 64.5 (51.3–76.3) | 60.5 (44.4–75.0) | 66.7 (52.5–78.9) | 60.5 (43.4–76.0) | 50 (15.7–84.3) |
| Neuraminidase inhibition | ||||||
| GMT (95% CI) | 844.2 (508.1–1402.7) | 614.8 (458.4–824.4) | 565.4 (396.0–807.2) | 584.6 (424.7–804.7) | 506.1 (345.4–741.6) | 863.3 (358.3–2080.3) |
Numbers of participants in the modified intent-to-treat population with available results are shown in the column heading.
Abbreviations: CI, confidence interval; GMT, geometric mean titer; MMID, mild-to-moderate influenza disease; RT-PCR, qualitative reverse-transcription polymerase chain reaction.
Including at least 1 influenza A detection (primary clinical endpoint).
Including at least 2 influenza A detections (secondary clinical endpoint).
We defined seroprotection for hemagglutination inhibition and microneutralization titers by a titer ≥40, reflecting a putative cut point commonly used to define susceptible versus seroprotective titers. We did not define a seroprotective titer for neuraminidase inhibition.
Influenza Virus Detection
Among participants receiving the influenza virus challenge, 62 (81.6%) had at least 1 influenza A virus detection and 43 (56.6%) had at least 2 influenza A detections through study day 8 (Table 2). The frequency of influenza A detections peaked on study day 2 and decreased rapidly (Supplementary Figure 1). Sixty (78.9%) participants had an initial influenza A detection on study day 2, and 2 (2.6%) had an initial influenza A detection on study day 3. By study day 8, 6 (7.9%) participants still had an influenza A detection and were treated with baloxavir. One participant (1.3%) had influenza A detections through study day 13, and 1 (1.3%) left the challenge unit prior to study day 8 after several negative influenza A tests.
Baseline HAI and MN were negatively associated with influenza A detection postchallenge (Table 2). Participants without any influenza A detection (n = 14) had baseline HAI GMT of 117.9 (95% CI, 61.3–227.0), MN GMT of 276.7 (95% CI, 124.8–613.1), and NAI GMT of 844.2 (95% CI, 508.1–1402.7). Participants with an influenza A detection (n = 62) had baseline HAI GMT of 34.0 (95% CI, 24.7–47.0), MN GMT of 60.1 (95% CI, 42.3–85.4), and NAI GMT of 614.8 (95% CI, 458.4–824.4). The proportions of participants with baseline titers ≥40 by HAI and MN among those with any influenza A detection were 41.9% (95% CI, 29.5%–55.2%) and 64.5% (95% CI, 51.3%–76.3%), respectively, while the proportions of participants with baseline titers ≥40 by HAI and MN among those without influenza A detections was 78.6% (95% CI, 49.2%–95.3%) and 92.9% (95% CI, 66.1%–99.8%). Using the more stringent definition for viral shedding of ≥2 days showed an even greater difference between these groups (Table 2).
The mean duration of viral shedding by qualitative RT-PCR for the 61 participants with at least 1 influenza A detection who were followed for the entire challenge period was 3.1 days (95% CI, 2.5–3.8 days). The duration tended to be higher in participants with baseline HAI titer <40 (n = 35), with a mean of 3.8 days (95% CI, 2.9–4.7 days), compared to participants with baseline HAI titer ≥40 (n = 26), with a mean of 2.3 days (95% CI, 1.6–3.0 days), with overlapping 95% CIs. Participants with baseline MN titer <40 (n = 23) shed virus longer on average (mean, of 3.9 days [95% CI, 2.6–5.2 days]) than participants with baseline MN titer ≥40 (n = 39) (mean, 2.7 days [95% CI, 2.1–3.3 days]), also with overlapping 95% CIs. The frequency of influenza A detection by study day was consistently higher among those with baseline titer <40 for both HAI and MN.
Peak and total viral load by quantitative RT-PCR were both significantly lower in participants with baseline HAI titer ≥40 versus <40 (difference in mean peak viral load of 2.6 log10 copies/mL [95% CI, .8–4.4]; difference in mean area under the curve, 2.8 log10 copies/mL per day observed [95% CI, 1.0–4.6]). The differences between MN seroprotection groups were slightly smaller and did not meet statistical significance.
Influenza Illness
Fifty-four (71.1%) participants met the MMID case definition, while 38 (50.0%) met the MMID-2 case definition (Table 1). For the primary analysis of the association of baseline HAI titer and the subsequent development of MMID, univariable logistic regression estimated 19% decrease in the odds of MMID for every 2-fold increase in baseline HAI titer (ie, 1-unit increase in log2 titer) (OR, 0.81 [95% CI, .62–1.06]; P = .126) (Table 3). In a secondary analysis, there was a stronger association between baseline HAI titer and development of MMID-2—an estimated 32% decrease in the odds of MMID-2 for every 2-fold increase in baseline HAI titer (OR, 0.68 [95% CI, .52–.89]; P = .006). Univariable logistic regression estimated a 23% decrease in the odds of MMID for every fold increase in baseline MN titer (OR, 0.77 [95% CI, .60–.98]; P = .035) and an estimated 33% decrease in the odds of MMID-2 for every fold increase in baseline MN titer (OR, 0.67 [95% CI, .52–.86]; P = .002) (Table 3). There was a nonsignificant association between baseline NAI titer and development of MMID (OR, 0.80 [95% CI, .57–1.11]; P = .183), but there was evidence of an association between baseline NAI titer and development of MMID-2 (OR, 0.74 [95% CI, .55–1.00]; P = .050) (Table 3).
Table 3.
Univariable Logistic Regression Modelsa Evaluating the Relationship of Baseline Antibodies With Mild-to-Moderate Influenza Disease Including Influenza A Detection
| Antibody | Model Parameterb | Parameter Estimate | Standard Error | P Value | Odds Ratio | (95% CI) |
|---|---|---|---|---|---|---|
| MMIDc | ||||||
| Hemagglutination inhibitiond | Intercept | 2.06 | 0.82 | .012 | … | … |
| Baseline log2 titer | −0.21 | 0.14 | .126 | 0.81 | (.62–1.06) | |
| Microneutralization | Intercept | 2.65 | 0.90 | .003 | … | … |
| Baseline log2 titer | −0.27 | 0.13 | .035 | 0.77 | (.60–.98) | |
| Neuraminidase inhibition | Intercept | 3.02 | 1.64 | .065 | … | … |
| Baseline log2 titer | −0.22 | 0.17 | .183 | 0.80 | (.57–1.11) | |
| MMID-2e | ||||||
| Hemagglutination inhibition | Intercept | 2.05 | 0.78 | .008 | … | … |
| Baseline log2 titer | −0.38 | 0.14 | .006 | 0.68 | (.52–.89) | |
| Microneutralization | Intercept | 2.56 | 0.85 | .003 | … | … |
| Baseline log2 titer | −0.41 | 0.13 | .002 | 0.67 | (.52–.86) | |
| Neuraminidase inhibition | Intercept | 2.80 | 1.45 | .054 | … | … |
| Baseline log2 titer | −0.30 | 0.15 | .050 | 0.74 | (.55–1.00) | |
Abbreviations: CI, confidence interval; MMID, mild-to-moderate influenza disease.
Seventy-six subjects in the modified intent-to-treat population are included in the models.
Baseline log2 titer calculated based on subject-specific geometric mean titer values for each assay, then log2 transformed.
Including at least 1 influenza A detection (primary clinical endpoint).
Primary study analysis.
Including at least 2 influenza A detections (secondary clinical endpoint).
Based on a univariable logistic regression, each 2-fold increase in baseline MN titer was associated with a 42% (OR, 0.58 [95% CI, .41–.82]) decrease in the odds of becoming RT-PCR positive and symptomatic (ie, meeting MMID criteria) compared to remaining RT-PCR negative, and a 49% (OR, 0.51 [95% CI, .31–.83]) decrease in odds of being RT-PCR positive and asymptomatic compared to remaining RT-PCR negative.
Predicted probabilities of MMID according to baseline HAI, MN, or NAI from univariable models are in Supplementary Figures 2–4. Kaplan-Meier curves with cumulative infection probabilities by HAI and MN baseline titers ≥40 are in Supplementary Figures 5 and 6.
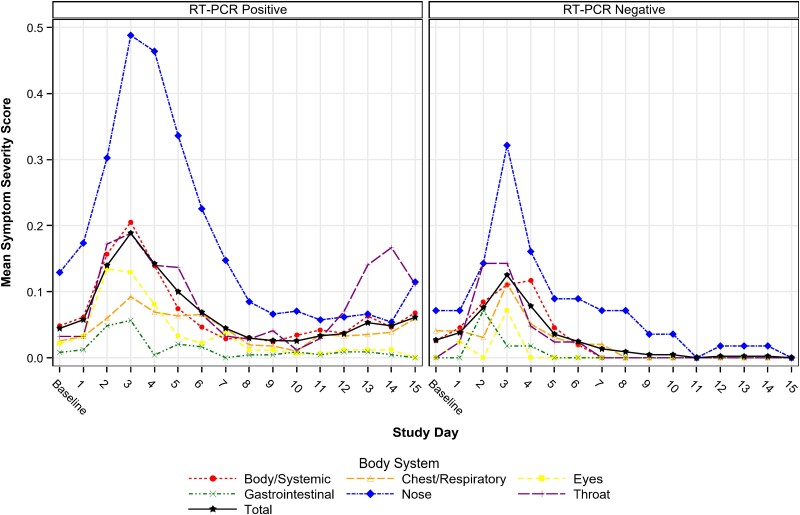
Symptom scores were low overall, with study day 3 mean scores of 0.25 (95% CI, .16–.36) among participants with at least 1 influenza A virus detection and 0.10 (95% CI, .06–.15) among participants without influenza A virus detection. The most common symptoms were nasal congestion/rhinorrhea, sore throat, and sinus congestion. Only 3 (4.8%) had a postchallenge fever (>38.0°C). Symptom scores by body system/domain peaked on study day 3, with generally higher scores observed among persons with any influenza A detection compared to those without influenza A detection (Figure 3), with statistically significant differences on study days 3, 5, and 6.
Figure 3.
Mean symptom score by body system, study day, and viral shedding status. Abbreviation: RT-PCR, qualitative reverse-transcription polymerase chain reaction.
Participants with higher baseline antibody titers had qualitatively fewer MMID cases when infected than participants with lower titers (Table 2). MMID cases (n = 54) had baseline HAI GMT of 36.8 (95% CI, 26.1–52.0), MN GMT of 62.9 (95% CI, 43.1–91.9), and NAI GMT of 584.6 (95% CI, 424.7–804.7). Asymptomatic participants with influenza A detections (n = 8) had baseline HAI GMT of 20.0 (95% CI, 6.9–58.0), MN GMT of 44.3 (95% CI, 13.8–142.6), and NAI GMT of 863.3 (95% CI, 358.3–2080.3). Baseline seroprotection by HAI and MN among MMID cases was 44.4% (95% CI, 30.9%–58.6%) and 66.7% (95% CI, 52.5%–78.9%). Baseline seroprotection by HAI and MN among asymptomatic participants with influenza A detection was 25.0% (95% CI, 3.2%–65.1%) and 44.3% (95% CI, 13.8%–142.6%).
Immune Response to Challenge
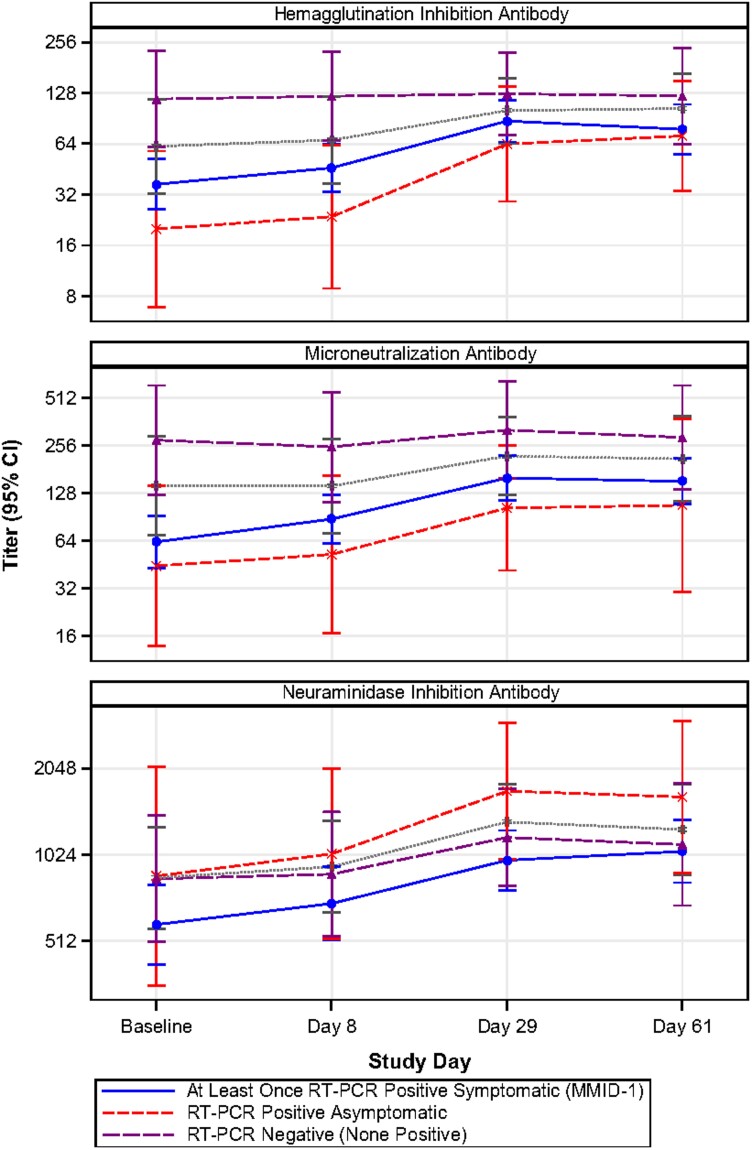
GMFR from baseline and seroconversion proportions over time (baseline and days 8, 29, and 61 postchallenge) by postchallenge infection status are in Figure 4, Supplementary Tables 3–5, and Supplementary Figure 7. HAI and MN titers tended to peak at day 29 and level off through day 61 for participants with any influenza A detection, while participants without influenza A detection maintained similar levels to baseline. NAI titers also peaked at day 61 among participants with any influenza A detection. No participants without influenza A detection (n = 14) seroconverted for HAI, MN, or NAI. Among MMID cases (n = 54) and asymptomatic participants with at least 1 influenza A RT-PCR detection (n = 8), the estimated probability of seroconversion by HAI was similar at day 8 (3.8% [95% CI, .5%–13.0%] vs 0.0% [.0%–36.9%], respectively) and at day 29 (28.6% [3.7%–71.0%] vs 28.3% [16.0%–43.5%], respectively), but it was qualitatively higher in the small group of asymptomatic participants than MMID cases at day 61 (33.3% [95% CI, 4.3%–77.7%] vs 18.6% [95% CI, 8.4%–33.4%], respectively; not significant). The same pattern was observed in seroconversion for MN and NAI over time. For NAI, participants without influenza A detection and asymptomatic participants with at least 1 influenza A detection tended to have higher titers than MMID cases at each study timepoint, although this pattern was less distinct than for the other assays.
Figure 4.
Hemagglutination inhibition, microneutralization, and neuraminidase inhibition antibody geometric mean titers (with 95% confidence intervals), by infection status and study day. Mild-to-moderate influenza disease (MMID) was defined as 1 or more positive polymerase chain reaction (PCR) tests and meeting the symptom threshold at any point during the challenge period. The gray dotted line displays the trajectory of the combined qualitative reverse-transcription PCR (RT-PCR)–positive asymptomatic and RT-PCR–negative groups (ie, participants who did not meet the definition of MMID).
Safety
Of the 76 participants in the safety population, 46 (60.5%) experienced an unsolicited adverse event (AE) of any attribution, including 26 participants (34.2%) who experienced at least 1 challenge-related unsolicited AE (Supplementary Table 6). Twenty-four (31.6%) participants had challenge-related unsolicited AEs of mild severity, 2 (2.6%) participants had challenge-related unsolicited AEs of moderate severity (ear pain and pyrexia), and no participants had severe unsolicited AEs.
The most common unsolicited AE was lymphadenopathy, occurring in 10 (13.2%) participants; all were mild in severity. No noninfluenza respiratory viruses were detected postchallenge. There were no postchallenge electrocardiographic changes or cardiac abnormalities. There were no deaths or other SAEs, and no participants discontinued due to AEs. Laboratory abnormalities graded as AEs were all mild in severity (hematology: 13/76 [17.1%]; chemistry: 7/76 [9.2%]).
DISCUSSION
In this multicenter study, intranasal administration of 1 × 107 TCID50 influenza A (H1N1)pdm09 virus resulted in overall attack rates of 71%, 50%, and 10%, respectively of MMID, MMID-2, and asymptomatic influenza virus infection. Clinical illness postchallenge was mild. Participants were diverse, unvaccinated, and almost evenly divided by baseline HAI titers <40 and ≥40. We chose ≥40 as a cutoff, recognizing that this HAI titer has been associated with a 50%–70% reduction in influenza illness in prior studies. The HAI GMT at baseline was higher in those individuals who did not subsequently develop MMID than in those who did, although not all these differences were statistically significant. Likewise, higher baseline HAI was generally associated with a reduction in overall viral detection and mean duration of viral detection. Corresponding results for analysis of baseline MN titers may have been more discerning.
Previously, NIAID researchers validated the influenza A(H1N1)pdm09 CHIM on which this study was modeled [25]. That study achieved a 68% attack rate of MMID using 1 × 107 TCID50 dose [25]. Similarly, we found that baseline HAI titer was associated with influenza outcomes, though the statistical significance of these associations was variable. The NIAID CHIM study reported that baseline NAI titer was more strongly correlated with clinical protection postchallenge than HAI [25], while we observed a weaker association between baseline NAI titer and influenza outcomes. A key difference between the NIAID study and ours was that the former excluded persons with challenge virus–specific HAI titers >40 assessed at approximately 8 weeks prior to challenge. Some participants likely had subsequent, prechallenge influenza exposures, as HAI titers were higher by the time of inpatient admission [25]. Nevertheless, excluding participants with baseline HAI titers ≥40 likely limited the ability of a study to evaluate associations of baseline HAI titers and infection or clinical influenza outcomes. Other immune responses that correlate with HAI may also be similarly affected.
Symptom severity in our study was consistent with previous influenza CHIMs [26], including the NIAID study with the same virus [25]. On study day 3, we measured peak average symptom score of 0.25 among participants with influenza A detections and 0.10 among participants without influenza A detection. This is equivalent to participants with influenza A detection reporting 8 mild symptoms on the 32-item FLU-PRO instrument and participants without influenza A detection reporting 3 mild symptoms on the instrument.
Symptom scores in our study are lower than naturally acquired, medically attended influenza illness [3, 26, 32]. Among 221 patients with laboratory-confirmed influenza illness seeking care in ambulatory clinics, peak total symptom scores were 1.6 (equivalent to 51 points on the 32-item FLU-PRO scale), with more predominant chest/respiratory symptoms than we identified in the CHIM study [33]. The influenza virus infections that we observed in the CHIM study may be more consistent with asymptomatic or pauci-symptomatic influenza virus infections that are common in community settings [34, 35]. For example, in a South African community study with routine, prospective testing for influenza virus infection, approximately 60% of adults aged 19–44 years with influenza detections were asymptomatic [34].
There are several strengths to this study. In closely monitored environments using standardized procedures and a GMP challenge virus, our multicenter influenza virus CHIM study was conducted safely and yielded consistent, high attack rates of MMID. Our participant population was diverse and had nearly equal numbers with and without baseline HAI titers ≥40. We corroborated previous observations about the role of HAI in protection from MMID and in modifying viral shedding [25]. We also assessed an alternative clinical outcome definition of MMID-2, which was more stringent and more strongly associated baseline with HAI and MN.
There were also limitations to our approach. We included only healthy adult participants, as is standard for CHIM studies, limiting the generalizability of our findings to children, older adults, and persons with chronic medical conditions. Further limiting the generalizability of influenza viral CHIM studies is the nature of inoculation, which may not recapitulate how persons become infected in the community and may contribute to the mild illnesses observed. As noted previously, HAI is a relative, and not absolute, correlate of protection. While about 50% of our participants did not have baseline HAI titers ≥40, it is possible that most had been exposed previously to (H1N1)pdm09 and developed some protective immunity via cellular or mucosal mechanisms. Our use of highly sensitive RT-PCR laboratory diagnostics did not allow us to determine whether the assay was detecting viable virus or nonviable, residual RNA from the challenge inoculum. The more stringent MMID-2 outcome definition, in which we required 2 or more positive RT-PCR tests, may be more likely to identify participants with true infection. The inclusion of double-blind placebo inoculations could have decreased bias regarding symptom self-reports and clinical assessments. Finally, future influenza vaccine efficacy CHIM trials may seek to assess prevention of influenza illness of greater public health and clinical importance. For influenza vaccines that lessen the severity of illness or have greater efficacy against more severe rather than mild illness, the use of this current challenge model might underestimate the product efficacy.
This trial has advanced NIAID goals to expand the capacity for conducting influenza virus CHIMs. In addition to the results in this report, we collected substantial data and specimens for future exploratory analyses that may help to refine study procedures and better understand the immunology of influenza virus CHIMs. We will explore exploratory objectives related to cellular and mucosal immune response, transcriptomics, and alternative clinical case definitions in subsequent analyses. Future influenza CHIMs should include more up-to-date influenza viruses, include placebo inoculations to minimize bias in outcome assessment, explore alternative methods to deliver the challenge virus to more closely mimic natural acquisition, and evaluate alternative clinical outcome definitions. There is a robust pipeline of next-generation influenza vaccines and therapeutics in preclinical development [36, 37], and we anticipate that influenza virus CHIMs will help to move them forward to licensure.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Influenza Controlled Human Infection Study Group. Azra Blazevic, Christopher S. Eickhoff, Karla Mosby, and Janice Tennant (Division of Infectious Diseases, Allergy and Immunology and Center for Vaccine Development, Department of Internal Medicine, Saint Louis University School of Medicine, Missouri); Kristen Buschle and Susan Parker (Cincinnati Children’s Hospital Medical Center, Ohio); Wilbur Chen, Lisa Chrisley, Marcelo B. Sztein, and Franklin R. Toapanta (Center for Vaccine Development and Global Health, University of Maryland School of Medicine, Baltimore); Michelle Dickey (Cincinnati Children’s Hospital Medical Center, University of Cincinnati, Ohio); Hana M. El Sahly (Department of Molecular Virology and Microbiology and Department of Medicine, Baylor College of Medicine, Houston, Texas); Esther Fevrier, Ge Li, and Rachel Tsong (The Emmes Company, Rockville, Maryland); Sarah George (Department of Internal Medicine and Department of Molecular Microbiology and Immunology, Division of Infectious Diseases, Allergy and Immunology, Saint Louis University School of Medicine and Saint Louis Veterans Affairs Medical Center, Missouri); Lynn S. Harrington (Duke Vaccine and Trials Unit, Duke University Medical Center, Durham, North Carolina); Sonnie Kim, M. Chelsea Lane, Catherine Luke, Rhonda Pikaart-Tautges, and Diane J. Post (Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases [NIAID], Bethesda, Maryland); Karen Kotloff (Department of Pediatrics, Center for Vaccine Development and Global Health, University of Maryland School of Medicine, Baltimore); Bradly P. Nicholson (Institute for Medical Research, Durham Veterans Affairs Health Care System, North Carolina).
Acknowledgments. The authors thank the members of the data and safety monitoring board for their oversight. From NIAID, we thank Brooke Bozick, Sonja Crandon, Mohamed Elsafy, Binh Hoang, Tena Knudsen, Robin Mason, Kathy Ormanoski, Rhonda Pikaart-Tautges, Christine Oshansky-Weilnau, and Tammy Yokum. From University of Cincinnati, we thank Kristen Buschle, Margie Huron, Jesse LePage, and Monica McNeal. From University of Maryland, we thank Paula Bernal, Melissa Billington, Colleen Boyce, Anna Carmack, Nancy Greenberg, Panagiota Komninou, Hanna LeBuhn, and Jennifer Marron. From Duke University, we thank Jack Anderson, Luis Ballon, Thomas Burke, Kathlene Chmielewski, Min Gao, Thad Gurley, Maria Miggs, and Tim Veldman. From Emory University, we thank Evan Anderson, Colleen Kraft, G. Marshall Lyon, Michele McCullough, and Aneesh Mehta. From Baylor College of Medicine, we thank Connie Rangel. From Emmes, we thank Peter Dubyoski, Alexander Noll, David Santos, Dan Sinnett, Karineh Tarpinian, Julia Weiss, and Shu Yang.
Financial support. This project has been funded in whole or part with federal funds from NIAID, National Institutes of Health (NIH), under contract numbers HHSN272201300016I (to Cincinnati Children's Hospital Medical Center), 75N93021C00012 (The Emmes Company), HHSN272201300022I (University of Maryland, Baltimore), HHSN2722013000017I (Duke University), HHSN272201300018I (Emory University), HHSN272200800003C (Saint Louis University), and HHSN272201300015I (Baylor College of Medicine). Partial support of the Institute for Clinical and Translational Research at the University of Maryland, Baltimore, was provided by the National Center for Advancing Translational Sciences Clinical Translational Science Award (grant number 1UL1TR003098).
Supplementary Material
Contributor Information
Justin R Ortiz, Center for Vaccine Development and Global Health, University of Maryland School of Medicine, Baltimore.
David I Bernstein, Cincinnati Children’s Hospital Medical Center, University of Cincinnati, Ohio; Departments of.
Daniel F Hoft, Internal Medicine and; Molecular Microbiology and Immunology, Division of Infectious Diseases, Allergy and Immunology and Center for Vaccine Development, Saint Louis University School of Medicine, Missouri.
Christopher W Woods, Division of Infectious Diseases, Duke University Medical Center, Durham, North Carolina.
Micah T McClain, Division of Infectious Diseases, Duke University Medical Center, Durham, North Carolina.
Sharon E Frey, Internal Medicine and.
Rebecca C Brady, Cincinnati Children’s Hospital Medical Center, University of Cincinnati, Ohio; Departments of.
Christopher Bryant, Vaccine and Infectious Disease Therapeutic Research Unit, The Emmes Company, Rockville, Maryland.
Ashley Wegel, Vaccine and Infectious Disease Therapeutic Research Unit, The Emmes Company, Rockville, Maryland.
Robert W Frenck, Jr, Cincinnati Children’s Hospital Medical Center, University of Cincinnati, Ohio; Departments of.
Emmanuel B Walter, Duke Human Vaccine Institute, Duke University School of Medicine, Durham, North Carolina.
Getahun Abate, Internal Medicine and.
Sarah R Williams, Division of Pulmonary and Critical Care Medicine, University of Maryland School of Medicine, Baltimore.
Robert L Atmar, Section of Infectious Diseases, Department of Medicine, Baylor College of Medicine, Houston, Texas.
Wendy A Keitel, Departments of Molecular Virology & Microbiology and Medicine, Baylor College of Medicine, Houston, Texas.
Nadine Rouphael, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia.
Mathew J Memoli, Laboratory of Infectious Diseases.
Mamodikoe K Makhene, Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases, Bethesda, Maryland.
Paul C Roberts, Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases, Bethesda, Maryland.
Kathleen M Neuzil, Center for Vaccine Development and Global Health, University of Maryland School of Medicine, Baltimore.
References
- 1. Anderson MJ, Heath RB. Cell mediated immunity in experimental influenza and parainfluenza infection. Dev Biol Stand 1977; 39:379–83. [PubMed] [Google Scholar]
- 2. Brown TA, Murphy BR, Radl J, Haaijman JJ, Mestecky J. Subclass distribution and molecular form of immunoglobulin A hemagglutinin antibodies in sera and nasal secretions after experimental secondary infection with influenza A virus in humans. J Clin Microbiol 1985; 22:259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carrat F, Vergu E, Ferguson NM, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol 2008; 167:775–85. [DOI] [PubMed] [Google Scholar]
- 4. Clements ML, Snyder MH, Buckler-White AJ, Tierney EL, London WT, Murphy BR. Evaluation of avian-human reassortant influenza A/Washington/897/80 x A/Pintail/119/79 virus in monkeys and adult volunteers. J Clin Microbiol 1986; 24:47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cretescu L, Beare AS, Schild GC. Formation of antibody to matrix protein in experimental human influenza A virus infections. Infect Immun 1978; 22:322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dolin R, Richman DD, Murphy BR, Fauci AS. Cell-mediated immune responses in humans after induced infection with influenza A virus. J Infect Dis 1977; 135:714–9. [DOI] [PubMed] [Google Scholar]
- 7. Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, Straus SE. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J Clin Invest 1998; 101:643–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jameson J, Cruz J, Ennis FA. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J Virol 1998; 72:8682–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kasel JA, Alford RH, Knight V, Waddell GH, Sigel MM. Experimental infection of human volunteers with equine influenza virus. Nature 1965; 206:41–3. [DOI] [PubMed] [Google Scholar]
- 10. Kasel JA, Couch RB. Experimental infection in man and horses with influenza A viruses. Bull World Health Organ 1969; 41:447–52. [PMC free article] [PubMed] [Google Scholar]
- 11. Rudenko LG, Shadrin AS, Geiker VI, Zibina EA, Zykov MP. Associated seroconversions to respiratory viruses in volunteers with experimental influenza infection. Acta Virol 1976; 20:135–41. [PubMed] [Google Scholar]
- 12. Scheinberg M, Blacklow NR, Goldstein AL, Parrino TA, Rose FB, Cathcart ES. Influenza: response of T-cell lymphopenia to thymosin. N Engl J Med 1976; 294:1208–11. [DOI] [PubMed] [Google Scholar]
- 13. Snyder MH, Clements ML, Herrington D, London WT, Tierney EL, Murphy BR. Comparison by studies in squirrel monkeys, chimpanzees, and adult humans of avian-human influenza A virus reassortants derived from different avian influenza virus donors. J Clin Microbiol 1986; 24:467–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sherman AC, Mehta A, Dickert NW, Anderson EJ, Rouphael N. The future of flu: a review of the human challenge model and systems biology for advancement of influenza vaccinology. Front Cell Infect Microbiol 2019; 9:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smorodintseff A, Tushinsky M, Drobyshevskaya A, Korovin A, Osetroff A. Investigation on volunteers infected with the influenza virus. Am J Med Sci 1937; 194:159–70. [Google Scholar]
- 16. Keitel WA, Couch RB, Cate TR, Six HR, Baxter BD. Cold recombinant influenza B/Texas/1/84 vaccine virus (CRB 87): attenuation, immunogenicity, and efficacy against homotypic challenge. J Infect Dis 1990; 161:22–6. [DOI] [PubMed] [Google Scholar]
- 17. US Food and Drug Administration . https://www.fda.gov/regulatory-information/search-fda-guidance-documents/clinical-data-needed-support-licensure-seasonal- inactivated-influenza-vaccines.
- 18. Hobson D, Curry RL, Beare AS, Ward-Gardner A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg (Lond) 1972; 70:767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hayden FG. Experimental human influenza: observations from studies of influenza antivirals. Antivir Ther 2012; 17:133–41. [DOI] [PubMed] [Google Scholar]
- 20. Lambkin-Williams R, Noulin N, Mann A, Catchpole A, Gilbert AS. The human viral challenge model: accelerating the evaluation of respiratory antivirals, vaccines and novel diagnostics. Respir Res 2018; 19:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen WH, Cohen MB, Kirkpatrick BD, et al. Single-dose live oral cholera vaccine CVD 103-HgR protects against human experimental infection with Vibrio cholerae O1 El Tor. Clin Infect Dis 2016; 62:1329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Erbelding EJ, Post DJ, Stemmy EJ, et al. A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis 2018; 218:347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Centers for Disease Control and Prevention . Outbreak of swine-origin influenza A (H1N1) virus infection—Mexico, March–April 2009. MMWR Morb Mortal Wkly Rep 2009; 58:467–70. [PubMed] [Google Scholar]
- 24. Centers for Disease Control and Prevention . FluView interactive.https://www.cdc.gov/flu/weekly/fluviewinteractive.htm. Accessed 3 January 2023.
- 25. Memoli MJ, Czajkowski L, Reed S, et al. Validation of the wild-type influenza A human challenge model H1N1pdMIST: an A(H1N1)pdm09 dose-finding investigational new drug study. Clin Infect Dis 2015; 60:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han A, Poon JL, Powers JH 3rd, Leidy NK, Yu R, Memoli MJ. Using the Influenza Patient-Reported Outcome (FLU-PRO) diary to evaluate symptoms of influenza viral infection in a healthy human challenge model. BMC Infect Dis 2018; 18:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Powers JH, Guerrero ML, Leidy NK, et al. Development of the Flu-PRO: a patient-reported outcome (PRO) instrument to evaluate symptoms of influenza. BMC Infect Dis 2016; 16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Powers JH, Bacci ED, Leidy NK, et al. Evaluation of the performance properties of the influenza patient-reported outcomes instrument (Flu-Pro). Value Health 2016; 19:A220–1. [Google Scholar]
- 29. Wan H, Gao J, Xu K, et al. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol 2013; 87:9290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Potter CW, Oxford JS. Determinants of immunity to influenza infection in man. Br Med Bull 1979; 35:69–75. [DOI] [PubMed] [Google Scholar]
- 31. Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol 2010; 17:1055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han A, Czajkowski LM, Donaldson A, et al. A dose-finding study of a wild-type influenza A(H3N2) virus in a healthy volunteer human challenge model. Clin Infect Dis 2019; 69:2082–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Powers JH 3rd, Bacci ED, Guerrero ML, et al. Reliability, validity, and responsiveness of InFLUenza Patient-Reported Outcome (FLU-PRO) scores in influenza-positive patients. Value Health 2018; 21:210–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen C, Kleynhans J, Moyes J, et al. Asymptomatic transmission and high community burden of seasonal influenza in an urban and a rural community in South Africa, 2017–18 (PHIRST): a population cohort study. Lancet Glob Health 2021; 9:e863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ip DK, Lau LL, Leung NH, et al. Viral shedding and transmission potential of asymptomatic and paucisymptomatic influenza virus infections in the community. Clin Infect Dis 2017; 64:736–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Biomedical Advanced Research and Development Authority . Influenza therapeutic landscape. https://www.isirv.org/site/images/Influenza%20Antiviral%20Landscape%202022.pdf. Accessed 24 July 2022.
- 37. Center for Infectious Disease Research and Policy . Universal influenza vaccine technology landscape. https://ivr.cidrap.umn.edu/universal-influenza-vaccine-technology-landscape. Accessed 24 July 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.