Abstract
To develop a better understanding of the interaction between retroviruses and their hosts, we have investigated the polymorphism in endogenous murine leukemia proviruses (MLVs). We used genomic libraries of wild mouse DNAs and PCR to analyze genetic variation in the proviruses found in wild mouse species, including Mus musculus (M. m. castaneus, M. m. musculus, M. m. molossinus, and M. m. domesticus), Mus spretus, and Mus spicelegus, as well as some inbred laboratory strains. In this analysis, we detected several unique forms of sequence organization in the U3 regions of the long terminal repeats of these proviruses. The distribution of the proviruses with unique U3 structures demonstrated that xenotropic MLV-related proviruses were present only in M. musculus subspecies, while polytropic MLV-related proviruses were found in both M. musculus and M. spretus. Furthermore, one unique provirus from M. spicelegus was found to be equidistant from ecotropic provirus and nonecotropic provirus by phylogenetic analysis. This provirus, termed HEMV, was thus likely to be related to the common ancestor of these MLVs. Moreover, an ancestral type of polytropic MLV-related provirus was detected in M. spretus species. Despite their “ancestral” phylogenetic position, proviruses of these types are not widespread in mice, implying more-recent spread by infection rather than inheritance. These results imply that recent evolution of these proviruses involved alternating periods of replication as virus and residence in the germ line.
All inbred laboratory strains of mice contain numerous endogenous proviruses, of which those related to murine leukemia virus (type C MLVs) are the best-characterized group. Endogenous MLVs are divided into two major groups, ecotropic and nonecotropic, classified by their potential host ranges (4). Ecotropic proviruses are present in one to five copies in some, but not all, common laboratory mouse strains (26, 30, 40). Nonecotropic viruses are subdivided into three major groups, termed xenotropic, polytropic, and modified polytropic, and are present in about 20 copies each in the genome of inbred mice (21, 30, 50). Many of these proviruses have been chromosomally mapped in laboratory strains, and several have been molecularly cloned and sequenced (3, 7, 19, 20, 21, 23, 27, 28, 32, 37). These studies demonstrated that the nonecotropic MLV proviruses are highly polymorphic in their insertion sites and exhibit limited genetic variation from one provirus to the next. Our previous studies have shown that the members of each group of nonecotropic proviruses share a set of linked polymorphisms in env and the long terminal repeat (LTR) regions that distinguishes them from the members of other groups (12, 49). Most usefully, the polymorphisms allowed us to develop a set of oligonucleotide probes that unambiguously detect members of each nonecotropic group in the mouse genome (18, 50).
The nonecotropic proviruses are more widely distributed than the ecotropic proviruses and are also found in wild mouse species, especially Mus musculus, which are the progenitors of common inbred laboratory strains (7, 25, 26, 30, 54, 55, 63). The distribution of the nonecotropic proviruses in taxonomically distinct wild mouse species indicates that these germ line sequences were acquired independently and have remained largely segregated in Mus (30, 55). Either interbreeding between different subspecies or cross-species infection could have contributed to this spread. Some of the nonecotropic proviruses in wild mice have been cloned and analyzed (7, 55). Recently, we have demonstrated that, although in common laboratory strains the linkage of group-specific sequences of the proviruses is strict, proviruses that combine env and LTR sequences from different groups are commonly observed in M. musculus subspecies (55). Furthermore, we have found extensive genetic variation of nonecotropic proviruses in the wild mice (55). These characteristics of the endogenous nonecotropic MLV proviruses provide better understanding not only of the host-retrovirus interaction but also of coevolution of MLVs and their murine host. Furthermore, because MLVs have survived as both viruses and endogenous proviruses in their murine hosts, analysis of the polymorphism of the nonecotropic MLV proviruses could give us a good way to understand adaptation of the MLVs to the hosts. In fact, the presence of endogenous proviruses that have undergone recombination with other endogenous viruses in wild mice implies that the endogenous proviruses have also adapted as viruses in their hosts (55).
We describe here a systematic investigation of polymorphism of nonecotropic MLV proviruses in wild mice, including M. musculus subspecies (M. m. castaneus, M. m. musculus, M. m. molossinus, and M. m. domesticus), Mus spretus, and Mus spicelegus (formerly known as Mus hortulanus), as well as some inbred laboratory strains. Using genomic libraries of wild mouse DNA and specific oligonucleotide probes, we detected several unique U3 regions of nonecotropic proviruses and determined their detailed distribution in the wild mice. Although most of the structures were found only in M. musculus subspecies, some were distributed in both M. musculus and other distinct species. Furthermore, we could detect possible ancestral forms of the nonecotropic MLVs. This paper reports a possible evolutionary relationship between MLVs and wild mouse species.
MATERIALS AND METHODS
DNAs.
In addition to four common inbred laboratory strains (AKR/J, HRS/J, C3H/HeJ, and C57BL/6J), DNAs from several species of Mus were used in this study. The DNAs of CAST/Ei, CASA/Rk (M. m. castaneus), CZECH II/Ei (M. m. musculus), MOLC/Rk, MOLF/Ei, MOLG/Dn (M. m. molossinus), WSB/Ei, ZALENDE/Ei (M. m. domesticus), SF/CamEi, PERA/Rk, PERC/Ei (M. musculus), SPRET/Ei (M. spretus), and PANCEVO/Ei (M. spicelegus) strains were obtained from the Mouse DNA Resource of The Jackson Laboratory, Bar Harbor, Maine. Samples of M. spicelegus (Halbturn), Mus cervicolor, Mus caroli, and Mus cookii DNA were kindly supplied by Christine A. Kozak of the National Institute of Allergy and Infectious Diseases, Bethesda, Maryland.
Preparation of genomic libraries.
DNAs from the CZECH II/Ei (M. m. musculus) and MOLC/Rk (M. m. molossinus) strains were partially digested with the restriction enzyme BamHI. The DNAs were then ligated into the BamHI site of lambda EMBL3 vector (Stratagene), packaged, and subsequently amplified in XL1-blue MRA-P2 cells. The library was plated out and then screened by using a 5′-end 32P-labeled xenotropic MLV-related or polytropic MLV-related (KT-69) oligonucleotide probe. The xenotropic virus-related probe (Xltr) is described elsewhere (18). Positive clones were selected and purified. Some of the unique clones were subcloned into the BamHI site of the pBluescript SK(−) plasmid vector (Stratagene) and sequenced.
Dried gel hybridization and oligonucleotide probes.
Hybridization in dried agarose gels (unblotting) was described previously (51). Briefly, genomic DNA digested with appropriate restriction enzymes was electrophoresed in a 0.8% agarose gel. After staining with ethidium bromide (EtBr), the DNA in the gel was denatured. After drying, the gel was hybridized for 16 h with a 5′-end 32P-labeled oligonucleotide probe (0.5 × 106 cpm/ml). The dried gel was then washed, briefly air dried, and exposed to X-ray film for 1 to 5 days by using an intensifying screen at −70°C. Sequences of oligonucleotide probes specific for each nonecotropic provirus LTR region (Xltr, Pltr, and Mltr) and the hybridization temperatures are described elsewhere (18). Sequences of oligonucleotide probes specific for other types of the nonecotropic proviruses (and the hybridization temperatures) are as follows: KT-51, 5′-TAC TAG GAC AAG GGC CAA ACA GG-3′ (62°C); KT-53, 5′-GAC AAG GGC CAA GAA CCG ATG GTA C-3′ (62°C); KT-55, 5′-GGA TAT CTG TGG TCG AGC ACC TGG-3′ (60°C); KT-58, 5′-GGC TGA ATA GGT ATC GGT GGT-3′ (58°C); KT-59/60, 5′-GGA AGT TCA GTT A(G/A)A GAT CAA GGC TG-3′ (60°C); KT-61, 5′-GCC ATA AGC AAG CTA GCA ATA GTA AC-3′ (60°C); KT-69, 5′-GAA CCA GCA ACA GAC ACA GAA G-3′ (59°C); and KT-76, 5′-GCT GCC ATT TTG CAA GGC ATA G-3′ (59°C).
Synthetic oligonucleotide primers and PCR analysis.
To detect nonecotropic MLV proviruses in mouse genomes by PCR, several amplification primers were generated in this study. The nucleotide sequences of the primers are as follows: Uniltr-4, 5′-CGG GCG ACT CAG TCT ATC GG-3′; KS-50, 5′-CAG TAT CAC CAA CTC AAA TC-3′; Unienv-3, 5′-GGA TAC ACG CCG CTC ACG TA-3′; HE-8, 5′-CTA CAG AAC CGT AGA GGA CT-3′; and KA-59C, 5′-CAG CCT TGA TCT CTA ACT GAA CTT CC-3′. The locations of these sequences on the proviruses are shown in the relevant figures. PCRs were carried out in a total volume of 50 μl containing 0.5 μg of genomic DNA, 50 pmol each of sense and antisense primers, and 2.5 U of Thermus aquaticus DNA polymerase (Taq polymerase; Perkin-Elmer Cetus). The reaction mixtures for amplification were incubated at appropriate temperatures, and the cycle was repeated 30 times in a GeneAmp PCR system 2400 (Perkin-Elmer Cetus).
Cloning and sequencing analysis.
The endogenous provirus fragments detected by PCR were purified from agarose gels and blunt ended by T4 DNA polymerase (New England Biolabs, Inc.). The fragments were then cloned into the SmaI site of the pUC119 vector. DNA sequences were determined in accordance with the double-stranded dideoxy-chain termination method (45) by using the Sequenase version 2.0 kit (United States Biochemical Co.).
Sequence alignments and phylogenetic analysis.
Sequences were aligned by using the algorithm of Needleman and Wunsch (39) as implemented in the PILEUP program in the Genetics Computer Group (GCG) program (14). For phylogenetic analysis, genetic distances between pairs of the sequences were calculated by the DNADIST or PROTDIST program in PHYLIP version 3.5 (16), using Kimura’s two-parameter model (29). A neighbor-joining tree was estimated by NEIGHBOR program and a bootstrap analysis (15) using 1,000 bootstrap replications to assess the support at each of the internal nodes of the neighbor-joining tree.
Nucleotide sequence accession numbers.
Provirus sequences reported in this study have been deposited in GenBank under accession no. AF070719 to AF070732.
RESULTS
Our previous study demonstrated that nonecotropic MLV proviruses show extensive genetic variation in wild mice (55). In particular, U3 regions of the proviruses are highly polymorphic, reflecting evolution of the MLVs in the mice. To investigate genetic variation of U3 regions in the endogenous nonecotropic MLV proviruses in more detail, we generated genomic libraries from M. m. musculus and M. m. molossinus DNAs. To analyze unique U3 forms of the nonecotropic proviruses, the libraries were screened by using xenotropic or other type-specific oligonucleotide probes, and individual fragments were selected and subcloned (see Materials and Methods). A number of clones containing either the 5′ or the 3′ LTR sequences of nonecotropic MLV proviruses were selected for analysis.
In addition to genomic libraries, U3 regions of the proviruses were also cloned following PCR amplification. For the PCR, we designed an antisense primer, Uniltr-4, in the R region of the LTR that annealed to all groups of nonecotropic proviruses and a sense primer, KS-50, derived from a conserved region near the 5′ end of the LTR. We used this primer pair for amplification of U3 regions from wild mouse DNAs. The exact sequences and locations of the primers are shown in Materials and Methods and Fig. 1, respectively. Amplified products were cloned into a plasmid vector and then sequenced. We have analyzed a total of 55 U3 clones from M. musculus and M. spretus species, designated by the prefix Mxv, Mcv, or SPR (Table 1). U3 sequences of unique clones were aligned with those of previous isolates of MLVs. Although U3 sequences of the nonecotropic proviruses are highly conserved, reflecting their recent evolutionary relationship, the proviruses could be clearly classified by unique structural features resulting from duplication, deletion, or point mutation of the sequences.
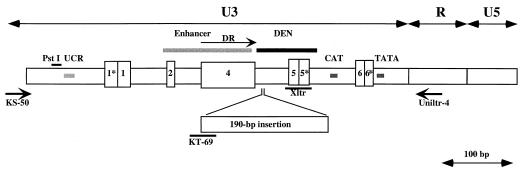
FIG. 1.
Structure of the MLV LTR. The unique sequences in the U3 region are indicated. The locations of primers and probes used in this study are also shown. The numbering of the boxed regions corresponds to that in our previous study (55). UCR, upstream conserved region (17); DR, direct repeat.
TABLE 1.
Summary of nonecotropic MLV U3 clones from wild mice
| Species | Clone | Type |
|---|---|---|
| M. m. musculus | Mxv2 | X-II |
| Mxv3 | X-II | |
| Mxv4 | P-III | |
| Mxv7 | X-II | |
| Mxv8 | X-II | |
| Mxv9 | X-I | |
| Mxv10 | X-II | |
| Mxv11 | X-IV | |
| Mxv16 | X-II | |
| Mxv28 | X-IV | |
| Mxv36 | P-IV | |
| M. m. molossinus | Mcv3 | X-III |
| Mcv4 | X-IIIa | |
| Mcv6 | X-I/X-II, X-IIIb | |
| Mcv7 | X-III | |
| Mcv11 | X-III | |
| Mcv15 | X-IIIa | |
| Mcv16 | X-IIIa | |
| Mcv17 | X-I | |
| Mcv18 | X-I | |
| Mcv20 | X-IIIa | |
| M. spretus | SPR1 | P-I |
| SPR2 | P-II | |
| SPR5 | P-IV | |
| SPR6 | P-I | |
| SPR8 | P-II | |
| SPR9 | P-I | |
| SPR14 | P-V |
Type X-III proviruses that have an additional region 4 enhancer sequence.
Possible mosaic provirus between different xenotropic types.
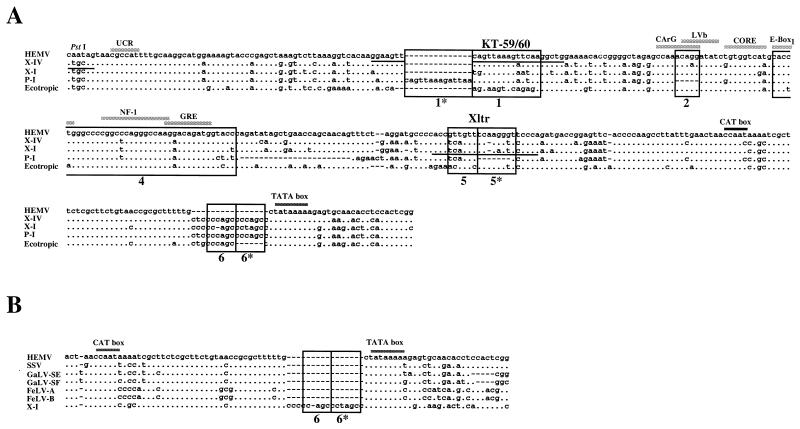
The sequence regions in U3 useful for classification of the structures are indicated in Fig. 1. The numbers of the boxed regions correspond to those in our previous study (55). The 5′ end is the most conserved site among known MLV U3 regions. It includes a PstI recognition site and the upstream conserved region (UCR) motif, CGCCAT (17). The approximately 100-bp sequence following region 1 contains a number of core-enhancer binding sequences that are well conserved among nonecotropic proviruses. Some MLVs contain a direct repeat of the enhancer-rich region region 4. The 93-bp sequence immediately 3′ to region 4 includes several transcriptional factor binding sites and is called the downstream-of-the-enhancer (DEN) region (57). This region has been shown to play an important role in transcriptional activation of the LTR (10, 57). Characteristically, the polytropic MLV-related proviruses contain a unique 190-bp insertion in the 5′ end of the DEN region. The U3 regions of MLVs also contain both CAT and TATA promoter-associated motifs in the 3′ ends of the sequences.
In the following paragraphs, we describe some specific features of these regions, which are useful for classifying the various provirus types and understanding their evolutionary relationships.
Structure of endogenous xenotropic MLV-related provirus U3 regions.
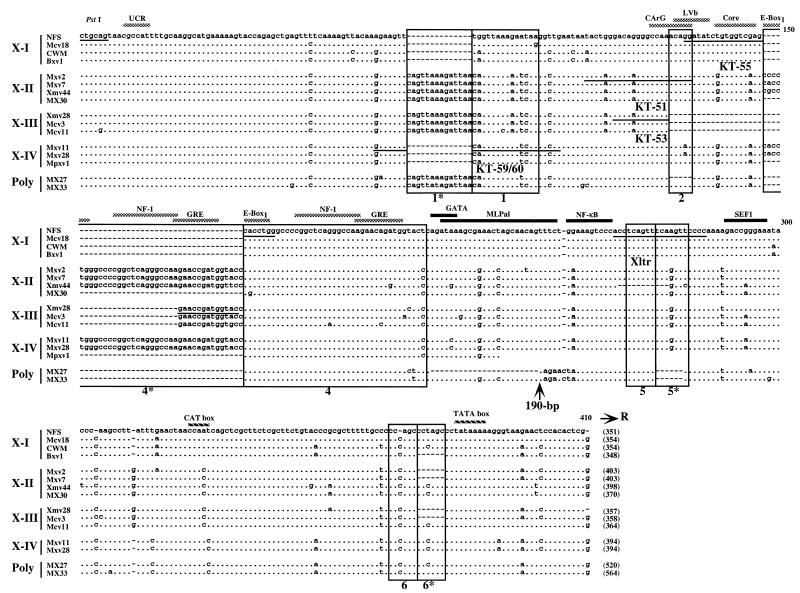
The xenotropic MLV-related proviruses could be divided into four subgroups, termed X-I to X-IV (Fig. 2). The type X-I proviruses include the previously identified proviruses Bxv1, NFS, and CWM, which encode infectious xenotropic virus (28, 35). Bxv1 is also involved in the generation of oncogenic mink cell focus-forming (MCF) viruses in certain mice (23). The X-I proviruses differed from the others by the presence of only one copy of region 1 as well as the enhancer-rich region 4. This group was also distinguished from the others by a number of single-base differences, some of which affected enhancer motifs.
FIG. 2.
U3 regions of xenotropic MLV-related proviruses. U3 sequences were cloned from M. musculus DNAs following amplification with the KS-50 and Uniltr-4 primers. Sequences which were potentially reactive the Xltr probe and which lacked the 190-bp insert are aligned with those of several previously sequenced provirus isolates: NFS (NFS-Th-1) (28), CWM (CWM-S-5X) (35), Bxv1 (23), Xmv44 (52), MX30 (49), Xmv28 (6), MX27, and MX33 (49). The sequence of NFS xenotropic provirus was used as a standard. Dots indicate nucleotide identity. Dashes indicate absence of a nucleotide. Direct repeats and unique sequences present in the proviruses are boxed. Potential enhancer sequence regions and transcriptional factor binding sites are indicated by the shaded and black bars, respectively. Locations of two promoter-associated motifs, the CAT and TATA boxes, are also indicated. The position of the 190-bp insertion in polytropic virus-related proviruses, MX27 and MX33, is shown by an arrow. The sequences of oligonucleotide probes used for unblotting analysis are underlined. The conserved PstI recognition site is also shown. The size of the U3 region of each provirus is indicated at the end of the sequence in parentheses.
Type X-II and X-III xenotropic MLV-related proviruses were isolated from M. m. musculus and M. m. molossinus, respectively, as well as inbred laboratory strains. The structures of X-II and X-III were virtually identical to each other (Fig. 2). Both proviruses contained a 14-bp duplication of region 1 in addition to region 1 (1* and 1), and both types included common mutations in several regions, as well as a common insertion located 12 bp upstream of the CAT box. The X-III proviruses were distinguished by a 45-bp deletion in the middle of the enhancer region, resulting in the absence of CArG, LVb, and core-binding enhancer motifs. The deletion probably occurred by recombination between the 7-bp direct repeats, GGGCCAA, found at both ends of the deleted sequence in the X-II proviruses. Furthermore, half of the type X-III clones we examined contained an additional direct repeat of region 4 (data not shown). In addition, one X-II sequence, Xmv44 (52), contained an 11-bp deletion in the small region (region 5) repeat.
A few clones from the M. m. musculus library were classified as type X-IV. The U3 regions of these proviruses had the same structure as type A Poly/Xeno recombinant proviruses isolated from M. m. musculus and M. m. domesticus (55). Although the U3 structure of this type of provirus was similar to that of the X-I proviruses, several common mutations found in the X-I proviruses were not conserved in type X-IV proviruses. For example, the region 1 sequence was different from that of the X-I provirus (Fig. 2). Furthermore, the sequences of both enhancer and DEN regions clearly distinguished the X-IV proviruses from the X-I proviruses. Interestingly, this type of provirus was very similar to polytropic proviruses in the 3′ end of the U3 region, including the CAT box.
Structure of endogenous polytropic MLV-related provirus U3 regions in wild mice.
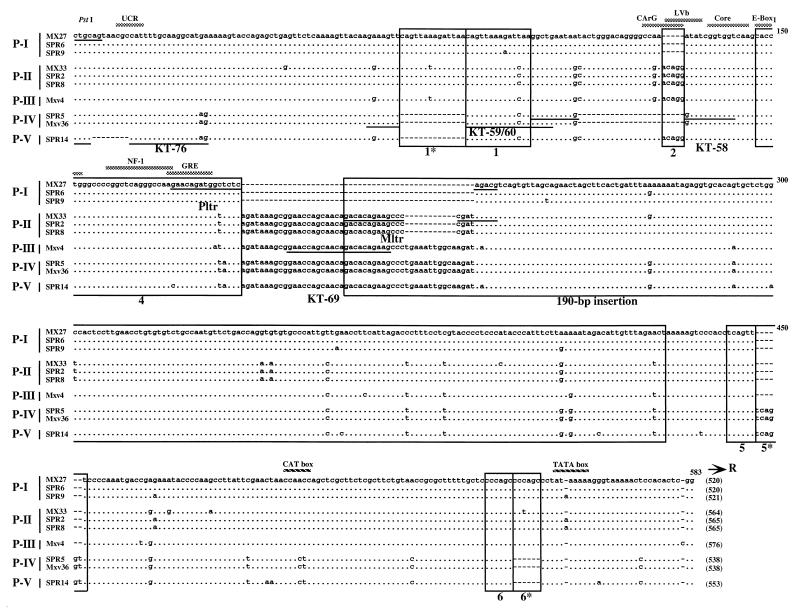
We next analyzed the U3 regions of polytropic MLV-related proviruses (Fig. 3). Since previous studies showed that M. spretus species also carry polytropic MLV-related proviruses (7, 30), we also isolated proviruses from M. spretus as well as M. musculus DNAs. U3 clones from M. spretus species were named with designations beginning with SPR. All proviruses contained a unique 190-bp insertion characteristic of this group (49). Despite the similarity of these proviruses, they could readily be subdivided into several types.
FIG. 3.
Alignment of U3 sequences of polytropic MLV-related proviruses. Sequences of polytropic MLV-related U3 regions cloned from M. musculus and M. spretus DNAs as described in the previous legend are aligned with those of polytropic (MX27) and modified polytropic (MX33) proviruses (49). The MX27 sequence was used as a standard. Dots indicate nucleotide identity. Dashes indicate absence of a nucleotide. Direct repeats and unique sequences present in the proviruses are boxed (55). The 190-bp inserted region is also boxed. Potential enhancer sequence regions are indicated by the shaded bar. Locations of two promoter-associated motifs, the CAT and TATA boxes, are also indicated. The sequences of oligonucleotide probes are underlined. The conserved PstI recognition site is also shown. The size of the U3 region of each provirus is indicated at the end of the sequence in parentheses.
The structures of type P-I and P-II proviruses were identical to those of proviruses isolated from inbred laboratory strains and previously classified as polytropic (MX27) and modified polytropic (MX33), respectively (49). Both types were also found in M. spretus. The P-I and P-II proviruses shared a 14-bp duplication of region 1. P-I proviruses also differed by two deletions from other sequences. The lack of region 2 resulted in deletion of the CArG and LVb enhancer sequences. Furthermore, there was a 49-bp deletion in the variable 5′ boundary region between the 190-bp insertion and original viral sequences where the P-II proviruses had an 11-bp deletion relative to the apparently complete P-III, -IV, and -V sequences. Both types lacked the region 5* duplication but contained a complete duplication of region 6 (Fig. 3).
Mxv4 was isolated from the M. musculus library and classified as a type P-III provirus, although its U3 region was virtually identical to that of the P-II proviruses, since it lacked some nucleotide changes commonly found in the P-II proviruses and it carried an “intact” 190-bp inserted sequence. Type P-IV and P-V proviruses were detected in both M. musculus and M. spretus DNAs. The 5′ half of the P-IV U3 sequence had the same structure as type B Xeno/mPoly recombinant proviruses from M. m. domesticus (55). The sequences of the P-IV and P-V U3 regions were almost identical to each other but contained distinguishing deletions in the 5′ portion. As with the P-I proviruses, the 23-bp deletion in the P-IV proviruses resulted in the absence of CArG and LVb enhancer motifs. Both types also contained intact 190-bp insertions in the sequences. Interestingly, unlike other polytropic virus-related proviruses, both proviruses lacked the 1* and 6* duplications but contained the region 5* duplication. Moreover, the CAT boxes of these types resembled those of type X-II and X-III proviruses rather than the other polytropic MLV-related proviruses.
Distribution of nonecotropic MLV proviruses.
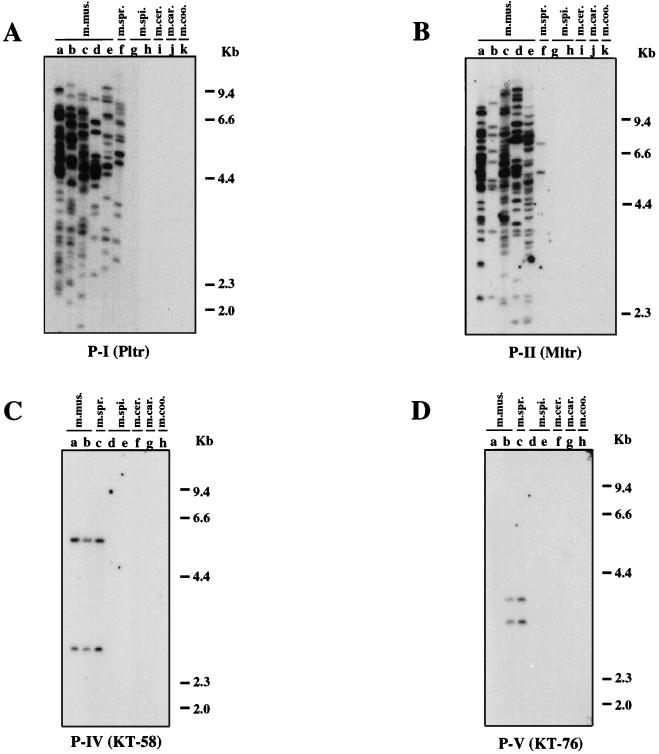
To investigate in detail the distribution of nonecotropic MLV proviruses in wild mice, we used structural features unique to each U3 type to design specific oligonucleotide probes. The locations of the probes are shown in Fig. 2 and 3, and their exact sequences are listed in Materials and Methods. Using these probes, we analyzed PvuII-digested genomic DNAs from the wild and laboratory mouse strains listed in Table 2. The approximate number of fragments reactive with each probe is also indicated in Table 2. Because of the similarity of the U3 sequences of some proviruses, some probes detected more than one type. For example, a probe used for detection of X-IV proviruses, KT-59/60, could also hybridize to the P-IV and P-V proviruses (Fig. 2 and 3). In this case, we determined the number of the X-IV provirus by subtracting the numbers of P-IV and P-V proviruses from those of KT-59/60 reactive fragments. As shown in Table 2, wild mice contained large and variable numbers of proviruses.
TABLE 2.
Distribution of nonecotropic proviral sequences in wild micea
| Species | Strain | No. of fragments for:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Xenotropic MLV-related provirus
|
Polytropic MLV-related provirus
|
||||||||||
| X-I (KT-55) | X-II (KT-51) | X-III (KT-53) | X-IV (KT-59-60) | Xltr | P-I (Pltr) | P-II (Mltr) | P-III (KT-69b) | P-IV (KT-58) | P-V (KT-76) | ||
| M. musculus (laboratory strains) | AKR/J | 6 | 4 | 19 | 2 | 25 | 38 | 24 | >1 | 0 | 0 |
| HRS/J | 10 | 2 | 7 | 2 | 19 | 46 | 23 | 0 | 0 | 0 | |
| C3H/HeJ | 6 | 3 | 12 | 0 | 16 | 34 | 17 | 0 | 0 | 0 | |
| C57BL/6J | 19 | 13 | 20 | 2 | 36 | 46 | 26 | >2 | 0 | 0 | |
| M. musculus | SF/CamEi | 4 | 4 | 4 | 0 | 13 | 41 | 40 | >1 | 0 | 0 |
| PERA/Rk | 1 | 0 | 2 | 4 | 6 | 21 | 30 | >1 | 0 | 0 | |
| PERC/Ei | 0 | 0 | 0 | 5 | 5 | 20 | 34 | >1 | 2 | 2 | |
| M. m. castaneus | CAST/Ei | 2 | 8 | 6 | 0 | 35 | 30 | 5 | 0 | 0 | 0 |
| CASA/Rk | 6 | 9 | 6 | 0 | 33 | 37 | 11 | 0 | 0 | 0 | |
| M. m. musculus | CZECH II/Ei | 31 | >60 | 12 | 19 | >100 | 23 | 1 | >50 | 2 | 0 |
| M. m. molossinus | MOLC/Rk | >50 | >50 | >100 | 2 | >100 | 14 | 6 | 0 | 0 | 0 |
| MOLF/Ei | >50 | >50 | >100 | 2 | >100 | 14 | 3 | 0 | 0 | 0 | |
| MOLG/Dn | >50 | >50 | >100 | 2 | >100 | 14 | 5 | 0 | 0 | 0 | |
| M. m. domesticus | WSB/Ei | 2 | 0 | 0 | 2 | 9 | 39 | 35 | >5 | 0 | 2 |
| ZALENDE/Ei | 0 | 0 | 0 | 0 | 11 | 23 | >50 | >3 | 2 | 2 | |
| M. spretus | SPRET/Ei | 0 | 0 | 0 | 0 | 5 | 15 | 2 | 0 | 2 | 2 |
| M. hortulanus | PANCEVO/Ei | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Halbturn | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| M. cervicolor | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| M. caroli | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| M. cookii | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Numbers of fragments after PvuII digestion and hybridization of dried agarose gels with the probes are shown. Note that an intact provirus gives rise to two such fragments.
This probe is not strictly specific (see text).
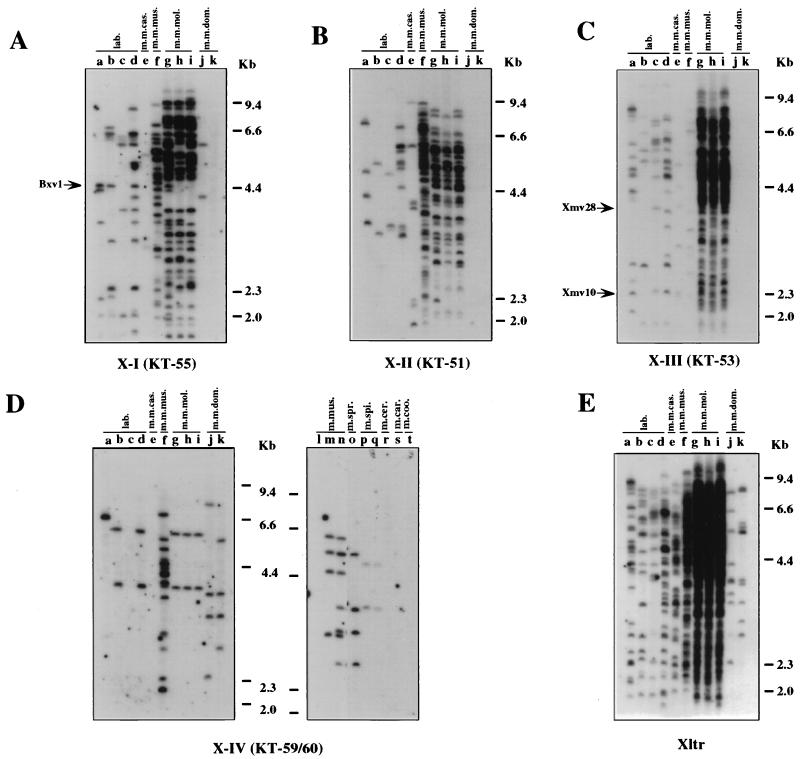
The X-I, X-II, and X-III proviruses showed similar distributions in the wild mouse species. These proviruses were found only in M. musculus subspecies (Fig. 4A to C and Table 2). Although the probe specific for the X-I provirus detected a few fragments in M. m. domesticus DNAs (Fig. 4A, lanes j, and Table 2), the dominant carriers of the X-I and X-II proviruses were M. m. musculus and M. m. molossinus (Fig. 4A and B, lanes f to i, and Table 2) with more than 50 fragments each in the latter DNA. M. m. molossinus DNAs also had a large number of X-III fragments, but only 12 fragments weakly reactive with the probe were found in M. m. musculus DNA (Fig. 4C, lane f, and Table 2). By contrast, X-IV-reactive proviruses were found in only two distinct species, M. musculus and M. spicelegus (Fig. 4D and Table 2). The X-IV probe (KT-59/60) also detected fragments in M. spretus DNA (Fig. 4D, lanes o), but these bands belonged to either type P-IV or P-V proviruses (see below, Fig. 5C and D, lanes a to c). Comigration of the fragments found in the two different M. spicelegus strains (PANCEVO/Ei strain and Halbturn) (Fig. 4D, lanes p and q) implies that this endogenous provirus is widespread in this species. Furthermore, the two X-IV fragments found in M. m. molossinus species comigrated with those of two laboratory strains (Fig. 4D, lanes b, d, and g to i).
FIG. 4.
Distribution of xenotropic MLV-related proviruses in wild mice. Analysis of PvuII-digested mouse DNAs was performed by hybridization of dried agarose gels with an X-I-specific oligonucleotide probe (KT-55) (A), an X-II-specific oligonucleotide probe (KT-51) (B), an X-III-specific oligonucleotide probe (KT-53) (C), and the X-IV-reactive oligonucleotide probe (KT-59/60) (D). Lanes (a to d contain laboratory [lab.] strains): a, AKR/J; b, HRS/J; c, C3H/HeJ; d, C57BL/6J; e, CAST/Ei (M. m. castaneus) (m.m.cas.); f, CZECH II/Ei (M. m. musculus) (m.m.mus.); g, MOLC/Rk (M. m. molossinus); h, MOLF/Ei (M. m. molossinus); i, MOLG/Dn (M. m. molossinus) (m.m.mol.); j, WSB/Ei (M. m. domesticus); k, ZALENDE/Ei (M. m. domesticus) (m.m.dom.); l, SF/CamEi (M. musculus); m, PERA/Rk (M. musculus); n, PERC/Ei (M. musculus) (m.mus.); o, SPRET/Ei (M. spretus) (m.spr.); p, PANCEVO/Ei (M. spicelegus); q, Halbturn (M. spicelegus) (m.spi.); r, M. cervicolor (m.cer.); s, M. caroli (m.car.); t, M. cookii (m.coo.). Some known provirus loci, Bxv1 (23), Xmv10, and Xmv28 (19), are shown by arrows. The approximate positions of molecular markers are also shown.
FIG. 5.
Distribution of polytropic MLV-related proviruses in wild mice. Analysis of PvuII-digested mouse DNAs was performed by hybridization of the DNA in dried agarose gels with the P-I-specific oligonucleotide probe (Pltr) (A), the P-II-specific oligonucleotide probe (Mltr) (B), the P-IV-specific oligonucleotide probe (KT-58) (C), and the P-V-specific oligonucleotide probe (KT-76) (D). (A and B) Lanes: a, C57BL/6J; b, CASA/Rk (M. m. castaneus); c, SF/CamEi (M. musculus); d, PERA/Rk (M. musculus); e, PERC/Ei (M. musculus) (m.mus.); f, SPRET/Ei (M. spretus) (m.spr.); g, PANCEVO/Ei (M. spicelegus); h, Halbturn (M. spicelegus) (m.spi.); i, M. cervicolor (m.cer.); j, M. caroli (m.car.); k, M. cookii (m.coo.). (C and D) a, CZECH II/Ei (M. m. musculus); b, ZALENDE/Ei (M. m. domesticus) (m.mus.); c, SPRET/Ei (M. spretus) (m.spr.); d, PANCEVO/Ei (M. spicelegus); e, Halbturn (M. spicelegus) (m.spi.); f, M. cervicolor (m.cer.); g, M. caroli (m.car.); h, M. cookii (m.coo.). The approximate positions of molecular markers are also shown.
Almost all the fragments reactive with the xenotropic MLV-related provirus probes could be detected by the more generalized xenotropic MLV-related provirus LTR probe, Xltr (Fig. 4E) (55), because all types of the xenotropic MLV-related proviruses conserved region 5 (Fig. 2 and Table 2). The type-specific probes, however, allowed us to investigate the detailed distribution of each type of xenotropic MLV-related proviruses. For example, using the X-I probe we could see that the Bxv1 provirus fragment comigrated among three laboratory strains and one M. m. molossinus strain (Fig. 4A, arrow). Furthermore, although we detected many xenotropic LTR-reactive fragments in both M. m. musculus and M. m. molossinus subspecies with the Xltr probe (Fig. 4E and Table 2), X-II and X-III are the dominant types in M. m. musculus and M. m. molossinus, respectively (Fig. 4B and C and Table 2). Laboratory strains contained roughly equal numbers of the three types of proviruses, together accounting for most or all of the Xltr-reactive fragments. By contrast, fewer fragments altogether were detected by the type-specific probes in M. m. castaneus, M. m. domesticus, and M. spretus than by the Xltr probe (Table 2), indicating that these mice contain still another type(s) of xenotropic MLV-related provirus in the genomes. Note that the P-IV and P-V proviruses should also be detected by the Xltr probe (Fig. 3).
Our previous study demonstrated that polytropic MLV-related proviruses are widely distributed in M. musculus subspecies and also suggested that the proviruses existed in the Mus germ line before subspeciation of M. musculus (55). To verify and extend this observation, we next analyzed the distribution of the polytropic MLV-related proviruses using the type-specific probes (Fig. 5). The P-I and P-II specific probes (Pltr and Mltr) detected specific fragments in M. musculus and M. spretus DNAs but not in others (Fig. 5A and B). The M. spretus DNA contained 15 and 2 fragments specific for the P-I and P-II probes, respectively (Fig. 5A and B, lanes f, and Table 2).
The distribution of the P-IV and P-V proviruses was also examined by using specific probes. The P-IV provirus-specific probe, KT-58, detected two comigrating bands, most likely representing a single copy of this type of provirus, in the M. musculus and M. spretus species DNAs (Fig. 5C). A similar observation was made by using the P-V specific probe, KT-76 (Fig. 5D), with which fragments common to M. spretus and one strain of M. musculus were also observed. These observations suggested that these proviruses were integrated into the Mus germ line before separation between M. musculus and M. spretus species. Unfortunately, we could not generate a P-III provirus-specific probe because it lacks a sequence feature to distinguish it from the other polytropic MLV-related proviruses. However, hybridization with the probe, KT-69, that detected all types of polytropic MLV-related proviruses except for P-I provirus implied that the P-III provirus was found only in M. musculus subspecies and distributed predominantly in M. m. musculus subspecies (Table 2).
Sequence and phylogenetic analyses of M. spicelegus endogenous MLV provirus.
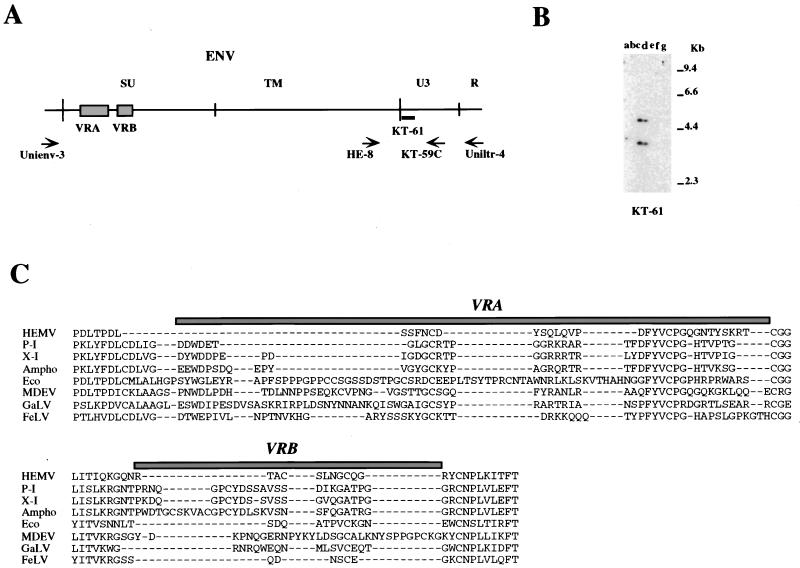
Analyses using type-specific probes suggested the possibility that the X-IV-related provirus might represent the oldest type of nonecotropic provirus among those examined, since it was the only one detected in the relatively distantly related M. spicelegus. Thus, we cloned an X-IV-reactive fragment from this species in two steps. First, we used a PCR primer that recognized a conserved MLV group env sequence, Unienv-3 (Fig. 6A), and an antisense primer, KA-59C, complementary to the X-IV-specific probe (Fig. 2 and 6A) to amplify and clone the whole env region. To determine whether this cloned fragment was the same X-IV-related provirus detected in the analysis presented Fig. 4D, we hybridized PvuII-digested M. spicelegus DNAs with a specific U3 region probe, KT-61, designed from the cloned product. As shown in Fig. 6B, KT-61 detected two fragments identical to those seen with KT-59/60 (Fig. 4D), indicating that the cloned fragment was indeed derived from the X-IV-related provirus. This provirus was named hortulanus endogenous MLV (HEMV).
FIG. 6.
Detection of fragments of the M. spicelegus endogenous provirus (HEMV). (A) The locations of PCR primers and the oligonucleotide probes used. The approximate positions of the SU and TM regions of the env gene and the U3 and R regions of the LTR are indicated. Two hypervariable regions (VRA and VRB) in the SU region are also shown by boxes. (B) Detection of the HEMV provirus. Analysis of PvuII-digested mouse DNAs was performed by using the HEMV-specific oligonucleotide probe KT-61. Lanes: a, CZECH II/Ei (M. musculus); b, SPRET/Ei (M. spretus); c, PANCEVO/Ei (M. spicelegus); d, Halbturn (M. spicelegus); e, M. cervicolor; f, M. caroli; g, M. cookii. The approximate positions of molecular markers are also shown. (C) Amino acid sequences of the VRA and VRB regions of the HEMV env gene are aligned with the analogous regions of several type C retroviruses. Abbreviations and strains of the viruses included in this alignment are as follows: P-I (MX27) (49); X-I (NZB) (41); Ampho, amphotropic MLV (4070A) (42); Eco, ecotropic MLV (Akv) (58); MDEV, M. dunni endogenous virus (62); GalV, (U20589); FeLV, feline leukemia virus subgroup A (Glasgow-1) (48).
To infer the host range of HEMV, we determined the nucleotide sequence of part of its env gene. The host range of MLVs is specified by two variable regions (VRA and VRB) in the 5′ portion of the surface (SU) protein (Fig. 6A) (1). The analysis indicated that the amino acid sequences of VRA and VRB of HEMV were quite different from those of other type C leukemia viruses (Fig. 6C), although other parts of the Env protein were highly conserved. Most strikingly, HEMV encoded a very short VRA sequence in the genome, implying a host range distinct from that of other known MLVs.
For the second step, we designed an HEMV-specific sense primer located in the env region, HE-8 (Fig. 6A), and performed PCR amplification with it and the Uniltr-4 antisense primer. The PCR product was purified and sequenced. By using a specific probe, the PCR product was confirmed to encode the HEMV U3 region (data not shown). The sequence of the HEMV U3 region is shown in Fig. 7. Although this sequence was distinct from the other proviruses, it was still closely related to both ecotropic and nonecotropic MLV proviruses. A highly conserved 24-bp region, including the UCR motif, was present just 3′ of the absent PstI site (Fig. 7A). HEMV encoded only one region 1 and the sequence showed a 1-bp difference from that of the X-IV provirus. This difference probably accounts for the relatively weak hybridization signals seen with M. spicelegus DNAs by using the KT-59/60 probe (Fig. 4D). Interestingly, the enhancer regions, including region 4, were well conserved among all MLV proviruses (Fig. 7A). The core-binding and E-box1 motifs of HEMV were found in ecotropic and nonecotropic types, respectively. The DEN region of HEMV was relatively close to that of ecotropic proviruses. Furthermore, HEMV had a complete CAT box, CCAAT, as was the case for the X-I and ecotropic proviruses. Moreover, unlike the U3 sequences of known MLV proviruses (22), HEMV lacked both region 6 and region 6* in the 3′ end of U3. Interestingly, simian sarcoma virus (SSV) (13), gibbon ape leukemia virus (GaLV) (56), and feline leukemia virus (48) U3 sequences also lack these regions (Fig. 7B).
FIG. 7.
Alignment of HEMV U3 sequences. (A) The U3 region of the HEMV provirus was obtained as described in the text and is aligned with the corresponding sequences from the X-IV (Mxv11), X-I (NFS-Th-1) (28), P-I (MX27) (49), and ecotropic (Akv) (58) proviruses. Dots indicate nucleotide identity. Dashes indicate absence of a nucleotide. The sequence of an oligonucleotide probe, KT-59/60, reactive with X-IV proviruses is underlined. Direct repeats in the enhancer regions found in ecotropic proviruses are not shown. The PstI recognition site is also shown. (B) Lack of the region 6 direct repeat in HEMV. The 3′ portion of the HEMV U3 sequence is aligned with the analogous regions of the several type C retroviruses. Abbreviations and strains of the viruses included in this alignment are as follows: GaLV-SE and GaLV-SF, SEATO and San Francisco isolates (56); FeLV-A and FeLV-B, feline leukemia virus, subgroup A (Glasgow-1) and subgroup B (Gardner-Arnstein) (48); X-I, NFS-Th-1 (28).
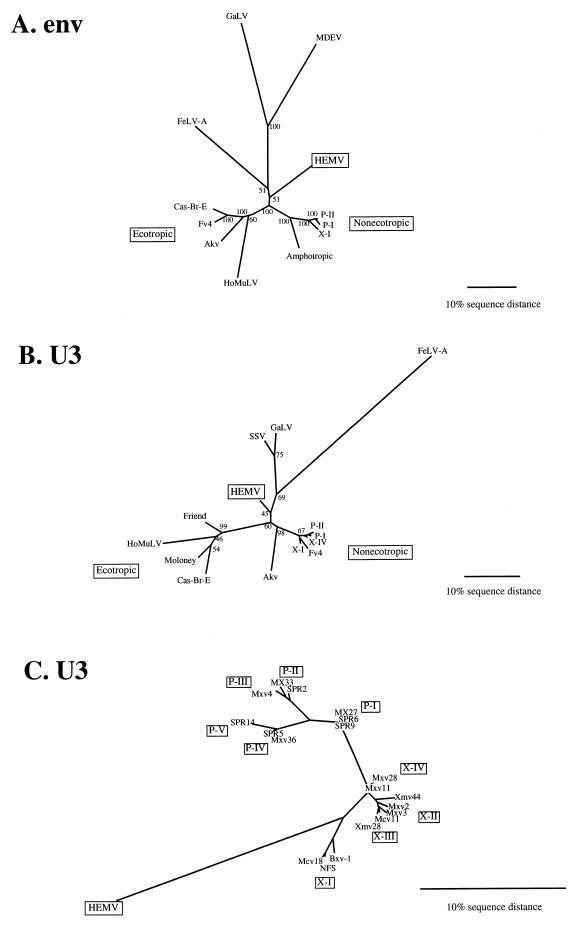
To assess the phylogenetic relationship between HEMV and the other proviruses, we generated unrooted neighbor-joining phylogenetic trees for the env and U3 sequences of the proviruses presented here (Fig. 8). In the env tree (Fig. 8A), each group of proviruses was clearly separated from the others. Interestingly, HEMV was found to be equidistant from the ecotropic and nonecotropic proviruses as well as from the viruses of various species. The U3 tree also supports the phylogenetic position of HEMV and is also supported by bootstrap analysis (Fig. 8B and C). These observations support the possibility that HEMV might resemble a common ancestor of recent MLVs.
FIG. 8.
Phylogenetic analysis of the nonecotropic U3 proviruses. Unrooted phylogenetic trees for the env (A) and U3 (B and C) regions of HEMV and several type C leukemia viruses were estimated by neighbor joining. Branch lengths are drawn to scale. To illustrate consistency all bootstrap values obtained with 1,000 replications of bootstrap sampling are shown. Viruses used for this analysis are as follows: ecotropic viruses, Akv (58), Moloney (47), Friend (FE29 strain) (43), Fv4 (M33884), Cas-Br-E (P08360), and HoMuLV (60); nonecotropic viruses, P-I (MX27) (49), P-II (MX33) (49), and X-I (NFS-Th-1) (28); amphotropic viruses, (4070A) (42), MDEV (62), GaLV (SEATO) (56), and feline leukemia virus subgroup A (FeLV-A) (Glasgow-1) (48).
DISCUSSION
Genetic variation and distribution of nonecotropic MLV proviruses in wild mice.
Endogenous nonecotropic MLV proviruses are stable genetic elements that were fixed recently in the Mus germ line and are therefore highly polymorphic. Endogenous ecotropic MLV proviruses are also polymorphic but are neither widely distributed nor greatly amplified in mice (4, 12), indicative of much more recent origin. Study of the nonecotropic MLV proviruses should provide a tool to obtain a better understanding of the association between retroviruses and their host during their evolutionary history. Here, we analyzed the extended genetic variation of nonecotropic endogenous MLV proviruses in wild mice including M. musculus, M. spretus, and M. spicelegus. We report here several new and distinct classes of nonecotropic provirus U3 sequences and their relationships to one another and to known endogenous proviruses.
The proviruses we studied showed a high degree of sequence similarity in U3, consistent with a recent evolutionary relationship. The 5′ portion including the UCR motif, which is highly conserved among a large number of type C retroviruses (22), was also the most conserved among the wild-mouse proviruses. This motif has been reported to function as a negative regulatory element in several murine type C viruses (17). Other regions of U3 are less well conserved and allowed us to group the proviruses into a number of subtypes, using polymorphisms arising from duplication, deletion, insertion, or point mutation. A schematic view of the relationship among the various groups is shown in Fig. 9, in the form of a possible scheme for their evolution. For example, type X-I proviruses contain distinctive enhancer-promoter sequences in the U3 region (Fig. 2). All X-I proviruses include the consensus MLV core-enhancer binding motif and the factor binding sites in region 4 but not the direct repeats of region 4*, and almost all contain a unique set of factor binding sequences in the DEN region. Furthermore, the promoter-associated CAT box of these proviruses was a perfect match to the consensus.
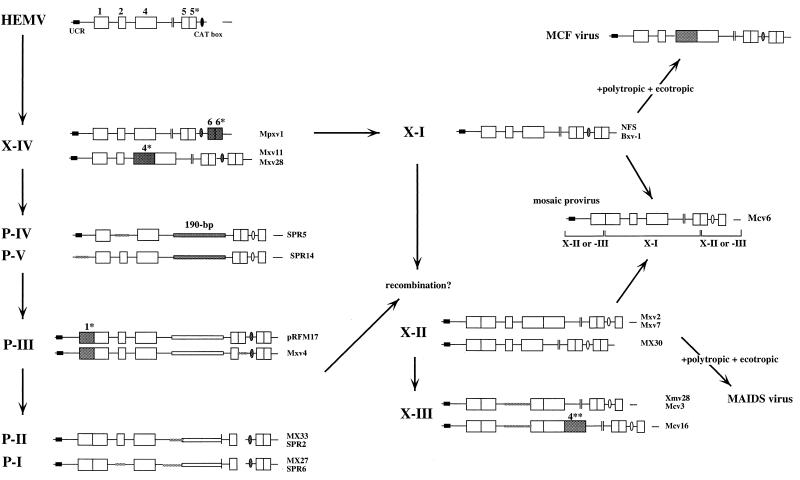
FIG. 9.
A possible evolutionary scheme relating nonecotropic MLV U3 regions. Schematic representations of the U3 structures are shown. Locations of the UCR and CAT box are also shown. Corresponding clones are shown on the right of the structures. Clone pRFM17 was described previously (8). Note that this scheme is parsimonious with respect to insertions and deletions. Discrepancies relative to the tree shown in Fig. 8 probably stem from these events having occurred multiple times.
Although we have not studied the functions of the enhancer-promoter sequences in the wild-mouse proviruses, the U3 regions of retroviruses, in general, are intimately involved in viral replication and pathogenicity (24, 33, 35, 36, 44, 53, 57, 64). Among all known nonecotropic proviruses, only the X-I group includes proviruses capable of yielding infectious virus (28, 35, 41). In this regard, lack of region 4* of the X-I proviruses could be critical. Duplication of core-enhancer regions of MLVs is associated with increases in transcriptional activity (9, 10, 35, 38, 57), and strong enhancer activity in retroviruses often results in increased pathogenicity for the host (33, 35, 44, 53, 57). Indeed, leukemogenicity of MCF viruses in certain mice is associated with and appears to be dependent on the acquisition (by recombination) of an X-I-type LTR followed by duplication of region 4 enhancer sequences (53). In fact, MCF viruses harboring such repeated sequences have not been detected as endogenous elements in the mice (55). Considering these observations, it is likely that X-I proviruses containing duplicated enhancer sequences could not remain in the mouse genome as endogenous elements on account of their strong enhancer activity.
The X-II and X-III proviruses were virtually identical to each other, and X-III was most likely generated by a deletion in the X-II provirus (Fig. 2 and 9). Members of these two groups contained several unique point differences or deletions in their U3 sequences that differentiate them from others. Both X-II and X-III proviruses have been isolated in previous studies, although the sequence characteristics were not examined. Two provirus clones harboring the X-II-type U3 structure, pGP24 and pGP68 (46), were cloned from a cDNA library from the liver of NZB mice and found to encode the env glycoprotein (gp70) found in serum and expressed as an acute-phase protein in mice. Sequences related to these clones are highly expressed in the liver and kidney tissues of NZB mice injected with lipopolysaccharide (46). Furthermore, two independent isolates, MRV (11) and EDV (31), which are related to the murine AIDS (MAIDS) virus, were identified as proviruses containing the X-II-type U3 in their genomes. These two MAIDS-related viruses are endogenous in laboratory strains and have donated the gag sequences characteristic of MAIDS viruses to the related exogenous viruses by recombination (11, 31). The X-III proviruses have also been cloned before. Insertion of Xmv28, an X-III provirus, into the mouse genome is associated with the rd mutation (6).
The distribution of the proviruses also has important implications for the evolutionary relationship of the xenotropic MLVs. First, most of the xenotropic relatives were found only in M. musculus subspecies (Fig. 4 and Table 2). In particular, M. m. musculus and M. m. molossinus were the predominant carriers of the X-I, X-II, and X-III proviruses, indicating that they entered the mouse germ line after separation between M. musculus and M. spretus, probably around the time of subspeciation between M. m. musculus and M. m. domesticus. Despite their presence, the dissimilarities of the distributions and numbers of the proviruses among the subspecies imply that a variety of amplification and germ line integration events occurred after separation of the subspecies. Furthermore, the type-specific probes revealed introgression of some proviruses involved in pathogenicity or mutation into certain laboratory mice. The inbred strains could have inherited the Bxv1 MCF-related provirus from the M. m. molossinus subspecies (Fig. 4A). The MAIDS-related proviruses could have been generated after subspeciation of M. musculus and then acquired in laboratory strains from either M. m. musculus or M. m. molossinus (Fig. 4B). Two mutation-associated proviruses, Xmv10 and Xmv28 (6, 61), were probably inherited from the M. m. molossinus subspecies (Fig. 4C). Further study of the distribution among mice of provirus integrated at specific sites will be required to determine the relationship between integration and speciation events.
Among xenotropic MLV-related proviruses, the X-IV type showed unique genetic features and a unique distribution pattern in the mice. Despite a xenotropic MLV-related U3 structure, the X-IV proviruses shared some common features with polytropic MLV-related proviruses, including identical region 1 and CAT box sequences. Furthermore, some X-IV proviruses and Poly/Xeno recombinant proviruses from M. m. musculus and M. m. domesticus contain polytropic virus-type env sequences (55). Sequence analysis of a clone from our genomic library, Mxv11, indicated that the SU region of the provirus was more similar to that of polytropic viruses than to that of xenotropic viruses, although it contained a large deletion (data not shown). These observations imply that this type of provirus could be related to a recent common ancestor of the nonecotropic proviruses. It could be necessary to investigate larger numbers of samples from M. spretus or the related species to verify this hypothesis.
Polytropic MLV-related proviruses showed less diversity than xenotropic MLV-related proviruses in the wild mice (Fig. 3). Sequence analysis demonstrated that proviruses analogous to polytropic (type P-I) and modified polytropic (P-II) proviruses in laboratory strains (49) were also present and widely distributed in M. spretus. This result was also confirmed by unblotting (Fig. 5 and Table 2). Among the polytropic MLV-related proviruses, the predominant proviruses were of the P-I and P-II types, while P-IV and P-V were not widespread, although some proviruses yielded comigrating bands from DNAs from two distinct species, M. musculus and M. spretus (Fig. 5C and D). This result implies that polytropic MLV-related proviruses were integrated into the Mus germ line before separation between these species, but that only the P-I and P-II proviruses were able to spread within the Mus genomes. Furthermore, the sequence analysis revealed that both the P-I and P-II proviruses contain an internal deletion in the 5′ portion of the 190-bp unique insertion (Fig. 3), while the P-III, P-IV and P-V proviruses contained intact forms of the inserted sequence in their genomes. This observation suggests that the P-I and P-II proviruses were derived from other polytropic MLV-related proviruses with an intact 190-bp insertion. It is likely that P-II was generated from the P-III provirus by simple deletion in the inserted sequence (see below).
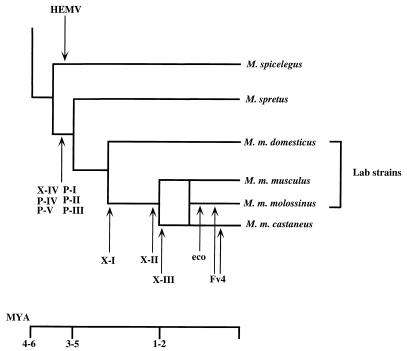
One of the interesting findings of this study was obtained from analysis of the provirus from M. spicelegus, HEMV. Although originally detected with the type X-IV probe, the U3 region of this provirus does not belong with either the xenotropic or the polytropic group. HEMV junction fragments did comigrate in two different strains of M. spicelegus, indicating that it is indeed endogenous in these mice. In a previous study, a distantly related pathogenic ecotropic virus (Hortulanus MLV or HoMLV) was isolated from M. spicelegus species (59, 60). It is believed, however, that HoMLV is exogenous and was acquired from Asian wild mice (59, 60). The sequence and phylogenetic analyses of HEMV clearly demonstrate its close relationship to MLVs, but it is equidistant from ecotropic and nonecotropic provirus groups (Fig. 7B and 8). These results suggest that HEMV is likely to be related to the common ancestor of MLVs. Despite the ancestral phylogenetic position of HEMV, however, the provirus is not widespread in wild mice (Fig. 6B). This result implies a more recent spread by infection of the provirus rather than inheritance. Furthermore, the phylogenetic position of HEMV was also found to be equidistant from simian leukemia viruses (GaLV and SSV) and MLVs. Indeed, the sequence analysis also revealed that the 3′ end of its U3 region is more closely related to the simian viruses than to the murine proviruses (Fig. 7B). It is now believed that GaLV and SSV originated subsequent to a transspecies infection of primates by xenotropic viruses of murine origin derived from M. caroli, M. cervicolor, or other related mouse species (2, 34). Although sequence analysis of the xenotropic viruses from M. caroli and M. cervicolor has not been carried out and the HEMV sequence was not detected in these species (Fig. 6B), there is a possibility that HEMV is the origin of the GaLV/SSV group in primates. Moreover, an endogenous virus (MDEV) that shows a unique host range has recently been isolated from M. dunni (5). The sequences of the vrA and vrB regions of both MDEV (62) and HEMV are consistent with a host range different from that of all other known viruses of this genus (Fig. 6C). However, they are also quite different from each other and occupy distinct phylogenetic positions (Fig. 8A).
Possible evolutionary relationships among MLVs in wild mice.
The unique structural features and detailed distribution of the nonecotropic provirus U3 regions raise several interesting possibilities regarding their evolutionary relationships. In the scheme proposed in Fig. 9, HEMV could represent the progenitor of the LTRs found in other nonecotropic proviruses. The HEMV U3 sequence could have evolved into the nonecotropic-type U3 and given rise to proviruses like the X-IV type following several mutations, including insertion of regions 6 and 6* and a number of duplication and recombination events. The X-IV sequences could generate P-IV and P-V proviruses by insertion of the 190-bp sequence and deletion of region 6*. The polytropic virus-like env sequence of the X-IV proviruses supports this evolutionary step. The P-IV and P-V proviruses seem to be ancestral forms of the polytropic MLV-related proviruses. The P-III sequence could have been derived from P-IV and P-V following duplication of region 1, and it could have given rise to P-I and P-II proviruses by deletions including the 5′ side of the 190-bp insertion. The apparent presence of type P-IV and P-V proviruses in both M. musculus and M. spretus suggests that the separation of these viruses occurred before the separation of M. musculus and M. spretus about 5 million years ago. Type X-I proviruses could have been derived from type X-IV proviruses by several mutations. The X-II and X-III proviruses could have been generated by duplication of region 1 from the X-I proviruses or recombination with polytropic MLV-related proviruses. These events could have occurred after the separation of M. musculus and M. spretus.
In order to accomplish these evolutionary steps, it would be necessary for the proviruses to have extended periods of replication as viruses and involve numerous mutational and recombinational events. Both the nature of the mutational events (duplications, insertions, and deletions, particularly involving enhancer regions) and their rapid accumulation over short evolutionary periods are inconsistent with mutations occurring during residence in the mouse germ line. These events are consistent with those commonly found in the generation of the recombinant MCF and MAIDS viruses derived in part from the X-I and X-III proviruses, respectively. Furthermore, a mosaic provirus, which has the type X-I enhancer-DEN region on the X-II- or X-III-based sequence, was found in a strain of M. m. molossinus, Mcv6 (Fig. 9), directly implicating recombination during virus replication.
It is important to keep in mind that the scheme shown in Fig. 9 is the simplest, involving the smallest number of insertion, duplication, and deletion events. It shares many features with the point mutation-based tree shown in Fig. 8, but the two patterns are not perfectly concordant, particularly in the polytropic lineage. This discordance implies that some events may have occurred more than once during the evolution of these viruses. Given the high frequency of recombination and duplication events observed during infection of a single animal (53), it is likely that some of the duplication and deletion events have occurred multiple times during the evolution of these viruses.
Figure 10 shows a schematic representation of the coevolution of MLV proviruses and the wild-mouse species we included in this study. The observation that the ancestral-type proviruses are not widespread in mice implies that these proviruses only recently expanded into these mice by infection rather than inheritance. Thus, rather than serving as either a static repository for ancient viruses or a reservoir from which exogenous viruses appear from time to time, endogenous proviruses should be viewed as a snapshot of a complex dynamic process involving long periods of replication as viruses, during which recombination and mutation events accumulate, and relatively recent germ line insertion. These proviruses could provide new genetic markers for the strain identification or evolutionary study of both MLVs and their murine host. The approach taken in this study will serve as the basis for further examination of the evolution and involvement of retroviral sequences in tumorigenicity in the murine host.
FIG. 10.
Coevolution of MLVs and wild mice. A schematic phylogenetic relationship of wild-mouse species is shown. Possible periods of provirus integration are represented by arrows. The bar under the tree indicates the approximate time scale. MYA, million years ago.
ACKNOWLEDGMENTS
We are grateful to Jonathan Stoye and Christopher Tipper for helpful comments, to Mary Bostic-Fitzgerald for preparing the manuscript, and to Christine Kozak for the generous gift of wild-mouse DNAs.
This work was supported by National Cancer Institute award R35CA44385 to J.M.C. and a Leukemia Society of America Special Fellowship to K.T. J.M.C. was a research professor of the American Cancer Society.
REFERENCES
- 1.Battini J-L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoprotein of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benveniste R E, Callahan R, Sherr C J, Chapman V, Todaro G J. Two distinct endogenous type C viruses isolated from the Asian rodent Mus cervicolor: conservation of virogene sequences in related rodent species. J Virol. 1977;21:849–862. doi: 10.1128/jvi.21.3.849-862.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blatt C, Mileham K, Haas M, Nesbitt M N, Harper M E, Simon M I. Chromosomal mapping of the mink cell focus-inducing and xenotropic env gene family in the mouse. Proc Natl Acad Sci USA. 1983;80:6298–6302. doi: 10.1073/pnas.80.20.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 5.Bonham L, Wolgamot G, Miller A D. Molecular cloning of Mus dunni endogenous virus: an unusual retrovirus in a new murine viral interference group with a wide host range. J Virol. 1997;71:4663–4670. doi: 10.1128/jvi.71.6.4663-4670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowes C, Li T, Frankel W N, Danciger M, Coffin J M, Applebury M L, Farber D B. Localization of a retroviral element within the rd gene coding for the b subunit of cGMP-phosphodiesterase. Proc Natl Acad Sci USA. 1993;90:2955–2959. doi: 10.1073/pnas.90.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ch’ang L-Y, Yank W K, Myer F E, Koh C K, Boone L R. Specific sequence deletions in two classes of murine leukemia virus-related proviruses in the mouse genome. Virology. 1989;168:245–255. doi: 10.1016/0042-6822(89)90264-x. [DOI] [PubMed] [Google Scholar]
- 8.Ch’ang L-Y, Yang W K, Myer F E, Yang D-M. Negative regulatory element associated with potentially functional promoter and enhancer elements in the long terminal repeats of endogenous murine leukemia virus-related proviral sequences. J Virol. 1989;63:2746–2757. doi: 10.1128/jvi.63.6.2746-2757.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chattopadhyay S K, Morse H C I, Makino M, Ruscetti S K, Hartley J W. Defective virus is associated with induction of murine retrovirus-induced immunodeficiency syndrome. Proc Natl Acad Sci USA. 1989;86:3862–3866. doi: 10.1073/pnas.86.10.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Yoshimura F K. Identification of a region of a murine leukemia virus long terminal repeat with novel transcriptional regulatory activities. J Virol. 1994;68:3308–3316. doi: 10.1128/jvi.68.5.3308-3316.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho B C, Shaughnessy J J, Largaespada D A, Bedigian H G, Buchberg A M, Jenkins N A, Copeland N G. Frequent disruption of the Nf1 gene by a novel murine AIDS virus-related provirus in BXH-2 murine myeloid lymphomas. J Virol. 1995;69:7138–7146. doi: 10.1128/jvi.69.11.7138-7146.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffin J M, Stoye J P, Frankel W N. Genetics of endogenous murine leukemia viruses. Ann N Y Acad Sci. 1989;567:39–49. doi: 10.1111/j.1749-6632.1989.tb16457.x. [DOI] [PubMed] [Google Scholar]
- 13.Devare S G, Reddy E P, Law J D, Robbins K C, Aaronson S A. Nucleotide sequence of the simian sarcoma virus genome: demonstration that its acquired cellular sequences encode the transforming gene product p28sis. Proc Natl Acad Sci USA. 1983;80:731–735. doi: 10.1073/pnas.80.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein J. PHYLIP: phylogeny inference package, version 3.5. Seattle: University of Washington; 1995. [Google Scholar]
- 17.Flanagan J R, Krieg A M, Max E E, Khan A S. Negative control region at the 5′ end of murine leukemia virus long terminal repeats. Mol Cell Biol. 1989;9:739–746. doi: 10.1128/mcb.9.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankel W N, Coffin J M. Endogenous nonecotropic proviruses mapped with oligonucleotide probes from the long terminal repeat region. Mamm Genome. 1994;5:275–281. doi: 10.1007/BF00389541. [DOI] [PubMed] [Google Scholar]
- 19.Frankel W N, Stoye J P, Taylor B A, Coffin J M. Genetic analysis of endogenous xenotropic murine leukemia viruses: association with two common mouse mutations and the viral restriction locus Fv-1. J Virol. 1989;63:1763–1774. doi: 10.1128/jvi.63.4.1763-1774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frankel W N, Stoye J P, Taylor B A, Coffin J M. Genetic identification of endogenous polytropic proviruses by using recombinant inbred mice. J Virol. 1989;63:3810–3821. doi: 10.1128/jvi.63.9.3810-3821.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frankel W N, Stoye J P, Taylor B A, Coffin J M. A linkage map of endogenous murine leukemia proviruses. Genetics. 1990;124:221–236. doi: 10.1093/genetics/124.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golemis E A, Speck N A, Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990;64:534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoggan M D, O’Neill R R, Kozak C A. Nonecotropic murine leukemia viruses in BALB/c and NFS/N mice: characterization of the BALB/c Bxv-1 provirus and the single NFS endogenous xenotrope. J Virol. 1986;60:980–986. doi: 10.1128/jvi.60.3.980-986.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland C A, Thomas C Y, Chattopadhyay S K, Koehne C, O’Donnell P V. Influence of enhancer sequences on thymotropism and leukemogenicity of mink cell focus-forming viruses. J Virol. 1989;63:1284–1292. doi: 10.1128/jvi.63.3.1284-1292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inaguma Y, Miyashita N, Moriwaki K, Huai W C, Mei-Lei J, Zinqiao H, Ikeda H. Acquisition of two endogenous ecotropic murine leukemia viruses in distinct Asian wild mouse populations. J Virol. 1991;65:1796–1802. doi: 10.1128/jvi.65.4.1796-1802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins N A, Copeland N G, Taylor B A, Lee B K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan A S. Nucleotide sequence analysis establishes the role of endogenous murine leukemia virus DNA segments in formation of recombinant mink cell focus-forming murine leukemia viruses. J Virol. 1984;50:864–871. doi: 10.1128/jvi.50.3.864-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan A S, Martin M A. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc Natl Acad Sci USA. 1983;80:2699–2703. doi: 10.1073/pnas.80.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 30.Kozak C A, O’Neill R R. Diverse wild mouse origins of xenotropic, mink-cell focus-forming, and two types of ecotropic proviral genes. J Virol. 1987;61:3082–3088. doi: 10.1128/jvi.61.10.3082-3088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubo Y, Nakagawa Y, Kakimi K, Matsui H, Higo K, Wang L, Kobayashi H, Hirama T, Ishimoto A. Molecular cloning and characterization of a murine AIDS virus-related endogenous transcript expressed in C57BL/6 mice. J Gen Virol. 1994;78:881–888. doi: 10.1099/0022-1317-75-4-881. [DOI] [PubMed] [Google Scholar]
- 32.Lamont C, Culp P, Talbott R L, Phillips T R, Trauger R J, Frankel W N, Wilson M C, Coffin J M, Elder J H. Characterization of endogenous and recombinant proviral elements of a highly tumorigenic AKR cell line. J Virol. 1991;65:4619–4628. doi: 10.1128/jvi.65.9.4619-4628.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenz J, Celander D, Crowther R L, Patarca R, Perkins D W, Haseltine W A. Determination of the leukemogenicity of a murine retrovirus by sequences within the long terminal repeat. Nature. 1984;308:467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- 34.Lieber M M, Sherr C J, Todaro G J, Benveniste R E, Callahan R, Coon H G. Isolation from the Asian mouse Mus caroli of an endogenous type C virus related to infectious primate type C viruses. Proc Natl Acad Sci USA. 1975;72:2315–2319. doi: 10.1073/pnas.72.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massey A C, Coppola M A, Thomas C Y. Origin of pathogenic determinants of recombinant murine leukemia viruses: analysis of Bxv-1-related xenotropic viruses from CWD mice. J Virol. 1990;64:5491–5499. doi: 10.1128/jvi.64.11.5491-5499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Massey A C, Lawrenz S S, Innes D J, Thomas C Y. Origins of enhancer sequences of recombinant murine leukemia viruses from spontaneous B- and T-cell lymphomas of CWD mice. J Virol. 1994;68:3773–3783. doi: 10.1128/jvi.68.6.3773-3783.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meruelo D, Rossomando A, Offer M, Buxbaum J, Pellicer A. Association of endogenous viral loci with genes encoding murine histocompatibility and lymphocyte differentiation antigens. Proc Natl Acad Sci USA. 1983;80:5032–5036. doi: 10.1073/pnas.80.16.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison H L, Soni B, Lenz J. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J Virol. 1995;69:446–455. doi: 10.1128/jvi.69.1.446-455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 40.O’Brien S J, Moore J L, Martin M A, Womack J E. Evidence for the horizontal acquisition of murine AKR virogenes by recent horizontal infection of the germ line. J Exp Med. 1982;155:1120–1123. doi: 10.1084/jem.155.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Neill R R, Buckler C E, Theodore T S, Martin M A, Repaske R. Envelope and long terminal repeat sequences of a cloned infectious NZB xenotropic murine leukemia virus. J Virol. 1985;53:100–106. doi: 10.1128/jvi.53.1.100-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perryman S, Nishio J, Chesebro B. Complete nucleotide sequence of Friend murine leukemia virus, strain FB29. Nucleic Acids Res. 1991;19:6950. doi: 10.1093/nar/19.24.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosen C A, Haseltine W A, Lenz J, Ruprecht R, Cloyd M W. Tissue selectivity of murine leukemia virus infection is determined by long terminal repeat sequences. J Virol. 1985;55:862–866. doi: 10.1128/jvi.55.3.862-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shigemoto K, Kubo S, Itoh Y, Tate G, Handa S, Maruyama N. Expression and structure of serum gp70 as an acute phase protein in NZB mice. Mol Immunol. 1992;29:573–582. doi: 10.1016/0161-5890(92)90193-2. [DOI] [PubMed] [Google Scholar]
- 47.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 48.Stewart M A, Warnock M, Wheeler A, Wilkie N, Mullins J I, Onions D E, Neil J C. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol. 1986;58:825–834. doi: 10.1128/jvi.58.3.825-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoye J P, Coffin J M. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol. 1987;61:2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoye J P, Coffin J M. Polymorphism of murine endogenous proviruses revealed by using virus class-specific oligonucleotide probes. J Virol. 1988;62:168–175. doi: 10.1128/jvi.62.1.168-175.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoye J P, Frankel W N, Coffin J M. DNA hybridization in dried gels with fragmented probes: an improvement over blotting techniques. Technique. 1991;3:123–128. [Google Scholar]
- 52.Stoye J P, Kaushik N, Jeremiah S, Best S. Genetic map of the region surrounding the retrovirus restriction locus, Fv1, on mouse chromosome 4. Mamm Genome. 1995;6:31–36. doi: 10.1007/BF00350890. [DOI] [PubMed] [Google Scholar]
- 53.Stoye J P, Moroni C, Coffin J. Virological events leading to spontaneous AKR thymomas. J Virol. 1991;65:1273–1285. doi: 10.1128/jvi.65.3.1273-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor B A. Recombinant inbred strains: use in gene mapping. In: Morse III H C, editor. Origins of inbred mice. New York, N.Y: Academic Press; 1978. pp. 423–438. [Google Scholar]
- 55.Tomonaga K, Coffin J M. Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J Virol. 1998;72:8289–8300. doi: 10.1128/jvi.72.10.8289-8300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trainor C D, Scott M L, Josephs S F, Fry K E, Reitz M J. Nucleotide sequence of the large terminal repeat of two different strains of gibbon ape leukemia virus. Virology. 1984;137:201–205. doi: 10.1016/0042-6822(84)90025-4. [DOI] [PubMed] [Google Scholar]
- 57.Tupper J C, Chen H, Hays E F, Bristol G C, Yoshimura F K. Contributions to transcriptional activity and to viral leukemogenicity made by sequences within and downstream of the MCF13 murine leukemia virus enhancer. J Virol. 1992;66:7080–7088. doi: 10.1128/jvi.66.12.7080-7088.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Beveren C, Rands E, Chattopadhyay S K, Lowy D R, Verma I M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982;41:542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voytek P, Kozak C. HoMuLV: a novel pathogenic ecotropic virus isolated from the European mouse, Mus hortulanus. Virology. 1988;165:469–475. doi: 10.1016/0042-6822(88)90590-9. [DOI] [PubMed] [Google Scholar]
- 60.Voytek P, Kozak C A. Nucleotide sequence and mode of transmission of the wild mouse ecotropic virus, HoMuLV. Virology. 1989;173:58–67. doi: 10.1016/0042-6822(89)90221-3. [DOI] [PubMed] [Google Scholar]
- 61.Winkes B M, Ollmann M M, Barsh G S. Association of Xmv-10 and the non-agouti (a) mutation explained by close linkage instead of causality. Mamm Genome. 1994;5:3–10. doi: 10.1007/BF00360560. [DOI] [PubMed] [Google Scholar]
- 62.Wolgamot G, Bonham L, Miller A D. Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope. J Virol. 1998;72:7459–7466. doi: 10.1128/jvi.72.9.7459-7466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yonekawa H, Gotoh O, Tagashira Y, Matsushima Y, Shi L I, Cho W S, Miyashita N, Moriwaki K. A hybrid origin of Japanese mice “Mus musculus molossinus”. Curr Top Microbiol Immunol. 1986;127:62–67. [PubMed] [Google Scholar]
- 64.Yoshimura F K, Davison B, Chaffin K. Murine leukemia virus long terminal repeat sequences can enhance gene activity in a cell-type-specific manner. Mol Cell Biol. 1985;5:2832–2835. doi: 10.1128/mcb.5.10.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]