Abstract
The use of benign stimuli to control the binding and release of labile biologics for their isolation from complex feedstocks is a key goal of modern biopharmaceutical technology. This study introduces cyclic azobenzene-peptide (CAP) hybrid ligands for the rapid and discrete photo-responsive capture and release of blood coagulation Factor VIII (FVIII). A predictive method – based on amino acid sequence and molecular architecture of CAPs – was developed to correlate the conformation of cis/trans CAP photo-isomers to FVIII binding and release. The combined in silico and in vitro analysis of FVIII:peptide interactions guided the design of a rational approach to optimize isomerization kinetics and biorecognition of CAPs. A photoaffinity adsorbent, prepared by conjugating selected CAP G-cycloAZOB[Lys-YYKHLYN-Lys]-G on translucent chromatographic beads, featured high binding capacity (> 6 mg of FVIII per mL of resin) and rapid photo-isomerization kinetics (τ < 30s) when exposed to 420-450 nm light at the intensity of 0.1 W·cm−2. The adsorbent purified FVIII from a recombinant harvest using a single mobile phase, affording high product yield (>90%), purity (>95%), and blood clotting activity. The CAPs introduced in this report demonstrate a novel route integrating gentle operational conditions in a rapid and efficient bioprocess for the purification of life-saving biotherapeutics.
Keywords: azobenzene-peptide hybrids, affinity ligands, photo-affinity chromatography, biomolecular recognition, Factor VIII
1. Introduction.
The purification of biotherapeutics for treating life-threatening diseases relies on the controlled partitioning of the target molecules between a complex solution and an adsorbent substrate1–3. In this context, chromatographic technology has undergone continuous development through the past four decades to deliver porous media and affinity ligands with excellent binding selectivity and capacity4–5. Current chromatographic methods rely on multiple mobile phases with varying composition, concentration, and pH to control the binding and release of the target product6–7. While adequate for purifying robust biomolecules, such as monoclonal antibodies, this approach is not suitable for labile products, such as blood factors and therapeutic enzymes, which are stable in narrow intervals of ionic strength, pH, and temperature8–9. These biologics require ligands whose biorecognition activity can be controlled using benign external stimuli, enabling the entire chromatographic process to be performed under a constant mobile phase that ensures the stability and bioactivity of the product.
A representative example of labile, lifesaving biotherapeutics is Factor VIII (FVIII). To date, the purification of recombinant Factor VIII (rFVIII) for the episodic and prophylactic treatment of uncontrolled bleeding10–11 has relied on immuno-affinity and ion exchange chromatography12–16. Of recent introduction are affinity adsorbents that enable eluting rFVIII under mild conditions: for example, an adsorbent functionalized with a camelid antibody targeting the B-domain depleted rFVIII affords high yield and purity using elution buffers formulated with polypropylene glycol and arginine17. Among synthetic affinity ligands18–21 is a peptide mimetic whose rFVIII binding is controlled by the ionic strength and electrostatic conditions of the solution. Using a sharp variation in ionic strength to trigger release, however, is unideal for rFVIII, whose integrity relies on non-covalent interactions between its heavy and light chains22.
The cyclic azobenzene-peptide (CAP) hybrid ligands and design framework developed in this study introduce a new paradigm of purification, where irradiation at defined wavelengths is used as a capture/release switch (Figure 1). The method of hybridization of peptides with azobenzene linkers is generalizable and offers a new route for biological purifications. An ensemble of rFVIII-targeting CAPs was discovered through initial library screening and optimized by (i) adjusting the architecture of the azobenzene-peptide cycle using flexible amino acid linkers, and (ii) evolving the peptide segment interacting with rFVIII via sequence mutations. This optimization was conducted through a concerted in silico - in vitro approach to yield a set of adsorbents for rFVIII purification from recombinant sources via photo-affinity chromatography. By employing both preliminary screening and in silico structural optimization, we demonstrate a complete pipeline for predicting CAP ligands with optimal performance.
Figure 1. Photo-affinity chromatography of labile therapeutics.
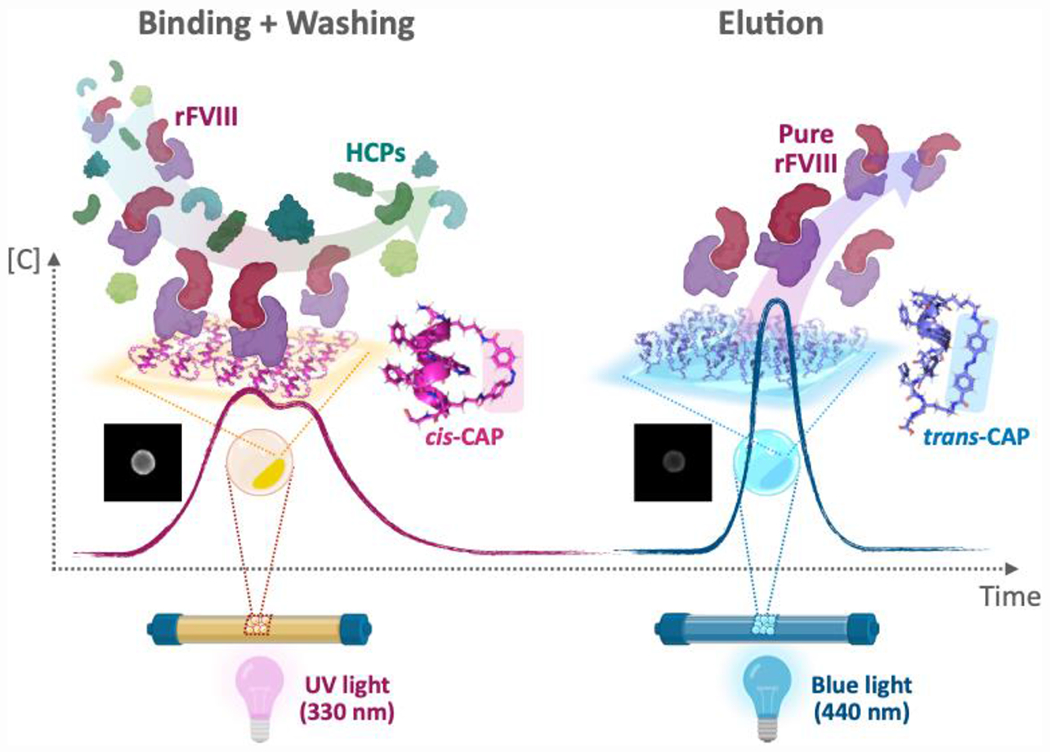
The target recombinant human blood coagulation Factor VIII (rVIII) – a labile therapeutic protein – is captured by the CAPs displayed on translucent ChemMatrix beads, while the impurities are cleared, and subsequently released at high purity under benign conditions (< 10 mW·cm−2). Specifically, (i) the pore surface of the beads is functionalized with cyclic azobenzene-peptides (CAPs), each comprising a peptide segment, which interacts with the target rFVIII, and a photo-responsive azobenzene linker, which switches reversibly the peptide segment between a rFVIII-binding (cis) and a rFVIII-releasing (trans) conformation; (ii) when rFVIII in solution contacts the beads, it diffuses into their pores, and reaches the inner surface of the CAP-functionalized beads, where it is captured by the affinity binding of cis-CAPs § (note: the CAPs developed in this study mainly target the A1 domain of rFVIII); conversely, the other species present in the feedstock do not interact with the CAPs and are therefore removed by washing; (iii) following the adsorption of rFVIII, the beads are washed and exposed to blue light (440 nm) to isomerize cis-CAPs into trans-CAPs, thus triggering the dissociation of the rFVIII:CAP complex and the release of rFVIII from the resin beads; finally, (iv) the beads can subsequently be exposed to UV light (330 nm), which switches the trans-CAPs back into cis-CAPs, thus preparing the beads for a subsequent round of photo-affinity purification of rFVIII.
2. Results
2.1. Selection of rFVIII-targeting CAPs.
Human FVIII is a large glycoprotein (2332 amino acids, 330 kDa) comprising six domains, namely A1-A2-B-A3-C1-C2 23–26. The light chain (LC), formed by domains A3, C1, and C2, and the heavy chain (HC), formed by domains A1, A2, and part of B, are linked by divalent metal cations (Ca2+ and Mg2+) into a non-covalent heterodimer27–28. Relevant to this work, the A1 domain features a large surface area and does not entertain biomolecular interactions with other protein members of the coagulation cascade29. This makes A1 domain an ideal target site for the purification of FVIII-related products, such as full rFVIII, FVIIIa, and B-domain deleted rFVIII.
Informed by the in silico druggability study of the FVIII-A1 domain (Figure S1A), the G-cycloAZOB[Dap-X1X2X3X4X5X6X7-Dap]-G framework, with hydrodynamic radius of ~ 10 - 17 Å for the cyclic 7-mer combinatorial segment (X1-X7) (Figure S1B), was selected to fit the putative binding pockets. The length of 7 amino acids has been chosen to balance binding strength and selectivity, and cost and scalability. The combinatorial positions Xi were randomized using amino acids capable of forming non-covalent interactions with the putative binding pockets, namely asparagine (N), glutamic acid (E), glycine (G), histidine (H), leucine (L), lysine (K), serine (S), and tyrosine (Y). The CAPs were synthesized on ChemMatrix beads following the “split-couple-and-recombine” technique described by Lam et al.30; porous, hydrophilic, translucent ChemMatrix microparticles are an excellent substrate for conventional and photo-affinity chromatographic applications31–37.
Primary screening of FVIII-targeting, light-responsive CAPs was performed with a high-throughput (350 beads·hr−1) dual-fluorescence assay (Figure S2) from prior work33, 38–39. In this study, the ensemble of CAP-ChemMatrix beads was screened against a screening mix comprising fluorescently labelled rFVIII-A1 and Chinese hamster ovary host cell proteins (CHO HCPs); the mix was formulated to mimic the bioreactor harvest fluids utilized in the biomanufacturing of recombinant blood clotting factors for human therapy. The identified 25 sequences showed (i) a significant enrichment in tyrosine (Y) and lysine (K), specifically arranged in the recurring dimer KY, (ii) a leucine dimer (LL) consistently located at X3X4, and (iii) asparagine (N) found frequently at the X3 position (Table S2). Nine sequences, selected based upon the sequence homology analysis in Figure S2, and one sequence (CAP.T) designed using the amino acids with top positional frequency were adopted for further investigation.
During the primary library screening, a single CAP-ChemMatrix bead was used to evaluate each candidate. While the emergence of sequence homology and the enrichment of specific amino acids (N, L, K, and Y) gives confidence in the outcome of the initial selection, further evaluation of the selected sequences was sought to provide statistical rigor. Furthermore, ChemMatrix beads feature polydispersity in particle diameter and morphology, pore size, and ligand density, which can impact their fluorescence signature; additional variability during library screening is caused by the different time spent by beads in aqueous suspension prior to imaging, during which protein dissociation and quenching of the fluorophores can occur.
A secondary screening was carried out by evaluating each candidate CAP on multiple ChemMatrix beads (60 for each candidate) for quantitative assessment using full-length rFVIII. The resulting values of binding strength and selectivity, and elution efficiency are reported in Figure 2A. Most of the candidate sequences, including the CAP.T, featured high binding strength and selectivity, but limited rFVIII elution. Conversely, CAP.14 (G-cycloAZOB[Dap-KYYGSYY-Dap]-G) and CAP.16 (G-cycloAZOB[Dap-YYKHLYN-Dap]-G) showed appreciable photo-responsive biorecognition activity and were most promising for subsequent evaluation.
Figure 2. Secondary screening of candidate and variant rFVIII-targeting photo-responsive CAP ligands.
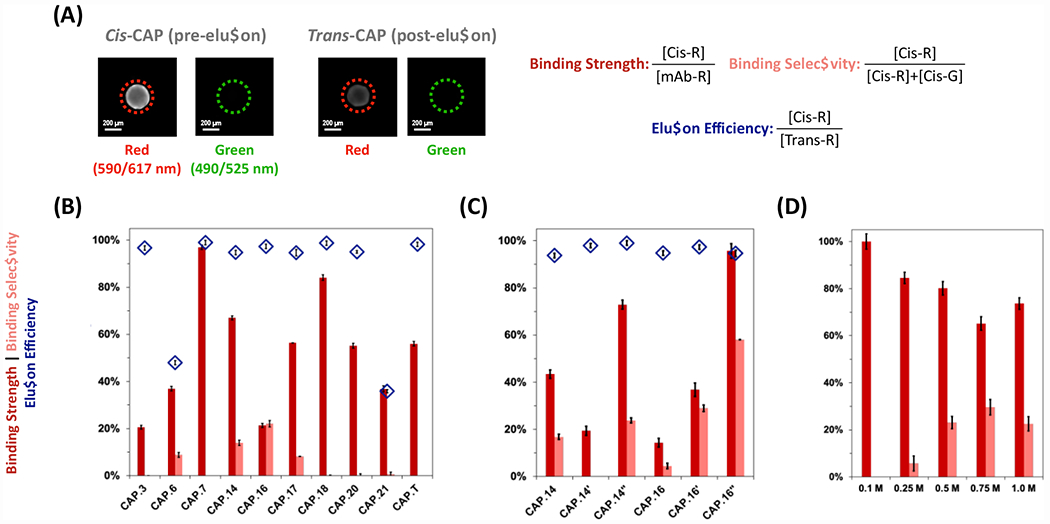
(A) The values of rFVIII binding strength were calculated as the ratio between the corrected intensity of red fluorescence (exc/em: 590/617 nm) of a cis-CAP-ChemMatrix bead and the corrected intensity of a control ChemMatrix bead (i.e. functionalized with anti-rFVIII antibodies) following AF594-rFVIII adsorption. The values of rFVIII binding selectivity were calculated as the ratio of the corrected intensity of red fluorescence of a cis-CAP-ChemMatrix bead after AF594-rFVIII adsorption against the total red and green (exc/em: 490/525 nm) fluorescence of a cis-CAP-ChemMatrix bead after AF594-rFVIII and AF488-HCPs adsorption. The values of rFVIII elution efficiency were calculated as the ratios of the corrected red fluorescence intensity of a cis-CAP-ChemMatrix bead after AF594-rFVIII adsorption and the corrected red fluorescence intensity of the same trans-CAP-ChemMatrix bead after AF594-rFVIII desorption upon exposure to visible light (λex = 420-450 nm) at ~220 mW·cm−2 for 2 mins at room temperature (25°C). Scale bar = 200 μm. (B) Sequences selected for in vitro analysis of rFVIII binding strength (dark red), elution efficiency (light red), and selectivity (blue diamonds), (C) their structural variants, and (C) performance of at different values of ionic strength of the mobile phase – namely, X M NaCl (X: 0.1, 0.25, 0.5, 0.75, and 1) added to 0.1 M HEPES buffer, 5 mM CaCl2, and 0.01% v/v Tween20 at pH 7.4. Error bars represent one standard deviation of 30 beads. The raw values of average fluorescence and are in Figure S4.
2.2. Engineering the architecture of CAP cyclization.
The proposed CAPs are engineered to bind a target biomolecule in their cis conformation and release when isomerized to the trans conformation. While trans-CAPs are more thermodynamically stable than cis-CAPs, spontaneous cis-to-trans conversion occurs rather slowly (~ 12 hrs); conversely, the photo-isomerization of resin-bound cis-CAPs to trans-CAPs is rapid39–40. This behavior is key to the success of photo-affinity chromatography: during the binding phase, cis-CAPs maintain their conformation without additional irradiation with UV light, thus avoiding the risk of damaging the target biomolecule; when exposed to blue light, cis-CAPs rapidly convert to trans-CAPs and release the product, thus minimizing the impact of photo-isomerization kinetics on recovery. Accordingly, the characteristic time of product release is determined primarily by the kinetics of transport phenomena in the pores and the interstitial space around the packed beads.
To our surprise, none of the CAPs identified from library screening – except for CAP.20 – presented the expected absorbance spectra10,11 (Figure S5) nor did they exhibit a satisfactory light-triggered elution of rFVIII (Figure 2A). The photo-isomerization kinetics of CAPs are governed by the molecular flexibility of the azobenzene-peptide construct and the excitation light source39, 41–43. The in silico modeling of the selected CAPs revealed a network of π-π interactions between azobenzene and at least one tyrosine (Y) as well as hydrogen bonds formed by polar (K, N, and S) residues. Conversely, CAP.20 (G-cycloAZOB[Dap-GGGKYGK-Dap]-G), which exhibits the expected absorbance spectra, contains a single tyrosine not interacting with the azobenzene linker and a flexible tri-glycine (GGG) motif. We therefore hypothesized that replacing Dap with lysine (Lys) as the azobenzene-linker provides flexibility to the CAP constructs, thus promoting their photo-isomerization yield.
Accordingly, we prepared and characterized variants CAP.14’ (G-cycloAZOB[Dap-KYYGSYY-Lys]-G), CAP.14” (G-cycloAZOB[Lys-KYYGSYY-Lys]-G), CAP.16’ (G-cycloAZOB[Dap-YYKHLYN-Lys]-G), and CAP.16” (G-cycloAZOB[Lys-YYKHLYN-Lys]-G). The secondary screening of variants (Figure 2B) showed a defined increase in rFVIII binding and elution. The absorbance profiles of variants CAP.14’ and CAP.16’ are more in line with those observed in prior work, albeit they present a rather limited cis-to-trans inversion (Figure S6B and S6E). Conversely, CAP.14” and CAP.16” displayed the canonical CAP behavior, namely (i) the 330 nm and a 440 nm signature and (ii) reversible cis-trans photo-isomerization (Figure S6C and S6F). These results show that linker editing provides an effective route to improve the photo-responsive behavior of CAPs.
2.3. In silico study of CAP structure and biorecognition.
The photo-isomerization kinetics studies and the preliminary chromatographic results indicate a strong influence of the CAP architecture on the biorecognition activity (Figures 2A and 2B, and supplementary information Sections S.3 and S.4) and suggested a strategy for kinetic tuning. To gather quantitative, molecular-level insight into the isomerization-dependent rFVIII:CAP interaction, we performed an in silico analysis of the free energy of isomers and of the paired interactions in FVIII:CAP complexes via molecular docking and dynamics simulations. The values of free energy (Table S4) of the trans isomers of CAP.14, CAP.14’, CAP.16, and CAP.16’ were significantly lower than of the corresponding cis isomers, the latter featuring unrealistically high values; conversely, the values of free energy of both isomers of CAP.14” and CAP.16”, and therefore their ΔECis-Trans, were comparable to those found in earlier work39, 44–45. This suggests that the lack of photo-isomerization of CAPs constructed with Dap (Figures S6A and S6D) is due to an untraversable energy barrier, which locks the peptide-azobenzene constructs in the trans conformation, irrespective of the energy provided by the incident light. The values of ΔECis-Trans may point at CAP.14” as a better candidate for photo-affinity applications; however, CAP.16” exhibits a secondary α-helix structure in cis and a lack of secondary structure in trans (Table S4), which helps modulating target binding upon isomerization.
The trans and cis isomers of CAP.14” and CAP.16” were docked in silico against human FVIII to evaluate the free energy of binding (ΔGb) and the paired interactions between FVIII and CAP residues. The results in Figure 3 are consistent with the library design criteria and the outcomes of secondary screening. Specifically, both CAP.14” and CAP.16” targeted the A1 domain of human FVIII (residues 1 – 336, in pink) preferentially and with high affinity (KD,cis < 10−6 M), albeit low-affinity (KD,cis > 10−5 M) binding poses of CAP.14” were identified on the A2 and C2 domains and of CAP.16” at the interface of the C1 and C2 domains. Notably, the trans isomers of both variants displayed a much lower binding affinity than their cis counterparts, with a loss of binding energy upon isomerization (ΔΔGb,cis-trans) of 2.38 and 3.84 kcal/mol for CAP.14” and CAP.16”, respectively.
Figure 3. In silico evaluation of binding interaction between candidate CAPs and FVIII.
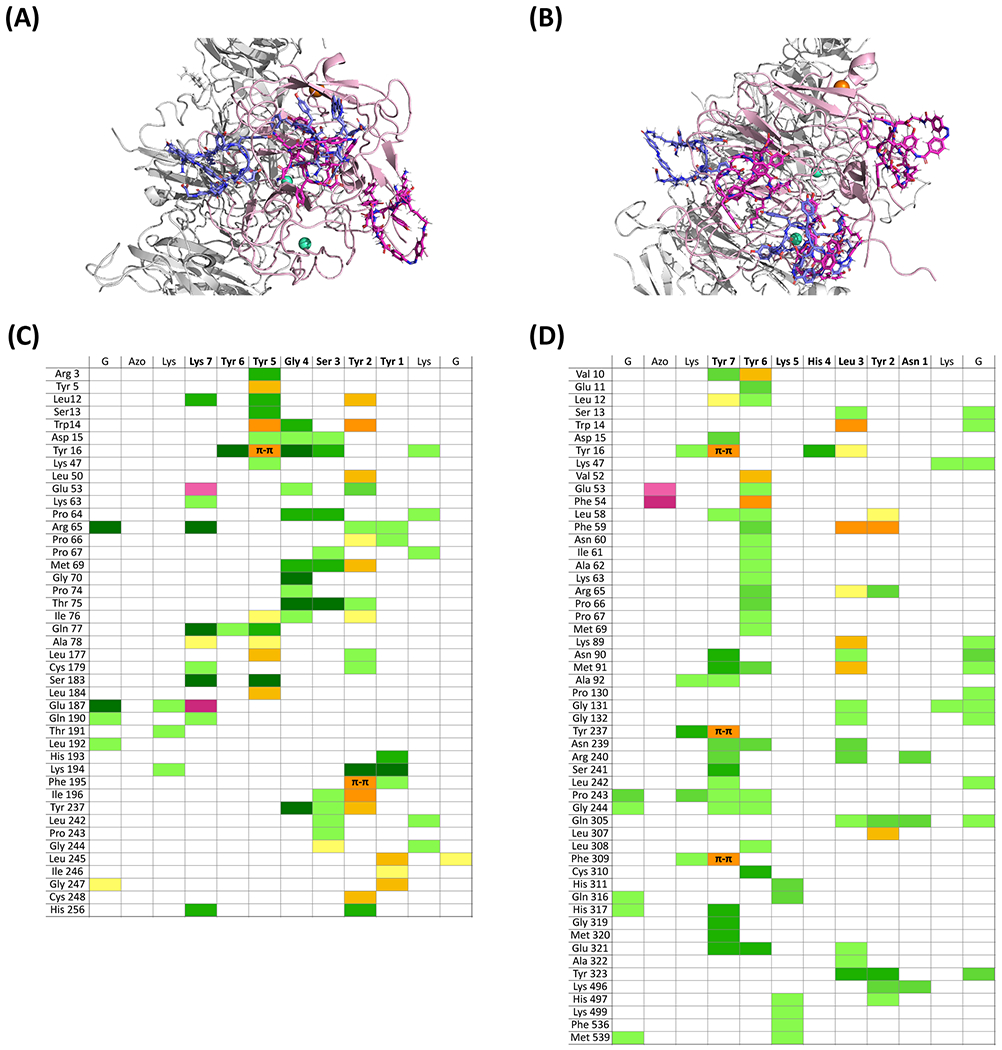
Structures of the complexes formed by human FVIII and the trans and cis isomers of (A) CAP.14” (G-cycloAZOB[Lys-KYYGSYY-Lys]-G) and (B) CAP.16” (G-cycloAZOB[Lys-YYKHLYN-Lys]-G); the trans-CAP isomers are in blue, the cis-CAP isomers are in magenta, the A1 domain of human FVIII (PDB ID: 3CDZ) is in pink, while all other FVIII domains are in grey. Paired interactions between FVIII and (C) cis-CAP.14” and (D) cis-CAP.16”; hydrogen bonds and polar contacts are denoted in green; hydrophobic and π-π interactions are denoted in yellow; electrostatic interactions are denoted in purple; the color intensity denotes the contribution of the paired interaction to the ΔGb.
The interplay between binding pattern and ΔGb is helpful to explain the behavior of these CAPs during secondary screening. First, cis-CAP.14” targets BP.1 (ΔGb ~ −8.89 kcal/mol) and BP.2 (−7.62 kcal/mol) of the A1 domain of FVIII; following photo-isomerization, trans-CAP.14” loses binding of BP.1 completely and BP.2 substantially (−5.07 kcal/mol) but may form a new interaction with BP.3 (5.71 kcal/mol). Similarly, cis-CAP.16” targets BP.1 (ΔGb ~ −8.01 kcal/mol), BP.2 (−7.62 kcal/mol), and the interface between BP.1 and BP.2 (−7.16 kcal/mol), while trans-CAP.16” loses binding of BP.1 and BP.2 (−4.03 kcal/mol) and forms a new interaction with BP.3; the latter, however, is significantly weaker (3.98 kcal/mol) than that formed thereupon by trans-CAP.14”. Therefore, for CAP.16”, photo-isomerization translates in a >650-fold increase of average KD upon photo-isomerization, which is sufficient to trigger release of bound FVIII.
2.4. Optimizing the FVIII:CAP interaction environment.
Next, we resolved to explore the composition of the mobile phase continuously fed during the photo-affinity chromatographic process. The pH and the basal levels of CaCl2 and Tween20 were held constant at the values recommended for FVIII stability46,47. The in silico analysis of FVIII:CAP interactions was consulted to devise a mobile phase promoting efficient release upon photo-isomerization (Figure 3C and 3D). The cis isomer of CAP.14” forms a complex network of hydrogen bonds as well as several ionic and hydrophobic interactions: of note were the dominant roles of (i) Tyr5 and Tyr2, which formed π-π interactions with Tyr and Phe residues on FVIII; (ii) Gly4 and Ser3, which formed a dense network of hydrogen bonds with polar residues on FVIII; and (iii) Lys7, which, together with strong hydrogen bonding, formed ionic interactions with Glu53 and Glu187 on FVIII. Conversely, FVIII binding by the cis isomer of CAP.16” was almost exclusively reliant on hydrogen bonding and 3 π-π interactions formed by Tyr7 with Tyr16, Tyr237, and Phe309 on FVIII; hydrogen bonds encompassed both sidechain-to-sidechain (57%) and sidechain-to-backbone (43%) interactions. The hydrogen bond-dominated makeup of FVIII:CAP.16” ΔGb is different from that of most peptide ligands in the literature and may be a unique feature of the photo-responsive biorecognition activity of CAPs. The binding mechanism characteristic of linear peptides is governed by electrostatic and hydrophobic interactions, which require strong variations in ionic strength and pH to trigger protein elution48–49. Hydrogen bonds are weaker and shorter-ranged than electrostatic interactions50 and can therefore be more easily disrupted by variations in the composition of the surrounding aqueous environment.
We therefore propose that photo-responsive biorecognition is uniquely engendered by protein:CAP binding that relies mostly on weak and short-range interactions. Accordingly, we resolved to investigate the role of ionic strength of the mobile phase, since added ions can interfere with the hydrogen bonding network, providing a means to temper the FVIII:CAP interaction and promote light-triggered dissociation51. To avoid disruption of the FVIII’s non-covalent dimeric structure, we adopted NaCl – an intermediate between chaotropic and kosmotropic salts. Increasing the ionic strength of the mobile phase produced a notable increase in FVIII elution efficiency for CAP.16” without affecting adsorption (Figure 2C). Specifically, 0.75 M offered the best balance of binding capacity and yield of rFVIII elution.
2.5. Photo-affinity purification of FVIII.
The spectroscopic and in silico evaluation of CAP variants converged in the design of a photo-affinity chromatographic protocol for purifying rFVIII from a clarified Chinese Hamster Ovary (CHO-S) cell culture fluid. The timed exposure of CAP-ChemMatrix beads to blue light was the only action implemented during the chromatographic protocol, which was entirely conducted at room temperature (25°C) and under constant flow of 0.1 M HEPES buffer (added with 5 mM CaCl2 and 0.01% v/v Tween20 at pH 7.4) to safeguard the stability and activity of rFVIII. The resulting chromatogram in Figure 4 collates the temporal profiles of rFVIII and CHO-S HCP titer in the effluent, and the corresponding values of rFVIII yield and purity. The absence of rFVIII in the flow-through fraction (0 – 0.4 mL) indicates complete binding of the rFVIII load (30 μg) by the CAP.16”-ChemMatrix packed bed (5 μL). This corresponds to a binding capacity above 6 mg of rFVIII per mL of resin, in line with commercial affinity adsorbents. The CAP.16”-ChemMatrix resin afforded high values of cumulative product yield (89.6%) and purity (92.6%) starting from a complex and diluted feedstock (initial rFVIII purity ~4%), corresponding to a purification ratio > 23-fold.
Figure 4. Photo-affinity chromatographic purification of rFVIII from using CAP.16”-ChemMatrix beads and blood clotting activity of purified rFVIII.
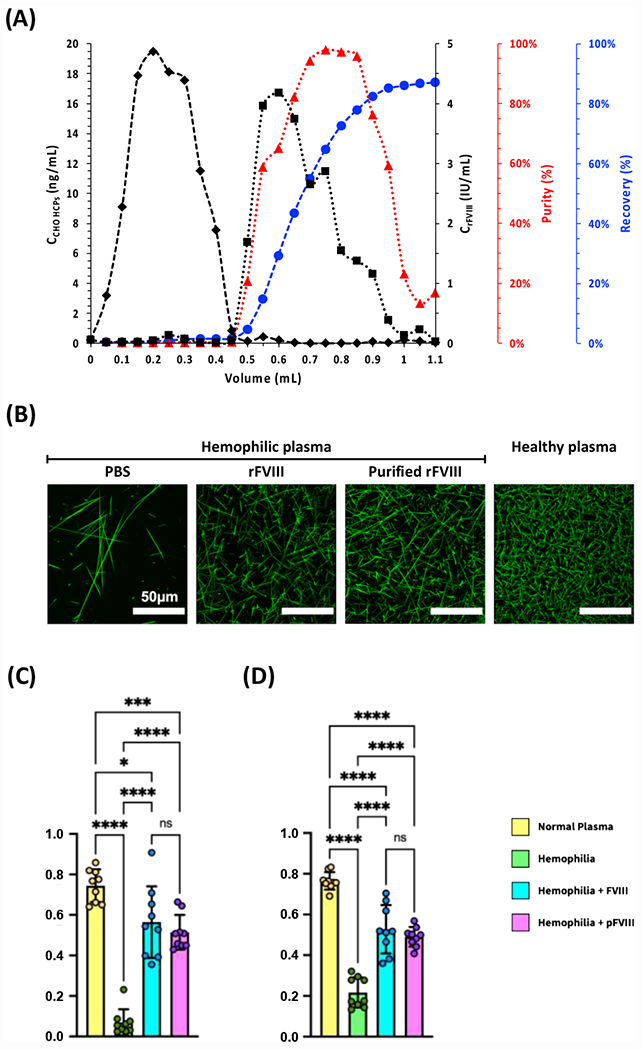
(A) Values of concentration of rFVIII (■) and CHO HCPs (◆);rFVIII purity (▲) and recovery (●) vs. volume of effluent collected from the measurement cell; the chromatographic operation was entirely conducted at room temperature (25°C) and under constant flow of 0.1 M HEPES buffer added with 5 mM CaCl2 and 0.01% v/v Tween20 at pH 7.4. (B) Clotting images generated by mixing platelet-poor plasma isolated from two Hemophilia A patients with aqueous 500 mM CaCl2, a solution of AF488-Fibrinogen at 50 μg·mL−1 in Binding Buffer, and either no FVIII or native FVIII, or FVIII purified via photo-affinity chromatography (fractions collected between 0.6 – 0.9 mL); clotting image generated by mixing healthy plasma with aqueous 500 mM CaCl2 and a solution of AF488-Fibrinogen at 50 μg·mL−1 in Binding Buffer. The images were generated from 5 μm z-stacks via confocal microscopy and the pixel density was measured in ImageJ. (C) Values of relative fiber density, calculated as the ratio of fiber pixels to background pixels after binarization of the images in panel (B). (D) Values of intersection density, calculated as the ratio of skeleton intersections post-erosion to the total number of fiber pixels pre-erosion of the images in panel (B). Plots show mean ± standard deviation. (**** p < 0.0001; two-way ANOVA).
The ability of the rFVIII purified via photo-affinity chromatography to restore the blood clotting activity of hemophilic plasma was evaluated against native rFVIII, using healthy plasma as a control (Table S6). The confocal fluorescence images of the resulting clots (Figure 4B) were analyzed to determine two key metrics of clotting efficiency: (i) relative fiber density and (ii) fiber intersection density, or network crosslinking. Thick fibrin fibers and low network density are known features of leaky, hemophilic fibrin networks52; conversely, in healthy plasma, the fibers are thinner, more branched, and more intersected/crosslinked. The analysis of the imaged clots shows that the rFVIII purified from the CHO harvest via photo-affinity chromatography equals native rFVIII in producing clots whose fiber density and crosslinking approximates those recorded in healthy plasma (Figure 4C and 4D).
2.6. Improving the light-controlled biorecognition of CAPs via sequence engineering.
Together with the cyclization architecture, the peptide sequence represents a key design parameter governing the isomerization kinetics and purification performance of CAP ligands. The amino acid sequences selected from library screening and the in silico analysis of pairwise interactions highlighted the role of hydrogen bonding in FVIII:CAP interactions. Furthermore, the trajectories of cis-to-trans isomerization portrayed by the simulations indicate that the interactions between the azobenzene and the hydroxybenzyl groups of tyrosine residues determine the energy of the intermediate conformations and thus the characteristic time and yield of photo-isomerization. Finally, the docking studies showed that the tyrosine residues – in particular Tyr5 on CAP.14” and Tyr6 on CAP.16” – participate in hydrogen bond, hydrophobic, and π-π interactions with FVIII. Collectively, these observations suggest replacing tyrosine residues with aromatic analogues as an alternative route to modulate the photo-responsive affinity of CAPs and improve the outcome of photo-affinity chromatography.
Accordingly, we designed variants CAP.14A (tyrosine to alanine), CAP.14F (tyrosine to phenylalanine), CAP.14mY (tyrosine to O-methyl-tyrosine), CAP.16A, CAP.16F, and CAP.16mY to evaluate the role of hydrogen bonding and π-π interactions through amino acid mutation. The trans and cis isomers of these variants were modeled in silico to evaluate photo-isomerization kinetics, which were compared against the spectroscopic measurements.
All variants of CAP.14 and CAP.16 displayed the signature absorbance spectra characteristic of CAPs, in particular the variation at 330 and 440 nm upon cis-trans isomerization (Figure S7). Replacing tyrosine with alanine obliterates the interactions between the azobenzene linker and the peptide segment, thus producing the largest variations in ΔEcis-trans among all variants. Additional mutations with phenylalanine and O-methyl-tyrosine explored the effect of removing of hydrogen bonding contribution of tyrosine, while maintaining its steric hindrance and ability to form π-π interaction. Collectively, the spectroscopic data (Figures S8 and S9) and the in silico values of ΔEcis-trans (Figure 5A) of the sequence-based variants indicate that the network of hydrogen bonds formed by tyrosines with the peptide sequence – at both backbone and side chain levels – greatly outweighs the effect of π-π interaction between tyrosines and the azobenzene linker in determining the energy landscape – and consequently the isomerization kinetics – of the isomers produced by CAP.14 and CAP.16 variants. As observed in prior work,39 we noticed that the normalized cis-trans energy difference obtained in silico (ΔEcis-trans/RT) correlated to the ratio of the kinetic constant of photo-isomerization vs. irradiance (κ/I). Notably, the data points collected in this study aligned on the plot originally derived for VCAM1-targeting CAPs, corroborating the hypothesis of design-driven photo-switching kinetics (Figure 5A).
Figure 5.
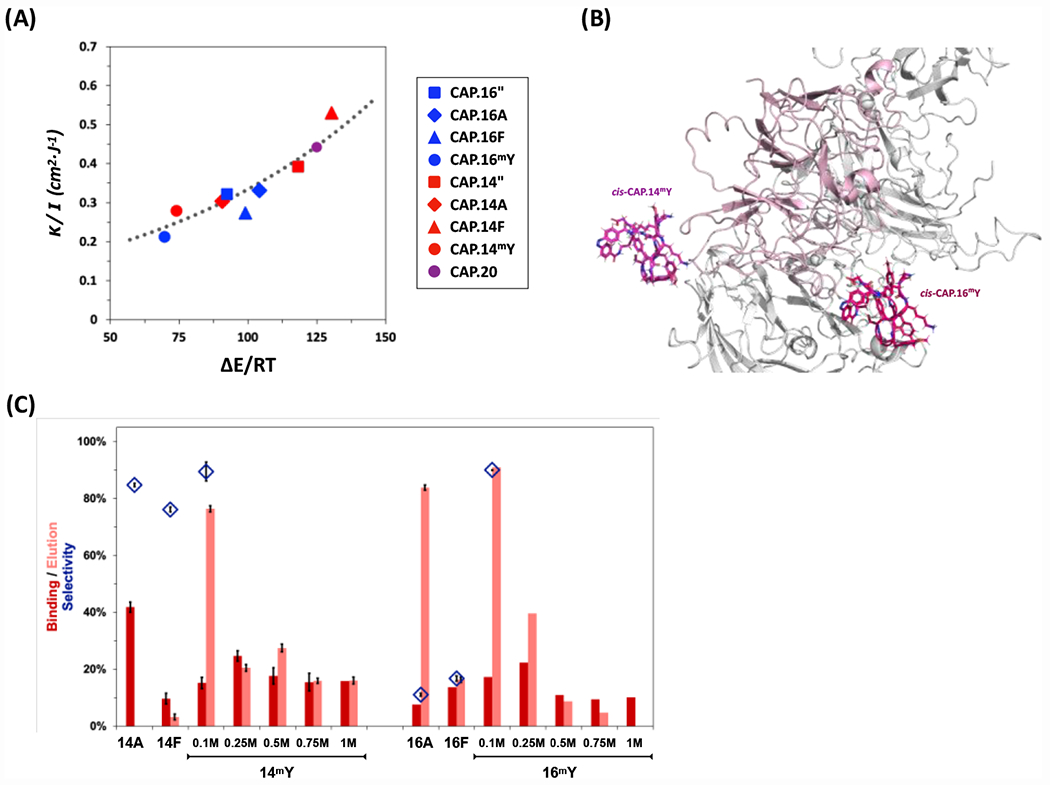
(A) Correlation of CAP photo-isomerization kinetics (κ/I) and energetics (ΔEcis-trans). The values of κ/I ratio of ChemMatrix-bound CAPs are plotted against the normalized values of ΔEcis-trans derived in silico via MD modeling of the corresponding CAPs; the dotted line data are reported from prior work on CAPs39. (B) Structures of the complexes formed by human FVIII and the cis isomers of CAP.14mY and CAP.16mY; the cis-CAP isomers are in magenta, the A1 domain of human FVIII (PDB ID: 3CDZ) is in pink, while all other FVIII domains are in grey. (C) Values of rFVIII binding strength (dark red), elution efficiency (light red), and selectivity (blue diamonds) of sequence-based variants of CAP.14 and CAP.16 conjugated on ChemMatrix beads at different values of ionic strength of the mobile phase - namely X M NaCl (X: 0.1, 0.25, 0.5, 0.75, and 1) added to 0.1 M HEPES buffer, 5 mM CaCl2, and 0.01% v/v Tween20 at pH 7.4. The values of rFVIII binding strength, selectivity, and elution, and error bars were calculated as described in Figure 2.
Secondary screening showed that, for a subset of sequence-based variants, photo-isomerization caused a significant variation in binding strength, resulting in efficient rFVIII release (Figure 5B); not all variants, however, afforded a high elution yield or possessed sufficient binding selectivity to qualify as effective ligands. CAP variants built with O-methyl-tyrosine (mY) were noticed for their rFVIII binding strength, selectivity, and light-controlled release: the hydrogen bond-acceptor ether group and the benzene ring of mY maintain most of the contacts formed by cis-CAP.14” and cis-CAP.16” with FVIII (Figure 5B); at the same time, the rapid photo-isomerization kinetics enabled by a weaker intramolecular hydrogen bonding network promotes efficient release of rFVIII.
Upon screening the effect of ionic strength of the mobile phase (Figure 5C), ligands CAP.14mY (operated at 0.5 M NaCl) and CAP.16mY (0.25 M NaCl) were adopted for rFVIII purification via photo-affinity chromatography (Figure 6). No rFVIII was recorded in the flow-through fractions, confirming the high binding capacity of the photo-affinity adsorbents; CAP.14mY-ChemMatrix resin afforded rFVIII yield of 91.2% and purity of 90.1%, while CAP.16mY-ChemMatrix resin afforded 90.5% and 90.8%, corresponding to a ~20-fold purification ratio.
Figure 6. Photo-affinity chromatographic purification of rFVIII from using (A) CAP.14mY- and (B) CAP.16mY-ChemMatrix beads.
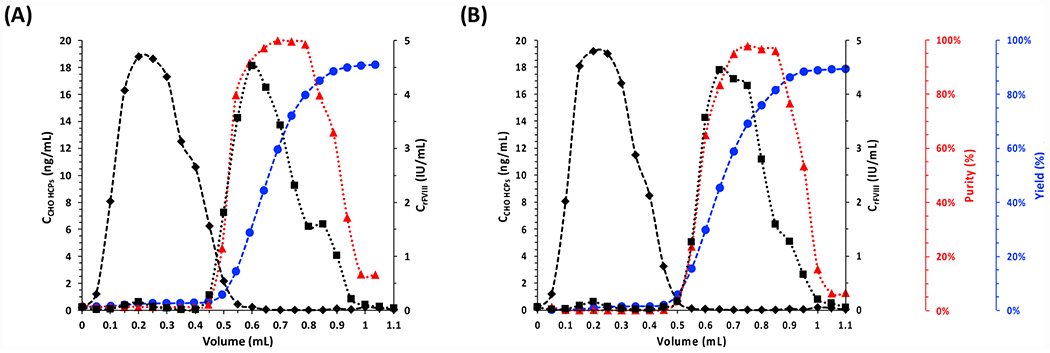
Values of concentration of rFVIII (■) and CHO HCPs (◆); rFVIII purity (▲) and cumulative yield (●) vs. volume of effluent collected from the measurement cell. the chromatographic operation was entirely conducted at room temperature (25°C) and under constant flow of 0.1 M HEPES buffer added with 5 mM CaCl2 and 0.01% v/v Tween20 at pH 7.4.
Notably, the CAP-ChemMatrix beads were not exposed to UV light during the adsorption step (5 min). As observed in prior work39–40, the proposed CAPs do not exhibit any spontaneous cis-to-trans conversion at room temperature (25°C) and in the dark, thus allowing to conduct the capture step without the need of additional irradiation, which avoids the risk of compromising the stability of the target product (note: cis-CAPs can maintain their conformation for up to 6 hrs; for reference, the duration of the product capture step in affinity chromatography ranges from 2 – 10 min for small scale applications to 120 – 150 min for large-scale applications)53–54. The broad elution peaks in Figure 4A and Figure 6 may in principle be attributed to kinetic limitations of CAP photo-isomerization and the diffusional transport of rFVIII through the pores of ChemMatrix beads. The spectroscopic profiles reported in Figure S8, however, indicate that ChemMatrix-bound CAP.16”, CAP.14mY and CAP.16mY photo-isomerize completely within 20 – 40 seconds. Conversely, the characteristic time of rFVIII diffusion from the core to the surface of the beads is estimated to be 10-fold higher (~ 250 – 400 seconds, see Section S.6). This suggests that the rates of rFVIII release – and thus the elution profiles – are kinetically controlled by pore diffusion and that increasing the intensity of blue light may improve only marginally the release of rFVIII.
3. Conclusions.
The purification of labile biologics via chromatographic technology has not yet reached a level of maturity sufficient to reduce costs and broaden the access to high-value therapeutics to patients. Accomplishing this goal will critically rely on next-generation affinity adsorbents that combine high binding capacity and selectivity with the ability to release the product under mild conditions, thus ensuring structural and functional product stability. The framework presented in this study for developing CAPs as photo-affinity ligands holds true promise to attain next-generation downstream bioprocessing. To that end, our workflow integrates (i) a modular CAP architecture enabling target-agnostic tuning of light-controlled biorecognition activity; (ii) high-throughput quantitative assessment of CAPs for rapid identification of bespoke ligands with effective photo-affinity activity; (iv) in silico modeling aiding the design of CAPs and their behavior to reduce benchwork and accelerate discovery; and (iv) spectroscopic evaluation of the kinetics of photo-isomerization and photo-affinity chromatography on chromatographic substrates for rigorous assessment of performance in actual bioprocess conditions. The correlations between CAP design, photo-isomerization, and protein binding and release developed in this study provides a robust and effective platform for the future design of CAPs and strategies to overcome bioprocess roadblocks. Particularly relevant towards the success of photo-affinity chromatography is the conformational stability of cis-CAPs (binding isomer) and rapid cis-to-trans photo-isomerization that triggers the release of the target protein. Further work will explore the use of translucent beads that minimize the limitations of mass transfer kinetics and promote rapid product recovery. The success of this case study demonstrated that this technology provides a pathway towards next-generation purification strategies tailored to labile biotherapeutics, improving the availability of life-saving medicines to patients.
4. Methods
4.1. Materials.
Alexa Fluor 488 (AF488) and NHS-Alexa Fluor 594 (NHS-AF594), N,N’-dimethylformamide (DMF), dichloromethane (DCM), HPLC-grade acetonitrile, HPLC-grade water, HPLC-grade dimethylsulfoxide (DMSO), hydrochloric acid, sodium chloride, sodium hydroxide, and calcium chloride were obtained from ThermoFisher Scientific (Waltham, MA). Fmoc/tBu-protected amino acids, hexafluorophosphate azabenzotriazole tetramethyl uronium (HATU), hexafluorophosphate chlorobenzotriazole tetramethyl uronium (HCTU), sodium diethyldithiocarbamate trihydrate, palladium tetrakis(triphenylphosphine) (Pd(PPh3)4), diisopropylethylamine (DIPEA), N-methyl-2-pyrrolidone (NMP), piperidine, trifluoroacetic acid (TFA), were purchased from ChemImpex Inc. (Wood Dale, IL). Nitrogen gas was obtained from Airgas National Welders (Raleigh, NC). Azobenzene-4,4′-dicarbonyl dichloride, triethylamine (TEA), triisopropylsilane (TIPS), Tween 20, sodium hydroxide, HEPES, phosphate-buffered saline (PBS) pH 7.4, and a Kaiser test kit were from MilliporeSigma (St. Louis, MO). Aminomethyl ChemMatrix and HMBA-ChemMatrix (HMBA, loading: 0.6 mmol amine per g resin) resins were sourced from PCAS Biomatrix, Inc. (Saint-Jean-sur-Richelieu, Quebec, Canada). The recombinant A1-domain of human Factor VIII (rFVIII-A1, 17 kDa) and the full-length recombinant human Factor VIII (rFVIII) were from Biomatick (Wilmington, DE) and CreativeBioMart (Shirley, NY). Silicon wafers were from University Wafers (Boston, MA). AF488-conjugated fibrinogen (AF488-fibrinogen) was from ThermoFisher Scientific (Waltham, MA). Hemophilia A patient plasmas was obtained from George King Bio-Medical, Inc. (Overland Park, KS), while non-hemophilic adult plasma was from the New York Blood Center (New York, NY). The SU-8 2150 epoxy-based photoresist and the SU-8 Developer were obtained from MicroChem (Round Rock, TX). Dow SYLGARD™ 184 Silicone Encapsulant Clear Kit was obtained from Ellsworth Adhesives (Germantown, WI).
4.2. Synthesis of a library of 7-mer CAPs on ChemMatrix beads.
The linear combinatorial 7-mer peptide G-Dap(Alloc)-X1-X2-X3-X4-X5-X6-X7-Dap(Mtt)-G, flanked by glycine (G) and Nγ-Mtt diaminopropionic acid (Dap) on the N-terminus and Nγ-Alloc Dap and G on the C-terminus, was initially synthesized on 1 g of HMBA-ChemMatrix resin (loading: 0.6 mmol amine per g resin) as previously described39, 55–56. All steps of amino acid conjugation and Fmoc deprotection were performed using a Syro I peptide synthesizer (Biotage, Uppsala, Sweden). Briefly, each amino acid coupling step was performed at 50°C for 20 min using 5 equivalents (eq.) of amino acid dissolved in dry DMF at a concentration of 0.5 M, 5 eq. of HATU in DMF at 0.5 M (HCTU was used for Dap coupling only), and 6 eq. of DIPEA in NMP at 0.5 M. A Kaiser test was performed after each amino acid coupling step to ensure complete conjugation. Fmoc deprotection was performed at room temperature (25°C) using 5 mL of 20% piperidine in DMF. The combinatorial portion of the library (X1-X7) was produced using the “divide-couple-recombine” technique37, 57 using the following Fmoc/tBu-protected amino acids: Fmoc-Gly-OH, Fmoc-His(Trt)-OH, Fmoc-Leu-OH, Fmoc-Glu(OtBu)-OH, Fmoc-Tyr(tBu)-OH, Fmoc-Asn(Trt)-OH, Fmoc-Lys-OH and Fmoc-Ser(tBu)-OH. The cyclization of the library peptides using the azobenzene linker was then performed as described in our previous work39–40. Briefly, Alloc deprotection was initially performed using 5 eq. Pd(PPh3)4 at 3 mM in a 7:1 (v/v) solution of DCM:TIPS for 5 min at 40°C under microwave heating and mixing. Following copious rinsing with DMF, DCM, a solution of sodium diethyldithiocarbamate at 0.5% w/w in DMF, and DMF, the resin was incubated with 5 eq. of a 50 mM solution of azobenzene-4,4’-dicarbonyl dichloride in 1:19 (v/v) anhydrous TEA:DMF for 3 hrs at room temperature, in dark and under mild agitation. Following copious rinsing with DMF, water, DMF, DCM, and DMF, the Mtt group was removed from C-terminal Dap using a cleavage cocktail of TFA/TIPS/DCM (2/6/92) for 15 min at room temperature, under mild agitation. Following rinsing with DCM, DMF, peptide cyclization was performed by incubating the library beads with a solution of HATU (5 eq.) and DIPEA (10 eq.) at 0.5 M in dry NMP for 20 min at 45°C; the resins were washed with DMF, and the completion of the cyclization reaction was confirmed by Kaiser test. The peptides were then deprotected by acidolysis using a cleavage cocktail of TFA/TIPS/H2O (93/5/2) for 3 hrs at room temperature, in dark and under mild agitation. The resins were finally washed with DCM, DMF, and stored in 20% v/v aqueous methanol at 4°C.
4.3. Fluorescent labeling of the A1-domain of rFVIII, full-length rFVIII, and CHO host cell proteins.
The A1-domain of rFVIII (rFVIII-A1, 17 kDa) and full-length rFVIII were individually labeled with Alexafluor 594 (AF594, red), whereas the host cell proteins (HCP) present in a clarified CHO-S cell culture fluid were collectively labeled with Alexafluor 488 (AF488, green). Each fluorescent dye was initially dissolved in DMSO at a concentration of 10 mg·mL−1. A volume of 2 μL of AF594 solution was added to 100 μL of a solution of either α-rFVIII or rFVIII at 1 mg·mL−1 in PBS pH 7.4, whereas a volume of 1.3 μL of AF488 solution was added to 50 μL of CHO HCP solution at 1.3 mg·mL−1. The labeling reactions were allowed to proceed for 1 hour at room temperature (25°C), under dark and gentle agitation. The unreacted dyes were removed using 0.5 mL Zeba™ Dye and Biotin Removal Spin Columns (ThermoFisher Scientific, Waltham, MA). The concentration of the labeled proteins in solution was determined by Bradford assay. The absorbance of the solutions of AF488-labeled CHO HCPs and AF594-labeled α-rFVIII and rFVIII was measured by UV spectrophotometry at the wavelength of 490 and 590 nm, respectively, using a Synergy H1 plate reader (Biotek, Winooski, VT). The values of the dye-to-protein (D/P) ratio were calculated using Equation 1:
| (1) |
Wherein Abs, ε, and C are the UV absorbance, the molar extinction coefficient, and the concentration of the protein solution, while DF is the dilution factor. An average of 2.9 AF594 molecules were conjugated to each rFVIII and 2.1 AF488 molecules were conjugated to each HCP.
4.4. Primary dual-fluorescence screening of the CAP library against rFVIII-A1.
The primary selection of the CAP -ChemMatrix library was performed against a screening mix prepared by spiking AF594-labeled rFVIII-A1 at 0.10 mg·mL−1 in a solution of AF488-labeled CHO HCPs at 1.0 mg·mL−1. An aliquot of 10 μL of library beads were initially rinsed with 0.10% v/v Tween 20 in PBS at pH 7.4, and exposed to UV light (λex = 305-390 nm) at ~ 60 mW·cm−2 for 20 mins at room temperature (25°C) to equilibrate all library CAPs in the cis photo-isomer. The library beads were then incubated with 50 μL of screening mix for 2 hours at room temperature and under dark, rinsed with 0.10% v/v Tween 20 in PBS, and sorted using the microfluidic library screening device38–39. Beads displaying strong red-only fluorescence were selected as positive leads (Figure S2), incubated with 100 μL of 0.1 M glycine buffer pH 2.5 for 1 hr at room temperature and under dark to elute the bound AF594-labeled rFVIII, rinsed with Milli-Q water and acetonitrile. Finally, the selected beads were analyzed via Edman degradation using a PPSQ-33A protein sequencer (Shimadzu, Kyoto, Japan) to sequence the peptides displayed thereupon39.
4.5. Secondary screening of selected ChemMatrix-bound CAPs against full-length rFVIII
CAPs selected from library screening G-cycloAZOB[Dap-YKLYKYK-Dap]-G, G-cycloAZOB[Dap-KYHYYYK-Dap]-G, G-cycloAZOB[Dap-KYSLLGY-Dap]-G, G-cycloAZOB[Dap-KYYGSYY-Dap]-G, G-cycloAZOB[Dap-YYKHLYN-Dap]-G, G-cycloAZOB[Dap-YHYSKHY-Dap]-G, G-cycloAZOB[Dap-GKSKNNY-Dap]-G, G-cycloAZOB[Dap-GGGKYGK-Dap]-G, and G-cycloAZOB[Dap-YNNKHYK-Dap]-G; the homology-based sequence G-cycloAZOB[Dap-KYNLLGK-Dap]-G; and the variants G-cycloAZOB[Dap-KYYGSYY-Lys]-G, G-cycloAZOB[Dap-YYKHLYN-Lys]-G, G-cycloAZOB[Lys-KYYGSYY-Lys]-G, G-cycloAZOB[Lys-YYKHLYN-Lys]-G, G-cycloAZOB[Dap-KAAGSAA-Dap]-G, G-cycloAZOB[Dap-KFFGSFF-Dap]-G, G-cycloAZOB[Dap-KHGSH-Dap]-G, G-cycloAZOB[Dap-KAAGSAA-Dap]-G, G-cycloAZOB[Dap-KmYmYGSmYmY-Dap]-G (mY: O-methyl-tyrosine), G-cycloAZOB[Dap-AAKHLAN-Dap]-G, G-cycloAZOB[Dap-HKHLHN-Dap]-G, G-cycloAZOB[Dap-FFKHLFN-Dap]-G, and G-cycloAZOB[Dap-mYmYKHLmYN-Dap]-G were synthesized on 50 mg of aminomethyl-ChemMatrix resin as described in Section M.2.2. Full-length rFVIII contains calcium in the binding pocket and requires calcium in solution; 0.1 M HEPES buffer added with 0.1M NaCl, 5 mM CaCl2, and 0.01% v/v Tween20 at pH 7.4 was used for screening and photo physics experiments, unless specified otherwise. The resulting beads were rinsed with 0.10% v/v Tween 20 in HEPES, exposed to UV light (λex = 305-390 nm) at ~60 mW·cm−2 for 10 mins at room temperature (25°C) to equilibrate the CAPs to the cis photo-isomer, and incubated with 50 μL of a solution of AF594-labeled rFVIII at 1500 IU·mL−1 or (0.3 mg·mL−1) in AF488-labeled CHO HCPs at 0.2 mg·mL−1. Aliquots of 30 beads carrying each cis-CAP were then fed to the microfluidic screening device and imaged in both red (exc/em: 590/617 nm) and green (exc/em: 490/525 nm) channels. The remaining beads were exposed to visible light (λex = 420-450 nm) at ~220 mW·cm−2 for 2 mins at room temperature to trigger the cis-to-trans photo-isomerization of the CAPs (note: photo-isomerization is expected to trigger the elution of bound rFVIII). Aliquots of 30 beads carrying each trans-CAP were then fed to the microfluidic screening device and imaged in both red and green channels. The images of the cis-CAP and trans-CAP beads were analyzed using a custom MATLAB algorithm that determines the location of the pixels in the image, and measures and stores the values of brightness of each of the pixels38–39: using these data, the algorithm returns the values in the 90th percentile of the brightest pixels and calculates the values of average and standard deviation of intensity of the pixels that exceed the 90th percentile. The mean intensity of the top 10% of pixels was utilized as it circumvents the morphological variability among beads. The averaged value serves as the primary indicator of the fluorescence intensity of the bead displaying either cis-CAP or trans-CAP.
4.6. Photo-isomerization kinetics of ChemMatrix-bound CAPs
The spectroscopic device39 comprised (i) a PDMS measurement cell that holds 5 μL of CAP -ChemMatrix beads and maintains them hydrated and thermostatted through a steady flow of aqueous buffer; (ii) a rotating light mask that alternates the exposure of the beads to either the excitation beam generated by the light source or the measurement beam from the spectrophotometer with controlled frequency; and (iii) a custom holder that orients the measurement cell to maximize the exposure of the sample to the excitation beam and ensure ample sampling of the CAP -ChemMatrix beads. The holder was installed in a Cary 60 spectrophotometer (Agilent, Santa Clara, CA) to maintain a 65 mm pathlength at a 60° angle with respect to the measurement light beam for UV-Vis spectroscopy and a 100 mm pathlength at a 30° angle with respect to the light beam generated by the lamp for irradiance. A UV bandpass filter (BP305-390, Thorlabs, Newton, NJ) was used for the trans-to-cis isomerization, whereas a 420 nm longpass (LP420, Edmund Optics, Barrington, NJ) filter was used for the cis-to-trans isomerization. The incident light to induce photo-isomerization was generated using a BlueWave 200 lamp (Dymax, Torrington, CT) whose irradiance power was calculated by measuring the output power using an Accu-Cal 50 radiometer (Dymax, Torrington, CT) and computing the corresponding incident irradiance for the known optical filters. The kinetics of trans-to-cis and cis-to-trans photo-isomerization were measured by monitoring the absorbance at 440 nm; the absorbance at 600 nm was also monitored to detect disturbances during the measurements caused by mechanical perturbations. All measurements were performed in triplicate using the “kinetics” function of the Cary 60 instrument at the speed of acquisition of 30 readings per minute. Prior to (10 min) and during the photo-kinetic measurements, a steady flow of 0.1 mL·min−1 of 0.1 M HEPES buffer added with 0.1 M NaCl, 5 mM CaCl2, and 0.01% v/v Tween 20 at pH 7.4 (Binding Buffer) was maintained in the measurement chamber to keep the beads at constant temperature and hydrated. The kinetics of the cis-to-trans isomerization were of primary interest as its light stimulus (λex >420 nm) is expected to cause minimal or no damage to the target protein. The values of average calculated irradiance and measurement times are listed in Table S3.
The spectroscopic data were initially curated by maintaining only the data collected while the excitation light beam was screened by the rotating light mask. The temporal profiles of absorbance were fit against a 1st order kinetics model (Equation 2).
| (2) |
Wherein A, B, and C are fitting parameters, x and xo are respectively the actual and initial times of measurement, Abs420nm is the value of absorbance of the beads packed in the measurement cell at time x, and τ is the characteristic time of photo-isomerization.
4.7. Design of the CAP structure and amino acid composition as informed by the in silico druggability study of human FVIII
In silico “druggability” study of the A1 surface (PDB ID: 3CDZ)58–59 was performed using SiteMap60–61 and identified 4 putative pockets, of which 3 presented the appropriate morphological features (i.e., pocket depth (PD), solvent-accessible surface area (SASA), and pocket volume (PV)) and biophysical properties (i.e., isoelectric point (pI), polarity, and grand average hydropathy (GRAVY) index) to harbor peptide ligands. The structure and properties of the selected binding sites are reported in Figure S1 and Table S1, respectively. The analysis of the prospective binding sites informed the design of an ensemble of CAPs tailored to target FVIII-A1 domain. The putative binding sites feature a similar depth (32.3 – 36.7 Å) and volume (2614 – 3374 Å3) as well as amphiphilic character (GRAVY index: −0.4 to −0.9), but vary widely in electrostatic charge (isoelectric point, pI: 5.1 – 10.1).
4.8. In silico evaluation of CAPs as free and FVIII-bound photo-isomers
The cis or trans isomers of the CAPs used in architecture and sequence optimizations were constructed using the molecular editor Avogadro62. The equilibration and production steps were performed in GROMACS using the GROMOS 43a1 force field, using the parameters for the azobenzene linker obtained by Nguyen et al.44, 63–64. Briefly, every CAP photo-isomer was initially placed in a simulation box with periodic boundary containing 800 water molecules (TIP3P model), and equilibrated with 10,000 steps of steepest gradient descent; the CAP was then heated to 300 K in an NVT ensemble for 250 ps with 1 fs time steps, and equilibrated to 1 atm with a 500-ps NPT simulation with 2 fs time steps. The production runs were performed in the NPT ensemble at T = 300 K and P = 1 atm using the Nosé-Hoover thermostat and the Parrinello-Rahman barostat, respectively65–66. The leap-frog algorithm was used to integrate the equations of motion, with integration steps of 2 fs, and the atomic coordinates were saved every 2 ps. All covalent bonds were constrained using the LINCS algorithm, the short‐range electrostatic and Lennard-Jones interactions were calculated using cut-off values of 1.0 nm and 1.4 nm, and the particle-mesh Ewald method was utilized for long-range electrostatic interactions [67]; the list of non-bonded interactions was updated every 5 fs using a cutoff of 1.4 nm. The conformational space of the trans and cis isomers of the CAPs was then sampled by starting with the structures obtained from the molecular dynamics simulations. Briefly, the distance between the terminal glycine residues was varied within a range of 0.5 nm to 1.5 nm, divided in 20 windows of 0.05 nm each, by applying a harmonic biasing potential with a force constant of 2000 kJ/mol/nm2; the elongated structure was then simulated for up to 500 ns while tracking the variation in potential of mean force (PMF).
The structures of the trans and cis isomers of CAP variants were initially derived by exploring the energetic landscape of the various conformations and identifying the structures with absolute energy minima as representative of the CAP isomers displayed on ChemMatrix resins. The resultant structures, the corresponding values of structural description and free energy of the isomers and their difference (ΔECis-Trans), together with the root-mean-square deviation of the atomic positions of trans vs. cis isomers (RMSD) are reported in Table S4 and S5.
The crystal structure of human FVIII (PDB ID: 3CDZ)58–59 was prepared using Protein Prep Wizard (PPW, Schrödinger, New York, NY)68 by correcting missing atoms and/or side chains (PRIME), removing salts and ligands, adding explicit hydrogens, and optimizing the hydrogen-bonding network; the ionization state at pH 7.4 and minimization of the protein structure were finally performed using PROPKA69–70. The adjusted structure was then analyzed using SiteMap to identify sites for CAP binding60–61, 71, and the sites with high S-score (> 0.8) and D-score (> 0.9) were selected for CAP docking.
Finally, the CAP isomers corresponding to the absolute energy minima were docked in silico against the putative binding sites on human FVIII (PDB ID: 3CDZ) using the docking software HADDOCK (High Ambiguity Driven Protein-Protein Docking, v.2.4)72–73. The residues on the selected binding sites of FVIII and the X1-X2-X3-X4-X5-X6-X7 residues on the CAPs were marked as “active”, and the surrounding residues were marked as “passive”. The docked FVIII:CAPstructures were grouped in clusters of up to 20 complexes based on Cα RMSD < 7.5 Å and ranked using the dMM-PBSA score74. Finally, the top FVIII:trans/cis-CAP complexes were refined via 100-ns atomistic MD simulations and evaluated to estimate the free energy of binding (ΔGB).
4.9. Photo-affinity chromatographic purification of rFVIII from CHO cell culture harvest.
A clarified CHO-S cell culture supernatant and rFVIII were initially buffer-exchanged into Binding Buffer (5 mM CaCl2 and 0.10% v/v Tween20 in 0.1 M HEPES buffer at pH 7.4 ) using Amicon™ Ultra centrifugal filters with 3 kDa MWCO (MilliporeSigma, Burlington, MA). The feedstock for all photo-chromatography runs was then prepared as a solution of rFVIII at 700 IU·mL−1 (~ 150 μg·mL−1) in CHO fluid with HCP titer of 0.25 mg·mL−1. A volume of 5 μL of G-cycloAZOB[Lys-YYKHLYN-Lys]-G-ChemMatrix resin was loaded in the measurement cell, equilibrated with Binding Buffer at the flow rate of 0.1 mL·mL−1, and exposed to UV light (λex = 305-390 nm) for 20 mins to induce photo-isomerization of all CAPs to the cis conformation. Upon reaching a steady UV absorbance at 440 nm (azobenzene) and 600 nm (background), a volume of 20 μL of feedstock was loaded onto the measurement cell at the flow rate of 0.1 mL·mL−1, at which point the collection of the effluent in 50 μL fraction began. Following loading, the CAP -ChemMatrix beads were washed in Binding Buffer for 2 mins and exposed to visible light (λex = 420-450 nm) for 3 mins to trigger the cis-to-trans photoisomerization. The effluent was collected for the subsequent 1 min, until UV absorbance at 280 nm regained the baseline value. The collected fractions were analyzed using an anti-CHO HCP ELISA kit (Cygnus Technologies, Southport, NC) and a rFVIII-specific ELISA kit (Abcam, Cambridge, UK) to calculate the values of HCP and rFVIII concentration in the chromatographic fractions. These values were ultimately utilized to calculate rFVIII yield and purity.
4.10. Blood Clotting Efficiency of FVIII purified via photo-affinity chromatography.
The ability of FVIII – either commercial or purified via photo-affinity chromatography - to restore fibrin networks to standard branching densities was evaluated via clotting tests. The samples listed in Table S6 were prepared in triplicate by mixing either healthy or hemophilic plasma with aqueous 500 mM CaCl2, a solution of FVIII at approximately 1 IU·mL−1 in Binding Buffer, and a solution of AF488-Fibrinogen at 50 μg·mL−1 in Binding Buffer. A volume of 20 μL of each sample was pipetted onto glass slides, covered with a thin cover glass, and incubated for 2 hours at room temperature (25°C); the slides were sealed to prevent evaporation. Three 5 μm z-stacks with 0.5 μm sectioning and 16-bit 1024 x 1024 pixel resolution were generated for each slide. The relative fiber density was calculated as the ratio of fiber (green) pixels to background (black) pixels after binarization using ImageJ (National Institutes of Health, Bethesda, MD); binarized images were also eroded and skeletonized in ImageJ. The intersection density, or network crosslinking, was calculated as the ratio of skeleton intersections post-erosion to the total number of fiber pixels pre-erosion. The erosion function was applied to limit the number of false intersections imposed in images.
Supplementary Material
Acknowledgments.
The authors acknowledge support from the National Science Foundation (CBET 1743404 and CBET 1653590) and the Novo Foundation (AIM-Bio Grant NNF19SA0035474). The authors also wish to acknowledge the contribution of Daniel Midkiff in the San Miguel group and Michael Mantini group (Chemical and Biomolecular Engineering, NC State), Dr. John D. Schneible, and Miguel Lorenzo Roncal. The microfluidic devices for kinetics and photo-affinity studies were fabricated in part with the Chapel Hill Analytical and Nanofabrication Laboratory, CHANL, a member of the North Carolina Research Triangle Nanotechnology Network, RTNN, which is supported by the National Science Foundation, Grant ECCS-2025064, as part of the National Nanotechnology Coordinated Infrastructure, NNCI.
Footnotes
Conflict of interest disclosure. No conflict of interest to declare.
References
- 1.Arora S; Saxena V; Ayyar BV, Affinity chromatography: A versatile technique for antibody purification. Methods (San Diego, Calif.) 2017, 116, 84–94. [DOI] [PubMed] [Google Scholar]
- 2.Zhao M; Vandersluis M; Stout J; Haupts U; Sanders M; Jacquemart R, Affinity chromatography for vaccines manufacturing: Finally ready for prime time? Vaccine 2019, 37 (36), 5491–5503. [DOI] [PubMed] [Google Scholar]
- 3.Clonis YD, Separation processes in biotechnology. Process affinity chromatography. Bioprocess technology 1990, 9, 401–45. [PubMed] [Google Scholar]
- 4.Chu W; Prodromou R; Day KN; Schneible JD; Bacon KB; Bowen JD; Kilgore RE; Catella CM; Moore BD; Mabe MD; Alashoor K; Xu Y; Xiao Y; Menegatti S, Peptides and pseudopeptide ligands: a powerful toolbox for the affinity purification of current and next-generation biotherapeutics. Journal of chromatography. A 2021, 1635, 461632. [DOI] [PubMed] [Google Scholar]
- 5.Li Y; Stern D; Lock LL; Mills J; Ou SH; Morrow M; Xu X; Ghose S; Li ZJ; Cui H, Emerging biomaterials for downstream manufacturing of therapeutic proteins. Acta biomaterialia 2019, 95, 73–90. [DOI] [PubMed] [Google Scholar]
- 6.Geuijen KPM; van Wijk-Basten D; Egging DF; Schasfoort RBM; Eppink MH, Rapid Buffer and Ligand Screening for Affinity Chromatography by Multiplexed Surface Plasmon Resonance Imaging. Biotechnology journal 2017, 12 (9). [DOI] [PubMed] [Google Scholar]
- 7.Arruda G; Mello M; Cavalcante R; Gama M; Marcellos C; Barreto A Jr, Technological Bottlenecks in purification of monoclonal antibodies: prospective study and proposals for improved process design. 2020. [Google Scholar]
- 8.Bratt J, Angela Linderholm G David Green, and Chamow Steven M., Buffers in Biologics Manufacturing. BioProcess International 2017, 15. [Google Scholar]
- 9.Lobo SA; Bączyk P; Wyss B; Widmer JC; Jesus LP; Gomes J; Batista AP; Hartmann S; Wassmann P, Stability liabilities of biotherapeutic proteins: Early assessment as mitigation strategy. Journal of Pharmaceutical and Biomedical Analysis 2021, 192, 113650. [DOI] [PubMed] [Google Scholar]
- 10.Cafuir LA; Kempton CL, Current and emerging factor VIII replacement products for hemophilia A. Ther Adv Hematol 2017, 8 (10), 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aledort L; Mannucci PM; Schramm W; Tarantino M, Factor VIII replacement is still the standard of care in haemophilia A. Blood transfusion = Trasfusione del sangue 2019, 17 (6), 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vehar GA; Davie EW, Preparation and properties of bovine factor VIII (antihemophilic factor). Biochemistry 1980, 19 (3), 401–10. [DOI] [PubMed] [Google Scholar]
- 13.Burnouf T; Burnouf-Radosevich M; Huart JJ; Goudemand M, A highly purified factor VIII:c concentrate prepared from cryoprecipitate by ion-exchange chromatography. Vox sanguinis 1991, 60 (1), 8–15. [DOI] [PubMed] [Google Scholar]
- 14.Addiego JE Jr.; Gomperts E; Liu SL; Bailey P; Courter SG; Lee ML; Neslund GG; Kingdon HS; Griffith MJ, Treatment of hemophilia A with a highly purified factor VIII concentrate prepared by anti-FVIIIc immunoaffinity chromatography. Thrombosis and haemostasis 1992, 67 (1), 19–27. [PubMed] [Google Scholar]
- 15.Thim L; Vandahl B; Karlsson J; Klausen NK; Pedersen J; Krogh TN; Kjalke M; Petersen JM; Johnsen LB; Bolt G; NØRby PL; Steenstrup TD, Purification and characterization of a new recombinant factor VIII (N8). Haemophilia 2010, 16 (2), 349–359. [DOI] [PubMed] [Google Scholar]
- 16.Cheng E; Jinzenji D; Lorthiois AP; de Carvalho RR; Tanaka-Azevedo AM; Raw I; Martins EA, Purification of coagulation factor VIII using chromatographic methods. Direct chromatography of plasma in anion exchange resins. Biotechnology letters 2010, 32 (9), 1207–14. [DOI] [PubMed] [Google Scholar]
- 17.Drouin LM; Agbandje-McKenna M, Adeno-associated virus structural biology as a tool in vector development. Future Virol 2013, 8 (12), 1183–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley BD; Tannatt M; Magnusson R; Hagelberg S; Booth J, Development and validation of an affinity chromatography step using a peptide ligand for cGMP production of factor VIII. Biotechnology and Bioengineering 2004, 87 (3), 400–412. [DOI] [PubMed] [Google Scholar]
- 19.Kelley BD; Booth J; Tannatt M; Wu Q-L; Ladner R; Yu J; Potter D; Ley A, Isolation of a peptide ligand for affinity purification of factor VIII using phage display. Journal of Chromatography A 2004, 1038 (1), 121–130. [DOI] [PubMed] [Google Scholar]
- 20.Pflegerl K; Hahn R; Berger E; Jungbauer A, Mutational analysis of a blood coagulation factor VIII-binding peptide. The journal of peptide research : official journal of the American Peptide Society 2002, 59 (4), 174–82. [DOI] [PubMed] [Google Scholar]
- 21.Amatschek K; Necina R; Hahn R; Schallaun E; Schwinn H; Josić D; Jungbauer A, Affinity Chromatography of Human Blood Coagulation Factor VIII on Monoliths with Peptides from a Combinatorial Library. Journal of High Resolution Chromatography 2000, 23 (1), 47–58. [Google Scholar]
- 22.Wakabayashi H; Varfaj F; Deangelis J; Fay PJ, Generation of enhanced stability factor VIII variants by replacement of charged residues at the A2 domain interface. Blood 2008, 112 (7), 2761–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toole JJ; Knopf JL; Wozney JM; Sultzman LA; Buecker JL; Pittman DD; Kaufman RJ; Brown E; Shoemaker C; Orr EC; et al. , Molecular cloning of a cDNA encoding human antihaemophilic factor. Nature 1984, 312 (5992), 342–7. [DOI] [PubMed] [Google Scholar]
- 24.Vehar GA; Keyt B; Eaton D; Rodriguez H; O’Brien DP; Rotblat F; Oppermann H; Keck R; Wood WI; Harkins RN; Tuddenham EG; Lawn RM; Capon DJ, Structure of human factor VIII. Nature 1984, 312 (5992), 337–42. [DOI] [PubMed] [Google Scholar]
- 25.Kane WH; Davie EW, Blood coagulation factors V and VIII: structural and functional similarities and their relationship to hemorrhagic and thrombotic disorders. Blood 1988, 71 (3), 539–55. [PubMed] [Google Scholar]
- 26.Lenting PJ; van Mourik JA; Mertens K, The life cycle of coagulation factor VIII in view of its structure and function. Blood 1998, 92 (11), 3983–96. [PubMed] [Google Scholar]
- 27.Stoilova-McPhie S; Lynch GC; Ludtke S; Pettitt BM, Domain organization of membrane-bound factor VIII. Biopolymers 2013, 99 (7), 448–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakabayashi H; Koszelak ME; Mastri M; Fay PJ, Metal ion-independent association of factor VIII subunits and the roles of calcium and copper ions for cofactor activity and inter-subunit affinity. Biochemistry 2001, 40 (34), 10293–300. [DOI] [PubMed] [Google Scholar]
- 29.Ngo JCK; Huang M; Roth DA; Furie BC; Furie B, Crystal Structure of Human Factor VIII: Implications for the Formation of the Factor IXa-Factor VIIIa Complex. Structure (London, England : 1993) 2008, 16 (4), 597–606. [DOI] [PubMed] [Google Scholar]
- 30.Lam KS; Liu R; Miyamoto S; Lehman AL; Tuscano JM, Applications of One-Bead One-Compound Combinatorial Libraries and Chemical Microarrays in Signal Transduction Research. Accounts of Chemical Research 2003, 36 (6), 370–377. [DOI] [PubMed] [Google Scholar]
- 31.Menegatti S; Ward KL; Naik AD; Kish WS; Blackburn RK; Carbonell RG, Reversible Cyclic Peptide Libraries for the Discovery of Affinity Ligands. Analytical Chemistry 2013, 85 (19), 9229–9237. [DOI] [PubMed] [Google Scholar]
- 32.Lavoie RA; di Fazio A; Carbonell RG; Menegatti S, Multiplexed Competitive Screening of One-Bead-One-Component Combinatorial Libraries Using a ClonePix 2 Colony Sorter. Int J Mol Sci 2019, 20 (20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Day K; Prodromou R; Saberi Bosari S; Lavoie A; Omary M; Market C; San Miguel A; Menegatti S, Discovery and Evaluation of Peptide Ligands for Selective Adsorption and Release of Cas9 Nuclease on Solid Substrates. Bioconjugate Chemistry 2019, 30 (12), 3057–3068. [DOI] [PubMed] [Google Scholar]
- 34.Lavoie RA; Williams TI; Blackburn RK; Carbonell RG; Menegatti S, Development of Peptide Ligands for Targeted Capture of Host Cell Proteins from Cell Culture Production Harvests. Methods in molecular biology (Clifton, N.J.) 2021, 2261, 489–506. [DOI] [PubMed] [Google Scholar]
- 35.Lavoie RA; di Fazio A; Blackburn RK; Goshe MB; Carbonell RG; Menegatti S, Targeted Capture of Chinese Hamster Ovary Host Cell Proteins: Peptide Ligand Discovery. Int J Mol Sci 2019, 20 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kish WS; Sachi H; Naik AD; Roach MK; Bobay BG; Blackburn RK; Menegatti S; Carbonell RG, Design, selection, and development of cyclic peptide ligands for human erythropoietin. Journal of Chromatography A 2017, 1500, 105–120. [DOI] [PubMed] [Google Scholar]
- 37.Barozzi A; Lavoie RA; Day KN; Prodromou R; Menegatti S, Affibody-Binding Ligands. International Journal of Molecular Sciences 2020, 21 (11), 3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saberi-Bosari S; Omary M; Lavoie A; Prodromou R; Day K; Menegatti S; San-Miguel A, Affordable Microfluidic Bead-Sorting Platform for Automated Selection of Porous Particles Functionalized with Bioactive Compounds. Scientific Reports 2019, 9 (1), 7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prodromou R; Day KN; Saberi-Bosari S; Schneible JD; Mabe MD; San Miguel A; Daniele MA; Pozdin V; Menegatti S, Engineering Next Generation Cyclized Peptide Ligands for Light-Controlled Capture and Release of Therapeutic Proteins. Advanced Functional Materials 2021, 31 (27), 2101410. [Google Scholar]
- 40.Day K; Schneible JD; Young AT; Pozdin VA; Van Den Driessche G; Gaffney LA; Prodromou R; Freytes DO; Fourches D; Daniele M; Menegatti S, Photoinduced reconfiguration to control the protein-binding affinity of azobenzene-cyclized peptides. Journal of Materials Chemistry B 2020, 8 (33), 7413–7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wachtveitl J; Spörlein S; Satzger H; Fonrobert B; Renner C; Behrendt R; Oesterhelt D; Moroder L; Zinth W, Ultrafast Conformational Dynamics in Cyclic Azobenzene Peptides of Increased Flexibility. Biophysical Journal 2004, 86 (4), 2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Amorós J; Díaz-Lobo M; Nonell S; Velasco D, Fastest Thermal Isomerization of an Azobenzene for Nanosecond Photoswitching Applications under Physiological Conditions. Angewandte Chemie International Edition 2012, 51 (51), 12820–12823. [DOI] [PubMed] [Google Scholar]
- 43.Renner C; Moroder L, Azobenzene as conformational switch in model peptides. Chembiochem : a European journal of chemical biology 2006, 7 (6), 868–78. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen PH; Stock G, Nonequilibrium molecular dynamics simulation of a photoswitchable peptide. Chemical Physics 2006, 323 (1), 36–44. [Google Scholar]
- 45.Nguyen PH; Mu Y; Stock G, Structure and energy landscape of a photoswitchable peptide: A replica exchange molecular dynamics study. Proteins: Structure, Function, and Bioinformatics 2005, 60 (3), 485–494. [DOI] [PubMed] [Google Scholar]
- 46.Unesaki T; Nakagawa O, studies of the stability of Factor VIII especially on the pH, time and temperature dependency. Journal of the Japan Society of Blood Transfusion 1972, 19 (1), 1–8. [Google Scholar]
- 47.Oh SH; Kim HW; Kim YJ, Structure of heavy and light chains of blood coagulation factor VIII (FVIII) involved in the activation of FVIII. Experimental & Molecular Medicine 1997, 29 (2), 97–102. [Google Scholar]
- 48.Menegatti S; Naik A; Carbonell R, The hidden potential of small synthetic molecules and peptides as affinity ligands for bioseparations. Pharmaceutical Bioprocessing 2013, 1, 467–485. [Google Scholar]
- 49.Reese HR; Shanahan CC; Proulx C; Menegatti S, Peptide science: A “rule model” for new generations of peptidomimetics. Acta Biomaterialia 2020, 102, 35–74. [DOI] [PubMed] [Google Scholar]
- 50.Berg JM TJ, Stryer L, Section 1.3Chemical Bonds in Biochemistry. In Biochemistry. 5th edition., W. H. Freeman and Company: New York, 2002. [Google Scholar]
- 51.Beladjine S; Amrani M; Zanoun A; Belaidi A; Vergoten G, Structure and hydrogen bonding in aqueous sodium chloride solutions using theoretical water model AB4: Effects of concentration. Computational and Theoretical Chemistry 2011, 977 (1), 97–102. [Google Scholar]
- 52.Bucay I; O’Brien ET 3rd; Wulfe SD; Superfine R; Wolberg AS; Falvo MR; Hudson NE, Physical determinants of fibrinolysis in single fibrin fibers. PLoS One 2015, 10 (2), e0116350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelley B, Very Large Scale Monoclonal Antibody Purification: The Case for Conventional Unit Operations. Biotechnology Progress 2007, 23 (5), 995–1008. [DOI] [PubMed] [Google Scholar]
- 54.Brechmann NA; Eriksson P-O; Eriksson K; Oscarsson S; Buijs J; Shokri A; Hjälm G; Chotteau V, Pilot-scale process for magnetic bead purification of antibodies directly from non-clarified CHO cell culture. Biotechnology Progress 2019, 35 (3), e2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bordelon T; Bobay B; Murphy A; Reese H; Shanahan C; Odeh F; Broussard A; Kormos C; Menegatti S, Translating antibody-binding peptides into peptoid ligands with improved affinity and stability. Journal of chromatography. A 2019, 1602, 284–299. [DOI] [PubMed] [Google Scholar]
- 56.Kish WS; Roach MK; Sachi H; Naik AD; Menegatti S; Carbonell RG, Purification of human erythropoietin by affinity chromatography using cyclic peptide ligands. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences 2018, 1085, 1–12. [DOI] [PubMed] [Google Scholar]
- 57.Day K; Prodromou R; Saberi Bosari S; Lavoie A; Omary M; Market C; San Miguel A; Menegatti S, Discovery and Evaluation of Peptide Ligands for Selective Adsorption and Release of Cas9 Nuclease on Solid Substrates. Bioconjug Chem 2019, 30 (12), 3057–3068. [DOI] [PubMed] [Google Scholar]
- 58.Ngo JC; Huang M; Roth DA; Furie BC; Furie B, Crystal structure of human factor VIII: implications for the formation of the factor IXa-factor VIIIa complex. Structure (London, England : 1993) 2008, 16 (4), 597–606. [DOI] [PubMed] [Google Scholar]
- 59.Smith IW; d’Aquino AE; Coyle CW; Fedanov A; Parker ET; Denning G; Spencer HT; Lollar P; Doering CB; Spiegel PC Jr., The 3.2 Å structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain. Journal of thrombosis and haemostasis : JTH 2020, 18 (1), 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Halgren T, New Method for Fast and Accurate Binding-site Identification and Analysis. Chemical Biology & Drug Design 2007, 69 (2), 146–148. [DOI] [PubMed] [Google Scholar]
- 61.Halgren TA, Identifying and Characterizing Binding Sites and Assessing Druggability. Journal of Chemical Information and Modeling 2009, 49 (2), 377–389. [DOI] [PubMed] [Google Scholar]
- 62.Hanwell MD; Curtis DE; Lonie DC; Vandermeersch T; Zurek E; Hutchison GR, Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. Journal of Cheminformatics 2012, 4 (1), 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen PH; Hamm P; Stock G, Nonequilibrium molecular dynamics simulation of photoinduced energy flow in peptides: theory meets experiment. Proteins: Energy, Heat and Signal Flow 2009, 149–168. [Google Scholar]
- 64.Nguyen PH; Mu Y; Stock G, Structure and energy landscape of a photoswitchable peptide: a replica exchange molecular dynamics study. Proteins 2005, 60 (3), 485–94. [DOI] [PubMed] [Google Scholar]
- 65.Kleinerman DS; Czaplewski C; Liwo A; Scheraga HA, Implementations of Nosé-Hoover and Nosé-Poincaré thermostats in mesoscopic dynamic simulations with the united-residue model of a polypeptide chain. The Journal of chemical physics 2008, 128 (24), 245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parrinello M; Rahman A, Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics 1981, 52 (12), 7182–7190. [Google Scholar]
- 67.Cheatham TE III; Miller JL; Fox T; Darden TA; Kollman PA, Molecular Dynamics Simulations on Solvated Biomolecular Systems: The Particle Mesh Ewald Method Leads to Stable Trajectories of DNA, RNA, and Proteins. Journal of the American Chemical Society 1995, 117 (14), 4193–4194. [Google Scholar]
- 68.Madhavi Sastry G; Adzhigirey M; Day T; Annabhimoju R; Sherman W, Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design 2013, 27 (3), 221–234. [DOI] [PubMed] [Google Scholar]
- 69.Rostkowski M; Olsson MHM; Søndergaard CR; Jensen JH, Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Structural Biology 2011, 11 (1), 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olsson MHM; Søndergaard CR; Rostkowski M; Jensen JH, PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. Journal of Chemical Theory and Computation 2011, 7 (2), 525–537. [DOI] [PubMed] [Google Scholar]
- 71.Sharma A; Tiwari V; Sowdhamini R, Computational search for potential COVID-19 drugs from FDA-approved drugs and small molecules of natural origin identifies several anti-virals and plant products. Journal of Biosciences 2020, 45 (1), 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Honorato RV; Koukos PI; Jiménez-García B; Tsaregorodtsev A; Verlato M; Giachetti A; Rosato A; Bonvin AMJJ, Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Frontiers in Molecular Biosciences 2021, 8 (708). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Zundert GCP; Rodrigues JPGLM; Trellet M; Schmitz C; Kastritis PL; Karaca E; Melquiond ASJ; van Dijk M; de Vries SJ; Bonvin AMJJ, The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. Journal of Molecular Biology 2016, 428 (4), 720–725. [DOI] [PubMed] [Google Scholar]
- 74.Spiliotopoulos D; Kastritis PL; Melquiond ASJ; Bonvin AMJJ; Musco G; Rocchia W; Spitaleri A, dMM-PBSA: A New HADDOCK Scoring Function for Protein-Peptide Docking. Frontiers in Molecular Biosciences 2016, 3 (46). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


