Abstract
Programmed constitutive heterochromatin silencing is essential for eukaryotic genome regulation, yet the initial step of this process is ambiguous. A large proportion of R-loops (RNA:DNA hybrids) had been unexpectedly identified within Arabidopsis pericentromeric heterochromatin with unknown functions. Through a genome-wide R-loop profiling screen, we find that DDM1 (decrease in DNA methylation 1) is the primary restrictor of pericentromeric R-loops via its RNA:DNA helicase activity. Low levels of pericentromeric R-loops resolved by DDM1 cotranscriptionally can facilitate constitutive heterochromatin silencing. Furthermore, we demonstrate that DDM1 physically excludes histone H2A variant H2A.Z and promotes H2A.W deposition for faithful heterochromatin initiation soon after R-loop clearance. The dual functions of DDM1 in R-loop resolution and H2A.Z eviction are essential for sperm nuclei structure maintenance in mature pollen. Our work unravels the cotranscriptional R-loop resolution coupled with accurate H2A variants deposition is the primary step of constitutive heterochromatin silencing in Arabidopsis, which might be conserved across eukaryotes.
A chromatin remodeling protein removes R-loops and rejects H2A.Z to prime heterochromatin silencing.
INTRODUCTION
Constitutive heterochromatin is the major composition of the eukaryotic genome and consists of repetitive elements, such as satellite repeats and transposable elements (TEs) near centromeres and telomeres, which is associated with crucial cellular functions including genome integrity, chromosome segregation, DNA recombination, transposition inhibition, transcription silencing, and maintenance of nuclear territories (1–4). The conserved contributors to constitutive heterochromatin formation in eukaryotes involve DNA and histone methylation, histone variants, chromatin remodeling complexes, polycomb proteins, transcription factors, and short and long RNAs (2, 5, 6). However, the early steps of the constitutive heterochromatin formation are still unclear.
In plants, decrease in DNA methylation 1 (DDM1), a putative SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeler, has long been known to maintain DNA and histone methylation of constitutive heterochromatin (7–10), although DDM1 is not a DNA methyltransferase (8), and is unlikely to be a histone methyltransferase (based on its domain organization). Previous evidences support a model that DDM1 is capable of remodeling nucleosomes and facilitating access for DNA methyltransferases to H1-containing heterochromatin or nucleosome-wrapped DNA (11–13). It was shown that DDM1 can interact with histone H2A variants H2A.W and deposit H2A.W into the pericentromeric region for silencing transposons (14). However, the h2a.w mutant does not mirror the phenotype of ddm1 mutant, as in h2a.w, there is a subtle impact on constitutive heterochromatin formation, TE silencing, and DNA methylation (15). These genetically inconsistent results imply the molecular functions of DDM1 in constitutive heterochromatin formation remain largely mysterious.
R-loops are three-stranded structures where the newly transcribed RNA invades into the double-stranded DNA (dsDNA) to pair with the template strand DNA and form an RNA:DNA hybrid, thus leaving the other strand as a single-stranded DNA. R-loops have been implicated in a multitude of biological processes, including transcriptional regulation, transcription termination, DNA replication, DNA damage and repair, and genome maintenance (16–21). Prevention and resolution of R-loops can occur through RNA processing factors, DNA topoisomerases, RNA:DNA helicases, ribonuclease H (RNase H) enzymes, as well as chromatin remodelers [reviewed in (21)]. We had developed an efficient R-loop profiling method ssDRIP-seq (single-strand DNA ligation–based library construction from RNA:DNA hybrid immunoprecipitation, followed by sequencing), which had been applied in multiple species to characterize R-loop formation and dynamic pattern genome-widely [(22–30), reviewed in (31)]. Unexpectedly, we found that a large proportion of R-loops is correlated with H3K9me2 and H3K27me1 heterochromatic marks in Arabidopsis (22); however, the biological relevance of these observations was not clear.
In this study, through a ssDRIP-seq-based screen, we show that DDM1 is the primary regulator that restricts R-loops in pericentromeric heterochromatin in Arabidopsis. Similarly, pericentromeric RNA:DNA hybrids are highly accumulated in the absence of the DDM1 ortholog in mammals (32). DDM1 can resolve RNA from RNA:DNA hybrids or R-loops in vitro through its adenosine triphosphate (ATP)–dependent helicase activity. In addition, our data show that DDM1 have higher affinities to the heterochromatic variant H2A.W than to the variant H2A.Z. In the absence of DDM1, the variant H2A.Z could replace H2A.W at pericentromeric regions through the SWR1 complex (SWR1-C), thus promoting cotranscriptional R-loop formation. Our results revealed that DDM1 protein has dual functions: restricts the initial cotranscriptionally formed R-loops and excludes the H2A.Z deposition in pericentromeric heterochromatin, to allow heterochromatic histone H2A.W deposition taking place, thus ensuring accurate heterochromatin initiation and transcriptional gene silencing. Furthermore, we found that this process is important for structure maintenance of sperm nuclei (SN), in where there is the enrichment of DDM1 and H2A.W, and in absence of H2A.Z.
RESULTS
The R-loops profiling screen identifies DDM1 as the primary restrictor of pericentromeric R-loops
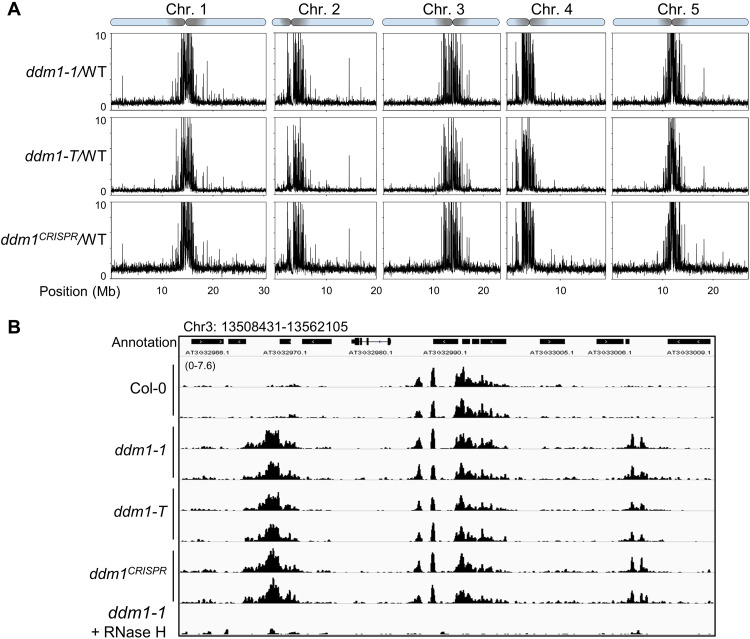
To understand the functions of, as well as the components that might participate in regulating pericentromeric R-loops formation, we performed an R-loop profiling (ssDRIP-seq) screen in a group of mutants defective for heterochromatin formation (Fig. 1, fig. S2, and data S1; including the detailed information of the mutants used for ssDRIP-seq). By analyzing the ssDRIP-seq in wild-type Col-0 and the mutants, while there is modest to almost no differences of R-loops at pericentromeric regions in most of the mutants (Fig. 1 and fig. S2), however, we found a marked R-loop accumulation at pericentromeric regions in ddm1 (Fig. 1 and fig. S2), a mutant that greatly decreases global DNA methylation (7). Slot blot assay in two different allelic mutants of ddm1 (ddm1-1 and ddm1-2) confirmed elevated R-loop levels in ddm1 (fig. S1, A to C). ssDRIP-seq analysis of the collected allelic mutants of ddm1 (fig. S1A), including a T-DNA insertion line ddm1-T and a CRISPR-Cas9–based genomic DNA deletion mutant, shows the increased R-loops at pericentromeric regions relative to wild-type (Fig. 1 and fig. S1D). Together, these results suggest that DDM1 is a major negative regulator of R-loops specifically at the pericentromeric heterochromatin.
Fig. 1. DDM1 is a major restrictor for pericentromeric R-loop homeostasis.
(A) ssDRIP-seq data from different allelic mutants of ddm1. Mutant/wild-type (WT) values are plotted in 10-kb bins on chromosomes. Pericentromeric regions are shown as shaded area on each chromosome. (B) Snapshots of Integrative Genomics Viewer show R-loop levels (ssDRIP-seq) of ddm1 mutants in the chromosome 3 region: 13508431-13562105. “ddm1-1 + RNase H” was the control experiment for ddm1 ssDRIP-seq with commercial RNase H treatment before DRIP.
Together with the data from screened mutants (Fig. 1 and fig. S2) and the facts that DDM1 affects DNA and histone methylation [(7–10), see also below], our findings imply that pericentromeric heterochromatin modifications have no impact on R-loop clearance by DDM1, while DDM1 functions as a key factor to link R-loop clearance with heterochromatin marks deposition.
Cotranscriptional R-loop accumulation disturbs pericentromeric heterochromatin formation in the ddm1 mutant
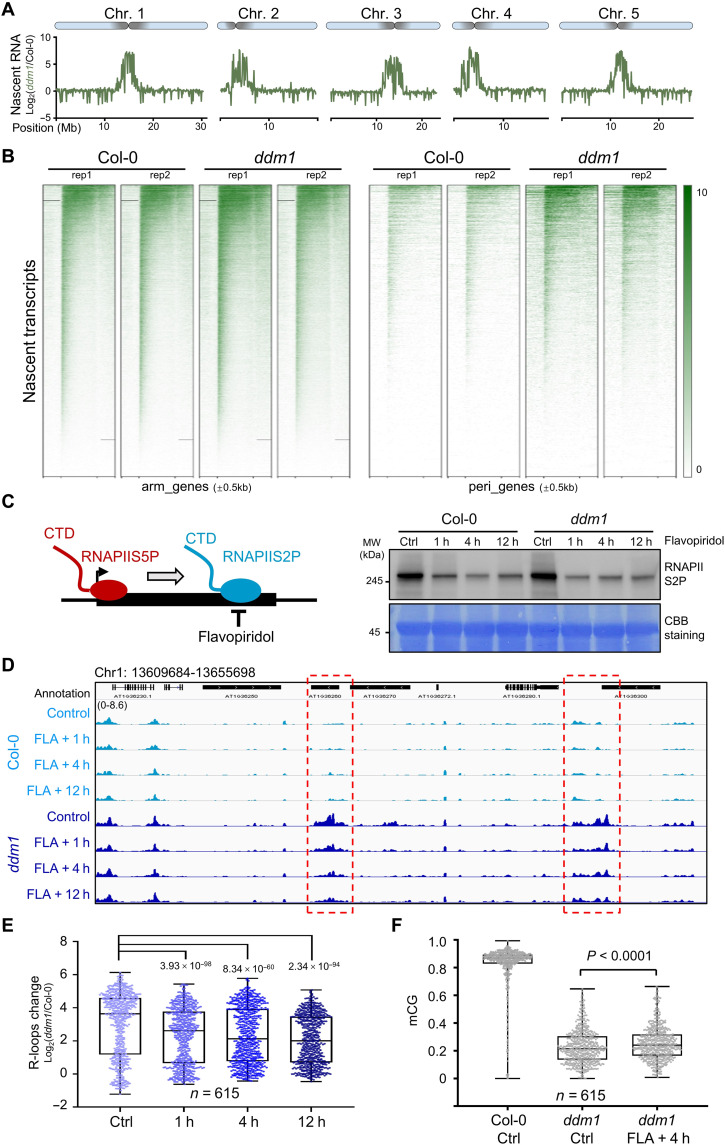
To test whether the cotranscriptional R-loops occur in ddm1 mutant, we analyzed the changes in transcription efficiency in ddm1 via the pNET-seq (plant native elongating transcript sequencing) experiment by using the antibody specific for the RNAPII C-terminal domain (CTD) (33, 34). When compared to Col-0, there is no substantial difference of transcription in arm regions; however, a marked transcription burst was observed in ddm1, which was highly and peculiarly enriched at the pericentromeric heterochromatin (Fig. 2, A and B) and colocalized with accumulated R-loops (Fig. 1A). To determine whether the accumulated R-loops in ddm1 derive from transcription burst, we characterized R-loop levels using an inhibition analysis with flavopiridol (FLA), a chemical that is widely used to inhibit RNAPII transcription elongation (Fig. 2C) (35, 36). FLA treatment indeed decreased phosphorylation levels of RNAPII CTD at serine 2 (RNAPIIS2P), starting within 1 hour after treatment (Fig. 2C). Using ssDRIP-seq, we found that FLA treatments significantly reduced the R-loop levels in ddm1 (Fig. 2, D and E). On the basis of these data, we concluded that cotranscriptional R-loops generated by RNAPII are eliminated by DDM1 at pericentromeric heterochromatin.
Fig. 2. Cotranscriptional R-loop accumulation disturbs pericentromeric heterochromatin formation in ddm1 mutant.
(A) Nascent RNA enrichment [log2NET-seq(ddm1/Col-0)] plotted on 5 chromosomes. (B) Heat maps showing pNET-seq of WT and ddm1 at genes on chromosome arms (n = 26,106) and from pericentromeric regions (n = 7217). The color bar shows RPGC (reads per genomic content, 1× normalization) values. (C) Left: Cartoon showing the chemical FLA is used to inhibit phosphorylation of RNAPII CTD at serine 2 (RNAPIIS2P). Right: Western blot showing RNAPIIS2P abundance after FLA treatment. Plants treated with dimethyl sulfoxide for 12 hours were used as controls. Membrane was stained with CBB (Coomassie Brilliant Blue) as a loading control. MW, molecular weight. (D) Snapshots from Integrative Genomics Viewer showing R-loop levels after FLA treatment. Regions with decreased R-loop levels during FLA treatment are highlighted in red boxes. (E) Box and whisker plot showing R-loop changes Log2(ddm1/Col-0) after FLA treatment at regions with increased R-loops in ddm1 (data from Fig. 1A, n = 615). Minimum to maximum and interquartile range are shown. (F) Box and whisker plot showing CG DNA methylation levels after 4 hours of FLA treatment. Min to max and interquartile range are shown.
To further investigate whether changes in R-loops are correlated with changes in histone and DNA modifications in ddm1, we performed native histone chromatin immunoprecipitation and sequencing (ChIP-seq) and bisulfite sequencing in Col-0 and ddm1 (fig. S4A). When comparing the alterations of R-loops (ssDRIP-seq data; Fig. 1A) and heterochromatic marks H3K9me2 and H3K27me1 (fig. S4A), we found that pericentromeric regions with elevated levels of R-loops in ddm1 showed a marked decrease in H3K9me2 and H3K27me1 levels in ddm1 (fig. S4A). In addition, ddm1 had considerably lower levels of DNA methylation in all sequence contexts, especially in CG and CHG at pericentromeric regions (fig. S4A). We also investigated the differences in chromatin accessibility between Col-0 and ddm1 using ATAC-seq (transposase-accessible chromatin with high-throughput sequencing) (37). Our results showed that the chromatin state of ddm1 is much less compacted than that of Col-0 in pericentromeric regions (fig. S4C). These results suggest that accumulated R-loops in ddm1 are inversely related to the deposition of heterochromatic marks such as H3K9me2, H3K27me1, DNA methylation, and high nucleosome density (fig. S4F). The R-loop depression by FLA treatment could partially recover DNA methylation (but not H3K9me2) where there are accumulated R-loops in ddm1 (Fig. 2F and fig. S4, D and E), together with the fact that ddm1atrxr5/6 has comparable R-loop levels to ddm1 single mutant on the tested loci (fig. S3), indicating that DDM1 could be epistasis to the heterochromatin modifiers (such as H3K27me1 methyltransferases ATXR5/ATXR6) on R-loop clearance, and the R-loop accumulation could disturb pericentromeric heterochromatin marks deposition such as DNA methylation, thus disturbing pericentromeric heterochromatin formation.
DDM1 associates with pericentromeric heterochromatin and restricts R-loop accumulation
To investigate the direct chromatin localization of DDM1, we generated FLAG-tagged complementation lines (DDM1-FLAG/ddm1; fig. S5A). ChIP-seq data with FLAG antibody showed that DDM1-enriched reads were particularly localized at pericentromeric regions of all five chromosomes (fig. S4B), in line with the subnuclear localization of DDM1–green fluorescent protein (GFP) (38). Moreover, DDM1-associated regions exhibit higher R-loop levels in ddm1 mutant compared to Col-0 (fig. S5B). RNA immunoprecipitation (RIP) and quantitative polymerase chain reaction (qPCR) results showed that DDM1 could bind to the RNAs from pericentromeric loci where the R-loop levels are highly accumulated in the ddm1 mutant (fig. S5C). These results suggest that DDM1 physically binds to constitutive pericentromeric regions and might restrict R-loops locally.
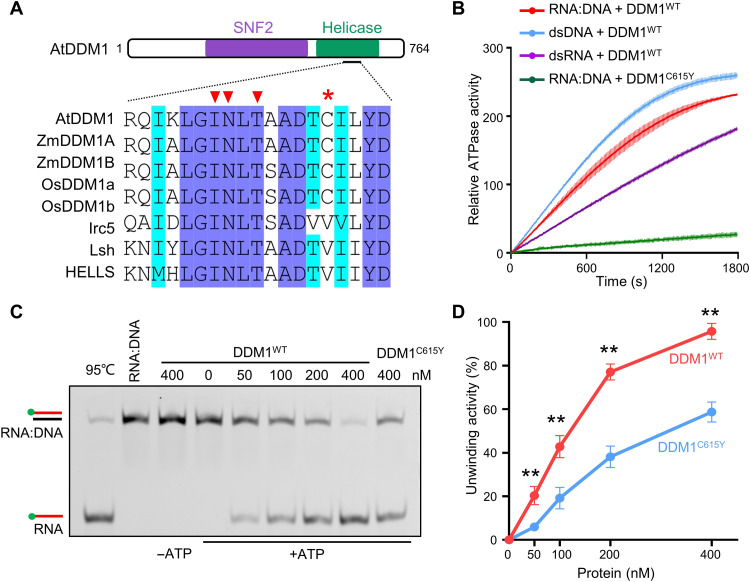
DDM1 is a chromatin remodeler and contains two domains, an N-terminal SNF2 and a C-terminal helicase region (Fig. 3A and fig. S6) (9, 13, 39). Given that chromatin remodelers have been reported to be associated with R-loop abundance in vivo (40–43) [reviewed in (44)], we decided to investigate the possibility of DDM1 in removing RNA:DNA hybrids and R-loops. First, we performed ATP hydrolysis assays to examine adenosine triphosphatase (ATPase) activity of DDM1 using different forms of nucleic acid as substrate. Results showed that the ATPase activity of recombinant DDM1 was stimulated by RNA:DNA, and this stimulation is slightly lower than dsDNA but higher than double-stranded RNA (dsRNA) substrate (Fig. 3B). While the protein DDM1C615Y, the mutant that mimics the ddm1-1 plant carrying a C615Y mutation at a conserved position in DDM1 orthologs among land plants (Fig. 3A and fig. S6D), showed less ATPase activity in the presence of RNA:DNA hybrid substrate, suggesting a possibility that C615 residue is essential for ATP hydrolysis of DDM1, consistent with the fact that C615 is very close to the conserved ATP binding sites of the protein (Fig. 3A).
Fig. 3. DDM1 resolves RNA:DNA hybrids in vitro.
(A) Alignment of amino acid sequences nearby C615Y mutation site among DDM1 orthologs. The asterisk indicates the mutation site (C615Y) in the ddm1-1 mutant. The triangles refer to the predicted ATP binding sites, which is conserved within DDM1 orthologs. (B) ATPase activities of DDM1WT under the substrate of RNA:DNA, dsDNA and ssDNA, respectively, and ATPase activities of mutant DDM1C615Y under the substrate of RNA:DNA. All data points are shown by mean ± SD with two replicates. (C) Helicase assay showing unwinding of RNA:DNA hybrids by WT and mutant DDM1. (D) A graph showing quantification of helicase activity of WT and mutant DDM1. Values from two replicates (fig. S7B) are plotted. **P < 0.05 by two-tailed Student's t test.
Sequentially, we detected that DDM1 could unwind RNA:DNA hybrid in an ATP-dependent manner (Fig. 3, C and D, and fig. S7B), while the helicase activity of DDM1C615Y showed less than half efficiency relative to wild-type DDM1 (Fig. 3, C and D, and fig. S7B). We then purified the truncated DDM1N and DDM1C and performed the helicase assay, and results showed that DDM1N is responsible for resolving RNA:DNA hybrids (fig. S7C). Parallelly, using a cotranscriptional formed R-loop system (fig. S8, A and B), we found that purified His-tagged DDM1 was able to release RNAs from the cotranscriptional R-loops (fig. S8C). Consistently, the C615Y mutation in the C-terminal region markedly slowed down the resolving process (fig. S8C). Together with the results of DDM1 ChIP-seq, ssDRIP-seq, and bisulfite sequencing upon FLA treatment, our findings suggested that DDM1 can remove the RNA from RNA:DNA structures and clear the cotranscriptional R-loops for further pericentromeric marks formation, such as DNA methylation.
DDM1 interacts with histone H2A variants but blocks H2A.Z deposition in pericentromeric heterochromatin
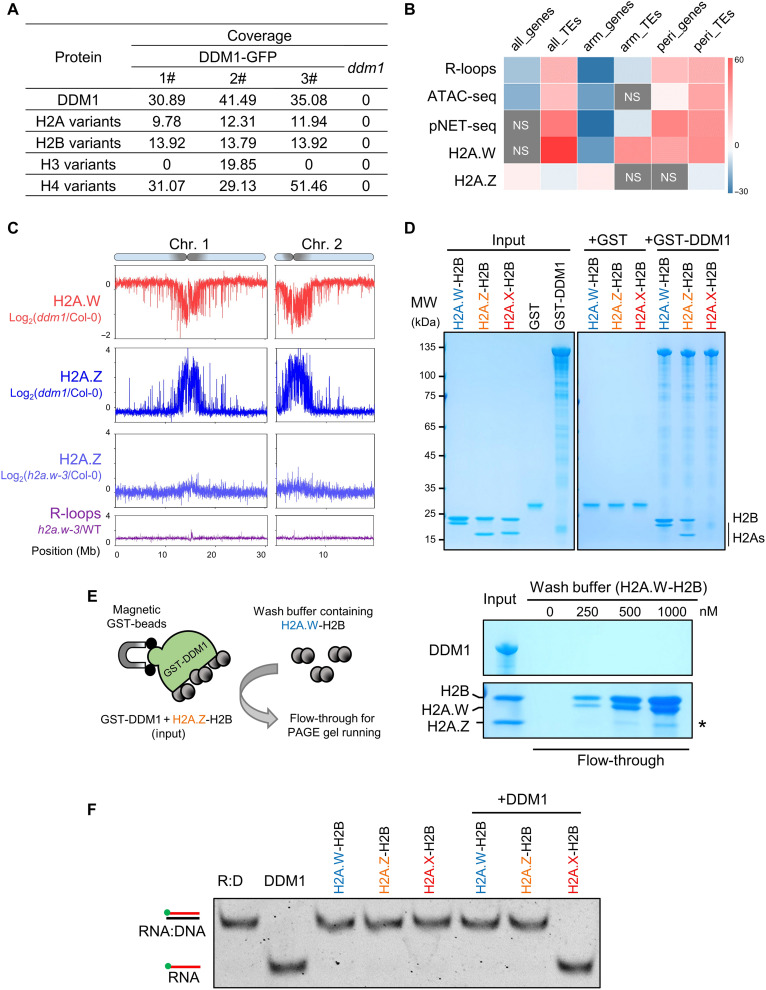
To dissect the role of DDM1 in promoting constitutive heterochromatin formation at pericentromeres, we sought to identify the proteins that interact with DDM1 using mass spectrometry (MS) after GFP immunoprecipitation of DDM1-GFP complementation plants (fig. S5, D to G). We noticed that peptides from histone proteins were enriched in multiple DDM1-GFP coimmunoprecipitation experiments but not in control experiments (Fig. 4A and data S2 to S5), which is consistent with the function of DDM1 in remodeling nucleosomes in vitro (13). Given that histone variant H2A.W is associated with heterochromatin in Arabidopsis (45), we focus on H2A variants among DDM1 interactome. Histone 2A has four variants in Arabidopsis (H2A, H2A.Z, H2A.W, and H2A.X) (45), and we could not determine which H2A variants directly interacts with DDM1 through the identified peptides from MS (red color boxes in fig. S9). We confirmed that as shown previously (15, 45), H2A, H2A.X, and H2A.Z are mainly present in chromosome arms, while H2A.W is exclusively present in pericentromeric heterochromatin (fig. S10, A and B), similar to the pattern of DDM1 and pericentromeric R-loops (Fig. 4B and fig. S4B). H2A.Z showed a relative decline in H2A.W-enriched regions (Fig. 4B and fig. S9B), and results of reciprocal coimmunoprecipitation in plants verified the interaction between DDM1 and H2A.W (fig. S11), implying that DDM1 is linked with the inverse localization of H2A.W and H2A.Z on chromatin.
Fig. 4. DDM1 interacts with H2A.W to promote pericentromeric heterochromatin maintenance.
(A) Peptides from histone 2A variants are detected after coimmunoprecipitation with DDM1-GFP followed by mass spectrometry (IP-MS). Three independent DDM1-GFP transgenic lines were used for GFP–IP-MS. ddm1 was used as a negative control. (B) Permutation test of colocalization among indicated sequencing peaks and annotated total/chromosome arm-located/pericentromeric genes and/or TEs. H2A.W ChIP-seq is from the previous data (45). The color bar scale indicates permutation z-score. NS, not significant. (C) The top shows H2A.W changes in ddm1 relative to WT, H2A.W.6-FLAG/ddm1, and H2A.W.6-FLAG/Col-0 transgenic plants that were used for H2A.W ChIP-seq. The second and third show H2A.Z changes in ddm1 and h2a.w triple mutants, respectively, relative to WT. Values in ChIP-seq are log2(fold change). The bottom shows R-loop changes (h2a.w-3/WT) from ssDRIP-seq. All data are averages of two independent biological replicates. (D) GST pull-down assays showing GST-DDM1 has binding affinities to H2A.W-H2B and H2A.Z-H2B dimers. (E) Left: Schematic representation showing the procedures of competitive pull-down experiment: H2A.Z-H2B dimer was incubated with GST-DDM1 to form DDM1-H2A.Z-H2B complex as input, then wash buffer containing different amounts of H2A.W-H2B dimers was used to be incubated with DDM1-H2A.Z-H2B complex. After wash, flow-through was collected for PAGE gel running and staining. Right: Wash buffer containing H2A.W-H2B was used to dissociate DDM1-H2A.Z-H2B complexes. Asterisk indicates the released H2A.Z. (F) RNA:DNA helicase activities of DDM1 were inhibited in the presence of H2A.W-H2B or H2A.Z-H2B dimers.
With the fact that H2A.W is not essential for Arabidopsis development and loss of H2A.W has a modest effect on DNA methylation or transcription of pericentromeric TEs (15), where the marked decrease of DNA methylation occurs in ddm1 mutant (fig. S4A), the functions of DDM1 in pericentromeric heterochromatin formation could not be explained by the DDM1-H2A.W interaction. On the basis of the results that (i) DDM1 is enriched at pericentromeric regions (fig. S4B), (ii) there is little-to-no pericentromeric enrichment of H2A.Z observed in the H2A.W fully depleted mutant h2a.w-3 (Fig. 4C and figs. S10D and S12), which is consistent with published data in h2a.w-2 mutant (fig. S10, B and C) (15), and (iii) H2A.X and H2A could replace the H2A.W at pericentromeric regions in h2a.w-2 (fig. S10, B and C) (15), we assume that the bona fide function of DDM1 could be to block H2A.Z deposition at pericentromeric regions in Col-0 and h2a.w-2 mutant, thus to inhibit the H2A.Z-instigated transcription burst (shown in Fig. 2, A and B). To evaluate the function of DDM1 in depositing H2A.W and H2A.Z on chromatin, we performed H2A.W-FLAG (using FLAG antibody) and H2A.Z [using native H2A.Z antibody (46)] ChIP-seq analysis in Col-0 and ddm1. Consistent with previous data (45), H2A.W was enriched in pericentromeric regions (fig. S13, A and B), and the enrichment of H2A.W in pericentromeric regions was reduced markedly in ddm1 (Fig. 4C and figs. S10D and S13, A and B). H2A.Z was not localized at the pericentromeric regions in Col-0 (fig. S13, A and C). However, in ddm1 mutant, H2A.Z was deposited into the same pericentromeric regions where H2A.W had decreased (Fig. 4C and fig. S13, B to E). Together, these results genetically proved our hypothesis that DDM1 promotes H2A.W but blocks H2A.Z deposition at pericentromeric regions.
H2A.Z eviction relies on the affinities of DDM1 and H2A variants
Our pull-down assay showed that DDM1 could bind to H2A.W and H2A.Z (Fig. 4D). To further determine the binding affinities of H2A.W and H2A.Z to DDM1, we performed a competitive pull-down experiment (Fig. 4E, see Methods). We found that the dissociated H2A.Z from DDM1-H2A.Z complex was increased when increasing the amounts of H2A.W in wash buffer (Fig. 4E and fig. S14A), while H2A.Z could not affect the binding of DDM1 to H2A.W (fig. S14B), suggesting that DDM1 preferably binds to H2A.W. These results confirmed that H2A.W was able to disrupt the interaction of DDM1-H2A.Z, which was corresponding to the chromosomal distributions of these proteins that DDM1 and H2A.W were enriched in, while H2A.Z was excluded out of the pericentromeric regions (Fig. 4B and fig. S13A). Together with the genetic evidence that the loss of DDM1, rather than loss of H2A.W, results in aberrant enrichment of H2A.Z in pericentromeric regions (Fig. 4C and fig. S10, C and D), we concluded that the H2A.Z eviction from pericentromeric regions was in a DDM1-dependent manner.
To further dissect the genetic relationship between R-loop resolution and H2A.W/Z deposition by DDM1, we added recombinant histone variants into the RNA:DNA helicase assay (see Methods). The RNA:DNA helicase activity of DDM1 was significantly blocked by either H2A.W or H2A.Z (Fig. 4F), while the control proteins H2A.X and GST could not affect the helicase activity of DDM1 (Fig. 4F and fig. S14D). In vivo, we quantified the changes in pericentromeric R-loops in h2a.w-3 compared to that in Col-0 through ssDRIP-seq, and the results showed there were almost no differences of R-loop levels in h2a.w-3 (Fig. 4C and fig. S10D, bottom). Together with the FLA inhibition results (Fig. 2, D and E), these data indicated that DDM1-facilitated cotranscriptional R-loop resolution is strictly separated with and genetically upstream of H2A variants deposition.
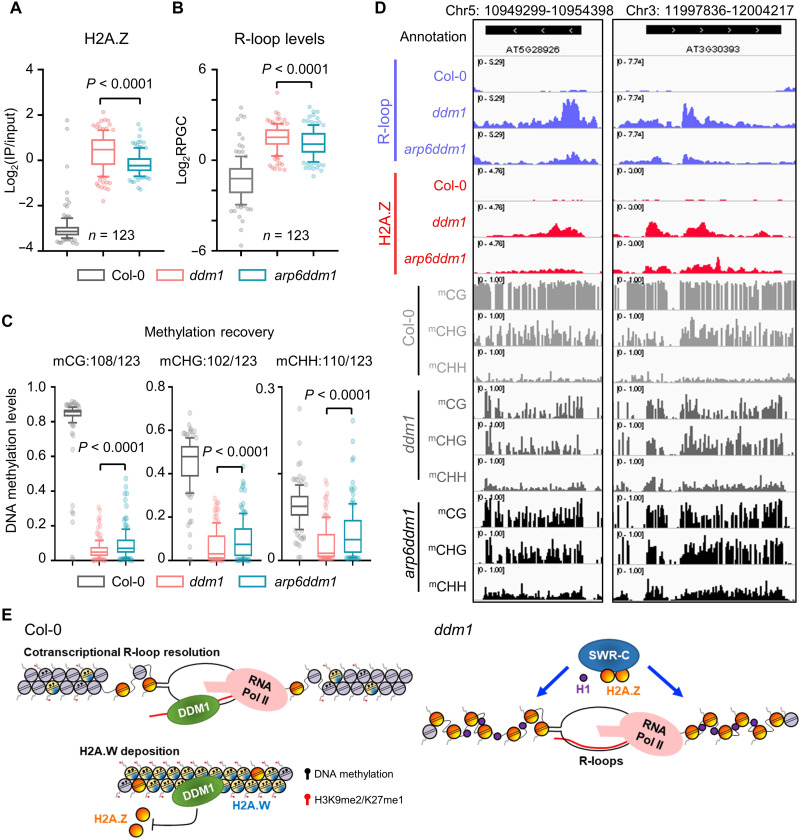
Suppression of H2A.Z deposition in pericentromeric regions rescues the R-loops and DNA methylation defects in ddm1 mutant
The deposition of H2A.Z is mediated by the chromatin remodeler SWR1-C (47). To test whether disruption of SWR1-C–mediated H2A.Z incorporation could alleviate the R-loop accumulation and DNA methylation defects in ddm1 mutant, we respectively introduced the mutation of three key components of SWR1-C (ARP6, SWC6, and PIE1) into ddm1 by either crossing with arp6 or CRISPR-Cas9–mediated deletion of the SWC6 and PIE1 genes (fig. S15, A and B) and performed H2A.Z ChIP-seq, ssDRIP-seq, and bisulfite sequencing. We identified that 123 up-regulated pericentromeric R-loop peaks in ddm1 (total 615 up-regulated R-loop peaks, shown in Fig. 2E) showed decreased H2A.Z abundance in arp6ddm1 double mutant (Fig. 5A). The global decreased R-loop levels on these peaks were observed (Fig. 5B), which is accompanied by gain of DNA methylation levels (Fig. 5, C and D). Similar dynamic patterns can be detected in swc6ddm1 and pie1ddm1 (fig. S16). In addition, several tested pericentromeric loci displayed decreased R-loops and partially recovered DNA methylation in the H2A.Z fully depleted mutant ddm1h2a.z (ddm1 hta8/9/11 quadruple mutant; fig. S15C) compared to ddm1 single mutant (fig. S17). These results clearly showed that inhibition of H2A.Z incorporation at pericentromeric region can rescue the defects of R-loops and DNA methylation in pericentromeric heterochromatin in ddm1 mutant, genetically consistent with our results (fig. S4, A and C).
Fig. 5. Suppression of H2A.Z deposition in pericentromeric regions rescues the R-loops and DNA methylation defects in ddm1 mutant.
(A) One hundred twenty-three up-regulated pericentromeric R-loop peaks of ddm1 showed decreased H2A.Z abundance in arp6ddm1. Ten to 90% and interquartile range are shown. (B) R-loop levels of 123 decreased H2A.Z peaks in Col-0, ddm1, and arp6ddm1. Ten to 90% and interquartile range are shown. (C) DNA methylation levels of 123 decreased H2A.Z peaks in Col-0, ddm1, and arp6ddm1. Ten to 90% and interquartile range are shown. (D) Snapshots from Integrative Genomics Viewer showing R-loop, H2A.Z, and DNA methylation levels in Col-0, ddm1, and arp6ddm1. (E) Constitutive heterochromatin formation starts from DDM1-mediated cotranscriptional R-loop clearance. The low but detectable levels of cotranscriptional R-loops formed at pericentromeric heterochromatin stimulate the RNA:DNA helicase activity of DDM1, and DDM1 promotes the deposition of H2A.W but excludes the H2A.Z, thus promoting constitutive heterochromatin formation. In the absence of DDM1, SWR complex–mediated anomalous H2A.Z deposition introduces enormous cotranscriptional R-loops and disturbs the pericentromeric heterochromatin. Histone Linker H1 could help the SWR complex to deposit H2A.Z at constitutive heterochromatin in the ddm1 mutant.
Previous results had shown that the depletion of linker histone H1 could partially rescue the defects of DNA methylation in ddm1 (11). When comparing the H2A.Z abundance in h1ddm1 to ddm1, we detected the reduced H2A.Z abundance in pericentromeric regions (fig. S18, A and B). Similar to the swr1ddm1 and ddm1h2a.z mutants (Fig. 5 and figs. S16 and S17), reduced H2A.Z in h1ddm1 results in decreased R-loops and recovery of DNA methylation (fig. S18, C to E). These results further enhanced our model of DDM1 and raised a possibility that the linker histone H1 could help pericentromeric H2A.Z deposition in the ddm1 mutant (Fig. 5E). It had also been reported that mutation of MET1, the de novo DNA methyltransferase, could increase the H2A.Z deposition (48). Our H2A.Z ChIP-seq data also showed that H2A.Z levels were increased across the genome in the absence of MET1, not only restrictedly in pericentromeric regions (fig. S18A, black lines), which is different from the patterns of increased pericentromeric H2A.Z in ddm1. Because de novo DNA methylation is the fasten step to firmly brake transcription and maintain heterochromatin silencing, thus genome-wide depletion of DNA methylation could disrupt the transcription silencing and introduce anomalous H2A.Z deposition (fig. S18B) (48). Genome-wide R-loop profile in met1 mutant has shown a much higher abundance of R-loop to all the tested mutants except ddm1 (about three- to fivefold lower than that in ddm1 mutant; fig. S2). These data are in line with the RNA:DNA helicase activity of DDM1: As in met1, the DDM1 is still functional; however, without the brake action of DNA methylation in the initial transcription step of heterochromatin silencing, the helicase activity of DDM1 could not cope with these massive transcription bursting caused R-loops (Fig. 5E). The function of DDM1 in R-loops removal is noneffective in the case of disruption of de novo DNA methylation, thus leaving a noticeable amount of R-loops in met1 (fig. S2).
Dual functions of DDM1 in R-loop resolution and H2A.Z eviction are important for tricellular nuclei development in pollen
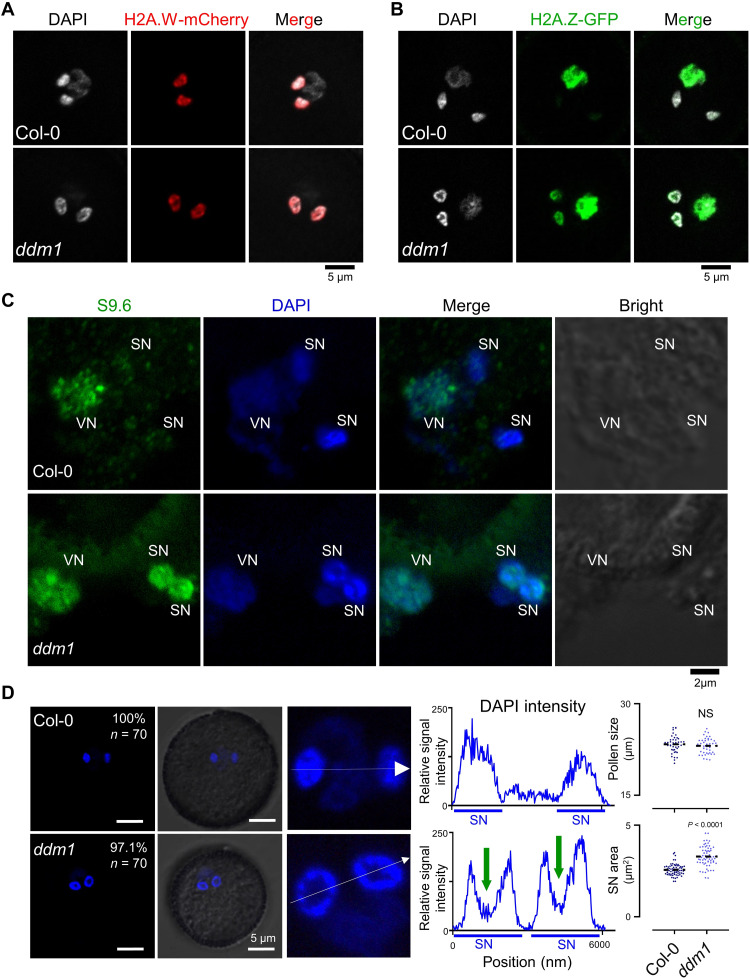
In pollen development, the tricellular nuclei stage contains one vegetative nucleus (VN) and two SN (49), and DDM1 was only accumulated in SN (fig. S19A) (50). Thus, the mature pollen is the ideal model to dissect the biological functions of DDM1 in heterochromatin regulation. After introducing constructs expressing the H2A.W or H2A.Z fused with fluorescent protein into Col-0 and ddm1, we observed that H2A.W was present in the SN but excluded in the VN, similar to the localization of DDM1 (Fig. 6A and fig. S19A), while H2A.Z was enriched in the VN where there is less abundance of DDM1 (Fig. 6B). Unexpectedly, in the absence of functional DDM1, H2A.Z were present both in the SN and VN (Fig. 6B); however, the cellular localization of H2A.W was not changed in ddm1 pollens (Fig. 6A). These results of single-cell analysis reinforce the function of DDM1 in blocking the H2A.Z deposition. Next, we sought to determine whether any R-loop differences could be found in maturated pollen when comparing Col-0 with ddm1. By staining the nuclei with 4′,6-diamidino-2-phenylindole (DAPI) and R-loops with antibody S9.6, we found that, in Col-0, R-loop signals were enriched in the VN but absent in SN (Fig. 6C and fig. S19, B and C). However, in the ddm1 pollen, the R-loop signals were present in all three nuclei (Fig. 6C). Thus, we concluded that DDM1 is required in mature pollen for maintaining the proper exclusion of R-loops and H2A.Z from SN.
Fig. 6. DDM1 facilitates R-loop resolution and H2A.Z eviction during tricellular nuclei development in pollen.
(A) Microscopic images showing H2A.W.6-mCherry in intact pollen from Col-0 and ddm1-1. Scale bar, 5 μM. (B) Microscopic images showing H2A.Z.9-GFP in intact pollen from Col-0 and ddm1-1. Scale bar, 5 μM. (C) Immunostaining detection of R-loops in tricellular nuclei of Arabidopsis pollen. At least 10 tricellular nuclei were observed for each sample. R-loops in nuclei were stained with S9.6 antibodies (green). Nuclear DNA was stained with DAPI (blue). Scale bar, 2 μM. (D) Left: DAPI staining of tricellular nuclei in intact pollen. Scale bars, 5 μM. Middle: Relative DAPI intensity of SN (white lines on left) was quantified by Zeiss Zen software, and a nuclear valley could be observed in each nucleus. Green arrows indicate regions with less DAPI staining in ddm1 SN. Right: Quantitative analysis of pollen size (diameter) and SN area of WT and ddm1 pollen. All data are plotted and means of value are shown.
While preparing the nuclei from mature pollen for the staining assays, we observed that the shape and morphology of SN in the ddm1 were markedly different from Col-0 (Fig. 6, A and B, DAPI panels). We further characterized this observation and found that the SN in intact pollen was significantly larger in ddm1 than in Col-0 (Fig. 6D). A “nuclear valley” could be seen in almost all SN from ddm1 pollen, which was absent in wild-type Col-0 (Fig. 6D). Calculation of the “nuclei valleys” showed a range from 25 to 45% of the total SN nuclei area (Fig. 6D). In addition, the h2a.w-3 mutant did not show the abnormal nuclear valley phenotype (fig. S19D), genetically providing another layer of the clue that the DDM1-promoted exclusion of H2A.Z and R-loops from SN, rather than the H2A.W localization into SN, has the biological roles to ensure the tricellular nuclei development.
DISCUSSION
A model for DDM1 in helping DNA methyltransferases access nucleosomes for DNA methylation and transcriptional silencing had been proposed (11, 12). Here, we reported the additional function of DDM1 in R-loop clearance and linked it to heterochromatin formation. On the basis of our R-loop profiling screen data, we proposed that, in pericentromeric heterochromatin, the low abundance of H2A.Z could produce low levels of cotranscriptional R-loops, which then could be eliminated by the primary function of DDM1 via the helicase activity (Fig. 5E). Links between R-loops/RNA:DNA hybrids and heterochromatin have been revealed in some biological processes. For instance, R-loop–induced heterochromatin marks are efficient for transcriptional termination (51); major satellite repeat transcript–mediated RNA:DNA hybrids stabilize chromatin retention of histone methylases Suv39h for heterochromatin formation (52); suppression of R-loops ensures proper DNA replication of highly condensed heterochromatin region (41). RNA:DNA hybrids could help the RNA interference–directed heterochromatin assembly in fission yeast (53). Here, considering the marked elevated pericentromeric R-loop levels in the ddm1 mutant (Fig. 1 and fig. S2), together with the results that DDM1-mediated H2A.Z eviction from pericentromeric regions (Fig. 4C and fig. S13A), we propose that DDM1 could function in early stage of heterochromatin silencing in Arabidopsis (see below; Fig. 5E).
Compared to the dsDNA substrate, there is almost no activity of plant DNA methyltransferases on single-stranded DNA or RNA:DNA hybrids (54). Our data showed that DDM1 has the RNA:DNA helicase activity in vitro and thus ensures the cotranscriptional pericentromeric R-loops clearance, further providing the fine substrate for DNA methyltransferases. The model of DDM1 promoting R-loops clearance also explains most of the phenotypic observations in the ddm1 mutant. For example, loss of function of DDM1 in Arabidopsis and its orthologs in other species causes DNA damage, defects in DNA repair, as well as increases in DNA recombination (55–59), and all these phenomena are similar to those reported mutants that contain an overabundance of R-loops (16, 18–20, 60–62). DDM1 was first isolated with the function of maintaining DNA methylation, and it is conserved among different species (fig. S6). The loss of helicase, lymphoid specific (HELLS), the ortholog of DDM1 in mammals, results in RNA:DNA hybrids accumulation at pericentromeric repeats (32), suggesting the similar functions of HELLS in R-loop resolution as the DDM1 in Arabidopsis. However, the DDM1 homolog in fungus Neurospora crassa mutagen sensitive-30 (MUS-30) is not responsible for DNA methylation maintenance (58). Instead, MUS-30 is required for genome stability, and loss of function of MUS-30 presented similar phenomena (e.g., hypersensitivity to DNA damage) to mutants with R-loop accumulation.
With the results of protein-protein interactions and ChIP-seq, we proposed that DDM1 is required for the histone H2A variant H2A.Z exclusion from pericentromeric regions, as well as for the heterochromatic H2A.W deposition. Our findings showed that both H2A.Z and H2A.W inhibit the helicase function of DDM1 (Fig. 4), while there are no R-loop differences in the h2a.w (Fig. 4C and fig. S10D) but R-loop and H2A.W enrichment are both altered in ddm1 (Figs. 1 and 4C and fig. S10D), implying that the R-loop clearance and histone variants deposition by DDM1 could not occur at the same time. In the absence of H2A.W, the histone H2A and H2A.X, but not H2A.Z, could be enriched in pericentromeric regions (fig. S10, B and C) which is regulated by unclear mechanisms, suggesting that the bona fide function of DDM1 is to evict the H2A.Z from pericentromeric heterochromatin (Fig. 5E). On the basis of these facts, our results revealed a model whereby DDM1 has functions in facilitating the constitutive pericentromeric heterochromatin formation in Arabidopsis: Initially, a low abundance of H2A.Z could generate modest but detectable levels of cotranscriptional R-loops at pericentromeric heterochromatin, which further stimulates the RNA:DNA helicase activity of DDM1 and then promotes DDM1-mediated H2A.W deposition, following with justified nucleosome density and facilitates the heterochromatic modifications (DNA and histone methylation), with the results of established constitutive pericentromeric heterochromatin formation and evicted H2A.Z-instigated transcription (Fig. 5E).
Together, the results presented in this study highlight the essential role of cotranscriptional R-loop clearance in promoting constitutive heterochromatin silencing and patterning the heterochromatin landscape. The findings suggest an evolved model of constitutive heterochromatin formation, in which nascent RNAs directly pair with templated DNA to trigger heterochromatin packing and silencing (Fig. 5E). DDM1 clears the low levels of R-loops that cotranscriptional formed in pericentromeric heterochromatin through its RNA:DNA removing activity, followed by depositing proper histone H2A variants. This in turn allows DNA methyltransferases and other histone modifiers to access pericentromeric heterochromatin for transcriptional silencing and heterochromatin maintenance (Fig. 5E). The notable interplays that we have uncovered between R-loop resolution and histone variants deposition/eviction in heterochromatin formation could be considered as a paradigm for other eukaryotic organisms.
MATERIALS AND METHODS
Plant material and growth conditions
All mutants used in this study are Columbia ecotype (Col-0), including ddm1-1 (7), ddm1-2 (9), ddm1-T (SALK_000590), hta6 hta7 (45), arp6-1 (SAIL_599_G03), h1.1 (SALK_128430), h2a.z (FLAG_593B04 for hta8, SALK_054814 for hta9, and SALK_017235 for hta11). Mutants for ssDRIP-seq screening are listed in data S1. We used CRISPR-Cas9 genome editing system to obtain genomic DNA deletion mutants, including ddm1CRISPR, hta12, pie1, swc6, and h1.2. We used second-generation homozygous ddm1-1 mutants obtained from the segregated ddm1/DDM1 heterozygous for all the experiments. Surface sterilized seeds were grown on Murashige and Skoog media (Sigma-Aldrich, M5519) for 12 days at 22°C with 16-hour light/8-hour dark cycle. For FLA treatment, 10-day-old seedlings were transferred to liquid MS media containing 10 μM FLA and incubated for 1, 4, and 12 hours.
Transgenic plants
To generate the DDM1-FLAG and DDM1-GFP complementation lines, DDM1 genomic sequence containing native promoter and terminator was amplified from genomic DNA and inserted into pCambia1301. For DDM1-FLAG, FLAG-tag was inserted at both the N and C terminus of DDM1 coding regions, and the C-terminal FLAG-tag was replaced with GFP-tag for DDM1-GFP. Constructs were transformed into ddm1/DDM1 heterozygotes by using the floral dip method, and transgenic plants containing the transgene were screened by selective resistance.
For H2A.W-FLAG/mCherry lines, H2A.W.6 (HTA6 and AT5G59870) genomic DNA containing native promoter and terminator was amplified and cloned into pCambia1300. FLAG-tag and mCherry-tag were inserted at the C terminus of the H2A.W.6 coding region, respectively. Construct was transformed into ddm1/DDM1, and segregated wild-type and ddm1 lines from T1 generation were used for H2A.W-FLAG ChIP-seq and H2A.W.6-mCherry observation in pollen nuclei. For H2A.Z-GFP lines, H2A.W.9 (HTA9 and AT1G52740) genomic DNA containing native promoter and terminator was amplified and cloned into pBINPLUS, and transformation was performed as described above.
Single-strand DNA ligation–based library construction from RNA:DNA hybrid immunoprecipitation, followed by sequencing
ssDRIP-seq was performed as previously described (22, 25). Briefly, nuclei were extracted from 12-day-old seedlings with Honda buffer [0.44 M sucrose, 1.25% Ficoll, 2.5% Dextran T40, 20 mM Hepes-KOH (pH 7.4), 10 mM MgCl2, 0.5% Triton X-100, and protease inhibitors]. DNA was recovered with phenol:chloroform extraction after digesting the nuclei with 0.5% SDS and proteinase K at 37°C overnight. Purified DNA was then digested with Mse I, Nla III, Dde I, and Mbo I [New England Biolabs (NEB)] by following the manufacturer’s instructions. Qubit was used to measure the DNA concentration, and 3 μg of DNA was used to perform the immunoprecipitation with S9.6 overnight. DNA-antibody complexes were incubated with Protein G beads (Invitrogen, 10004D) at 4°C for 5 hours with rotation. DNA was then eluted from washed beads with elution buffer [50 mM tris-HCl (pH 8.0) and 10 mM EDTA] and proteinase K at 55°C for 1 hour. After phenol:chloroform extraction, DNA was recovered by ethanol precipitation. The eluted DNA was used for sequencing library constructions using an ssDNA library preperation kit (Vazyme, ND620).
Chromatin immunoprecipitation and sequencing
For histone ChIP-seq (H3K9me2, H3K27me1, H2A.W-FLAG, and H2A.Z), nuclei were extracted from 12-day-old seedlings with Honda buffer [0.44 M sucrose, 1.25% Ficoll, 2.5% Dextran T40, 20 mM Hepes-KOH (pH 7.4), 10 mM MgCl2, 0.5% Triton X-100, 5 mM dithiothreitol (DTT), and protease inhibitors]. Nuclei were suspended in 1× micrococcal nuclease (MNase) buffer [50 mM tris-HCl (pH 7.6), 5 mM CaCl2, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF)] containing 1× protease inhibitor cocktail. After RNase A treatment at 37°C for 30 min, chromatin was digested with micrococcal nuclease (NEB, M0247S) for 6 min, and the reaction was stopped with EDTA (10 mM). Then, the nucleosomes were released with the addition of 0.005% SDS while rotating at 4°C for 2 to 3 hours. Next, the supernatant was separated and diluted with dilution buffer (0.1% Triton X-100, 50 mM NaCl, 0.1 mM PMSF, 17 mM tris-HCl, 3.33 mM EDTA, and 1× protease inhibitor cocktail). Chromatin was precleaned with protein G beads and immunoprecipitated with H3 (ABclonal, A2348), H3K9me2 (Abcam, ab1220), H3K27me1 (Millipore, 07-448), FLAG M2 beads (Sigma-Aldrich, M8823), and H2A.Z (46) antibodies at 4°C overnight. The complexes were incubated with protein G beads (Invitrogen, 10004D) at 4°C overnight with rotation and washed three times with wash buffer having different concentrations of NaCl in each wash [50 mM tris-HCl (pH 7.6), 10 mM EDTA, 50/100/150 mM NaCl, 0.1 mM PMSF, and protease inhibitors]. Then, DNA was eluted with elution buffer (0.1% SDS and 0.1 M NaHCO3) at 65°C for 15 min. Proteinase K treatment was performed at 55°C for 2 hours. DNA was recovered after phenol:chloroform extraction for sequencing library construction.
For DDM1-FLAG ChIP-seq, the samples were cross-linked with formaldehyde. Then, nuclei extraction, sonication, immunoprecipitation with anti-FLAG beads (Sigma-Aldrich, M8823), washing, and chromatin immunoprecipitated DNA extraction were performed as previously described (63). Eluted DNA was used to make the sequencing libraries as previously described (22) or used for qPCR.
RNA immunoprecipitation
The DDM1-FLAG RIP-qPCR was performed as previously described (64) with some modifications. Because of the low abundance of input RNAs from pericentromeric loci in Col-0 and DDM1-FLAG/ddm1, the genomic DNA from input samples was purified to act as a normalized control. The qPCR primers for the pericentromeric and chromosome arm-located loci are listed in data S7.
Bisulfite sequencing
Nuclear DNA was extracted from 12-day-old seedlings as described in the ssDRIP-seq section and sonicated to 250-bp fragmentation with COVARIS. Next, bisulfite conversion and DNA recovery were performed with the EZ DNA Methylation-Gold Kit (Zymo Research, D5005) by following the manufacturer’s instructions. The sequencing library was made using the EpiArt DNA Methylation Library Kit for Illumina V3 (Vazyme, NE103) by following the manufacturer’s instructions.
Plant native elongating transcript sequencing
pNET-seq was performed as previously described (34). Briefly, 12-day-old seedlings were grounded in liquid nitrogen and resuspended in ice-cold lysis buffer [50 mM Hepes (pH 7.5), 150 mM NaCl, 1 mM EDTA (pH 8.0), 1% Triton X-100, 10% glycerol, 5 mM β-mercaptoethanol, 1 mM PMSF, aprotinin (2 μg/μl), and pepstatin A (2 μg/μl)]. Lysis mixture was filtered and centrifuged to discard the supernatant. Nuclear pellet was washed once with homogenization buffer B (HBB) [25 mM tris-HCl (pH 7.6), 0.44 M sucrose, 10 mM MgCl2, 0.10% Triton X-100, and 10 mM β-mercaptoethanol] and once with homogenization buffer C (HBC) [20 mM tris-HCl (pH 7.6), 0.352 M sucrose, 8 mM MgCl2, 0.08% Triton X-100, 8 mM β-mercaptoethanol, and 20% glycerol] buffers and then resuspended in MNase buffer [20 mM tris-HCl (pH 8.0), 5 mM NaCl, and 2.5 mM CaCl2], supplemented with 20 U of micrococcal nuclease (Takara, 2910A). The reaction mixture was incubated at 37°C for 5 min, and the reaction was stopped with EDTA. After MNase digestion, sonication was performed with 1 s on/off for 10 cycles, and the supernatant was obtained by centrifugation. The supernatant was incubated with antibody (Abcam, ab817)–coated beads for 2 hours at 4°C, and the beads containing the complex were washed eight times with precooled NET-2 buffer [50 mM tris-HCl (pH 7.4), 150 mM NaCl, and 0.05% NP-40]. Last, the beads were resuspended in T4 polynucleotide kinase (T4 PNK) reaction mixture containing 75 μl of T4 PNK buffer, 10 μl of T4 PNK enzymes (NEB, M0236), and 15 μl of 10 mM ATP and incubated at 37°C for 10 min on a Thermomixer (1400 rpm). After that, the supernatant was removed, and beads were washed with NET-2 buffer. The beads were again resuspended in 500 μl of TRIzol and incubated for 10 min at room temperature. After chloroform purification, RNA was precipitated with 1/10 volume of ammonium acetate, 3 volumes of ethanol, and 1 μl of GlycoBlue. The precipitated RNA was run on an 8% polyacrylamide gel electrophoresis (PAGE) gel, and the gel corresponding to 35- to 100-bp RNA was excised. After RNA purification from the gel, the NEXTflex small RNA-Seq Kit V3 was used to construct the libraries. Last, the libraries were recovered from a 6% PAGE gel corresponding to 140 to 250 bp.
Transposase-accessible chromatin with high-throughput sequencing
The ATAC-seq method was modified from previous publications (65, 66). Briefly, 0.3 g of 12-day-old seedlings was freshly collected and ground with a MACHMA-100 Laboratory Homogenizer in ice-cold nuclei purification buffer [20 mM Mops (pH 7.0), 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, 0.2 mM spermine, and protease inhibitor cocktail]. The homogenate was filtered with a 40-μm nylon mesh and then centrifuged to discard the supernatant. The nuclear pellet was sequentially purified with NEB2 [0.25 M sucrose, 10 mM tris-HCl (pH 8.0), 10 mM MgCl2, 1% Triton X-100, and protease inhibitor cocktail], NEB3 [1.7 M sucrose, 10 mM tris-HCl (pH 8.0), 2 mM MgCl2, 0.15% Triton X-100, and protease inhibitor cocktail], and nuclei purification buffer. The nuclear pellet was resuspended in diethyl pyrocarbonate–treated (DEPC-treated) H2O and then was immediately subjected to transposition reaction. Nuclei were fragmented and incubated with 5 μl TTE Mix V5 (Vazyme, TD502) in 10 μl of 2× dimethylformamide (DMF) buffer [66 mM tris-acetate (pH 7.8), 132 mM K-acetate, 20 mM Mg-acetate, and 32% DMF] at 37°C for 30 min. Fragmented DNA was purified with the ChIP DNA Clean & Concentrator Kit (Zymo Research Corporation, D5205). The purified DNA was used for library construction.
Expression and purification of recombinant proteins
The coding sequence of DDM1 was amplified from Columbia (Col-0) cDNA. The amplified PCR fragments were cloned into a pET-28a or pGEX-4 T-1 vector and expressed in Rosetta (DE3) cells. Briefly, Rosetta (DE3) cells were grown at 37°C until optical density at 600 nm reached 0.4 to 0.6, and then isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.5 mM. The cells were then held at 16°C and incubated overnight with shaking. The harvested cells were resuspended in 1× phosphate-buffered saline (PBS) and sonicated (30 s on and 60 s off in cold) until the suspension became transparent). The supernatant was collected and incubated with HisSep Ni-NTA agarose resin (Yeasen, 20502ES50) or GSTSep Glutathione agarose resin (Yeasen, 20507ES50) at 4°C for 2 hours. The agarose resin-protein complexes were washed four times with 1× PBS, and the protein was eluted with elution buffer (250 mM imidazole or 10 mM glutathione in 1× PBS). His-tagged Arabidopsis histones H2B.9, H2A.X.3, H2A.Z.9, and H2A.W.6 were expressed using pET-28a vector and were purified as previously described (14) but without His tag removal. H2B-H2As heterodimers were constructed as previously described (14).
ATPase assays
The ATPase activity measurement was conducted using the EnzChek Phosphate Assay Kit (Thermo Fisher Scientific, E6646). First, three kinds of nucleic acid substrates were prepared (dsRNA, dsDNA, and RNA:DNA; see sequences information in data S6). In the reaction, 200 nM His-DDM1 and 250 nM nucleic acid substrates were added in the buffer of 50 mM tris (pH 7.5), 1 mM MgCl2, 0.2 mM DTT, and 150 mM NaCl. The product of ATP hydrolysis was measured per 10 s.
RNA:DNA unwinding assay
The 23-nt of 6-carboxyfluorescein (6-FAM)–labeled RNA (5′-6-FAM-AAAACAAAAUAGCACCGUAAAGC-3′) and 23-nt of cDNA were annealed in the annealing buffer [20 mM tris-HCl (pH 7.0), 100 mM NaCl, and 1 mM EDTA]. For the helicase reactions, the increasing amount of recombinant protein (0, 50, 100, 200, and 400 nM) was added into each reaction system containing 10 nM RNA:DNA hybrids, and the reaction was incubated at 37°C for 1 hour. Reactions were stopped by adding 5 μl of stop buffer [20 mM Hepes (pH 7.4), 300 mM KCl, 10 mM EDTA, and proteinase K (200 ng/μl)] for 1 hour at 37°C. The product was separated by 15% PAGE gel running, and the FAM signal was detected by Typhoon FLA9500 with a LPR660 FAM filter (GE Healthcare). For Fig. 4F, 400 nM recombinant heterodimers were added into the reactions, and then the assays were performed as described above.
R-loop resolution assay
Cotranscriptional R-loop formation was performed as previously described (67). (TTTAGGG) repeats (~1 kb) were amplified from Col-0 genomic DNA with specific primers (see data S7), and then PCR products were cloned into pEASY-T5-zero (Transgen, CT501). In vitro transcription was performed using T7 polymerase (Promega, P1300) with 0.3 mM ribonucleoside adenosine/cytosine/guanine triphosphate (rATP/rCTP/rGTP), 0.1 mM uridine triphosphate (UTP) and 0.3 mM cyanine5 (Cy5)–labeled UTP (GeneCopoeia, C422B) by adding 5 μg (TTTAGGG) repeat containing plasmid, and the reaction was incubated at 37°C for 60 min and heated at 65°C for 10 min. Transcription products were treated with RNase A in 330 mM NaCl and 30 mM MgCl2 and then were purified by phenol:chloroform extraction. R-loop–containing products were treated by Apyrase (NEB, M0398L) to remove excess ribonucleoside adenosine triphosphate (rATP) and then were purified by phenol:chloroform. After purification though S-400 columns (GE Healthcare, 27514001), R-loop–containing products (R-loop substrate) were dissolved in 100 μl of 100 mM tris-HCl (pH 8.0) and stored at 4°C in the dark.
For R-loop resolving reactions, 1 μl of R-loop substrate was incubated with indicated proteins in 10 μl of system [6.6 mM tris-HCl (pH 7.5), 3% glycerol, 0.1 mM EDTA (pH 8.0), 1 mM DTT, 0.5 mM MgCl2, and 1 mM ATP] at 37°C, and then the reaction was stopped by adding 2 μl of stop buffer [proteinase K (10 mg/ml) and 1% SDS] incubating at 37°C for 15 min. Samples were run on 0.9% agarose tris-borate-EDTA (TBE) gels in 0.5× TBE buffer for 2 hours in the dark. R-loop signal was detected using Typhoon FLA9500 with a LPR660 Cy5 filter (GE Healthcare).
Coimmunoprecipitation and mass spectrometry
For coimmunoprecipitation assays in protoplasts, 9FLAG-DDM1 and 9MYC-H2A.W.6-GFP were cloned into the pUC19-35S construct. Transfected protoplasts were incubated in extraction buffer [50 mM tris-HCl (pH 7.4), 154 mM NaCl, 10% glycerol, 5 mM MgCl2, 1% Triton X-100, 0.3% NP-40, 5 mM DTT, 1 mM PMSF, and protease inhibitor cocktail] and then centrifuged to collect the supernatant for protein extraction. The supernatant was then incubated with anti-FLAG beads (Sigma-Aldrich, M8823) overnight. After incubation, the beads were washed with extraction buffer (without DTT, PMSF, and protease inhibitor cocktail) for four times and were boiled in SDS loading buffer. Samples were subjected to Western blot.
For coimmunoprecipitation assays in Arabidopsis, native promoter-drived DDM1-FLAG/H2A.W-mCherry in ddm1 and H2A.W-mCherry transgenic lines were used. Total proteins were extracted with buffer lysis buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% NP-40, 1 mM PMSF, and protease inhibitor cocktail]. The supernatant was incubated with anti-FLAG beads (Sigma-Aldrich, M8823) overnight.
For DDM1-GFP coimmunoprecipitation-MS, inflorescence tissues from DDM1-GFP transgenic plants and ddm1 mutants were collected for total protein extraction with lysis buffer. After centrifugation, the supernatant was incubated with anti-GFP agarose beads (Chromteck, gta-20) at 4°C for 4 hours. After washing with lysis buffer, beads were boiled in SDS loading buffer. Samples were separated on the SDS-PAGE gel and excised for MS.
Competitive pull-down assay
The competitive pull-down assay was conducted as previously described (14) with some modifications. Briefly, GST-DDM1 was incubated with glutathione magnetic beads in the binding buffer at 30°C for 1 hour, and then the GST-DDM1-bead complexes were sequentially incubated with H2As-H2B dimers in the binding buffer at 30°C for 1 hour. Then, the complexes were washed in the binding buffer three times to remove the unbound H2As-H2B dimers. Then, the complexes were incubated with wash buffer [20 mM tris-HCl (pH 8.0), 500 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM PMSF, and 2 mM DTT] containing the indicated amounts of competitive dimers in Fig. 4E and fig. S14 (A and B) at room temperature for 1 hour. The washed buffer was obtained for PAGE gel running and staining or Western blot analysis. For normal pull-down assay, the GST–DDM1–bead–histone dimer complexes were washed four times with the binding buffer and then were eluted for Western blot analysis.
Western blot
Total protein was extracted from Arabidopsis seedlings with protein extraction buffer [150 mM NaCl, 0.5% Triton X-100, 50 mM tris-HCl (pH 7.5), and protease inhibitor cocktail]. Anti-Pol II Ser2P (Medical & Biological Laboratories Co., Ltd., MABI0602), anti-FLAG (Sigma-Aldrich, F1804), anti-Myc (EASYBIO, BE2010), anti-GST (Yeasen, 30903ES50), and anti-His (Yeasen, 30403ES40) were used in the assays.
Slot blot
Different amounts of nuclear DNA (isolated using Honda buffer) were treated with or without RNase H. Then, the DNA was slotted onto a nylon membrane (Amersham Hybond-N+) and detected using S9.6.
Pollen nuclei immunostaining
Pollen collection and nuclei extraction were performed as previously described (68). S9.6 or dsRNA antibody rJ2 (Millipore, MABE1134) was used for immunostaining.
Chop-qPCR
The genomic DNA was extracted using the hexadecyltrimethylammonium bromide (CTAB) method from 12-day-old seedlings and was digested with or without McrBC endonuclease (NEB, M0272) at 37°C overnight. The digested as well as nondigested DNA was used to perform qPCR, and qPCR values were normalized to nondigested samples.
Data analysis
ssDRIP-seq/ChIP-seq/ATAC-seq analysis
The analysis of ssDRIP-seq was performed as previously described (22) with minor modifications. Briefly, trimmed reads were aligned to the TAIR10 genome with Bowtie 2 (version 2.3.0) using the default settings, and samtools 1.3.1 was used to remove reads with more than three mismatches and non-uniquely mapped reads. Then, total mapped reads were further divided into forward and reverse reads. MACS2 was used to identify peaks, and normalized coverage files (bigWig) with 5-bp bin sizes were obtained using bamCoverage from deeptools (v2.26.0) to visualize on the Integrative Genomics Viewer. Furthermore, bamCompare or bigwigCompare from deeptools was used to compare immunoprecipitated and input samples for ChIP-seq analysis.
pNET-seq analysis
R2 reads were trimmed for Illumina adaptors, and then reads < 35 bp were filtered using the Cutadapt (v1.9.1) based on the reads distribution density. Filtered reads were aligned to the Arabidopsis reference genome (TAIR10) using Bowtie 2 (version 2.3.0) with default settings. Reads mapped to specific strands (forward or reverse) were divided as described above.
Whole-genome bisulfite sequencing analysis
Bismark (v0.19.0) was used to map the trimmed reads and extract the methylation level. The UCSC application tool wigToBigWig was used to get the bigwig file.
Acknowledgments
We thank E. J. Richards from Boyce Thompson Institute, New York, for sharing ddm1-1 seed, F. Berger from Gregor Mendel Institute, Vienna, for sharing hta6 and hta7 single mutant seeds, and D. Jiang from Chinese Academy of Sciences, Beijing, for sharing hta8/hta9/hta11 triple mutant seed. We thank the Core Facility of Center of Biomedical Analysis at Tsinghua University for assistance with Confocal Microscopy and mass spectrometry analysis.
Funding: This work was funded by grants from the National Natural Science Foundation of China (grant nos. 31822028, 91940306, 91740105, and 32261133529 to Q.S., 32100428 to J.Zhou, and 32070651 to W.Z.) and the Ministry of Science and Technology of China (2016YFA0500800). J.Zhou, S.S., and W.Z. are supported by postdoctoral fellowships from Tsinghua-Peking Center for Life Sciences.
Author contributions: Q.S. conceptualized the study. Q.S. and J.Zhou designed the experiments. J.Zhou performed most of the experiments and data analysis. X.L. performed a portion of helicase assays. S.S. performed H3K9me2, H3K27me1, and DDM1-FLAG ChIP-seq. W.Z. performed coimmunoprecipitation assay with protoplasts. Q.L. contributed to the ssDRIP-seq of mom1, morc6, and smc4. K.L. performed a portion of bioinformatics analysis. J.Zhu and Z.D. provided help for pNET-seq and data analysis. X.H. provided help for H2A.Z ChIP-seq. J.Zhou and Q.S. wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. High-throughput sequencing data have been deposited in the Gene Expression Omnibus database (accession no. GSE218148).
Supplementary Materials
This PDF file includes:
Figs S1 to S19
Legends for data S1 to S7
Other Supplementary Material for this manuscript includes the following:
Data S1 to S7
REFERENCES AND NOTES
- 1.A. Janssen, S. U. Colmenares, G. H. Karpen, Heterochromatin: Guardian of the genome. Annu. Rev. Cell Dev. Biol. 34, 265–288 (2018). [DOI] [PubMed] [Google Scholar]
- 2.N. Saksouk, E. Simboeck, J. Dejardin, Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 8, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.W. Feng, S. D. Michaels, Accessing the inaccessible: The organization, transcription, replication, and repair of heterochromatin in plants. Annu. Rev. Genet. 49, 439–459 (2015). [DOI] [PubMed] [Google Scholar]
- 4.J. Wang, S. T. Jia, S. Jia, New insights into the regulation of heterochromatin. Trends Genet. 32, 284–294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.B. Loppin, F. Berger, Histone variants: The nexus of developmental decisions and epigenetic memory. Annu. Rev. Genet. 54, 121–149 (2020). [DOI] [PubMed] [Google Scholar]
- 6.W. L. Johnson, A. F. Straight, RNA-mediated regulation of heterochromatin. Curr. Opin. Cell Biol. 46, 102–109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. Vongs, T. Kakutani, R. A. Martienssen, E. J. Richards, Arabidopsis thaliana DNA methylation mutants. Science 260, 1926–1928 (1993). [DOI] [PubMed] [Google Scholar]
- 8.T. Kakutani, J. A. Jeddeloh, E. J. Richards, Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 23, 130–137 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. A. Jeddeloh, T. L. Stokes, E. J. Richards, Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97 (1999). [DOI] [PubMed] [Google Scholar]
- 10.W. J. Soppe, Z. Jasencakova, A. Houben, T. Kakutani, A. Meister, M. S. Huang, S. E. Jacobsen, I. Schubert, P. F. Fransz, DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21, 6549–6559 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. Zemach, M. Y. Kim, P. H. Hsieh, D. Coleman-Derr, L. Eshed-Williams, K. Thao, S. L. Harmer, D. Zilberman, The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153, 193–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D. B. Lyons, D. Zilberman, DDM1 and Lsh remodelers allow methylation of DNA wrapped in nucleosomes. eLife 6, e30674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.J. Brzeski, A. Jerzmanowski, Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 278, 823–828 (2003). [DOI] [PubMed] [Google Scholar]
- 14.A. Osakabe, B. Jamge, E. Axelsson, S. A. Montgomery, S. Akimcheva, A. L. Kuehn, R. Pisupati, Z. J. Lorković, R. Yelagandula, T. Kakutani, F. Berger, The chromatin remodeler DDM1 prevents transposon mobility through deposition of histone variant H2A.W. Nat. Cell Biol. 23, 391–400 (2021). [DOI] [PubMed] [Google Scholar]
- 15.P. Bourguet, C. L. Picard, R. Yelagandula, T. Pélissier, Z. J. Lorković, S. Feng, M. N. Pouch-Pélissier, A. Schmücker, S. E. Jacobsen, F. Berger, O. Mathieu, The histone variant H2A.W and linker histone H1 co-regulate heterochromatin accessibility and DNA methylation. Nat. Commun. 12, 2683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J. M. Santos-Pereira, A. Aguilera, R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 16, 583–597 (2015). [DOI] [PubMed] [Google Scholar]
- 17.L. A. Sanz, S. R. Hartono, Y. W. Lim, S. Steyaert, A. Rajpurkar, P. A. Ginno, X. Xu, F. Chédin, Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.T. Garcia-Muse, A. Aguilera, R loops: From physiological to pathological roles. Cell 179, 604–618 (2019). [DOI] [PubMed] [Google Scholar]
- 19.C. Niehrs, B. Luke, Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 21, 167–178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.F. Chedin, Nascent connections: R-loops and chromatin patterning. Trends Genet. 32, 828–838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.E. Petermann, L. Lan, L. Zou, Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat. Rev. Mol. Cell Biol. 23, 521–540 (2022). [DOI] [PubMed] [Google Scholar]
- 22.W. Xu, H. Xu, K. Li, Y. Fan, Y. Liu, X. Yang, Q. Sun, The R-loop is a common chromatin feature of the Arabidopsis genome. Nat. Plants 3, 704–714 (2017). [DOI] [PubMed] [Google Scholar]
- 23.W. Yuan, J. Zhou, J. Tong, W. Zhuo, L. Wang, Y. Li, Q. Sun, W. Qian, ALBA protein complex reads genic R-loops to maintain genome stability in Arabidopsis. Sci. Adv. 5, eaav9040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.X. Yang, Q. L. Liu, W. Xu, Y. C. Zhang, Y. Yang, L. F. Ju, J. Chen, Y. S. Chen, K. Li, J. Ren, Q. Sun, Y. G. Yang, m(6)A Promotes R-loop formation to facilitate transcription termination. Cell Res. 29, 1035–1038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.W. Xu, K. Li, S. Li, Q. Hou, Y. Zhang, K. Liu, Q. Sun, The R-loop atlas of arabidopsis development and responses to environmental stimuli. Plant Cell 32, 888–903 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Y. Li, Y. Song, W. Xu, Q. Li, X. Wang, K. Li, J. Wang, Z. Liu, S. Velychko, R. Ye, Q. Xia, L. Wang, R. Guo, X. Dong, Z. Zheng, Y. Dai, H. Li, M. Yao, Y. Xue, H. R. Schöler, Q. Sun, H. Yao, R-loops coordinate with SOX2 in regulating reprogramming to pluripotency. Sci. Adv. 6, eaba0777 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.P. Yan, Z. Liu, M. Song, Z. Wu, W. Xu, K. Li, Q. Ji, S. Wang, X. Liu, K. Yan, C. R. Esteban, W. Ci, J. C. I. Belmonte, W. Xie, J. Ren, W. Zhang, Q. Sun, J. Qu, G. H. Liu, Genome-wide R-loop Landscapes during cell differentiation and reprogramming. Cell Rep. 32, 107870 (2020). [DOI] [PubMed] [Google Scholar]
- 28.L. Chen, W. Xu, K. Liu, Z. Jiang, Y. Han, H. Jin, L. Zhang, W. Shen, S. Jia, Q. Sun, A. Meng, 5' Half of specific tRNAs feeds back to promote corresponding tRNA gene transcription in vertebrate embryos. Sci. Adv. 7, eabh0494 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Y. Liu, Q. Liu, H. Su, K. Liu, X. Xiao, W. Li, Q. Sun, J. A. Birchler, F. Han, Genome-wide mapping reveals R-loops associated with centromeric repeats in maize. Genome Res. 31, 1409–1418 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Q. Xiao, X. Huang, Y. Zhang, W. Xu, Y. Yang, Q. Zhang, Z. Hu, F. Xing, Q. Sun, G. Li, X. Li, The landscape of promoter-centred RNA-DNA interactions in rice. Nat. Plants 8, 157–170 (2022). [DOI] [PubMed] [Google Scholar]
- 31.J. Zhou, W. Zhang, Q. Sun, R-loop: The new genome regulatory element in plants. J. Integr. Plant Biol. 64, 2275–2289 (2022). [DOI] [PubMed] [Google Scholar]
- 32.M. Unoki, J. Sharif, Y. Saito, G. Velasco, C. Francastel, H. Koseki, H. Sasaki, CDCA7 and HELLS suppress DNA:RNA hybrid-associated DNA damage at pericentromeric repeats. Sci. Rep. 10, 17865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. Nojima, T. Gomes, M. Carmo-Fonseca, N. J. Proudfoot, Mammalian NET-seq analysis defines nascent RNA profiles and associated RNA processing genome-wide. Nat. Protoc. 11, 413–428 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. Zhu, M. Liu, X. Liu, Z. Dong, RNA polymerase II activity revealed by GRO-seq and pNET-seq in Arabidopsis. Nat. Plants 4, 1112–1123 (2018). [DOI] [PubMed] [Google Scholar]
- 35.S. H. Chao, D. H. Price, Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276, 31793–31799 (2001). [DOI] [PubMed] [Google Scholar]
- 36.C. Chen, J. Shu, C. Li, R. K. Thapa, V. Nguyen, K. Yu, Z. C. Yuan, S. E. Kohalmi, J. Liu, F. Marsolais, S. Huang, Y. Cui, RNA polymerase II-independent recruitment of SPT6L at transcription start sites in Arabidopsis. Nucleic Acids Res. 47, 6714–6725 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. D. Buenrostro, P. G. Giresi, L. C. Zaba, H. Y. Chang, W. J. Greenleaf, Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. Zemach, Y. Li, B. Wayburn, H. Ben-Meir, V. Kiss, Y. Avivi, V. Kalchenko, S. E. Jacobsen, G. Grafi, DDM1 binds Arabidopsis methyl-CpG binding domain proteins and affects their subnuclear localization. Plant Cell 17, 1549–1558 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.E. J. Cho, S. H. Choi, J. H. Kim, J. E. Kim, M. H. Lee, B. Y. Chung, H. R. Woo, J. H. Kim, A mutation in plant-specific SWI2/SNF2-Like chromatin-remodeling proteins, DRD1 and DDM1, delays leaf senescence in Arabidopsis thaliana. PLOS ONE 11, e0146826 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D. T. Nguyen, H. P. J. Voon, B. Xella, C. Scott, D. Clynes, C. Babbs, H. Ayyub, J. Kerry, J. A. Sharpe, J. A. Sloane-Stanley, S. Butler, C. A. Fisher, N. E. Gray, T. Jenuwein, D. R. Higgs, R. J. Gibbons, The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 18, 914–928 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.N. Taneja, M. Zofall, V. Balachandran, G. Thillainadesan, T. Sugiyama, D. Wheeler, M. Zhou, S. I. S. Grewal, SNF2 family protein Fft3 suppresses nucleosome turnover to promote epigenetic inheritance and proper replication. Mol. Cell 66, 50–62.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L. Prendergast, U. L. McClurg, R. Hristova, R. Berlinguer-Palmini, S. Greener, K. Veitch, I. Hernandez, P. Pasero, D. Rico, J. M. G. Higgins, A. Gospodinov, M. Papamichos-Chronakis, Resolution of R-loops by INO80 promotes DNA replication and maintains cancer cell proliferation and viability. Nat. Commun. 11, 4534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Bayona-Feliu, S. Barroso, S. Munoz, A. Aguilera, The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat. Genet. 53, 1050–1063 (2021). [DOI] [PubMed] [Google Scholar]
- 44.S. Uruci, C. S. Y. Lo, D. Wheeler, N. Taneja, R-loops and its chro-mates: The strange case of Dr. Jekyll and Mr. Hyde. Int. J. Mol. Sci. 22, 8850 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R. Yelagandula, H. Stroud, S. Holec, K. Zhou, S. Feng, X. Zhong, U. M. Muthurajan, X. Nie, T. Kawashima, M. Groth, K. Luger, S. E. Jacobsen, F. Berger, The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell 158, 98–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Y. X. Luo, X. M. Hou, C. J. Zhang, L. M. Tan, C. R. Shao, R. N. Lin, Y. N. Su, X. W. Cai, L. Li, S. Chen, X. J. He, A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding. EMBO J. 39, e102008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. A. Jarillo, M. Pineiro, H2A.Z mediates different aspects of chromatin function and modulates flowering responses in Arabidopsis. Plant J. 83, 96–109 (2015). [DOI] [PubMed] [Google Scholar]
- 48.D. Zilberman, D. Coleman-Derr, T. Ballinger, S. Henikoff, Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125–129 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.V. V. Ashapkin, L. I. Kutueva, N. I. Aleksandrushkina, B. F. Vanyushin, Epigenetic Regulation of Plant Gametophyte Development. Int. J. Mol. Sci. 20, 3051 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R. K. Slotkin, M. Vaughn, F. Borges, M. Tanurdžić, J. D. Becker, J. A. Feijó, R. A. Martienssen, Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.K. Skourti-Stathaki, K. Kamieniarz-Gdula, N. J. Proudfoot, R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516, 436–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O. Velazquez Camacho, C. Galan, K. Swist-Rosowska, R. Ching, M. Gamalinda, F. Karabiber, I. de la Rosa-Velazquez, B. Engist, B. Koschorz, N. Shukeir, M. Onishi-Seebacher, S. van de Nobelen, T. Jenuwein, Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. eLife 6, e25293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. Nakama, K. Kawakami, T. Kajitani, T. Urano, Y. Murakami, DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 17, 218–233 (2012). [DOI] [PubMed] [Google Scholar]
- 54.X. Zhong, J. du, C. J. Hale, J. Gallego-Bartolome, S. Feng, A. A. Vashisht, J. Chory, J. A. Wohlschlegel, D. J. Patel, S. E. Jacobsen, Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157, 1050–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. Ogrocká, P. Polanská, E. Majerová, Z. Janeba, J. Fajkus, M. Fojtová, Compromised telomere maintenance in hypomethylated Arabidopsis thaliana plants. Nucleic Acids Res. 42, 2919–2931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.H. Shaked, N. Avivi-Ragolsky, A. A. Levy, Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173, 985–994 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.X. Xie, D. E. Shippen, DDM1 guards against telomere truncation in Arabidopsis. Plant Cell Rep. 37, 501–513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.E. Y. Basenko, M. Kamei, L. Ji, R. J. Schmitz, Z. A. Lewis, The LSH/DDM1 homolog MUS-30 is required for genome stability, but not for DNA methylation in Neurospora crassa. PLOS Genet. 12, e1005790 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. Burrage, A. Termanis, A. Geissner, K. Myant, K. Gordon, I. Stancheva, The SNF2 family ATPase LSH promotes phosphorylation of H2AX and efficient repair of DNA double-strand breaks in mammalian cells. J. Cell Sci. 125, 5524–5534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.L. Costantino, D. Koshland, The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 34, 39–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.D. Zong, P. Oberdoerffer, P. J. Batista, A. Nussenzweig, RNA: A double-edged sword in genome maintenance. Nat. Rev. Genet. 21, 651–670 (2020). [DOI] [PubMed] [Google Scholar]
- 62.M. P. Crossley, M. Bocek, K. A. Cimprich, R-loops as cellular regulators and genomic threats. Mol. Cell 73, 398–411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.S. Shafiq, A. Berr, W. H. Shen, Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time regulation. New Phytol. 201, 312–322 (2014). [DOI] [PubMed] [Google Scholar]
- 64.M. J. Rowley, G. Bohmdorfer, A. T. Wierzbicki, Analysis of long non-coding RNAs produced by a specialized RNA polymerase in Arabidopsis thaliana. Methods 63, 160–169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.M. Bajic, K. A. Maher, R. B. Deal, Identification of open chromatin regions in plant genomes using ATAC-seq. Methods Mol. Biol. 1675, 183–201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M. E. Potok, Y. Wang, L. Xu, Z. Zhong, W. Liu, S. Feng, B. Naranbaatar, S. Rayatpisheh, Z. Wang, J. A. Wohlschlegel, I. Ausin, S. E. Jacobsen, Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A.Z deposition. Nat. Commun. 10, 3352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.B. Silva, R. Pentz, A. M. Figueira, R. Arora, Y. W. Lee, C. Hodson, H. Wischnewski, A. J. Deans, C. M. Azzalin, FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops. Nat. Commun. 10, 2253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.M. Borg, D. Buendia, F. Berger, A simple and robust protocol for immunostaining Arabidopsis pollen nuclei. Plant Reprod. 32, 39–43 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs S1 to S19
Legends for data S1 to S7
Data S1 to S7