Abstract
Adeno-associated virus (AAV) replication depends on two viral components for replication: the AAV nonstructural proteins (Rep) in trans, and inverted terminal repeat (ITR) sequences in cis. AAV type 5 (AAV5) is a distinct virus compared to the other cloned AAV serotypes. Whereas the Rep proteins and ITRs of other serotypes are interchangeable and can be used to produce recombinant viral particles of a different serotype, AAV5 Rep proteins cannot cross-complement in the packaging of a genome with an AAV2 ITR. In vitro replication assays indicated that the block occurs at the level of replication instead of at viral assembly. AAV2 and AAV5 Rep binding activities demonstrate similar affinities for either an AAV2 or AAV5 ITR; however, comparison of terminal resolution site (TRS) endonuclease activities showed a difference in specificity for the two DNA sequences. AAV2 Rep78 cleaved only a type 2 ITR DNA sequence, and AAV5 Rep78 cleaved only a type 5 probe efficiently. Mapping of the AAV5 ITR TRS identified a distinct cleavage site (AGTG TGGC) which is absent from the ITRs of other AAV serotypes. Comparison of the TRSs in the AAV2 ITR, the AAV5 ITR, and the AAV chromosome 19 integration locus identified some conserved nucleotides downstream of the cleavage site but little homology upstream.
A key step in the infectious cycle of adeno-associated virus (AAV) is the replication of the viral genome. Two viral components are required for replication: the AAV nonstructural proteins (Rep), and the inverted terminal repeat (ITR) sequences at the end of the viral genome. The Rep open reading frame (ORF) of AAV encodes four related proteins that are transcribed from one of two promoters along with a splice variant of each. These four proteins are referred to by their apparent molecular sizes of 78, 68, 52, 40 kDa. Rep78 and Rep68 are necessary for viral DNA replication (3, 14, 25, 35), whereas Rep52 and Rep40 function as DNA helicases (2) and facilitate the accumulation of single-stranded progeny virus (30).
Two of the Rep proteins (Rep78 and Rep68) bind DNA in a sequence-specific manner and can nick duplex ITRs in a site- and strand-specific manner (6, 15, 33). This cleavage reaction occurs at the terminal resolution site (TRS) within the ITR and permits the replication of the ends of the linear genome. DNA binding requires a tandem repeat of a GAGY motif which is present in the ITR structures at the ends of the viral genome (4, 6, 26, 28, 32). Furthermore, DNA binding and terminal resolution are central to the preferential integration of the AAV genome into a region on the q arm of human chromosome 19 (18, 29, 35, 37).
In addition to the role of the Rep proteins in the life cycle of AAV, their expression results in a characteristic phenotype. Overexpression of Rep68 and Rep78 has been shown to either negatively or positively affect transcription, to inhibit cellular transformation, (11–13, 17, 20, 21), and to inhibit progression through the cell cycle (10, 39). The importance of the Rep proteins is emphasized by their high degree of conservation among four of the five sequenced AAV serotypes (5, 8, 24, 27, 34).
The ITRs can fold into T-shaped hairpin structures and serve as the origins of viral DNA replication. Two elements within the ITR are central to its function: a GAGY repeat motif and the TRS (RGTTGG). The ITRs of AAV serotypes 2, 3, 4, 5, and 6 (AAV2 to -6) exhibit sequence differences, but the ability to fold into a hairpin conformation and the existence of a Rep binding motif are conserved. The TRS is conserved only in AAV2, -3, -4, and -6.
The Rep proteins of AAV2, -3, -4, and -6 are approximately 90% identical, with most of the changes being conservative amino acid substitutions. This high degree of conservation of the Rep gene and ITR among the serotypes results in cross-complementation among the Rep genes and ITRs of different serotypes (8).
In contrast, the Rep gene and ITR of AAV5 are only 60% similar to those of the other serotypes and fail to cross-complement with other serotypes (5); i.e., no AAV2 ITR-containing genomes can be packaged by using AAV5 Rep. However, viral particles are produced if the AAV2 Rep ORF is included. From these data, we hypothesize that the lack of complementation is not at the level of viral assembly—that is, viral DNA packaging—but is rather at the level of viral DNA replication. Comparison of the ITR sequence of AAV5 with those of the other serotypes identified a Rep binding motif; however, only a partial consensus TRS could be identified in the AAV5 ITR. Thus, AAV5 Rep proteins may have an endonuclease different from those of the other AAV serotypes.
In this study, we confirmed our hypothesis that the lack of cross-complementation occurs at the level of DNA replication. AAV5 and AAV2 Rep proteins were shown to bind the ITRs of the other serotype; however, efficient TRS endonuclease activity was detected only in their respective ITRs. Mapping of the AAV5 Rep terminal resolution endonuclease activity identified a novel recognition sequence that is present only in the AAV5 ITR.
MATERIALS AND METHODS
Plasmids and DNA.
All oligonucleotide probes were purified on denaturing polyacrylamide gels before being annealed and on native gels after being annealed. Construction of recombinant AAV2 (AAV2RnLacZ) and AAV5 (AAV5RnLacZ) vector plasmids expressing nucleus-localized β-galactosidase is described elsewhere (5, 7). The P1 element from the AAV integration locus was cloned into pUC19, producing pMAT50 (37). The Rep ORF of AAV5 was cloned into the maltose binding protein (MBP) expression plasmid pMAL-c2 by PCR amplification of AAV5 DNA (nucleotides [nt] 359 to 3600) with Pfu polymerase (Statagene, La Jolla, Calif.). The PCR product was digested with XhoI, which cuts at nt 3589. The 3.2-kb fragment was cloned into the XmnI and SalI sites of pMAL-c2 (New England Biolabs, Beverly, Mass.). The construct was verified by sequencing. The recombinant AAV5 MBP-Rep78 fusion protein was also expressed in Escherichia coli and purified by using an amylose affinity resin (New England Biolabs). The MBP-Rep78 fusion protein had a molecular mass of 115 kDa as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. For in vitro translation of AAV5 Rep78, AAV5 DNA was cut with NruI and a 1.9-kb fragment was isolated and cloned into the SmaI site of pGem3Zf. AAV2 MBP-Rep78 and in vitro-translated AAV2 Rep78 expression plasmids are described elsewhere (6, 31).
In vitro replication assay.
HeLa S3 cell extracts were prepared as described elsewhere (36). The replication assays were performed as described previously (35), with the following modifications: the total dCTP concentration was 10 μM, and the reaction mixtures were preincubated for 1.5 h before the addition of [α-32P]dCTP (5,000 Ci/mmol; Amersham). Each assay mixture contained 0.1 mg of cellular protein in a final volume of 15 μl; 4 mM ATP; 200 μM (each) CTP, GTP, and UTP; 100 μM (each) dATP, dGTP, and dTTP; 10 μCi of [α-32P]dCTP; 2 mM dithiothreitol (DTT); 2 μg of creatine phosphokinase; 300 ng of plasmid DNA; and, when indicated, 1 μg of recombinant Rep protein. Reaction mixtures were incubated at 34°C for 16 to 18 h. The level of stimulation of replication was determined by the amount of 32P in the acid-precipitable material and assayed by thin-layer chromotography (TLC). Aliquots of each reaction mixture were assayed by spotting them on silica gel TLC plates (Baker-flex Silica Gel 1B; J. T. Baker, Phillipsburg, N.J.). The unincorporated nucleotides were separated from acid-precipitable DNA by development of the plates with 1 M HCl. The level of 32P was determined with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). For samples analyzed by agarose gel electrophoresis, unincorporated nucleotides were removed from reaction mixtures by the use of a nucleotide removal kit (Qiagen, Valencia, Calif.), and the DNA was resolved on 0.6% agarose gels, dried, and autoradiographed.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (6). Briefly, gel-purified radiolabeled probes (50,000 cpm) were incubated with 100 to 200 ng of purified AAV2 MBP-Rep78 or AAV5 MBP-Rep78 in a 15-μl reaction mixture containing shift buffer (40 mM Tris-HCl [pH 8], 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 4% glycerol, 0.1 mg of bovine serum albumin per ml) at 30°C for 15 min. The reaction mixtures were then cooled to 4°C, and 5 μl of sample loading dye was added to each. The DNA-protein complexes were then resolved on 4% nondenaturing polyacrylamide gels, dried, and autoradiographed.
Terminal resolution endonuclease assays.
Site- and strand-specific endonuclease assays were performed as described previously (6) with the following modifications. A 2-μl aliquot of in vitro-translated AAV2 Rep78 or AAV5 Rep78 was added, along with radioactively 5′-end-labeled probe (50,000 cpm) to a 15-μl reaction volume containing TRS buffer [25 mM piperazine-N,N′-bis(2-ethanosulfonic acid) [PIPES], 4 mM MgCl2, 1 mM DTT, 1 mM ATP, 0.1 mg of bovine serum albumin/ml]. Reaction mixtures were incubated for 1 h at 37°C, and then the reactions were terminated by addition of 5 μl of formamide gel loading buffer followed by heating to 80°C for 5 min. The reaction products were resolved on 10% polyacrylamide–urea gels (Novex). The gels were fixed in 10% methanol and 10% acetic acid followed by gel drying solution (Novex) prior to being dried and subjected to autoradiography. The TRS mapping experiments were performed with the same reaction mixture, but the reactions were terminated with stop buffer (10 mM Tris-HCl [pH 7.9], 10 mM NaCl, 0.5% sodium dodecyl sulfate, 0.2 mg of yeast tRNA/ml, 20 mM EDTA, 2 mg of proteinase K/ml). The reaction mixtures were incubated in stop buffer for 30 min at 37°C, and the nucleic acid was purified by phenol-chloroform extraction and ethanol precipitation. The DNA was resuspended in formamide gel loading buffer and resolved on a 6% polyacrylamide sequencing gel, which was subsequently dried and subjected to autoradiography. The sizes of the cleavage products were determined by comparison to the products of a purine-specific sequencing reaction of the labeled probe (Sigma Chemical Co.) (23).
RESULTS
Cross-complementation refers to the ability of a protein of one serotype to replicate and package the genome of an alternative serotype. To determine whether cross-complementation is occurring at the level of DNA synthesis with heterologous Rep proteins, AAV2 or AAV5 ITR-containing genomes were tested in an in vitro replication assay that included either AAV2 or AAV5 Rep proteins.
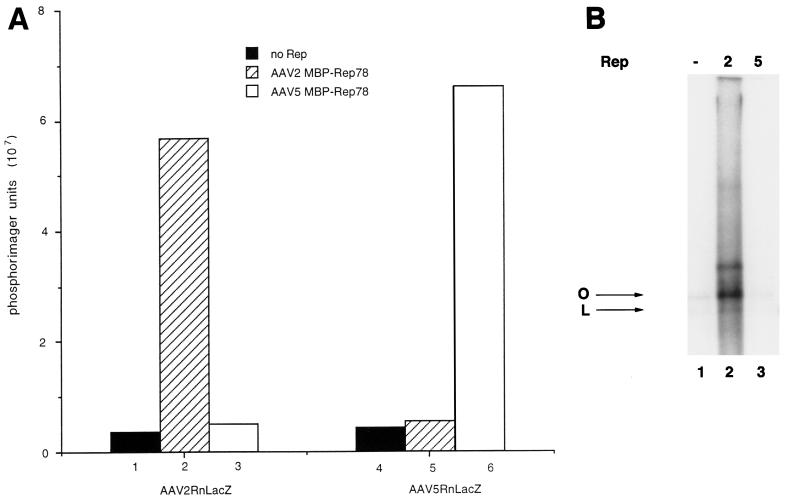
The DNA templates (AAV2RnLacZ and AAV5RnLacZ) tested in the replication assay were the recombinant genomes used in the packaging experiments and contained the ITR of either AAV2 or AAV5 flanking a nucleus-localized β-galactosidase gene with a Rous sarcoma virus promoter. These plasmids lack the Rep and Cap ORFs of AAV. Addition of the AAV2RnLacZ DNA to HeLa cell lysate supplemented with recombinant AAV2 MBP-Rep78 resulted in a 12-fold increase in DNA replication compared to control reactions (lysate minus Rep) (Fig. 1A, bar 1) as measured by radiolabel incorporation (Fig. 1A, bar 2). A similar increase in DNA replication was measured when the AAV5RnLacZ DNA was added to HeLa cell lysate supplemented with AAV5 MBP-Rep78 (Fig. 1A, bar 6). In contrast, little or no DNA replication was detected if AAV5 Rep was added to AAV2RnLacZ DNA or AAV2 Rep was added to AAV5RnLacZ DNA (Fig. 1A, bars 3 and 5, respectively).
FIG. 1.
In vitro replication. (A) The effect of the MBP-Rep fusion protein from AAV-2 or AAV-5 on DNA replication was measured by TLC (see Materials and Methods). One of two templates was tested in the in vitro replication assay: AAV2RnLacZ or AAV5RnLacZ (bars 1 to 3 and 4 to 6, respectively). Background incorporation was defined as the amount of incorporation measured in the absence of Rep protein (bars 1 and 4). (B) Effects of AAV2 and AAV5 MBP-Rep78 with pMAT50 on replication. Replication reactions were performed as described in Materials and Methods, and the replication products were resolved by agarose gel electrophoresis. AAV2 and AAV5 MBP-Rep78 proteins were included in lanes 2 and 3, respectively; lane 1, extract only, no Rep. The open-circular (O) and linear (L) forms of pMAT50 are indicated.
AAV2 Rep protein can also initiate DNA replication from a cellular DNA fragment isolated from the AAV integration locus (35, 37). This fragment (P1) contains a Rep binding motif as well as a TRS, both of which are necessary for P1 to function as an origin of DNA replication (35). As previously described, the addition of a P1-containing plasmid to HeLa cell lysate supplemented with AAV2 MBP-Rep78 resulted in a 10-fold increase in DNA replication (Fig. 1B, lane 2) (35). However, only background levels of radiolabel incorporation were measured if no Rep or AAV5 MBP-Rep78 was added to the HeLa cell lysate (Fig. 1B, lanes 1 and 3).
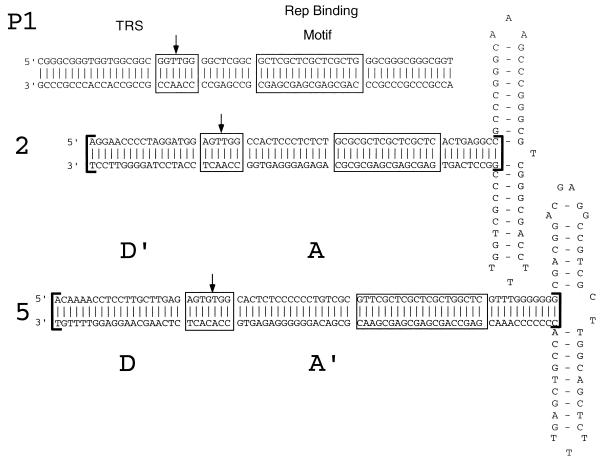
The above results support our hypothesis that the lack of cross-complementation occurs at the level of DNA replication. AAV DNA replication requires both a Rep binding motif and an adjacent TRS motif. Comparison of the ITR sequence of AAV5 with those of the AAV2 ITR and the P1 fragment identified repeats of the GAGY Rep binding motif in all three. However, the AAV5 ITR contained only a weak consensus TRS compared to the AAV2 ITR and the P1 fragment (Fig. 2).
FIG. 2.
Sequence and alignment of ITR and probes. (Top) Sequence of the P1 element contained within the SmaI subfragment of AAVS1 (nt 354 to 468) (37). (Middle) Sequence of the AAV2 ITR as previously described (34). (Bottom) Sequence of the AAV5 ITR as previously described (5). Linear probes used in the EMSA and TRS assays are indicated by brackets flanking the AAV2 and AAV5 ITR stem regions. Rep binding motifs are boxed and labeled. TRSs are boxed and labeled, and the site of cleavage in each is indicated by a vertical arrow.
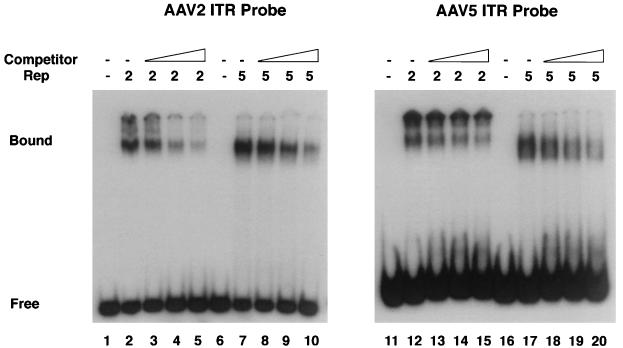
To compare the DNA binding activities of AAV5 MBP-Rep78 and AAV2 MBP-Rep78 with either the AAV2 ITR or AAV5 ITR, we performed EMSAs using truncated ITR probes (Fig. 3). Both Rep proteins formed complexes with either probe, producing slightly different shift patterns with the different Rep proteins. The AAV5 MBP-Rep78 complex migrated slightly faster than the AAV2 Rep complex with either the type 2 or type 5 ITR probe (Fig. 3). This mobility difference most likely results from the reduced size and more acidic charge of AAV5 Rep78 compared to AAV2 Rep78. Addition of a 20-fold excess of cold competitor DNA inhibited all four shift complexes three- to fourfold, suggesting that AAV2 and AAV5 Rep proteins have similar binding affinities for either probe (Fig. 3, lanes 5, 10, 15, and 20).
FIG. 3.
EMSAs. Gel-purified radiolabeled probes (50,000 cpm each) were incubated with 100 ng of purified AAV2 MBP-Rep78 or AAV5 MBP-Rep78 as indicated. In the absence of protein, no shift complex was detected (lanes 1, 6, 11, and 16). The addition of purified AAV2 MBP-Rep78 (lanes 2 and 12) or AAV5 MBP-Rep78 (lanes 7 and 17) resulted in the formation of a shift complex. Binding competition studies were done in the presence of a 5-fold (lanes 3, 8, 13, and 18), 10-fold (lanes 4, 9, 14, and 19), or 20-fold (lanes 5, 10, 15, and 20) molar excess of unlabeled probe. Quantitation of the shift complex was done with a scanning densitometer (Molecular Dynamics). Bound, bound probe; free, free probe.
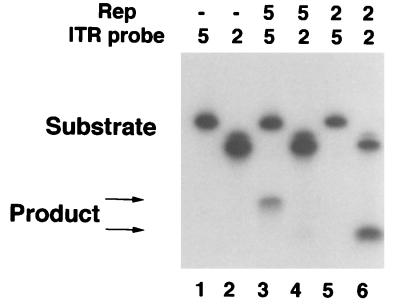
Characterization of the TRS endonuclease activities of AAV2 and AAV5 Rep proteins on either the AAV2 or AAV5 ITR showed a difference in specificity for the two probes. Incubation of in vitro-translated AAV2 Rep78 with the AAV2 ITR probe radiolabeled on the 5′ end of the D′A strand generated a cleavage product of the expected size (Fig. 4, lane 6). In contrast, no cleavage product was detected with AAV2 Rep78 and the AAV5 ITR (Fig. 4, lane 5). In vitro-translated AAV5 Rep78 demonstrated a similar specificity for its own ITR compared to the AAV2 ITR. AAV5 Rep78 produced a major cleavage when AAV5 Rep78 was incubated with the AAV5 ITR probe; in contrast, a very faint band was detected when AAV5 Rep78 was incubated with the AAV2 ITR probe (Fig. 4, lanes 3 and 4, respectively). The inefficient cleavage of the AAV2 ITR by AAV5 Rep78 indicates that in the absence of its cognate TRS, AAV5 Rep78 may utilize a nonnative TRS. Thus, AAV2 Rep78 and AAV5 Rep78 have similar DNA binding affinities for the two probes but lack efficient TRS endonuclease cross-reactivity.
FIG. 4.
TRS specificity. In vitro-translated AAV2 Rep78 or AAV5 Rep78 was incubated with radiolabeled probe (50,000 cpm), and the products were resolved on denaturing gels. The positions of the full-length substrate and cleavage products are indicated by arrows. Lanes: 1 and 2, probe alone; 3 and 4, AAV5 Rep78 with the AAV5 and AAV2 ITRs, respectively; 5 and 6, AAV2 Rep78 with the AAV5 and AAV2 ITRs, respectively. The upper arrow identifies the AAV5 cleavage product (lane 3), and the lower arrow indicates the AAV2 cleavage product (lane 6).
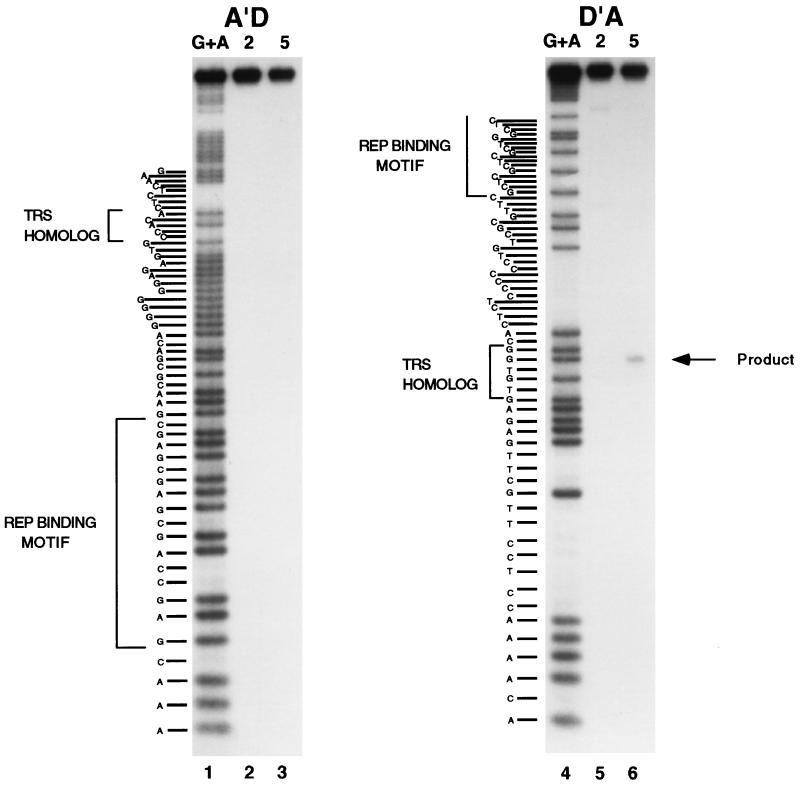
Previous research mapped the AAV2 TRS to a unique site (AGTTGG) on the D′A strand (15, 33). To map the AAV5 TRS, the cleavage reactions were repeated with the AAV5 ITR probe derived from the stem region of the ITR (Fig. 2). 5′-End-labeled D′A or A′D duplexed oligonucleotide probes were incubated with either AAV2 or AAV5 MBP-Rep and resolved on a sequencing gel (Fig. 5). The site of cleavage was determined by comparison with a G+A chemical sequencing reaction and is diagrammatically indicated in Fig. 2. AAV5 Rep78-directed cleavage was detected only on the coding strand (D′A) and not on the noncoding strand (A′D) (Fig. 5, lanes 6 and 3, respectively). As in Fig. 4, some minor cleavage products were also detected with AAV2 Rep78 on the D′A probe (Fig. 5, lane 5). As stated above, this may represent an ability of Rep78 to cleave at a suboptimal site when a preferred site is not present.
FIG. 5.
TRS mapping of the AAV5 ITR. 32P-5′-end-labeled oligonucleotide probes (A′D or D′A) were annealed with unlabeled complementary oligonucleotides and incubated with either recombinant MBP-AAV2 Rep78 (lanes 2 and 5) or MBP-AAV5 Rep78 (lanes 3 and 6). The position of the cleavage product is indicated by an arrow. The sizes of the cleavage products were determined by comparison to the products of purine-specific sequencing reactions of the labeled probe (lanes 1 and 4). The sequence of the labeled oligonucleotide probe is on the left of each panel, with the Rep binding motif and TRS homolog indicated by brackets.
The cleavage site is positioned 21 to 25 bases upstream of the GAGY Rep binding motif, compared to the 16 to 20 bases in the AAV2 ITR. Whereas the AAV5 DNA upstream of the cleavage site has little homology with the AAV2 DNA, some homology exists downstream of the TRS. In both AAV2 and AAV5, the TGGC motif immediately downstream of the cleavage site is conserved. This sequence is followed by a pyrimidine-rich region which abuts the Rep binding motif (Fig. 2). The sequence surrounding the TRS is conserved proximal to the Rep binding sites in P1, AAV2, and AAV5 (TGG); there is a lack of homology upstream. However, the spacing between the GAGY motif and the TRS is reduced, and the pyrimidine-rich region present in the AAV ITRs is absent.
DISCUSSION
AAV5 is the only dependent parvovirus that was originally isolated from a patient sample instead of from laboratory stocks of adenovirus (1). In this study, the discrete TRS endonuclease activity of type 5 Rep in comparison to that of type 2 Rep further defines AAV5 as a unique dependovirus. The ITR and Rep protein of AAV5 are distinct and do not complement those of AAV2. In vitro replication experiments indicate that the lack of cross-reactivity occurs at the level of DNA replication. Although further biochemical analysis demonstrated that AAV2 Rep78 and AAV5 Rep78 will bind to either AAV2 or AAV5 ITR probes with similar affinities, comparison of their TRS endonuclease activities showed a difference in specificity for the two DNA sequences. AAV2 Rep78 would only cleave an AAV2 ITR DNA sequence, and AAV5 Rep78 had a very strong preference for cleaving the AAV5 ITR DNA sequence.
Despite the differences in TRS specificity, there are conserved sequence elements within the probes. The sequence immediately downstream of the cleavage site (TGG) is common to all three probes. Further downstream of the cleavage site, both the AAV2 ITR and the AAV5 ITR contain a pyrimidine tract between the TRS site and the Rep binding motif. In contrast, in P1, the spacing is reduced and the pyrimidine tract is absent. Because of its absence from the P1 fragment, the pyrimidine tract may not be important for Rep binding or TRS activity; instead, this region may promote stability of the ITR hairpin structure, which cannot form in the P1 fragment.
Rep-mediated cleavage of the P1 fragment is central to the integration of AAV2 at the AAVS1 cellular locus on chromosome 19 (22). Initiation of DNA synthesis results from Rep cleavage of these DNA substrates. The type 5 Rep proteins are unable to initiate DNA synthesis in vitro from these substrates as a result of their unique TRS endonuclease activity. By extension of the model for type 2 integration, AAV5 integration may occur at a unique locus compared to the other serotypes of AAV.
The Rep consensus binding motif consists of at least two repeats of GAGY (9). These repeats have been identified in a number of cellular loci (9, 38) and are reported to affect gene transcription (19). Based on analysis of the AAVS1 integration locus, a TRS adjacent to the Rep binding motif is also required for the site to function in targeted integration (22). For AAV2, the requirement of a properly positioned TRS and a Rep binding motif may further reduce the probability of the sequence’s occurrence to that of a unique site in the human genome (35). This is supported by the preferential integration of AAV2 at the chromosome 19 locus (16). Whereas AAV5 Rep recognizes a TRS distinct from that recognized by AAV2 Rep, the probability of this sequence occurring adjacent to a Rep binding site will be similar to that for AAV2.
Sequence analysis, capsid interactions, packaging efficiency, and biochemical activities define AAV5 as being unique among the dependoviruses. Several of these features will have applications for the field of gene therapy and the development of recombinant and chimeric vectors. Comparison of the amino acid sequences and biochemical activities of AAV5 Rep and AAV2 Rep will also be useful for defining the Rep DNA binding domain, as well as amino acids that are involved in TRS endonuclease activity.
ACKNOWLEDGMENTS
We thank Richard H. Smith and Michael Schmidt for review of the manuscript.
REFERENCES
- 1.Bantel-Schall U, zur Hausen H. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology. 1984;134:52–63. doi: 10.1016/0042-6822(84)90271-x. [DOI] [PubMed] [Google Scholar]
- 2.Chejanovsky N, Carter B J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989;173:120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 3.Chejanovsky N, Carter B J. Replication of a human parvovirus nonsense mutant in mammalian cells containing an inducible amber suppressor. Virology. 1989;171:239–247. doi: 10.1016/0042-6822(89)90531-x. [DOI] [PubMed] [Google Scholar]
- 4.Chiorini J A, Wiener S M, Owens R A, Kyöstiö S R M, Kotin R M, Safer B. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J Virol. 1994;68:7448–7457. doi: 10.1128/jvi.68.11.7448-7457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiorini J A, Kim F, Yang L, Kotin R M. Cloning and characterization of adeno-associated virus type 5. J Virol. 1999;73:1309–1319. doi: 10.1128/jvi.73.2.1309-1319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiorini J A, Wendtner C M, Urcelay E, Safer B, Hallek M, Kotin R M. High-efficiency transfer of the T cell co-stimulatory molecule B7-2 to lymphoid cells using high-titer recombinant adeno-associated virus vectors. Hum Gene Ther. 1995;6:1531–1541. doi: 10.1089/hum.1995.6.12-1531. [DOI] [PubMed] [Google Scholar]
- 8.Chiorini J A, Yang L, Lui Y, Safer B, Kotin R M. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J Virol. 1997;71:6823–6833. doi: 10.1128/jvi.71.9.6823-6833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiorini J A, Yang L, Safer B, Kotin R M. Determination of adeno-associated virus Rep68 and Rep78 binding sites by random sequence oligonucleotide selection. J Virol. 1995;69:7334–7338. doi: 10.1128/jvi.69.11.7334-7338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermanns J, Schulze A, Jansen-Dürr P, Kleinschmidt J A, Schmidt R, zur Hausen H. Infection of primary cells by adeno-associated virus type 2 results in a modulation of cell cycle-regulating proteins. J Virol. 1997;71:6020–6027. doi: 10.1128/jvi.71.8.6020-6027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermonat P L. The adeno-associated virus Rep78 gene inhibits cellular transformation induced by bovine papillomavirus. Virology. 1989;172:253–261. doi: 10.1016/0042-6822(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 12.Hermonat P L. Inhibition of H-ras expression by the adeno-associated virus Rep78 transformation suppressor gene product. Cancer Res. 1991;51:3373–3377. [PubMed] [Google Scholar]
- 13.Hermonat P L. Inhibition of bovine papillomavirus plasmid DNA replication by adeno-associated virus. Virology. 1992;189:329–333. doi: 10.1016/0042-6822(92)90710-7. [DOI] [PubMed] [Google Scholar]
- 14.Hong G, Ward P, Berns K I. In vitro replication of adeno-associated virus DNA. Proc Natl Acad Sci USA. 1992;89:4673–4677. doi: 10.1073/pnas.89.10.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Im D-S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 16.Kearns W G, Afione S A, Fulmer S B, Pang M C, Erikson D, Egan M, Landrum M J, Flotte T R, Cutting G R. Recombinant adeno-associated virus (AAV-CFTR) vectors do not integrate in a site-specific fashion in an immortalized epithelial cell line. Gene Ther. 1996;3:748–755. [PubMed] [Google Scholar]
- 17.Khleif S N, Myers T, Carter B J, Trempe J P. Inhibition of cellular transformation by the adeno-associated virus rep gene. Virology. 1991;181:738–741. doi: 10.1016/0042-6822(91)90909-u. [DOI] [PubMed] [Google Scholar]
- 18.Kotin R M, Menninger J C, Ward D C, Berns K I. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 19.Kyöstiö S R M, Wonderling R S, Owens R A. Negative regulation of the adeno-associated virus (AAV) P5 promoter involves both the P5 Rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J Virol. 1995;69:6787–6796. doi: 10.1128/jvi.69.11.6787-6796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labow M A, Berns K I. The adeno-associated virus rep gene inhibits replication of an adeno-associated virus/simian virus 40 hybrid genome in cos-7 cells. J Virol. 1988;62:1705–1712. doi: 10.1128/jvi.62.5.1705-1712.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labow M A, Graf L H, Jr, Berns K I. Adeno-associated virus gene expression inhibits cellular transformation by heterologous genes. Mol Cell Biol. 1987;7:1320–1325. doi: 10.1128/mcb.7.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linden R M, Winocour E, Berns K I. The recombination signals for adeno-associated virus site-specific integration. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 24.Muramatsu S I, Mizukami H, Young N S, Brown K E. Nucleotide sequence and generation of an infectious clone of adeno-associated virus 3. Virology. 1996;221:208–217. doi: 10.1006/viro.1996.0367. [DOI] [PubMed] [Google Scholar]
- 25.Ni T-H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owens R A, Trempe J P, Chejanovsky N, Carter B J. Adeno-associated virus rep proteins produced in insect and mammalian expression systems: wild-type and dominant-negative mutant proteins bind to the viral replication origin. Virology. 1991;184:14–22. doi: 10.1016/0042-6822(91)90817-u. [DOI] [PubMed] [Google Scholar]
- 27.Rutledge E A, Halbert C L, Russell D W. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan J H, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. . (Erratum, 11:1228, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith R H, Kotin R M. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith R H, Spano A J, Kotin R M. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J Virol. 1997;71:4461–4471. doi: 10.1128/jvi.71.6.4461-4471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder R O, Im D-S, Ni T, Xiao X, Samulski R J, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder R O, Im D-S, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) Rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srivastava A, Lusby E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R M. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward P, Berns K I. In vitro rescue of an integrated hybrid adeno-associated virus/simian virus 40 genome. J Mol Biol. 1991;218:791–804. doi: 10.1016/0022-2836(91)90267-a. [DOI] [PubMed] [Google Scholar]
- 37.Weitzman M D, Kyöstiö S R M, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wonderling R S, Owens R A. Binding sites for adeno-associated virus Rep proteins within the human genome. J Virol. 1997;71:2528–2534. doi: 10.1128/jvi.71.3.2528-2534.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Q, Chen F, Trempe J P. Characterization of cell lines that inducibility express the adeno-associated virus Rep proteins. J Virol. 1994;68:4847–4856. doi: 10.1128/jvi.68.8.4847-4856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]