Abstract
Interactions between Rev and the Rev-responsive element (RRE) control the order, rate, and extent of gene expression in human immunodeficiency virus type 1. Rev decoys may therefore prove to be useful RNA therapeutics for the treatment of AIDS. To improve upon the current generation of Rev decoys that bind single Rev molecules, it would be useful to generate polyvalent Rev decoys that could bind multiple Rev molecules. J. Kjems and P. A. Sharp (J. Virol. 67:4769–4776, 1993) originally constructed functional polyvalent Rev decoys, but the structural context of these polyvalent decoys remains unclear, and it has been argued that the individual decoys were either structurally discrete (Kjems and Sharp, J. Virol. 67:4769–4776, 1993) or were part of an extended helix (R. W. Zemmel et al., Mol. Biol. 258:763–777, 1996). To resolve the differences between these models, we have designed and synthesized concatemers of Rev-binding elements (RBEs) that fold to form multiple, discrete, high-affinity Rev-binding sites. We find that the concatenated RBEs can facilitate the cytoplasmic transport of viral mRNAs and therefore likely bind multiple Rev molecules. These artificial RREs may simultaneously sequester Rev and hinder access to the cellular transport machinery.
The Rev protein of human immunodeficiency virus type 1 (HIV-1) interacts with a Rev-responsive element (RRE) on retroviral mRNAs to regulate the expression of viral proteins (reviewed in references 5 and 43). Early in the HIV-1 life cycle, fully spliced (∼2-kb) viral messages accumulate in the cytoplasm and produce regulatory proteins such as Rev, Tat, and Nef. Rev then enters the nucleus and facilitates the nucleocytoplasmic transport of incompletely spliced (∼4-kb) and unspliced (∼9-kb) viral messages that encode viral structural proteins (33; reviewed in reference 20).
The mechanism of Rev-dependent mRNA transport has been elucidated in some detail. An N-terminal arginine-rich motif (ARM) within a single Rev protein first interacts with a 30-nucleotide Rev-binding element (RBE) (4, 23, 28) on a 234-nucleotide RRE. The RRE is in turn located within an intron that spans the env gene (14, 34, 56). An oligomerization domain that flanks and overlaps the ARM guides the formation of Rev tetramers and higher-order Rev oligomers (42, 57). After Rev binds to the RBE, additional Rev molecules accumulate on the RRE (6, 36, 57). The specific RNA-protein complex interacts with the nuclear export machinery to promote the transport of incompletely spliced mRNAs that contain the RRE (7, 9, 37, 49). A C-terminal leucine-rich activation domain has been shown to be critical for nucleocytoplasmic transport (33, 36), to act as a nuclear export signal (8, 53; reviewed in reference 11), and to bind to proteins involved in the nuclear pore complex (3, 10). The Rev activation domain redirects RRE-containing mRNAs to the non-mRNA export pathway used by 5S rRNA and U snRNAs (8, 46, 47; reviewed in reference 20). The Rev ARM includes a nuclear localization signal that allows Rev to reenter the nucleus and transport additional mRNA molecules (18). Rev thus shuttles between the nucleus and cytoplasm and regulates the timing and level of structural protein expression by kinetically competing with the splicing machinery (40, 45).
Rev and the RBE have both been targets for the development of antiviral therapies. Mutants of the Rev protein that inhibit oligomerization can transdominantly disrupt Rev function and inhibit viral replication (19, 37, 42, 57). Antisense oligonucleotides (26, 48) and ribozymes (59) directed against the RRE have also been used to interrupt the viral life cycle. Rev decoys based on the RBE have been shown to inhibit RNA-protein interactions and viral replication (30, 31, 54, 55). Similarly, aptamers that can bind tightly and selectively to the Rev protein have been selected from random sequence nucleic acid population (2, 12, 51). These anti-Rev aptamers have also been shown to bind Rev in vivo (50) and to inhibit viral replication (13, 22).
Just as single Rev molecules bind to the RBE whereas multiple Rev molecules accumulate on the RRE, the efficacies of individual Rev decoys can potentially be augmented by building polyvalent Rev decoys in which multiple Rev-binding sites are present on a single RNA transcript. However, it is unclear how polyvalent Rev decoys should be constructed. Kjems and Sharp have suggested that simple concatenation of multiple high-affinity sites may successfully sequester multiple Rev molecules (29). In contrast, Zemmel and coworkers have suggested that Rev accumulates on extended helices adjacent to a single high-affinity site (38, 58). Although these models are not necessarily mutually exclusive, Zemmel et al. (58) have argued that the constructs originally designed by Kjems and Sharp (29) did not in fact fold to form discrete high-affinity sites but instead formed an extended helix. To clearly distinguish between these different structural models, we designed a polyvalent Rev decoy in which multiple, high-affinity Rev-binding sites were presented in a nonhelical structural context. The polyvalent decoy functions as an artificial RRE and efficiently supports mRNA transport in tissue culture cells. These findings have important implications not only for the design of antiviral therapies but also for understanding the mechanism of Rev-dependent viral mRNA transport.
MATERIALS AND METHODS
Materials.
Plasmids pDM128 (17, 18), pDM138 (21), and pRSVRev (17) were generous gifts from T. Hope. Plasmids pDM128 and pDM138 contain the HIV-1 intron that splits the rev gene and flanking exonic sequences; a chloramphenicol acetyltransferase (CAT) reporter gene has been inserted into the intron. The HIV-1 intron and CAT gene are under the control of the simian virus 40 promoter and enhancer (44). In pDM138, a unique ClaI restriction site was introduced into pDM128 in place of the RRE. Plasmid pRSVbgal (2), generously supplied by M. Zapp, contains the β-galactosidase gene under the control of the strong Rous sarcoma virus (RSV) promoter. Plasmids to be used in cellular transfection experiments were purified by using Qiagen (Chatsworth, Calif.) columns. Media, reduced-serum media, supplements, phosphate-buffered saline, and Lipofectamine were purchased from Gibco BRL (Gaithersburg, Md.). Fetal bovine serum was purchased from Summit Biotechnologies (Fort Collins, Colo.).
Cell lines.
African green monkey kidney (CV-1) cells were grown according to standard procedures in high-glucose Dulbecco’s modified Eagle medium supplemented with fetal bovine serum (11%), penicillin (2.5 U/ml), streptomycin (2.5 mg/ml), and l-glutamine (2 mM) in 24-well cell culture plates (Costar, Cambridge, Mass.).
Construction of RBE concatemers.
A modular synthesis scheme was devised for the construction of RBE concatemers. Three overlapping pairs of oligonucleotides encoded a 5′ T7 RNA polymerase promoter, an RBE-linker monomer, and a 3′ cap, respectively. The paired oligonucleotides corresponding to the RBE-linker monomer contained overhangs that facilitated their oligomerization. These overhangs also facilitated addition of a T7 RNA polymerase promoter to the 5′ end of RBE concatemers and a constant sequence cap to the 3′ end. Both constant sequences also contained ClaI restriction sites to facilitate cloning. The sequences of the oligonucleotides are shown in Table 1.
TABLE 1.
Oligonucleotides used
| Oligonucleotide | Sequencea |
|---|---|
| 5′ T7 promoter | 5′ AAT TCT AAT ACG ACT CAC TAT AGG GAG ATC GAT |
| 5′ AGA GAA GAG ATC GAT CTC CCT ATA GTG AGT CGT ATT A | |
| RBE-linker monomer | 5′ CTC TTC TCT GGT GGG CGC AGC TTC GGC TGA CGG TAC ACC |
| 5′ AGA GAA GAG GGT GTA CCG TCA GCC GAA GCT GCG CCC ACC | |
| 3′ cap | 5′ CTC TTC TCT ATC GAT ACC CAA AGT CGT GAC TGG GAA AAC G |
| 5′ GAT CCG TTT TCC CAG TCA CGA CTT TGG GTA TCG AT |
ClaI restriction sites are italicized, and the T7 RNA polymerase promoter is shown in boldface.
All oligonucleotides were phosphorylated by using T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) for 30 min at room temperature. The oligonucleotides encoding the RBE linker monomers were denatured for 1 min at 95°C, annealed for 3 min at 45°C, and incubated at room temperature for at least 10 min. The oligonucleotide concatemers that were formed were ligated with T4 DNA ligase (New England Biolabs). The T7 promoter and 3′ cap were then ligated to the RBE concatemers. PCR amplification yielded a mixture of products which served as templates for Ampliscribe in vitro transcription reactions (Epicentre Technologies, Madison, Wis.). Transcribed RNAs were treated with DNase I and gel purified on a 6% polyacrylamide denaturing gel. Individual bands were eluted from the gel, ethanol precipitated, and reverse transcribed by using avian myeloblastosis virus reverse transcriptase (Seikagaku America, Ijamsville, Md.) for 1 h at 42°C. The reverse transcription reaction products corresponding to individual concatemers were then PCR amplified and cloned into the pCRII vector from the TA cloning kit (Invitrogen, San Diego, Calif.). Individual colonies were screened for the presence of RBE-linker inserts. Clones containing concatemers corresponding to one to five tandem copies of the RBE (T1 to T5) were identified, and the sequences of the concatemers were confirmed by standard dideoxy sequencing methods. The RBE concatemers were PCR amplified from the pCRII vector, digested with ClaI, and cloned into ClaI-digested pDM138 to generate plasmids pDM138-T1 to pDM138-T5. The sequences and orientations of the concatemers in pDM138 were verified by dideoxy sequencing. The insert in plasmid pDM138-T5 contained point mutations in two of the linker regions (one linker lacked a U, while the second had an additional C).
Nuclease mapping.
A DNA template corresponding to T5 was transcribed in vitro by using an Ampliscribe transcription kit according to the manufacturer’s directions. The transcribed RNA was purified on a 6% denaturing polyacrylamide gel, ethanol precipitated, and dephosphorylated with alkaline phosphatase (Boehringer Mannheim, Indianapolis, Ind.). The dephosphorylated RNA was phenol-chloroform extracted and treated with T4 polynucleotide kinase (New England Biolabs) in the presence of 5 fmol of [γ-32P]ATP (7,000 Ci/mmol; ICN, Costa Mesa, Calif.). The radiolabeled RNA was again purified on a 6% polyacrylamide gel and precipitated. Radiolabeled T5 was digested with various amounts of RNase A (1, 0.1, 0.01, 0.001, and 0.0001 U) or RNase T1 (10, 5, 2.5, 1.25, 0.63, and 0.31 U) in 6 μl of Hanks balanced salt solution (1) at 37°C for 5 min. The digestion reactions were quenched with 2 μg of yeast tRNA (Gibco Life Technologies, Gaithersburg, Md.) in 4 μl of 0.5 M EDTA, immediately phenol-chloroform extracted, and precipitated. To determine the spacings between hydrolysis products, 0.5 μg of radiolabeled T5 RNA was hydrolyzed in the presence of 25 mM sodium bicarbonate, 1 mM EDTA, and 10 μg of yeast tRNA. This alkaline hydrolysis reaction mixture was heated to 90°C for 10 min, neutralized with 2 μl of 0.5 M acetic acid, and ethanol precipitated. Hydrolysis products were separated on a 6% denaturing polyacrylamide gel, and the digestion patterns were analyzed with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Cellular assays of RBE concatemers.
To determine if RBE concatemers supported Rev function, the reporter plasmids were cotransfected with a Rev expression plasmid, pRSVRev, and CAT activities were assessed. One day prior to transfection, CV-1 cells were counted with a Coulter (Hialeah, Fla.) cell counter, and 55,000 cells were seeded into each well of a 24-well cell culture plate. The next day, the cells (at 70 to 80% confluency) were transiently transfected in duplicate with either a Rev-responsive reporter plasmid (pDM128; 0.2 μg) or derivatives of pDM138 containing RBE concatemers (0.2 μg), pRSVbgal (0.5 μg) as a control for transfection efficiency, and a Rev expression plasmid (pRSVRev). The amount of pRSVRev used in each transfection was varied between limiting pRSVRev (0.01 μg), saturating pRSVRev (0.2 μg), or a titration of pRSVRev (0.002 μg to 0.2 μg). Cells were prepared for transfection by preincubation for 30 min in 1 ml of OptiMEM reduced-serum medium. Plasmid DNAs (1.2 μg in total, including various amounts of a carrier plasmid, pUC118) in 100 μl of OptiMEM were mixed with 1.5 μl of Lipofectamine in 100 μl of OptiMEM. Lipid amalgams (200 μl) were incubated at ambient temperature for 30 min to allow complex formation to occur; the mixture was added to cells in a total volume of 0.5 ml of OptiMEM (7.2 ng of liposome:2.4 ng of DNA per ml [final concentration]) and incubated for an additional 4.5 to 5 h at 37°C. The transfection medium was removed from the wells and replaced with complete medium. Forty-eight hours posttransfection, the cells were washed with 1 ml of phosphate-buffered saline and lysed in 0.25 M Tris-Cl (pH 7.6)–0.5% Triton X-100 (150 μl). Harvested cellular extracts (100 μl) were centrifuged for 5 min, and the supernatant was used for CAT assays and β-galactosidase assays.
CAT activity was measured according to standard procedures (32). Cellular extracts (40 μl) were mixed with acetyl coenzyme A (0.45 mM [final concentration]; Pharmacia, Piscataway, N.J.), glycerol (1.8% final), and 14C-labeled chloramphenicol (0.1 μCi; 50 mCi/mmol; DuPont NEN, Boston, Mass.) in 176 μl of 0.14 M Tris-Cl (pH 7.6). The reaction mix was incubated at 37°C for 1.5 h; control experiments indicated that this was within the linear range of the assay. Chloramphenicol and less polar acetylated products were ethyl acetate extracted and separated by thin-layer chromatography in 5% methanol–95% chloroform on silica gel IB2 sheets (J. T. Baker, Phillipsburg, N.J.). The labeled chloramphenicol products were quantitated with a Phosphorimager (Molecular Dynamics). The amount of CAT activity present in each extract was defined as the percentage of total chloramphenicol that had been converted to acetylated products. CAT activities were normalized to the amount of β-galactosidase activity detected in each extract. The β-galactosidase levels were determined by incubating 15 μl of extract with 0.1 mM MgCl2, 0.35% β-mercaptoethanol (Sigma, St. Louis, Mo.), and 0.88 mg of o-nitrophenyl-β-d-galactopyranoside (Sigma) per ml in a final volume of 150 μl of 0.1 M NaH2PO4-Na2HPO4. After incubation at 37°C for 45 min, β-galactosidase activities were determined with a microtiter plate reader (Cambridge Technology, Cambridge, Mass.) fitted with a 450-nm filter.
RESULTS AND DISCUSSION
The structural context of polyvalent Rev decoys.
Rev decoys have been developed as potential therapeutics to combat HIV infection. The RBE mounted on a retroviral vector sequesters Rev and inhibits viral replication (30, 31, 54, 55). Anti-Rev aptamers that have higher affinities for Rev than the wild-type RBE have been selected from random-sequence populations (2, 12, 51) and have been shown to be efficient Rev decoys both in vitro and in vivo (13, 22, 50).
To further improve the efficacies of Rev decoys, we wished to design RNA molecules that would capture multiple Rev molecules. The fact that Rev responsiveness is potentiated by the accumulation of multiple Rev molecules on the RRE (33, 36) suggested that polyvalent decoys might prove to be especially effective at blocking viral replication. Biochemical analyses have previously revealed that 7 to 8 Rev molecules form a stable complex with the RRE in vitro (6), and up to 11 to 12 Rev molecules may contribute to full Rev responsiveness in vivo (38). Moreover, the accumulation of Rev on the RRE is assisted by the tetramerization or oligomerization of Rev monomers (15, 36, 42, 57), and mutations in the multimerization domain of Rev inhibit in vivo Rev function (36, 57).
The simplest model for the design of polyvalent Rev decoys would be the concatenation of multiple high-affinity Rev-binding sites. In support of this model, fusion proteins between Rev and the bacteriophage MS2 coat protein directed transport of intron-containing RNAs when multiple MS2 phage operator sites were present (39, 52). These studies found that three to four MS2 operator sites were required for efficient Rev-dependent mRNA transport (39). Similarly, Kjems and Sharp (29) designed constructs that contained one, three, or six copies of RBEs. RNAs containing only one RBE fostered no Rev-dependent mRNA transport, while RNAs expressing three copies of the RBE had an intermediate Rev response (55% of the wild-type RRE level), and six copies produced an almost full Rev response (74% of the wild-type RRE level). A hexamer RBE has also been shown to mediate Rev-dependent suppression of RNA splicing in vitro (27).
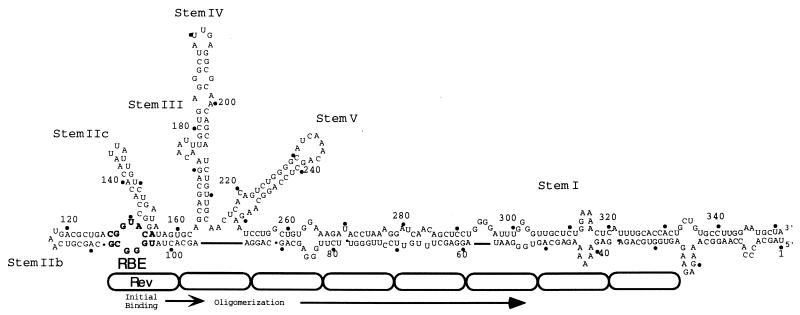
However, a competing model suggested that the simple concatenation of high-affinity Rev-binding sites might not direct the accumulation of multiple Rev molecules (38, 58). In this model, the high-affinity RBE within stem IIB of the RRE binds the first Rev molecule, while the adjacent stem I, a long and imperfect RNA duplex, mediates the subsequent accumulation of additional Rev molecules (23, 38, 58) (Fig. 1). Nuclease protection studies and gel shift analyses have shown that initial Rev binding to the high-affinity RBE is required for additional binding to adjacent, lower-affinity RNA duplexes (38, 58). Deletion analyses originally revealed that the presence of stem I was important for Rev responsiveness (35) and that truncation of stem I reduces the number of Rev molecules bound in vitro and Rev responsiveness in vivo (38). Taken together, these results support the notion that coupled high- and low-affinity sites on the RRE function as a molecular rheostat that is sensitive to the concentration of Rev in a cell (38).
FIG. 1.
The molecular rheostat model for the accumulation of Rev on the RRE. One model for Rev function suggests that Rev initially binds to the RBE whereas subsequent Revs oligomerize along an adjacent, imperfectly paired duplex in stem I (38, 58). In this model, the overall architecture of the RRE complex plays a critical role in sequestering multiple Rev molecules. The RRE is drawn and numbered according to Mann et al. (38). Rev protein molecules are represented as oblongs; the actual number and alignment of Revs on the RRE is unknown.
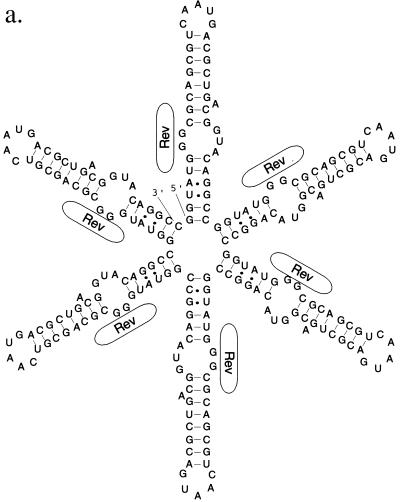
While these two models are not of necessity mutually exclusive, Zemmel et al. (58) have argued that the RBE concatemers constructed by Kjems and Sharp (29) (Fig. 2a) could be folded into an alternative structure in which one or more RBEs was adjacent to a long and imperfect RNA duplex (Fig. 2b). An energetic analysis (60) of the Kjems and Sharp (29) RBE concatemers suggests that the alternative conformation predicted by Zemmel et al. (58) was in fact the more favorable conformation. In this view, increasing numbers of RBEs promoted greater levels of mRNA transport not because multiple, high-affinity Rev-binding sites were sequentially introduced into a transcript but rather because a proportionately longer, albeit unplanned, stem structure was formed. Before design strategies for polyvalent RBEs could be established it was first necessary to resolve these disparate interpretations.
FIG. 2.
Alternative structures of RBE concatemers. (a) Design of an RBE hexamer by Kjems and Sharp (29). The desired structure is shown. Rev protein molecules interacting with the internal loop of the RBE are again represented as oblongs. (b) Predicted secondary structure of the RBE hexamer. When the secondary structure of the RBE hexamer shown in panel a is modeled with the program Mulfold (24, 25, 60), an elongated RNA originally predicted by Zemmel et al. (58) is observed.
Concatenation of discrete RBEs into polyvalent Rev decoys.
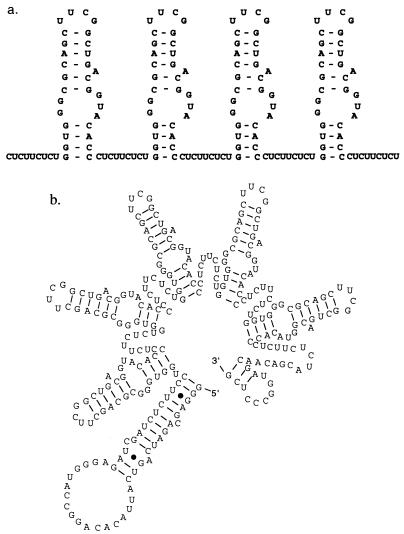
A minimal, 30-nucleotide, stem-internal loop-stem RBE has previously been shown to interact with Rev monomers both in vitro (23, 28) and in vivo (16, 35). We therefore attempted to link together several RBEs to form polyvalent Rev decoys (Fig. 3a). The RBEs were topped with stable tetraloops (UUCG [41]) that should have guided the formation of discrete stem-internal loop-stem structures within the cell, and they were flanked by pyrimidine spacers that should not have allowed the formation of an extended helix similar to that shown in Fig. 2b. An energetic analysis of the RBE concatemers suggested that the structure shown in Fig. 3b was likely the most favorable conformation.
FIG. 3.
Design and construction RBE concatemers. (a) Design of RBE concatemers. Individual RBEs were capped with a stable UUCG tetraloop sequence and separated by nine-nucleotide pyrimidine linkers (CUCUUCUCU). The tetramer T4 is shown. (b) Predicted secondary structure of a RBE pentamer. When the secondary structure of the RBE concatemer T5 (this report) is modeled with the program Mulfold, a discrete series of unit-length RBE structures is observed.
The length (>200 nucleotides) of the larger RBE concatemers prevented their direct synthesis as a single oligonucleotide, and a modular synthetic scheme was instead devised. Individual oligonucleotides encoding RBE-linker monomers were synthesized. The RBE monomers were ligated to form concatemers, which were then capped with oligonucleotides that allowed PCR amplification; one of the capping oligonucleotides also contained a T7 RNA polymerase promoter to further biochemical analysis. Following amplification, a ladder of bands corresponding to concatemers of different lengths was observed on an agarose gel. The concatemer pool was cloned, and concatemers T1, T2, T3, T4, and T5 were recovered and sequenced. To further buttress our contention that the concatemers folded into discrete Rev-binding sites, the secondary structure of the T5 concatemer was mapped in vitro (Fig. 4). The nuclease sensitivity of T5 is consistent with the formation of unit-length RBEs and inconsistent with the formation of an extended secondary structure.
FIG. 4.
Mapping the structure of T5. The secondary structure of T5 was mapped with RNase T1 and RNase A as described in Materials and Methods. Lanes 1 to 5, digestion of T5 with decreasing concentrations of RNase A; lanes 7 to 12, digestion of T5 with decreasing concentrations of RNase T1; lanes 6 and 13, alkaline hydrolysis ladder of T5. A repeating pattern is observed with both nucleases; elements of this pattern are indicated by arrows stretching between the proposed secondary structure of the RBE concatemer and the digests. The sizes and spacings of the digestion products that make up this pattern are consistent with the formation of unit-length RBEs that contain single-stranded pyrimidines (in the case of RNase A) or single-stranded guanosines (in the case of RNase T1). The digestion patterns are inconsistent with the formation of a long, extended helix.
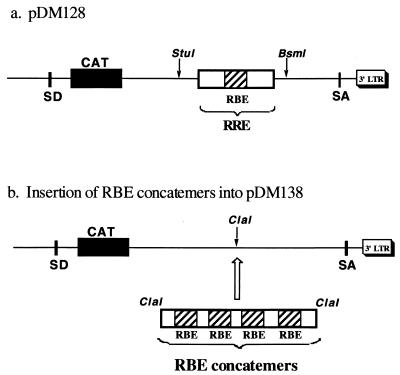
The RBE concatemers were subcloned into pDM138, a well-characterized Rev-dependent CAT reporter for use with tissue culture cells (17, 18, 21). Plasmid pDM138 is derived from pDM128 (Fig. 5a), which contains the RRE and a CAT reporter gene within an HIV-1 intron. In the absence of Rev, RNAs transcribed from pDM128 are spliced and negligible CAT activity is observed. When Rev is present and interacts with the RRE, Rev-mediated mRNA transport competes with splicing and CAT activity is detected. Plasmid pDM138 contains a unique ClaI site in place of the RRE (Fig. 5b). By placing the RBE concatemers into the pDM138 reporter, constructs that were superficially similar to pDM128 were formed. If the RBE concatemers can in fact bind multiple Rev molecules and thereby be transported from the nucleus, then higher amounts of CAT activity should be present in transfected cellular extracts in the presence of Rev than in its absence.
FIG. 5.
Reporter plasmids. (a) Plasmid pDM128 (17, 18), which serves as a positive control and contains the wild-type RRE within an HIV-1 intron adjacent to a CAT reporter gene. SD, splice donor; SA, splice acceptor; LTR, long terminal repeat. (b) Insertion of RBE concatemers into pDM138. Plasmid pDM138 (21), a derivative of pDM128, contains a unique ClaI site in place of the RRE. RBE concatemers T1, T2, T3, T4, and T5 were inserted into the ClaI site of pDM138.
Optimization of assay conditions.
To determine how much reporter plasmid should be transfected into cells to obtain a robust Rev response, we first carried out a series of ranging experiments. Plasmid pRSVRev (driver) expresses the Rev protein under the control of the strong RSV promoter. Various amounts of the pRSVRev driver were transfected into CV-1 African green monkey kidney cells along with the parental reporter plasmid, pDM128. A plasmid that contained the β-galactosidase gene was transfected in parallel with the reporter and driver plasmids as a control for transfection efficiencies (2). At 48 h posttransfection, the cells were harvested and levels of CAT activity were determined and normalized to levels of β-galactosidase activity. As shown in Fig. 6, when 0.2 μg of pDM128 is transfected into tissue culture cells the cotransfection of small amounts (e.g., 0.01 μg) of pRSVRev leads to the production of small amounts of CAT, while the cotransfection of large amounts of pRSVRev (e.g., 0.2 μg) saturates the Rev-dependent CAT signal. These results are similar to those previously observed in our lab with pDM128 and mutant derivatives of pDM128 (50).
FIG. 6.
Rev responsiveness as a function of pRSVRev concentration. The relative activities of the wild-type RRE in pDM128 are shown as a function of the amount of pRSVRev included in transfection experiments. CV-1 cells were transiently transfected with constant amounts of pDM128 (0.2 μg) and constant amounts of pRSVbgal (0.5 μg). The amounts of pRSVRev included in each transfection varied (0.002, 0.01, 0.02, 0.05, and 0.2 μg). CAT activities (arbitrary units) were determined and normalized to β-galactosidase levels (β-gal) (arbitrary units). The results of eight separate determinations (four repetitions in duplicate) were averaged. Bars indicate standard deviations of the data.
Polyvalent Rev decoys function as artificial RREs.
RBE concatemers in pDM138 were cotransfected with pRSVRev, and CAT signals were assayed at 48 h posttransfection. Since the Rev-dependent CAT signal from the pDM128 reporter was readily observed and reproducible in the presence of both limiting and saturating amounts of cotransfected pRSVRev driver, the RBE concatemer reporter constructs were assayed under both conditions. A representative sample of the data is shown in Fig. 7; a graphical summary of all of the data is shown in Fig. 8.
FIG. 7.
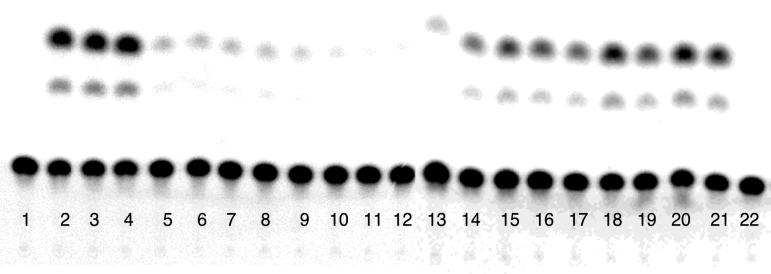
RBE concatemers are Rev responsive. A representative sample of results of CAT assays carried out with the RBE concatemer series is shown. Lane 1, no-plasmid control; lanes 2 to 4, pDM128 plus pRSVREV (positive control; triplicate determinations); lanes 5, 6, and 22, pDM138 plus pRSVRev (negative control); lanes 7 to 11, RBE concatemers pDM138-T1 through pDM138-T5, respectively; lanes 12 to 21, RBE concatemers pDM138-T1 through pDM138-T5 plus pRSVRev, respectively (duplicate determinations).
FIG. 8.
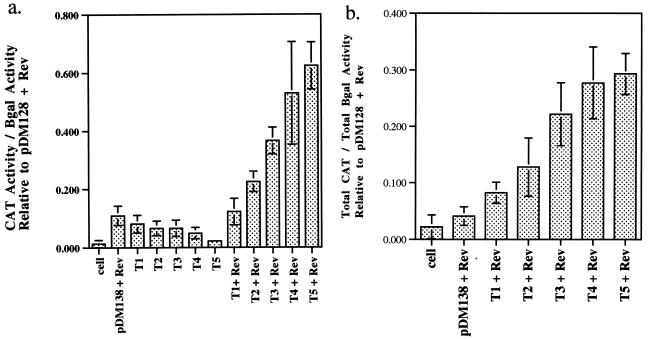
Rev responsiveness of RBE concatemers at limiting and saturating concentrations of pRSVRev. Graphical summaries of the data from Fig. 4 and other determinations. CV-1 cells were transiently transfected with constant amounts of RBE concatemer (0.2 μg), pRSVbgal (0.5 μg), and limiting (0.01 μg) (a) or saturating (0.2 μg) (b) levels of pRSVRev. The CAT activities of RBE concatemers were normalized to β-galactosidase (βgal) activities and to the CAT activity of the positive control (pDM128 plus pRSVRev). Results of six separate determinations (three repetitions in duplicate) were averaged. Bars indicate standard deviations of the data.
In the absence of Rev, the RBE concatemer reporters (pDM138-T1, -T2, -T3, -T4, and -T5) produced only negligible levels of CAT activity, comparable to results for a no-transfection control and to pDM138 in the presence of Rev. However, when the Rev expression plasmid was included in the transfection experiment, substantially greater amounts of CAT activity were observed. The levels of Rev responsiveness increased in a graded fashion; each RBE that was added to a concatemer led to the production of additional CAT activity. At limiting concentrations of the driver plasmid, the amount of Rev-dependent CAT activity observed ranged from 1.3% of wild-type activity (pDM128) for pDM138-T1 to 51.5% of wild-type activity for pDM138-T5 (background activity subtracted relative to Fig. 8). At saturating concentrations of the driver plasmid, the values ranged from 4.2% above background for pDM138-T1 to 25.2% for pDM138-T5.
Rev responsiveness of artificial RREs as a function of Rev concentration.
The experiments at limiting and saturating concentrations of the Rev expression plasmid gave some indication of how the artificial RREs differed from their natural counterparts. To derive a fuller understanding of how the Rev responses of the artificial and natural elements differed or overlapped, Rev titration experiments similar to those originally carried out with pDM128 were also carried out with pDM138-T2 and -T5 (Fig. 9). The RBE concatemer reporter pDM138-T2 was chosen rather than pDM138-T1 because the extremely small amounts of signal produced by pDM138-T1 would have been difficult to follow at low concentrations of pRSVRev. The amounts of RBE concatemer reporter plasmids used for transfection were held constant (0.2 μg), while the amounts of pRSVRev were varied from 0.002 to 0.2 μg.
FIG. 9.
Rev responsiveness of artificial RREs as a function of pRSVRev concentration. The relative CAT activities of pDM138-T2 and pDM138-T5 are shown as a function of Rev concentration. CV-1 cells were transiently transfected with constant amounts of T2, T5, or pDM128 (0.2 μg), constant amounts of pRSVbgal (0.5 μg), and various amounts of pRSVRev (0.002, 0.01, 0.02, 0.05, and 0.2 μg). CAT activities were normalized to β-galactosidase (β-gal) levels. Results of eight separate determinations (four repetitions in duplicate) were averaged.
In consonance with the results shown in Fig. 8, the RBE concatemer reporter pDM138-T5 was again more Rev responsive than pDM138-T2 and elicited higher levels of Rev-dependent CAT activity. Interestingly, the level of Rev-responsive mRNA transport exhibited by pDM138-T5 at saturating concentrations of the driver plasmid was roughly 2.5 times the level exhibited by pDM138-T2 at saturating concentrations of the driver plasmid. This result is consistent with a model in which the number of Rev-binding sites on a multivalent decoy is the sole determinant of the level of mRNA transport. However, both pDM138-T2 and pDM138-T5 reached maximal levels of Rev responsiveness when approximately 0.01 μg of pRSVRev was added, while the RRE did not reach maximal levels of Rev responsiveness until approximately 0.05 μg of pRSVRev had been added (Fig. 6). The results presented in Fig. 6, 8, and 9 indicate that there is not only a quantitative difference between the natural and artificial RREs but a qualitative one as well. While concatenated Rev-binding sites allow a transcript to be transported in a Rev-dependent fashion, they do not necessarily respond to Rev or accumulate Rev in the same manner as the natural RRE does.
Toward design principles for polyvalent decoys and artificial RREs.
By eliminating heterologous (i.e., MS2) components (39, 52) and by clarifying the structures of the RNA substrates (29), our studies are the first to fully delineate and delimit Rev-RNA interactions in a functional (albeit artificial) RRE. The graded increase in Rev-dependent mRNA transport activity observed with increasing numbers of RBEs confirms the findings of Kjems and Sharp (29) and is consistent with the model in which multiple, discrete Rev-RNA complexes can mediate Rev responsiveness. The use of discrete Rev-binding sites greatly simplifies the analysis of where and how many Rev molecules are bound by a RNA molecule. For example, Huang et al. (21) demonstrated that a single copy of an 88-nucleotide RRE fragment encompassing the RBE supported a partial Rev response (26% of the wild-type RRE response), but it was unclear how many Rev-binding sites may have been present on this fragment. Based on our results, a similar level of Rev-dependent mRNA transport would have required approximately five independent, high-affinity Rev-binding sites (see also Fig. 8). By simplifying the structural context for the presentation of high-affinity Rev-binding sites, it should now be possible to directly compare the efficacies of polyvalent Rev decoys in which the number, spacing, and affinities of the binding sites are varied.
However, it should be noted that our results are also in accord with the rheostat model for the accumulation of Rev on the RRE, since artificial RREs with either two or five RBEs saturated at roughly the same concentration of the driver plasmid, while the natural RRE saturated at a higher concentration. Thus, it is likely that the discrete-binding-site and rheostat models may be simultaneously correct. To accumulate sufficient Rev molecules to mediate mRNA transport, it is sufficient either to have multiple, independent high-affinity Rev-binding sites or to have multiple, interdependent high- and low-affinity Rev-binding sites. Thus, in addition to varying the placement and type of Rev decoys in a polyvalent decoy, it would likely be fruitful to explore whether the inclusion of additional secondary structural elements can drive the cooperative accumulation of even more Rev molecules.
Overall, our results support the simple design principle that we originally set out to assess, that simple concatenation of Rev-binding sites can be used to generate polyvalent Rev decoys. Most importantly, since polyvalent Rev decoys can function as artificial RREs, they may be able to inhibit viral replication by additional mechanisms, such as binding cytoplasmic as well as nuclear Rev or competing for nuclear export pathways.
ACKNOWLEDGMENT
This work was supported by Department of Health and Human Services grant AI-36083 from the National Institutes of Health.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R, Moore D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 2.Bartel D P, Zapp M L, Green M R, Szostak J W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 3.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 4.Cook K S, Fisk G J, Hauber J, Usman N, Daly T J, Rusche J R. Characterization of HIV-1 Rev protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 1991;19:1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen B R. Regulation of HIV-1 gene expression. FASEB J. 1991;5:2361–2367. doi: 10.1096/fasebj.5.10.1712325. [DOI] [PubMed] [Google Scholar]
- 6.Daly T J, Doten R C, Rennert P, Auer M, Jaksche H, Donner A, Fisk G, Rusche J R. Biochemical characterization of binding of multiple HIV-1 Rev monomeric proteins to the Rev responsive element. Biochemistry. 1993;32:10497–10505. doi: 10.1021/bi00090a028. [DOI] [PubMed] [Google Scholar]
- 7.Fischer U, Meyer S, Teufel M, Heckel C, Luhrmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 9.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 10.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 11.Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 12.Giver L, Bartel D, Zapp M, Pawul A, Green M, Ellington A D. Selective optimization of the Rev-binding element of HIV-1. Nucleic Acids Res. 1993;21:5509–5516. doi: 10.1093/nar/21.23.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Good P D, Krikos A J, Li S X L, Bertrand E, Lee N S, Giver L, Ellington A D, Zaia J A, Rossi J J, Engelke D R. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 1997;4:45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
- 14.Hadzopoulou-Cladaras M, Felber B, Cladaras C, Athanassopoulos A, Tse A, Pavlakis G N. The Rev (Trs/Art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the Env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heaphy S, Finch J T, Gait M J, Karn J, Singh M. Human immunodeficiency virus type 1 regulator of virion expression, rev, forms nucleoprotein filaments after binding to a purine-rich “bubble” located with the rev response region of viral mRNAs. Proc Natl Acad Sci USA. 1991;88:7366–7370. doi: 10.1073/pnas.88.16.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holland S M, Chavez M, Gerstberger S, Venkatesan S. A specific sequence with a bulged guanosine residue(s) in a stem-bulge-stem structure of Rev-responsive element RNA is required for trans activation by human immunodeficiency virus type 1 Rev. J Virol. 1992;66:3699–3706. doi: 10.1128/jvi.66.6.3699-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hope T J, McDonald D, Huang X, Low J, Parslow T G. Mutational analysis of the human immunodeficiency virus type 1 Rev transactivator: essential residues near the amino terminus. J Virol. 1990;64:5360–5366. doi: 10.1128/jvi.64.11.5360-5366.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hope T J, Huang X, McDonald D, Parslow T G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hope T J, Klein N P, Elder M E, Parslow T G. Trans-dominant inhibition of human immunodeficiency virus type 1 Rev occurs through formation of inactive protein complexes. J Virol. 1992;66:1849–1855. doi: 10.1128/jvi.66.4.1849-1855.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hope T J. Viral RNA export. Chem Biol. 1997;4:335–344. doi: 10.1016/s1074-5521(97)90124-1. [DOI] [PubMed] [Google Scholar]
- 21.Huang X, Hope T J, Bond B L, McDonald D, Gahl K, Parslow T G. Minimal Rev-response element for type 1 human immunodeficiency virus. J Virol. 1991;65:2131–2134. doi: 10.1128/jvi.65.4.2131-2134.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inouye R T, Du B, Boldt-Houle D, Ferrante A, Park I-W, Hammer S M, Duan L, Groopman J E, Pomerantz R J, Terwilliger E F. Potent inhibition of human immunodeficiency virus type 1 in primary T cells and alveolar macrophages by a combination anti-Rev strategy delivered in an adeno-associated virus vector. J Virol. 1997;71:4071–4078. doi: 10.1128/jvi.71.5.4071-4078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwai S, Pritchard C, Mann D A, Karn J, Gait M J. Recognition of the high affinity binding site in rev-response element RNA by the human immunodeficiency virus type 1 rev protein. Nucleic Acids Res. 1992;20:6465–6472. doi: 10.1093/nar/20.24.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 26.Junker U, Rittner K, Homann M, Bevec D, Bohnlein E, Sczakiel G. Reduction in replication of the human immunodeficiency virus type 1 in human T cell lines by polymerase III-drived transcription of chimeric tRNA-antisense RNA genes. Antisense Res Dev. 1994;4:165–172. doi: 10.1089/ard.1994.4.165. [DOI] [PubMed] [Google Scholar]
- 27.Kjems J, Frankel A D, Sharp P A. Specific regulation of mRNA splicing in vitro by a peptide from HIV-1. Cell. 1991;67:169–178. doi: 10.1016/0092-8674(91)90580-r. [DOI] [PubMed] [Google Scholar]
- 28.Kjems J, Calnan B J, Frankel A D, Sharp P A. Specific binding of basic peptide from HIV-1 Rev. EMBO J. 1992;11:1119–1129. doi: 10.1002/j.1460-2075.1992.tb05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kjems J, Sharp P A. The basic domain of Rev from human immunodeficiency virus type 1 specifically blocks the entry of U4/U6-U5 small nuclear ribonucleoprotein in spliceosome assembly. J Virol. 1993;67:4769–4776. doi: 10.1128/jvi.67.8.4769-4776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S-W, Gallardo H F, Gilboa E, Smith C. Inhibition of human immunodeficiency virus type 1 in human T cells by a potent Rev response element decoy consisting of the 13-nucleotide minimal Rev-binding domain. J Virol. 1994;68:8254–8264. doi: 10.1128/jvi.68.12.8254-8264.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee T C, Sullenger B A, Gallardo H F, Ungers G E, Gilboa E. Overexpression of RRE-derived sequences inhibits HIV-1 replication in CEM cells. New Biol. 1992;4:66–74. [PubMed] [Google Scholar]
- 32.Lin Y S, Green M R. Similarities between prokaryotic and eukaryotic cyclic AMP-responsive promoter elements. Nature. 1989;340:656–659. doi: 10.1038/340656a0. [DOI] [PubMed] [Google Scholar]
- 33.Malim M H, Bohnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev trans-activator: derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 34.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 35.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 36.Malim M H, Cullen B R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 37.Malim M H, McCarn D F, Tiley L S, Cullen B R. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mann D A, Mikaelian I, Zemmel R W, Green S M, Lowe A D, Kimura T, Singh M, Butler J G, Gait M J, Karn J. Co-operative rev binding to stem I of the rev-response element modulates human immunodeficiency virus type 1 late gene expression. J Mol Biol. 1994;241:193–207. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- 39.McDonald D, Hope T J, Parslow T G. Posttranscriptional regulation by the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type 1 Rex proteins through a heterologous RNA binding site. J Virol. 1992;66:7232–7238. doi: 10.1128/jvi.66.12.7232-7238.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 41.Molinaro M, Tinoco I J. Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: thermodynamic and spectroscopic applications. Nucleic Acids Res. 1995;23:3056–3063. doi: 10.1093/nar/23.15.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen H S, Cochrane A W, Dillon P J, Nalin C M, Rosen C A. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 1990;4:1357–1364. doi: 10.1101/gad.4.8.1357. [DOI] [PubMed] [Google Scholar]
- 43.Parslow T G. Post-transcriptional regulation of human retroviral gene expression. In: Cullen B R, editor. Human retroviruses. New York, N.Y: Oxford University Press; 1993. pp. 101–136. [Google Scholar]
- 44.Peterlin B M, Luciw P A, Barr P J, Walker M D. Elevated levels of mRNA can account for the trans-activation of human immunodeficiency virus. Proc Natl Acad Sci USA. 1986;83:9734–9738. doi: 10.1073/pnas.83.24.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richard N, Iacampo S, Cochrane A. HIV-1 Rev is capable of shuttling between the nucleus and cytoplasm. Virology. 1994;204:123–131. doi: 10.1006/viro.1994.1516. [DOI] [PubMed] [Google Scholar]
- 46.Ruhl M, Himmelspach M, Bahr G M, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington G K, Probst H, Bevec D, Hauber J. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating transactivation. J Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schatz O, Oft M, Dascher C, Schebesta M, Rosorius O, Jaksche H, Dobrovnik M, Bevec D, Hauber J. Interaction of the HIV-1 Rev cofactor eukaryotic initiation factor 5A with ribosomal protein L5. Proc Natl Acad Sci USA. 1998;95:1607–1612. doi: 10.1073/pnas.95.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sczakiel G, Oppenlander M, Rittner K, Pawlita M. Tat- and Rev-directed antisense RNA expression inhibits and abolishes replication of human immunodeficiency virus type 1: a temporal analysis. J Virol. 1992;66:5576–5581. doi: 10.1128/jvi.66.9.5576-5581.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stutz F, Izaurralde E, Mattaj I W, Rosbash M. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol Cell Biol. 1996;16:7144–7150. doi: 10.1128/mcb.16.12.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Symensma T L, Giver L, Zapp M, Takle G B, Ellington A D. RNA aptamers selected to bind human immunodeficiency virus type 1 Rev in vitro are Rev responsive in vivo. J Virol. 1996;70:179–187. doi: 10.1128/jvi.70.1.179-187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuerk C, MacDougal-Waugh S. In vitro evolution of functional nucleic acids: high-affinity RNA ligands of HIV-1 proteins. Gene. 1993;137:33–39. doi: 10.1016/0378-1119(93)90248-2. [DOI] [PubMed] [Google Scholar]
- 52.Venkatesan S, Gerstberger S M, Park H, Holland S M, Nam Y S. Human immunodeficiency virus type 1 Rev activation can be achieved without Rev responsive element RNA if Rev is directed to the target as a Rev/MS2 fusion protein which tethers the MS2 operator RNA. J Virol. 1992;66:7469–7480. doi: 10.1128/jvi.66.12.7469-7480.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 54.Yamada O, Kraus G, Luznik L, Yu M, Wong-Staal F. A chimeric human immunodeficiency virus type 1 (HIV-1) minimal Rev response element-ribozyme molecule exhibits dual antiviral function and inhibits cell-cell transmission of HIV-1. J Virol. 1996;70:1596–1601. doi: 10.1128/jvi.70.3.1596-1601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuyama N, Ohkawa J, Koguma T, Shirai M, Taira K. A multifunctional expression vector for an anti-HIV-1 ribozyme that produces a 5′- and 3′-trimmed trans-acting ribozyme, targeted against HIV-1 RNA, and cis-acting ribozymes that are designed to bind to and thereby sequester trans-activator proteins such as Tat and Rev. Nucleic Acids Res. 1994;22:5060–5067. doi: 10.1093/nar/22.23.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zapp M L, Green M R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- 57.Zapp M L, Hope T J, Parslow T G, Green M R. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc Natl Acad Sci USA. 1991;88:7734–7738. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zemmel R W, Kelley A C, Karn J, Butler P J G. Flexible regions of RNA structure facilitate co-operative rev assembly on the rev-response element. Mol Biol. 1996;258:763–777. doi: 10.1006/jmbi.1996.0285. [DOI] [PubMed] [Google Scholar]
- 59.Zhou C, Bahner I C, Larson G P, Zaia J A, Rossi J J, Kohn D B. Inhibition of HIV-1 in human T-lymphocytes by retrovirally transduced anti-tat and rev hammerhead ribozymes. Gene. 1994;149:33–39. doi: 10.1016/0378-1119(94)90409-x. [DOI] [PubMed] [Google Scholar]
- 60.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]